Dysregulation of innate immune- and inflammatory-related pathways is implicated in hematopoietic defects associated with aging, clonal hematopoiesis, and MDS. We review the current state of this field and emerging concepts that reveal critical biology and novel therapeutic opportunities.
Abstract
With a growing aged population, there is an imminent need to develop new therapeutic strategies to ameliorate disorders of hematopoietic aging, including clonal hematopoiesis and myelodysplastic syndrome (MDS). Cell-intrinsic dysregulation of innate immune- and inflammatory-related pathways as well as systemic inflammation have been implicated in hematopoietic defects associated with aging, clonal hematopoiesis, and MDS. Here, we review and discuss the role of dysregulated innate immune and inflammatory signaling that contribute to the competitive advantage and clonal dominance of preleukemic and MDS-derived hematopoietic cells. We also propose how emerging concepts will further reveal critical biology and novel therapeutic opportunities.
Introduction
Myelodysplastic syndromes (MDSs) are hematopoietic stem cell (HSC) disorders defined by myeloid cell dysplasia and ineffective hematopoiesis (Cazzola, 2020). MDSs are propagated by defective HSC(s) and are genetically defined by somatic mutations and chromosomal abnormalities (Shastri et al., 2017). In certain cases, MDS can be preceded by an asymptomatic phase characterized by clonal expansion of HSCs harboring mutations found in hematologic malignancies, termed clonal hematopoiesis of indeterminate potential (CHIP; Jaiswal and Ebert, 2019). Moreover, chronic inflammatory diseases associated with activated innate immune signaling pathways often precede MDS; thus, a role for dysregulated innate immune and inflammatory signaling in hematopoietic cells as well as in the microenvironment is associated with preleukemic states, such as CHIP and MDS (Barreyro et al., 2018). The innate immune system utilizes evolutionarily conserved host cell receptors that recognize commensal and pathogenic microorganisms and cellular by-products. Genomic sequencing of organisms from early-branching phyla has revealed that the prototypical innate immune receptors, namely the TLR family and NF-κB pathway components, are found in organisms that predate bilaterians (Brennan and Gilmore, 2018). The biological function of these innate immune- and inflammatory-related pathways not only serves to sense and defend against foreign pathogens but is also critical for adapting to stressed environments, developmental processes, and directing cell survival and self-renewal programs. Thus, the complexity and consequences of dysregulated innate immune and inflammatory signaling pathways in the emergence of preleukemic states and in the pathogenesis of MDS are becoming appreciated. It has been nearly 10 yr since the first comprehensive review was published addressing the role of dysregulated innate immune signaling as a driver of the MDS phenotype (Starczynowski and Karsan, 2010). The field has reached a critical inflection point, as it is evident that cell-intrinsic dysregulation of innate immune- and inflammatory-related pathways as well as systemic inflammation contribute to hematopoietic cell aging, CHIP, and MDS. Here, we review and discuss the current state of data supporting the role of dysregulated innate immune and inflammatory signaling that contributes to the competitive advantage and clonal dominance of preleukemic and MDS-derived hematopoietic cells. We also propose how emerging concepts will further reveal critical biology and novel therapeutic opportunities.
Overview of premalignancies
Aging and risk of hematopoietic premalignancies
The greatest risk factor for developing MDS is advanced age. It is well understood that aging immune and hematopoietic systems undergo dysregulation and functional decline (Dorshkind et al., 2009). In elderly populations, these changes result in increased risk of infection, poor vaccination response, anemias, autoimmunity, and a higher risk of hematologic malignancies (Dorshkind et al., 2009). While the molecular mechanisms involved in immune and hematopoietic aging are well understood to include accumulation of DNA damage, oxidative stress, shortening of telomeres, activation of tumor-suppressor genes, and epigenetic dysregulation, what triggers these alterations is a key question. Increasing evidence supports that environmental stimuli and stressors, including infection and inflammatory processes, play a key role in aging and risk of preleukemic states (Barreyro et al., 2018). Understanding these processes is timely and imperative because, at a global level, the population aged 65 yr and over is growing faster than all other age groups (United Nations, Department of Economic and Social Affairs, Population Dynamics, 2019). By 2050, one in six people in the world will be over age 65 yr (16%), up from 1 in 11 in 2019 (9%), which has significant implications for the incidence of CHIP and MDS (United Nations, Department of Economic and Social Affairs, Population Dynamics, 2019).
Clonal hematopoiesis and CHIP
The recent World Health Organization’s definitions of myeloid neoplasms consider CHIP and clonal cytopenia of undetermined significance (CCUS) as precursor conditions, while MDSs are considered myeloid neoplasms with myelodysplasia (Arber et al., 2016; DeZern et al., 2019). Clonal hematopoiesis was initially discovered in aging females by nonrandom patterns of X inactivation (Ayachi et al., 2020) and was subsequently found to be driven by recurrent somatic mutations, including DNMT3A, TET2, and ASXL1, along with mosaic chromosomal alterations (Gao et al., 2021; Jaiswal et al., 2014; Kwok et al., 2015). CHIP is defined in a subset of individuals with clonal hematopoiesis based on a variant allele frequency ≥2% of a somatic mutation in a hematologic malignancy-associated gene. Individuals with CHIP have a modestly increased long-term risk of development of hematologic malignancy (Abelson and Wang, 2018; Desai et al., 2018; Young et al., 2019; Genovese et al., 2014), which is associated with the size of the mutant clone, although the absolute risk of leukemic transformation in individuals with CHIP is very low. Similar to CHIP, individuals with CCUS are defined by clonal expansion of mutant HSCs (referred to herein as preleukemic HSCs); however, individuals with CCUS exhibit cytopenia and a higher risk of developing a hematologic malignancy. Strikingly, many clonal hematopoiesis-associated somatic mutations have now been detected at low levels at the earliest stages of life (Williams et al., 2020 Preprint; Van Egeren et al., 2021). These landmark findings have shifted focus from efforts to understand how these somatic mutations are acquired toward understanding how these mutations are selected for in the context of aging and inflammation. Beyond increased risk of hematologic malignancies, CHIP is associated with higher morbidity and mortality of atherosclerotic cardiovascular disease and ischemic stroke (Jaiswal et al., 2017; Jaiswal et al., 2014; Dorsheimer et al., 2019), as well as increased incidence of pulmonary disease (Buscarlet et al., 2017; Zink et al., 2017) and type 2 diabetes (Bonnefond et al., 2013). Experimental evidence in murine models supports that this increased risk relates to activation of the NLRP3 inflammasome and IL-1β production by CHIP mutant macrophages (Fuster et al., 2020; Sano et al., 2018). Understanding the innate immune and inflammatory processes dysregulated in the context of CHIP and CCUS will be critical for developing methods to intercept hematologic malignancies and other associated pathologies.
Pathogenesis of MDSs
MDSs are characterized by a clonal advantage of HSCs, cytopenias, bone marrow (BM) cell dysplasia, and ineffective hematopoiesis (Corey et al., 2007; Nimer, 2008). MDSs are propagated by defective HSC(s) and are defined by somatic mutations and chromosomal abnormalities affecting several critical cellular functions, including epigenetic plasticity, DNA repair, cohesion complexes, ribosome and spliceosome function, and immune signaling (Nilsson et al., 2007; Nilsson et al., 2000; Pang et al., 2013). The most common recurrently mutated genes occur in ASXL1, DNMT3A, RUNX1, SRSF2, SF3B1, and TET2, which are considered early genetic events and are thought to contribute to clonal dominance. In addition, chromosomal abnormalities including del(5q), +8, −7/del(7q), del(20q), and a complex karyotype are commonly observed in MDS patients. The gradual outgrowth of these mutant hematopoietic clones in the BM coincides with the clinical manifestation of MDS. At the onset of disease, the median number of somatic mutations is two to three per patient (Ogawa, 2019). In a subset of patients, evolution to acute myeloid leukemia (AML) may occur through a linear and/or branching evolutionary process (Chen et al., 2019). Given the older age of MDS patients, only a portion are potential candidates for allogeneic stem cell transplantation, which remains the only curative option.
A central paradox of MDS pathogenesis is that the initiating mutations are thought to provide a competitive advantage and clonal dominance at the stem cell level; however, MDS HSCs along with the hematopoietic progenitors and mature cells are inherently impaired, as demonstrated by a differentiation defect, inefficient growth ex vivo, and poor engraftment in immune-compromised mice (Côme et al., 2020). For instance, MDS hematopoietic stem and progenitor cells (HSPCs) grow poorly in vitro and generally exhibit limited engraftment in xenograft models compared with normal HSPCs (Côme et al., 2020). Moreover, the majority of molecular alterations observed in MDSs are not sufficient to increase the self-renewal potential of human HSPCs as demonstrated in mouse models (Zhou et al., 2015; Basheer and Vassiliou, 2021). Understanding the discrepancy between clonal dominance of MDS HSCs and their functional impairment in the BM niche is necessary to improve the outcome of MDSs.
The role of systemic inflammation in hematopoietic premalignancies
Inflamm-aging and CHIP
With aging in both humans and mouse models, increases in systemic inflammation, termed “inflamm-aging,” are widely reported. This includes elevated circulating levels of the pro-inflammatory cytokines TNFα, IL-1β, IL-6, RANTES, and numerous others (Rea et al., 2018). Chronic inflammatory challenge is well understood to be harmful to HSCs. For example, mimicking TLR3-mediated type I IFN signaling by repeated injection with polyI:C results in progressive depletion of functional HSCs in young mice, with no sign of later recovery (Bogeska et al., 2020 Preprint). As these mice were aged, they developed anemia, thrombocytopenia, BM hypocellularity, and increased adipocyte density, as observed in elderly humans. Similar results reporting attrition of HSC function have been described following other types of chronic inflammatory challenge, including LPS (Esplin et al., 2011; Mann et al., 2018) and mycobacterial infection (Matatall et al., 2016). These results support that low-grade, chronic inflammation observed in the context of aging is not conducive for sustaining the long-term function of normal aged HSCs.
Low-grade, chronic inflammation and other microenvironmental factors have been posited to favor the expansion of HSCs with CHIP mutations, providing an explanation for the clonal dominance of mutant HSCs over the normal HSC population in the context of aging. A clonogenic advantage for Tet2-knockout murine and TET2 mutant human HSPCs is observed in an in vitro environment that contains the proinflammatory cytokine TNFα (Abegunde et al., 2018). These findings imply that TET2 mutations promote clonal dominance with aging by conferring cell-intrinsic TNFα resistance to sensitive hematopoietic cell progenitors while also propagating such an inflammatory environment. Along these lines, it was also revealed that an increase in Tet2 mutant mature myeloid cells and HSPCs in response to inflammatory stress results in enhanced production of inflammatory cytokines, including IL-6, and concurrent resistance to apoptosis. IL-6 induces hyperactivation of the Shp2–Stat3 signaling axis, resulting in increased expression of an anti-apoptotic long noncoding RNA, Morrbid, in Tet2 mutant myeloid cells and HSPCs (Cai et al., 2018). TET2 deficiency in HSPCs results in increased self-renewal and myeloid proliferation in humans and mice. These phenotypes are variable and occur only in a subset of humans with TET2 mutations, suggesting that extrinsic non–cell-autonomous factors could also contribute to disease onset. In support of this concept, bacterial translocation and increased IL-6 production, resulting from dysfunction of the small-intestinal barrier, contribute to the myeloid cell expansion of TET2 mutant HSPCs (Meisel et al., 2018). In symptom-free TET2 mutant mice, myeloid cell expansion can be induced by disrupting intestinal barrier integrity or in response to systemic bacterial stimuli such as a TLR2 agonist. Pointing to a causal link, myeloid cell expansion of TET2 mutant cells was reversed by antibiotic treatment, which illustrates the importance of microbial signals in the development of CHIP.
