Inflammation can activate hematopoietic stem and progenitor cells (HSPCs) to proliferate and differentiate, signal the specification of HSPCs during embryonic development, and maintain adult HSPC homeostasis. Collins et al. review the role of inflammatory signaling in these diverse hematopoietic scenarios.
Abstract
Inflammation exerts multiple effects on the early hematopoietic compartment. Best studied is the role of proinflammatory cytokines in activating adult hematopoietic stem and progenitor cells to dynamically replenish myeloid lineage cells in a process known as emergency myelopoiesis. However, it is increasingly appreciated that the same proinflammatory signaling pathways are used in diverse hematopoietic scenarios. This review focuses on inflammatory signaling in the emergence of the definitive hematopoietic compartment during embryonic life, and tonic inflammatory signals derived from commensal microbiota in shaping the adult hematopoietic compartment in the absence of pathogenic insults. Insights into the unique and shared aspects of inflammatory signaling that regulate hematopoietic stem and progenitor cell function across the lifespan and health span of an individual will enable better diagnostic and therapeutic approaches to hematopoietic dysregulation and malignancies.
Introduction
The cells of the blood system provide oxygen-carrying capacity, hemostasis, and innate and adaptive immunity and contribute to the regeneration and repair of other tissues. In the adult, these mature effector cells are generated from hematopoietic stem cells (HSCs) that reside in the bone marrow (BM), sit at the top of the hematopoietic hierarchy, and self-renew to maintain themselves for the lifetime of the organism. HSCs are normally found in a dormant state of functional quiescence that shelters them from insult while sustaining their ability to respond to a range of signals and produce various types of progenitor cells, starting with lineage-biased multipotent progenitors (MPPs; Olson et al., 2020). MPPs and their lineage-restricted progeny are the workhorses of the blood system, rapidly proliferating to drive the production of different mature cell types according to the demands of the organism (Cabezas-Wallscheid et al., 2014; Pietras et al., 2015). In particular, inflammation exerts a profound effect across the entire hematopoietic hierarchy, activating and mobilizing effector cells to respond to the organismal challenge and stimulating hematopoietic stem and progenitor cells (HSPCs) to replenish the depleted lineages before returning to homeostasis (Baldridge et al., 2011; Mirantes et al., 2014; Pietras, 2017). In this current understanding of hematopoietic function, a continuum exists between quiescent HSCs that are protected from damage associated with replication and metabolic activation as a consequence of their dormancy, and activated HSCs that respond to various signals including acute inflammation to properly tailor blood cell production. Positioned along this continuum, tonic inflammation is crucial for shaping the development and homeostasis of the HSPC compartment even in the absence of pathogenic inflammation (e.g., injury, infection, or malignancy). Indeed, work within the past decade has revealed that many of the same inflammatory pathways controlling adult emergency myelopoiesis also control the emergence of definitive HSCs during embryonic development and shape the adult hematopoietic compartment throughout the lifespan. In this context, the commensal microbiome is increasingly appreciated as a critical regulator of many aspects of adult vertebrate biology (Schroeder and Bäckhed, 2016), with the hematopoietic system being no exception. The molecular mechanisms underlying the spaciotemporal execution of these inflammatory pathways in hematopoietic ontogeny are the focus of the first part of this review, and the second part concentrates on the role of nonpathogenic microbial inflammation in regulating the HSPC compartment during homeostasis in adults.
Inflammatory signaling in hematopoietic ontogeny
Reference to adult hematopoiesis
Young adult HSCs reside in specialized niches in the BM microenvironment, where they are maintained in a quiescent state (Schepers et al., 2015; Crane et al., 2017; Wei and Frenette, 2018), protected from genotoxic stress, and are characterized by the dual “stemness” properties of self-renewal (experimentally measured by transplantation capacity) and multilineage differentiation potential. HSCs give rise to several MPP populations, each with distinct biases toward different lineages of the blood system, which altogether form the HSPC compartment. At steady-state, the erythroid/megakaryocyte-biased MPP2 and granulocyte/macrophage-biased MPP3 are the smallest produced MPP subsets, while lymphoid-biased MPP4s are abundantly made by a mostly quiescent HSC population (Cabezas-Wallscheid et al., 2014; Pietras et al., 2015). Importantly, substantial plasticity exists such that in the face of inflammation, the organization of the HSPC compartment is dynamically shifted, with HSCs activating and transiently decreasing their self-renewal capacity as they overproduce MPP2/MPP3 and redirect MPP4 output toward the myeloid lineages, hence driving the downstream formation of granulocyte-macrophage progenitor (GMP) “clusters” that serve as production hubs for myeloid differentiation (Pietras et al., 2015; Hérault et al., 2017). This process is orchestrated by various inflammatory cytokines acting at multiple levels of the early hematopoietic hierarchy. HSCs are readily activated to proliferate in response to IL-1β (Ueda et al., 2009; Pietras et al., 2016; Weisser et al., 2016), G-CSF (Wilson et al., 2008; Schuettpelz et al., 2014), and type I and II IFNs (IFNα/β and IFNγ, respectively; Essers et al., 2009; Sato et al., 2009; Baldridge et al., 2010; Pietras et al., 2014). Other inflammatory cytokines such as TNFα exert a prosurvival effect on HSCs, shielding them from its apoptotic effects on many downstream progenitors, while activating a myeloid differentiation program via induction of the master regulator of myeloid commitment, the transcription factor Pu.1 (Etzrodt et al., 2019; Yamashita and Passegué, 2019). Indeed, the common result of HSC activation by inflammatory cytokines is myeloid differentiation, although divergent signaling pathways are used by discrete cytokines. As with TNFα, IL-1β and M-CSF induce myeloid differentiation via up-regulation of Pu.1 (Mossadegh-Keller et al., 2013; Pietras et al., 2016), while IFNs stimulate expression of Batf2 and Cebpβ (Matatall et al., 2014, 2016), two key transcription factors also promoting myeloid lineage differentiation. Other cytokines such as IL-6 act primarily at the level of MPP4s, redirecting their output from lymphoid to myeloid lineages (Reynaud et al., 2011), while cytokines such as G-CSF modulate the formation of GMP clusters (McLemore et al., 2001; Panopoulos et al., 2006; Zhang et al., 2010; Hérault et al., 2017). The combined effect of these inflammatory signaling pathways results in a dramatic reshaping of the HSPC compartment to transiently meet organismal needs followed by a rapid return to homeostasis, when another set of cytokines such as TGF-β and CXCL4 kick in to reestablish quiescence in HSCs and terminate the acute regenerative response (Hérault et al., 2017). These same signaling pathways are coopted in a range of other hematopoietic scenarios, both adaptive, as in HSPC emergence and microbial tuning of hematopoiesis that are discussed here, and maladaptive, as in aging (“inflamm-aging”) and cancer, which are the topics of accompanying reviews.
