Abstract
The aerial portion of a plant, namely the leaf, is inhabited by pathogenic and non-pathogenic microbes. The leaf’s physical and chemical properties, combined with fluctuating and often challenging environmental factors, create surfaces that require a high degree of adaptation for microbial colonization. As a consequence, specific interactive processes have evolved to establish a plant leaf niche. Little is known about the impact of the host immune system on phyllosphere colonization by non-pathogenic microbes. These organisms can trigger plant basal defenses and benefit the host by priming for enhanced resistance to pathogens. In most disease resistance responses, microbial signals are recognized by extra- or intracellular receptors. The interactions tend to be species specific and it is unclear how they shape leaf microbial communities. In natural habitats, microbe–microbe interactions are also important for shaping leaf communities. To protect resources, plant colonizers have developed direct antagonistic or host manipulation strategies to fight competitors. Phyllosphere-colonizing microbes respond to abiotic and biotic fluctuations and are therefore an important resource for adaptive and protective traits. Understanding the complex regulatory host–microbe–microbe networks is needed to transfer current knowledge to biotechnological applications such as plant-protective probiotics.
Keywords: Biofilm, innate immunity, microbe–microbe interaction, microbial colonization, phyllosphere, quorum sensing
Microbial colonization of above-ground parts of plants is a dynamic and interactive process that requires a high degree of adaptation. Understanding complex host–microbe–microbe interactions is key to new strategies for plant protection.
Introduction
This review examines how aerial parts of plants, particularly leaves, are colonized by microbes. The first section (‘Colonizing leaf surfaces’) dissects biotic and abiotic factors that shape leaf microbial communities and determine the quality and quantity of colonization. We discuss pre-formed barriers, such as the cuticle, that restrict plant host colonization, and environmental conditions that enhance selection pressures. As a consequence of the extreme conditions on leaves, the properties and generation of biofilms through quorum sensing (QS) are considered. Leaf colonization by microbes is not only impacted by host and environmental factors but also by resident microbes, including pathogens that can severely perturb microbial communities. The second section (‘Microbe–microbe–host interactions’) discusses effects of microbial communities on host susceptibility to pathogens and the impact of plant pathogens and endophytes on microbial host colonization and community composition. A particular focus is on microbe–microbe interactions that are often mediated via the plant host. Individual plant cells also have the capacity to steer microbial activities by pre-formed or induced structures and compounds that influence microbial growth on the leaf surface. The third section (‘Role of the plant immune system in shaping the phyllosphere microbiome’) considers the role of the plant immune system on microbial host interactions with a focus on plant leaf colonization, leaf–microbe outputs, and microbial diversity.
Colonizing leaf surfaces
The leaf environment
All terrestrial plants are inhabited by diverse, complex, and interactive communities of microorganisms. With this intimate association, the host plant and its associated microbiota are regarded as a close knit entity and are collectively defined as the holobiont. The holobiont concept implies that evolutionary selection takes place between the host and its associated microbes, and within microbe–microbe members (Vandenkoornhuyse et al., 2015; Hassani et al., 2018; Teixeira et al., 2019). The phenotype of a plant host is the collective outcome of numerous interactions with its microbiota in a particular environment at a time (Vorholt et al., 2017). The ‘phyllosphere’ is referred to as the above-ground portion of plants, dominated by leaves. Its surface represents one of the most abundant habitats on earth (Lindow and Brandl, 2003). Leaves create a fluctuating and unstable environment exposed to multiple stresses and relatively devoid of nutrient sources (Bringel and Couée, 2015). The study of microbial communities inhabiting this stressful leaf habitat and their collective contribution to plant growth, development, and protection has gained intense interest over the last decade. The leaf harbors diverse microorganisms that inhabit the surface and the interior, and are known as epiphytes and endophytes, respectively (Beattie and Lindow, 1999; Lindow and Brandl, 2003). These microorganisms include bacteria as the most common inhabitants, followed by filamentous fungi and yeast strains (Stone et al., 2018), protists (Sapp et al., 2018), and bacteriophages (Balogh et al., 2018). The bacterial titer accounts for ~106–107 cells cm–2 of leaf area (Lindow and Brandl, 2003), whereas a typical yeast titer ranges from 10 to 104 cells cm–2 of leaf (Shivas and Brown, 1984). The origin of leaf microbial communities is not restricted to a single source. Microbes can colonize the plant leaf vertically through seeds or pollen and horizontally from the air, soil, and insects (Vorholt, 2012; Bodenhausen et al., 2013; Maignien et al., 2014; Bai et al., 2015; Frank et al., 2017).
A stressed and nutrient-poor condition of the leaf surface makes this environment selective to certain microorganisms. Hence, different microbial mechanisms such as ability to extract nutrients, produce hormones and surfactants, as well as motility and biofilm formation can be key to colonization success (Nadakuduti et al., 2012; Ueda et al., 2018; Leveau, 2019; Oso et al., 2019; Streletskii et al., 2019). Most epiphytes survive on the leaf surface by forming large aggregates which help them to cope with the surrounding milieu and maintain a hydrated surface by production of extracellular polymeric substances (EPSs) (Morris and Kinkel, 2002; Lindow and Brandl, 2003; Baldotto and Olivares, 2008; Vorholt, 2012). Other microbes, which are not considered as part of common leaf microbiota, are human commensal or pathogenic bacteria. These can survive and proliferate on the plant leaf, as documented by numerous outbreak studies of human infections on leafy vegetables (Beuchat, 2002; Lindow and Brandl, 2003; Naimi et al., 2003; Islam et al., 2004; Melotto et al., 2006; Munther et al., 2020). Such microbes are able to colonize and survive in an unfavorable leaf environment if they are teamed up with aggregates of pre-colonized leaf microbiota (Monier and Lindow, 2005).
Host-adapted microbial colonizers are more tolerant to abiotic stresses such as harmful UV radiation (Kamo et al., 2018), oxidative stress, and desiccation (Vorholt, 2012), and can utilize nutrients (Crombie et al., 2018) and vitamins (Yoshida et al., 2019) available on the leaf surface. By mitigating biotic and/or abiotic stress(es) and influencing plant growth and fitness, microbes develop adaptive traits and intimate associations with leaves (Vorholt, 2012; Helfrich et al., 2018). Plant host–microbiota interactions are built on the transfer of molecular and genetic information. Important colonization factors, such as secondary metabolites, QS systems, biofilm formation, and cell signaling, are responsible for this exchange of information (Braga et al., 2016; Leveau, 2019; Flores-Núñez et al., 2020). Leaf microbial communities influence plant fitness by modulating the host plant immune system and promoting plant growth in above-ground tissues (Stone et al., 2018).
Leaf surface structure and chemistry relevant to microbiota assembly
Leaf structure and its surface chemistry create a peculiar microenvironment. During evolution, the formation of a leaf cuticle layer was a prerequisite for land plants to survive out of water. The composition and function of the cuticle is summarized in a review (Müller and Riederer, 2005). The cuticle also covers preferential sites for microbiota colonization, such as the surface of leaf epidermal cells, stomata, and trichomes (Teplitski et al., 2011; Peredo and Simmons, 2018). The cuticle layer is composed of structurally and chemically heterogeneous compounds primarily made of biopolyester cutin, wax, and more minor compounds such as phenolics, cutan, and polysaccharides (Gniwotta et al., 2005; Nawrath et al., 2013). Under constant exposure to abiotic and biotic factors, the epidermal layer of leaf tissues performs its primary function as a protective barrier by preventing seepage of water from the leaf surface as well as external water and solutes from entering the plant. Moreover, the cuticle plays a critical role in mediating interactions with leaf microbiota, including commensal, beneficial, and pathogenic microorganisms (Schönherr, 2006; Vorholt, 2012; Vacher et al., 2016).
Leaf microbiota utilize a number of strategies to enter and penetrate the leaf cuticle. A major route is through natural stomatal openings and wounds resulting from lytic enzymes and osmotic pressure (Frank et al., 2017). Stomata are enclosed by two guard cells to regulate gas exchange and transpiration from the leaf epidermis. Movement of microbes between the external and internal parts of the phyllosphere via stomata has been generally regarded as a passive process, in which the microorganism and plant leaf do not engage in active dialog to permit and/or restrict microbe entry (Underwood et al., 2007). Studies have demonstrated the role of signal transduction cascades in bacterial regulation of stomatal aperture (Zeng et al., 2010; Zheng et al., 2012).
Stomatal aperture is regulated by biotic and abiotic environmental conditions. In general, successful microbial colonization of the leaf depends on stomatal aperture (Ou et al., 2014). Decades of research have shown that phytopathogenic bacteria and fungi exploit stomata as a point of entry for invasion. To breach surface barriers via stomata, host-adapted bacteria subvert plant abiotic stress signaling to suppress stomatal closure during infection (Melotto et al., 2006; Okamoto et al., 2009; Zeng et al., 2010; Xin et al., 2018). To counter pathogen invasion, stomatal guard cells recognize diverse pathogen-/microbe-associated molecular patterns (PAMPs/MAMPs) such as flagellin, chitin, and chitosan (Arnaud and Hwang, 2015). These recognitions and downstream signaling processes lead to closing of stomatal pores and hence prevent bacterial entry as part of the plant immune response. Suppressing the stomatal defense system is an important adaptation mechanism for switching from an epiphytic to an endophytic lifestyle, leading to bacterial disease (Melotto et al., 2017).
Structural and chemical heterogeneity of the leaf cuticle is detected within and between plant genotypes, organs, and even developmental stages (Müller and Riederer, 2005). A role for leaf surface microbiota, together with leaf cuticle mechanisms, was observed in Arabidopsis thaliana in resistance against Botrytis cinerea, a broad host-range necrotrophic fungal pathogen (Ritpitakphong et al., 2016). Analyses reveal important effects of variation in cuticle chemical and physical composition modulating associations between plants and microbiota, including beneficial and pathogenic microorganisms (Aragón et al., 2017).
Apoplastic spaces inside leaves are large intercellular spaces which mediate gas exchange between cells and are essential for most plant species to achieve efficient photosynthesis (Chen et al., 2020). Humidity controls occupancy of pathogens in apoplastic spaces and is an important initial determinant of leaf colonization (Xin et al., 2016, 2018). Indeed, water availability in the leaf apoplast is a key factor determining successful colonization by neutral and beneficial, but also pathogenic microbes that compete with the host for water (Aung et al., 2018; Chen et al., 2020). It is therefore not surprising that the leaf apoplast has emerged as a decisive environment for host–microbe communication during colonization and for the mobilization of active defense mechanisms to counter pathogen infection.
Diversity of leaf-colonizing microbiota
Distinct microbiota interactions are found in the leaf compartment which influence the plant host, shaping the microbial community and colonization success. The microbiota in the leaf is not composed of a single species but rather intraspecies, interspecies, and cross-kingdom microbial assemblies of bacteria, yeast, fungi, and protists, establishing the leaf environment (Hardoim et al., 2015). The establishment and abundance of these leaf microbial communities and their distinct effects on the host plant—whether this is commensal, beneficial, or detrimental—are the outcome of numerous interactive processes. These processes are, in turn, influenced by incoming and outgoing microorganisms to and from the leaf habitat and their rate of multiplication, dispersal, and decline in a particular niche (Vorholt, 2012; Wagner et al., 2014, 2016; Lebeis et al., 2015; Vacher et al., 2016; Remus-Emsermann and Schlechter, 2018; Stone et al., 2018; Laforest-Lapointe and Whitaker, 2019).
To gain a better understanding of plant leaf–microbe interactions and outcomes, it is crucial to identify and characterize the microbial community that has evolved and adapted to the leaf environment. There are now ample studies describing the diversity and community structure of leaf-associated microbes, their characterization by next-generation sequencing, culture-independent and culture-dependent methods based on taxonomic markers, and their roles in host development and protection against stress (Romero et al., 2014; Harsonowati et al., 2017; Wallace et al., 2018; Dong et al., 2019). Notably, the leaf-associated microbial community in plants such as common bean (Phaseolus vulgaris), lettuce (Lactuca sativa), and neotropical forest and poplar trees consists of four major bacterial phyla, namely Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria (de Oliveira Costa et al., 2012; Rastogi et al., 2012; Kembel et al., 2014; Durand et al., 2018). Rastogi et al. (2012) demonstrated variability in bacterial community profiles on field-grown lettuce leaves with respect to time, space, and environment. In characterizing a tropical tree microbiome, Kembel et al. (2014) showed that leaf bacterial communities are dominated by Actinobacteria, Alpha-, Beta-, and Gammaproteobacteria, and Sphingobacteria. The Kembel study also identified microbial correlations with host growth, mortality, and function. In A. thaliana, there is a taxonomic and functional overlap between bacterial communities in the leaves and roots, and evidence that soil is the main driver of leaf bacterial community structure (Bai et al., 2015). Phyla belonging to Proteobacteria, Actinobacteria, and Bacteroidetes were found to be most abundant in A. thaliana, common ash (Fraxinus excelsior), and other tree leaves (Redford et al., 2010; Bodenhausen et al., 2013; Bai et al., 2015; Griffiths et al., 2020; Ulrich et al., 2020).