The studies described above heavily rely on models in which TET2 is inactivated, and far less is known about the relative competitive advantage of HSCs with DNMT3A, ASXL1, and other CHIP mutations. Early studies support similar general principles; for example, IgE-stimulated mast cells from Dnmt3a-knockout mice produce higher amounts of inflammatory cytokines including IL-6 and TNFα (Leoni et al., 2017), and IFN-γ is sufficient to drive expansion of Dnmt3a-knockout HSCs (Abegunde et al., 2018). Elevated pro-inflammatory molecules are not equally observed across all individuals with CHIP. Initial reports find that TET2 mutant CHIP is more highly associated with elevated IL-6, while DNMT3A mutant CHIP is more highly associated with elevated TNFα (Cook et al., 2019). In a cohort of patients with ulcerative colitis, an autoimmune disease characterized by increased levels of pro-inflammatory cytokines, CHIP was associated with a specific mutational spectrum, notably positive selection of clones with DNMT3A and PPM1D mutations (Zhang et al., 2019). This study observed a specific association between elevated levels of serum IFN-γ and DNMT3A mutations. Together, this suggests that inflamm-aging as a general principle is not sufficiently refined to predict or explain elevated risk of CHIP across aged individuals or those with autoimmune disorders. Rather, detailed mechanistic studies will be needed to understand the contextual basis for synergy between specific inflammatory molecules, dysregulation of particular innate immune pathways, and favorable selection of HSCs with precise CHIP mutations.
Preleukemia and MDS
A relationship between microenvironment-associated systemic inflammation and progression of MDS has been established; however, the molecular and cellular mechanisms of how preleukemic or MDS HSCs selectively acquire a clonal advantage over normal HSCs and the distinction of these mechanisms from those promoting CHIP are less understood (Barreyro et al., 2018). Despite overwhelming evidence of chronic inflammation in the BM and peripheral blood (PB) of MDS patients, the effects of systemic inflammatory signals on MDS HSCs and disease progression remain uncharacterized and are under ongoing investigations. As described above during aging and in CHIP, the MDS BM niche exhibits increased inflammatory signaling, including elevated cytokine, chemokine, and alarmin levels. Microenvironmental alterations might simply be related to aging, but there is also evidence that MDS hematopoietic cells themselves may alter the stem cell niche through activation of the innate immune system and related inflammatory signaling. For example, elevated expression of IL-6 in MDSs is directly linked to dysregulation of innate immune pathways, particularly the myddosome. Loss of the 5q genes miR-145 and miR-146a results in TRAF6-mediated overexpression of IL-6 in HSPCs (Starczynowski et al., 2010). The observed increase in IL-6 contributes to non–cell-autonomous effects on PB cytopenias. These findings suggest that the dysplastic hematopoiesis, cytopenias, and thrombocytosis as a result of TRAF6 overexpression in HSPCs are due to a myddosome-mediated increase in IL-6 expression and resulting paracrine, cell-nonautonomous effects. Such alterations in expression of microenvironmental factors may favor the expansion of preleukemic or MDS HSCs over aged normal HSCs and may provide an explanation for the discrepancy between clonal dominance of MDS HSCs and their functional impairment.
In contrast to the above, clonal expansion and the propensity to develop acute leukemia are not mediated by IL-6 and are due to cell-autonomous effects of TRAF6-mediated activation. While deletion of IL-6 ameliorated the PB cytopenias in TRAF6-expressing mice, these mice continued on to develop overt BM failure or AML (Starczynowski et al., 2010). A similar observation was reported in AML. Human AML suppresses erythroid differentiation and causes anemia via paracrine effects of IL-6 (Zhang et al., 2020). Building on these prior observations, it was recently shown that miR-146a expression declined in old wild-type mice, and loss of miR-146a promoted premature HSC aging and inflammation (i.e., through increased expression of IL-6 and TNF) in young miR-146a–null mice, before onset of myeloid malignancy (Grants et al., 2020). Reducing inflammation by targeting IL-6 or TNF was sufficient to restore miR-146a–deficient HSC function and reduced the incidence of hematological malignancy in miR-146a–deficient mice. Thus, loss of miR-146a regulates cell-extrinsic and cell-intrinsic mechanisms linking HSC inflamm-aging to the development of myeloid malignancy. Thus, changes in systemic cytokine expression in MDSs and preleukemia have pleotropic effects on the etiology of the disease.
The emerging model of how dysregulated inflammatory and innate immune signaling contribute to preleukemia and MDSs perhaps can be viewed as a two-step process (Fig. 1). The first step results in dysregulation of key signaling hubs that control innate immune and inflammatory pathways in preleukemic or MDS HSPCs. This occurs through the various genetic and molecular changes affecting innate immune- and inflammatory-related supramolecular organizing centers (SMOCs; discussed below and summarized in Fig. 2), which disrupt the stoichiometry of key signaling hubs in a way that alters the circuitry of the response. Thus, dysregulated expression of key signaling hubs within innate immune and inflammatory pathways in preleukemia and MDSs does not necessarily result in increased pathway activation, but rather short-circuits evolutionarily conserved responses to the microenvironment in a way that favors the competitive advantage of preleukemic and MDS clones. Based on this logic, the second step in pathogenesis relies on inflammatory changes in the BM microenvironment (Fig. 1). There is substantive evidence of age- and disease-related microenvironment changes within the MDS BM niche, associated with inflammatory states resulting not only from cell-intrinsic but also cell-extrinsic innate immune signaling activation. These observations suggest that MDS HSPCs persist over normal HSPCs in an environment associated with chronic inflammation.
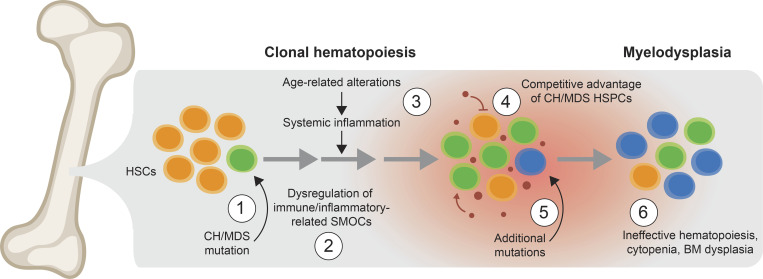
Figure 1.
Proposed model of the step-wise progression of clonal hematopoiesis to MDS. (1) An initiating mutation, such as TET2, DNMT3A, or ASXL1, occurs within an HSC (green cell). (2) The mutant (“preleukemic”) HSPCs exhibit dysregulation of key SMOCs that control innate immune and inflammatory pathways. (3) Certain diseases and conditions, such as aging, autoimmune disorders, and chronic infections, can result in systemic inflammation characterized by increased alarmins and/or cytokines. (4) Preleukemic and MDS HSPCs (green cells), which have altered their response to the systemic effects of inflammation as a result of dysregulated SMOCs that control innate immune and inflammatory pathways, gain a competitive advantage over normal HSPCs (orange cells) in an environment associated with chronic inflammation (small red circles represent inflammatory mediators). In contrast, the inflammatory environment suppresses the normal HSPCs. (5) Over time, the mutant HSPCs acquire additional mutations that may lead to MDS (blue cells). (6) At the MDS stage, the mutant HSPCs gain further competitive advantage and exhibit impaired hematopoiesis. CH, clonal hematopoiesis.
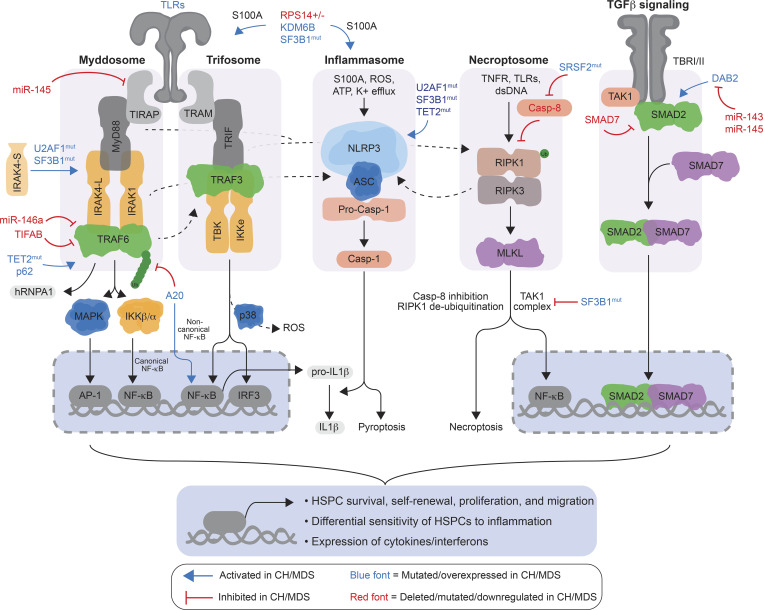
Figure 2.
Cell-intrinsic dysregulation of SMOCs involved in innate immune, inflammasome, and inflammatory-related pathways in preleukemic and MDS HSPCs. TLRs recruit the adaptors TIRAP and MyD88, along with IRAK kinases and TRAF6, to form the myddosome complex. TLR3 and TLR4 can recruit the adaptors TRIF and TRAM, along with TBK and IKKe kinases and TRAF3, to form the trifosome complex. The inflammasome serves as a platform to activate Casp-1 through pyrin domain–containing receptors (i.e., NLRPs, AIM2, and Pyrin) and the adaptor proteins ASC or NLRC4. Activated Casp-1 cleaves signaling substrates, such as pro–IL-1β, leading to pyroptosis. Initiation of necroptosis is mediated by inflammatory ligands leading to RIPK1- and RIPK3-mediated activation of MLKL (“necroptosome”), which disrupts the plasma membrane integrity. The TGF-β superfamily signals through a dual receptor system of type I (ALK1, 2, and 5) and type II (TbRII) transmembrane serine/threonine kinases, leading to activation of the SMAD transcription factors. Although these SMOCs are characterized by distinct receptors and assembling adaptors and enzymes, they converge on critical downstream effectors (i.e., NF-κB, AP-1, IRFs, and SMADs) that affect preleukemic and MDS HSPC survival, self-renewal, proliferation, and migration. In addition, dysregulation of these pathways results in a differential sensitivity of HSPCs to systemic inflammation and the competitive advantage of preleukemia and MDS HSPCs. Genes/proteins in blue font are mutated and/or overexpressed in preleukemic and MDS HSPCs. Genes/proteins in red font are deleted, mutated, and/or down-regulated in preleukemic and MDS HSPCs. Casp-8, Caspase 8; dsDNA, double-stranded DNA; mut, mutant.