Overview of fetal hematopoiesis
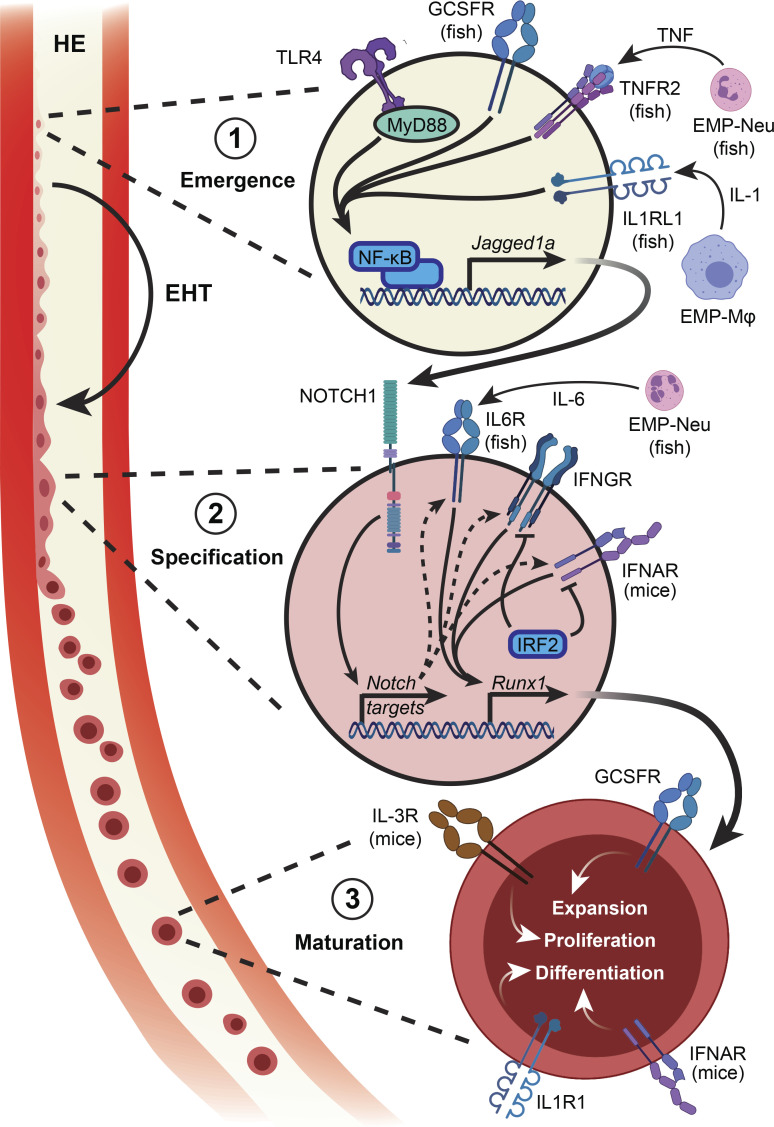
Hematopoietic cells such as macrophages and erythroid cells are present in the developing embryo before the appearance of HSCs with adult self-renewal capacity. These cells support the rapidly developing embryo and arise from both primitive progenitors and definitive erythroid-myeloid progenitors that are transiently present in the yolk sac during this window of development (Bertrand et al., 2007; Frame et al., 2013; Palis, 2014; McGrath et al., 2015). These early waves of hematopoiesis are replaced by a subsequent wave that includes pre-HSCs, which are already multipotent but have poor engraftment capability in adult recipient mice and mature to become definitive HSCs around embryonic day 10.5–11.5 (E10.5–E11.5; Kieusseian et al., 2012; Taoudi et al., 2008). Definitive HSPCs are variably identified as Runx1+/c-Kit+/Procr+/vascular endothelial cadherin (VE-Cad)+/CD45+ cells at E10.5–E11.5 in the mouse (Sánchez et al., 1996; North et al., 1999, 2002; Taoudi et al., 2005; Zheng et al., 2019) and as cMyb+ or Runx1+ cells 24–48 h postfertilization (hpf) in the zebrafish (Kobayashi, 2018). While bona fide HSCs can be functionally defined in the mouse using transplantation assays, a comparable functional assay does not exist in zebrafish, where stem cell function is inferred from interventions that modulate the number of cells expressing cMyb or Runx1, but this does not distinguish between stem and progenitor populations. In both models, HSPCs arise from endothelial cells in a process known as endothelial-to-hematopoietic transition (EHT), which takes place in the hemogenic endothelium (HE) in the dorsal portion of the aorta within the aorta-gonad-mesonephros (AGM) region (de Bruijn et al., 2000; Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010). EHT and HSC specification revolve around Notch signaling and the function of the Runx1 transcription factor, with both zebrafish and mice lacking key components of the Notch pathway or deficient for Runx1 being devoid of definitive hematopoietic cells (Müller et al., 1994; Cai et al., 2000; North et al., 2002; Kumano et al., 2003; Hadland et al., 2004; Burns et al., 2005; Robert-Moreno et al., 2008; Chen et al., 2009; Bigas et al., 2010; Lam et al., 2010; Kim et al., 2014; Kobayashi et al., 2014). The development of definitive HSPCs involves three discrete steps: emergence, specification, and maturation (Fig. 1), and inflammatory signaling has been implicated in each of these steps in both mice and zebrafish, with some indications of its involvement in humans (Crosse et al., 2020).
Figure 1.
Tonic inflammatory signaling in the establishment of definitive hematopoiesis. The development of HSCs involves three distinct steps. (1) Hemogenic endothelial cells respond to inflammatory signals secreted by primitive myeloid cells that converge on NF-κB to up-regulate Notch ligand. (2) Neighboring endothelial cells expressing Notch are stimulated to up-regulate a number of target genes required for HSC emergence; this signal is relayed in part through inflammatory pathways. (3) Mobilization of newly specified HSCs from the AGM into the circulation, migration to subsequent hematopoietic organs, and further functional maturation is regulated by cytokines. EMP, erythroid-myeloid progenitor; Mϕ, macrophage; Neu, neutrophil.
Inflammatory signals in HSPC emergence
The first indication that inflammatory signaling plays a role in HSPC specification followed from the observation that TNFα signaling is essential for endothelial emergence (Espín et al., 2013). Further work in zebrafish detailed this mechanism, by which endothelial cells expressing Tnfr2 respond to TNFα by up-regulating expression of the Notch ligand Jagged1a via an NF-κB-dependent pathway (Espín-Palazón et al., 2014). These Jagged1a-expressing endothelial cells within the dorsal aorta then bind Notch1a on neighboring endothelial cells, triggering cleavage of the Notch intracellular domain and licensing activation of a number of genes required for HSPC specification. Morphant fishes lacking Tnfa or Tnfr2 generate fewer cMyb+ HSPCs, and this phenotype is rescued by ectopic expression of the Notch intracellular domain in the endothelium. Furthermore, genetic inhibition of NF-κB in endothelial cells results in a depletion of cMyb+ HSPCs. The centrality of NF-κB, upstream of Notch and downstream of inflammatory signaling, was confirmed when an unbiased screening approach identified the TLR4-MyD88-NF-κB axis as required for HSPC emergence in both mice and zebrafish (He et al., 2015). Emerging Runx1+ HSPCs in the zebrafish HE express a number of inflammation-related genes, notably the cytokine receptors Tnfr2 and Gcsfr, Tlr4bb, and the NF-κB family member p65. Morphant fishes lacking Tlr4bb or Myd88 have decreased HSPC and T lymphoid marker expression, suggesting a functional HSPC defect, as T lymphopoiesis is exclusively HSPC dependent in zebrafish. Similarly, the AGM region from Tlr4−/− mouse embryos harbors fewer Runx1+ cells; decreased Runx1, cMyb, and Notch target gene expression; and diminished multilineage repopulating activity. More recently, inflammatory signaling through the RIG-I–like receptors Mda5, Rig-1, and Lgp2 has been shown to modulate HSPC numbers in zebrafish and mice. These receptors are at least partially stimulated by repetitive element expression, which triggers NF-κB activation via Traf6, leading to HSPC specification (Lefkopoulos et al., 2020). Together, these data demonstrate that inflammatory signals converging on NF-κB activation and Notch up-regulation are the initiating signal for the emergence of definitive HSPCs.
Inflammatory signals in HSPC specification
Endothelial cells that experience Notch signaling respond by up-regulating Runx1, which is required for specification of definitive HSPCs. Recent work has demonstrated the role of inflammatory signaling pathways in relaying the Notch-to-Runx1 signal. In zebrafish, Notch signaling results in Il6r up-regulation, rendering those endothelial cells receptive to IL-6 secreted by primitive neutrophils. IL-6–IL6R signaling in turn stimulates Runx1 expression, and enforced expression of Il6r in Notch-inhibited or Tnfa-deficient embryos restores HSPC numbers (Tie et al., 2019). IL-6 is also implicated downstream of Hif1α-induced Pdgfrβ regulation of zebrafish HSPC production (Lim et al., 2017). Notch signaling also positively regulates the expression of Ifng and its receptor Crfb17 in the zebrafish embryo, and Ifng1-2 overexpression restores HSPC numbers following Notch inhibition (Sawamiphak et al., 2014). Zebrafish in which either Ifng1-2 or expression of the IFNγ receptors Crfb6, Crfb13, or Crfb17 is abrogated have diminished numbers of HSPCs (Sawamiphak et al., 2014; Li et al., 2014), while knockdown of Irf2, a negative regulator of IFN signaling, overactivates Runx1 expression in the AGM and increases the number of HSPCs and lymphocytes (Li et al., 2014). The involvement of IFN signaling in HSC emergence has also been described in mice. A GFP reporter transgene driven by the Ly6a/Sca-1 locus marks cells with lymphoid potential in the murine AGM region, distinguishing them as definitive HSCs (de Bruijn et al., 2002; Ma et al., 2002). Ly6a-GFP+ AGM cells have an innate immune/inflammatory signature, with TLRs, histocompatibility antigens, lymphocyte-associated genes, and IFN-regulated genes being particularly enriched. Type I and type II IFNs, and to a lesser extent TNFα, also induce Ly6a-GFP expression in murine embryo explant cultures, in contrast to IL-1β, IL-4, and IL-6. Stromal cultures with dissociated E10.5 AGM cells from Ifng−/−, Ifngr−/−, and Ifnar1−/− embryos yield fewer progenitors with lymphoid potential, and HSC repopulation capacity is significantly reduced in E11.5 Ifngr−/− mice (Li et al., 2014). As with stimulation of Notch ligand expression, these studies illustrate the convergence of multiple pathways on Runx1 expression. Combinatorial knockdown of both Tnfa and Ifng in zebrafish causes a greater reduction in Runx1 expression than knockdown of either gene alone (Li et al., 2014), but it remains to be determined how the interplay between different related pathways controls HSPC specification.