Redford et al. (2010) in a study of leaves of 56 tree species also concluded that interspecies variation is more prevalent than intraspecies variation and that there is a correlation between tree phylogeny and bacterial community composition. Using 16S rRNA, ammonia oxidation (amoA), and nitrogen fixation (nifH) gene markers, Bao et al. (2020) characterized phyllosphere bacteria and established differences in the diversity and composition of bacteria, including diazotrophic communities, over two seasons in three different tree species. An abundance of season-specific bacterial genera highlighted that there might be particular mechanisms of leaf adaptation in different seasons (Bao et al., 2020). However, Methylobacterium and Sphingomonas species were highly abundant in the plant leaf environment of three species, namely A. thaliana, Trifolium repens, and Glycine max (Delmotte et al., 2009). Furthermore, Durand et al. (2018) reported bacterial community members belonging to Methylobacterium, Kineococcus, Sphingomonas, and Hymenobacter on the leaf surface of poplar trees. Apart from these, members of the genus Pseudomonas are also predominantly found in the phyllosphere of a wide range of plant species (Rastogi et al., 2013).
Numerous studies revealed the association of diverse leaf epiphytic and endophytic filamentous fungi and yeasts with plant host (Arnold et al., 2007; Kharwar et al., 2010; Porras-Alfaro and Bayman, 2011; Sun et al., 2014; Wang et al., 2016; Qian et al., 2018; Yao et al., 2019; Into et al., 2020). Huge diversity, spatial structure, and host association were observed among leaf endophytes and a role in protecting the plant against the devastating foliar oomycete pathogen, Phytophthora sp. (Arnold et al., 2003). Qian et al. (2018) found that Dothideomycetes and Eurotiomycetes are dominant members in Mussaenda pubescens and identified intraspecific host genetics as primary drivers in shaping regional phyllosphere fungal communities. Yao et al. (2019) reported that Dothideomycetes and Tremellomycetes are dominant members in the mangrove ecosystem in six mangrove species, namely Aegiceras corniculatum, Avicennia marina, Bruguiera gymnorrhiza, Kandelia candel, Rhizophora stylosa, and Excoecaria agallocha, and obtain ecosystem insights for species co-existence and community stability. Using a culture-independent approach, Agler et al. (2016) extracted yeast genera belonging to Protomyces, Dioszegia, Leucosporidium, and Rhodotorula in the phyllosphere of wild A. thaliana populations from Germany. In another study, Dhayanithy et al. (2019) characterized fungal endophytes from the leaves and stems of Catharanthus roseus and reported Colletotrichum, Alternaria, and Chaetomium genera as common members. Cladosporium and Alternaria filamentous fungi, Cryptococcus and Sporobolomyces yeasts, and Pseudomonas spp. and Erwinia herbicola bacteria were commonly found colonizing leaves of Beta vulgaris (Thompson et al., 1993). In another study, by Glushakova and Chernov (2004), changes in epiphytic yeast populations were observed over the year in evergreen common wood sorrel Oxalis acetosella L., revealing that species diversity was high in autumn and low in spring. In contrast, Rhodotorula glutinis and Sporobolomyces roseus species were abundant throughout the year. Interestingly, leaves also harbor certain suppressive bacteria that can restrict phyllosphere bacterial diversity and increase resistance, for example in maize, to Southern leaf blight (SLB) fungal infection (Balint-Kurti et al., 2010). Numerous studies showed the diversity and abundance of yeast in the leaf environment; however, there is a need for more in-depth understanding of the biological mechanisms that showed their role towards host growth and protection.
Sometimes, serious human pathogens such as Salmonella enterica serovar Typhimurium 14028s (S. typhimurium 14028s) and Escherichia coli O157:H7 (EcO157) colonize fresh leafy vegetables such as lettuce (Lactuca sativa) via damaged leaf tissue and can cause food-borne disease outbreaks (Saldaña et al., 2011; Roy et al., 2013). At the site of injury, lettuce leaf tissue provides substrates for proliferation, and choline which helps the pathogen combat osmotic stress (Scott et al., 2017). It is not clearly understood how human pathogenic/commensal bacteria survive in the extreme environmental conditions encountered by plants. Nevertheless, human pathogens can stay as persister cells also known as cells in a transient dormant state on the plant and cause disease once they encounter a new environment (Munther et al., 2020). In a recent study, Jacob and Melotto (2020) discovered genetic diversity among lettuce genotypes and resident human pathogenic S. Typhimurium 14028s and E. coli O157:H7, and found a link between genetic diversity and differences in plant immune responses to these bacteria. However, in comparison with human pathogens, less is studied on persister cells in phytopathogen associations with the leaf (Martins et al., 2018). Mechanisms preventing invasion by plant pathogenic microorganisms and plant-induced defense responses are discussed below.
Role of metabolites in leaf preferential colonization by microbiota
Although present as epiphytes on plant hosts, not all microbes are able to colonize and establish themselves inside leaves. Initial colonization and entry of microbes as a community into a leaf is not a random process in which arbitrary communities adhere and grow, but an organized series of events. Steps involve attachment, movement, and cellular interactions. These steps are facilitated by the leaf surface structure (see above) which regulates colonization as an important priming event in microbial community interactions with the plant (Lebeis et al., 2015; Flemming and Wuertz, 2019). Research hypothesizes that a small community of established microbes associated with the host are inherited vertically through the seed (Nelson, 2018). These microbes are thought to influence recruitment, structuring, and stabilizing of microbiota throughout the plant life cycle (Newcombe et al., 2018).
While several studies have uncovered a role for microbiota at the site of leaf colonization, the functional relationship between leaves and their associated microbial community is poorly understood. Some studies showed production of biosurfactants by epiphytic bacteria on the leaf and the role of these molecules in movement and nutrient acquisition aiding leaf surface adaptation (Bunster et al., 1989; Neu et al., 1990; Hutchison and Johnstone, 1993; Schreiber et al., 2005; Burch et al., 2011, 2012, 2014). Analysis of gain- and loss-of-biosurfactant (Syringafactin) Pseudomonas syringae pv. syringae B728a strains on bean (Phaseolus vulgaris) leaves indicated that this hygroscopic biosurfactant increases diffusion of water across a waxy leaf cuticle surface which attracts moisture and nutrients to benefit the bacteria (Burch et al., 2014). As the initial microbe–leaf contact point, a role for cuticle wax biosynthesis genes in phyllosphere bacterial community composition was observed in A. thaliana (Reisberg et al., 2013).
As an abundant genus of leaf microbiota, culturable isolates assigned to Sphingomonas sp. were found to provide protection in A. thaliana against the foliar pathogen P. syringae pv. tomato strain DC3000 (Innerebner et al., 2011). Combinatorial metagenome and metaproteome studies conducted on the leaf microbiota of three plant species, namely A. thaliana, T. repens, and G. max, offer clues to leaf microbiota functions and suggest an important role for one-carbon metabolism and transport processes in the microbiota (Delmotte et al., 2009). Methanol is a common one-carbon substrate available to leaf microbiota as a result of the diurnal metabolic cycle and is a pectin methylesterase by-product processed by plants in large amounts during cell wall degradation for growth and development (Fall and Benson, 1996). Methanol-utilizing microorganisms assigned mostly to the genus Methylobacterium consume methanol during leaf colonization of numerous plant species, which enhances fitness (Sy et al., 2005; Delmotte et al., 2009; Knief et al., 2010; Sanjenbam et al., 2020). Together with phyllosphere-specific metabolites, leaf-colonizing microbiota offer a unique pool of bioactive metabolites and traits to counter stresses such as UV rays, reactive oxygen species (ROS), and dehydration (Delmotte et al., 2009; Vorholt, 2012; Helfrich et al., 2018). Such traits might become useful in developing probiotics for agriculture.
Biofilm formation by leaf microbiota
Microbes colonize leaves as complex multicellular communities. Long-term co-evolution of communities that have co-adapted and specialized results in distinct associations which further facilitate mutualistic, symbiotic, competitive, antagonistic, and indeed pathogenic microbial lifestyles with the host (Braga et al., 2016). Association between communities starts with initial adhesion to the leaf surface and ends with a complex network of interactions. Most research on leaf microbiota has focused on bacterial communities which assemble in aggregates of up to 104 cells (Monier and Lindow, 2004). These bacterial clusters are the result of aggregation between multiple cell types or clonal reproduction of a single cell (Tecon and Leveau, 2012). Bacterial surfaces play a critical role in aggregation, biofilm formation, adherence, and survival on leaf surfaces. As a part of a survival strategy, human pathogens also formed aggregates with other bacteria on the leaf, probably affording some protection during their limited survival span (Brandl and Mandrell, 2002).
Biofilms are aggregates of microbial communities in which cells adhere to each other and to a surface enveloped in a matrix of extracellular polymeric compounds, which protects the community under adverse conditions (Davey and O’toole, 2000). In nature, ~70% of bacteria on leaves are found in aggregates which confer a survival and colonization selective advantage over solitary cells on leaf surfaces (Morris and Kinkel, 2002; Monier and Lindow, 2003). Bacterial cell aggregates need to reach a minimum size to gain protection in unfavorable environments. For instance, Monier and Lindow (2003) showed that aggregates of ~100 or more cells are essential for protection against desiccation on plant leaf surfaces. A large pool of microbial communities on the leaf is protected in stress-tolerant aggregates, and dispersal of single cells leads to new microcolonies (Danhorn and Fuqua, 2007). From the attachment of cells to generation of mature biofilms, specific traits such as motility and adhesion are necessary to move and disperse on the leaf, for optimal resilience to biotic and abiotic stresses (Grinberg et al., 2019).
Using atomic force microscopy (AFM), Mittelviefhaus et al. (2019) quantified high magnitude differences in adhesion forces of leaf bacteria. Biofilm formation can also be observed in symbiotic and pathogenic lifestyles on plants, and linked with the disease cycle of phytopathogenic bacteria (Bogino et al., 2013). A role for the biofilm in colonization, disease development, and biocontrol activity was established in different studies for Xanthomonas axonopodis pv. citri, Xanthomonas vesicatoria, and Bacillus amyloliquefaciens (Malamud et al., 2011; Felipe et al., 2018; Salvatierra-Martinez et al., 2018). Nevertheless, the precise mechanism(s) by which plants regulate biofilm-associated communities are unclear (Ference et al., 2018; Kyrkou et al., 2018). The phytopathogen X. axonopodis pv. citri forms biofilms on leaves of citrus species during the development of citrus canker disease (Brunings and Gabriel, 2003; Rigano et al., 2007; Malamud et al., 2011). Notably, microbial biofilms are also important for pathogenesis by Xylella fastidiosa, the causal agent of devastating Pierce’s disease of grapes, olives, and citrus fruits, which culminates in blockage of the host vascular system (Hopkins, 1989; Marques et al., 2002; Thorne et al., 2006; Rudrappa et al., 2008; Kyrkou et al., 2018). Bacterial brown spot disease of bean leaves caused by P. syringae pv. syringae was also found to require biofilm formation (Monier and Lindow, 2004). Similarly, motility in biofilm formation was essential for host colonization by phytopathogens such as Ralstonia solanacearum, Pantoea stewartii, and Dickeya dadantii (Tans-Kersten et al., 2001; Herrera et al., 2008; Jahn et al., 2008). Much biofilm research has concentrated on specific groups of microorganisms with emphasis on bacteria. These studies emphasize the importance of biofilms in the survival and colonization of leaves by both damaging and potentially beneficial microbe communities under unfavorable conditions.
Quorum sensing in leaf microbiota
Microbial colonization to plants is regulated by the density-dependent QS phenomenon, a strategy to survive in the challenging leaf habitat. QS mechanisms involve intra- and also interspecies bacterial communication to share information and regulate their physiological activities and coordinate gene expression of factors such as motility, biofilm, host colonization, and virulence (Ng and Bassler, 2009; Elias and Banin, 2012). Different bacterial groups synthesize and use particular chemical signals or QS molecules for communication. For example, Gram-negative bacteria employ N-acyl-l-homoserine lactone [AHL; also called autoinducer-1 (AI-1)] and quinolones as QS molecules, whereas modified oligopeptides (autoinducer peptides, AIPs) are commonly used by Gram-positive bacteria for communication between cells (Taga and Bassler, 2003). Other autoinducers belonging to boron furan-derived QS molecules or AI-2 are specifically for interspecies communication (Federle, 2009). Diffusible signal factor (DSF) is another family of conserved QS signals utilized for the regulation of virulence factor in numerous Gram-negative bacterial pathogens (Li et al., 2019).
Interestingly, these QS molecules can also be recognized by cells of eukaryotes, including plants and fungi (Dudler and Eberl, 2006; Irie and Parsek, 2008). QS signal information during initial leaf surface colonization is highly localized and the quorum area can be as low as 10 cells (Gantner et al., 2006; Dulla and Lindow, 2008). AHL QS signaling molecules occur naturally in the leaf environment and might impact leaf–bacteria interactions (Enya et al., 2007). Lv et al. (2012) screened a number of AHLs produced by Gram-negative Proteobacteria as QS signals in the tobacco phyllosphere and monitored bacterial community composition. Here, Pseudomonas and other AHL-producing Gammaproteobacteria were found to use QS signals for survival and protection against other epiphytic members in the nutrient-limited phyllosphere environment. It is therefore likely that AHL QS signaling can also limit pathogenic microbes on leaves. On the other hand, in some phytopathogens, intraspecies QS was studied; for instance in Xanthomonas associated with grapevines, QS molecules control the expression of virulence factor as well as biofilm formation (Danhorn and Fuqua, 2007). For Pseudomonas syringae in tobacco and bean interaction, QS mediated control of motility and exopolysaccharide synthesis was observed for their role in biofilm formation and colonization of bacteria on leaf (Quiñones et al., 2005).
In this section, we covered microbial community composition and diversity, and colonization, survival, and adaptation in a leaf habitat. There are still significant gaps in knowledge of the types of microbial interaction, and mechanisms of competition and cooperation between leaf microbiota members, that facilitate microbial community stability and structure.