One of the questions emerging from these observations is how are MDS HSPCs protected from the inflammatory milieu? Namely, what are mechanisms for the differential response and competitive advantage of MDS HSPCs during low-grade inflammation? MDS mouse models and patient samples exhibit a significant enrichment of NF-κB activation (Muto et al., 2020). Although this observation is expected as NF-κB activation is observed in MDS and normal HSPCs exposed to an inflammatory stimulus, the MDS HSPCs (human and mouse) exhibited preferential activation of noncanonical NF-κB while normal HSPCs exhibited activation of canonical NF-κB activation (Muto et al., 2020). This distinction is important, as canonical RelA/p50 and noncanonical RelB/p52 NF-κB transcriptional factors both recognize similar κB-site DNA sequences and share an overlapping set of target genes; however, the transcription factors can activate a unique set of target genes in a ligand-, cell type–, and promoter-dependent manner, resulting in distinct cellular outcomes. MDS HSPCs are preferentially sensitive to inhibition of noncanonical NF-κB signaling compared with normal HSPCs. These findings suggest that one mechanism by which MDS HSCs may persist in an inflammatory environment is by activating the noncanonical NF-κB pathway at the expense of canonical NF-κB signaling, which is the typical response of HSPCs to inflammatory stimuli.
What contributes to the shift from canonical NF-κB signaling to noncanonical NF-κB signaling in MDS HSPCs? Based on mouse models and MDS patient samples, noncanonical NF-κB activation is exclusively mediated by TNFAIP3 (i.e., A20), a dual-ubiquitin editing enzyme that suppresses TRAF6-mediated canonical NF-κB activation while simultaneously having the ability to induce noncanonical NF-κB activation by stabilization of NIK (Muto et al., 2020). This finding was initially surprising, as previous reports linked A20 to suppression of canonical NF-κB by inhibiting TRAF6; however, its effects on noncanonical NF-κB are less well understood. Notwithstanding, A20 expression in human and mouse MDS HSPCs is responsible for activating the noncanonical NF-κB pathway. More importantly, genetic loss of A20 in the MDS HSPCs resulted in suppression of noncanonical NF-κB signaling and impaired the competitive advantage of the MDS HSPCs during chronic low-grade inflammation.
The collective studies suggest that key innate immune and inflammatory pathways are rewired, not simply activated, in CHIP and MDS HSPCs, which alters their response to the systemic effects of inflammation and supports their persistence over normal HSPCs in an environment associated with chronic inflammation. The precise contribution of individual cytokines, chemokines, and bacterial metabolites on this process requires further investigation and resolution.
Dysregulation of immune-related pathways in hematopoietic premalignancies
Overview of implicated innate immune pathways
Systemic inflammation and alterations in the immune microenvironment are implicated in hematopoietic premalignancies; moreover, HSPCs with mutations in CHIP and/or MDS-related genes also exhibit differential activation of innate immune and inflammatory pathways compared with age-matched HSPCs. Microbial and cellular by-product sensing by immune effector cells is achieved by a variety of structurally unrelated pattern recognition receptors (PRRs; Janeway and Medzhitov, 2002). The majority of PRRs belong to one of the following groups: TLRs, leucine rich repeat–containing proteins (NLRs), AIM2-like receptors, and C-type lectin receptors. Despite their ability to detect diverse microbial and cellular by-product signals, the PRRs can engage distinct but also overlapping signal transduction pathways. The PRRs seed the formation of and concentrate modular oligomeric protein complexes that include a receptor, adaptors, and effector enzymes (Kagan et al., 2014; Qiao and Wu, 2015), which are referred to as SMOCs. The detailed aspects of innate immune-related signaling have been recently reviewed (Kagan et al., 2014). Briefly, SMOCs are characterized by their composition and modular nature of assembling adaptor and effector enzymes (Fig. 2). TLRs (except for TLR3), which reside on the plasma and endosomal membranes, recruit the adaptors TIRAP and MyD88, along with IRAK kinases and TRAF6, to form the myddosome complex (Fitzgerald and Kagan, 2020). Alternatively, TLR3 and TLR4 can recruit the adaptors TRIF and TRAM, along with TBK and IKKe kinases and TRAF3, to form the trifosome complex (Rubin et al., 1988; Kawasaki and Kawai, 2014).
The inflammasome serves as a platform to activate caspase 1 (Casp-1) through pyrin-domain containing receptors (i.e., NLRPs, AIM2, and Pyrin) and the adaptor proteins ASC or NLRC4 (Kaczmarek et al., 2013). Activated Casp-1 mediates cleavage of signaling substrates, such as pro-IL1β, and can lead to inflammatory-mediated cell death called pyroptosis. An alternative form of inflammatory cell death, necroptosis, has also been implicated in MDS. Initiation of necroptosis is mediated by inflammatory ligands leading to RIPK1 and RIPK3-mediated activation of MLKL (“necroptosome”), which disrupts the plasma membrane integrity (Kaczmarek et al., 2013). Lastly, the TGF-β family comprises multifunctional growth factors that regulate a range of cellular processes and immune responses. The TGF-β superfamily signals through a dual receptor system of type I (ALK1, 2, and 5) and type II (TbRII) transmembrane serine/threonine kinases, leading to transmission of signals from the receptors to target gene expression by a family of SMAD transcription factors (Vaidya and Kale, 2015). Although the SMOCs are characterized by distinct receptors and assembling adaptors and enzymes, they converge on critical pathways such as NF-κB, MAPK/ERK, AKT serine/threonine kinases, TGFβ/SMAD, and interferon regulatory factors (IRFs), resulting in changes in cellular states due to activation of downstream effectors that impact cell survival, differentiation, self-renewal, migration, and metabolism.
HSC aging and CHIP
HSCs express innate immune receptors (King and Goodell, 2011) such as TLRs and IFN receptors and directly respond to many inflammatory mediators, including LPS (Nagai et al., 2006), IFN-γ (Baldridge et al., 2010), and macrophage-CSF (Mossadegh-Keller et al., 2013). This response influences not only their division rate but also their lineage fate (Hormaechea-Agulla et al., 2020) and serves an important function in coordinating and tailoring defense responses. Intrinsic alterations in aged HSCs affect their response to inflammation and signaling through innate immune-related pathways. In human HSCs, age-associated epigenetic reprogramming, including de-repression of the transcriptional regulator KLF6, modifies the transcriptional response to immune signaling (Adelman et al., 2019). This general principle is also observed in mice, where epigenetic alterations in aged HSCs occur at loci/genes associated with inflammation (Sun et al., 2014; Chambers et al., 2007; Rossi et al., 2005); furthermore, aged HSCs demonstrate myeloid-biased “memory” of inflammatory challenge that can persist for several months, involving alterations in the transcriptional regulators KLF5, IKZF1, and STAT3 (Mann et al., 2018). In the context of aging, mitochondrial stress initiates aberrant activation of the NLRP3 inflammasome in HSCs, mediating their functional decline and impaired regenerative capacity (Luo et al., 2019), and dysregulated TGF-β signaling is strongly implicated in HSC lineage bias (Valletta et al., 2020; Challen et al., 2010). Aged HSCs also express lower levels of TNFAIP3 (A20), a negative regulator of innate immune signaling impacting myddosome, trifosome, and necroptosome effectors (Fig. 2). Heterozygous deletion of A20 in young HSCs results in characteristic features of hematopoietic aging, including expansion of the HSC pool, reduced HSPC fitness, and myeloid-biased hematopoiesis due to increased NF-κB signaling (Smith et al., 2020). Interestingly, a recent analysis identified frequent loss of one copy of TNFAIP3/A20 in aged healthy individuals with clonal hematopoiesis of Japanese descent (Smith et al., 2020), and mutations in the myddosome adaptor MyD88 have been reported in previous studies of clonal hematopoiesis (Genovese et al., 2014; Zink et al., 2017), linking innate immune dysregulation in aged HSCs and CHIP.
Dysregulation of numerous innate immune-related pathways has been implicated as a contributing factor, conferring selective advantage of HSCs and immune effector cells in CHIP, particularly in the context of TET2 mutations. For example, Tet2 mutant mouse HSPCs exhibit increased protein expression of the myddosome effector Traf6 (Muto et al., 2020), and expression of wild-type Tet2 can repress Traf6-mediated signaling (Zhang et al., 2015). Inflammasome dysregulation is also observed in the Tet2 mutant context. Macrophages in Tet2-knockout mice increase expression of NLRP3 and Casp-1 activity, which correlates with increased expression and production of IL-1β (Fuster et al., 2017). While Dnmt3a- and Asxl1-mutant HSPCs exhibit altered expression of several mediators of innate immune signaling and mice deficient for Dnmt3a have higher mortality rates after challenge with RNA viruses (Li et al., 2016), far less is understood regarding the mechanistic basis for dysregulation of innate immune signaling by these and other CHIP-associated mutations. The consequences of dysregulated innate immune and inflammatory signaling on the competitive advantage of CHIP mutant HSCs remain under intense investigation.
MDS
As observed in CHIP, dysregulation of innate immune and inflammatory signaling is a hallmark of MDS. Despite mutations in a spectrum of driver genes, MDS HSPCs exhibit differential activation of innate immune and inflammatory pathways compared with age-matched HSPCs. Several recent reviews have described in great detail the genetic and molecular evidence of cell-intrinsic dysregulation of innate immune and inflammatory pathways in MDS HSPCs (Paracatu and Schuettpelz, 2020; Ratajczak et al., 2020; Sallman and List, 2019; Barreyro et al., 2018; Ivy and Brent Ferrell, 2018; Monlish et al., 2016; Varney et al., 2015a; Gañán-Gómez et al., 2015). Dysregulation of immune-related genes in purified human HSPCs likely occurs in the majority of MDS patients; however, the implicated innate immune and inflammatory signaling pathways widely differ within MDS subtypes and disease stage.
Myddosome dysregulation in MDS
Evidence that innate immune pathway dysregulation related to the myddosome contributes to MDS resulted from work related to the del(5q) MDS genes, miR-145, miR-146a, and TIFAB (Fig. 2). As reviewed above, in normal immune cells, binding of ligand to TLRs recruits intracellular adaptors (TIRAP, MYD88, TRIF, TRAM), kinases (IRAKs, TBK), and effector molecules (TRAFs). However, deletion of miR-146a increases TRAF6 and IRAK1 mRNA and translation, while loss of TIFAB increases TRAF6 protein stability, thus resulting in overexpression and activation of TRAF6 and IRAK1 in MDS HSCs in the absence of ligand-mediated activation of the TLRs (Varney et al., 2015b; Starczynowski et al., 2010). Haploinsufficiency of miR-146a in mouse hematopoietic cells results in myeloid expansion in the BM and then progression to a BM failure and/or MDS-like disease (Zhao et al., 2011; Lu et al., 2010; Boldin et al., 2011), in part due to TRAF6 signaling (Su et al., 2020). TRAF6 is an E3 ubiquitin ligase that catalyzes the formation of lysine (K) 63-linked ubiquitin chains on itself and substrates and a central signal transducer of several innate immune receptors, including the myddosome (Fig. 2). Overexpression of TRAF6 at disease-relevant levels in hematopoietic cells is sufficient to induce HSC defects in mice that are cell intrinsic and associated with myeloid-biased differentiation (Fang et al., 2017). Unexpectedly, the hematopoietic phenotype following TRAF6 overexpression is not simply due to activation of canonical NF-κB signaling and/or chronic inflammation (see below for a detailed perspective). TRAF6-mediated ubiquitination of the RNA binding protein hnRNPA1 leads to aberrant RNA splicing of Rho family guanosine triphosphatases, Arhgap1 in mice or Arhgap17 in humans, and subsequent Cdc42 activation (Fang et al., 2017). IRAK1, a serine-threonine kinase complexed with TRAF6, is not only a target of miR-146a but is also overexpressed and activated in MDS HSPCs (Rhyasen et al., 2013). Furthermore, small-molecule inhibitors targeting IRAK1 and IRAK4 block TRAF6-mediated signaling and suppress MDS HSCPs (Rhyasen et al., 2013).