Inflammatory signals in HSPC maturation
The aforementioned studies focus on inflammatory signaling in definitive HSPC specification, but inflammatory cues continue to provide maturation and expansion signals as HSCs leave the AGM and transit to the next hematopoietic compartment. IFNα and Jak/Stat1-associated genes are significantly enriched in murine fetal liver HSCs relative to their AGM HSC counterparts. AGM HSCs are responsive to type I IFN signaling, as shown in ex vivo culture with IFNα that partially matures AGM HSCs and enhances their long-term repopulation capacity upon transplantation (Kim et al., 2016). Interleukins such as IL-1 and IL-3 have also been implicated in HSC maturation downstream of Runx1. In particular, IL-3 expression in the AGM is diminished in Runx1+/− and absent in Runx1−/− embryos, and Il3−/− AGM transplantations exhibit significantly reduced overall chimerism, thereby implicating IL-3 as an HSC proproliferative and prosurvival factor downstream of Runx1. However, IL-3 does not specify HSCs, as treatment with the cytokine does not rescue HSC activity in Runx1−/− AGM explants (Robin et al., 2006). Similarly, IL-1 signaling enhances AGM or fetal liver HSC repopulation activity, but absence of IL-1 or its receptor does not prevent HSC emergence in mice (Orelio et al., 2008, 2009). IL-1β plays an important role in HSPC specification in zebrafish, where inflammasome activation in primitive macrophages in response to metabolic cues results in IL-1β activation and modulation of HSPC numbers (Frame et al., 2020). IL-1 receptor signaling in the HE also plays a role in specifying HSPCs in response to macrophage-secreted IL-1β, with knockdown of Il1rl1 significantly reducing Runx1/cMyb expression in emerging Flk1+ HSPCs (Frame et al., 2020). In the same study, Il1b was also found to be expressed in HSPCs in response to metabolic cues and with ectopic expression of Il1b in emerging Flk1+ HSPCs directly increasing Runx1 expression. Thus, in zebrafish, IL-1β appears to act both upstream of HSPC specification and within emerging HSPCs themselves to amplify and promote their number and/or function.
Another cytokine with species-specific action is G-CSF. The zebrafish has two copies of the G-CSF ligand, Gcsfa and Gcsfb, with overlapping and nonoverlapping roles in hematopoiesis (Stachura et al., 2013). Overexpression of either ligand causes HSPC expansion in the dorsal aorta at 24 hpf and in the caudal hematopoietic tissue (CHT) at 36 hpf, while knockdown of the receptor Gcsfr or either Gcsfa or Gcsfb ligands causes significant reduction in HSPCs in the dorsal aorta and CHT (Stachura et al., 2013). In contrast, mice lacking either Gcsf or Gcsfr have reductions in myeloid progenitors and further differentiated myeloid cells but relatively normal HSC numbers, as measured by frequency of long-term culture-initiating cells in the adult BM (Lieschke et al., 1994; Liu et al., 1996; Richards et al., 2003). As discussed above, these differences may also reflect heterogeneity in the cell populations being studied, HSPCs in zebrafish and HSCs in mice. Overall, it is clear that several inflammatory pathways continue to be used to signal HSPC maturation after specification, and further work may uncover additional cytokines involved in this process.
Sources of inflammatory signals during development
A key element in understanding how inflammatory signals contribute to HSPC emergence and specification is identifying the source of those signals. Consensus dictates that this inflammation is sterile and not derived from microbes, either commensal or pathogenic. Whether or not there are commensal microbes present at different stages of prenatal development and what their impact might be is an area of considerable interest and controversy (Aagaard et al., 2014; de Goffau et al., 2019; Perez-Muñoz et al., 2017). Still, most studies agree that early embryonic development, at the time of HSC emergence, is sterile. The inflammatory signals that specify definitive HSCs appear to originate at least in part from hematopoietic cells derived from the earlier waves of hematopoiesis, primarily myeloid cells. In zebrafish, depletion of neutrophils consistently results in decreased HSPCs, while the effect of macrophage depletion varies depending on methodology (Li et al., 2014; Espín-Palazón et al., 2014; He et al., 2015; Tie et al., 2019; Frame et al., 2020; Table 1). While most studies using Irf8 morpholinos report a skewing of myeloid differentiation with a decrease in macrophages and concomitant excess of neutrophils, resulting in increased cytokine release and increased HSPC numbers, one study reported instead a decrease in emerging HSPCs (Li et al., 2014). In line with this result, another work using genetic ablation of macrophages as well as macrophage-specific inhibition of NF-κB signaling convincingly showed decreased HSPC numbers with those approaches (Frame et al., 2020), hence validating the idea that both primitive neutrophils and macrophages are important for proper HSC emergence.
Table 1. Summary of results from zebrafish studies evaluating the contribution of primitive myeloid cells to inflammatory cytokines involved in HSC emergence.
| Intervention | Cells affected | Effect on HSPC | Effect on cytokines | Reference |
|---|---|---|---|---|
| Irf8a morpholino | ↓ Csf1ra+ macrophages | ↓ Runx1+ in AGM | Li et al., 2014 | |
| ↓ Cd41:Gfp+ in CHT | ||||
| ↓ Mpeg1+ macrophages | ↑ Runx1+ in AGM | ↑ TNFα | Espín-Palazón et al., 2014 | |
| ↑ Kdrl+;cMyb+ in AGM | ||||
| ↓ Mfap4+ macrophages | No effect | ↓ IL-1β; ↓ G-CSF; ↑ TNFα | He et al., 2015 | |
| ↑ Lyz+ neutrophils | ||||
| ↑ Mpx+ neutrophils | ↑ Runx1+ in AGM | ↑ IL-6 | Tie et al., 2019 | |
| Cebp1b morpholino | ↓ Lyz+ neutrophils | ↓ Runx1+ in AGM | ↓ IL-1β; ↓ G-CSF; ↓ TNFα | He et al., 2015 |
| Spi1bc morpholino | ↓ Mfap4+ macrophages | ↓ Runx1+ in AGM | ↓ IL-1β; ↓ G-CSF; ↓ TNFα | He et al., 2015 |
| ↓ Lyz+ neutrophils | ||||
| Spi1bd morpholino | ↓ l-Plastin+ leukocytes | ↓ Kdrl+;cMyb+ in AGM | ↓ TNFα | Espín-Palazón et al., 2014 |
| ↓ Mpx+ neutrophils | ||||
| ↓ l-Plastin+ leukocytes | ↓ Runx1+ in AGM | ↓ IL-6 | Tie et al., 2019 | |
| Spi1ae/Spi1bd morpholino | ↓ Irf8+ macrophages | ↓ Runx1+ in AGM | Unknown | Li et al., 2014 |
| ↓ Mpo+ granulocytes | ↓ Cd41:Gfp+ in CHT | |||
| ↓↓ Mfap4+ macrophages | ↓ Runx1+ in AGM | Unknown | Frame et al., 2020 | |
| ↓ Mpx+ neutrophils | ↓ Cd41:Gfp+ in CHT | |||
| Mpeg:NTRf + MTZ | ↓ Mpeg1+ macrophages | ↓ cMyb+ in CHT | Unknown | Frame et al., 2020 |
| Mpeg:Dnikbaag | ↓ Mpeg1+ macrophages | ↓ cMyb+ in CHT | Unknown | Frame et al., 2020 |
Irf8: 5′-AATGTTTCGCTTACTTTGAAAATGG-3′ and 5′-TCAGTCTGCGACCGCCCGAGTTCAT-3′.
Cebp1: 5′-GTCAGACACCGACATGGCTGTGTGT-3′, 5′-GGAGCTGCTGAACTCTACTCGATCT-3′, and 5′-GTCTGACTCCGTCATCGCTGAGTGT-3′.
Pu.1: 5′-CCTCCATTCTGTACGGATGCAGCAT-3′ and 5′-GGTCTTTCTCCTTACCATGCTCTCC-3′.
