Abstract
Cyanobacteria produce a plethora of compounds with unique chemical structures and diverse biological activities. Importantly, the increasing availability of cyanobacterial genome sequences and the rapid development of bioinformatics tools have unraveled the tremendous potential of cyanobacteria in producing new natural products. However, the discovery of these compounds based on cyanobacterial genomes has progressed slowly as the majority of their corresponding biosynthetic gene clusters (BGCs) are silent. In addition, cyanobacterial strains are often slow-growing, difficult for genetic engineering, or cannot be cultivated yet, limiting the use of host genetic engineering approaches for discovery. On the other hand, genetically tractable hosts such as Escherichia coli, Actinobacteria, and yeast have been developed for the heterologous expression of cyanobacterial BGCs. More recently, there have been increased interests in developing model cyanobacterial strains as heterologous production platforms. Herein, we present recent advances in the heterologous production of cyanobacterial compounds in both cyanobacterial and noncyanobacterial hosts. Emerging strategies for BGC assembly, host engineering, and optimization of BGC expression are included for fostering the broader applications of synthetic biology tools in the discovery of new cyanobacterial natural products.
Keywords: Cyanobacterial natural products, Biosynthetic gene cluster, Heterologous expression
Introduction
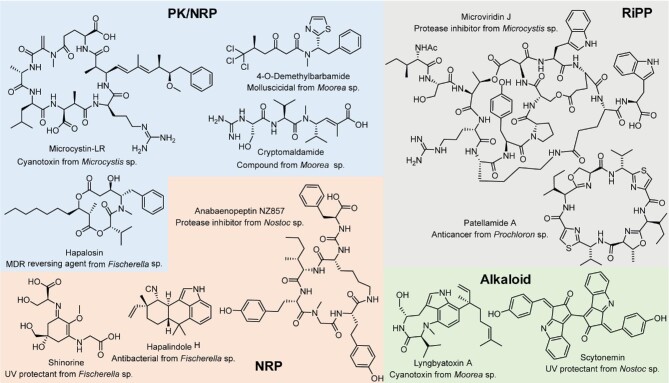
Cyanobacteria are a phylum of photosynthetic prokaryotes that possess exceptional adaptability in almost all environmental niches. They can survive in diverse conditions varying from Antarctic Dry Valleys to Atacama Desert (Friedmann & Kibler, 1980; Wierzchos et al., 2006) and hot springs near volcanic zones (Teece et al., 2020, 3). In addition, they can exist as symbionts or commensals associated with other animals and plants (Demay et al., 2019; Mazard et al., 2016; Thuan et al., 2019). Accompanying their ecological plasticity, cyanobacteria demonstrate notable metabolic versatility (Xiong et al., 2017), particularly the production of a number of structurally and functionally diverse metabolites (Fig. 1). These compounds possess a wide array of bioactivities, such as antibacterial, antifungal antiviral, and antitumor activities (Cai et al., 2017; Mason et al., 1982; Salvador-Reyes & Luesch, 2015; Shishido et al., 2015). In addition, they possess antimalarial (Linington et al., 2009), anti-inflammatory (Choi et al., 2012; Montaser et al., 2013), insecticidal (Becher & Jüttner, 2005), antimetabolic (Brilisauer et al., 2019), and allelopathic properties (Leão et al., 2010). On the other hand, due to the toxicity to humans, plants, and animals, the toxins produced by cyanobacteria, named cyanotoxins (Carmichael, 1992), have attained increasing public attention. The broad bioactivity spectrum of cyanobacterial natural products (NPs) highlights the importance to further explore the cyanobacterial resource for applications.
Fig. 1.
Chemical structures of representative cyanobacterial compounds that have been heterologously produced. Compounds are grouped and colored according to structural families.
Cyanobacterial NP drug discovery and development has focused primarily on three aspects, the discovery of novel compounds, the assessment of bioactivities through in vitro and in vivo studies, and the detailed exploration of the mechanism of action on the molecular level (Liang et al., 2019; Salvador-Reyes & Luesch, 2015; Tan & Phyo, 2020). Cyanobacterial NPs possess novel bioactivities of biomedical relevance (Al-Awadhi et al., 2018, 2020; Lee et al., 2011; Pereira et al., 2012). In addition to the exploration of un- or less-studied resources, the metabolomics-based approach that is based on reference compounds as potential biomarkers for the identification of molecular fingerprints determining particular bioactivities has recently become useful in finding new chemical diversities with versatile acti-vities (Kuehnbaum & Britz-McKibbin, 2013). More recently, the genome centric approach, or the bottom-up approach, which that combines functional genomics and bioinformatics to associate genomic context called biosynthetic gene clusters (BGCs) to potential chemical entities, has been increasingly used to unravel the biosynthetic capabilities of cyanobacteria (Dittmann et al., 2015). Along with the rapid growth of cyanobacterial genome data, diverse bioinformatics tools such as antiSMASH (Blin et al., 2019), PRISM (Skinnider et al., 2015), and RODEO (Tietz et al., 2017) have enabled identifying the cyanobacterial BGCs of different compound families. Furthermore, the availability of a global repository of microbial BGCs provides another useful resource for accurate prediction and analysis of the cyanobacterial BGCs (Kautsar et al., 2020; Navarro-Muñoz et al., 2020). These resources open new opportunities to cyanobacterial NP research using the bottom-up approach.
Cyanobacterial BGCs encode mainly polyketides (PKs) and peptides, but also pharmaceutically important compounds of other families, for example, terpenes, alkaloids, and lipids (Fig. 1) (Demay et al., 2019; Dittmann et al., 2015). Among known cyanobacterial NPs, the majority (66 per cent) belong to the peptide family. Peptide molecules are biosynthesized from both nonribosomal and ribosomal origins through the actions of nonribosomal peptide synthetases (NRPSs) and ribosomally synthesized and post-translationally modified peptides (RiPPs) biosynthetic machinery, respectively (Sieber & Marahiel, 2005; Ortega & van der Donk, 2016; Hudson & Mitchell, 2018). Similarly, acyl-CoA building blocks are processed through sequential decarboxylative condensations by polyketide synthases (PKSs) to generate PKs (Dhakal et al., 2019; Smith & Tsai, 2007; Mishra et al., 2019). Furthermore, some cyanobacterial NPs are derived from hybrid biosynthetic systems. The increased understanding of cyanobacterial NP biosynthesis facilitates the discovery with the bottom-up approach.
One major challenge in cyanobacterial NP research using the bottom-up approach is that the majority of identified BGCs are silent. Many engineering approaches, such as traditional strain mutagenesis and screening and rational metabolic engineering, have been developed to activate the silent BGCs of other bacterial phyla, for example, actinomycetes. However, these approaches have barely been used for the discovery of cyanobacterial NPs as most of cyanobacterial strains are slow growers and/or genetically less amenable (Berla et al., 2013; Santos-Merino et al., 2019). As such, a large number of potentially novel and bioactive compounds are yet to be characterized from cyanobacteria. For example, comparative genome analysis of diverse Moorea species revealed 30–45 BGCs in each genome, whereas only a few compounds have been isolated from each strain (Leao et al., 2017). On the other hand, the mobilization of one BGC into a suitable heterologous host has been an effective means for not only producing the encoded compound but also redesigning genetic contents for generating NP analogs or optimizing production yield (Huo et al., 2019; Zhang et al., 2019). Herein, we present recent advances in heterologous expression of cyanobacterial NPs in both noncyanobacterial hosts, including Escherichia coli, yeast and Streptomyces, and model cyanobacterial strains. Furthermore, we discuss different metabolic engineering and synthetic biology tools for assembling BGCs, crafting production platforms, and optimizing productivity at the transcriptional, post-transcriptional, and translational levels.
Heterologous Expression of Cyanobacterial NPs
So far, a limited number of cyanobacterial NPs of several families have been produced in non-native hosts (Fig. 1, Table 1). These hosts include the Gram-negative bacterium E. coli (Ongley et al., 2013; Schmidt et al., 2005), the Gram-positive bacterium Streptomyces venezuelae (Kim et al., 2012), genetically amenable model cyanobacterial strains (Taton et al., 2020; Videau et al., 2016, 2020; Yang et al., 2018), and yeast (Park et al., 2019).