TIFAB is a fork-head–associated domain protein that is also within the deleted region on chromosome 5 (5q31.1). TIFAB binds and induces lysosomal degradation of TRAF6 protein, while deletion of TIFAB results in increased TRAF6 protein expression (Varney et al., 2015b). TIFAB deletion also results in diminished USP15 de-ubiquitinase function and consequently p53 activation in hematopoietic cells (Niederkorn et al., 2020). Deletion of TIFAB in mouse hematopoietic cells results in modest hematopoietic defects and occasionally BM failure (Varney et al., 2015b). However, concurrent hematopoietic-specific deletion of TIFAB and miR-146a results in higher levels of TRAF6 expression and innate immune pathway activation, which coincides with a highly penetrant BM failure, a diseased phenotype that more faithfully recapitulates human del(5q) MDS (Varney et al., 2017). Loss of another 5q gene, miR-145, results in de-repression of Mal/TIRAP, which is important for an initial step of myddosome formation (Kumar et al., 2011; Starczynowski et al., 2010). DIAPH1 encodes mDia1, another 5q gene within the deleted segment in del(5q) MDS. mDia1-deficient mice either alone or when codeleted with miR-146a exhibit an age-dependent granulocytopenia and myeloid dysplasia, in part through increased TLR4-IL6 signaling (Mei et al., 2018). A gene overexpressed from the intact 5q allele, SQSTM1/p62, that binds and activates TRAF6 is essential for miR-146a–deficient human and mouse HSPCs (Fang et al., 2014).
As an indirect mechanism of innate immune activation in del(5q) MDS, loss of RPS14 contributes to a p53-dependent increase of the TLR ligands S100A8/A9 (Schneider et al., 2016). However, combined Rps14, Csnk1a1, miR-145/146a deficiency recapitulates the cardinal features of the 5q− syndrome, including more severe anemia with faster kinetics than Rps14 haploinsufficiency alone and pathognomonic megakaryocyte morphology (Ribezzo et al., 2019). Macrophages are significantly increased in Rps14/Csnk1a1/miR-145/146a–deficient mice as well as in 5q− syndrome patient BM and exhibit increased expression of S100A8 and decreased phagocytic function (Ribezzo et al., 2019). Collectively, these studies highlight the critical role of dysregulated myddosome signaling in del(5q) MDS as a result of alterations in the function of several key regulators of this pathway.
Although independent studies have shown dysregulation of innate immune signaling in MDS HSPCs, the precise genetic and mutational alterations that drive innate immune signaling in MDS HSCs beyond del(5q) MDS only recently became more evident. Several recent studies have implicated dysregulation of myddosome signaling in various models of MDS. KDM6b (JMJD3), which encodes a histone demethylase that removes methyl groups on H3K27, is overexpressed in MDS CD34+ cells (Wei et al., 2018). In mouse HSPCs, KDM6B overexpression results in mild hematopoietic defects; however, chronic innate immune stimulation of KDM6B-overexpressing HSPCs with the TLR ligand LPS results in significant hematopoietic defects, including leukopenia, dysplasia, and compromised repopulating function of BM HSPCs (Wei et al., 2018). KDM6B overexpression alone results in activation of disease-relevant genes such as S100a9 in BM HSPCs; when combined with innate immune stimulation, KDM6B overexpression results in more profound overexpression of innate immune and disease-relevant genes, indicating that KDM6B was involved in a feed-forward activation of innate immune signaling in BM HSPCs. Deletion of the 20q gene STK4 (encoding Hippo kinase MST1) results in thrombocytopenia, megakaryocytic dysplasia, and a propensity for chronic granulocytosis. These studies linked the MDS/myeloproliferative neoplasm phenotype to IRAK1 signaling. MST1 interacted with IRAK1, which resulted in innate immune activation. Moreover, inhibition of IRAK1 signaling with a small-molecule IRAK1/4 inhibitor was sufficient to restore innate immune signaling in these cells (Stoner et al., 2019). Recently, induced pluripotent stem cells with ASXL1 mutations modeling distinct preleukemia (i.e., CHIP and MDS) and AML stages revealed cell-autonomous dysregulation and dependency on innate immune signaling via IRAK1/4 and TRAF6 (Wang et al., 2021).
The first direct link of an MDS-associated mutation with dysregulation of innate immune signaling was demonstrated in MDS with U2AF1 mutations (Smith et al., 2019). Mutation in splicing factor U2AF1 leads to retention of exon 4 and overexpression of a longer hyperactive isoform of IRAK4 that contributes to the function of leukemic HSPCs (Fig. 2). IRAK4 is a serine/threonine kinase that forms the myddosome complex with IRAK1 and MyD88 and mediates signaling downstream of the TLR superfamily, resulting in NF-κB and MAPK activation (Li et al., 2002). IRAK4 protein consists of the death domain (aa 1–125), a hinge domain (aa 140–150), and a kinase domain (aa 150–460). IRAK4 in MDS and AML is encoded by at least two major protein isoforms: a long isoform (IRAK4-L), which contains the three major domains, and a shorter isoform (IRAK4-S), which lacks the death domain (Fig. 2). Primary MDS/AML HSPCs express IRAK4-L, while normal BM-derived CD34+ hematopoietic cells and mononuclear cells predominantly express IRAK4-S. These findings demonstrated that a significant proportion of MDS/AML patients primarily express the longer IRAK4 RNA and protein isoforms. IRAK4-L protein isoforms retain the N-terminal death domain and therefore can directly bind MyD88 and IRAK1. Death domain interactions between MyD88 and IRAK4 initiate myddosome formation, which promotes IRAK4 trans-autophosphorylation and activation (Balka and De Nardo, 2019). IRAK4 then activates IRAK1, enabling recruitment of TRAF6 and NF-κB activation. IRAK4-S protein isoforms, which lack the N-terminal death domain, are not able to bind MyD88 and participate in seeding the myddosome. In contrast, IRAK4-L mediates maximal activation of NF-κB signaling through assembly of the myddosome complex in leukemic cells, even in the absence of receptor activation with ligands (Smith et al., 2019).
Of all MDS primary samples examined, the majority of MDS patients with U2AF1 mutations express IRAK4-L. Interestingly, ∼50% of MDS patients without splicing factor mutations also express IRAK4-L. In contrast, only a minority of healthy age-matched BM CD34+ cells express IRAK4-L (<20%). A similar phenomenon of innate immune signaling dysregulation was observed in SF3B1 mutant MDS. SF3B1 mutations promote expression of IRAK4-L but also induce mis-splicing and nonsense-mediated decay of MAP3K7 (Smith et al., 2019). MAP3K7 encodes the kinase TAK1, which, under certain cellular conditions, is required for restricting NF-κB signaling; therefore, altered splicing of MAP3K7 results in hyperactivation of NF-κB signaling in HSPCs from SF3B1 mutant mice and patient samples (Lee et al., 2018). These observations were confirmed in an orthogonal validation of MDS patients harboring SF3B1 K700E mutations. Genes in proinflammatory signaling pathways were substantially up-regulated by the SF3B1-K700E mutation in patient blast cells, most prominently the expression of S100A8. There is a growing body of evidence that spliceosome gene mutations directly and/or indirectly enhance inflammatory signaling in MDS HSPCs.
Although dysregulation of innate immune signaling MDSs as well as CHIP HSPCs can occur in the absence of ligand-receptor activation, there are numerous reports of aberrant expression of immune- and inflammatory-related receptors. For instance, in a large cohort of MDS cases, TLR1, TLR2, and TLR6 (Wei et al., 2013; Zeng et al., 2016; Maratheftis et al., 2007) are significantly overexpressed in MDS BM CD34+ cells. Furthermore, deep sequencing uncovered a recurrent genetic variant, TLR2-F217S, in ∼10% of patients (Wei et al., 2013). Functionally, TLR2-F217S results in enhanced activation of downstream signaling, including NF-κB activity after TLR2 agonist treatment, suggesting that aberrant ligand-receptor activation in MDS HSPCs can also contribute to disease. TLR2 forms a heterodimer with TLR1 or TLR6, and while overexpression of TLR2 is associated with lower-risk disease, increased expression of TLR6, but not TLR1, correlates with higher-risk disease (Monlish et al., 2020). In a mouse model of MDS driven by the NUP98-HOXD13 fusion (NHD13), TLR2 ligand exposure results in cell-autonomous TLR2 signaling that promotes the cell death of premalignant NHD13-HSPCs. In contrast, deletion of TLR2 in the NHD13 MDS mouse model accelerated transformation to AML (Monlish et al., 2018). These findings are consistent with patient data demonstrating that higher TLR2 expression is associated with increased HSPC apoptosis, lower-risk disease, and longer survival. In support of these clinical observations, chronic stimulation of TLR2/6, but not TLR1/2, results in a more rapid transformation from MDS to overt leukemia of the NHD13 mouse model. In contrast, deletion of TLR6, but not TLR1, delays transformation from MDS to AML. Mechanistically, TLR2/6 stimulation leads to cell-intrinsic and cell-autonomous expansion of premalignant HSPCs (Monlish et al., 2020).
Despite evidence that myddosome signaling is dysregulated in MDS, there is no documentation of somatic mutations involving any of the signaling components. As discussed above, gain-of-function mutations in MyD88, such as the ones commonly detected in lymphoma and Waldenstrom macroglobulinemia, are occasionally observed in CHIP (Genovese et al., 2014; Zink et al., 2017). However, these gain-of-function MyD88 mutations are rarely observed in myeloid malignancies, suggesting that the MyD88 mutant protein may contribute to clonal hematopoiesis but not to the development of MDS or AML.
Trifosome dysregulation in MDS
The trifosome complex has been implicated in the response of HSCs to chronic inflammation. In vivo stimulation of HSCs with LPS induces proliferation of dormant HSCs directly via TLR4, and sustained LPS exposure impairs HSC self-renewal and competitive repopulation activity. This process is mediated via TLR4-TRIF-ROS-p38, but not MyD88 signaling, and can be inhibited pharmacologically (Takizawa et al., 2017). Based on these observations and the modular nature of innate immune signaling proteins, it can be inferred that TRIF-dependent activation may play a role in impaired function of MDS HSCs (Fig. 2). Although there hasn’t been documentation directly implicating TRIF in the pathogenesis of MDS, dysregulation of TRIF-dependent pathways has been observed in MDS, such as IFN signaling. A global gene expression analysis of CD34+ BM cells from a large cohort of MDS patients revealed that several of the genes commonly up-regulated in the MDS patients were IFN-stimulated genes. IFIT1, IFITM1, IFI44L, and IFIT3 were up-regulated by more than twofold in at least 60% of the patients (Pellagatti et al., 2010).