Microbe–microbe–host interactions
Communication between pathogenic microbes
Infection by pathogens can have a significant impact on the resident leaf microbial community. For example, severe SLB disease was correlated with reduced species richness in the epiphytic bacterial population of maize (Manching et al., 2014). Pathogenic microbes can also increase the susceptibility of their host plant to colonization by other microbes, which would not normally be invasive. For example, Albugo candida (white rust) enhanced susceptibility of various Brassicaceae species to fungal mildew pathogens (Cooper et al., 2002, 2008). In turn, white rust disease symptoms caused by A. candida in Brassica juncea were elevated by subsequent inoculation with the downy mildew pathogen Hyaloperenospora parasitica, which normally colonizes B. juncea asymptomatically and thereby increases its susceptibility towards white rust (Kaur et al., 2011). A similar mutual infectivity relationship was found in A. thaliana, in which an adapted oomycete pathogen, Albugo laibachii, induced susceptibility to the non-host pathogen Phytophthora infestans (Belhaj et al., 2017). Therefore, Albugo infections in Brassicaceae in some way promote a host jump by certain pathogens (Thines, 2014).
While several reports highlight the importance of interspecies microbial communication in disease development, it should be emphasized that different cells of a single pathogenic microorganism behave distinctly to orchestrate successful colonization of the host. This was shown for hyphal cells of the fungal pathogen Sclerotinia sclerotiorum, which have differential gene expression patterns and metabolic heterogeneity during successful colonization of host plants (Peyraud et al., 2019).
QS between pathogenic microbes leads to an increase in virulence and pathogenicity in the host plant. QS-based systems of the Pseudomonas savastanoi pv. savastanoi (olive knot pathogen) and Erwinia toletana (olive knot cooperator) stabilize the community and exchange QS signals, and this cooperation results in a more aggressive disease on olive plants (Olea europaea) (Caballo-Ponce et al., 2018). An intriguing example of QS mediating pathogen infection in eukaryotes was reported for the oomycete pathogen Phytophthora nicotinae. Zoospore-derived extracellular fluids contain QS components which induced zoospore aggregation that increased pathogen infectivity (Kong et al., 2010).
Interactions of foliar pathogens with endophytes
Endophytes are microorganisms which colonize the internal organs of the plant without causing visible symptoms. In several cases, endophytic microbes were reported to impact plant stress protection and development. Plant pathogens can be inhibited by a number of mechanisms, for example hyperparasitism, competition, and/or antibiosis (Busby et al., 2016). Fungal endophytes promoted induction of phenolic compounds in perennial ryegrass, thereby providing resistance against pathogenic growth (Pańka et al., 2013). Direct interactions are also observed between endophytes and pathogens. Fungal endophytes of oak tree were found to be possible antagonists of Erysiphe alphitoides, the causal agent of powdery mildew disease (Jakuschkin et al., 2016). Metarhizium robertsii colonizes insect larvae present in the plant root tissue and transfers nutrients from the insect to the host (Branine et al., 2019). Biosynthetic gene clusters including non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) genes were identified in endophytes (Miller et al., 2012; Ludlow et al., 2019), which might contribute to their biocontrol potential.
Busby et al. (2015) reported that disease modification is an ecological function shared by common foliar fungi of Populus trichocarpa. Species of Cladosporium and Trichoderma were identified to be antagonists of Melampsora rust pathogen in wild P. tricocarpa populations. These results differ from previous studies by Raghavendra and Newcombe (2013) where Stachybotrys sp., Trichoderma atroviride, Ulocladium atrum, and Truncatella angustata have been reported to induce quantitative disease resistance in P. trichocarpa against Melampsora rust pathogen under controlled experimental conditions. On the contrary, the above fungi were found to be quite rare in wild P. trichocarpa (Busby et al., 2015) and this hints at the disparity between disease-modifying action of foliar fungi under wild and experimental conditions.
Endophytes utilize QS to act against pathogenic microbes by expressing QS inhibitors (QSIs) to attenuate the activity of AIs, or quorum quenching (QQ) enzymes to disrupt signaling molecules. For example, AHL lactonase enzyme (a potent quorum quencher) present in endophytic bacteria has been reported to inhibit the plant pathogens Erwinia carotovora (Dong et al., 2000, 2001), Bacillus sp., subspecies of Bacillus thuringiensis (Lee et al., 2002; Ulrich, 2004), and Enterobacter asburiae (Rajesh and Ravishankar Rai, 2014). Ma et al. (2013) explored the diversity of tobacco (Nicotiana tabacum) leaf-associated strains with QQ activity for disruption of AHL-mediated QS, by using the biosensor reference strain Chromobacterium violaceum CV026. These bacterial quorum quenchers can be used as effective biocontrol agents against plant pathogens (Ma et al., 2013). More research is needed to understand how these interactive chemical processes impact plant microbiota community structure and function on plant hosts, and their consequences for plant health.
Microbial succession in host interactions
There is a constant struggle between different microorganisms residing inside the plant for nutrients, space, and survival. In this arena, the order of arrival of microbes can be a decisive factor between host disease resistance and facilitation. In planta experiments on Phaseolus lunatus have shown that, if a pathogen was introduced to the plant on the same day or before inoculation with an endophyte, disease resistance was more strongly reduced than when the endophyte had already colonized the host (Adame-Alvarez et al., 2014).
The situation is reversed in the case of the biotrophic maize smut fungus Ustilago maydis which is inhibited by the endophyte Fusarium verticillioides when both organisms are co-inoculated to the plant. Pre-inoculation with the endophyte had no impact on disease severity, whereas post-inoculation caused greater disease progression and decreased plant growth (Lee et al., 2009). This result suggests that F. verticillioides can inhibit U. maydis by direct interaction and not by induction of host defense responses. In line with this, the presence of U. maydis does not result in a significant difference in the diversity of the endophytic community, causing small localized differences in the community structure because of infection (Pan et al., 2008). Also, variation of the endophytic community does not correlate with levels of resistance to U. maydis in different maize lines (Pan et al., 2008).
Microbial lifestyle in host interactions
While so far there is no direct evidence for modulation of U. maydis infection with endophytic microbes, the group of smut fungi themselves represents an interesting example of organisms which exhibit different lifestyles in different niches. Generally, basidiomycete yeasts are present abundantly in the leaf microbial community of A. thaliana, along with other endophytic bacteria, as well as oomycetes (Agler et al., 2016). The anamorphic yeast Moesziomyces albugensis was recently found to antagonize Albugo laibachii infection and reduce disease development on plants (Eitzen et al., 2020, Preprint). This happens to differ from previous studies by Agler et al. (2016), where the presence of the basidiomycete yeast, Dioszegia sp., was positively correlated with the oomycete, A. laibachii.
Moesziomyces sp. (classified as Pseudozyma sp. until phylogenetic reconstruction by Wang et al., 2015) belong to the order of Ustilaginales, and have been reported to act as biocontrol agents in a number of cases (Barda et al., 2014; Gafni et al., 2015). Comparative transcriptomics identified a secreted hydrolase of M. albugensis being induced on the Arabidopsis leaf surface in the presence of A. laibachii, and reverse genetics demonstrated that the antagonism of M. albugensis towards A. laibachii depends on the expression of this enzyme (Eitzen et al., 2020, Preprint).
Such insights into the functional basis of microbial interactions of the Ustilaginales, where members of the same species can either be plant pathogens or beneficial epiphytes, show us that there might be no clearly demarcated barrier between organisms which behave as a pathogen and a plant-protecting microbe (i.e. a pathogen’s antagonist). Similarly, different strains of the pathogenic fungus Fusarium oxysporum can act as microbial antagonists against other F. oxysporum strains (van Dam et al., 2016). The differences in their lifestyle have traced back to the effector repertoire, with the epi-/endophytic strains having fewer or no host-specific effectors (de Lamo and Takken, 2020). Moesziomyces sp., however, encodes a fully equipped set of effector genes (Eitzen et al., 2020, Preprint), including a functional homolog of the U. maydis core virulence effector Pep1 (Sharma et al., 2019). This evidence suggests that anamorphic Ustilaginales yeasts have the potential to form infectious filamentous structures (Kruse et al., 2017) and at the same time raises the question of which factors drive the adaptation of these organisms to either a pathogenic or an epiphytic lifestyle.
Knowledge of the roles of microbe–microbe–host interactions in determining microbial invasiveness will aid understanding of the cross-domain interactions in pathogenicity. Nevertheless, more fundamental research is needed to disentangle microbe–microbe and microbe–host interactions at the level of individual strains to determine what underpins functional microbial assemblies in nature.
Role of the plant immune system in shaping the leaf microbiome
The plant innate immune system comprises a large repertoire of plasma membrane-localized (surface) and intracellular receptors which recognize microbial or modified host molecular signatures and retain plant health and secure plant propagation. Surface immune receptors (often referred to as pattern recognition receptors, or PRRs) are members of a diverse family of ligand-binding proteins that sense microbial, environmental, developmental, and nutritional cues (Saijo et al., 2018; Cheng et al., 2019). In terms of shaping microbial communities, it is the PRR activities that are thought to gate microbial entry into leaf tissues, and effectively ward off colonization by host non-adapted strains (Boutrot and Zipfel, 2017). The intracellular receptor panels [consisting mostly of nucleotide-binding/leucine-rich repeat (NLR) proteins] are similarly diverse and are selected as triggers of strain-specific resistance to host-adapted pathogens (Eitas and Dangl, 2010; Jones et al., 2016; Wu et al., 2017; Burdett et al., 2019; de Weyer et al., 2019).
The activation of plant immune responses by mobilizing a network of defense and stress hormone pathways has been extensively characterized in binary plant–pathogen interactions (Noman et al., 2019; Zhao et al., 2019). Little is known about the impact of plant immunity signaling networks on host–microbe interactions in leaf microbial communities (Fig. 1D). High-throughput DNA and RNA sequencing of leaf samples from natural environments have enabled examination of complex microbial communities in plant-specific niches in time and space (Agler et al., 2016). Analysis of microbial metadata and their integration with experimental testing should provide a clearer picture of the role of plant immunity signaling in shaping leaf microbial community structure and, in turn, how resident microbes influence host immunity.
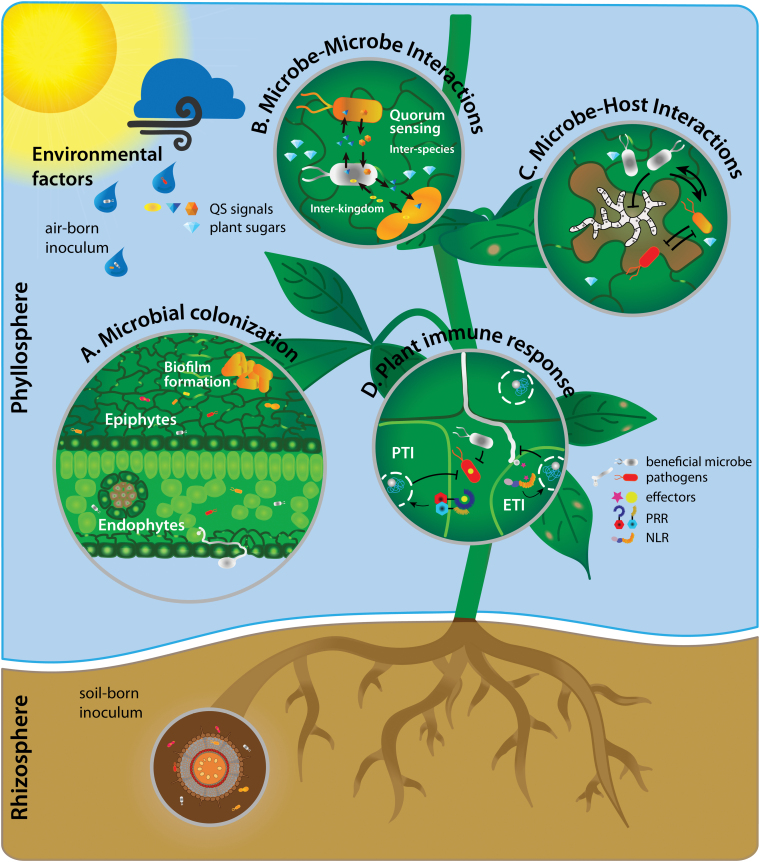
Fig. 1.
Microbial colonization of the above-ground part of the plant (phyllosphere), as well as the below-ground part (rhizosphere). (A) The microbial colonization on the leaf takes place on the leaf surface (epiphytes) from air-borne and soil-borne inocula and the inner leaf part (endophytes). Microbial colonization can lead to exogenous intraspecies biofilm formation on the leaf surface. (B) Microbe–microbe interactions occur between interspecies and interkingdoms, referred to as quorum sensing. Quorum-sensing molecules impacting microbial recognition and biofilm formation on leaves. (C) Pathogenic microbes colonize host plants by means of their virulence. The genetic make-up of both the host and pathogen contributes to disease progression. However, other microbes in the host phyllosphere can influence this plant–pathogen interaction by either facilitation or antagonism. (D) Plant immune responses are of specific interest as host–microbe interactions shaping the phyllosphere microbiome. Non-host-adapted pathogens are involved in PAMP-triggered immunity (PTI) and recognized via pattern-recognition receptors (PRRs). Host-adapted microbes are recognized via nucleotide-binding leucine-rich repeat receptors (NLRs), summarized in effector-triggered immunity (ETI).
In this section, we consider evidence that abiotic and biotic stress responses modulate microbial consortia on leaves and discuss the consequences for plant fitness. It is becoming clear that microbial community structure throughout a plant host’s life cycle is dynamic and modulated by the innate immune system, which itself is tuned to environmental changes.