Table 1.
List of cyanobacterial compounds that have been heterologously produced
| Name | Types | Native producer | Heterologous host | Cloning method/BGC size (kb) | Yield in heterologous host | References |
|---|---|---|---|---|---|---|
| Hapalosin | PK/NRP | Fischerella sp. PCC 9431 | E. coli BAP1 | DiPAC/23 | ∼0.45-fold of native producer | D'Agostino Gulder (2018) |
| 4-O-Demethylbarbamide | PK/NRP | M. producens | S. venezuelae DHS2001 | RDL/26 | <1 μg/l | Kim et al. (2012) |
| Anabaenopeptins | NRP | N. punctiforme PCC 73102 | E. coli | DiPAC/28.1 | >100-fold of native producer | Greunke et al. (2018) |
| Patellamide, ulithiyacyclamide and eptidemnamide | RiPP | Prochloron spp. | E. coli Rosetta (DE3) | RDL/- | DM | Donia et al. (2006) |
| Patellamide A and C | RiPP | P. didemnid | E. coli BL21(DE3) pLys | GLBC/11 | DM | Schmidt et al. (2005) |
| Patellamide D and ascidiacyclamide | RiPP | P. didemnid | E. coli DH10B | RDL/30 | 80–100 ng/ml | Long et al. (2005) |
| Trukamide | RiPP | Prochloron spp. | E. coli TOP10 | RDL/11 | DM | Donia et al. (2008) |
| Anacyclamides | RiPP | Anabaena sp. 90 | E. coli TOP10 | RDL/11 | 0.5-fold of native producer | Leikoski et al. (2010) |
| Microviridins | RiPP | Microcystis UOWOCC MRC | E. coli Epi300 | GLBC/- | 20–7280 ng/ml | Ziemert et al. (2008) |
| Microviridin L | RiPP | M. aeruginosa NIES843 | E. coli BL21 | RDL/6.5 | DM | Weiz et al. (2011) |
| Perchlorosins | RiPP | Prochlorococcus MIT9313 | E. coli BL21 Gold | RDL/- | DM | Tang & van der Donk (2012) |
| Lyngbyatoxin A | NRP | M. producta | Anabaena sp. PCC 7120 | TAR/11.3 | ∼ 3.2 mg/l | Videau et al. (2016) |
| E. coli GB05-MtaA | RDL/11.3 | ∼25.6 mg/l | Ongley et al. (2013) | |||
| M. producta, | Anabaena sp. PCC 7120 | RDL/- | ∼313 ng/mg DCW | Videau et al. (2020) | ||
| Pendolmycin | NRP | M. producta, M. thermotolerans SCSIO 00652 | Anabaena sp. PCC 7120 | RDL/- | ∼200 ng/mg DCW | Videau et al. (2020) |
| Teleocidins | NRP | M. producta, S. blastmyceticus NBRC 12747 | Anabaena sp. PCC 7120 | RDL/- | ∼1 mg/mg DCW | Videau et al. (2020) |
| Cryptomaldamide | PK/NRP | M. producens JHB | Anabaena sp. PCC 7120 | TAR/28.7 | ∼26 mg/l | Taton et al. (2020) |
| Shinorine | NRP | Fischerella sp. PCC9339 | Synechocystis sp. PCC6803 | RDL/4.3 | ∼ 0.71 mg/l | Yang et al. (2018) |
| N. punctiforme | S. cerevisiae CEN.PK2−1C | RDL/6.5 | 31.0 mg/l | Park et al. (2019) | ||
| Hapalindole H, and 12-epi hapalindole U | Alkaloids | Fischerella ambigua UTEX 1903 | Synechococcus sp. UTEX 2973 | RDL/42 | ∼3 mg/l | Knoot et al. (2019) |
| Mycosporine-ornithine/mycosporine-lysine | NRP | C. stagnale PCC 7417 | E. coli BL21(DE3) | RDL/6.2 | DM | Katoch et al. (2016) |
| Mycosporine‐ycospdeoxygadusolyl‐ornithine) | NRP | Nostoc flagelliforme | Anabaena PCC 7120 | RDL/8.2 | DM | Shang et al. (2018) |
| [d-Asp3] microcystin-LR | PK/NRP | M. aeruginosa PCC 7806 | E. coli GB05-MtaA | GLBC/55 | ∼65 μg/l | Liu et al. (2017) |
| [d-Asp3, DMAdda5] microcystin-LR | PK/NRP | M. aeruginosa PCC 7806 | E. coli GB05-MtaA | GLBC/NA | DM | Liu et al. (2019) |
| Scytonemin | Alkaloid | N. punctiforme ATCC 29133 | E. coli BL21(DE3) | RDL/20kb | 8.9 mg/l | Malla & Sommer (2014) |
M producta: Moorea producens; M thermotolerans: Marinactinospora thermotolerans; M. aeruginosa: Microcystis aeruginosa; RDL: restriction digestion and ligation; GLBC: genomic library-based cloning; DCW: dry cell weight; DM: detected by mass spectrometry or NMR spectroscopy; -: no information available.
Metabolic Engineering and Synthetic Biology Toolkits for Heterologous Production of Cyanobacterial Compounds
Recent advances in metabolic engineering approaches and applications of synthetic biology tools have revolutionized the discovery of NPs from both native and heterologous hosts (Smanski et al., 2016). The major thrust for the heterologous production of cyanobacterial NPs can be directed to the cloning and assembly of BGCs, selection and optimization of heterologous hosts, and transcriptional and translational tuning of BGC expression.
Cloning and Assembly of BGCs
An initial and fundamental aspect for the heterologous production of any NP is to transfer genetic materials into one heterologous host. The assembly and cloning steps include the insertion of one desired BGC in one suitable vector. The vector utilizing the homologous recombination for integrating the desired DNA fragment into the neutral site I (NSI) has been a robust and most popular approach for cyanobacterial genetic engineering (Hitchcock et al., 2020). The NSI is a functionally neutral location in cyanobacterial genomes where the integration of heterologous DNA causes no noticeable phenotypic change at least under the relevant growth conditions (Ng et al., 2015). But recently there is growing interest in developing new vectors for cyanobacterial engineering particularly for transferring cyanobacterial BGCs for heterologous expression. Hence, replicative plasmids based on two major origins of replication RSF1010 (Scherzinger et al., 1984) or pMB1 (Rosano & Ceccarelli, 2014) have been widely used for cyanobacterial genetic engineering, while different integration plasmids based on Tn5-1063 and Tn5-1058 have also been developed (Cohen et al., 1998). Furthermore, chromosomal integration vectors carrying standard prefix (e.g., promoters and ribosome binding sites, RBS) and suffix sequences (e.g., transcriptional terminators) suitable for BioBrick-based cloning have been designed (Kim et al., 2017; Vogel et al., 2017).
Generally, shuttle vectors (replicative plasmids) that include E. coli plasmid replicons (pMB1) and endogenous plasmid segments have commonly been used for cyanobacterial genetic engineering. However, endogenous plasmids are often large, difficult for further engineering, and specific only to certain cyanobacterial species, limiting their wide use (Jin et al., 2018; Taton et al., 2014). In contrast, one non-native replicon RSF1010 exhibits a broad host range (Bishe et al., 2019). The first RSF1010-based shuttle vector pPMQAK1 enabled the generation of different recombinant constructs for biochemical characterization of cyanobacterial enzymes (Huang et al., 2010). The pPMQAK1 vector has a low copy number (∼ 10–20 copies per cell in E. coli), and a high copy number variant pSCB-YFP was later developed with the putative replicon of the plasmid pCC5.2 and the origin of replication of pMB1 from E. coli (Jin et al., 2018). This vector was used for genetic manipulation of Synechocystis sp. PCC6803, but its wide applications in engineering different cyanobacterial strains have not been validated (Jin et al., 2018). Furthermore, the CYANO-VECTOR assembly portal was developed to organize various module elements of one vector and facilitate the in silico construction of plasmids (Taton et al., 2014). Similarly, a versatile system called CyanoGate based on the Plant Golden Gate MoClo kit was recently developed and the resulting constructs were used to engineer two cyanobacterial species, Synechocystis sp. PCC 6803 and Synechococcus elongatus UTEX 2973 (Vasudevan et al., 2019).