Pu.1/Spi1b: 5′-GATATACTGATACTCCATTGGTGGT-3′.
Spi1a: 5′-AGCGACTCACGCTGTGGAGGAACT-3′.
Mpeg:NTR: Tg(mpeg1:GAL4;UAS:NTRmCherry) + metronidazole.
Mpeg:Dnikbaa: Tg(mpeg1:GAL4;UAS:nfsB-mCherry;UAS:dnikbaa).
These genetic studies have been nicely complemented by in situ imaging approaches, which show primitive myeloid cells in close proximity to emerging HSCs in the HE of zebrafish, mice, and humans (Travnickova et al., 2015; Yuan et al., 2019; Mariani et al., 2019). In mice, Cx3cr1-expressing macrophages are recruited to the AGM and physically associate with cKit+ cells in intra-aortic hematopoietic clusters at E10.5 (Mariani et al., 2019). In this work, chemical depletion of macrophages in AGM explants with either the Csf1r inhibitor BLZ945 or clodronate liposomes decreases colony-forming capacity and reduces donor chimerism in transplantation assays, suggesting that macrophages positively regulate AGM hematopoietic output. Profiling of these macrophages also reveals a proinflammatory signature with specific enrichment of Tnf, Mmp9, and Mmp13 (Mariani et al., 2019). Similarly, in zebrafish, macrophages accumulate along the ventral wall of the dorsal aorta in close proximity to emerging HSPCs (Travnickova et al., 2015). Depletion of these macrophages results in an accumulation of HSPCs in the AGM and concomitant decrease in the CHT, suggesting that macrophages facilitate HSPC mobilization from the endothelial layer and into the circulation, permitting them to seed other hematopoietic organs. As in mice, zebrafish macrophages express Mmp9 and Mmp13a, and matrix metalloproteinase inhibition phenocopies the accumulation of HSPCs in the AGM at the expense of CHT colonization (Travnickova et al., 2015). However, these studies are not without controversy. For instance, macrophage-deficient zebrafish embryos engineered by disruption of Cepba show normal numbers and distribution of HSPCs in the AGM and CHT, challenging the conclusion that AGM macrophages are necessary for HSPC mobilization (Yuan et al., 2019). Furthermore, clodronate liposome treatment in macrophage-deficient zebrafish resulted in AGM HSPC accumulation and CHT depletion, suggesting a macrophage-independent mechanism. Thus, while these studies clearly implicate primitive myeloid cells in zebrafish, mice, and humans in the emergence of the definitive hematopoietic compartment, unraveling which cell types are required and what signals they are providing remains an area of active investigation.
Beyond specification and emergence, the developing blood system exhibits unique properties specific to the fetal and early neonatal period, resulting in HSC function that is quite different from adult HSCs. Unlike quiescent adult HSCs, fetal/neonatal HSCs exhibit enhanced proliferative and self-renewal capacities as well as distinct lineage biases (Pietras and Passegué, 2013). In mice, the switch from fetal to adult HSCs appears to occur at 3–4 wk postnatal age (Bowie et al., 2006, 2007), and recent work suggests that inflammatory signaling again plays a role in the maturation of the perinatal HSPC compartment. In particular, perinatal HSCs gradually acquire adult-like features, and this maturation is accompanied by a late fetal (just before birth) spike in type I IFNs that modulates the HSPC compartment and reinforces an adult transcriptional program in both HSCs and MPPs (Li et al., 2020). The source of this IFN surge appears to be the fetal skin and, as in HSC specification, is sterile, occurring even in germ-free (GF) mice. Whether inflammatory signaling, either sterile or bacteria induced, is involved in the transition from fetal to adult HSC function at 3–4 wk postnatal age remains to be determined.
Another major hurdle for the field is determining the initial trigger for primitive myeloid cells or other organs to secrete inflammatory signals. Where measured, the concentration of these cytokines during fetal life is orders of magnitude lower than what is generated in response to extrinsic inflammatory stimuli, consistent with tonic secretion being sufficient to support normal development. Inflammasome-driven macrophage IL-1β production has already been linked to the growing metabolic cues of the maturing embryo (Frame et al., 2020), but whether other metabolic signals are linked to inflammation-induced hematopoietic development, and whether these cues can be harnessed to improve in vitro therapeutic applications, including generation of adult-like HSCs from induced pluripotent stem cells, remains an area of active research. Although the transcription factor networks underlying inflammatory signaling in embryonic hematopoiesis are now known with some certainty, it is less clear what the precise impact of stage-specific inflammatory signaling on the epigenetic state of HSCs may be, and which of these changes stay imprinted into adulthood. Additionally, though recent work using an FLT3-ITD mouse model of perinatal leukemia indicated a genetic requirement for type I IFN signaling in malignant MPP overexpansion at this stage (Li et al., 2020), it remains largely to be determined what the connections are between activating inflammatory signaling in hematopoietic ontogeny with the oncogenesis of pediatric leukemias. This is perhaps a particularly apt line of investigation going forward, given the well-documented relationship between cancer and inflammation in the adult.
Tonic inflammatory signaling in adult hematopoietic homeostasis
Commensal microbiome
Once specified, expanded, and migrated to the BM cavity, HSCs complete their transition to an adult phenotype that includes functional quiescence, a state in which HSCs are protected from genotoxic insults by their dormant proliferative and metabolic state (Olson et al., 2020). However, quiescent HSCs remain attuned to their environment, as BM niche signals exert a crucial role in maintaining HSC dormancy. In this vein, tonic inflammatory signals originating from commensal microbes play a key role in steady-state hematopoiesis. It has been appreciated for decades that intestinal commensal bacteria are needed to shape the hematopoietic compartment and myeloid generative capacity of adult mice (Fig. 2), as reductions in commensal bacteria, achieved either in GF mice or by oral administration of antibiotics, result in severe dysregulation of BM myelopoiesis (Chang and Pollard, 1973; Staber et al., 1978; MacVittie and Walker, 1978; Joshi et al., 1979; Goris et al., 1985). In contrast, the cellularity of the spleen, thymus, and peripheral blood as well as the distribution of B and T lymphocytes in all organs remains unaffected (Tada et al., 1996). More recently, the microbiome’s effect on the myeloid compartment has been traced to myeloid progenitors in the BM. Antibiotic-treated or GF pups have an impaired wave of postnatal granulopoiesis and diminished GMP numbers in the BM (Deshmukh et al., 2014), and weaning is associated with a shift in gut microbiome complexity, with the corresponding immune response being critical for acquisition of immunopathological resistance (Al Nabhani et al., 2019). Furthermore, adult GF mice have reduced GMP and HSPC numbers and impaired myeloid colony formation (Balmer et al., 2014; Khosravi et al., 2014). These defects are associated with increased susceptibility to infection, which can be rescued by bacterial colonization or transfer of heat-killed serum from specific pathogen–free mice in a MyD88/Trif-dependent manner (Balmer et al., 2014). Intriguingly, microbial tuning of myelopoiesis is not an all-or-nothing phenomenon, as the size of the myeloid compartment is proportional to the complexity of the intestinal microbiome. GF mice colonized with three-species flora or more populous but “low-complexity” flora have incremental increases in HSPCs, GMPs, and mature myeloid cells in their BM (Balmer et al., 2014). These results are commensurate with studies in zebrafish showing that certain low-abundance microbes have an outsized effect on myelopoiesis (Rolig et al., 2015), emphasizing the complexity and composition of the microbiome as an additional layer of regulation in host myelopoiesis.
Figure 2.
Intestinal commensal microbes shape the hematopoietic compartment through systemic and niche effects. Commensal bacteria exert effects on type 3 innate lymphoid cells (ILC3s) within the intestinal lamina propria to produce IL17A, which stimulates circulating levels of G-CSF and acts distally on GMPs in the BM cavity. The intestinal microbiome also exerts distant effects on HSPCs by releasing bacterial products such as Nod1L that interact with the Nod1 receptor expressed on marrow MSCs and extracellular vesicles containing bacterial DNA that are taken up by Cx3cr1+ MNCs to stimulate the production of a whole range of cytokines that act locally within the BM microenvironment. The source of most of those circulating cytokines remains under investigation.
