Cells constantly sense their environment to adapt their behavior to changing needs. Kull and Schroeder discuss the requirements, potential, and limitations of current technologies for quantification and manipulation of cellular signaling in the context of inflammatory signaling in hematopoietic cells.
Abstract
Cells constantly sense their environment, allowing the adaption of cell behavior to changing needs. Fine-tuned responses to complex inputs are computed by signaling pathways, which are wired in complex connected networks. Their activity is highly context-dependent, dynamic, and heterogeneous even between closely related individual cells. Despite lots of progress, our understanding of the precise implementation, relevance, and possible manipulation of cellular signaling in health and disease therefore remains limited. Here, we discuss the requirements, potential, and limitations of the different current technologies for the analysis of hematopoietic stem and progenitor cell signaling and its effect on cell fates.
Cellular signaling
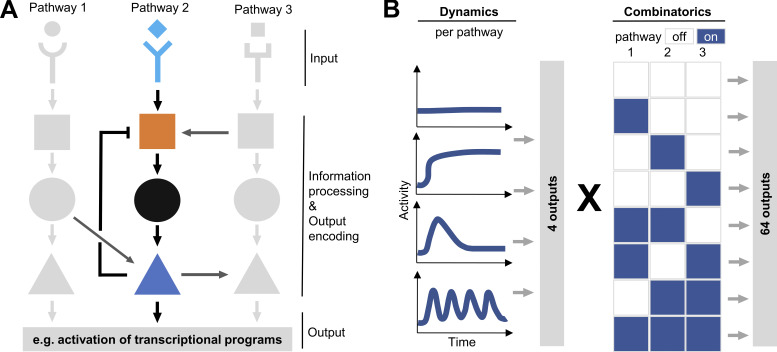
Cells must adapt their behavior to the needs of the organism. These needs are always changing, not only during development and aging, but throughout life even in seemingly homeostatic conditions. Cells constantly sense their environment through receptors for different chemical and physical components. These receptors activate signaling pathways, which activate molecular target programs initiating the cells’ responses (Fig. 1 A). Relevant environmental stimuli are highly diverse, complex, and dynamic, and cells must be able to compute finely tuned output responses, both as individual cells and coordinated, as a population, from these combined and changing inputs. This response computation happens at the level of signaling pathways, which translate the different combinations of inputs into output programs (Landry et al., 2015). While many receptors exist in our genomes, individual cells express far fewer receptors than the number of different environmental situations they can react to. In addition, an even more limited number of cell-intrinsic signaling pathways are available. As a result, although the activation of different receptors leads to different cellular responses, they often activate a highly overlapping set of pathways. To enable the required high number of fine-tuned responses with the limited set of existing pathways, these are wired in complex interconnected networks. Their activity is nonlinear, context-dependent, dynamic, and heterogeneous, even between closely related individual cells (Tay et al., 2010; Lane et al., 2017; Mitra et al., 2017; Wang et al., 2021), enabling a much-increased number of combinatoric and dynamic outputs (Fig. 1 B). A further increase in output possibilities is achieved by modulating the quantity and quality of the input ligands or receptors (Ho et al., 2017; Purvis et al., 2012). For example, a change in the topology of erythropoietin dimers by variation of distance and angle between monomers changes how the receiving pathways are activated (Kim et al., 2017; Moraga et al., 2015).
Figure 1.
Complex signaling pathway network wiring increases output encoding possibilities. (A) Ligand-receptor binding activates specific signaling pathways. Variation of ligand and receptor configurations and concentrations increase the number of possible outputs. Feedback/forward loops and inter-pathway crosstalk lead to complex pathway activity patterns and make the comprehensive analysis of signal transduction mechanisms extremely challenging. (B) Individual receptors can activate combinations of multiple signaling pathways, each with different possible activity dynamics. This increases the number of distinct possible outputs encoded by the limited number of available signaling pathways, allowing cells to fine-tune their responses to numerous different external signaling inputs.
Inflammatory signaling in hematopoietic stem and progenitor cells (HSPCs)
Inflammatory signaling in HSPCs is activated upon detection of injury or infection and regulates the appropriate cellular responses. Various pathways, most prominently NF-κB, JAK/STAT, protein kinase B (AKT), and p38, are triggered by pathogen-associated molecular patterns or pro-inflammatory cytokines like TNF-α, IFNs, and ILs (Hemmati et al., 2017). Pathways lead to the expression of an array of target genes involved in pathogen elimination, immune cell recruitment, and a general adaptation of cell state and fate (Newton and Dixit, 2012). Inflammatory signaling controls HSPC fates (Endele et al., 2014; Rieger et al., 2009; Takizawa and Manz, 2017; Trumpp et al., 2010), and misregulation is associated with stem cell exhaustion, malignant transformation, and other disorders (Hemmati et al., 2017; Takizawa et al., 2012). For example, both IFN-γ and IFN-α activate quiescent hematopoietic stem cells (HSCs) and trigger cycling (Baldridge et al., 2010; Essers et al., 2009) and emergency megakaryopoiesis (Chicha et al., 2004; Haas et al., 2015; Manz, 2008). LPS-induced NF-κB activation triggers apoptosis in T cells but proliferation in the common lymphoid progenitor population (Chandra et al., 2008; Welner et al., 2008). TNF-α promotes HSC survival but induces apoptosis in granulocyte monocyte progenitors (Yamashita and Passegué, 2019), and a correct NF-κB response seems crucial for ex vivo HSC expansion (Chagraoui et al., 2019). Inflammatory signaling is used as a target for therapeutic manipulation, most successfully by small molecules or peptides. For example, JAK2 inhibitors treat JAK2-induced myeloproliferative neoplasms (Kiladjian et al., 2016), and the p38 inhibitor ralimetinib reduced the proliferative effects of IL-1 in acute myeloid leukemia (AML) in preclinical studies (Carey et al., 2017). Inhibition of NF-κB signaling by parthenolide shows activity in AML stem cells (Guzman et al., 2005). Differentiation in myelodysplastic syndrome and AML can be induced by TLR agonists (Hemmati et al., 2017). For a more exhaustive discussion of the many effects of signaling, please refer to the other reviews of this series.
Thus, signaling is a core regulator of HSPC fate choices in health and disease, and its comprehensive understanding also holds the potential for the development of drugs for improved therapeutic manipulation. However, much remains unknown about exact signal transduction mechanisms, crosstalk and combinatorics of pathways, and how specific fate changes are encoded by these signals. This is due to the complexity of signaling networks and the required theoretical and experimental approaches for their analysis. Here, we discuss some of the requirements, potential, and limitations of current technologies for the quantification of signaling components and activity and their relevance for HSPC fate control.
Technological requirements
Analyzing signaling networks and their functional implementation is notoriously difficult. As discussed above, individual activated receptors usually activate multiple parallel and interconnected pathways, each with complex regulation through cascades of biochemical layers and feed-forward and feedback loops (Fig. 1 A). This wiring leads to many possible dynamics and combinations, which massively increase the amount of information that can be encoded from the limited number of available signal transduction pathways (Fig. 1 B). Short- and long-range feedback signaling between cells of a tissue or even the whole organism makes this even more complex (Charest and Firtel, 2006). This increases both the signalings’ potential for accurately controlling biology and the experimental requirements for its comprehensive analysis. How information is encoded, how specificity is achieved (Behar and Hoffmann, 2010), and how it is possible to integrate many input combinations and their spatiotemporal dynamics over time to generate accurate spatiotemporal outputs remain some of the big central mysteries in life sciences. This is due not only to the limitations in mathematical and computational tools for understanding nonlinear dynamic control systems but also to the lack of technologies for the quantification of the many signaling components and associated cell fate choices with the required spatiotemporal resolution, dimensionality, and throughput.