The role of pattern-triggered immunity in shaping the leaf microbiota
Most microorganisms on plant leaves are non-pathogenic. However, a broad range of microbes are able to prime innate plant immunity to counter subsequent pathogen attacks (Ritpitakphong et al., 2016; Vogel et al., 2016). Many microbes are recognized by terrestrial plants through their MAMPs initiating pattern-triggered immunity (PTI) responses. PTI is an induced and often low-level but broadly effective resistance response involving phytohormone signaling, secretion of antimicrobial compounds, generation of ROS and mitogen-activated proein (MAP) kinase cascades, and stomatal closure (Bigeard et al., 2015; Bi and Zhou, 2017). Notably, the phytohormone ethylene is required for ROS production in PTI, for example in Arabidopsis resistance to P. syringae bacteria and rice resistance to the rice blast fungus, Magnapothe oryzae (Mersmann et al., 2010; Guan et al., 2015; Helliwell et al., 2016; Yang et al., 2017). In Arabidopsis, an ethylene-insensitive2 (ein2) mutant displayed an altered bacterial leaf community compared with wild-type plants, suggesting that ethylene signaling is important for modulating the leaf microbiota (Bodenhausen et al., 2014; Nascimento et al., 2018).
A recent study by Chen et al. (2020) provided experimental evidence that PTI signaling controls the diversity of endophytic leaf microbiota in microorganism-rich environments. An Arabidopsis quadruple mutant [min7 fls2 efr cerk1 (mfec)] that is defective in PTI and the MIN7 vesicle trafficking pathway (affecting the aqueous apoplastic microenvironment) and a constitutively activated cell death1 (cad1) mutant had altered endophytic bacterial leaf diversity (Chen et al., 2020). In particular, the relative abundance of the bacterial phyla Firmicutes was significantly reduced, whereas Proteobacteria became the dominating bacterial community members in the mutant plants. The occurrence of PTI components MIN7 and CAD1 across major plant lineages suggests that a number of common pathways might govern endophytic microbial proliferation of certain taxa in leaves.
Further research has revealed the importance of resident Pseudomonas sp. (Proteobacteria) in protecting Arabidopsis against infection by a fungal necrotrophic pathogen, B. cinerea (Ritpitakphong et al., 2016). Notably, prominent bacterial clades from soil microbiota such as filamentous Actinobacteria (Strepotmycetes sp.) are able to activate plant biosynthesis of salicylic acid (SA) and promote leaf defense responses against fungal pathogens (Vergnes et al., 2020). These findings highlight actions of soil-borne microbial inocula of leaves on immunity (Bakker et al., 2013; Haney et al., 2018; Vannier et al., 2019).
The above studies emphasize the role of both commensal and pathogenic microbes in priming PTI as a barrier to colonization of the leaf compartment by host non- or poorly adapted pathogens. Nevertheless these host–microbe interactions were examined mostly under controlled laboratory conditions. Further research is needed to gain an understanding of how PTI shapes plant immune responses and microbiota communities in nature.
Leaf effector-triggered immunity as a potential microbial gateway
Strain-specific resistance, known as effector-triggered immunity (ETI), is often mediated by intracellular NLR receptors which recognize certain pathogen-delivered virulence factors (effectors) to induce immunity (Monteiro and Nishimura, 2018; Seong et al., 2019; Feehan et al., 2020). Pathogen effector-activated NLRs accelerate and amplify many PTI responses, often resulting in host-localized cell death (a hypersensitive response) and rapid pathogen containment (Peng et al., 2018). Expressed NLR genes in roots are observed in dicot plant species such as the legume Lotus (Lai and Eulgem, 2018). This in contrast to tested Brassicaceae species including A. thaliana and the crop oilseed rape (Brassica napus), which favor NLR expression in the phyllosphere (Munch et al., 2018). Although NLR activation and downstream signaling mechanisms are becoming resolved, the extent to which this layer of protection against pathogens shapes plant microbial communities is hardly understood.
Diverse microbial communities in leaves can be controlled directly through pathogen colonization on the host or indirectly by host–microbe interactions involving the innate immunity network (Agler et al., 2016). Thus, pathogenic microbes can act as highly interconnected community members (so-called ‘hub microbes’) that dominate microbial community assemblies. For example, the causal agent of white rust on Arabidopsis, Albugo sp., appears to act as a hub which alters epiphytic and endophytic bacterial colonization of leaves (Agler et al., 2016; Ruhe et al., 2016). Perturbations of microbial communities by host-adapted biotrophic pathogens such as Albugo and Hyaloperonospora arabidopsidis (Hpa) reduce microbial diversity within leaf habitats and stabilize microbial communities among wild plants (Karasov et al., 2019, Preprint). Hence, microbial diversity can be used as an indicator for microbial community imbalance (Chen et al., 2020).
Whether ETI reactions directly lead to defense priming is not well studied, although in Arabidopsis one important ETI branch leads to a reinforcement and spread of pathogen resistance (so-called basal immunity) in leaf tissues (Lapin et al., 2020). A recent study by Levy et al. (2018) analyzed >3800 genomes of plant-associated (pathogenic and non-pathogenic) bacteria. The analysis identified plant-mimicking protein domains (named PREPARADOS) that carry non-canonical ‘embedded’ NLR domains. An increasing number of NLR-fused domains are related to authentic effector targets. PREPARADOS are highly abundant in the bacterial families Bacteroides and Xanthomonadaceae (Frank, 2019). These findings point to potential interactions between commensal and or pathogenic bacteria with intracellular receptors in host plants. Additional studies are needed to test this hypothesis and dissect functional relationships between NLR panels and the leaf microbiota.
Stability of microbial consortia against pathogen perturbation
The plant and its associated microbiota is not a static environment but is altered by numerous factors including host genotype, environmental fluctuations, surrounding macro- and microorganisms, and geographical location and associated local variables such as climate (Laforest-Lapointe et al., 2016; Poudel et al., 2016; Wagner et al., 2016; Singh et al., 2018). The stability of a leaf microbial community is measured as the ability to maintain a stable equilibrium state (homeostasis) under biotic or abiotic perturbations (Thébault and Fontaine, 2010). Generally, higher community complexity in a network reflects a more stable community structure (Mougi and Kondoh, 2012). Stable microbial communities or consortia have greater ability to resist perturbation (Ives et al., 2000; Luo et al., 2019; Morella et al., 2020). Studies using culture-independent DNA sequencing revealed similar microbial community patterns in successive year samplings (Copeland et al., 2015). In the phyllosphere, microbial communities can often undergo drastic changes and establish a distinctive and less diverse community (Manching et al., 2014; Copeland et al., 2015). Different computational and experiment-based approaches have been used to capture microbial community homeostasis or deviations over time. Computational microbial network analysis and mining of core microbes are valuable in understanding the factors underlying microbial resilience to controlled perturbations (Astudillo-García et al., 2017; Lemanceau et al., 2017). Much less is known about the dynamics and stability of leaf microbiomes in the field since there is a lack of high-resolution experimental data linked to plant disease and health with respect to time, space, and environmental scale. In recent studies, leaf diseases were linked to disruption of microbial community network stability, resulting in ecosystem dysfunction (Kerdraon et al., 2019; Luo et al., 2019; Leopold and Busby, 2020, Preprint). Understanding how a microbial community corrects itself under conditions of environmental stress is crucial to harness its potential in probiotic applications against aggressive plant pathogens and to track plant-associated human pathogen outbreaks.
Does immunity priming affect microbial leaf communities?
Various abiotic and biotic factors impact dynamic changes on microbial leaf communities as depicted in the modes of microbial colonization, microbe–microbe, and microbe–host interactions (see Fig. 1 and Table 1). Nevertheless, fundamental mechanisms of microbial community assembly remain barely understood. One major goal of current microbiome research is to understand how microbial consortia in nature secure plant protection during pathogen perturbation. Immunity priming (IP) effects through abiotic (applied chemical compounds) and biotic (biocontrol agents) stimuli seem to play an important role in managing abiotic stress tolerance and disease resistance (Kumar and Verma, 2018). IP has been described as a ‘positive cost–benefit balance in times of stress’ (Martinez-Medina et al., 2016). IP induction involves the phytohormones SA and jasmonic acid (JA), and pipecolic acid-derived signaling molecules that are known to mediate systemic acquired resistance, as well as the non-protein amino acid defense primer β-aminobutyric acid (BABA) (Martinez-Medina et al., 2016). BABA was found naturally in Arabidopsis experiencing abiotic stress (high salinity) and biotic stress, induces broad-spectrum pathogen resistance (Thevenet et al., 2017; Buswell et al., 2018). Another interesting IP compound, (R)-β-homoserine (RBH), primes ethylene and JA pathways and is effective against necrotrophic pathogens such as B. cinerea in tomato and Plectosphaerella cucumerina (Buswell et al., 2018). Also, brassinosteroids (BRs) have been discussed as factors in an IP mechanism that balances the trade-off between immunity and growth (Yu et al., 2018). These findings highlight the potential utility of chemical compounds for IP. They also prompt studies of how IP impacts leaf microbial diversity under conditions of abiotic and biotic stress.
Table 1.
Summary of important studies associated with the leaf microbiome
| Host plant | Leaf microbiota/leaf microbe under study | Perturbation | Key findings | Reference |
|---|---|---|---|---|
| Microbial colonization | ||||
| Arabidopsis thaliana | Bacteria | – | Phyllosphere community profile of A. thaliana wild-type Landsberg erecta (Ler) and eceriferum (cer) mutants (cer1, cer6, cer9, and cer16) involved in cuticle biosynthesis. Plant cuticular wax composition affects the phyllosphere bacterial community. | Reisberg et al. (2013) |
| Faba bean (Vicia faba L.) and Arabidopsis thaliana | Pseudomonas syringae DC3118, a coronatine-deficient mutant of Pseudomonas syringae DC3000 | – | In a specific environmental setting, leaf surface colonization by bacteria correlated with stomatal aperture regulation. | Ou et al. (2014) |
| Bean (Phaseolus vulgaris L.) | P. syringae pv. syringae B728a | – | Biosurfactant, syringafactin, produced by P. syringae pv. syringae B728a on leaves adsorbed on waxy leaf cuticle surface. Provide benefit to bacteria by attracting moisture and aid in nutrient availability. | Burch et al. (2014) |
| Arabidopsis thaliana | Pseudomonas syringae DC3000 | – | Humidity-controlled, pathogen-guided establishment of an aqueous intercellular space (apoplast) as an important step in leaf bacterial infection. | Xin et al. (2016) |
| Microbial composition and diversity | ||||
| Sugar beet (Beta vulgaris) | Bacteria, yeasts, and filamentous fungi | – | Seasonal dynamics over a growing season. Fungi: Cladosporium and Alternaria sp. | Thompson et al. (1993) |
| Yeast: Cryptococcus and Sporobolomyces Bacteria: Pseudomonas sp. and Erwinia herbicola |
||||
| Cacao (Theobroma cacao) | Fungi (endophytes) | Phytophthora sp. | High diversity, spatial structure, and host affinity among foliar endophytes.