The effect of tonic inflammatory signaling derived from commensal microbiota has been traced to the level of the HSPC compartment, including HSCs themselves (Burberry et al., 2014; Fiedler et al., 2013; Iwamura et al., 2017; Josefsdottir et al., 2017; Lee et al., 2019). However, there is significant variability in the reported effect of commensal microbiota or impaired inflammatory signaling on the number of phenotypic HSCs (Table 2). This likely reflects a combination of differences in surface markers used to identify HSCs, variations in the methods used to deplete commensal bacteria or block inflammatory signaling pathways, the redundancy in those signaling pathways, and facility-specific microbiological variations. Another plausible explanation is that functional changes in HSCs are not always revealed by simple enumeration. Accordingly, most studies suggest that GF, antibiotic-treated, or inflammatory signaling pathway–deficient mice (i.e., Myd88−/−, Unc93b1−/−, and Nod1−/−) have altered distribution of MPPs, which is a direct consequence of HSC activation, in contrast to single TLR-deficient mice (i.e., Tlr3−/−, Tlr4−/−, Tlr5−/−, and Tlr9−/−) that have no alterations in their HSPC compartment cellularity (Table 2). Currently, there are limited studies directly examining the consequences of tonic microbial exposure on HSC function. However, a recent work showed that acute lipopolysaccharide challenge had only a transient effect on HSC numbers but caused a marked opening of chromatin around myeloid lineage priming factors that persisted for 4 wk and lent a Cebpβ-dependent epigenetic “memory” of prior infection to the HSCs, which were then able to more rapidly respond to a subsequent bacterial challenge (de Laval et al., 2020). This work provides support for trained immunity at the HSC level, lending credence to the idea that tonic inflammatory signals from commensal microbiota can epigenetically prime HSCs to respond to activating signals.
Table 2. Summary of results from studies looking at effects of microbiome depletion or inflammatory signaling pathway deletion on numbers of cells in the HSPC compartment.
| HSPC definition | Intervention | HSPC number | Reference |
|---|---|---|---|
| HSC | |||
| LSK CD48−/CD150+ | GF mice | No effect | Lee et al., 2019 |
| MyD88−/− | No effect | ||
| MyD88−/− | Depleted | Fiedler et al., 2013 | |
| Ripk2−/− | No effect | Burberry et al., 2014 | |
| Tlr3−/− | No effect | Lee et al., 2019 | |
| Tlr4−/− | |||
| Tlr5−/− | |||
| Tlr9−/− | |||
| Unc93b1−/− | No effect | ||
| LSK CD48−/CD150+/CD34−/Flk2− | Antibioticsa | Depleted | Josefsdottir et al., 2017 |
| LSK CD127−/Flk2−/CD34− | GF mice | Depleted | Iwamura et al., 2017 |
| Nod1−/− | Depleted | ||
| Short-term HSC | |||
| LSK CD48−/CD150− | MyD88−/− | No effect | Fiedler et al., 2013 |
| GF mice | Increased | Lee et al., 2019 | |
| LSK CD127−/Flk2−/CD34+ | GF mice | Depleted | Iwamura et al., 2017 |
| MPP | |||
| LSK CD48+/CD150− | GF mice | Depleted | Lee et al., 2019 |
| MyD88−/− | No effect | ||
| Unc93b1−/− | Depleted | ||
| Tlr3−/− | No effect | ||
| Tlr4−/− | |||
| Tlr5−/− | |||
| Tlr9−/− | |||
| LSK CD48+/−/CD150+/−/CD34+/Flk2+/− | Antibioticsa | Depleted | Josefsdottir et al., 2017 |
| LSK CD127−/Flk2+ | GF mice | Depleted | Iwamura et al., 2017 |
| Nod1−/− | Depleted | ||
| LSK CD48+/CD150+ | GF mice | Depleted | Lee et al., 2019 |
| MyD88−/− | Depleted | ||
| Unc93b1−/− | Depleted | ||
| Tlr3−/− | No effect | ||
| Tlr4−/− | |||
| Tlr5−/− | |||
| Tlr9−/− |
LSK, Lin−/Sca-1+/c-Kit+.
Antibiotics in drinking water: vancomycin, neomycin, ampicillin, and metronidazole.
Cell-extrinsic microbial signals
Although HSCs are competent to respond directly to microbial signals (Nagai et al., 2006; Zhao et al., 2014), the major effect of commensal microbes on HSCs seems to be cell-extrinsic via regulation of other cell types that act as intermediaries. As neonatal mice undergo initial commensal colonization, intestinal microbiota stimulate local type 3 innate lymphoid cells to produce IL-17A, resulting in increased plasma G-CSF and granulopoiesis (Deshmukh et al., 2014). GF mice have decreased bacterial cell wall component Nod1L (lauroyl-γ-D-glutamyl-meso-diaminopimelic acid; C12-iE-DAP) in their serum, correlating with significant decreases in IL-7, Flt3-L, thrombopoietin, and stem cell factor levels in peripheral blood (Iwamura et al., 2017). Serum cytokine levels can be restored by oral gavage with Nod1L, which also rescues HSPC numbers in the BM (Iwamura et al., 2017), supporting an instructive role for this ligand in controlling hematopoiesis via its effect on the BM microenvironment. Accordingly, BM mesenchymal stromal cells (MSCs) secrete IL-7, Flt3-L, thrombopoietin, and stem cell factor in response to Nod1L, and supernatant from wild type but not Nod1−/− MSCs stimulates HSPCs to proliferate (Iwamura et al., 2017). TNFα, IL-1β, and IL-6 levels are also lower in BM fluid but not peripheral blood of GF mice, and simultaneous injection of TNFα, IL-1β, and IL-6 blocking antibodies decreases both HSC and MPP numbers in the BM (Lee et al., 2019). That study pinpointed BM Cx3cr1+ mononuclear cells (MNCs) as the primary producers of TNFα, IL-1β, and IL-6 in the BM, with specific abrogation of either MyD88 or Unc93b1 in Cx3cr1+ MNCs phenocopying GF mice with respect to HSPC cellularity. BM Cx3cr1+ MNCs take up bacterial DNA in extracellular vesicles, which stimulates TNFα, IL-1β, and IL-6 secretion in a TLR-dependent manner.
These observations draw interesting parallels to the role that Cx3cr1+ AGM macrophages play in HSC emergence, and the partial dependence on TNFα in both scenarios (Mariani et al., 2019). It now remains to be determined whether there is correspondence between these cell populations at developmentally and anatomically disparate times and locations. As with many microbiome studies, standardizing experimental design and promoting greater transparency in reporting the many nongenetic factors (animal facility variations, mouse chow, disease models, and sequencing methods) that contribute to variation in microbiome composition and function will be indispensable to allow for optimal comparisons of findings across laboratories.
Hematopoietic imbalance
Commensal microbes play a crucial role in regulating steady-state myelopoiesis in the BM, which suggests that the hematopoietic compartment has coevolved with the microbiome so as to be appropriately tuned to the environment. It is also clear that dysfunction of each goes hand in hand with the other. Both clinical studies and experimental models indicate that dysbiosis of the gut in inflammatory bowel disorders is associated with abnormal, clonal, and even malignant hematopoiesis (Griseri et al., 2012; Zhang et al., 2019; Askling et al., 2005). The maladapted intestinal microbiome of obese mice directly causes hematopoietic stress, inducing depletion of HSCs and expansion of downstream MPPs, with stool transfer to healthy mice recapitulating features of dysregulated hematopoiesis (Luo et al., 2015). Pediatric leukemia patients exhibit alterations to their oral microbiome (Wang et al., 2014), and a direct role for barrier dysfunction and commensal-induced IL-6 in driving preleukemic myeloproliferation was also identified in a Tet2−/− model of clonal hematopoiesis, with the effect being abrogated in GF mice (Meisel et al., 2018). Conversely, microbial depletion enhances disease penetrance in a Pax5+/− mouse model of acute lymphoblastic leukemia (Vicente-Dueñas et al., 2020), underscoring the importance of a healthy and appropriately tuned microbiome in regulating hematopoiesis. Aging is also associated with a drop in microbiome diversity, expanded pathobiont colonization, and loss of gut barrier integrity, with resultant bacterial leakage triggering chronic multitissue inflammation (Nagpal et al., 2018). Further studies will also clarify whether HSCs themselves are the functional mediators of these phenomena or whether more active progenitors such as MPPs are instead the main targets. Lastly, dysregulation of the intestinal flora has significant clinical consequences in allogeneic HSC transplantation, which has recently been reviewed in detail (Shono and van den Brink, 2018). Further insight into the complex relationships between the microbiome and hematopoiesis is likely to lead to improved outcomes for patients in these scenarios.
Concluding remarks
Inflammatory signaling functions not only as an activator of emergency myelopoiesis in the context of inflammation, infection, and malignancy, but also as a core shaper of the hematopoietic compartment during development and homeostasis. Understanding the unique features of each of these hematopoietic scenarios that allows the same signaling pathways to yield disparate outcomes will be a crucial step to a better understanding of normal development and dysregulation and disease at each of these time points in the lifespan.