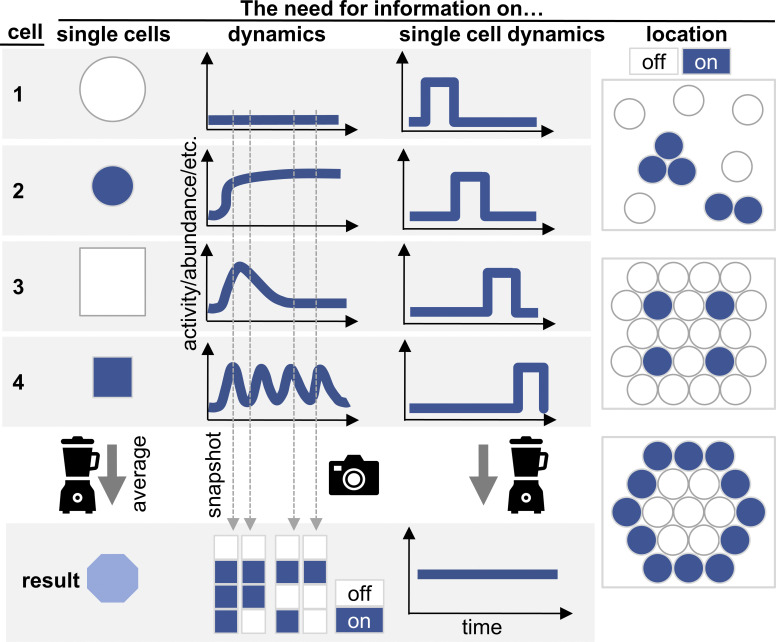
Ideally, signaling is quantified (1) at the single-cell level, (2) continuously, (3) with spatial information of the cells in their environment, and (4) linked to past and future molecular and cellular behavior of the analyzed cells (Fig. 2).
Figure 2.
Continuous multiplexed spatial single-cell quantification is required for a comprehensive analysis of signaling activities. Ideally, signaling is quantified at the single-cell level (left), continuously (middle) and with spatial information of the cells in their environment (right). Analysis of population, temporal and/or spatial averages can obscure true signaling activities (bottom).
Since signaling is usually heterogeneous between individual cells even in seemingly homogeneous and closely related cells, the analysis of population averages is insufficient (Etzrodt et al., 2014; Hoppe et al., 2014; Loeffler and Schroeder, 2019; Schroeder, 2005, 2008; Fig. 2). For example, there is a linear correlation between the TNF concentration used for stimulation and the NF-κB response amplitude of the whole stimulated 3T3 fibroblast population, suggesting an analogue NF-κB response. However, analysis at the single-cell level demonstrated an entirely different regulation: Individual cells respond in a digital all-or-nothing on/off fashion, and only the frequency of responding cells is correlating with the TNF concentrations (Tay et al., 2010). Furthermore, the current geometric state of individual cells can influence their transcriptional responses to TNF stimulation (Mitra et al., 2017).
In addition, signaling pathways are active not only in an on/off type fashion but also with different dynamics (e.g., sustained, transient, oscillatory; Fig. 1 B; Behar and Hoffmann, 2010; Purvis and Lahav, 2013). If done only at individual time points, even single-cell analyses will miss this crucial information (Fig. 2; Hoppe et al., 2014; Loeffler and Schroeder, 2019; Purvis and Lahav, 2013; Schroeder, 2005, 2011). These signaling activity dynamics can be specific for the target cell type or stimulus used but are typically heterogeneous between individual cells. Importantly, these different dynamics seem to activate different molecular target programs (Hoffmann et al., 2002), and to cause specific cell fates, even though only one pathway is activated (Marshall, 1995; Nandagopal et al., 2018; Purvis et al., 2012). For inflammatory signaling, different ligands trigger NF-κB signaling, but with different dynamics, and different NF-κB activity dynamics are followed by the activation of distinct transcriptional target programs in the RAW 264.7 cell line and fibroblasts (Hoffmann et al., 2002; Lane et al., 2017; Werner et al., 2005). Even if all cells exhibited the same response dynamics, but in an asynchronous way, time course quantification of population averages would lead to wrong conclusions (Fig. 2).
Also, cellular responses are typically dependent on their location within a tissue or a culture dish (Battich et al., 2015), thus requiring analysis technologies to also provide that spatial information (Fig. 2). The generation and effects of spatiotemporally layered patterns of signaling activation and its coordinated propagation through 2D cultures and 3D tissues are even more complex, fascinating, crucial for normal biology, and ill-understood. The best in vivo examples come from embryology in simpler model organisms as well as vertebrates and mammals (Eldar et al., 2002; Golan et al., 2018; Hashimura et al., 2019; Mongera et al., 2019; Pohl and Bao, 2010; Schier, 2001; Chen and Schier, 2001).
In vitro assays, while providing better cell accessibility, throughput, and well-defined chemical conditions, only partly reflect conditions in vivo, and adjacent niche cells as well as surrounding factors typically differ substantially between the two (Juno et al., 2019). Therefore, in vitro findings have to be validated by in vivo analyses of cells in their native 3D environments. However, in vivo analyses are extremely complex, with many unknown and usually confounding influences on cells. Cell and molecular systems in vivo will be redundant and quickly adapt to, e.g., chemical or genetic manipulation, making it almost impossible to analyze the precise immediate effect of a defined manipulation. The combination of simpler and chemically defined in vitro assays with in vivo studies will therefore often be required.
In vivo analysis is especially difficult for the hematopoietic system, where most tissues are (semi)liquid, are constantly mixing, and lack the obvious compartmentalized and stratified 3D organization of other tissues. The location and morphology of a cell in its tissue can reliably predict its type, environment, and signaling inputs and responses in many solid tissues (Barker et al., 2007; Quiroz et al., 2020; Sato et al., 2011). However, this is not the case, or not yet understood, for most of the highly motile hematopoietic cell types, which also share very similar in vivo morphologies. Indeed, the location of HSPCs in hematopoietic tissues, as well as the location and cellular and molecular composition of their niches, remain disputed in mice and unknown in humans (Acar et al., 2015; Hérault et al., 2017; Kokkaliaris et al., 2020).
Available technologies
Snapshot analyses of population averages
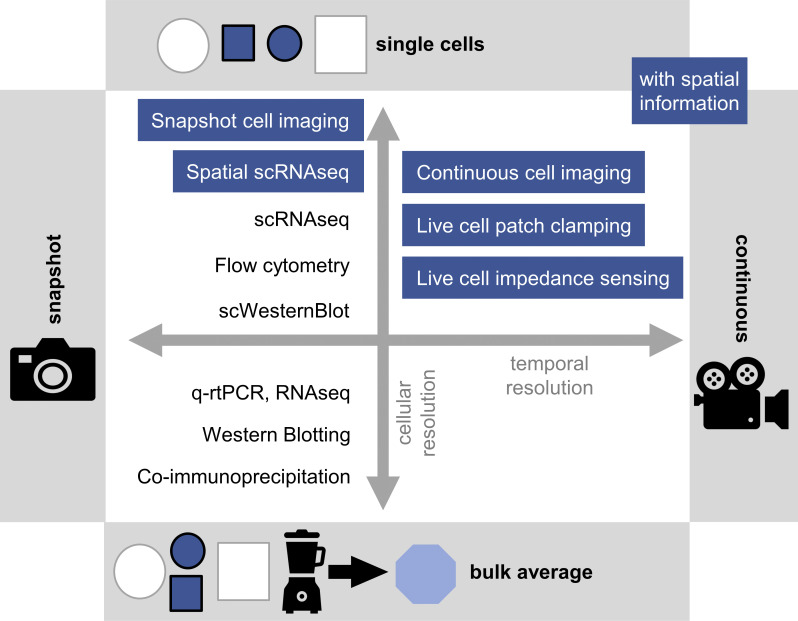
Quantitative PCR and RNA sequencing technologies enable detection of signaling target genes, but not of the activity of protein pathway components (Wu et al., 2014; Ziegenhain et al., 2017). While direct proteome analyses can analyze averages of large cell populations (Blagoev et al., 2003), specific signaling proteins are typically detected by antibodies (ABs). Western blotting identified expression and activity of signaling proteins (Janes, 2015; Renart et al., 1979), e.g., by detecting their phosphorylation (Li et al., 2006), and coimmunoprecipitation can detect their molecular interactions (Bonifacino et al., 1999; Laird et al., 2009; Lee, 2007). These approaches provided fundamental insights about signaling pathways (Danial et al., 1995; Zhang et al., 1997). However, while single-cell Western blot technology with limited sensitivity exists (Hughes et al., 2014), these analyses of bulk populations are not suitable for rare cell populations and miss crucial information about individual cells (Fig. 3).