Endophyte-mediated protection against foliar pathogen. |
Arnold et al. (2003) |
| Common wood sorrel (Oxalis acetosella L.) | Yeast (epiphytes) | – | Seasonal dynamics of yeasts. Species diversity—maximum in autumn; minimum in spring. |
Glushakova and Chernov (2004) |
| Rhodotorula glutinis and Sporobolomyces roseus species abundant throughout the year. | ||||
| Loblolly pine (Pinus taeda) | Fungi (endophytes) | – | High diversity of foliar fungal endophytes. | Arnold et al. (2007) |
| Arabidopsis thaliana, Trifolium repens, and Glycine max | Bacteria | – | Metaproteogenomic analysis found consistency in three plant species. | Delmotte et al. (2009) |
| High abundance of Sphingomonas sp. and Methylobacterium sp. | ||||
| Important role of the one-carbon metabolism and transport processes in the microbiota. | ||||
| Tree species | Bacteria (epiphytes) | – | In trees, interspecies variation is more than intraspecies variation in bacterial communities. | Redford et al. (2010) |
| Correlation between tree phylogeny and bacterial community composition. | ||||
| Maize | Bacteria (epiphytes) | Southern leaf blight (SLB) | A specific set of epiphytic bacteria can restrict phyllosphere bacterial diversity and increase resistance to Southern leaf blight (SLB) fungal infection. | Balint-Kurti et al. (2010) |
| Eucalyptus citriodora Hook | Fungi (epiphytes and edophytes) | – | Total 33 fungal species assigned to 33 taxa (endophytes, 20; epiphytes, 22). | Kharwar et al. (2010) |
| Difference in frequency of colonization. Antagonism against human and plant pathogen. | ||||
| Lettuce | Bacteria | – | Bacterial community composition by pyrosequencing. Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria—most abundant phyla. Insights on variability in bacterial community profile with respect to time, space, and environment. | Rastogi et al. (2012) |
| Common bean (Phaseolus vulgaris) | Bacteria (endophytes) | – | 158 culturable endophytic bacteria. Phyla distribution 36.7% Proteobacteria, 32.9% Firmicutes, 29.7% Actinobacteria, and 0.6% Bacteroidetes | de Oliveira Costa et al. (2012) |
| Arabidopsis thaliana | Bacteria (epiphytes and endophytes) | – | Proteobacteria, Actinobacteria, and Bacteroidetes were found most abundant. Massilia and Flavobacterium are prevalent genera | Bodenhausen et al. (2013) |
| Tomato (Solanum lycopersicum L.) | Bacteria (epiphytes) | – | Members of endophytic bacterial communities of tomato leaves exert multiple effects on growth and health of tomato plants. | Romero et al. (2014) |
| Neotropical forest | Bacteria | – | Dominated bacterial communities: Actinobacteria, Alpha-, Beta-, Gammaproteobacteria, and Sphingobacteria. Correlation of bacterial community with host growth, mortality, and function. | Kembel et al. (2014) |
| Arabidopsis thaliana | Bacteria | – | Taxonomic and functional overlap of leaf and root bacterial communities. Soil as main driver for bacterial members.. | Bai et al. (2015) |
| Rice (Oryza sativa L.) | Actinomycetes | Pyricularia oryzae (syn. Magnaporthe oryzae) | Rice phyllosphere-associated actinomycetes produce bioactive compounds and control leaf blast disease caused by Pyricularia oryzae. | Harsonowati et al. (2017) |
| Sugar maple (Acer saccharum) | Bacteria and fungi (epiphytes and endophytes) | – | Microbial communities at the edge of the species’ elevational range differ from those within the natural range. | Wallace et al. (2018) |
| Poplar tree | Bacteria and fungi (epiphytes and endophytes) | Mercury | Methylobacterium, Kineococcus, Sphingomonas, and Hymenobacter on the leaf surface. | Durand et al. (2018) |
| Mussaenda pubescens var. alba | Fungi | – | Dothideomycetes and Eurotiomycetes are dominant members. Intraspecific host genetic identity, primary driver in shaping regional phyllosphere fungal communities. | Qian et al. (2018) |
| Arabidopsis thaliana | Bacteria | – | Determined biosynthetic potential of 224 bacterial strains from Arabidopsis leaf microbiome. Phyllosphere as a valuable resource for the identification and characterization of antibiotics and natural products. | Helfrich et al. (2018) |
| Tomato (Solanum lycopersicum L.) | Bacteria (epiphytes) | – | Comprehensive view of the tomato-associated bacterial community. | Dong et al. (2019) |
| Isolation of beneficial bacterial for future functional studies. | ||||
| Mangrove | Fungi (epiphytes and endophytes) | – | Dothideomycetes and Tremellomycetes are dominant members. Plant identity significantly affects endophytic but not epiphytic fungi. | Yao et al. (2019) |
| Catharanthus roseus | Fungi (Endophytes) | – | Colletotrichum, Alternaria, and Chaetomium are common genera. | Dhayanithy et al. (2019) |
| Biofilm | ||||
| Common bean (Phaseolus vulgaris) | P. syringae pv. syringae | – | Cause of brown spot disease of bean leaves was the result of biofilm formation of P. syringae. | Monier and Lindow (2004) |
| Citrus limon ‘Eureka’ | Xanthomonas axonopodis pv. citri | – | Motility and role of flagellum is required for mature biofilm and canker development. | Malamud et al. (2011) |
| Tomato (Solanum lycopersicum L.) | Xanthomonas vesicatoria | – | Aggressiveness of Xv strains correlated with their ability to move by flagella or type IV pili, adherence to leaves and form well-developed biofilms, help in improved phyllosphere colonization. | Felipe et al. (2018) |
| Tomato (Solanum lycopersicum L.) | Bacillus amyloliquefaciens | Botrytis cinerea | Reduction of biocontrol of BBC 023 on leaves due to its limited ability to generate robust biofilms and colonization in the phylloplane. | Salvatierra-Martinez et al. (2018) |
| Quorum sensing | ||||
| Tomato (Solanum lycopersicum L.) | Bacteria | – | Culturable leaf-associated bacteria community with BCA activity against tomato disease have the ability to produce AHL and IAA. | Enya et al. (2007) |
| Tobacco (Nicotiana tobacum) | Epiphytes | – | AHLs induced variation in the bacterial community composition. Pseudomonas and other AHL-producing Gammaproteobacteria use QS signals for their survival and protection. | Lv et al. (2012) |
| Tobacco (Nicotiana tobacum), common bean (Phaseolus vulgaris) | Pseudomonas syringae | – | QS-mediated control of motility and exopolysaccharide synthesis was observed for their role in biofilm formation and colonization of bacteria on leaf. | Quiñones et al. (2005) |
| Microbe–microbe–host interactions | ||||
| Arabidopsis thaliana | Hyaloperonospora parasitica subsp., Arabidopsis thaliana, H. parasitica subsp. Brassica oleracea, Bremia lactucae, and Albugo candida | – | Albugo candida suppressed defense signaling pathways in the host, facilitating sporulation by the incompatible downy mildews | Cooper et al., (2002) |
| Quercus robur L. | Foliar fungi and bacteria | Erysiphe alphitoides | Direct interaction between E. alphitoides and 13 fungal and bacterial operational taxonomic units (OTUs). Fungal endophytes Mycosphaerella punctiformis and Monochaetia kansensis could be possible antagonists of E. alphitoides. | Jakuschkin et al. (2016) |
| Arabidopsis thaliana | - | Phytophthora infestans: Albugo laibachii | Prior colonization of host by A. laibachii, helps P. infestans to infect an essentially non-host plant. | Belhaj et al., (2017) |
| Phaseolus lunatus | Endophytic fungi for e.g. Rhizopus, Fusarium, Penicillium, Cochliobolus, and Artomyces spp. | Pseudomonas syringae pv. syringae, Enterobacter sp. strain FCB1, and the fungus Colletotrichum lindemuthianum | Order of arrival of fungal endophytes and pathogens on the plant surface can determine disease resistance or facilitation. | Adame-Alvarez et al. (2014) |
| Zea mays | Endophyte Fusarium verticillioides | Ustilago maydis | F. verticillioides can inhibit U. maydis disease progression by direct interaction. | Lee et al. (2009) |
| Olive plants (Olea europaea) | Pseudomonas savastanoi pv. savastanoi (olive knot pathogen) and Erwinia toletana (olive knot cooperator). | The bacteria stabilize the community, exchange QS signals, and this cooperation results in disease aggression. | Caballo-Ponce et al. (2018) | |
| Arabidopsis thaliana | Basidiomycete yeast, Dioszegia sp. | Albugo laibachii | Construction of an extensive phyllosphere microbial network encompassing bacterial, fungal, and oomycetal communities. Presence of Dioszegia sp. is positively correlated with that of A. laibachii. | Agler et al. (2016) |
| Arabidopsis thaliana | Basidiomycete yeast, Moesziomyces albugensis | Albugo laibachii | Moesziomyces albugensis antagonizes A. laibachii on the host leaf surface. | Eitzen et al. (2020) |
| Innate immunity interaction | ||||
| Arabidopsis thaliana | Bacteria | – | The author showed evidence of ethylene signaling (ein2) affecting the abundance of Variovorax. | Bodenhausen et al, (2014) |
| Arabidopsis thaliana | Bacteria | – | Affected diversity of Firmicutes sp. and Proteobacteria sp. in min7 fls2 efr cerk1 (mfec) and constitutively activated cell death1 (cat1) mutants (involving PTI, MIN7 vesicle trafficking, or cell death pathways). | Chen et al. (2020) |
| Arabidopsis thaliana | Streptomyces AgN23. | Alternaria brassicicola | The bacteria Streptomyces induces defense responses, which prevents Alternaria infection. | Vergnes et al. (2020) |
| Tomato (Solanum lycopersicum, Solanum pimpinellifolium) | Bacteria | – | Host resistance shapes leaf microbiota under environmental fluctuations and is time dependent. | Morella et al. (2020) |
| Cucumber Cucumis sativus (Suyan 10) | Bacteria and fungi | Pseudomonas syringae pv. Lachrymans | Plant-specific microbes such as Sphingomonas, Methylobacterium, Pseudomonas, and Alternaria are significantly affected by the causal agent of angular leaf-spot of cucumber at different infection stages. | Luo et al. (2019) |
| Pepper (Capsicum annuum L.) | Bacillus thuringiensis | – | Significant changes of phyllosphere microbiota in Firmicutes and Gammaproteobacteria. | Zhang et al. (2008) |
| Grapevine (Vitis vinifera) | Bacteria | Botrytis cinerea, Phytophthora infestans | Potential biocontrol agents (Bacillus, Variovorax, Pantoea, Staphylococcus, Herbaspirillum, Sphingomonas) from leaf microbiome acting against phytopathogens. | Bruisson et al. (2019) |
| Wheat (Triticum aestivum) | Bacteria and fungi | Zymoseptoria tritici | Microbial dynamics upon infection | Kerdraon et al. (2019 |
| Tobacco (Nicotiana sp.) | Bacteria | Pseudomonas syringae pv. tabaci | The application of two BCAs changed the bacterial phyllosphere community and decreased bacterial wildfire outbreak. | Qin et al. (2019) |
Effects of biocontrol agents (BCAs) on crops such as potato against biotrophic (P. infestans) and grapevine against necrotrophic (B. cinerea) fungi have been studied extensively in vitro (Bailly and Weisskopf, 2017; De Vrieze et al., 2018; Bruisson et al., 2019). In contrast, applying P. syringae pathovar tomato (Pst) to Arabidopsis roots attracted Bacillus subtilis and led to IP upon Pst infection (Rudrappa et al., 2008; Vannier et al., 2019). The ecological impact of BCAs on the leaf microbiome while controlling disease resistance remains an open research question. Current reports emphasize a linkage between certain bacterial taxa (Bacillus, Pantoea, Sphingomonas, Pseudomonas, and Trichoderma) affecting microbial diversity (Zhang et al., 2008; Bruisson et al., 2019; Ulrich et al., 2020) and IP induction on leaves (Cawoy et al., 2014; Ritpitakphong et al., 2016; Qin et al., 2019). In particular, highly diverse leaf communities are negatively correlated with pathogen invasion and colonization, and vice versa (Purahong et al., 2018; Qin et al., 2019). Other reports describe difficulties encountered in the application of biocontrol agents such as B. subtilis, which did not alter the microbial leaf community under rainy field conditions (Wei et al., 2016). Thus, use of biocontrol agents under natural conditions might be challenging and require further analysis. However, BCAs and IP-inducing compounds can potentially be used to monitor disease control to improve crop yield and production in new biological breeding strategies. There is clearly a need to increase efforts in this research field to explore the effects and underlying mechanisms of abiotic and biotic stress on IP and how they are transmitted to microbial leaf communities.
Conclusion and outlook
The plant phyllosphere is a highly competitive and challenging habitat for microbes to colonize. Pre-formed barriers such as the hydrophobic cuticle, stomata, or cell wall structures require specific adaptation for the microbes, and persistence of microbes strongly depends on their ability to interact with others. Thus understanding microbiota assembly and persistence in the plant phyllosphere requires investigating ecological factors that shape pre-formed plant structures and therefore directly act on host–microbe and indirectly microbe–microbe interactions. Microbe–microbe interactions in turn not only impact microbial behavior but can impact host fitness by antagonizing plant pathogens. Pathogen invasion generally has a significant negative effect on host fitness caused by tissue damage, nutrient loss to invaders, and reallocation of resources to immune activation. Successful pathogenesis on the other hand is a complex process that requires multiple steps of host colonization and reproduction, and is generally the result of long-term co-evolution (Hall et al., 2017). Considering the enormous gene pool and diversity of all non-pathogenic microbes that are associated with the phyllosphere and other parts of the plant, and considering this pool under constant selection to benefit plant fitness directly or indirectly, we can expect an enormous unexploited pool of beneficial microbes to antagonize pathogenicity processes. In addition, phyllosphere-colonizing microbes are highly adapted to abiotic and biotic fluctuations and are therefore an enormous pool for new adaptive traits.
The downside is, however, that this pool is highly dynamic and probably requires a stable co-existence of different microbial species in one habitat in order to express beneficial traits. Interconnected networks between organisms can be an important element in providing a buffer against perturbations since such links help to recruit microbes to fulfill specific functions in cases where another organism that, for example, provides important antimicrobial compounds or enzymes within the network is lost.
A main goal to develop strategies to protect the phyllosphere from pathogen invasion, such as wheat from rusts, is to identify probiotics that can either stabilize the natural community or become stable on its own. One major effort to develop such probiotics is therefore understanding the multistep process of establishing a niche and defending this niche. Only once we know how to combine traits for stability with our desired traits such as plant protection will we be able to develop products that can replace the majority of our current agrochemicals.