Inflammasome dysregulation in MDS
The inflammasome has a key role in normal hematopoiesis as well as in myeloid malignancies, including MDS and myeloproliferative neoplasms (Ratajczak et al., 2020). Several recent reviews have extensively summarized the role of the inflammasome in MDSs (Sallman and List, 2019; Sallman et al., 2016; Banerjee et al., 2019). Of the NLR family, NLRP3 has been implicated in pyroptosis of MDS cells. Once activated, NLRP3 recruits the adaptor apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC), which initiates ASC polymerization creating filamentous, helical clusters that recruit and cleave pro–Casp-1 (Fig. 2). Active Casp-1 then converts pro–IL-1β and pro–IL-18 to their active forms, as well as the pore-forming protein gasdermin D, which permits the release of IL-1β and IL-18 and serves as the executioner of pyroptosis (Liu et al., 2016). The NLRP3-pyroptosis axis is found to be activated in MDS BM cells independent of gene driver mutation (Basiorka et al., 2016). Although these findings are paradoxical to the observed competitiveness of MDS cells, these studies propose that activation of β-catenin in MDS HSCs may provide a competitive advantage, yet the relationship between NLRP3-pyropotosis and β-catenin as a driver of MDS expansion needs further exploration. Alternatively, it is also possible that activation of NLRP3 and Casp-1–mediated processing of IL-1β in MDS HSCs occurs in the absence of pyroptosis (Evavold et al., 2018). NLRP3 is activated by diverse damage-associated molecular pattern signals, including S100A8 and S100A9, which suggests that NLRP3 may be actively primed in MDS HSPCs (Basiorka et al., 2016). However, given that stimulation of inflammasome assembly requires IRAK1 and IRAK4 (Fernandes-Alnemri et al., 2013), it is possible that preleukemic and/or MDS HSPCs are chronically primed in the absence of an extracellular priming step in these cells. Although pyroptosis is one cellular endpoint following induction of NLRP3, it will be important to discern the relative contribution of pyroptosis versus the other functions of NLRP3-mediated Casp-1 activation in MDS HSPCs.
Necroptosome dysregulation in MDS
Unlike apoptosis, necroptosis results in membrane permeabilization and release of damage-associated molecular patterns, which are then recognized by PRRs, resulting in a feed-forward cycle of cell death and inflammation. Gene expression analysis of CD34+ BM cells from MDS patients revealed that expression of MLKL, but not RIPK1 or RIPK3, was significantly overexpressed in MDS compared with healthy control CD34+ cells (Ombrato et al., 2019). Moreover, overexpression of MLKL correlated with lower hemoglobin levels, which may indicate that necroptosis-inducing inflammation may contribute to anemia in some MDS patients. Mice that are deficient in the proapoptotic BCL2 family members BAX, BAK, and BID, which are prone to necroptosis, develop hematopoietic defects related to MDS, including aberrant differentiation of BM cells, PB cytopenias, and a propensity for developing leukemia (Wagner et al., 2019). Suppression of necroptosis in these mice by reducing RIPK1 expression partly restored the hematopoietic defects, suggesting that necroptosis may contribute to the pathogenesis of MDS. Initiation of necroptosis is mediated by strictly regulated stepwise activation of RIPK1, RIPK3, and MLKL (Fig. 2). However, for activation of necroptosis to occur, the cells first require inhibition of the Caspase 8 complex; otherwise, cells default to cell death by apoptosis (Kaczmarek et al., 2013). Interestingly, SRSF2 mutations in MDS promote aberrant splicing and expression of Caspase 8 (Lee et al., 2018). Thus, it is possible that lineage-restricted hematopoietic progenitors from certain subtypes of MDS are prone to cell death mediated by necroptosis and feed-forward cycle of inflammation.
TGF-β signaling in MDS
TGF-β signaling has pleotropic effects on hematopoiesis. Mouse models that target key TGF-β signaling molecules have a critical requirement of this pathway in developmental and adult hematopoiesis, and its dysregulation has been implicated in MDS. TGF-β receptor activation triggers a regulatory circuit of activating and inhibitory SMAD proteins and increased activation of the TGF-β signaling pathway either by a loss of negative feedback or constitutive activation has been associated with the myelosuppression and ineffective erythropoiesis in MDS (Fig. 2; Bewersdorf and Zeidan, 2019). Early studies demonstrated that SMAD2 is up-regulated and overactivated in MDS HSPCs, thereby inducing unrestrained TGF-β signal activation. Moreover, SMAD7, a negative regulator of TBRI kinase, is decreased in MDS, and this leads to overactivation of TGF-β signaling even in the absence of increased levels of extracellular TGF-β (Zhou et al., 2011). In more recent studies, it has been described that there is a direct link between the loss of miR-143 and miR-145, two microRNAs within the deleted segment of chr 5q in MDS, and the activation of TGF-β signaling in del(5q) MDS HSCs. Combined loss of miR-143 and miR-145 results in enhanced activation of the TGF-β pathway in MDS HSPCs (Lam et al., 2018). Moreover, inhibition of the TGF-β pathway by interference with Dab2 or Smad3, the targets of miR-143/145, in miR-143/145−/− HSPCs inhibited activity of more mature progenitors while concomitantly inhibiting more primitive HSPCs. These findings suggest that DAB2 overexpression cooperates with other genetic aberrations in MDS to drive the disease phenotype. Although activation of the DAB2/Smad-dependent TGF-β pathway inhibited LT-HSC activity early during the progression of the disease, there was expansion of myeloid progenitor cells concomitant with progression to myeloid malignancy (Lam et al., 2018). Although TGF-β signaling normally induces negative feedback, in early-stage MDS high levels of microRNA-21 (miR-21) contribute to chronic TGF-β signaling (Muench et al., 2018). Furthermore, a TGF-β–related gene signature is sufficient to identify an MDS patient population with abnormal RNA splicing, which is independent of splicing factor mutations, but rather is correlated with aberrant expression of the RNA binding protein HNRNPK. The mechanism by which miR-21 expression induces TGF-β activation in MDS was linked to one of its targets, SKI, a transcription factor and negative regulator of TGF-β signaling (Tecalco-Cruz et al., 2018). miR-21–mediated loss of SKI results in activation of TGF-β signaling and alternative splicing to impair the competitive advantage of normal HSCs. Based on the emerging role of TGF-β activation in MDSs, modulation of TGF-β signaling is being explored for therapeutic purposes in MDSs.
Therapeutic implications of targeting immune-related pathways in hematopoietic premalignancies
Reducing selection pressure favoring CHIP mutant HSCs
Dysregulation of innate immune-related pathways, in addition to low-grade, chronic inflammation and microenvironmental factors, is implicated in clonal dominance of CHIP mutant HSCs. Thus, in therapeutic strategies to restore proper regulation of innate immune signaling in CHIP mutant HSCs, reduce chronic inflammation or alter microenvironmental factors, and/or enhance regenerative function and competitive advantage of non-CHIP mutants, aged HSCs represent ideal concepts for preclinical and translational studies. Proof-of-concept studies in murine models support that NF-κB inhibitors can prevent Tet2-knockout HSCs from expanding in response to LPS (Cai et al., 2018), and blocking IL-6 can reverse expansion of Tet2-knockout myeloid cells (Meisel et al., 2018). However, systemic treatments that impair immune system function and pathogen clearance are not appropriate in the context of prophylactic treatment in otherwise healthy aged individuals. Other, more practical strategies include boosting tumor-suppressive functions of genes mutated or lost in CHIP. For example, administration of ascorbate (vitamin C) mimics TET2 function and can reduce preleukemic changes and risk of transformation to overt leukemia in Tet2-deficient mice (Cimmino et al., 2017; Agathocleous et al., 2017). The mechanisms by which—and extent to which—this treatment modifies innate immune signaling is unknown. Emerging strategies to enhance regenerative function and competitive advantage of aged HSCs (reviewed in Akunuru and Geiger, 2016; SanMiguel et al., 2020) are the subject of active investigation to determine whether these may tip the balance back toward oligoclonal hematopoiesis, reducing the competitive advantage of CHIP mutant and/or preleukemic HSCs.
Targeting disease-associated innate immune and inflammatory pathways in MDS
Treatment of MDS patients is based on their risk profile as calculated according to the Revised International Prognostic Scoring System (IPSS-R; Greenberg et al., 2012). The scoring system predicts the course of the patient’s disease by incorporating the percentage of leukemic blasts in the BM, the type of chromosomal changes, and detailed information on the presence and type of PB cytopenias. Although the IPSS-R provides prognostic value to the disease course, it is less effective at predicting response to disease-modifying therapies. Moreover, the predictive value of the current prognostic system for MDS may be further improved by incorporating information on the “immune status” of the patients (Winter et al., 2020). Information such as dysregulation of innate immune pathways and associated inflammation and perturbations to immune responses may provide vital clinical information and potential therapeutic applications for MDS patients. As we learn more about the relationship between MDS gene mutations, dysregulated innate immune and inflammatory signaling, and the microenvironment, the status of smoldering inflammation, immunosenescence, and the microbiome may provide further prognostic value. Immunomodulatory therapies have long been used for MDS. Immunosuppressive therapy with antithymocyte globulin and in combination with prednisone or cyclosporine provides a therapeutic option for selected lower-risk patients, particularly those with hypoplastic MDS (Stahl et al., 2018). At the time of this review, there have been recent advances in preclinical and clinical studies, demonstrating the potential of inhibiting dysregulated innate immune signaling in MDS by targeting key hub mediators, such as IRAK1 and IRAK4, as well as ligands and receptors, including S100A9, CD33, IL1RAP, and TGF-β. These early studies are revealing the potential of more selective strategies to target dysregulated innate immune and inflammatory pathways in MDS. Selective IRAK4 inhibitors are being evaluated in early-stage clinical studies for MDS and AML (Curis: NCT04278768). Several IRAK4 degraders have also been developed for other indications (Kymera: NCT04440410, Pfizer: NCT02996500), but these may also show promise in subtypes of MDS exhibiting dysregulation of these pathways. Given that IRAK1 and IRAK4 are nonredundant kinases and are both critical in formation of the myddosome complex, another strategy is to target both kinases simultaneously, an effort currently being employed (https://www.kurometherapeutics.com and https://www.rigel.com). CX-01, which disrupts TLR4 activation by blocking one of its endogenous ligands, HMGB1, is under investigation in MDS (NCT02995655). Luspatercept, a recombinant fusion protein that binds TGF superfamily ligands and reduces SMAD2 and SMAD3 signaling, was recently approved for low-risk MDS with refractory anemia. In a recent phase 2 study, Luspatercept reduced the severity of anemia in patients with lower-risk MDS with ring sideroblasts who were transfusion dependent and nonresponsive to erythropoiesis-stimulating agents (Fenaux et al., 2020). Given the complexity and redundancy of immune- and inflammatory-related pathways and their modular nature of signaling, it is unlikely that targeting a selective ligand or a specific upstream immune-related pathway will have sufficient therapeutic benefit. Thus, our emerging therapies will be only as effective as our knowledge of the signaling circuitries driven by the unique genetics and molecular alterations in MDS.
Emerging questions in the field
Over the last 10 yr, significant progress has been made in understanding the contribution of innate immune pathways and inflammation to age-related hematopoietic premalignancies. There are now multiple molecular and genetic examples showing that the function of innate immune- and inflammatory-related pathways is altered in preleukemic and MDS HSPCs. More recently, substantial effort has been devoted to understanding how the dysregulation of innate immune- and inflammatory-related pathways contributes to a competitive advantage of preleukemic and MDS HSPCs and development of overt disease. Despite the tremendous progress and expansion of this emerging field, there are pressing questions that need to be answered: How do common CHIP and MDS-associated mutations (i.e., TET2, DNMT3A, and ASXL1) contribute to dysregulation of innate immune and inflammatory signaling in HSPCs and mature immune cells? Which immune/inflammatory-related pathways are best to target to prevent the competitive advantage of CHIP/MDS HSPCs? How does the modular nature of the CHIP/MDS–associated SMOCs impact HSPC function and the response to the microenvironment? How do CHIP and/or MDS HSPCs outcompete normal cells during aging and under low-grade inflammation? What is the mechanistic explanation for the paradoxical signaling that leads to induction of cell death (apoptosis/pyroptosis/necroptosis) of MDS progenitors while permitting expansion of MDS HSCs? What are age-related changes and microenvironmental cues that favor expansion of CHIP/MDS HSCs? What is the contribution of the dysregulated innate immune/inflammatory signaling pathways as the diseases progress to overt AML?