Figure 3.
Technologies for analyzing signaling at different levels of resolution. Technologies often used for analysis of signaling, categorized by their capacity for providing single-cell, temporal, and spatial information. Continuous single-cell technologies yield most relevant information but are usually lower in throughput and/or dimensionality than other methods. Blue text boxes labels technologies that additionally retain spatial information. q-rtPCR, quantitative RT-PCR; RNAseq, RNA sequencing; scRNAseq, single-cell RNA sequencing; scWesternBlot, single-cell Western Blot.
Snapshot single-cell analyses
Flow cytometry is a core method for single-cell quantification, with high-throughput multiplexed protein detection. Fluorescent labels allow quantification of intracellular (cells are killed in the process; Firaguay and Nunès, 2009) and surface proteins, and sorting of living cells (Bonner et al., 1972; Herzenberg et al., 1976). Imaging flow cytometry adds useful information on cell morphology and subcellular molecule location (Doan et al., 2018; McGrath et al., 2008). However, fluorescent labels have limited multiplexing potential due to their spectral overlaps. Mass cytometry (cytometry by time of flight [CyTOF]) improves multiplexing by labeling ABs with heavy metal ions, distinguished by mass spectrometry with very high precision (Bandura et al., 2009; Forthun et al., 2019; Yao et al., 2014). CyTOF was used, for instance, to comprehensively quantify signaling behavior in different normal bone marrow HSPCs (Bendall et al., 2011), or to detect anti-tumor immune responses (Dempsey, 2017). However, CyTOF destroys cells and is thus blind to possible links between the cells’ current molecular state and future behavior. Labeling ABs by DNA sequences and their detection by high-throughput sequencing increases multiplexing and throughput potential even more (Peterson et al., 2017; Stoeckius et al., 2017; Todorovic, 2017). These approaches have the potential to replace most of the current flow cytometry experiments for snapshot cell analysis, but not for live-cell sorting, since cells get destroyed in the process. All these analyses of dissociated cells lose the information on cells’ locations in their tissue or culture, which is crucial to understand signaling regulation and cell–cell communication (Fig. 3).
Spatial single-cell analysis
Imaging provides this spatial information including molecules’ subcellular location. Fluorescent ABs are the most used label for imaging signaling components (Coons et al., 1942; Coons and Kaplan, 1950). For HSPCs, much remains to be done for the in situ quantification of their locations and signaling activities induced by their microenvironments in vivo. Despite numerous publications, the exact location of HSCs and their niches in mouse bone marrow remains disputed (Acar et al., 2015; Boulais and Frenette, 2015; Kokkaliaris et al., 2020; Méndez-Ferrer et al., 2010; Morrison and Scadden, 2014). The locations and niches of other HSPC types have not yet been well analyzed in mice, and almost no studies exist for human hematopoietic organs. This is mostly due to lacking technology. 3D imaging usually only allows a very limited number of fluorescent channels to be distinguished. Together with the lack of single markers for the identification of specific HSPC types—combinations of several markers are usually required for their reliable identification—this prevents the simultaneous identification of specific cell types, their microenvironment, and signaling components required for understanding the spatiotemporal distributions of signaling activities in specific HSPCs in vivo. In addition, due to the acquisition time requirements on expensive machinery and the huge amounts of data resulting from imaging approaches, only small spatial volumes are typically imaged, limiting the required sample numbers for representative data acquisition and statistically sound conclusions. Recently, imaging approaches for large 3D volumes, such as whole mouse bones with up to 10 simultaneous fluorescent channels, and custom software pipelines for the quantitative analysis of the resulting huge data volumes have been developed (Coutu et al., 2017, 2018; Nombela-Arrieta et al., 2013). This indeed demonstrated that previous conclusions about HSC location bone marrow have to be taken with caution (Kokkaliaris et al., 2020). Recently, the quantification of 3D distributions of secreted regulatory proteins in large tissue volumes with even single-molecule sensitivity was developed (Kunz and Schroeder, 2019), based on proximity ligation assay amplification of fluorescent signals (Söderberg et al., 2006). This will now also allow quantifying where signaling ligands actually locate in hematopoietic tissues, greatly improving our understanding of the spatiotemporal concentrations and availability of these signals to HSPCs in their native in vivo environments.
Cyclic immunostainings with several rounds of AB exposure increase multiplexing (Gut et al., 2018; Lin et al., 2015), and limited fluorescence multiplexing potential can be overcome by mass cytometry detection of metal-labeled ABs. Stepwise scanning of tissue sections by sequential laser ablation of AB-stained tissue voxels with subsequent mass spectrometry and computational image reconstruction can generate 2D images with highly multiplexed protein quantification per pixel (Giesen et al., 2014). This approach was used to map the immune system changes in type 1 diabetes (Wang et al., 2019) and to phenotype breast cancer (Ali et al., 2020), and is continuously improving in scanning speed, multiplexing ability, and possible 3D imaging (Bodenmiller, 2016; Lun and Bodenmiller, 2020). For the detection of mRNAs, single-molecule RNA fluorescence in situ hybridization imaging (Raj et al., 2008) and spatial transcriptomics (Ståhl et al., 2016) provide spatial information with subcellular resolution. Cyclic fluorescence in situ hybridization staining and imaging enable very high multiplexing and allow absolute quantification of mRNA molecule numbers per cell (Lubeck and Cai, 2012; Xia et al., 2019). Spatial transcriptomics physically barcode 3D tissue positions with subcellular resolution before dissociation and later match these barcodes to single-cell sequencing results (Vickovic et al., 2019). This approach has enabled the identification of cells’ previous tissue location and helped to identify a plaque-induced gene network in Alzheimer’s disease (Chen et al., 2020). All of these technologies provide snapshot observations at single time points. However, for quantification of the important dynamic behavior of signaling networks, continuous analyses are required (Fig. 3).
Continuous long-term single live-cell quantification
Patch clamping (Neher and Sakmann, 1992) or impedance sensing (Xu et al., 2016) can measure cellular electrical properties over time and are therefore able to detect concentrations of second messengers like Ca2+ in real time (Franks et al., 2005; Frey et al., 2010). However, fluorescent live-cell imaging is the most abundantly used technology for continuous single live-cell analysis, even for up to weeks (Kokkaliaris et al., 2016; Schroeder, 2011). Genetically encoded biosensors can be used to quantify even very fast signaling activity dynamics. They typically report signaling activity either by changing fluorescence resonance energy transfer (Fritz et al., 2013), leading to a spectral change of signaling reporter emission (Krause et al., 2013; Tuleuova et al., 2010), or by changing the subcellular location of the biosensor. Fluorescence resonance energy transfer sensors can also be modified to detect the abundance of small molecule secondary signaling messengers, like cyclic di–GMP, and therefore expand the detection limit beyond proteins (Anderson et al., 2020; Dippel et al., 2018). Translocation-based reporters often have a higher dynamic range and require only one fluorescent channel, thus improving multiplexing with other reporters. They are usually generated by adding a fluorescence tag to a key signaling protein, thus changing its intracellular location upon pathway activation (e.g., NF-κB [De Lorenzi et al., 2009], AKT [Gross and Rotwein, 2015], and STAT3 [Herrmann et al., 2004; Watanabe et al., 2004]). They have provided important insights into the dynamical encoding of signal information. For example, a GFP-p65 reporter showed that different NF-κB dynamics correlate with the expression of specific target gene programs (Lane et al., 2017) and that NF-κB signaling can be entrained in cell populations, which are also regulated by noise (Heltberg et al., 2016). An AKT translocation sensor was employed to screen for the effect of growth factors on signaling dynamics (Gross and Rotwein, 2016). A new generation of translocation biosensors is based on a fusion between fluorescent proteins and a protein fragment containing nuclear export and localization signals that change activity upon phosphorylation by kinases of signaling pathways. This approach can be applied to report the activity of essentially any kinase and thus many signaling pathways by adjusting the kinase docking site in the protein complex (Regot et al., 2014).