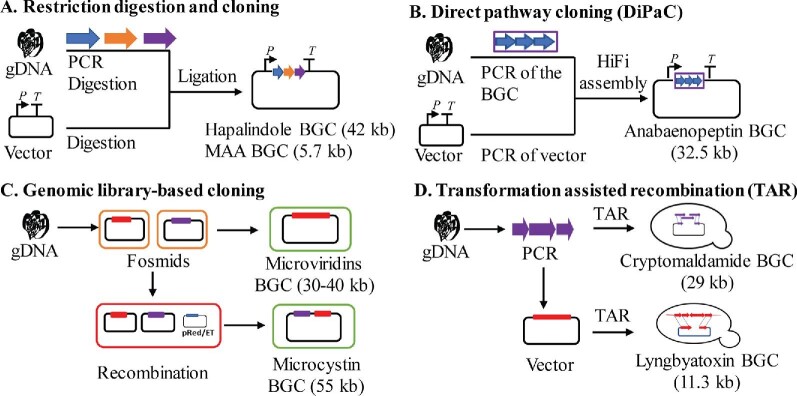
Generally, the expression of cyanobacterial compounds in a heterologous host is initiated by capturing the BGCs of cyanobacterial NPs from the genomic DNAs of native producers. This has been supported by the ease to obtain microbial whole-genome sequence, the accurate bioinformatic prediction of BGCs from genome sequences, and the availability of genetic tools (Albarano et al., 2020). Primary approaches for cloning and assembling BGCs of different sizes for heterologous expression are shown in Fig. 2. For the BGCs of small size, they can be directly cloned from native producers by conventional restriction digestion and ligation approach. This approach has demonstrated successes in the cloning of several cyanobacterial BGCs, for example, mycosporine-like amino acids (MAAs) from Cylindrospermum stagnale PCC7417 (Katoch et al., 2016), shinorine from Fischerella sp. PCC9339 (Yang et al., 2018), prochlorosins from Prochlorococcus sp. MIT9313 (Tang & van der Donk, 2012), and hapalindoles from F. ambigua UTEX 1903 (Knoot et al., 2019). Direct pathway cloning (DiPaC) is an effective homology-based assembly strategy for cloning small- to medium-sized BGCs (Fig. 2). This approach was utilized for the heterologous production of anabaenopeptins from Nostoc punctiforme (Greunke et al., 2018). Similarly, the combination of DiPaC and sequence- and ligation-independent cloning led to the efficient capture of the hapalosin BGC from Fischerella sp. PCC 9431 (D'Agostino & Gulder, 2018) (Fig. 2).
Fig. 2.
Primary approaches for cloning and assembling cyanobacterial BGCs of different sizes for heterologous expression. BGCs can be PCR amplified and ligated in a suitable vector (A) or directly assembled with the vector (B). The genomic library is generated and the clone carrying an entire BGC can be directly expressed. Alternatively, multiple clones can be recombined to generate a complete BGC by approaches such as Red-ET in E. coli (C) or TAR in yeast (D). gDNA: genomic DNA. In figure C, outside rectangles in orange, red, and green indicate different bacterial cells.
Another routine approach for cloning and assembling BGCs relies on the generation of genomic libraries of native producers using vectors of various capturing capacity such as fosmids (∼40 kb), cosmids (∼45 kb), bacterial artificial chromosomes BACs (∼200 kb) and P1 derived artificial chromosomes PACs (111–300 kb) (Fig. 2). Clones carrying the BGCs are selected from the libraries by polymerase chain reaction (PCR) or southern blotting. The positive clones are sequenced to provide concrete information about the putative BGCs. This approach has led to the characterization of the BGCs of diverse cyanobacterial NPs, such as nostophycin from Nostoc sp. strain 152 (Fewer et al., 2011), hectochlorin from Lyngbya majuscula (Ramaswamy et al., 2007), and microviridin from Microcystis aeruginosa NIES843 (Ziemert et al., 2010). Importantly, BGC-containing fosmids can be used directly for heterologous production, exemplified by the expression of the BGCs of lyngbyatoxin from Moorea producens (Ongley et al., 2013), microcystins from M. aeruginosa (Liu et al., 2017) and patellamides from Prochloron didemnid (Schmidt et al., 2005) (Fig. 2).
For the mobilization of a large DNA segment for heterologous expression, homologous recombination-based approaches such as lambda red (λ/Red) recombination in E. coli (Datsenko & Wanner, 2000) and transformation associated recombination (TAR) in Saccharomyces cerevisiae (Kouprina & Larionov, 2016) can be very efficient (Fig. 2). For example, the microcystin gene cluster from M. aeruginosa PCC 7806 was assembled by Red/ET recombineering and expressed in E. coli to yield a significant titer of [d-Asp3] microcystin-LR and microcystin-LR [66] (Fig. 2). Similarly, the TAR approach was used to mobilize the lyngbya-toxin BGC captured in the vector pPJAV633 into a customized and replicative vector pPJAV550 (optimized for the TAR-mediated capture of cyanobacterial BGCs) (Videau et al., 2016) (Fig. 2D). This same approach also assembled the cryptomaldamide BGC from M. producens JHB in a customized vector pAM5571 (Taton et al., 2020) (Fig. 2D), where different overlapping fragments of the cryptomaldamide BGC were first PCR amplified from an isolated chromosomal DNA template. On the other hand, direct capture of one entire cyanobacterial BGC from chromosomal DNA by Red/ET recombineering or TAR has not been reported yet, probably due to the challenge of attaining high quality and amount of large cyanobacterial genomic DNA fragments. Finally, as the cost of DNA synthesis is expected to continuously drop, the synthesis of entire BGCs will become economically viable for the heterologous production of cyanobacterial NPs in the coming years.
The assembled BGCs should be transferred into host cells for expression. For many conventional microbial strains, the transformation can be mediated by conjugation, protoplast transformation, or electroporation. On the other hand, some cyanobacterial hosts are naturally competent for the introduction of genetic materials, called natural transformation (Onai et al., 2004). In general, the conjugation and electroporation are facilitated by extrachromosomal genetic elements such as various replicative plasmids, whereas the natural transformation uptakes and integrates DNA fragments into the neutral sites of cyanobacterial genomes (Wijffels et al., 2013). In addition to gene overexpression, the natural transformation has been used for systematic gene inactivation in various cyanobacteria (Frigaard et al., 2004).
Selection of Heterologous Hosts for Producing Cyanobacterial NPs
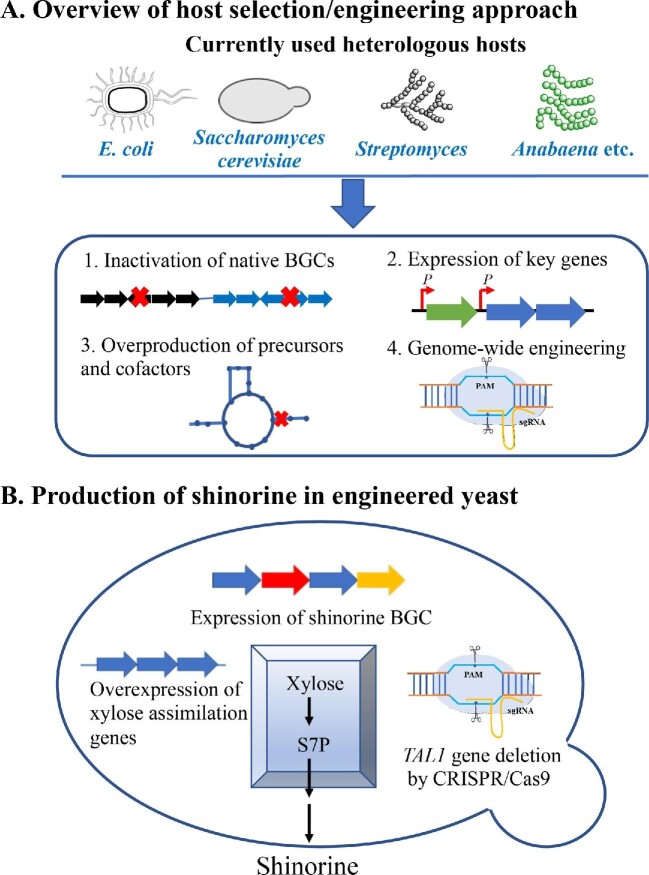
The selection of suitable production chassis is the most crucial aspect of successful heterologous production. A good host can be fast-growing and easy for genetic manipulation and handling in the laboratory or industrial processes. However, the successful expression of a BGC also requires effective protein expression and the availability of biosynthetic precursors and cofactors to accomplish all biosynthetic steps (Zhang et al., 2019). There have been significant efforts for selecting and optimizing suitable heterologous hosts for the successful expression of cyanobacterial NPs (Fig. 3).