Although there are many remaining questions that will inform our current understanding of preleukemic conditions, such as CHIP and MDS, the next 10 yr are likely to lay the foundation for novel therapies. In our perspective, ideal therapies will be defined based on knowledge of the innate immune and inflammatory signaling circuitries driven by the unique molecular alterations in CHIP and MDS. While a critical node integrating these signaling circuitries may be identified and targeted to reduce the selective advantage of CHIP and MDS clones, it is perhaps more likely that multiple pathways and processes will need to be targeted concurrently to achieve both reduction in the environmental changes driving selective advantage of CHIP and MDS mutant HSCs, therein supporting oligoclonal hematopoiesis, as well as restoring proper regulation of immune- and inflammatory-related SMOCs within CHIP and MDS mutant HSCs.
Acknowledgments
We thank members of the Trowbridge and Starczynowski laboratories for helpful suggestions and editing of the review. Figures and illustrations were inspired by BioRender.com.
The authors declare no competing financial interests.
References
- Abegunde, S.O., Buckstein R., Wells R.A., and Rauh M.J.. 2018. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp. Hematol. 59:60–65. 10.1016/j.exphem.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Abelson, S., and Wang J.C.Y.. 2018. Age-related clonal hematopoiesis: implications for hematopoietic stem cell transplantation. Curr. Opin. Hematol. 25:441–445. 10.1097/MOH.0000000000000465 [DOI] [PubMed] [Google Scholar]
- Adelman, E.R., Huang H.T., Roisman A., Olsson A., Colaprico A., Qin T., Lindsley R.C., Bejar R., Salomonis N., Grimes H.L., and Figueroa M.E.. 2019. Aging Human Hematopoietic Stem Cells Manifest Profound Epigenetic Reprogramming of Enhancers That May Predispose to Leukemia. Cancer Discov. 9:1080–1101. 10.1158/2159-8290.CD-18-1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous, M., Meacham C.E., Burgess R.J., Piskounova E., Zhao Z., Crane G.M., Cowin B.L., Bruner E., Murphy M.M., Chen W., et al. 2017. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 549:476–481. 10.1038/nature23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akunuru, S., and Geiger H.. 2016. Aging, Clonality, and Rejuvenation of Hematopoietic Stem Cells. Trends Mol. Med. 22:701–712. 10.1016/j.molmed.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., and Vardiman J.W.. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Ayachi, S., Buscarlet M., and Busque L.. 2020. 60 Years of clonal hematopoiesis research: From X-chromosome inactivation studies to the identification of driver mutations. Exp. Hematol. 83:2–11. 10.1016/j.exphem.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balka, K.R., and De Nardo D.. 2019. Understanding early TLR signaling through the Myddosome. J. Leukoc. Biol. 105:339–351. 10.1002/JLB.MR0318-096R [DOI] [PubMed] [Google Scholar]
- Banerjee, T., Calvi L.M., Becker M.W., and Liesveld J.L.. 2019. Flaming and fanning: The Spectrum of inflammatory influences in myelodysplastic syndromes. Blood Rev. 36:57–69. 10.1016/j.blre.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreyro, L., Chlon T.M., and Starczynowski D.T.. 2018. Chronic immune response dysregulation in MDS pathogenesis. Blood. 132:1553–1560. 10.1182/blood-2018-03-784116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheer, F., and Vassiliou G.. 2021. Mouse Models of Myeloid Malignancies. Cold Spring Harb. Perspect. Med. 11:a035535. 10.1101/cshperspect.a035535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basiorka, A.A., McGraw K.L., Eksioglu E.A., Chen X., Johnson J., Zhang L., Zhang Q., Irvine B.A., Cluzeau T., Sallman D.A., et al. 2016. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 128:2960–2975. 10.1182/blood-2016-07-730556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewersdorf, J.P., and Zeidan A.M.. 2019. Transforming growth factor (TGF)-β pathway as a therapeutic target in lower risk myelodysplastic syndromes. Leukemia. 33:1303–1312. 10.1038/s41375-019-0448-2 [DOI] [PubMed] [Google Scholar]
- Bogeska, R., Kaschutnig P., Fawaz M., Mikecin A.-M., Buchler-Schaff M., Paffenholz S., Asada N., Frauhammer F., Buettner F., Ball M., et al. 2020. Hematopoietic stem cells fail to regenerate following inflammatory challenge. bioRxiv. 10.1101/2020.08.01.230433. (Preprint posted August 3, 2020) [DOI]
- Boldin, M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., et al. 2011. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208:1189–1201. 10.1084/jem.20101823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond, A., Skrobek B., Lobbens S., Eury E., Thuillier D., Cauchi S., Lantieri O., Balkau B., Riboli E., Marre M., et al. 2013. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat. Genet. 45:1040–1043. 10.1038/ng.2700 [DOI] [PubMed] [Google Scholar]
- Brennan, J.J., and Gilmore T.D.. 2018. Evolutionary Origins of Toll-like Receptor Signaling. Mol. Biol. Evol. 35:1576–1587. 10.1093/molbev/msy050 [DOI] [PubMed] [Google Scholar]
- Buscarlet, M., Provost S., Zada Y.F., Barhdadi A., Bourgoin V., Lépine G., Mollica L., Szuber N., Dubé M.P., and Busque L.. 2017. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 130:753–762. 10.1182/blood-2017-04-777029 [DOI] [PubMed] [Google Scholar]
- Cai, Z., Kotzin J.J., Ramdas B., Chen S., Nelanuthala S., Palam L.R., Pandey R., Mali R.S., Liu Y., Kelley M.R., et al. 2018. Inhibition of Inflammatory Signaling in Tet2 Mutant Preleukemic Cells Mitigates Stress-Induced Abnormalities and Clonal Hematopoiesis. Cell Stem Cell. 23:833–849.e5. 10.1016/j.stem.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola, M. 2020. Myelodysplastic Syndromes. N. Engl. J. Med. 383:1358–1374. 10.1056/NEJMra1904794 [DOI] [PubMed] [Google Scholar]
- Challen, G.A., Boles N.C., Chambers S.M., and Goodell M.A.. 2010. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 6:265–278. 10.1016/j.stem.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, S.M., Shaw C.A., Gatza C., Fisk C.J., Donehower L.A., and Goodell M.A.. 2007. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5:e201. 10.1371/journal.pbio.0050201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Kao Y.R., Sun D., Todorova T.I., Reynolds D., Narayanagari S.R., Montagna C., Will B., Verma A., and Steidl U.. 2019. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat. Med. 25:103–110. 10.1038/s41591-018-0267-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino, L., Dolgalev I., Wang Y., Yoshimi A., Martin G.H., Wang J., Ng V., Xia B., Witkowski M.T., Mitchell-Flack M., et al. 2017. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell. 170:1079–1095.e20. 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côme, C., Balhuizen A., Bonnet D., and Porse B.T.. 2020. Myelodysplastic syndrome patient-derived xenografts: from no options to many. Haematologica. 105:864–869. 10.3324/haematol.2019.233320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, E.K., Izukawa T., Young S., Rosen G., Jamali M., Zhang L., Johnson D., Bain E., Hilland J., Ferrone C.K., et al. 2019. Comorbid and inflammatory characteristics of genetic subtypes of clonal hematopoiesis. Blood Adv. 3:2482–2486. 10.1182/bloodadvances.2018024729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey, S.J., Minden M.D., Barber D.L., Kantarjian H., Wang J.C., and Schimmer A.D.. 2007. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat. Rev. Cancer. 7:118–129. 10.1038/nrc2047 [DOI] [PubMed] [Google Scholar]
- Desai, P., Mencia-Trinchant N., Savenkov O., Simon M.S., Cheang G., Lee S., Samuel M., Ritchie E.K., Guzman M.L., Ballman K.V., et al. 2018. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24:1015–1023. 10.1038/s41591-018-0081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZern, A.E., Malcovati L., and Ebert B.L.. 2019. CHIP, CCUS, and Other Acronyms: Definition, Implications, and Impact on Practice. Am. Soc. Clin. Oncol. Educ. Book. 39:400–410. 10.1200/EDBK_239083 [DOI] [PubMed] [Google Scholar]
- Dorsheimer, L., Assmus B., Rasper T., Ortmann C.A., Ecke A., Abou-El-Ardat K., Schmid T., Brüne B., Wagner S., Serve H., et al. 2019. Association of Mutations Contributing to Clonal Hematopoiesis With Prognosis in Chronic Ischemic Heart Failure. JAMA Cardiol. 4:25–33. 10.1001/jamacardio.2018.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshkind, K., Montecino-Rodriguez E., and Signer R.A.. 2009. The ageing immune system: is it ever too old to become young again? Nat. Rev. Immunol. 9:57–62. 10.1038/nri2471 [DOI] [PubMed] [Google Scholar]
- Esplin, B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., and Kincade P.W.. 2011. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 186:5367–5375. 10.4049/jimmunol.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold, C.L., Ruan J., Tan Y., Xia S., Wu H., and Kagan J.C.. 2018. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity. 48:35–44.e6. 10.1016/j.immuni.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J., Barker B., Bolanos L., Liu X., Jerez A., Makishima H., Christie S., Chen X., Rao D.S., Grimes H.L., et al. 2014. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Rep. 8:1328–1338. 10.1016/j.celrep.2014.07.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J., Bolanos L.C., Choi K., Liu X., Christie S., Akunuru S., Kumar R., Wang D., Chen X., Greis K.D., et al. 2017. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 18:236–245. 10.1038/ni.3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux, P., Platzbecker U., Mufti G.J., Garcia-Manero G., Buckstein R., Santini V., Díez-Campelo M., Finelli C., Cazzola M., Ilhan O., et al. 2020. Luspatercept in Patients with Lower-Risk Myelodysplastic Syndromes. N. Engl. J. Med. 382:140–151. 10.1056/NEJMoa1908892 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri, T., Kang S., Anderson C., Sagara J., Fitzgerald K.A., and Alnemri E.S.. 2013. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J. Immunol. 191:3995–3999. 10.4049/jimmunol.1301681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, K.A., and Kagan J.C.. 2020. Toll-like Receptors and the Control of Immunity. Cell. 180:1044–1066. 10.1016/j.cell.2020.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster, J.J., MacLauchlan S., Zuriaga M.A., Polackal M.N., Ostriker A.C., Chakraborty R., Wu C.L., Sano S., Muralidharan S., Rius C., et al. 2017. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 355:842–847. 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster, J.J., Zuriaga M.A., Zorita V., MacLauchlan S., Polackal M.N., Viana-Huete V., Ferrer-Pérez A., Matesanz N., Herrero-Cervera A., Sano S., et al. 2020. TET2-Loss-of-Function-Driven Clonal Hematopoiesis Exacerbates Experimental Insulin Resistance in Aging and Obesity. Cell Rep. 33:108326. 10.1016/j.celrep.2020.108326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gañán-Gómez, I., Wei Y., Starczynowski D.T., Colla S., Yang H., Cabrero-Calvo M., Bohannan Z.S., Verma A., Steidl U., and Garcia-Manero G.. 2015. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 29:1458–1469. 10.1038/leu.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T., Ptashkin R., Bolton K.L., Sirenko M., Fong C., Spitzer B., Menghrajani K., Ossa J.E.A., Zhou Y., Bernard E., et al. 2021. Interplay between chromosomal alterations and gene mutations shapes the evolutionary trajectory of clonal hematopoiesis. Nat. Commun. 12:338. 10.1038/s41467-020-20565-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese, G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371:2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grants, J.M., Wegrzyn J., Hui T., O’Neill K., Shadbolt M., Knapp D.J.H.F., Parker J., Deng Y., Gopal A., Docking T.R., et al. 2020. Altered microRNA expression links IL6 and TNF-induced inflammaging with myeloid malignancy in humans and mice. Blood. 135:2235–2251. 10.1182/blood.2019003105 [DOI] [PubMed] [Google Scholar]