Importantly, the relevance of signaling dynamics for controlling cell fates can only be evaluated if the future fates of the same cells are known. Only if specific signaling outputs reliably lead to specific future fates of the same cell or its progeny can their role in inducing this fate be demonstrated. However, due to the slow implementation of fate programs, especially in mammals, it can take up to days or weeks until the lineage choice of a differentiating HSC can be detected (Hoppe et al., 2016; Kokkaliaris et al., 2016; Strasser et al., 2018). The same cells (and all of their progeny) in which signaling had been quantified at the start of the experiments have to be tracked without losing their identity over these long periods of time. This is technically much more demanding than the relatively short-term live-cell imaging for the identification of signaling activity or dynamics, which only has to span minutes to hours (Endele et al., 2014; Endele and Schroeder, 2012; Schroeder, 2005, 2011). Long-term single-cell imaging and fate quantification of mammalian HSPCs has been accomplished in vitro, for instance demonstrating the production of blood cells from hemogenic endothelium (Eilken et al., 2009), the lineage instruction of hematopoietic progenitor cells by inflammatory cytokine signaling (Endele et al., 2017; Rieger et al., 2009), the asymmetric cell division of HSCs (Loeffler et al., 2019), or the direct regulation of the core hematopoietic transcription factor PU.1 by TNF and IL-1 in HSCs (Etzrodt et al., 2019; Pietras et al., 2016). This approach also enables the identification and quantification of signaling dynamics in HSPCs (Endele et al., 2014; Wang et al., 2021). Combining protein analyses for detecting signaling activity with single-cell high-dimensional snapshot endpoint analysis methods like transcriptome sequencing (Wu et al., 2014; Ziegenhain et al., 2017) enables the comprehensive detection of molecular regulators and target programs of signaling pathways. It was used to show that specific NF-κB dynamics correlate with target gene expression (Lane et al., 2017). Detection of signaling differences even between closely related cells can further be used to distinguish cellular subpopulations that are not distinguishable by flow cytometry or other assays. In addition, it could contribute important insights to the ongoing discussion as to whether hematopoietic differentiation is a discrete process (with fast changes in relevant cell states) or a continuous one (with slow changes across the transcriptome and variable [de]activation sequences for individual genes), as suggested by recent transcriptome sequencing studies (Laurenti and Goettgens, 2018; Liggett and Sankaran, 2020).
While long-term single-cell imaging is possible in vitro, it remains even more demanding in vivo. In addition to the more difficult access to cells deeper in tissues, the required immobilization of animals or even patients typically prevents experiments longer than a day. Live HSPC in vivo imaging is possible (Lo Celso et al., 2009), but with much more limited duration and throughput, and signaling activity and links to future cell fates have not yet been analyzed. Due to its easier accessibility at the outside of the organism, single-cell imaging of skin (Rompolas et al., 2012; Sun et al., 2017) enables insights into the changing signaling activities of differentiating primary skin stem and progenitor cells in vivo (Quiroz et al., 2020).
Manipulation of cellular signaling
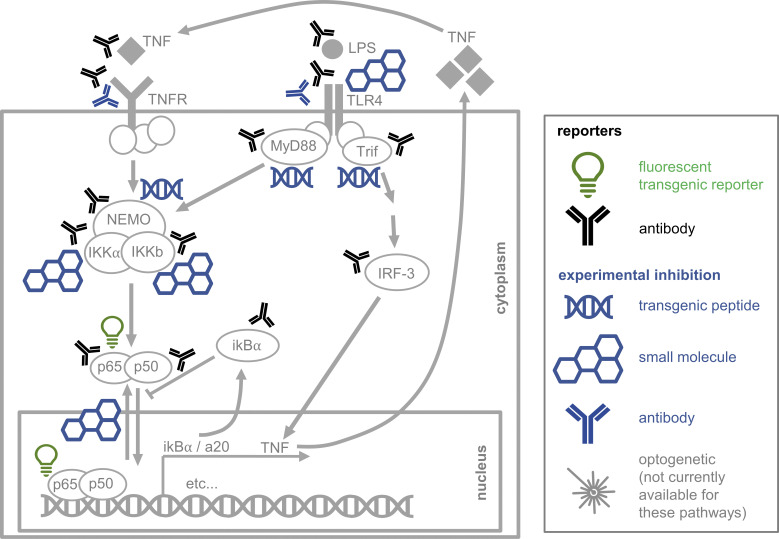
For a full understanding of signal transduction mechanisms, it is usually not enough to only observe. Instead, defined perturbations are necessary to reveal signaling properties that are otherwise hidden. Classic transgenic manipulations for knockdown, knockout, and overexpression of pathway components in animal models provide important insights (Gao et al., 2019; Yamamoto et al., 2003). In humans, insights into signaling regulation and relevance came from “natural” pathway perturbations, due to disease-causing germline or somatic mutations that alter signaling (Katoh, 2007; Pencik et al., 2016; Zhao et al., 2017). Signaling pathways can simply be stimulated with their ligands in vitro and, with much less control over concentrations, kinetics, and combinations, in vivo. In addition, pharmacologic activation or inhibition by small molecules, peptides, or ABs is an extremely successful addition, both for research and for clinical therapy (Fig. 4; Carey et al., 2017; Kiladjian et al., 2016; Nelson et al., 2004; Purvis et al., 2012). Technologies for efficient, automated, and time-controlled (micro)fluidic handling can be required here to enable automated high-throughput manipulation of media compositions, in particular with defined fast changes over time. For NF-κB, it was shown that the pathway entrains to external periodic TNF-α stimulations, which were achieved with microfluidic cell culture devices (Heltberg et al., 2016; Kellogg and Tay, 2015). However, most microfluidic devices are not suitable for use with rare nonadherent cells like primary HSPCs, which require efficient cell capture and flow-free media changes with sufficient speed (Dettinger et al., 2018, 2020). Optogenetic approaches are a very exciting approach allowing the experimental switching between signaling activity states by light exposure with single-cell resolution and extremely fast kinetics (Boyden et al., 2005; Bugaj et al., 2013). It has been used, e.g., to precisely control the activity of ERK and AKT pathways by light exposure (Johnson and Toettcher, 2019; Katsura et al., 2015), and will allow many important insights in the future.
Figure 4.
Simplified representation of the inflammatory NF-κB pathway with currently available detection and manipulation methods. After binding of TNF or LPS to their receptors, the pathway is activated and eventually releases the active transcription factor p65-p50, a heterodimer, into the nucleus, where transcription of many target genes is triggered. Tools for manipulation and quantification are available for various pathway components.
Data analysis
While it cannot be covered in more depth here, it is important to point out that the analysis of the data generated by these technologies, in particular by high-throughput and time-resolved single-cell approaches, is critically dependent on computational data analysis, modeling, and curation (Hilsenbeck et al., 2016; Skylaki et al., 2016). This, and not the data acquisition, often is the relevant bottleneck for comprehensive insights, and the required software is typically not readily available. Researchers must therefore be able to develop or adjust the required software to their needs.
Concluding remarks
Cellular signaling is central for controlling cell fates. While we already understand a lot, and targeted manipulation of signaling is successfully used even for clinical therapy, much more remains to be analyzed and understood. Recent technological developments, in particular for single-cell analyses and dynamics quantification and manipulation, now enable improved insights into the regulation and function of cellular signaling in controlling cell fates. While often developed in easy-to-use model systems, they are increasingly applied to also analyze primary cells including HSPCs and will lead to important novel insights and the development of improved targeted therapeutic manipulations.