Fig. 3.
(A) Currently available heterologous hosts as E. coli, yeast, Streptomyces, and cyanobacteria strains for cyanobacterial NP production and major approaches for host engineering. PAM: photospacer adjacent motif; SgRNA: synthetic guise RNA. (B) The overproduction of shinorine in yeast included multicopy delta integration of biosynthetic genes, the overexpression of xylose assimilation genes (XL1, XL2, and XL3) from Scheffersomyces stipites, and the modulation of the pentose phosphate pathway through deleting TAL1 and overexpressing STB5 and TKL1 to overproduce the biosynthetic precursor sedoheptulose 7-phosphate (S7P).
E. coli is among the most widely used hosts for the heterologous expression of BGCs of diverse NP families (Pfeifer et al., 2001; Watanabe et al., 2006; Zhang et al., 2018), due to its easy and flexible culturing, fast growth, cost-effectiveness, and well-established genetic manipulation tools (Kaur et al., 2018; Sanchez-Garcia et al., 2016). Not surprisingly, many cyanobacterial BGCs have been expressed in E. coli (Table 1, Figs 1 and 3), including multiple RiPPs (e.g., patellamide and microviridins), MAAs, the NRP lyngbyatoxin, and the NRP-PK hybrid microcystins (Liu et al., 2017; Long et al., 2005; Ongley et al., 2013; Schmidt et al., 2005; Ziemert et al., 2008). Similar to E. coli, Saccharomyces cerevisiae demonstrates many good features as the chassis for the hetero-logous expression of diverse NPs (Fig. 3) (Bond & Tang, 2019), such as artemisinic acid and taxadiene (Ding et al., 2014; Kung et al., 2018). However, only one cyanobacterial BGC, the shinorine BGC from N. punctiforme, has been expressed in yeast after multiple host engineering efforts (Fig. 3B) (Park et al., 2019). Actinomycetes are the most prominent source of NPs (Nguyen et al. 2020) [98], and multiple model Streptomyces strains (e.g., S. albus and S. coelicolor) have served as excellent hosts for the expression of BGCs from other actinomycete strains (Myronovskyi & Luzhetskyy 2019). However, the use of this type of hosts for the expression of cyanobacterial BGCs has so far succeeded only with the production of one barbamide analog at trace levels (Figs 1 and 3) (Kim et al., 2012).
Multiple model cyanobacterial species represent another type of promising chassis for the heterologous production of cyanobacterial NPs. Model cyanobacterial strains are considered as a sustainable platform for biotechnological applications due to their capacities of photosynthesis, N-fixation, and autotroph (Machado & Atsumi, 2012; Roulet et al., 2018). Recent advances in genomics and metabolomics of cyanobacterial strains along with the development of precise genetic engineering approaches provide additional opportunities for exploring them as a promising platform for chemical production (Lin & Pakrasi, 2019; Oliver et al., 2016). Importantly, the similar genetic backgrounds of cyanobacterial hosts can potentially lead to a higher success rate in the expression of cyanobacterial BGCs (Fig. 3). Indeed, the heterologous expression of lyngbyatoxin BGC has been accomplished in the cyanobacterial host Anabaena sp. PCC 7120 (Fig. 2), with a titer comparable to the native producer M. producens (Videau et al., 2016). Although the total yield of lyngbyatoxin A was higher in E. coli, this noncyanobacterial host led to the accumulation of significant amounts of biosynthetic intermediates N-methyl-l-valyl-l-tryptophan and indolactam V (Ongley et al., 2013; Videau et al., 2016). The complete conversion of biosynthetic precursors to the final product indicated that the lyngbyatoxin BGC has a balanced expression of individual genes in Anabaena sp. PCC 7120 (Videau et al., 2016). More recently, the heterologous expression of the cryptomaldamide BGC was attempted in two cyanobacterial strains Synechococcus elongatus PCC 7942 and Anabaena sp. PCC 7120 (Fig. 1). No targeted product was identified from the transformed S. elongatus PCC 7942, but a significant titer (∼15 mg/g biomass dry weight) was detected in engineered Anabaena sp. PCC 7120 (Taton et al., 2020).
Engineering Heterologous Hosts for Optimizing the Productivity
To enhance the productivity of target molecules, heterologous hosts can be engineered, mainly through the elimination of competitive pathways and/or the increased supply of appropriate precursors/cofactors (Fig. 3A). Common engineering strategies include the deletion of native BGCs, proteases and nucleases, and the addition of chromosomal integration elements for easing the introduction of genetic materials, precursor pathway genes and other key genes (Fig. 3A), for example, those encoding versatile phosphopantetheinyl transferases (PPTases). Of note, the PPTase is essential to produce the holo-thiolation domains of PKSs and NRPSs, which we will discuss more later. E. coli and S. cerevisiae do not encode any prominent secondary metabolite BGCs, leading to a clean NP background for easy detection of expressed foreign compounds. On the other hand, the optimal production of NPs in these two hosts often require the engineered production of essential building blocks and cofactors and the expression of key exogenous enzymes such as PPTases (Harvey et al., 2012; Pfeifer et al., 2001) (Fig. 3A). In contrast, extensive deletion of native BGCs in model Streptomyces hosts have created chassis with a clean background for heterologous production, for example, S. coelicolor (Gomez-Escribano & Bibb. 2011), S. avermitilis (Komatsu et al., 2010), S. chattanoogensis (Bu et al., 2019), and S. albus (Myronovskyi et al., 2018). However, these engineered Streptomyces hosts have not been tested for the expression of any cyanobacterial BGCs yet.
An excellent example of host engineering employed for overproduction of the cyanobacterial NP is that of shinorine in Saccharomyces cerevisiae (Park et al., 2019) (Fig. 3B). The shinorine BGC genes from N. punctiforme were cloned in a suitable expression vector and then subjected to the integration of multiple copies at the Ty retrotransposon delta sites in the yeast genome by homo-logous recombination (Shi et al., 2016). Subsequently, three xylose assimilation genes from Scheffersomyces stipites, including XYL1–3 encoding xylose reductase, xylose dehydrogenase, and xylose kinase, respectively, were introduced the yeast to convert xylose as a carbon source into xylulose-5-phosphate (X5P). X5P enters the pentose phosphate pathway (PPP), leading to the elevated level of sedoheptulose 7-phosphate (S7P) that is an important precursor for the shinorine biosynthesis. The cellular pool of S7P was further enhanced by overexpression of STB5 (a transcriptional activator of PPP) and TKL1 (a transketolase reversibly connecting PPP with the glycolysis) and deletion of TAL1 (a transaldolase mediating the interconversion of S7P and glyceraldehyde 3-phosphate in PPP) by the CRISPR/Cas9 approach (Park et al., 2019). The final engineered yeast produced 31.0 mg/l of shinorine in the optimized medium containing 8 g/l of xylose and 12 g/l of glucose (Park et al., 2019), demonstrating the power of host engineering for the heterologous production of cyanobacterial NPs.