- Greenberg, P.L., Tuechler H., Schanz J., Sanz G., Garcia-Manero G., Solé F., Bennett J.M., Bowen D., Fenaux P., Dreyfus F., et al. 2012. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 120:2454–2465. 10.1182/blood-2012-03-420489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla, D., Le D.T., and King K.Y.. 2020. Common Sources of Inflammation and Their Impact on Hematopoietic Stem Cell Biology. Curr. Stem Cell Rep. 6:96–107. 10.1007/s40778-020-00177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy, K.S., and Brent Ferrell P. Jr. 2018. Disordered Immune Regulation and its Therapeutic Targeting in Myelodysplastic Syndromes. Curr. Hematol. Malig. Rep. 13:244–255. 10.1007/s11899-018-0463-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, S., and Ebert B.L.. 2019. Clonal hematopoiesis in human aging and disease. Science. 366:eaan4673. 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371:2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, S., Natarajan P., Silver A.J., Gibson C.J., Bick A.G., Shvartz E., McConkey M., Gupta N., Gabriel S., Ardissino D., et al. 2017. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 377:111–121. 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway, C.A. Jr., and Medzhitov R.. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- Kaczmarek, A., Vandenabeele P., and Krysko D.V.. 2013. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 38:209–223. 10.1016/j.immuni.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Kagan, J.C., Magupalli V.G., and Wu H.. 2014. SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 14:821–826. 10.1038/nri3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., and Kawai T.. 2014. Toll-like receptor signaling pathways. Front. Immunol. 5:461. 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.Y., and Goodell M.A.. 2011. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11:685–692. 10.1038/nri3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M.S., Narla A., Nonami A., Mullally A., Dimitrova N., Ball B., McAuley J.R., Poveromo L., Kutok J.L., Galili N., et al. 2011. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood. 118:4666–4673. 10.1182/blood-2010-12-324715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, B., Hall J.M., Witte J.S., Xu Y., Reddy P., Lin K., Flamholz R., Dabbas B., Yung A., Al-Hafidh J., et al. 2015. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 126:2355–2361. 10.1182/blood-2015-08-667063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, J., van den Bosch M., Wegrzyn J., Parker J., Ibrahim R., Slowski K., Chang L., Martinez-Høyer S., Condorelli G., Boldin M., et al. 2018. miR-143/145 differentially regulate hematopoietic stem and progenitor activity through suppression of canonical TGFβ signaling. Nat. Commun. 9:2418. 10.1038/s41467-018-04831-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.C., North K., Kim E., Jang E., Obeng E., Lu S.X., Liu B., Inoue D., Yoshimi A., Ki M., et al. 2018. Synthetic Lethal and Convergent Biological Effects of Cancer-Associated Spliceosomal Gene Mutations. Cancer Cell. 34:225–241.e8. 10.1016/j.ccell.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni, C., Montagner S., Rinaldi A., Bertoni F., Polletti S., Balestrieri C., and Monticelli S.. 2017. Dnmt3a restrains mast cell inflammatory responses. Proc. Natl. Acad. Sci. USA. 114:E1490–E1499. 10.1073/pnas.1616420114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Strelow A., Fontana E.J., and Wesche H.. 2002. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. USA. 99:5567–5572. 10.1073/pnas.082100399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhang Q., Ding Y., Liu Y., Zhao D., Zhao K., Shen Q., Liu X., Zhu X., Li N., et al. 2016. Erratum: Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 17:1005. 10.1038/ni0816-1005d [DOI] [PubMed] [Google Scholar]
- Liu, X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., and Lieberman J.. 2016. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 535:153–158. 10.1038/nature18629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L.F., Boldin M.P., Chaudhry A., Lin L.L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., and Rudensky A.Y.. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 142:914–929. 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H., Mu W.C., Karki R., Chiang H.H., Mohrin M., Shin J.J., Ohkubo R., Ito K., Kanneganti T.D., and Chen D.. 2019. Mitochondrial Stress-Initiated Aberrant Activation of the NLRP3 Inflammasome Regulates the Functional Deterioration of Hematopoietic Stem Cell Aging. Cell Rep. 26:945–954.e4. 10.1016/j.celrep.2018.12.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, M., Mehta A., de Boer C.G., Kowalczyk M.S., Lee K., Haldeman P., Rogel N., Knecht A.R., Farouq D., Regev A., and Baltimore D.. 2018. Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli Are Altered with Age. Cell Rep. 25:2992–3005.e5. 10.1016/j.celrep.2018.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maratheftis, C.I., Andreakos E., Moutsopoulos H.M., and Voulgarelis M.. 2007. Toll-like receptor-4 is up-regulated in hematopoietic progenitor cells and contributes to increased apoptosis in myelodysplastic syndromes. Clin. Cancer Res. 13:1154–1160. 10.1158/1078-0432.CCR-06-2108 [DOI] [PubMed] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, Y., Zhao B., Basiorka A.A., Yang J., Cao L., Zhang J., List A., and Ji P.. 2018. Age-related inflammatory bone marrow microenvironment induces ineffective erythropoiesis mimicking del(5q) MDS. Leukemia. 32:1023–1033. 10.1038/leu.2017.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, M., Hinterleitner R., Pacis A., Chen L., Earley Z.M., Mayassi T., Pierre J.F., Ernest J.D., Galipeau H.J., Thuille N., et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 557:580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlish, D.A., Bhatt S.T., and Schuettpelz L.G.. 2016. The Role of Toll-Like Receptors in Hematopoietic Malignancies. Front. Immunol. 7:390. 10.3389/fimmu.2016.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlish, D.A., Bhatt S.T., Duncavage E.J., Greenberg Z.J., Keller J.L., Romine M.P., Yang W., Aplan P.D., Walter M.J., and Schuettpelz L.G.. 2018. Loss of Toll-like receptor 2 results in accelerated leukemogenesis in the NUP98-HOXD13 mouse model of MDS. Blood. 131:1032–1035. 10.1182/blood-2017-08-801944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlish, D.A., Greenberg Z.J., Bhatt S.T., Leonard K.M., Romine M.P., Dong Q., Bendesky L., Duncavage E.J., Magee J.A., and Schuettpelz L.G.. 2020. TLR2/6 signaling promotes the expansion of premalignant hematopoietic stem and progenitor cells in the NUP98-HOXD13 mouse model of MDS. Exp. Hematol. 88:42–55. 10.1016/j.exphem.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller, N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., and Sieweke M.H.. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 497:239–243. 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench, D.E., Ferchen K., Velu C.S., Pradhan K., Chetal K., Chen X., Weirauch M.T., Colmenares C., Verma A., Salomonis N., and Grimes H.L.. 2018. SKI controls MDS-associated chronic TGF-β signaling, aberrant splicing, and stem cell fitness. Blood. 132:e24–e34. 10.1182/blood-2018-06-860890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, T., Walker C.S., Choi K., Hueneman K., Smith M.A., Gul Z., Garcia-Manero G., Ma A., Zheng Y., and Starczynowski D.T.. 2020. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 21:535–545. 10.1038/s41590-020-0663-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., and Kincade P.W.. 2006. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 24:801–812. 10.1016/j.immuni.2006.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederkorn, M., Hueneman K., Choi K., Varney M.E., Romano L., Pujato M.A., Greis K.D., Inoue J.I., Meetei R., and Starczynowski D.T.. 2020. TIFAB Regulates USP15-Mediated p53 Signaling during Stressed and Malignant Hematopoiesis. Cell Rep. 30:2776–2790.e6. 10.1016/j.celrep.2020.01.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, L., Astrand-Grundström I., Arvidsson I., Jacobsson B., Hellström-Lindberg E., Hast R., and Jacobsen S.E.. 2000. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood. 96:2012–2021. 10.1182/blood.V96.6.2012 [DOI] [PubMed] [Google Scholar]
- Nilsson, L., Edén P., Olsson E., Månsson R., Astrand-Grundström I., Strömbeck B., Theilgaard-Mönch K., Anderson K., Hast R., Hellström-Lindberg E., et al. 2007. The molecular signature of MDS stem cells supports a stem-cell origin of 5q myelodysplastic syndromes. Blood. 110:3005–3014. 10.1182/blood-2007-03-079368 [DOI] [PubMed] [Google Scholar]
- Nimer, S.D. 2008. Myelodysplastic syndromes. Blood. 111:4841–4851. 10.1182/blood-2007-08-078139 [DOI] [PubMed] [Google Scholar]
- Ogawa, S. 2019. Genetics of MDS. Blood. 133:1049–1059. 10.1182/blood-2018-10-844621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ombrato, L., Nolan E., Kurelac I., Mavousian A., Bridgeman V.L., Heinze I., Chakravarty P., Horswell S., Gonzalez-Gualda E., Matacchione G., et al. 2019. Author Correction: Metastatic-niche labelling reveals parenchymal cells with stem features. Nature. 575:E8. 10.1038/s41586-019-1697-y [DOI] [PubMed] [Google Scholar]
- Pang, W.W., Pluvinage J.V., Price E.A., Sridhar K., Arber D.A., Greenberg P.L., Schrier S.L., Park C.Y., and Weissman I.L.. 2013. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc. Natl. Acad. Sci. USA. 110:3011–3016. 10.1073/pnas.1222861110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracatu, L.C., and Schuettpelz L.G.. 2020. Contribution of Aberrant Toll Like Receptor Signaling to the Pathogenesis of Myelodysplastic Syndromes. Front. Immunol. 11:1236. 10.3389/fimmu.2020.01236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellagatti, A., Cazzola M., Giagounidis A., Perry J., Malcovati L., Della Porta M.G., Jädersten M., Killick S., Verma A., Norbury C.J., et al. 2010. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 24:756–764. 10.1038/leu.2010.31 [DOI] [PubMed] [Google Scholar]
- Qiao, Q., and Wu H.. 2015. Supramolecular organizing centers (SMOCs) as signaling machines in innate immune activation. Sci. China Life Sci. 58:1067–1072. 10.1007/s11427-015-4951-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, M.Z., Bujko K., Cymer M., Thapa A., Adamiak M., Ratajczak J., Abdel-Latif A.K., and Kucia M.. 2020. The Nlrp3 inflammasome as a “rising star” in studies of normal and malignant hematopoiesis. Leukemia. 34:1512–1523. 10.1038/s41375-020-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, I.M., Gibson D.S., McGilligan V., McNerlan S.E., Alexander H.D., and Ross O.A.. 2018. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 9:586. 10.3389/fimmu.2018.00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyasen, G.W., Bolanos L., Fang J., Jerez A., Wunderlich M., Rigolino C., Mathews L., Ferrer M., Southall N., Guha R., et al. 2013. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 24:90–104. 10.1016/j.ccr.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribezzo, F., Snoeren I.A.M., Ziegler S., Stoelben J., Olofsen P.A., Henic A., Ferreira M.V., Chen S., Stalmann U.S.A., Buesche G., et al. 2019. Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia. 33:1759–1772. 10.1038/s41375-018-0350-3 [DOI] [PubMed] [Google Scholar]