Acknowledgments
T. Kull and T. Schroeder wrote the manuscript.
References
- Acar, M., Kocherlakota K.S., Murphy M.M., Peyer J.G., Oguro H., Inra C.N., Jaiyeola C., Zhao Z., Luby-Phelps K., and Morrison S.J.. 2015. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 526:126–130. 10.1038/nature15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, H.R., Jackson H.W., Zanotelli V.R.T., Danenberg E., Fischer J.R., Bardwell H., Provenzano E., Rueda O.M., Chin S.-F., Aparicio S., et al. 2020. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Can. 1:163–175. 10.1038/s43018-020-0026-6 [DOI] [PubMed] [Google Scholar]
- Anderson, W.A., Dippel A.B., Maiden M.M., Waters C.M., and Hammond M.C.. 2020. Chemiluminescent sensors for quantitation of the bacterial second messenger cyclic di-GMP. Methods Enzymol. 640:83–104. 10.1016/bs.mie.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura, D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., and Tanner S.D.. 2009. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81:6813–6822. 10.1021/ac901049w [DOI] [PubMed] [Google Scholar]
- Barker, N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., and Clevers H.. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Battich, N., Stoeger T., and Pelkmans L.. 2015. Control of Transcript Variability in Single Mammalian Cells. Cell. 163:1596–1610. 10.1016/j.cell.2015.11.018 [DOI] [PubMed] [Google Scholar]
- Behar, M., and Hoffmann A.. 2010. Understanding the temporal codes of intra-cellular signals. Curr. Opin. Genet. Dev. 20:684–693. 10.1016/j.gde.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall, S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I., et al. 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332:687–696. 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev, B., Kratchmarova I., Ong S.E., Nielsen M., Foster L.J., and Mann M.. 2003. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21:315–318. 10.1038/nbt790 [DOI] [PubMed] [Google Scholar]
- Bodenmiller, B. 2016. Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst. 2:225–238. 10.1016/j.cels.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Bonifacino, J. S., Dell’Angelica E. C., and Springer T. A.. 1999. Immunoprecipitation. Curr. Protoc. Mol. Biol. 48:10.16.1–10.16.29. 10.1002/0471142727.mb1016s48 [DOI] [PubMed] [Google Scholar]
- Bonner, W.A., Hulett H.R., Sweet R.G., and Herzenberg L.A.. 1972. Fluorescence activated cell sorting. Rev. Sci. Instrum. 43:404–409. 10.1063/1.1685647 [DOI] [PubMed] [Google Scholar]
- Boulais, P.E., and Frenette P.S.. 2015. Making sense of hematopoietic stem cell niches. Blood. 125:2621–2629. 10.1182/blood-2014-09-570192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden, E.S., Zhang F., Bamberg E., Nagel G., and Deisseroth K.. 2005. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8:1263–1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Bugaj, L.J., Choksi A.T., Mesuda C.K., Kane R.S., and Schaffer D.V.. 2013. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods. 10:249–252. 10.1038/nmeth.2360 [DOI] [PubMed] [Google Scholar]
- Carey, A., Edwards D.K. V, Eide C.A., Newell L., Traer E., Medeiros B.C., Pollyea D.A., Deininger M.W., Collins R.H., Tyner J.W., et al. 2017. Identification of Interleukin-1 by Functional Screening as a Key Mediator of Cellular Expansion and Disease Progression in Acute Myeloid Leukemia. Cell Rep. 18:3204–3218. 10.1016/j.celrep.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagraoui, J., Lehnertz B., Girard S., Spinella J.F., Fares I., Tomellini E., Mayotte N., Corneau S., MacRae T., Simon L., and Sauvageau G.. 2019. UM171 induces a homeostatic inflammatory-detoxification response supporting human HSC self-renewal. PLoS One. 14:e0224900. 10.1371/journal.pone.0224900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, R., Villanueva E., Feketova E., Machiedo G.W., Haskó G., Deitch E.A., and Spolarics Z.. 2008. Endotoxemia down-regulates bone marrow lymphopoiesis but stimulates myelopoiesis: the effect of G6PD deficiency. J. Leukoc. Biol. 83:1541–1550. 10.1189/jlb.1207838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest, P.G., and Firtel R.A.. 2006. Feedback signaling controls leading-edge formation during chemotaxis. Curr. Opin. Genet. Dev. 16:339–347. 10.1016/j.gde.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Chen, Y., and Schier A.F.. 2001. The zebrafish Nodal signal Squint functions as a morphogen. Nature. 411:607–610. 10.1038/35079121 [DOI] [PubMed] [Google Scholar]
- Chen, W.T., Lu A., Craessaerts K., Pavie B., Sala Frigerio C., Corthout N., Qian X., Laláková J., Kühnemund M., Voytyuk I., et al. 2020. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer’s Disease. Cell. 182:976–991.e19. 10.1016/j.cell.2020.06.038 [DOI] [PubMed] [Google Scholar]
- Chicha, L., Jarrossay D., and Manz M.G.. 2004. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J. Exp. Med. 200:1519–1524. 10.1084/jem.20040809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coons, A.H., and Kaplan M.H.. 1950. Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J. Exp. Med. 91:1–13. 10.1084/jem.91.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coons, A.H., Creech H.J., Norman J.R., and Berliner E.. 1942. The Demonstration of Pneumococcal Antigen in Tissues by the Use of Fluorescent Antibody. J. Immunol. 45:159–170. [Google Scholar]
- Coutu, D.L., Kokkaliaris K.D., Kunz L., and Schroeder T.. 2017. Three-dimensional map of nonhematopoietic bone and bone-marrow cells and molecules. Nat. Biotechnol. 35:1202–1210. 10.1038/nbt.4006 [DOI] [PubMed] [Google Scholar]
- Coutu, D.L., Kokkaliaris K.D., Kunz L., and Schroeder T.. 2018. Multicolor quantitative confocal imaging cytometry. Nat. Methods. 15:39–46. 10.1038/nmeth.4503 [DOI] [PubMed] [Google Scholar]
- Danial, N.N., Pernis A., and Rothman P.B.. 1995. Jak-STAT signaling induced by the v-abl oncogene. Science. 269:1875–1877. 10.1126/science.7569929 [DOI] [PubMed] [Google Scholar]
- De Lorenzi, R., Gareus R., Fengler S., and Pasparakis M.. 2009. GFP-p65 knock-in mice as a tool to study NF-kappaB dynamics in vivo. Genesis. 47:323–329. 10.1002/dvg.20468 [DOI] [PubMed] [Google Scholar]
- Dempsey, L.A. 2017. CyTOF analysis of anti-tumor responses. Nat. Immunol. 18:254. 10.1038/ni.3702 [DOI] [PubMed] [Google Scholar]
- Dettinger, P., Frank T., Etzrodt M., Ahmed N., Reimann A., Trenzinger C., Loeffler D., Kokkaliaris K.D., Schroeder T., and Tay S.. 2018. Automated Microfluidic System for Dynamic Stimulation and Tracking of Single Cells. Anal. Chem. 90:10695–10700. 10.1021/acs.analchem.8b00312 [DOI] [PubMed] [Google Scholar]
- Dettinger, P., Wang W., Ahmed N., Zhang Y., Loeffler D., Kull T., Etzrodt M., Lengerke C., and Schroeder T.. 2020. An automated microfluidic system for efficient capture of rare cells and rapid flow-free stimulation. Lab Chip. 20:4246–4254. 10.1039/d0lc00687d [DOI] [PubMed] [Google Scholar]
- Dippel, A.B., Anderson W.A., Evans R.S., Deutsch S., and Hammond M.C.. 2018. Chemiluminescent Biosensors for Detection of Second Messenger Cyclic di-GMP. ACS Chem. Biol. 13:1872–1879. 10.1021/acschembio.7b01019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan, M., Vorobjev I., Rees P., Filby A., Wolkenhauer O., Goldfeld A.E., Lieberman J., Barteneva N., Carpenter A.E., and Hennig H.. 2018. Diagnostic Potential of Imaging Flow Cytometry. Trends Biotechnol. 36:649–652. 10.1016/j.tibtech.2017.12.008 [DOI] [PubMed] [Google Scholar]
- Eilken, H.M., Nishikawa S., and Schroeder T.. 2009. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 457:896–900. 10.1038/nature07760 [DOI] [PubMed] [Google Scholar]
- Eldar, A., Dorfman R., Weiss D., Ashe H., Shilo B.Z., and Barkai N.. 2002. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 419:304–308. 10.1038/nature01061 [DOI] [PubMed] [Google Scholar]
- Endele, M., and Schroeder T.. 2012. Molecular live cell bioimaging in stem cell research. Ann. N. Y. Acad. Sci. 1266:18–27. 10.1111/j.1749-6632.2012.06560.x [DOI] [PubMed] [Google Scholar]