PPTases catalyze post-translational phosphopantetheinylation of thiolation domains of modular and iterative synthases, such as fatty acid synthases, PKSs and NRPSs (Beld et al., 2014), thereby producing catalytically active enzymes (Lambalot et al., 1996). Since the endogenous PPTases of one heterologous host may not be promiscuous toward noncognate thiolation substrates (Pfeifer et al., 2001), the expression of an exogenous catalytically versatile PPTase is crucial for the production of PKs, NRPs, and their hybrids (Bond & Tang, 2019; Siewers et al., 2009) (Fig. 3A). For example, the gene of a promiscuous PPTase from the myxobacterium Stigmatella aurantiaca (MtaA) was integrated into the chromosome of E. coli GB2005 to create E. coli GB05-MtaA, which successfully expressed the microcystin BGC to produce multiple analogs (Liu et al., 2017, 2019). To gain useful insights into the catalytic performance of cyanobacterial PPTases, we recently characterized the substrate scopes of 6 enzymes with 11 thiolation domains of known and silent BGCs from cyanobacterial and Streptomyces strains (Yang et al., 2017). Biochemical and genetic studies uncovered that the PPTase from Anabaena sp. PCC7120 (APPTase) rivals the widely used surfactin PPTase (Sfp) in terms of substrate flexibility. Furthermore, the coexpression of APPTase supported the heterologous expression of the shinorine BGC of Fischerella sp. PCC9339 in Synechocystis sp. PCC6803 (Yang et al., 2018).
The recent development of robust genome engineering approaches such as CRISPR-Cas further facilitates host engineering to improve the productivity of expressed NPs, including those from cyanobacterial BGCs (Wright et al., 2016; Mougiakos et al., 2018). The CRISPR-Cas9 approach has been developed primarily for creating markerless gene deletion, supporting genome engineering and transcriptional regulation. The use of CRISPR-Cas systems to engineering E. coli, yeast and Streptomyces strains have been reviewed previously (Alberti & Corre, 2019; Didovyk et al., 2016; Jakočiūnas et al., 2016; Stovicek et al., 2017). Particularly, the CRISPR/Cas9 based gene inactivation has been already utilized during heterologous production of shinorine in yeast (Park et al., 2019) as mentioned previously. However, their applications in engineering cyanobacterial strains remain poorly explored. One potential reason is that the off-target cleavage can generate a number of illegitimate colonies. There is a significant technical difficulty in screening and validating desired mutants from a high number of relatively slow-growing cyanobacterial transformants without a selection marker. However, it may be possible to tackle this limitation by encoding two spacers, instead of one, for targeting the desired genomic region, which can improve the success rate in markerless gene deletion as shown in engineering Anabaena sp. PCC 7120 (Niu et al., 2019). Another hindrance is that the expression of Cas9 appears to be toxic in some cyanobacteria, such as S. elongatus UTEX 2973 (Wendt et al., 2016). The mechanism by which Cas9 causes toxicity remains unclear, but it is suspected that CRISPR-Cas9 engineering has off-target effects and may negatively influence growth-essential genes. The Cas9 system requires two separate RNA strands, CRISPR RNA (crRNA) that encodes guide sequence, and trans-activating crRNA (tracrRNA). Interestingly, the use of Cas12a (formerly known as Cpf1), which requires only the tracrRNA, seemingly bypasses the Cas9-related toxicity issue in many species (Ungerer & Pakrasi, 2016). Indeed, the CRISPR/Cas12a engineering has been used to efficiently obtain segregated double recombinant Anabaena sp. PCC 7120 clones for the heterologous production of cryptomaldamide (Yang et al., 2018).
Transcriptional Tuning of BGC Expression
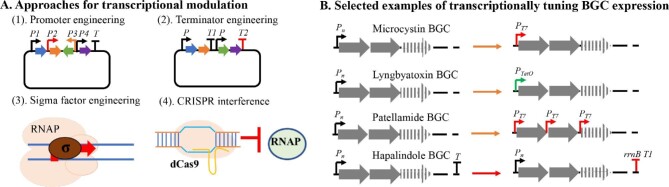
Fine-tuning of the expression level of biosynthetic genes is a proven strategy for improving the productivity of targeted NPs in native or heterologous hosts (Dhakal & Sohng, 2017; Xu et al., 2020). The use of different promoters can readily manipulate the gene expression at the transcription level (Fig. 4). For example, the T7 promoter has high strength and is most commonly used for driving gene expression in E. coli (Hawley & McClure, 1983). Indeed, the discovery, engineering and characterization of effective promoters have been the focus of metabolic engineering research for overproducing NPs in different microbes, which has been well-reviewed elsewhere (Blazeck & Alper, 2013; Jin et al., 2019; Tang et al., 2020).
Fig. 4.
(A) An overview of four major approaches for transcriptional of BGC expression, including the use of different promoters and terminators, sigma factor engineering and CRISPR interference with a deactivated Cas9 (dCas9). P: promoter; T: terminator; RNAP: RNA polymerase. (B) Selected examples that tuned the expression of cyanobacterial BGCs using promoters or terminators in E. coli or cyanobacterial strains. Pn: native promoter. Dashed arrows and lines indicate additional biosynthetic genes of the BGCs.
Cyanobacterial promoters can be classified into three major types. Type I promoters contain both −10 consensus element (5′-TATAAT-3′) and −35 element (5′-TTGACA-3′), while those of type II have only the −10 element. Type III promoters do not contain both consensus regions and are regulated by type III sigma (σ) factors in response to various stresses and stimuli. The σ factor is one critical component of the RNA polymerase (RNAP) holoenzyme and confers promoter selectivity. In E. coli, there are seven σ factors. Among them, the σ70 primarily controls the transcription of housekeeping genes, while the σ54 regulates nitrogen metabolism. Cyanobacteria have a conserved σ70 homolog, SigA, but no σ54, highlighting that the transcription of cyanobacterial genes in native hosts is controlled in different ways than E. coli (Srivastava et al., 2020). Furthermore, the cyanobacterial RNAP consists of α2ββ′γ subunits, different with most eubacterial counterparts that include α2ββ′ subunits (Kansara & Sukhodolets, 2011; Srivastava et al., 2020). The differences in core transcription machinery, as well as ancillary factors (e.g., the Crp binding sites), suggest the use of promoters compatible with selected hosts for the heterologous expression of cyanobacterial BGCs (Ebright & Busby, 1995; Gordon & Pfleger, 2018) (Fig. 4A, Table 1). For example, the production of microcystin-LR and [d-Asp3] microcystin-LR in E. coli GB05-MtaA was achieved by replacing the native promoter of the microcystin BGC from M. aeruginosa PCC 7806 with the T7 promoter (Liu et al., 2017, 2019), while the patellamide BGC was refactored by controlling the expression of each gene under the T7 promoter in E. coli Rosetta (DE3) (Donia et al., 2006) (Fig. 4B). Furthermore, the tetracycline-inducible promoter PtetO has been used to control the expression of the microcystin and lyngbyatoxin BGCs in E. coli (Liu et al., 2017; Ongley et al., 2013) (Fig. 4B). On the other hand, the Philmus group expressed all 12 sigma factors of Anabaena sp. PCC 7120 in E. coli and observed that four of them elevated the transcriptional rate from cyanobacterial promoters (Wells et al., 2018), suggesting that sigma factor engineering could be a strategy for tuning the heterologous expression of cyanobacterial BGCs in a heterologous host (Fig. 4A).
In cyanobacterial hosts, the transcriptional control of BGC expression can be achieved using constitutive and inducible promoters. Some notable inducible promoters include the high-light responsive psbA promoter, the nitrate/nitrite-inducible nirA promoter, the copper-regulated petE promoter, and the nickel-responsive nrsA promoter (Ma et al., 2014). PrnpB and Pcpc560 are two strong constitutive promoters in Synechocystis sp. PCC 6803 (Englund et al., 2016; Wang et al., 2018), while the Philmus group characterized the transcriptional performance of multiple constitutive promoters of cyanobacterial BGCs in Anabaena sp. PCC 7120 (Videau et al., 2016). In addition, diverse heterologous promoters have been tested for controlling gene expression in cyanobacteria. The cyanobacterial RNAP does not recognize the T7 promoter (Temme et al., 2012), making many widely used promoters in E. coli such as λPL, λPR, and PLac incompatible with cyanobacterial hosts (Huang and Lindblad, 2013). On the other hand, the IPTG inducible promoters (e.g., Ptrc, PLlacO1, PconII, PJ23101, and PJ23119) supported gene expression in cyanobacteria (Huang and Lindblad, 2013). However, one known major drawback of these promoters is their leaky expression (Geerts et al., 1995), making it important to characterize their performance in each cyanobacterial host. Nonetheless, these known native and heterologous promoters provide opportunities to tune the BGC expression in cyanobacteria (Fig. 4B). For example, three promoters with varied strengths, including PrnpB, Pcpc560 and the synthetic promoter Ptrc, were utilized for controlling the expression of the shinorine BGC in Synechocystis sp. PCC 6803 (Yang et al., 2018). Similarly, the constitutive promoter glnA (PglnA) and inducible promoters PpetE and PnirA have been used for the heterologous production of lyngbyatoxin A in Anabaena sp. PCC 7120 (Videau et al., 2016).