Acknowledgements
VC gratefully acknowledges support from the Alexander von Humboldt Foundation for a Humboldt Research Fellowship (2018–2020). PR acknowledges the financial support from the European Research Council (ERC) under the DeCoCt research program (grant agreement: ERC-2018-COG 820124). PS would like to acknowledge the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy EXC-2048/1, Project ID 390686111, and the DFG priority program SPP2125 ‘DECRyPT’, for their financial support. We thank the two anonymous reviewers for their suggestions and constructive comments.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Adame-Álvarez RM, Mendiola-Soto J, Heil M. 2014. Order of arrival shifts endophyte–pathogen interactions in bean from resistance induction to disease facilitation. FEMS Microbiology Letters 355, 100–107. [DOI] [PubMed] [Google Scholar]
- Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM. 2016. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biology 14, e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón W, Reina-Pinto JJ, Serrano M. 2017. The intimate talk between plants and microorganisms at the leaf surface. Journal of Experimental Botany 68, 5339–5350. [DOI] [PubMed] [Google Scholar]
- Arnaud D, Hwang I. 2015. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Molecular Plant 8, 566–581. [DOI] [PubMed] [Google Scholar]
- Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. 2007. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99, 185–206. [DOI] [PubMed] [Google Scholar]
- Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences, USA 100, 15649–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo-García C, Bell JJ, Webster NS, Glasl B, Jompa J, Montoya JM, Taylor MW. 2017. Evaluating the core microbiota in complex communities: a systematic investigation. Environmental Microbiology 19, 1450–1462. [DOI] [PubMed] [Google Scholar]
- Aung K, Jiang Y, He SY. 2018. The role of water in plant–microbe interactions. The Plant Journal 93, 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, et al. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. [DOI] [PubMed] [Google Scholar]
- Bailly A, Weisskopf L. 2017. Mining the volatilomes of plant-associated microbiota for new biocontrol solutions. Frontiers in Microbiology 8, 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker PA, Doornbos RF, Zamioudis C, Berendsen RL, Pieterse CM. 2013. Induced systemic resistance and the rhizosphere microbiome. The Plant Pathology Journal 29, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldotto LE, Olivares FL. 2008. Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Canadian Journal of Microbiology 54, 918–931. [DOI] [PubMed] [Google Scholar]
- Balint-Kurti P, Simmons SJ, Blum JE, Ballaré CL, Stapleton AE. 2010. Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Molecular Plant-Microbe Interactions 23, 473–484. [DOI] [PubMed] [Google Scholar]
- Balogh B, Nga NTT, Jones JB. 2018. Relative level of bacteriophage multiplication in vitro or in phyllosphere may not predict in planta efficacy for controlling bacterial leaf spot on tomato caused by Xanthomonas perforans. Frontiers in Microbiology 9, 2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Gu L, Sun B, Cai W, Zhang S, Zhuang G, Bai Z, Zhuang X. 2020. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiology Ecology 96, fiaa017. [DOI] [PubMed] [Google Scholar]
- Barda O, Shalev O, Alster S, Buxdorf K, Gafni A, Levy M. 2015. Pseudozyma aphidis induces salicylic-acid-independent resistance to Clavibacter michiganensis in tomato plants. Plant Disease 99, 621–626. [DOI] [PubMed] [Google Scholar]
- Beattie GA, Lindow SE. 1999. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89, 353–359. [DOI] [PubMed] [Google Scholar]
- Belhaj K, Cano LM, Prince DC, et al. 2017. Arabidopsis late blight: infection of a nonhost plant by Albugo laibachii enables full colonization by Phytophthora infestans. Cellular Microbiology 19, e12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchat LR. 2002. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes and Infection 4, 413–423. [DOI] [PubMed] [Google Scholar]
- Bi G, Zhou JM. 2017. MAP kinase signaling pathways: a hub of plant–microbe interactions. Cell Host & Microbe 21, 270–273. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Molecular Plant 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. 2014. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genetics 10, e1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8, e56329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogino PC, Oliva Mde L, Sorroche FG, Giordano W. 2013. The role of bacterial biofilms and surface components in plant–bacterial associations. International Journal of Molecular Sciences 14, 15838–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C. 2017. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annual Review of Phytopathology 55, 257–286. [DOI] [PubMed] [Google Scholar]
- Braga RM, Dourado MN, Araújo WL. 2016. Microbial interactions: ecology in a molecular perspective. Brazilian Journal of Microbiology 47 Suppl 1, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the Cilantro phyllosphere. Applied and Environmental Microbiology 68, 3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branine M, Bazzicalupo A, Branco S. 2019. Biology and applications of endophytic insect-pathogenic fungi. PLoS Pathogens 15, e1007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringel F, Couée I. 2015. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Frontiers in Microbiology 6, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruisson S, Zufferey M, L’Haridon F, Trutmann E, Anand A, Dutartre A, De Vrieze M, Weisskopf L. 2019. Endophytes and epiphytes from the grapevine leaf microbiome as potential biocontrol agents against phytopathogens. Frontiers in Microbiology 10, 2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunings AM, Gabriel DW. 2003. Xanthomonas citri: breaking the surface. Molecular Plant Pathology 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Bunster L, Fokkema NJ, Schippers B. 1989. Effect of surface-active Pseudomonas spp. on leaf wettability. Applied and Environmental Microbiology 55, 1340–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. 2011. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environmental Microbiology 13, 2681–2691. [DOI] [PubMed] [Google Scholar]
- Burch AY, Shimada BK, Mullin SW, Dunlap CA, Bowman MJ, Lindow SE. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. Journal of Bacteriology 194, 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AY, Zeisler V, Yokota K, Schreiber L, Lindow SE. 2014. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environmental Microbiology 16, 2086–2098. [DOI] [PubMed] [Google Scholar]
- Burdett H, Bentham AR, Williams SJ, Dodds PN, Anderson PA, Banfield MJ, Kobe B. 2019. The plant ‘resistosome’: structural insights into immune signaling. Cell Host & Microbe 26, 193–201. [DOI] [PubMed] [Google Scholar]
- Busby PE, Lamit LJ, Keith AR, Newcombe G, Gehring CA, Whitham TG, Dirzo R. 2015. Genetics-based interactions among plants, pathogens, and herbivores define arthropod community structure. Ecology 96, 1974–1984. [DOI] [PubMed] [Google Scholar]
- Busby PE, Peay KG, Newcombe G. 2016. Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. New Phytologist 209, 1681–1692. [DOI] [PubMed] [Google Scholar]
- Buswell W, Schwarzenbacher RE, Luna E, Sellwood M, Chen B, Flors V, Pétriacq P, Ton J. 2018. Chemical priming of immunity without costs to plant growth. New Phytologist 218, 1205–1216. [DOI] [PubMed] [Google Scholar]
- Caballo-Ponce E, Meng X, Uzelac G, Halliday N, Cámara M, Licastro D, da Silva DP, Ramos C, Venturi V. 2018. Quorum sensing in Pseudomonas savastanoi pv. savastanoi and Erwinia toletana: role in virulence and interspecies interactions in the olive knot. Applied and Environmental Microbiology 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy H, Mariutto M, Henry G, Fisher C, Vasilyeva N, Thonart P, Dommes J, Ongena M. 2014. Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Molecular Plant-Microbe Interactions 27, 87–100. [DOI] [PubMed] [Google Scholar]
- Chen T, Nomura K, Wang X, et al. 2020. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Zhang L, He SY. 2019. Plant–microbe interactions facing environmental challenge. Cell Host & Microbe 26, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJ, Latunde-Dada AO, Woods-Tör A, Lynn J, Lucas JA, Crute IR, Holub EB. 2008. Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Molecular Plant-Microbe Interactions 21, 745–756. [DOI] [PubMed] [Google Scholar]
- Cooper AJ, Woods-Tor A, Holub EB. 2002. Albugo candida (white rust) suppresses resistance to downy mildew pathogens in Arabidopsis thaliana. Plant Protection Sciences 38, 474–476. [Google Scholar]
- Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. 2015. Seasonal community succession of the phyllosphere microbiome. Molecular Plant-Microbe Interactions 28, 274–285. [DOI] [PubMed] [Google Scholar]
- Crombie AT, Larke-Mejia NL, Emery H, Dawson R, Pratscher J, Murphy GP, McGenity TJ, Murrell JC. 2018. Poplar phyllosphere harbors disparate isoprene-degrading bacteria. Proceedings of the National Academy of Sciences, USA 115, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annual Review of Microbiology 61, 401–422. [DOI] [PubMed] [Google Scholar]
- Davey ME, O’toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiology and Molecular Biology Reviews 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamo FJ, Takken FLW. 2020. Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Frontiers in Plant Science 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proceedings of the National Academy of Sciences, USA 106, 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Costa LE, de Queiroz MV, Borges AC, de Moraes CA, de Araújo EF. 2012. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Brazilian Journal of Microbiology 43, 1562–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vrieze M, Germanier F, Vuille N, Weisskopf L. 2018. Combining different potato-associated Pseudomonas strains for improved biocontrol of Phytophthora infestans. Frontiers in Microbiology 9, 2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weyer A-LV, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F. 2019. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178, 1260–1272.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayanithy G, Subban K, Chelliah J. 2019. Diversity and biological activities of endophytic fungi associated with Catharanthus roseus. BMC Microbiology 19, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CJ, Wang LL, Li Q, Shang QM. 2019. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS One 14, e0223847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817. [DOI] [PubMed] [Google Scholar]
- Dong YH, Xu JL, Li XZ, Zhang LH. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proceedings of the National Academy of Sciences, USA 97, 3526–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R, Eberl L. 2006. Interactions between bacteria and eukaryotes via small molecules. Current Opinion in Biotechnology 17, 268–273. [DOI] [PubMed] [Google Scholar]
- Dulla G, Lindow SE. 2008. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proceedings of the National Academy of Sciences, USA 105, 3082–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A, Maillard F, Alvarez-Lopez V, Guinchard S, Bertheau C, Valot B, Blaudez D, Chalot M. 2018. Bacterial diversity associated with poplar trees grown on a Hg-contaminated site: community characterization and isolation of Hg-resistant plant growth-promoting bacteria. The Science of the total environment 622–623, 1165–1177. [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. 2010. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Current Opinion in Plant Biology 13, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen K, Sengupta P, Kroll S, Kemen E, Doehlemann G. 2020. An antagonistic driver of the microbial phyllosphere suppresses infection of Arabidopsis thaliana by the oomycete pathogen Albugo laibachii via a secreted hydrolase. bioRxiv, 10.1101/2020.04.20.051367. [Preprint]. [DOI] [Google Scholar]
- Elias S, Banin E. 2012. Multi-species biofilms: living with friendly neighbors. FEMS Microbiology Reviews 36, 990–1004. [DOI] [PubMed] [Google Scholar]
- Enya J, Shinohara H, Yoshida S, Tsukiboshi T, Negishi H, Suyama K, Tsushima S. 2007. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microbial Ecology 53, 524–536. [DOI] [PubMed] [Google Scholar]
- Fall R, Benson AA. 1996. Leaf methanol—the simplest natural product from plants. Trends in Plant Science 1, 296–301. [Google Scholar]
- Federle MJ. 2009. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contributions to Microbiology 16, 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan JM, Castel B, Bentham AR, Jones JD. 2020. Plant NLRs get by with a little help from their friends. Current Opinion in Plant Biology 56, 99–108. [DOI] [PubMed] [Google Scholar]
- Felipe V, Romero AM, Montecchia MS, Vojnov AA, Bianco MI, Yaryura PM. 2018. Xanthomonas vesicatoria virulence factors involved in early stages of bacterial spot development in tomato. Plant Pathology 67, 1936–1943. [Google Scholar]
- Ference CM, Gochez AM, Behlau F, Wang N, Graham JH, Jones JB. 2018. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Molecular Plant Pathology 19, 1302–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wuertz S. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nature Reviews. Microbiology 17, 247–260. [DOI] [PubMed] [Google Scholar]
- Flores-Núñez VM, Fonseca-García C, Desgarennes D, Eloe-Fadrosh E, Woyke T, Partida-Martínez LP. 2020. Functional signatures of the epiphytic prokaryotic microbiome of agaves and cacti. Frontiers in Microbiology 10, 3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC. 2019. Molecular host mimicry and manipulation in bacterial symbionts. FEMS Microbiology Letters 366, fnz038. [DOI] [PubMed] [Google Scholar]
- Frank AC, Saldierna Guzmán JP, Shay JE. 2017. Transmission of bacterial endophytes. Microorganisms 5, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni A, Calderon CE, Harris R, Buxdorf K, Dafa-Berger A, Zeilinger-Reichert E, Levy M. 2015. Biological control of the cucurbit powdery mildew pathogen Podosphaera xanthii by means of the epiphytic fungus Pseudozyma aphidis and parasitism as a mode of action. Frontiers in Plant Science 6, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner S, Schmid M, Dürr C, Schuhegger R, Steidle A, Hutzler P, Langebartels C, Eberl L, Hartmann A, Dazzo FB. 2006. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiology Ecology 56, 188–194. [DOI] [PubMed] [Google Scholar]
- Glushakova AM, Chernov IYU. 2004. Seasonal dynamics in a yeast population on leaves of the common wood sorrel Oxalis acetosella L. Microbiology 73, 184–188. [PubMed] [Google Scholar]
- Gniwotta F, Vogg G, Gartmann V, Carver TLW, Riederer M, Jetter R. 2005. What do microbes encounter at the plant surface? Chemical composition of pea leaf cuticular waxes. Plant Physiology 139, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths SM, Galambao M, Rowntree J, Goodhead I, Hall J, O’Brien D, Atkinson N, Antwis RE. 2020. Complex associations between cross-kingdom microbial endophytes and host genotype in ash dieback disease dynamics. Journal of Ecology 108, 291–309. [Google Scholar]