- Endele, M., Etzrodt M., and Schroeder T.. 2014. Instruction of hematopoietic lineage choice by cytokine signaling. Exp. Cell Res. 329:207–213. 10.1016/j.yexcr.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Endele, M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Skylaki S., Hoppe P.S., Schambach A., Stanley E.R., and Schroeder T.. 2017. CSF-1-induced Src signaling can instruct monocytic lineage choice. Blood. 129:1691–1701. 10.1182/blood-2016-05-714329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, M.A.G., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Etzrodt, M., Endele M., and Schroeder T.. 2014. Quantitative single-cell approaches to stem cell research. Cell Stem Cell. 15:546–558. 10.1016/j.stem.2014.10.015 [DOI] [PubMed] [Google Scholar]
- Etzrodt, M., Ahmed N., Hoppe P.S., Loeffler D., Skylaki S., Hilsenbeck O., Kokkaliaris K.D., Kaltenbach H.M., Stelling J., Nerlov C., and Schroeder T.. 2019. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood. 133:816–819. 10.1182/blood-2018-02-832998 [DOI] [PubMed] [Google Scholar]
- Firaguay, G., and Nunès J.A.. 2009. Analysis of signaling events by dynamic phosphoflow cytometry. Sci. Signal. 2:pl3. 10.1126/scisignal.286pl3 [DOI] [PubMed] [Google Scholar]
- Forthun, R.B., Hellesøy M., Sulen A., Kopperud R.K., Sjøholt G., Bruserud Ø., McCormack E., and Gjertsen B.T.. 2019. Modulation of phospho-proteins by interferon-alpha and valproic acid in acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 145:1729–1749. 10.1007/s00432-019-02931-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, W., Schenker I., Schmutz P., and Hierlemann A.. 2005. Impedance characterization and modeling of electrodes for biomedical applications. IEEE Trans. Biomed. Eng. 52:1295–1302. 10.1109/TBME.2005.847523 [DOI] [PubMed] [Google Scholar]
- Frey, U., Sedivy J., Heer F., Pedron R., Ballini M., Mueller J., Bakkum D., Hafizovic S., Faraci F.D., Greve F., et al. 2010. Switch-matrix-based high-density microelectrode array in CMOS technology. IEEE J. Solid-State Circuits. 45:467–482. 10.1109/JSSC.2009.2035196 [DOI] [Google Scholar]
- Fritz, R.D., Letzelter M., Reimann A., Martin K., Fusco L., Ritsma L., Ponsioen B., Fluri E., Schulte-Merker S., van Rheenen J., and Pertz O.. 2013. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci. Signal. 6:rs12. 10.1126/scisignal.2004135 [DOI] [PubMed] [Google Scholar]
- Gao, L., Han H., Wang H., Cao L., and Feng W.H.. 2019. IL-10 knockdown with siRNA enhances the efficacy of Doxorubicin chemotherapy in EBV-positive tumors by inducing lytic cycle via PI3K/p38 MAPK/NF-kB pathway. Cancer Lett. 462:12–22. 10.1016/j.canlet.2019.07.016 [DOI] [PubMed] [Google Scholar]
- Giesen, C., Wang H.A.O., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B., Schüffler P.J., Grolimund D., Buhmann J.M., Brandt S., et al. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 11:417–422. 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- Golan, K., Kumari A., Kollet O., Khatib-Massalha E., Subramaniam M.D., Ferreira Z.S., Avemaria F., Rzeszotek S., García-García A., Xie S., et al. 2018. Daily Onset of Light and Darkness Differentially Controls Hematopoietic Stem Cell Differentiation and Maintenance. Cell Stem Cell. 23:572–585.e7. 10.1016/j.stem.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Gross, S.M., and Rotwein P.. 2015. Akt signaling dynamics in individual cells. J. Cell Sci. 128:2509–2519. 10.1242/jcs.168773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S.M., and Rotwein P.. 2016. Mapping growth-factor-modulated Akt signaling dynamics. J. Cell Sci. 129:2052–2063. 10.1242/jcs.183764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut, G., Herrmann M.D., and Pelkmans L.. 2018. Multiplexed protein maps link subcellular organization to cellular states. Science. 361:eaar7042. 10.1126/science.aar7042 [DOI] [PubMed] [Google Scholar]
- Guzman, M.L., Rossi R.M., Karnischky L., Li X., Peterson D.R., Howard D.S., and Jordan C.T.. 2005. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 105:4163–4169. 10.1182/blood-2004-10-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast Á.M., Schnell A., Hexel K., et al. 2015. Stem Cell-like Megakaryocyte Progenitors As Driving Forces of IFN-Induced Emergency Megakaryopooesis. Blood. 126:2391. 10.1182/blood.V126.23.2391.2391 [DOI] [PubMed] [Google Scholar]
- Hashimura, H., Morimoto Y.V., Yasui M., and Ueda M.. 2019. Collective cell migration of Dictyostelium without cAMP oscillations at multicellular stages. Commun. Biol. 2:34. 10.1038/s42003-018-0273-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltberg, M., Kellogg R.A., Krishna S., Tay S., and Jensen M.H.. 2016. Noise Induces Hopping between NF-κB Entrainment Modes. Cell Syst. 3:532–539.e3. 10.1016/j.cels.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati, S., Haque T., and Gritsman K.. 2017. Inflammatory signaling pathways in preleukemic and leukemic stem cells. Front. Oncol. 7:265. 10.3389/fonc.2017.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérault, A., Binnewies M., Leong S., Calero-Nieto F.J., Zhang S.Y., Kang Y.A., Wang X., Pietras E.M., Chu S.H., Barry-Holson K., et al. 2017. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 544:53–58. 10.1038/nature21693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, A., Sommer U., Pranada A.L., Giese B., Küster A., Haan S., Becker W., Heinrich P.C., and Müller-Newen G.. 2004. STAT3 is enriched in nuclear bodies. J. Cell Sci. 117:339–349. 10.1242/jcs.00833 [DOI] [PubMed] [Google Scholar]
- Herzenberg, L.A., Sweet R.G., and Herzenberg L.A.. 1976. Fluorescence-activated cell sorting. Sci. Am. 234:108–117. 10.1038/scientificamerican0376-108 [DOI] [PubMed] [Google Scholar]
- Hilsenbeck, O., Schwarzfischer M., Skylaki S., Schauberger B., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Hastreiter S., Skylaki E., Filipczyk A., et al. 2016. Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 34:703–706. 10.1038/nbt.3626 [DOI] [PubMed] [Google Scholar]
- Ho, C.C.M., Chhabra A., Starkl P., Schnorr P.J., Wilmes S., Moraga I., Kwon H.S., Gaudenzio N., Sibilano R., Wehrman T.S., et al. 2017. Decoupling the Functional Pleiotropy of Stem Cell Factor by Tuning c-Kit Signaling. Cell. 168:1041–1052.e18. 10.1016/j.cell.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A., Levchenko A., Scott M.L., and Baltimore D.. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245. 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- Hoppe, P.S., Coutu D.L., and Schroeder T.. 2014. Single-cell technologies sharpen up mammalian stem cell research. Nat. Cell Biol. 16:919–927. 10.1038/ncb3042 [DOI] [PubMed] [Google Scholar]
- Hoppe, P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N., et al. 2016. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 535:299–302. 10.1038/nature18320 [DOI] [PubMed] [Google Scholar]
- Hughes, A.J., Spelke D.P., Xu Z., Kang C.C., Schaffer D.V., and Herr A.E.. 2014. Single-cell western blotting. Nat. Methods. 11:749–755. 10.1038/nmeth.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes, K.A. 2015. An analysis of critical factors for quantitative immunoblotting. Sci. Signal. 8:rs2. 10.1126/scisignal.2005966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, H.E., and Toettcher J.E.. 2019. Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev. Cell. 48:361–370.e3. 10.1016/j.devcel.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno, J.A., Wragg K.M., Kristensen A.B., Lee W.S., Selva K.J., van der Sluis R.M., Kelleher A.D., Bavinton B.R., Grulich A.E., Lewin S.R., et al. 2019. Modulation of the CCR5 Receptor/Ligand Axis by Seminal Plasma and the Utility of In Vitro versus In Vivo Models. J. Virol. 93:e00242–e19. 10.1128/JVI.00242-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, M. 2007. Dysregulation of stem cell signaling network due to germline mutation, SNP, Helicobacter pylori infection, epigenetic change and genetic alteration in gastric cancer. Cancer Biol. Ther. 6:832–839. 10.4161/cbt.6.6.4196 [DOI] [PubMed] [Google Scholar]
- Katsura, Y., Kubota H., Kunida K., Kanno A., Kuroda S., and Ozawa T.. 2015. An optogenetic system for interrogating the temporal dynamics of Akt. Sci. Rep. 5:14589. 10.1038/srep14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, R.A., and Tay S.. 2015. Noise facilitates transcriptional control under dynamic inputs. Cell. 160:381–392. 10.1016/j.cell.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Kiladjian, J.J., Giraudier S., and Cassinat B.. 2016. Interferon-alpha for the therapy of myeloproliferative neoplasms: targeting the malignant clone. Leukemia. 30:776–781. 10.1038/leu.2015.326 [DOI] [PubMed] [Google Scholar]