In addition to the use of different promoters, transcriptional tuning can be achieved with transcriptional terminators. The use of high-capacity terminators has been shown to increase the flux in a heterologous metabolic pathway (Curran et al., 2013). In bacteria, the transcriptional termination is rho-dependent or -independent, and cyanobacteria utilize the latter (Nudler & Gottesman, 2002). Two different rho-independent terminators including the early transcription terminator of bacteriophage T7 (Brahamsha & Haselkorn, 1991) and the terminator of rrnB gene (Geerts et al., 1995) have been utilized individually or in combination to control gene transcription in cyanobacteria (Huang et al., 2010). For example, the combination of promoter refactoring and the E. coli rrnB T1 terminator was utilized for the heterologous production of hapalindoles in Synechococcus elongatus UTEX 2973 (Knoot et al., 2019) (Fig. 4B). On the other hand, the evaluation of 12 promoters, 20 RBSs, and 8 terminators in the endogenous plasmids of Synechocystis sp. PCC 6803 revealed a ∼8,000-fold induction range. Importantly, significant incompatibility was observed between promoters and noncognate RBSs (Liu & Pakrasi, 2018). These results demonstrated many future opportunities to optimize the metabolic flux and compound production by using different promoter-RBS-terminator sets.
Recently, CRISPR-Cas9 based approaches have been utilized to engineer diverse organisms at multiple levels. For example, the use of a catalytically inactive Cas9 (dCas9), which lacks endonucleolytic activity, can effectively suppress the transcription of any gene of interest when coexpressed with a target-specific sgRNA, named CRISPR interference (CRISPRi) (Qi et al., 2013) (Fig. 4A). When targeting the transcriptional activators/repressors, this new technique was further named as CRISPR activation (CRISPRa) or CRISPR repression (CRISPRr) (Tian et al., 2017). The CRISPRi approach has been used for dynamically regulating the metabolic flux in Synechococcus sp. PCC 7002 (Gordon et al., 2016) and partial repression of up to six genes in the acyl-ACP-consuming pathway in Synechocystis sp. PCC 6803 (Kaczmarzyk et al., 2018). We expect that these approaches can also be useful for tuning the heterologous expression of cyanobacterial BGCs.
Translational Tuning of BGC Expression
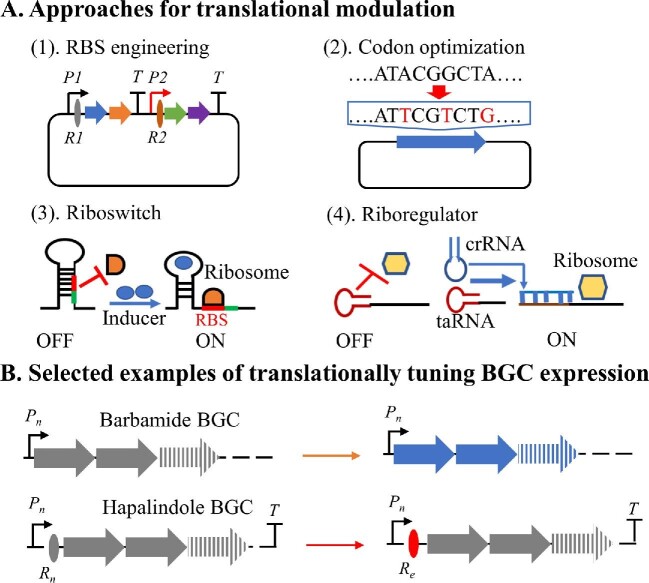
The rate of protein production from an mRNA transcript also depends on the strength of an RBS in recruiting ribosomes for translation (Fig. 5). The position and sequence of a given RBS significantly influence translational efficiency. The use of native and engineered RBSs for the control of BGC expression has been demonstrated in many organisms (e.g., E. coli) (Wang et al., 2012; Jeschek et al., 2017). In addition, several thermodynamic models have been developed to predict the translational efficiency of native and designed RBSs in diverse microorganisms (Espah Borujeni et al., 2014), such as the RBS calculator (Salis et al., 2009), the RBS designer (Na & Lee, 2010) and the UTR designer (Seo et al., 2015). Similarly, the RBS element can influence protein expression in cyanobacteria. For example, the use of an altered RBS for tuning the expression of limonene synthase led to a 13-fold production increase in Synechococcus elongatus PCC 7942 (Wang, Liu et al., 2016). Several RBS sequences from the BioBrick Registry of standard biological parts and 20 native cyanobacterial RBS elements have been characterized to control the translation in Synechocystis sp. PCC 6803 (Englund et al., 2016; Liu & Pakrasi, 2018). In addition, the application of designed RBS sequences using the RBS calculator enhanced the production of bisabolene in Synechocystis sp. PCC 6803 (Sebesta & Peebles, 2020), indicating the promise of this approach. However, in cyanobacteria, the RBS engineering for translational tuning remains less developed in comparison with other model organisms.
Fig. 5.
(A) An overview of major approaches for translational modulation of cyanobacterial BGC expression, including RBS engineering, codon optimization, and the use of riboswitch and riboregulators. crRNA: cis-repressing RNA; taRNA: trans-activating RNA; R: RBS. (B) Selected examples that tuned the expression of cyanobacterial BGCs by codon optimization and the use of engineered RBSs. Rn: native RBS; Re: engineered RBS.
In addition to the RBS engineering, the tuning of protein expression can further be achieved at the post-transcriptional level by RNA-based approaches (Fig. 5) such as riboswitch (Ma et al., 2014) and riboregulators (Sakamoto et al., 2018). Riboswitches allow the control of gene expression by forming secondary structures within an mRNA transcript. A riboswitch is composed of an aptamer sequence that imposes a secondary structure on the targeted mRNA, leading to the transcriptional activation or repression. The use of a modified theophylline-dependent synthetic riboswitch was first reported in S. elongatus PCC 7942, allowing a strict regulation of protein production (Nakahira et al., 2013), and later used in diverse cyanobacterial strains such as Synechocystis, Leptolyngbya, and Nostoc (Ma et al., 2014; Ohbayashi et al., 2016). The riboregulator system relies on the interactions of cis‐repressing (crRNA) and trans‐activating RNA (taRNA). When the taRNA is not expressed, the transcribed crRNA forms a loop structure at the 5′‐UTR of the targeted mRNA, preventing the binding of ribosomes for translation. On the other hand, when transcribed, the taRNA binds the crRNA to expose the RBS for translation (Isaacs et al., 2004). The use of riboregulators for controlling translation has been observed in Synechocystis sp. PCC 6803 (Abe et al., 2014; Sakai et al., 2015; Ueno et al., 2017). However, the practical applications of these two RNA based approaches for controlling the expression of cyanobacterial BGCs have not been reported yet.
Due to the different abundance of tRNAs in various hosts, each organism has its codon preference. Thus, codon optimization of biosynthetic genes proves to be a good strategy to improve the heterologous expression (Fig. 5). Indeed, the entire barbamide BGC was codon optimized and synthesized for heterologous expression in S. venezuelae (Kim et al., 2012). On the other hand, for optimal production of teleocidin and pendolmycin in the cyanobacterium host Anabaena sp. PCC 7120, the tleC and tleD from Streptomyces blastmyceticus NBRC 12747, and the mpnD from M. thermotolerans SCSIO0065 were also codon optimized and synthesized (Videau et al., 2020).