- Grinberg M, Orevi T, Kashtan N. 2019. Bacterial surface colonization, preferential attachment and fitness under periodic stress. PLoS Computational Biology 15, e1006815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S. 2015. Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiology 169, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MD, Bento G, Ebert D. 2017. The evolutionary consequences of stepwise infection processes. Trends in Ecology & Evolution 32, 612–623. [DOI] [PubMed] [Google Scholar]
- Haney CH, Wiesmann CL, Shapiro LR, et al. 2018. Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens. Molecular Ecology 27, 1833–1847. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiology and Molecular Biology Reviews 79, 293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsonowati W, Astuti RI, Wahyudi AT. 2017. Leaf blast disease reduction by rice-phyllosphere Actinomycetes producing bioactive compounds. Journal of General Plant Pathology 83, 98–108. [Google Scholar]
- Hassani MA, Durán P, Hacquard S. 2018. Microbial interactions within the plant holobiont. Microbiome 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich EJN, Vogel CM, Ueoka R, Schäfer M, Ryffel F, Müller DB, Probst S, Kreuzer M, Piel J, Vorholt JA. 2018. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nature Microbiology 3, 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell EE, Wang Q, Yang Y. 2016. Ethylene biosynthesis and signaling is required for rice immune response and basal resistance against Magnaporthe oryzae infection. Molecular Plant-Microbe Interactions 29, 831–843. [DOI] [PubMed] [Google Scholar]
- Herrera CM, Koutsoudis MD, Wang X, von Bodman SB. 2008. Pantoea stewartii subsp. stewartii exhibits surface motility, which is a critical aspect of Stewart’s wilt disease development on maize. Molecular Plant-Microbe Interactions 21, 1359–1370. [DOI] [PubMed] [Google Scholar]
- Hopkins DL. 1989. Xylella fastidiosa: xylem-limited bacterial pathogen of plants. Annual Review of Phytopathology 27, 271–290. [DOI] [PubMed] [Google Scholar]
- Hutchison ML, Johnstone K. 1993. Evidence for the involvement of the surface active properties of the extracellular toxin tolaasin in the manifestation of brown blotch disease symptoms by Pseudomonas tolaasii on Agaricus bisporus. Physiological and Molecular Plant Pathology 42, 373–384. [Google Scholar]
- Innerebner G, Knief C, Vorholt JA. 2011. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Applied and Environmental Microbiology 77, 3202–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Into P, Pontes A, Sampaio JP, Limtong S. 2020. Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms 8, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Parsek MR. 2008. Quorum sensing and microbial biofilms. Current Topics in Microbiology and Immunology 322, 67–84. [DOI] [PubMed] [Google Scholar]
- Islam M, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. Journal of Food Protection 67, 1365–1370. [DOI] [PubMed] [Google Scholar]
- Ives AR, Klug JL, Gross K. 2000. Stability and species richness in complex communities. Ecology Letters 3, 399–411. [Google Scholar]
- Jacob C, Melotto M. 2019. Human pathogen colonization of lettuce dependent upon plant genotype and defense response activation. Frontiers in Plant Science 10, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CE, Willis DK, Charkowski AO. 2008. The flagellar sigma factor fliA is required for Dickeya dadantii virulence. Molecular Plant-Microbe Interactions 21, 1431–1442. [DOI] [PubMed] [Google Scholar]
- Jakuschkin B, Fievet V, Schwaller L, Fort T, Robin C, Vacher C. 2016. Deciphering the pathobiome: intra- and interkingdom interactions involving the pathogen Erysiphe alphitoides. Microbial Ecology 72, 870–880. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Kamo T, Hiradate S, Suzuki K, Fujita I, Yamaki S, Yoneda T, Koitabashi M, Yoshida S. 2018. Methylobamine, a UVA-absorbing compound from the plant-associated bacteria Methylobacterium sp. Natural Product Communications 13, 141–143. [Google Scholar]
- Karasov TL, Neumann M, Duque-Jaramillo A, et al. 2019. The relationship between microbial biomass and disease in the Arabidopsis thaliana phyllosphere. bioRxiv, 828814. [Preprint]. [Google Scholar]
- Kaur P, Sivasithamparam K, Li H, Barbetti MJ. 2011. Pre-inoculation with Hyaloperonospora parasitica reduces incubation period and increases severity of disease caused by Albugo candida in a Brassica juncea variety resistant to downy mildew. Journal of General Plant Pathology 77, 101–106. [Google Scholar]
- Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL. 2014. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proceedings of the National Academy of Sciences, USA 111, 13715–13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdraon L, Barret M, Laval V, Suffert F. 2019. Differential dynamics of microbial community networks help identify microorganisms interacting with residue-borne pathogens: the case of Zymoseptoria tritici in wheat. Microbiome 7, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharwar RN, Gond SK, Kumar A, Mishra A. 2010. A comparative study of endophytic and epiphytic fungal association with leaf of Eucalyptus citriodora Hook., and their antimicrobial activity. World Journal of Microbiology and Biotechnology 26, 1941–1948. [Google Scholar]
- Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. 2010. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. The ISME Journal 4, 719–728. [DOI] [PubMed] [Google Scholar]
- Kong P, Tyler BM, Richardson PA, Lee BW, Zhou ZS, Hong C. 2010. Zoospore interspecific signaling promotes plant infection by Phytophthora. BMC Microbiology 10, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J, Doehlemann G, Kemen E, Thines M. 2017. Asexual and sexual morphs of Moesziomyces revisited. IMA Fungus 8, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Verma JP. 2018. Does plant–microbe interaction confer stress tolerance in plants: a review? Microbiological Research 207, 41–52. [DOI] [PubMed] [Google Scholar]
- Kyrkou I, Pusa T, Ellegaard-Jensen L, Sagot MF, Hansen LH. 2018. Pierce’s disease of grapevines: a review of control strategies and an outline of an epidemiological model. Frontiers in Microbiology 9, 2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest-Lapointe I, Messier C, Kembel SW. 2016. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest-Lapointe I, Whitaker BK. 2019. Decrypting the phyllosphere microbiota: progress and challenges. American Journal of Botany 106, 171–173. [DOI] [PubMed] [Google Scholar]
- Lai Y, Eulgem T. 2018. Transcript-level expression control of plant NLR genes. Molecular Plant Pathology 19, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Bhandari DD, Parker JE. 2020. Origins and immunity networking functions of EDS1 family proteins. Annual Review of Phytopathology 58, 253–276. [DOI] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, et al. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. [DOI] [PubMed] [Google Scholar]
- Lee K, Pan JJ, May G. 2009. Endophytic Fusarium verticillioides reduces disease severity caused by Ustilago maydis on maize. FEMS Microbiology Letters 299, 31–37. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Park SY, Lee JJ, Yum DY, Koo BT, Lee JK. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Applied and Environmental Microbiology 68, 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau P, Barret M, Mazurier S, Mondy S, Pivato B, Fort T, Vacher C. 2017. Plant communication with associated microbiota in the spermosphere, rhizosphere and phyllosphere. In: G Becard, ed. How plants communicate with their biotic environment. Advances in Botanical Research. Academic Press, 101–133. [Google Scholar]
- Leopold DR, Busby PE. 2020. Host genotype and colonist arrival order jointly govern plant microbiome composition and function. bioRxiv, 10.1016/j.cub.2020.06.011. [Preprint]. [DOI] [PubMed] [Google Scholar]
- Leveau JH. 2019. A brief from the leaf: latest research to inform our understanding of the phyllosphere microbiome. Current Opinion in Microbiology 49, 41–49. [DOI] [PubMed] [Google Scholar]
- Levy A, Salas Gonzalez I, Mittelviefhaus M, et al. 2018. Genomic features of bacterial adaptation to plants. Nature Genetics 50, 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li J, Zhang Y, Wang N. 2019. Diffusible signal factor (DSF)-mediated quorum sensing modulates expression of diverse traits in Xanthomonas citri and responses of citrus plants to promote disease. BMC Genomics 20, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Applied and Environmental Microbiology 69, 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow EJ, Vassiliadis S, Ekanayake PN, Hettiarachchige IK, Reddy P, Sawbridge TI, Rochfort SJ, Spangenberg GC, Guthridge KM. 2019. Analysis of the indole diterpene gene cluster for biosynthesis of the epoxy-janthitrems in Epichloë endophytes. Microorganisms 7, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Zhang Z, Wang P, et al. 2019. Variations in phyllosphere microbial community along with the development of angular leaf-spot of cucumber. AMB Express 9, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D, Ma A, Bai Z, Zhuang X, Zhuang G. 2012. Response of leaf-associated bacterial communities to primary acyl-homoserine lactone in the tobacco phyllosphere. Research in Microbiology 163, 119–124. [DOI] [PubMed] [Google Scholar]
- Ma A, Lv D, Zhuang X, Zhuang G. 2013. Quorum quenching in culturable phyllosphere bacteria from tobacco. International Journal of Molecular Sciences 14, 14607–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5, e00682–e00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud F, Torres PS, Roeschlin R, Rigano LA, Enrique R, Bonomi HR, Castagnaro AP, Marano MR, Vojnov AA. 2011. The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 157, 819–829. [DOI] [PubMed] [Google Scholar]
- Manching HC, Balint-Kurti PJ, Stapleton AE. 2014. Southern leaf blight disease severity is correlated with decreased maize leaf epiphytic bacterial species richness and the phyllosphere bacterial diversity decline is enhanced by nitrogen fertilization. Frontiers in Plant Science 5, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques LLR, Ceri H, Manfio GP, Reid DM, Olson ME. 2002. Characterization of biofilm formation by Xylella fastidiosa in vitro. Plant Disease 86, 633–638. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CMJ, Pozo MJ, Ton J, van Dam NM, Conrath U. 2016. Recognizing plant defense priming. Trends in Plant Science 21, 818–822. [DOI] [PubMed] [Google Scholar]
- Martins PMM, Merfa MV, Takita MA, De Souza AA. 2018. Persistence in phytopathogenic bacteria: do we know enough? Frontiers in Microbiology 9, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY. 2017. Stomatal defense a decade later. Plant Physiology 174, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. 2010. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiology 154, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KI, Qing C, Sze DM, Neilan BA. 2012. Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS One 7, e35953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelviefhaus M, Müller DB, Zambelli T, Vorholt JA. 2019. A modular atomic force microscopy approach reveals a large range of hydrophobic adhesion forces among bacterial members of the leaf microbiota. The ISME Journal 13, 1878–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proceedings of the National Academy of Sciences, USA 100, 15977–15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Applied and Environmental Microbiology 70, 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. 2005. Aggregates of resident bacteria facilitate survival of immigrant bacteria on leaf surfaces. Microbial Ecology 49, 343–352. [DOI] [PubMed] [Google Scholar]
- Monteiro F, Nishimura MT. 2018. Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annual Review of Phytopathology 56, 243–267. [DOI] [PubMed] [Google Scholar]
- Morella NM, Weng FC, Joubert PM, Metcalf CJE, Lindow S, Koskella B. 2020. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proceedings of the National Academy of Sciences, USA 117, 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CE, Kinkel LL. 2002. Fifty years of phyllosphere microbiology: significant contributions to research in related fields. In: Lindow SE, Hecht-Poinar EI, Elliott V, eds. Phyllosphere microbiology. St. Paul, MN: APS Press, 365–375. [Google Scholar]
- Mougi A, Kondoh M. 2012. Diversity of interaction types and ecological community stability. Science 337, 349–351. [DOI] [PubMed] [Google Scholar]
- Müller C, Riederer M. 2005. Plant surface properties in chemical ecology. Journal of Chemical Ecology 31, 2621–2651. [DOI] [PubMed] [Google Scholar]
- Munch D, Gupta V, Bachmann A, Busch W, Kelly S, Mun T, Andersen SU. 2018. The Brassicaceae family displays divergent, shoot-skewed NLR resistance gene expression. Plant Physiology 176, 1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munther DS, Carter MQ, Aldric CV, Ivanek R, Brandl MT. 2020. Formation of Escherichia coli O157:H7 persister cells in the lettuce phyllosphere and application of differential equation models to predict their prevalence on lettuce plants in the field. Applied and Environmental Microbiology 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen C Jr, Ohlrogge JB, Barry CS. 2012. Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiology 159, 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi TS, Wicklund JH, Olsen SJ, et al. 2003. Concurrent outbreaks of Shigella sonnei and enterotoxigenic Escherichia coli infections associated with parsley: implications for surveillance and control of foodborne illness. Journal of Food Protection 66, 535–541. [DOI] [PubMed] [Google Scholar]
- Nascimento FX, Rossi MJ, Glick BR. 2018. Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Frontiers in Plant Science 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Schreiber L, Franke RB, Geldner N, Reina-Pinto JJ, Kunst L. 2013. Apoplastic diffusion barriers in Arabidopsis. The Arabidopsis Book 11, e0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EB. 2018. The seed microbiome: origins, interactions, and impacts. Plant and Soil 422, 7–34. [Google Scholar]
- Neu TR, Härtner T, Poralla K. 1990. Surface active properties of viscosin: a peptidolipid antibiotic. Applied Microbiology and Biotechnology 32, 518–520. [Google Scholar]
- Newcombe G, Harding A, Ridout M, Busby PE. 2018. A hypothetical bottleneck in the plant microbiome. Frontiers in Microbiology 9, 1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annual Review of Genetics 43, 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman A, Aqeel M, Lou Y. 2019. PRRs and NB-LRRs: from signal perception to activation of plant innate immunity. International Journal of Molecular Sciences 20, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. 2009. High humidity induces abscisic acid 8'-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiology 149, 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oso S, Walters M, Schlechter RO, Remus-Emsermann MNP. 2019. Utilisation of hydrocarbons and production of surfactants by bacteria isolated from plant leaf surfaces. FEMS Microbiology Letters 366, fnz061. [DOI] [PubMed] [Google Scholar]