- Kim, A.R., Ulirsch J.C., Wilmes S., Unal E., Moraga I., Karakukcu M., Yuan D., Kazerounian S., Abdulhay N.J., King D.S., et al. 2017. Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation. Cell. 168:1053–1064.e15. 10.1016/j.cell.2017.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkaliaris, K.D., Drew E., Endele M., Loeffler D., Hoppe P.S., Hilsenbeck O., Schauberger B., Hinzen C., Skylaki S., Theodorou M., et al. 2016. Identification of factors promoting ex vivo maintenance of mouse hematopoietic stem cells by long-term single-cell quantification. Blood. 128:1181–1192. 10.1182/blood-2016-03-705590 [DOI] [PubMed] [Google Scholar]
- Kokkaliaris, K.D., Kunz L., Cabezas-Wallscheid N., Christodoulou C., Renders S., Camargo F., Trumpp A., Scadden D.T., and Schroeder T.. 2020. Adult blood stem cell localization reflects the abundance of reported bone marrow niche cell types and their combinations. Blood. 136:2296–2307. 10.1182/blood.2020006574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, C.D., Digioia G., Izotova L.S., Xie J., Kim Y., Schwartz B.J., Mirochnitchenko O.V., and Pestka S.. 2013. Ligand-independent interaction of the type I interferon receptor complex is necessary to observe its biological activity. Cytokine. 64:286–297. 10.1016/j.cyto.2013.06.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, L., and Schroeder T.. 2019. A 3D Tissue-wide Digital Imaging Pipeline for Quantitation of Secreted Molecules Shows Absence of CXCL12 Gradients in Bone Marrow. Cell Stem Cell. 25:846–854.e4. 10.1016/j.stem.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Laird, M.H.W., Rhee S.H., Perkins D.J., Medvedev A.E., Piao W., Fenton M.J., and Vogel S.N.. 2009. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J. Leukoc. Biol. 85:966–977. 10.1189/jlb.1208763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, B.D., Clarke D.C., and Lee M.J.. 2015. Studying Cellular Signal Transduction with OMIC Technologies. J. Mol. Biol. 427:3416–3440. 10.1016/j.jmb.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, K., Van Valen D., DeFelice M.M., Macklin D.N., Kudo T., Jaimovich A., Carr A., Meyer T., Pe’er D., Boutet S.C., and Covert M.W.. 2017. Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-κB Activation. Cell Syst. 4:458–469.e5. 10.1016/j.cels.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti, E., and Goettgens B.. 2018. From haematopoietic stem cells to complex differentiation landscapes. Nature. 553:418–426. 10.1038/nature25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. 2007. Coimmunoprecipitation assay. Methods Mol. Biol. 362:401–406. 10.1007/978-1-59745-257-1_31 [DOI] [PubMed] [Google Scholar]
- Li, Z., Theus M.H., and Wei L.. 2006. Role of ERK 1/2 signaling in neuronal differentiation of cultured embryonic stem cells. Dev. Growth Differ. 48:513–523. 10.1111/j.1440-169X.2006.00889.x [DOI] [PubMed] [Google Scholar]
- Liggett, L.A., and Sankaran V.. 2020. Unraveling Hematopoiesis through the Lens of Genomics. Cell. 182:1384–1400. 10.1016/j.cell.2020.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.R., Fallahi-Sichani M., and Sorger P.K.. 2015. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6:8390. 10.1038/ncomms9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso, C., Fleming H.E., Wu J.W., Zhao C.X., Miake-Lye S., Fujisaki J., Côté D., Rowe D.W., Lin C.P., and Scadden D.T.. 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 457:92–96. 10.1038/nature07434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler, D., and Schroeder T.. 2019. Understanding cell fate control by continuous single-cell quantification. Blood. 133:1406–1414. 10.1182/blood-2018-09-835397 [DOI] [PubMed] [Google Scholar]
- Loeffler, D., Wehling A., Schneiter F., Zhang Y., Müller-Bötticher N., Hoppe P.S., Hilsenbeck O., Kokkaliaris K.D., Endele M., and Schroeder T.. 2019. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature. 573:426–429. 10.1038/s41586-019-1531-6 [DOI] [PubMed] [Google Scholar]
- Lubeck, E., and Cai L.. 2012. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 9:743–748. 10.1038/nmeth.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun, X.K., and Bodenmiller B.. 2020. Profiling Cell Signaling Networks at Single-cell Resolution. Mol. Cell. Proteomics. 19:744–756. 10.1074/mcp.R119.001790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz, M.G. 2008. Bone Marrow Progenitors of Dendritic and Natural Interferon-producing Cells. In Handbook of Dendritic Cells. 10.1002/9783527619696.ch2 [DOI] [Google Scholar]
- Marshall, C.J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 80:179–185. 10.1016/0092-8674(95)90401-8 [DOI] [PubMed] [Google Scholar]
- McGrath, K.E., Bushnell T.P., and Palis J.. 2008. Multispectral imaging of hematopoietic cells: where flow meets morphology. J. Immunol. Methods. 336:91–97. 10.1016/j.jim.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer, S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., and Frenette P.S.. 2010. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 466:829–834. 10.1038/nature09262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, A., Venkatachalapathy S., Ratna P., Wang Y., Jokhun D.S., and Shivashankar G.V.. 2017. Cell geometry dictates TNFα-induced genome response. Proc. Natl. Acad. Sci. USA. 114:E3882–E3891. 10.1073/pnas.1618007114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera, A., Michaut A., Guillot C., Xiong F., and Pourquié O.. 2019. Mechanics of anteroposterior axis formation in vertebrates. Annu. Rev. Cell Dev. Biol. 35:259–283. 10.1146/annurev-cellbio-100818-125436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga, I., Wernig G., Wilmes S., Gryshkova V., Richter C.P., Hong W.J., Sinha R., Guo F., Fabionar H., Wehrman T.S., et al. 2015. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell. 160:1196–1208. 10.1016/j.cell.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., and Scadden D.T.. 2014. The bone marrow niche for haematopoietic stem cells. Nature. 505:327–334. 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal, N., Santat L.A., LeBon L., Sprinzak D., Bronner M.E., and Elowitz M.B.. 2018. Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell. 172:869–880.e19. 10.1016/j.cell.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher, E., and Sakmann B.. 1992. The patch clamp technique. Sci. Am. 266:44–51. 10.1038/scientificamerican0392-44 [DOI] [PubMed] [Google Scholar]
- Nelson, D.E., Ihekwaba A.E.C., Elliott M., Johnson J.R., Gibney C.A., Foreman B.E., Nelson G., See V., Horton C.A., Spiller D.G., et al. 2004. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 306:704–708. 10.1126/science.1099962 [DOI] [PubMed] [Google Scholar]