Conclusion and Future Perspectives
Cyanobacteria are a prolific resource of NPs with varied structural and bioactivity properties, which have drawn great attention in the isolation and characterization of new compounds. In particular, culture-independent approaches have uncovered the tremendous biosynthetic potential of cyanobacterial genomes for discovery. These approaches are supported by the increasing availability of the genome sequences of several hundred cyanobacteria. In addition, Kazusa DNA Research Institute (Japan) (http://www.kazusa.or.jp/) and DOE Joint Genome Institute (USA) (http://www.jgi.doe.gov/) have led efforts to sequence more cyanobacteria. Furthermore, advanced bioinformatics tools can accurately predict BGCs from cyanobacterial genome sequences. The information on the structure and functions of NPs from microbial sources including cyanobacteria can be accessed from different platforms such as Natural Product Atlas (Van Santen et al., 2019), NPASS (Zeng et al., 2018), and others. Furthermore, the information of secondary metabolites from cyanobacteria has recently been compiled and curated into one database, CyanoMetDB (Jones et al., 2020). In addition, the development of new analytic tools and databases, for example, GNPS (Wang, Carver et al., 2016) and MASST (Wang et al., 2020) for precise analysis and interpretation of molecular mass and automated NMR data analysis (Howarth et al., 2020), has eased structural predictions. These efforts have set the stage for the discovery of new cyanobacterial NPs, particularly using the bottom-up approaches. However, the biosynthetic potential of cyanobacteria has not been translated proportionally into chemical entities yet as the majority of cyanobacterial BGCs are cryptic or expressed at an extremely low level. Heterologous expression of cyanobacterial BGCs is becoming a viable solution to this critical problem in the discovery of new NPs. Diverse nonphotosynthetic hosts such as E. coli, yeast, and Streptomyces species have demonstrated successes in producing cyanobacterial NPs. In addition, there are growing interests in developing cyanobacterial platforms for heterologous production. When developed, these heterologous hosts can be suitable platforms for structural diversification by precursor-directed biosynthesis, mutasynthesis and combinatorial biosynthesis (Cummings et al., 2019; Dhakal & Sohng, 2017; Yan et al., 2018), expanding the applications of discovered cyanobacterial NPs. Moreover, many synthetic biology approaches can facilitate the development of multivariate combinational setups to further expand structural diversity (Tsukada et al., 2020). We anticipate that the above approaches will lead to the discovery of significantly more new cyanobacterial NPs and analogs for broad applications in the coming years.
Acknowledgement
We thank the current and past members of the Ding and Luesch groups for critical discussions on this topic.
Contributor Information
Dipesh Dhakal, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 31610, USA.
Manyun Chen, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 31610, USA.
Hendrik Luesch, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 31610, USA.
Yousong Ding, Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 31610, USA.
Funding
This work was supported by the cyanobacterial research from National Institutes of Health (NIH) R01CA172310 (H.L.), Debbie and Sylvia DeSantis Chair professorship (H.L.), a start-up fund from the University of Florida (Y.D.), and NIH R35GM128742 (Y.D.).
Conflict of Interest
The authors declare no conflict of interest.
References
- Abe K., Miyake K., Nakamura M., Kojima K., Ferri S., Ikebukuro K., Sode K. (2014). Engineering of a green-light inducible gene expression system in Synechocystis sp. PCC 6803. Microbial Biotechnology, 7, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H., Gao B., Rezaei M. A., Kwan J. C., Li C., Ye T., Ye T, Paul V. J., Luesch H. (2018). Discovery, synthesis, pharmacological profiling, and biological characterization of brintonamides A–E, novel dual protease and GPCR modulators from a marine cyanobacterium. Journal of Medicinal Chemistry, 61, 6364–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Awadhi F. H., Salvador-Reyes L. A., Elsadek L. A., Ratnayake R., Chen Q. Y., Luesch H. (2020). Largazole is a brain-penetrant class I HDAC inhibitor with extended applicability to glioblastoma and CNS Diseases. ACS Chemical Neuroscience, 11, 1937–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarano L., Esposito R., Ruocco N., Costantini M. (2020). Genome mining as new challenge in natural products discovery. Marine Drugs, 18, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti F., Corre C. (2019). Editing streptomycete genomes in the CRISPR/Cas9 age. Natural Product Reports, 36, 1237–1248. [DOI] [PubMed] [Google Scholar]
- Becher P. G., Jüttner F. (2005). Insecticidal compounds of the biofilm-forming cyanobacterium Fischerella sp. (ATCC 43239). Environmental Toxicology, 20, 363–372. [DOI] [PubMed] [Google Scholar]
- Beld J., Sonnenschein E. C., Vickery C. R., Noel J. P., Burkart M. D. (2014). The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Natural Product Reports, 31, 61–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berla B. M., Saha R., Immethun C. M., Maranas C. D., Moon T. S., Pakrasi H. (2013). Synthetic biology of cyanobacteria: Unique challenges and opportunities. Frontiers in Microbiology, 4, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishé B., Taton A., Golden J. W. (2019). Modification of RSF1010-based broad-host-range plasmids for improved conjugation and cyanobacterial bioprospecting. Iscience, 20, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck J., Alper H. S. (2013). Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnology Journal, 8, 46–58. [DOI] [PubMed] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., Medema M. H., Weber T. (2019). antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research, 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C. M., Tang Y. (2019) Engineering Saccharomyces cerevisiae for production of simvastatin. Metabolic Engineering, 51, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahamsha B., Haselkorn R. (1991). Isolation and characterization of the gene encoding the principal sigma factor of the vegetative cell RNA polymerase from the cyanobacterium Anabaena sp. strain PCC 7120. Journal of Bacteriology, 173 (8), 2442–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilisauer K., Rapp J., Rath P., Schöllhorn A., Bleul L., Weiß E., Stahl M., Grond S., Forchhammer K. (2019). Cyanobacterial antimetabolite 7-deoxy-sedoheptulose blocks the shikimate pathway to inhibit the growth of prototrophic organisms. Nature Communications, 10, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q. T., Yu P., Wang J., Li Z. Y., Chen X. A., Mao X. M., Li Y. Q. (2019). Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches. Microbial Cell Factories, 18, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Chen Q. Y., Dang L. H., Luesch H. (2017). Apratoxin S10, a dual inhibitor of angiogenesis and cancer cell growth to treat highly vascularized tumors. ACS Medicinal Chemistry Letters, 8, 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael W. W. (1992). Cyanobacteria secondary metabolites—the cyanotoxins. Journal of Applied Bacteriology, 72, 445–459. [DOI] [PubMed] [Google Scholar]
- Choi H., Mascuch S. J., Villa F. A., Byrum T., Teasdale M. E., Smith J. E., Preskitt L. B., Rowley D. C., Gerwick L., Gerwick W. H. (2012). Honaucins A−C, potent inhibitors of inflammation and bacterial quorum sensing: Synthetic derivatives and structure-activity relationships. Chemistry & Biology, 19, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. F., Meeks J. C., Cai Y. A., Wolk C. P. (1998). Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods in Enzymology, 297, 3–17. [Google Scholar]
- Cummings M., Peters A. D., Whitehead G. F., Menon B. R., Micklefield J., Webb S. J., Takano E. (2019). Assembling a plug-and-play production line for combinatorial biosynthesis of aromatic polyketides in Escherichia coli. PLoS Biology, 7, e3000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran K. A., Karim A. S., Gupta A., Alper H. S. (2013). Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metabolic Engineering, 19, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino P. M., Gulder T. A. (2018). Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression. ACS Synthetic Biology, 7, 1702–1708. [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L., (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demay J., Bernard C., Reinhardt A., Marie B. (2019). Natural products from cyanobacteria: Focus on beneficial activities. Marine Drugs, 17, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal D., Sohng J. K. (2017). Coalition of biology and chemistry for ameliorating antimicrobial drug discovery. Frontiers in Microbiology, 8, 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal D., Sohng J. K., Pandey R. P. (2019). Engineering actinomycetes for biosynthesis of macrolactone polyketides. Microbial Cell Factories, 18, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didovyk A., Borek B., Tsimring L., Hasty J. (2016). Transcriptional regulation with CRISPR-Cas9: Principles, advances, and applications. Current Opinion in Biotechnology, 40, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M. Z., Yan H. F., Li L. F., Zhai F., Shang L. Q., Yin Z., Yuan Y. J. (2014). Biosynthesis of taxadiene in Saccharomyces cerevisiae: Selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS One, 9, e109348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann E., Gugger M., Sivonen K., Fewer D. P. (2015). Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends in Microbiology, 23, 642–652. [DOI] [PubMed] [Google Scholar]