- Rossi, D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., and Weissman I.L.. 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 102:9194–9199. 10.1073/pnas.0503280102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, J.R., Pitluk H.C., King T.A., Hutton M., Kieger E.F., Plecha F.R., and Hertzer N.R.. 1988. Carotid endarterectomy in a metropolitan community: the early results after 8535 operations. J. Vasc. Surg. 7:256–260. 10.1016/0741-5214(88)90144-9 [DOI] [PubMed] [Google Scholar]
- Sallman, D.A., and List A.. 2019. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 133:1039–1048. 10.1182/blood-2018-10-844654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallman, D.A., Cluzeau T., Basiorka A.A., and List A.. 2016. Unraveling the Pathogenesis of MDS: The NLRP3 Inflammasome and Pyroptosis Drive the MDS Phenotype. Front. Oncol. 6:151. 10.3389/fonc.2016.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, J.M., Young K., and Trowbridge J.J.. 2020. Hand in hand: intrinsic and extrinsic drivers of aging and clonal hematopoiesis. Exp. Hematol. 91:1–9. 10.1016/j.exphem.2020.09.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, S., Oshima K., Wang Y., MacLauchlan S., Katanasaka Y., Sano M., Zuriaga M.A., Yoshiyama M., Goukassian D., Cooper M.A., et al. 2018. Tet2-Mediated Clonal Hematopoiesis Accelerates Heart Failure Through a Mechanism Involving the IL-1β/NLRP3 Inflammasome. J. Am. Coll. Cardiol. 71:875–886. 10.1016/j.jacc.2017.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R.K., Schenone M., Ferreira M.V., Kramann R., Joyce C.E., Hartigan C., Beier F., Brümmendorf T.H., Germing U., Platzbecker U., et al. 2016. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 22:288–297. 10.1038/nm.4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri, A., Will B., Steidl U., and Verma A.. 2017. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 129:1586–1594. 10.1182/blood-2016-10-696062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A., Choudhary G.S., Pellagatti A., Choi K., Bolanos L.C., Bhagat T.D., Gordon-Mitchell S., Von Ahrens D., Pradhan K., Steeples V., et al. 2019. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21:640–650. 10.1038/s41556-019-0314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.A., Culver-Cochran A.E., Adelman E.R., Rhyasen G.W., Ma A., Figueroa M.E., and Starczynowski D.T.. 2020. TNFAIP3 Plays a Role in Aging of the Hematopoietic System. Front. Immunol. 11:536442. 10.3389/fimmu.2020.536442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, M., DeVeaux M., de Witte T., Neukirchen J., Sekeres M.A., Brunner A.M., Roboz G.J., Steensma D.P., Bhatt V.R., Platzbecker U., et al. 2018. The use of immunosuppressive therapy in MDS: clinical outcomes and their predictors in a large international patient cohort. Blood Adv. 2:1765–1772. 10.1182/bloodadvances.2018019414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski, D.T., and Karsan A.. 2010. Innate immune signaling in the myelodysplastic syndromes. Hematol. Oncol. Clin. North Am. 24:343–359. 10.1016/j.hoc.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Starczynowski, D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., Hirst M., Hogge D., Marra M., Wells R.A., et al. 2010. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 16:49–58. 10.1038/nm.2054 [DOI] [PubMed] [Google Scholar]
- Stoner, S.A., Yan M., Liu K.T.H., Arimoto K.I., Shima T., Wang H.Y., Johnson D.T., Bejar R., Jamieson C., Guan K.L., and Zhang D.E.. 2019. Hippo kinase loss contributes to del(20q) hematologic malignancies through chronic innate immune activation. Blood. 134:1730–1744. 10.1182/blood.2019000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y.L., Wang X., Mann M., Adamus T.P., Wang D., Moreira D.F., Zhang Z., Ouyang C., He X., Zhang B., et al. 2020. Myeloid cell-targeted miR-146a mimic inhibits NF-κB-driven inflammation and leukemia progression in vivo. Blood. 135:167–180. 10.1182/blood.2019002045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K.F., Chen R., et al. 2014. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 14:673–688. 10.1016/j.stem.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, H., Fritsch K., Kovtonyuk L.V., Saito Y., Yakkala C., Jacobs K., Ahuja A.K., Lopes M., Hausmann A., Hardt W.D., et al. 2017. Pathogen-Induced TLR4-TRIF Innate Immune Signaling in Hematopoietic Stem Cells Promotes Proliferation but Reduces Competitive Fitness. Cell Stem Cell. 21:225–240.e5. 10.1016/j.stem.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Tecalco-Cruz, A.C., Ríos-López D.G., Vázquez-Victorio G., Rosales-Alvarez R.E., and Macías-Silva M.. 2018. Transcriptional cofactors Ski and SnoN are major regulators of the TGF-β/Smad signaling pathway in health and disease. Signal Transduct. Target. Ther. 3:15. 10.1038/s41392-018-0015-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations, Department of Economic and Social Affairs, Population Dynamics . 2019. World Population Prospects 2019: Highlights. https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf
- Vaidya, A., and Kale V.P.. 2015. TGF-β signaling and its role in the regulation of hematopoietic stem cells. Syst. Synth. Biol. 9:1–10. 10.1007/s11693-015-9161-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta, S., Thomas A., Meng Y., Ren X., Drissen R., Sengül H., Di Genua C., and Nerlov C.. 2020. Micro-environmental sensing by bone marrow stroma identifies IL-6 and TGFβ1 as regulators of hematopoietic ageing. Nat. Commun. 11:4075. 10.1038/s41467-020-17942-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Egeren, D., Escabi J., Nguyen M., Liu S., Reilly C.R., Patel S., Kamaz B., Kalyva M., DeAngelo D.J., Galinsky I., et al. 2021. Reconstructing the lineage histories and differentiation trajectories of individual cancer cells in myeloproliferative neoplasms. Cell Stem Cell. 28:514–523.e9. 10.1016/j.stem.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney, M.E., Melgar K., Niederkorn M., Smith M., Barreyro L., and Starczynowski D.T.. 2015a. Deconstructing innate immune signaling in myelodysplastic syndromes. Exp. Hematol. 43:587–598. 10.1016/j.exphem.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney, M.E., Niederkorn M., Konno H., Matsumura T., Gohda J., Yoshida N., Akiyama T., Christie S., Fang J., Miller D., et al. 2015b. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J. Exp. Med. 212:1967–1985. 10.1084/jem.20141898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney, M.E., Choi K., Bolanos L., Christie S., Fang J., Grimes H.L., Maciejewski J.P., Inoue J.I., and Starczynowski D.T.. 2017. Epistasis between TIFAB and miR-146a: neighboring genes in del(5q) myelodysplastic syndrome. Leukemia. 31:491–495. 10.1038/leu.2016.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, P.N., Shi Q., Salisbury-Ruf C.T., Zou J., Savona M.R., Fedoriw Y., and Zinkel S.S.. 2019. Increased Ripk1-mediated bone marrow necroptosis leads to myelodysplasia and bone marrow failure in mice. Blood. 133:107–120. 10.1182/blood-2018-05-847335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Pine A.R., Kotini A.G., Yuan H., Zamparo L., Starczynowski D.T., Leslie C., and Papapetrou E.P.. 2021. Sequential CRISPR gene editing in human iPSCs charts the clonal evolution of myeloid leukemia and identifies early disease targets. Cell Stem Cell. 28:P1074-1089.E7. 10.1016/j.stem.2021.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Dimicoli S., Bueso-Ramos C., Chen R., Yang H., Neuberg D., Pierce S., Jia Y., Zheng H., Wang H., et al. 2013. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 27:1832–1840. 10.1038/leu.2013.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Zheng H., Bao N., Jiang S., Bueso-Ramos C.E., Khoury J., Class C., Lu Y., Lin K., Yang H., et al. 2018. KDM6B overexpression activates innate immune signaling and impairs hematopoiesis in mice. Blood Adv. 2:2491–2504. 10.1182/bloodadvances.2018024166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N., Lee J., Moore L., Baxter E.J., Hewinson J., Dawson K.J., Menzies A., Godfrey A.L., Green A.R., Campbell P.J., and Nangalia J.. 2020. Phylogenetic reconstruction of myeloproliferative neoplasm reveals very early origins and lifelong evolution. bioRxiv. 10.1101/2020.11.09.374710 (Posted November 9, 2020). [DOI]
- Winter, S., Shoaie S., Kordasti S., and Platzbecker U.. 2020. Integrating the “Immunome” in the Stratification of Myelodysplastic Syndromes and Future Clinical Trial Design. J. Clin. Oncol. 38:1723–1735. 10.1200/JCO.19.01823 [DOI] [PubMed] [Google Scholar]
- Young, A.L., Tong R.S., Birmann B.M., and Druley T.E.. 2019. Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica. 104:2410–2417. 10.3324/haematol.2018.215269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Q., Shu J., Hu Q., Zhou S.H., Qian Y.M., Hu M.H., Hu L.Y., Wang Y.G., Zhou Y.M., and Lu J.H.. 2016. Apoptosis in human myelodysplastic syndrome CD34+ cells is modulated by the upregulation of TLRs and histone H4 acetylation via a β-arrestin 1 dependent mechanism. Exp. Cell Res. 340:22–31. 10.1016/j.yexcr.2015.12.008 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., Zhao D., Liu Y., Wang C., Zhang X., et al. 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 525:389–393. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.R.C., Nix D., Gregory M., Ciorba M.A., Ostrander E.L., Newberry R.D., Spencer D.H., and Challen G.A.. 2019. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp. Hematol. 80:36–41.e3. 10.1016/j.exphem.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T.Y., Dutta R., Benard B., Zhao F., Yin R., and Majeti R.. 2020. IL-6 blockade reverses bone marrow failure induced by human acute myeloid leukemia. Sci. Transl. Med. 12:eaax5104. 10.1126/scitranslmed.aax5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.L., Rao D.S., Boldin M.P., Taganov K.D., O’Connell R.M., and Baltimore D.. 2011. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA. 108:9184–9189. 10.1073/pnas.1105398108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L., McMahon C., Bhagat T., Alencar C., Yu Y., Fazzari M., Sohal D., Heuck C., Gundabolu K., Ng C., et al. 2011. Reduced SMAD7 leads to overactivation of TGF-beta signaling in MDS that can be reversed by a specific inhibitor of TGF-beta receptor I kinase. Cancer Res. 71:955–963. 10.1158/0008-5472.CAN-10-2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T., Kinney M.C., Scott L.M., Zinkel S.S., and Rebel V.I.. 2015. Revisiting the case for genetically engineered mouse models in human myelodysplastic syndrome research. Blood. 126:1057–1068. 10.1182/blood-2015-01-624239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, F., Stacey S.N., Norddahl G.L., Frigge M.L., Magnusson O.T., Jonsdottir I., Thorgeirsson T.E., Sigurdsson A., Gudjonsson S.A., Gudmundsson J., et al. 2017. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 130:742–752. 10.1182/blood-2017-02-769869 [DOI] [PMC free article] [PubMed] [Google Scholar]