Acknowledgments
The authors acknowledge the many investigators in the field whose primary data could not be cited in this review because of space limitations.
This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (K08-DK124657) and a Louis V. Gerstner Jr. Scholar Award to A. Collins, and a grant from the National Institutes of Health’s National Heart, Lung, and Blood Institute (R35-HL135763) to E. Passegué.
Author contributions: A. Collins with the help of C.A. Mitchell wrote the initial draft, prepared the tables and figures, and revised the manuscript. E. Passegué provided oversight and leadership responsibility for this paper and edited the manuscript.
References
- Aagaard, K., Ma J., Antony K.M., Ganu R., Petrosino J., and Versalovic J.. 2014. The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra65. 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Nabhani, Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., Sparwasser T., Bérard M., Cerf-Bensussan N., and Eberl G.. 2019. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 50:1276–1288.e5. 10.1016/j.immuni.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Askling, J., Brandt L., Lapidus A., Karlén P., Björkholm M., Löfberg R., and Ekbom A.. 2005. Risk of haematopoietic cancer in patients with inflammatory bowel disease. Gut. 54:617–622. 10.1136/gut.2004.051771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., and Goodell M.A.. 2011. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32:57–65. 10.1016/j.it.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer, M.L., Schürch C.M., Saito Y., Geuking M.B., Li H., Cuenca M., Kovtonyuk L.V., McCoy K.D., Hapfelmeier S., Ochsenbein A.F., et al. 2014. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193:5273–5283. 10.4049/jimmunol.1400762 [DOI] [PubMed] [Google Scholar]
- Bertrand, J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y.R., and Traver D.. 2010. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 464:108–111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, J.Y., Kim A.D., Violette E.P., Stachura D.L., Cisson J.L., and Traver D.. 2007. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 134:4147–4156. 10.1242/dev.012385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas, A., Robert-Moreno A., and Espinosa L.. 2010. The Notch pathway in the developing hematopoietic system. Int. J. Dev. Biol. 54:1175–1188. 10.1387/ijdb.093049ab [DOI] [PubMed] [Google Scholar]
- Boisset, J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., and Robin C.. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 464:116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Bowie, M.B., Kent D.G., Dykstra B., McKnight K.D., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. USA. 104:5878–5882. 10.1073/pnas.0700460104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, M.B., McKnight K.D., Kent D.G., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2006. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Invest. 116:2808–2816. 10.1172/JCI28310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burberry, A., Zeng M.Y., Ding L., Wicks I., Inohara N., Morrison S.J., and Núñez G.. 2014. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe. 15:779–791. 10.1016/j.chom.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C.E., Traver D., Mayhall E., Shepard J.L., and Zon L.I.. 2005. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19:2331–2342. 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid, N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Cai, Z., de Bruijn M., Ma X., Dortland B., Luteijn T., Downing R.J., and Dzierzak E.. 2000. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 13:423–431. 10.1016/S1074-7613(00)00042-X [DOI] [PubMed] [Google Scholar]
- Chang, C.F., and Pollard M.. 1973. Effects of microbial flora on levels of colonay stimulating factor in serums of irradiated CFW mice. Proc. Soc. Exp. Biol. Med. 144:177–180. 10.3181/00379727-144-37551 [DOI] [PubMed] [Google Scholar]
- Chen, M.J., Yokomizo T., Zeigler B.M., Dzierzak E., and Speck N.A.. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 457:887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, G.M., Jeffery E., and Morrison S.J.. 2017. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17:573–590. 10.1038/nri.2017.53 [DOI] [PubMed] [Google Scholar]
- Crosse, E.I., Gordon-Keylock S., Rybtsov S., Binagui-Casas A., Felchle H., Nnadi N.C., Kirschner K., Chandra T., Tamagno S., Webb D.J., et al. 2020. Multi-layered Spatial Transcriptomics Identify Secretory Factors Promoting Human Hematopoietic Stem Cell Development. Cell Stem Cell. 27:822–839.e8. 10.1016/j.stem.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn, M.F.T.R., Ma X., Robin C., Ottersbach K., Sánchez M.-J., and Dzierzak E.. 2002. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 16:673–683. 10.1016/S1074-7613(02)00313-8 [DOI] [PubMed] [Google Scholar]
- de Bruijn, M.F.T.R., Speck N.A., Peeters M.C.E., and Dzierzak E.. 2000. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19:2465–2474. 10.1093/emboj/19.11.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau, M.C., Lager S., Sovio U., Gaccioli F., Cook E., Peacock S.J., Parkhill J., Charnock-Jones D.S., and Smith G.C.S.. 2019. Human placenta has no microbiome but can contain potential pathogens. Nature. 572:329–334. 10.1038/s41586-019-1451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laval, B., Maurizio J., Kandalla P.K., Brisou G., Simonnet L., Huber C., Gimenez G., Matcovitch-Natan O., Reinhardt S., David E., et al. 2020. C/EBPβ-Dependent Epigenetic Memory Induces Trained Immunity in Hematopoietic Stem Cells. Cell Stem Cell. 26:657–674.e8. 10.1016/j.stem.2020.01.017 [DOI] [PubMed] [Google Scholar]
- Deshmukh, H.S., Liu Y., Menkiti O.R., Mei J., Dai N., O’Leary C.E., Oliver P.M., Kolls J.K., Weiser J.N., and Worthen G.S.. 2014. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20:524–530. 10.1038/nm.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín, R., Roca F.J., Candel S., Sepulcre M.P., González-Rosa J.M., Alcaraz-Pérez F., Meseguer J., Cayuela M.L., Mercader N., and Mulero V.. 2013. TNF receptors regulate vascular homeostasis in zebrafish through a caspase-8, caspase-2 and P53 apoptotic program that bypasses caspase-3. Dis. Model. Mech. 6:383–396. 10.1242/dmm.010249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín-Palazón, R., Stachura D.L., Campbell C.A., García-Moreno D., Del Cid N., Kim A.D., Candel S., Meseguer J., Mulero V., and Traver D.. 2014. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 159:1070–1085. 10.1016/j.cell.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, M.A.G., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Etzrodt, M., Ahmed N., Hoppe P.S., Loeffler D., Skylaki S., Hilsenbeck O., Kokkaliaris K.D., Kaltenbach H.M., Stelling J., Nerlov C., and Schroeder T.. 2019. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood. 133:816–819. 10.1182/blood-2018-02-832998 [DOI] [PubMed] [Google Scholar]
- Fiedler, K., Kokai E., Bresch S., and Brunner C.. 2013. MyD88 is involved in myeloid as well as lymphoid hematopoiesis independent of the presence of a pathogen. Am. J. Blood Res. 3:124–140. [PMC free article] [PubMed] [Google Scholar]
- Frame, J.M., Kubaczka C., Long T.L., Esain V., Soto R.A., Hachimi M., Jing R., Shwartz A., Goessling W., Daley G.Q., and North T.E.. 2020. Metabolic Regulation of Inflammasome Activity Controls Embryonic Hematopoietic Stem and Progenitor Cell Production. Dev. Cell. 55:133–149.e6. 10.1016/j.devcel.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, J.M., McGrath K.E., and Palis J.. 2013. Erythro-myeloid progenitors: “definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol. Dis. 51:220–225. 10.1016/j.bcmd.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris, H., de Boer F., and van der Waaij D.. 1985. Myelopoiesis in experimentally contaminated specific-pathogen-free and germfree mice during oral administration of polymyxin. Infect. Immun. 50:437–441. 10.1128/IAI.50.2.437-441.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griseri, T., McKenzie B.S., Schiering C., and Powrie F.. 2012. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity. 37:1116–1129. 10.1016/j.immuni.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland, B.K., Huppert S.S., Kanungo J., Xue Y., Jiang R., Gridley T., Conlon R.A., Cheng A.M., Kopan R., and Longmore G.D.. 2004. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 104:3097–3105. 10.1182/blood-2004-03-1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., Zhang C., Wang L., Zhang P., Ma D., Lv J., and Liu F.. 2015. Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood. 125:1098–1106. 10.1182/blood-2014-09-601542 [DOI] [PubMed] [Google Scholar]