- Newton, K., and Dixit V.M.. 2012. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4:a006049. 10.1101/cshperspect.a006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela-Arrieta, C., Pivarnik G., Winkel B., Canty K.J., Harley B., Mahoney J.E., Park S.Y., Lu J., Protopopov A., and Silberstein L.E.. 2013. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 15:533–543. 10.1038/ncb2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencik, J., Pham H.T.T., Schmoellerl J., Javaheri T., Schlederer M., Culig Z., Merkel O., Moriggl R., Grebien F., and Kenner L.. 2016. JAK-STAT signaling in cancer: From cytokines to non-coding genome. Cytokine. 87:26–36. 10.1016/j.cyto.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., Moore R., McClanahan T.K., Sadekova S., and Klappenbach J.A.. 2017. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35:936–939. 10.1038/nbt.3973 [DOI] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl, C., and Bao Z.. 2010. Chiral forces organize left-right patterning in C. elegans by uncoupling midline and anteroposterior axis. Dev. Cell. 19:402–412. 10.1016/j.devcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis, J.E., and Lahav G.. 2013. Encoding and decoding cellular information through signaling dynamics. Cell. 152:945–956. 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis, J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., and Lahav G.. 2012. p53 dynamics control cell fate. Science. 336:1440–1444. 10.1126/science.1218351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz, F.G., Fiore V.F., Levorse J., Polak L., Wong E., Pasolli H.A., and Fuchs E.. 2020. Liquid-liquid phase separation drives skin barrier formation. Science. 367:eaax9554. 10.1126/science.aax9554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., and Tyagi S.. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 5:877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot, S., Hughey J.J., Bajar B.T., Carrasco S., and Covert M.W.. 2014. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 157:1724–1734. 10.1016/j.cell.2014.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renart, J., Reiser J., and Stark G.R.. 1979. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc. Natl. Acad. Sci. USA. 76:3116–3120. 10.1073/pnas.76.7.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger, M.A., Hoppe P.S., Smejkal B.M., Eitelhuber A.C., and Schroeder T.. 2009. Hematopoietic cytokines can instruct lineage choice. Science. 325:217–218. 10.1126/science.1171461 [DOI] [PubMed] [Google Scholar]
- Rompolas, P., Deschene E.R., Zito G., Gonzalez D.G., Saotome I., Haberman A.M., and Greco V.. 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 487:496–499. 10.1038/nature11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., and Clevers H.. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469:415–418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier, A.F. 2001. Axis formation and patterning in zebrafish. Curr. Opin. Genet. Dev. 11:393–404. 10.1016/S0959-437X(00)00209-4 [DOI] [PubMed] [Google Scholar]
- Schroeder, T. 2005. Tracking hematopoiesis at the single cell level. Ann. N. Y. Acad. Sci. 1044:201–209. 10.1196/annals.1349.025 [DOI] [PubMed] [Google Scholar]
- Schroeder, T. 2008. Imaging stem-cell-driven regeneration in mammals. Nature. 453:345–351. 10.1038/nature07043 [DOI] [PubMed] [Google Scholar]
- Schroeder, T. 2011. Long-term single-cell imaging of mammalian stem cells. Nat. Methods. 8(4, Suppl):S30–S35. 10.1038/nmeth.1577 [DOI] [PubMed] [Google Scholar]
- Skylaki, S., Hilsenbeck O., and Schroeder T.. 2016. Challenges in long-term imaging and quantification of single-cell dynamics. Nat. Biotechnol. 34:1137–1144. 10.1038/nbt.3713 [DOI] [PubMed] [Google Scholar]
- Söderberg, O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Ståhl, P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M., et al. 2016. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 353:78–82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Stoeckius, M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., and Smibert P.. 2017. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 14:865–868. 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, M.K., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Schroeder T., Theis F.J., and Marr C.. 2018. Lineage marker synchrony in hematopoietic genealogies refutes the PU.1/GATA1 toggle switch paradigm. Nat. Commun. 9:2697. 10.1038/s41467-018-05037-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.Y., Haberman A.M., and Greco V.. 2017. Preclinical Advances with Multiphoton Microscopy in Live Imaging of Skin Cancers. J. Invest. Dermatol. 137:282–287. 10.1016/j.jid.2016.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, H., and Manz M.G.. 2017. Impact of inflammation on early hematopoiesis and the microenvironment. Int. J. Hematol. 106:27–33. 10.1007/s12185-017-2266-5 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Boettcher S., and Manz M.G.. 2012. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 119:2991–3002. 10.1182/blood-2011-12-380113 [DOI] [PubMed] [Google Scholar]
- Tay, S., Hughey J.J., Lee T.K., Lipniacki T., Quake S.R., and Covert M.W.. 2010. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 466:267–271. 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic, V. 2017. Single-cell RNA-seq—now with protein. Nat. Methods. 14:1028–1029. 10.1038/nmeth.4488 [DOI] [Google Scholar]
- Trumpp, A., Essers M., and Wilson A.. 2010. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 10:201–209. 10.1038/nri2726 [DOI] [PubMed] [Google Scholar]
- Tuleuova, N., Jones C.N., Yan J., Ramanculov E., Yokobayashi Y., and Revzin A.. 2010. Development of an aptamer beacon for detection of interferon-gamma. Anal. Chem. 82:1851–1857. 10.1021/ac9025237 [DOI] [PubMed] [Google Scholar]
- Vickovic, S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., Äijö T., Bonneau R., Bergenstråhle L., Navarro J.F., et al. 2019. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 16:987–990. 10.1038/s41592-019-0548-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Zhang Y., Dettinger P., Reimann A., Kull T., Loeffler D., Manz M., Lengerke C., and Schroeder T.. 2021. Cytokine combinations for human blood stem cell expansion induce cell type- and cytokine-specific signaling dynamics. Blood. 10.1182/blood.2020008386 [DOI] [PubMed] [Google Scholar]
- Wang, Y.J., Traum D., Schug J., Gao L., Liu C., Atkinson M.A., Powers A.C., Feldman M.D., Naji A., Chang K.M., and Kaestner K.H.. HPAP Consortium . 2019. Multiplexed In Situ Imaging Mass Cytometry Analysis of the Human Endocrine Pancreas and Immune System in Type 1 Diabetes. Cell Metab. 29:769–783.e4. 10.1016/j.cmet.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Saito K., Kinjo M., Matsuda T., Tamura M., Kon S., Miyazaki T., and Uede T.. 2004. Molecular dynamics of STAT3 on IL-6 signaling pathway in living cells. Biochem. Biophys. Res. Commun. 324:1264–1273. 10.1016/j.bbrc.2004.09.187 [DOI] [PubMed] [Google Scholar]
- Welner, R.S., Pelayo R., Nagai Y., Garrett K.P., Wuest T.R., Carr D.J., Borghesi L.A., Farrar M.A., and Kincade P.W.. 2008. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 112:3753–3761. 10.1182/blood-2008-04-151506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S.L., Barken D., and Hoffmann A.. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 309:1857–1861. 10.1126/science.1113319 [DOI] [PubMed] [Google Scholar]
- Wu, A.R., Neff N.F., Kalisky T., Dalerba P., Treutlein B., Rothenberg M.E., Mburu F.M., Mantalas G.L., Sim S., Clarke M.F., and Quake S.R.. 2014. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 11:41–46. 10.1038/nmeth.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, C., Babcock H.P., Moffitt J.R., and Zhuang X.. 2019. Multiplexed detection of RNA using MERFISH and branched DNA amplification. Sci. Rep. 9:7721. 10.1038/s41598-019-43943-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Xie X., Duan Y., Wang L., Cheng Z., and Cheng J.. 2016. A review of impedance measurements of whole cells. Biosens. Bioelectron. 77:824–836. 10.1016/j.bios.2015.10.027 [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S.. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- Yamashita, M., and Passegué E.. 2019. TNF-α Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell. 25:357–372.e7. 10.1016/j.stem.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y., Liu R., Shin M.S., Trentalange M., Allore H., Nassar A., Kang I., Pober J.S., and Montgomery R.R.. 2014. CyTOF supports efficient detection of immune cell subsets from small samples. J. Immunol. Methods. 415:1–5. 10.1016/j.jim.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Jones S., Hagood J.S., Fuentes N.L., and Fuller G.M.. 1997. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J. Biol. Chem. 272:30607–30610. 10.1074/jbc.272.49.30607 [DOI] [PubMed] [Google Scholar]
- Zhao, J., Cheng F., and Zhao Z.. 2017. Tissue-specific signaling networks rewired by major somatic mutations in human cancer revealed by proteome-wide discovery. Cancer Res. 77:2810–2821. 10.1158/0008-5472.CAN-16-2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain, C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., and Enard W.. 2017. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell. 65:631–643.e4. 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]