- Ou X, Gan Y, Chen P, Qiu M, Jiang K, Wang G. 2014. Stomata prioritize their responses to multiple biotic and abiotic signal inputs. PLoS One 9, e101587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JJ, Baumgarten AM, May G. 2008. Effects of host plant environment and Ustilago maydis infection on the fungal endophyte community of maize (Zea mays). New Phytologist 178, 147–156. [DOI] [PubMed] [Google Scholar]
- Pańka D, Piesik D, Jeske M, Baturo-Cieśniewska A. 2013. Production of phenolics and the emission of volatile organic compounds by perennial ryegrass (Lolium perenne L.)/Neotyphodium lolii association as a response to infection by Fusarium poae. Journal of Plant Physiology 170, 1010–1019. [DOI] [PubMed] [Google Scholar]
- Peredo EL, Simmons SL. 2018, Leaf-FISH: microscale imaging of bacterial taxa on phyllosphere. Frontiers in Microbiology 8, 2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, Zhang Y. 2018. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Molecular Plant-Microbe Interactions 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Peyraud R, Mbengue M, Barbacci A, Raffaele S. 2019. Intercellular cooperation in a fungal plant pathogen facilitates host colonization. Proceedings of the National Academy of Sciences, USA 116, 3193–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Alfaro A, Bayman P. 2011. Hidden fungi, emergent properties: endophytes and microbiomes. Annual Review of Phytopathology 49, 291–315. [DOI] [PubMed] [Google Scholar]
- Poudel R, Jumpponen A, Schlatter DC, Paulitz TC, Gardener BB, Kinkel LL, Garrett KA. 2016. Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology 106, 1083–1096. [DOI] [PubMed] [Google Scholar]
- Purahong W, Orrù L, Donati I, Perpetuini G, Cellini A, Lamontanara A, Michelotti V, Tacconi G, Spinelli F. 2018. Plant microbiome and its link to plant health: host species, organs and Pseudomonas syringae pv. actinidiae infection shaping bacterial phyllosphere communities of Kiwifruit plants. Frontiers in Plant Science 9, 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Duan T, Sun X, et al. 2018. Host genotype strongly influences phyllosphere fungal communities associated with Mussaenda pubescens var. alba (Rubiaceae). Fungal Ecology 36, 141–151. [Google Scholar]
- Qin C, Tao J, Liu T, Liu Y, Xiao N, Li T, Gu Y, Yin H, Meng D. 2019. Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Molecular Plant-Microbe Interactions 18, 682–693. [DOI] [PubMed] [Google Scholar]
- Raghavendra AK, Newcombe G. 2013. The contribution of foliar endophytes to quantitative resistance to Melampsora rust. New Phytologist 197, 909–918. [DOI] [PubMed] [Google Scholar]
- Rajesh PS, Ravishankar Rai V. 2014. Quorum quenching activity in cell-free lysate of endophytic bacteria isolated from Pterocarpus santalinus Linn., and its effect on quorum sensing regulated biofilm in Pseudomonas aeruginosa PAO1. Microbiological Research 169, 561–569. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Coaker GL, Leveau JH. 2013. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiology Letters 348, 1–10. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JH. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. The ISME Journal 6, 1812–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environmental Microbiology 12, 2885–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. 2013. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS One 8, e78613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus-Emsermann MNP, Schlechter RO. 2018. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytologist 218, 1327–1333. [DOI] [PubMed] [Google Scholar]
- Rigano LA, Siciliano F, Enrique R, et al. 2007. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Molecular Plant-Microbe Interactions 20, 1222–1230. [DOI] [PubMed] [Google Scholar]
- Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux JP, L’Haridon F. 2016. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytologist 210, 1033–1043. [DOI] [PubMed] [Google Scholar]
- Romero FM, Marina M, Pieckenstain FL. 2014. The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiology Letters 351, 187–194. [DOI] [PubMed] [Google Scholar]
- Roy D, Panchal S, Rosa BA, Melotto M. 2013. Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 103, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Biedrzycki ML, Bais HP. 2008. Causes and consequences of plant-associated biofilms. FEMS Microbiology Ecology 64, 153–166. [DOI] [PubMed] [Google Scholar]
- Ruhe J, Agler MT, Placzek A, Kramer K, Finkemeier I, Kemen EM. 2016. Obligate biotroph pathogens of the genus Albugo are better adapted to active host defense compared to niche competitors. Frontiers in Plant Science 7, 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S. 2018. Pattern recognition receptors and signaling in plant–microbe interactions. The Plant Journal 93, 592–613. [DOI] [PubMed] [Google Scholar]
- Saldaña Z, Sánchez E, Xicohtencatl-Cortes J, Puente JL, Girón JA. 2011. Surface structures involved in plant stomata and leaf colonization by shiga-toxigenic Escherichia coli O157:H7. Frontiers in Microbiology 2, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatierra-Martinez R, Arancibia W, Araya M, Aguilera S, Olalde V, Bravo J, Stoll A. 2018. Colonization ability as an indicator of enhanced biocontrol capacity—an example using two Bacillus amyloliquefaciens strains and Botrytis cinerea infection of tomatoes. Journal of Phytopathology 166, 601–612. [Google Scholar]
- Sanjenbam P, Buddidathi R, Venkatesan R, Shivaprasad PV, Agashe D. 2020. Phenotypic diversity of Methylobacterium associated with rice landraces in North-East India. PLoS One 15, e0228550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp M, Ploch S, Fiore-Donno AM, Bonkowski M, Rose LE. 2018. Protists are an integral part of the Arabidopsis thaliana microbiome. Environmental Microbiology 20, 30–43. [DOI] [PubMed] [Google Scholar]
- Schönherr J. 2006. Characterization of aqueous pores in plant cuticles and permeation of ionic solutes. Journal of Experimental Botany 57, 2471–2491. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Krimm U, Knoll D, Sayed M, Auling G, Kroppenstedt RM. 2005. Plant–microbe interactions: identification of epiphytic bacteria and their ability to alter leaf surface permeability. New Phytologist 166, 589–594. [DOI] [PubMed] [Google Scholar]
- Scott RA, Thilmony R, Harden LA, Zhou Y, Brandl MT. 2017. Escherichia coli O157:H7 converts plant-derived choline to glycine betaine for osmoprotection during pre- and post-harvest colonization of injured lettuce leaves. Frontiers in Microbiology 8, 2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K, Seo E, Witek K, Li M, Staskawicz B. 2020. Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species. New Phytologist 227, 1530–1543. [DOI] [PubMed] [Google Scholar]
- Sharma R, Ökmen B, Doehlemann G, Thines M. 2019. Saprotrophic yeasts formerly classified as Pseudozyma have retained a large effector arsenal, including functional Pep1 orthologs. Mycological Progress 18, 763–768. [Google Scholar]
- Shivas RG, Brown JF. 1984. Identification and enumeration of yeasts on Banksia collina and Callistemon viminalis leaves. Transactions of the British Mycological Society 83, 687–689. [Google Scholar]
- Singh P, Santoni S, This P, Péros J-P. 2018. Genotype–environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: an NGS approach. Microorganisms 6, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BWG, Weingarten EA, Jackson CR. 2018. The role of the phyllosphere microbiome in plant health and function. Annual Plant Reviews 1, 1–24. [Google Scholar]
- Streletskii RA, Kachalkin AV, Glushakova AM, Yurkov AM, Demin VV. 2019. Yeasts producing zeatin. PeerJ 7, e6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun PF, Fang WT, Shin LY, Wei JY, Fu SF, Chou JY. 2014. Indole-3-acetic acid-producing yeasts in the phyllosphere of the carnivorous plant Drosera indica L. PLoS One 9, e114196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Timmers AC, Knief C, Vorholt JA. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Applied and Environmental Microbiology 71, 7245–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Bassler BL. 2003. Chemical communication among bacteria. Proceedings of the National Academy of Sciences, USA 100 Suppl 2, 14549–14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans-Kersten J, Huang H, Allen C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. Journal of Bacteriology 183, 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Leveau JH. 2012. The mechanics of bacterial cluster formation on plant leaf surfaces as revealed by bioreporter technology. Environmental Microbiology 14, 1325–1332. [DOI] [PubMed] [Google Scholar]
- Teixeira PJP, Colaianni NR, Fitzpatrick CR, Dangl JL. 2019. Beyond pathogens: microbiota interactions with the plant immune system. Current Opinion in Microbiology 49, 7–17. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Warriner K, Bartz J, Schneider KR. 2011. Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends in Microbiology 19, 121–127. [DOI] [PubMed] [Google Scholar]
- Thébault E, Fontaine C. 2010. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856. [DOI] [PubMed] [Google Scholar]
- Thevenet D, Pastor V, Baccelli I, Balmer A, Vallat A, Neier R, Glauser G, Mauch-Mani B. 2017. The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytologist 213, 552–559. [DOI] [PubMed] [Google Scholar]
- Thines M. 2014. Phylogeny and evolution of plant pathogenic oomycetes—a global overview. European Journal of Plant Pathology 138, 431–447. [Google Scholar]
- Thompson IP, Bailey MJ, Fenlon JS, et al. 1993. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant and Soil 150, 177–191. [Google Scholar]
- Thorne ET, Stevenson JF, Rost TL, Labavitch JM, Matthews MA. 2006. Pierce’s disease symptoms: comparison with symptoms of water deficit and the impact of water deficits. American Journal of Enology and Viticulture 57, 1–11. [Google Scholar]
- Ueda H, Kurose D, Kugimiya S, Mitsuhara I, Yoshida S, Tabata J, Suzuki K, Kitamoto H. 2018. Disease severity enhancement by an esterase from non-phytopathogenic yeast Pseudozyma Antarctica and its potential as adjuvant for biocontrol agents. Scientific Reports 8, 16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich K, Becker R, Behrendt U, Kube M, Ulrich A. 2020. A comparative analysis of ash leaf-colonizing bacterial communities identifies putative antagonists of Hymenoscyphus fraxineus. Frontiers in Microbiology 11, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich RL. 2004. Quorum quenching: enzymatic disruption of N-acylhomoserine lactone-mediated bacterial communication in Burkholderia thailandensis. Applied and Environmental Microbiology 70, 6173–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W, Melotto M, He SY. 2007. Role of plant stomata in bacterial invasion. Cellular Microbiology 9, 1621–1629. [DOI] [PubMed] [Google Scholar]
- Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE. 2016. The phyllosphere: microbial jungle at the plant–climate interface. Annual Review of Ecology, Evolution, and Systematics 47, 1–24. [Google Scholar]
- van Dam P, Fokkens L, Schmidt SM, Linmans JHJ, Kistler HC, Ma L-J, Rep M. 2016. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environmental Microbiology 18, 4087–4102. [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. 2015. The importance of the microbiome of the plant holobiont. New Phytologist 206, 1196–1206. [DOI] [PubMed] [Google Scholar]
- Vannier N, Agler M, Hacquard S. 2019. Microbiota-mediated disease resistance in plants. PLoS Pathogens 15, e1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes S, Gayrard D, Veyssière M, Toulotte J, Martinez Y, Dumont V, Bouchez O, Rey T, Dumas B. 2020. Phyllosphere colonization by a soil Streptomyces sp. promotes plant defense responses against fungal infection. Molecular Plant-Microbe Interactions 33, 223–234. [DOI] [PubMed] [Google Scholar]
- Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. 2016. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytologist 212, 192–207. [DOI] [PubMed] [Google Scholar]
- Vorholt JA. 2012. Microbial life in the phyllosphere. Nature Reviews. Microbiology 10, 828–840. [DOI] [PubMed] [Google Scholar]
- Vorholt JA, Vogel C, Carlström CI, Müller DB. 2017. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host & Microbe 22, 142–155. [DOI] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Coleman-Derr D, Tringe SG, Dangl JL, Mitchell-Olds T. 2014. Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecology Letters 17, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. 2016. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications 7, 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J, Laforest-Lapointe I, Kembel SW. 2018. Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. PeerJ 6, e5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sipilä TP, Overmyer K. 2016. The isolation and characterization of resident yeasts from the phylloplane of Arabidopsis thaliana. Scientific Reports 6, 39403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Begerow D, Groenewald M, Liu XZ, Theelen B, Bai FY, Boekhout T. 2015. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilagino mycotina. Studies in Mycology 81, 55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Fanella B, Guo L, Wang X. 2016. Membrane glycerolipidome of soybean root hairs and its response to nitrogen and phosphate availability. Scientific Reports 6, 36172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S. 2017. NLR network mediates immunity to diverse plant pathogens. Proceedings of the National Academy of Sciences, USA 114, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Kvitko B, He SY. 2018. Pseudomonas syringae: what it takes to be a pathogen. Nature Reviews. Microbiology 16, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. 2016. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li W, Cao J, et al. 2017. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. The Plant Journal 89, 338–353. [DOI] [PubMed] [Google Scholar]
- Yao H, Sun X, He C, Maitra P, Li XC, Guo LD. 2019. Phyllosphere epiphytic and endophytic fungal community and network structures differ in a tropical mangrove ecosystem. Microbiome 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Iguchi H, Sakai Y, Yurimoto H. 2019. Pantothenate auxotrophy of Methylobacterium spp. isolated from living plants. Bioscience, Biotechnology, and Biochemistry 83, 569–577. [DOI] [PubMed] [Google Scholar]
- Yu M-H, Zhao Z-Z, He J-X. 2018. Brassinosteroid signaling in plant–microbe interactions. International Journal of Molecular Sciences 19, 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Melotto M, He SY. 2010. Plant stomata: a checkpoint of host immunity and pathogen virulence. Current Opinion in Biotechnology 21, 599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Bai Z, Hoefel D, Tang L, Yang Z, Zhuang G, Yang J, Zhang H. 2008. Assessing the impact of the biological control agent Bacillus thuringiensis on the indigenous microbial community within the pepper plant phyllosphere. FEMS Microbiology Letters 284, 102–108. [DOI] [PubMed] [Google Scholar]
- Zhao T, Liu W, Zhao Z, et al. 2019. Transcriptome profiling reveals the response process of tomato carrying Cf-19 and Cladosporium fulvum interaction. BMC Plant Biology 19, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Spivey NW, Zeng W, Liu P-P, Fu ZQ, Klessig DF, He SY, Dong X. 2012. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host & Microbe 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]