- Hérault, A., Binnewies M., Leong S., Calero-Nieto F.J., Zhang S.Y., Kang Y.-A., Wang X., Pietras E.M., Chu S.H., Barry-Holson K., et al. 2017. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 544:53–58. 10.1038/nature21693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura, C., Bouladoux N., Belkaid Y., Sher A., and Jankovic D.. 2017. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 129:171–176. 10.1182/blood-2016-06-723742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsdottir, K.S., Baldridge M.T., Kadmon C.S., and King K.Y.. 2017. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 129:729–739. 10.1182/blood-2016-03-708594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, J.H., Entringer M.A., and Robinson W.A.. 1979. Bacterial stimulation of serum colony-stimulating activity and neutrophil production in germ-free mice. Proc. Soc. Exp. Biol. Med. 162:44–47. 10.3181/00379727-162-40615 [DOI] [PubMed] [Google Scholar]
- Khosravi, A., Yáñez A., Price J.G., Chow A., Merad M., Goodridge H.S., and Mazmanian S.K.. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 15:374–381. 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieusseian, A., Brunet de la Grange P., Burlen-Defranoux O., Godin I., and Cumano A.. 2012. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 139:3521–3530. 10.1242/dev.079210 [DOI] [PubMed] [Google Scholar]
- Kim, A.D., Melick C.H., Clements W.K., Stachura D.L., Distel M., Panáková D., MacRae C., Mork L.A., Crump J.G., and Traver D.. 2014. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 33:2363–2373. 10.15252/embj.201488784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P.G., Canver M.C., Rhee C., Ross S.J., Harriss J.V., Tu H.-C., Orkin S.H., Tucker H.O., and Daley G.Q.. 2016. Interferon-α signaling promotes embryonic HSC maturation. Blood. 128:204–216. 10.1182/blood-2016-01-689281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa, K., and Herbomel P.. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 464:112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kobayashi, I. 2018. Development of Hematopoietic Stem Cells in Zebrafish. In Zebrafish, Medaka, and Other Small Fishes. Springer Singapore, Singapore. 37–57. 10.1007/978-981-13-1879-5_3 [DOI] [Google Scholar]
- Kobayashi, I., Kobayashi-Sun J., Kim A.D., Pouget C., Fujita N., Suda T., and Traver D.. 2014. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature. 512:319–323. 10.1038/nature13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano, K., Chiba S., Kunisato A., Sata M., Saito T., Nakagami-Yamaguchi E., Yamaguchi T., Masuda S., Shimizu K., Takahashi T., et al. 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 18:699–711. 10.1016/S1074-7613(03)00117-1 [DOI] [PubMed] [Google Scholar]
- Lam, E.Y.N., Hall C.J., Crosier P.S., Crosier K.E., and Flores M.V.. 2010. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood. 116:909–914. 10.1182/blood-2010-01-264382 [DOI] [PubMed] [Google Scholar]
- Lee, S., Kim H., You G., Kim Y.-M., Lee S., Le V.-H., Kwon O., Im S.-H., Kim Y.-M., Kim K.S., et al. 2019. Bone marrow CX3CR1+ mononuclear cells relay a systemic microbiota signal to control hematopoietic progenitors in mice. Blood. 134:1312–1322. 10.1182/blood.2019000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkopoulos, S., Polyzou A., Derecka M., Bergo V., Clapes T., Cauchy P., Jerez-Longres C., Onishi-Seebacher M., Yin N., Martagon-Calderón N.-A., et al. 2020. Repetitive Elements Trigger RIG-I-like Receptor Signaling that Regulates the Emergence of Hematopoietic Stem and Progenitor Cells. Immunity. 53:934–951.e9. 10.1016/j.immuni.2020.10.007 [DOI] [PubMed] [Google Scholar]
- Li, Y., Esain V., Teng L., Xu J., Kwan W., Frost I.M., Yzaguirre A.D., Cai X., Cortes M., Maijenburg M.W., et al. 2014. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28:2597–2612. 10.1101/gad.253302.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Kong W., Yang W., Patel R.M., Casey E.B., Okeyo-Owuor T., White J.M., Porter S.N., Morris S.A., and Magee J.A.. 2020. Single-Cell Analysis of Neonatal HSC Ontogeny Reveals Gradual and Uncoordinated Transcriptional Reprogramming that Begins before Birth. Cell Stem Cell. 27:732–747.e7. 10.1016/j.stem.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke, G.J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K.J., Basu S., Zhan Y.F., and Dunn A.R.. 1994. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 84:1737–1746. 10.1182/blood.V84.6.1737.1737 [DOI] [PubMed] [Google Scholar]
- Lim, S.-E., Esain V., Kwan W., Theodore L.N., Cortes M., Frost I.M., Liu S.Y., and North T.E.. 2017. HIF1α-induced PDGFRβ signaling promotes developmental HSC production via IL-6 activation. Exp. Hematol. 46:83–95.e6. 10.1016/j.exphem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Wu H.Y., Wesselschmidt R., Kornaga T., and Link D.C.. 1996. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 5:491–501. 10.1016/S1074-7613(00)80504-X [DOI] [PubMed] [Google Scholar]
- Luo, Y., Chen G.-L., Hannemann N., Ipseiz N., Krönke G., Bäuerle T., Munos L., Wirtz S., Schett G., and Bozec A.. 2015. Microbiota from Obese Mice Regulate Hematopoietic Stem Cell Differentiation by Altering the Bone Niche. Cell Metab. 22:886–894. 10.1016/j.cmet.2015.08.020 [DOI] [PubMed] [Google Scholar]
- Ma, X., Robin C., Ottersbach K., and Dzierzak E.. 2002. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells. 20:514–521. 10.1634/stemcells.20-6-514 [DOI] [PubMed] [Google Scholar]
- MacVittie, T.J., and Walker R.I.. 1978. Canine granulopoiesis: alterations induced by suppression of gram-negative flora. Exp. Hematol. 6:639–647. [PubMed] [Google Scholar]
- Mariani, S.A., Li Z., Rice S., Krieg C., Fragkogianni S., Robinson M., Vink C.S., Pollard J.W., and Dzierzak E.. 2019. Pro-inflammatory Aorta-Associated Macrophages Are Involved in Embryonic Development of Hematopoietic Stem Cells. Immunity. 50:1439–1452.e5. 10.1016/j.immuni.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Shen C.-C., Challen G.A., and King K.Y.. 2014. Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells. 32:3023–3030. 10.1002/stem.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, K.E., Frame J.M., Fegan K.H., Bowen J.R., Conway S.J., Catherman S.C., Kingsley P.D., Koniski A.D., and Palis J.. 2015. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 11:1892–1904. 10.1016/j.celrep.2015.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLemore, M.L., Grewal S., Liu F., Archambault A., Poursine-Laurent J., Haug J., and Link D.C.. 2001. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 14:193–204. 10.1016/S1074-7613(01)00101-7 [DOI] [PubMed] [Google Scholar]
- Meisel, M., Hinterleitner R., Pacis A., Chen L., Earley Z.M., Mayassi T., Pierre J.F., Ernest J.D., Galipeau H.J., Thuille N., et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 557:580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirantes, C., Passegué E., and Pietras E.M.. 2014. Pro-inflammatory cytokines: emerging players regulating HSC function in normal and diseased hematopoiesis. Exp. Cell Res. 329:248–254. 10.1016/j.yexcr.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller, N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., and Sieweke M.H.. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 497:239–243. 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A.M., Medvinsky A., Strouboulis J., Grosveld F., and Dzierzak E.. 1994. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1:291–301. 10.1016/1074-7613(94)90081-7 [DOI] [PubMed] [Google Scholar]
- Nagai, Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., and Kincade P.W.. 2006. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 24:801–812. 10.1016/j.immuni.2006.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, R., Mainali R., Ahmadi S., Wang S., Singh R., Kavanagh K., Kitzman D.W., Kushugulova A., Marotta F., and Yadav H.. 2018. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Healthy Aging. 4:267–285. 10.3233/NHA-170030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North, T., Gu T.L., Stacy T., Wang Q., Howard L., Binder M., Marín-Padilla M., and Speck N.A.. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 126:2563–2575. 10.1242/dev.126.11.2563 [DOI] [PubMed] [Google Scholar]
- North, T.E., de Bruijn M.F.T.R., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., and Speck N.A.. 2002. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 16:661–672. 10.1016/S1074-7613(02)00296-0 [DOI] [PubMed] [Google Scholar]
- Olson, O.C., Kang Y.-A., and Passegué E.. 2020. Normal Hematopoiesis Is a Balancing Act of Self-Renewal and Regeneration. Cold Spring Harb. Perspect. Med. 10:a035519. 10.1101/cshperspect.a035519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelio, C., Haak E., Peeters M., and Dzierzak E.. 2008. Interleukin-1-mediated hematopoietic cell regulation in the aorta-gonad-mesonephros region of the mouse embryo. Blood. 112:4895–4904. 10.1182/blood-2007-12-123836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelio, C., Peeters M., Haak E., van der Horn K., and Dzierzak E.. 2009. Interleukin-1 regulates hematopoietic progenitor and stem cells in the midgestation mouse fetal liver. Haematologica. 94:462–469. 10.3324/haematol.13728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis, J. 2014. Primitive and definitive erythropoiesis in mammals. Front. Physiol. 5:3. 10.3389/fphys.2014.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos, A.D., Zhang L., Snow J.W., Jones D.M., Smith A.M., El Kasmi K.C., Liu F., Goldsmith M.A., Link D.C., Murray P.J., and Watowich S.S.. 2006. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 108:3682–3690. 10.1182/blood-2006-02-003012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Muñoz, M.E., Arrieta M.-C., Ramer-Tait A.E., and Walter J.. 2017. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 5:48. 10.1186/s40168-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., and Passegué E.. 2013. Linking HSCs to their youth. Nat. Cell Biol. 15:885–887. 10.1038/ncb2817 [DOI] [PubMed] [Google Scholar]
- Pietras, E.M., Lakshminarasimhan R., Techner J.-M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262. 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.-M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Reynaud D., Kang Y.-A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud, D., Pietras E., Barry-Holson K., Mir A., Binnewies M., Jeanne M., Sala-Torra O., Radich J.P., and Passegué E.. 2011. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 20:661–673. 10.1016/j.ccr.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, M.K., Liu F., Iwasaki H., Akashi K., and Link D.C.. 2003. Pivotal role of granulocyte colony-stimulating factor in the development of progenitors in the common myeloid pathway. Blood. 102:3562–3568. 10.1182/blood-2003-02-0593 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno, A., Guiu J., Ruiz-Herguido C., López M.E., Inglés-Esteve J., Riera L., Tipping A., Enver T., Dzierzak E., Gridley T., et al. 2008. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 27:1886–1895. 10.1038/emboj.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, C., Ottersbach K., Durand C., Peeters M., Vanes L., Tybulewicz V., and Dzierzak E.. 2006. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev. Cell. 11:171–180. 10.1016/j.devcel.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Rolig, A.S., Parthasarathy R., Burns A.R., Bohannan B.J.M., and Guillemin K.. 2015. Individual Members of the Microbiota Disproportionately Modulate Host Innate Immune Responses. Cell Host Microbe. 18:613–620. 10.1016/j.chom.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez, M.-J., Holmes A., Miles C., and Dzierzak E.. 1996. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 5:513–525. 10.1016/S1074-7613(00)80267-8 [DOI] [PubMed] [Google Scholar]
- Sato, T., Onai N., Yoshihara H., Arai F., Suda T., and Ohteki T.. 2009. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 15:696–700. 10.1038/nm.1973 [DOI] [PubMed] [Google Scholar]
- Sawamiphak, S., Kontarakis Z., and Stainier D.Y.R.. 2014. Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev. Cell. 31:640–653. 10.1016/j.devcel.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers, K., Campbell T.B., and Passegué E.. 2015. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 16:254–267. 10.1016/j.stem.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, B.O., and Bäckhed F.. 2016. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22:1079–1089. 10.1038/nm.4185 [DOI] [PubMed] [Google Scholar]
- Schuettpelz, L.G., Borgerding J.N., Christopher M.J., Gopalan P.K., Romine M.P., Herman A.C., Woloszynek J.R., Greenbaum A.M., and Link D.C.. 2014. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia. 28:1851–1860. 10.1038/leu.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono, Y., and van den Brink M.R.M.. 2018. Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nat. Rev. Cancer. 18:283–295. 10.1038/nrc.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber, F.G., Tarcsay L., and Dukor P.. 1978. Modulations of myelopoiesis in vivo by chemically pure preparations of cell wall components from gram-negative bacteria: effects at different stages. Infect. Immun. 20:40–49. 10.1128/iai.20.1.40-49.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachura, D.L., Svoboda O., Campbell C.A., Espín-Palazón R., Lau R.P., Zon L.I., Bartůněk P., and Traver D.. 2013. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood. 122:3918–3928. 10.1182/blood-2012-12-475392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, T., Yamamura S., Kuwano Y., and Abo T.. 1996. Level of myelopoiesis in the bone marrow is influenced by intestinal flora. Cell. Immunol. 173:155–161. 10.1006/cimm.1996.0261 [DOI] [PubMed] [Google Scholar]
- Taoudi, S., Gonneau C., Moore K., Sheridan J.M., Blackburn C.C., Taylor E., and Medvinsky A.. 2008. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 3:99–108. 10.1016/j.stem.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Taoudi, S., Morrison A.M., Inoue H., Gribi R., Ure J., and Medvinsky A.. 2005. Progressive divergence of definitive haematopoietic stem cells from the endothelial compartment does not depend on contact with the foetal liver. Development. 132:4179–4191. 10.1242/dev.01974 [DOI] [PubMed] [Google Scholar]
- Tie, R., Li H., Cai S., Liang Z., Shan W., Wang B., Tan Y., Zheng W., and Huang H.. 2019. Interleukin-6 signaling regulates hematopoietic stem cell emergence. Exp. Mol. Med. 51:1–12. 10.1038/s12276-019-0320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova, J., Tran Chau V., Julien E., Mateos-Langerak J., Gonzalez C., Lelièvre E., Lutfalla G., Tavian M., and Kissa K.. 2015. Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 6:6227. 10.1038/ncomms7227 [DOI] [PubMed] [Google Scholar]
- Ueda, Y., Cain D.W., Kuraoka M., Kondo M., and Kelsoe G.. 2009. IL-1R type I-dependent hemopoietic stem cell proliferation is necessary for inflammatory granulopoiesis and reactive neutrophilia. J. Immunol. 182:6477–6484. 10.4049/jimmunol.0803961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Dueñas, C., Janssen S., Oldenburg M., Auer F., González-Herrero I., Casado-García A., Isidro-Hernández M., Raboso-Gallego J., Westhoff P., Pandyra A.A., et al. 2020. An intact gut microbiome protects genetically predisposed mice against leukemia. Blood. 136:2003–2017. 10.1182/blood.2019004381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Xue J., Zhou X., You M., Du Q., Yang X., He J., Zou J., Cheng L., Li M., et al. 2014. Oral microbiota distinguishes acute lymphoblastic leukemia pediatric hosts from healthy populations. PLoS One. 9:e102116. 10.1371/journal.pone.0102116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q., and Frenette P.S.. 2018. Niches for Hematopoietic Stem Cells and Their Progeny. Immunity. 48:632–648. 10.1016/j.immuni.2018.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser, M., Demel U.M., Stein S., Chen-Wichmann L., Touzot F., Santilli G., Sujer S., Brendel C., Siler U., Cavazzana M., et al. 2016. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J. Allergy Clin. Immunol. 138:219–228.e9. 10.1016/j.jaci.2015.11.028 [DOI] [PubMed] [Google Scholar]
- Wilson, A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Yamashita, M., and Passegué E.. 2019. TNF-α Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell. 25:357–372.e7. 10.1016/j.stem.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H., Gao S., Chen H., Liu X., Zhou J., de The H., and Zhu J.. 2019. Primitive macrophages are dispensable for HSPC mobilization and definitive hematopoiesis. Blood. 134:782–784. 10.1182/blood.2018893974 [DOI] [PubMed] [Google Scholar]
- Zhang, C.R.C., Nix D., Gregory M., Ciorba M.A., Ostrander E.L., Newberry R.D., Spencer D.H., and Challen G.A.. 2019. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp. Hematol. 80:36–41.e3. 10.1016/j.exphem.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Nguyen-Jackson H., Panopoulos A.D., Li H.S., Murray P.J., and Watowich S.S.. 2010. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 116:2462–2471. 10.1182/blood-2009-12-259630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.L., Ma C., O’Connell R.M., Mehta A., DiLoreto R., Heath J.R., and Baltimore D.. 2014. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell. 14:445–459. 10.1016/j.stem.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Zhang G., Gong Y., Ning X., Bai Z., He J., Zhou F., Ni Y., Lan Y., and Liu B.. 2019. Embryonic lineage tracing with Procr-CreER marks balanced hematopoietic stem cell fate during entire mouse lifespan. J. Genet. Genomics. 46:489–498. 10.1016/j.jgg.2019.10.005 [DOI] [PubMed] [Google Scholar]