- Donia M. S., Hathaway B. J., Sudek S., Haygood M. G., Rosovitz M. J., Ravel J., Schmidt E. W. (2006). Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nature Chemical Biology, 2, 729–735. [DOI] [PubMed] [Google Scholar]
- Donia M. S., Ravel J., Schmidt E. W. (2008). A global assembly line for cyanobactins. Nature Chemical Biology, 4, 341–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Busby S. (1995). The Escherichia coli RNA polymerase α subunit: Structure and function. Current Opinion in Genetics & Development, 5, 197–203. [DOI] [PubMed] [Google Scholar]
- Englund E., Liang F., Lindberg P. (2016). Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Scientific Reports, 6, 36640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espah Borujeni, Channarasappa A., A. S., Salis H. M. (2014). Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Research, 42, 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewer D. P., Österholm J., Rouhiainen L., Jokela J., Wahlsten M., Sivonen K. (2011). Nostophycin biosynthesis is directed by a hybrid polyketide synthase-nonribosomal peptide synthetase in the toxic cyanobacterium Nostoc sp. strain 152. Applied and Environmental Microbiology, 77, 8034–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann E.I., Kibler A.P. (1980). Nitrogen economy of endolithic microbial communities in hot and cold deserts. Microbial Ecology, 6, 95–108. [DOI] [PubMed] [Google Scholar]
- Frigaard N. U., Sakuragi Y., Bryant D. A. (2004). Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods in Molecular Biology, 274, 325–340. [DOI] [PubMed] [Google Scholar]
- Geerts D., Bovy A., de Vrieze G., Borrias M., Weisbeek P. (1995). Inducible expression of heterologous genes targeted to a chromosomal platform in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology, 141, 831–841. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Bibb M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microbial Biotechnology, 4, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G. C., Korosh T. C., Cameron J. C., Markley A. L., Begemann M. B., Pfleger B. F. (2016). CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metabolic Engineering, 38, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G. C., Pfleger B. F. (2018). Regulatory tools for controlling gene expression in cyanobacteria. Advances in Experimental Medicine and Biology, 1080, 281–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greunke C., Duell E. R., D'Agostino P. M., Glöckle A., Lamm K., Gulder T. A. M. (2018). Direct pathway cloning (DiPaC) to unlock natural product biosynthetic potential. Metabolic Engineering, 47, 334–345. [DOI] [PubMed] [Google Scholar]
- Harvey C. J., Puglisi J. D., Pande V. S., Cane D. E., Khosla C. (2012). Precursor directed biosynthesis of an orthogonally functional erythromycin analogue: Selectivity in the ribosome macrolide binding pocket. Journal of the American Chemical Society, 134, 12259–12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. (1983). Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Research, 11, 2237–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock A., Hunter C. N., Canniffe D. P. (2020). Progress and challenges in engineering cyanobacteria as chassis for light-driven Microbial Biotechnology, 13, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth A., Ermanis K., Goodman J. M. (2020). DP4-AI automated NMR data analysis: Straight from spectrometer to structure. Chemical Science, 11, 4351–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. H., Camsund D., Lindblad P., Heidorn T. (2010). Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Research, 38, 2577–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. H., Lindblad P. (2013). Wide-dynamic-range promoters engineered for cyanobacteria. Journal of Biological Engineering, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. A., Mitchell D. A. (2018). RiPP antibiotics: Biosynthesis and engineering potential. Current Opinion in Microbiology, 45, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L., Hug J. J., Fu C., Bian X., Zhang Y., Müller R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Natural Product Reports, 36, 1412–1436. [DOI] [PubMed] [Google Scholar]
- Isaacs F. J., Dwyer D. J., Ding C., Pervouchine D. D., Cantor C. R., Collins J. J. (2004). Engineered riboregulators enable post-transcriptional control of gene expression. Nature Biotechnology, 22, 841–847. [DOI] [PubMed] [Google Scholar]
- Jakočiūnas T., Jensen M. K., Keasling J. D. (2016). CRISPR/Cas9 advances engineering of microbial cell factories. Metabolic Engineering, 34, 44–59. [DOI] [PubMed] [Google Scholar]
- Jeschek M., Gerngross D., Panke S. (2017). Combinatorial pathway optimization for streamlined metabolic engineering. Current Opinion in Biotechnology, 47, 142–151. [DOI] [PubMed] [Google Scholar]
- Jin H., Wang Y., Idoine A., Bhaya D. (2018). Construction of a shuttle vector using an endogenous plasmid from the cyanobacterium Synechocystis sp. PCC6803. Frontiers Microbiology, 9, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. Q., Jin W. R., Ma Z. C., Shen Q., Cai X., Liu Z. Q., Zheng Y. G. (2019). Promoter engineering strategies for the overproduction of valuable metabolites in microbes. Applied Microbiology and Biotechnology, 103 (21–22), 8725–8736. [DOI] [PubMed] [Google Scholar]
- Jones M. R., Pinto E., Torres M. A., Doerr F., Mazur-Marzec H., Szubert K., Tartaglione L., Dell'Aversano C., Miles C. O., Beach D. G., McCarron P. (2020). Comprehensive database of secondary metabolites from cyanobacteria. bioRxiv. 10.1101/2020.04.16.038703. [DOI] [PubMed]
- Kaczmarzyk D., Cengic I., Yao L., Hudson E. P. (2018). Diversion of the long-chain acyl-ACP pool in Synechocystis to fatty alcohols through CRISPRi repression of the essential phosphate acyltransferase PlsX. Metabolic Engineering, 45, 59–66. [DOI] [PubMed] [Google Scholar]
- Kansara S. G., Sukhodolets M. V. (2011). Oligomerization of the E. coli core RNA polymerase: Formation of (α 2 ββ′ω) 2-DNA complexes and regulation of the oligomerization by auxiliary subunits. PLoS One, 6, e18990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoch M., Mazmouz R., Chau R., Pearson L. A., Pickford R., Neilan B. A. (2016). Heterologous production of cyanobacterial mycosporine-like amino acids mycosporine-ornithine and mycosporine-lysine in Escherichia coli. Applied and Environmental Microbiology, 82, 6167–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Kumar A., Kaur. J. (2018). Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. International Journal of Biological Macromolecules, 106, 803–822. [DOI] [PubMed] [Google Scholar]
- Kautsar S. A., Blin K., Shaw S., Navarro-Muñoz J. C., Terlouw B. R., van der Hooft J. J., Van Santen J. A., Tracanna V., Suarez Duran H. G., Pascal Andreu V., Selem-Mojica N. (2020). MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Research, 48, D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Lee J. H., Choi H., Pereira A. R., Ban Y. H., Yoo Y. J., Kim E., Park J.W., Sherman D. H., Gerwick W. H., Yoon Y. J. (2012). Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Organic Letters, 14 (23), 5824–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. J., Lee S. M., Um Y., Sim S. J., Woo H. M. (2017). Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Frontiers in Plant Science, 8, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoot C. J., Khatri Y., Hohlman R. M., Sherman D. H., Pakrasi H. B. (2019). Engineered production of hapalindole alkaloids in the cyanobacterium Synechococcus sp. UTEX 2973. ACS Synthetic Biology, 8, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Uchiyama T., Ōmura S., Cane D. E., Ikeda H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proceedings of the National Academy of Sciences of the USA, 107, 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Larionov V. (2016). Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma, 125, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnbaum N. L., Britz-McKibbin P. (2013) New advances in separation science for metabolomics: Resolving chemical diversity in a post-genomic era. Chemical Reviews, 113, 2437–2468. [DOI] [PubMed] [Google Scholar]
- Kung S. H., Lund S., Murarka A., McPhee D., Paddon C. J. (2018). Approaches and recent developments for the commercial production of semi-synthetic artemisinin. Frontiers in Plant Science, 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot R. H., Gehring A. M., Flugel R. S., Zuber P., LaCelle M., Marahiel M. A., Reid R., Khosla C., Walsh C. T. (1996). A new enzyme superfamily-the phosphopantetheinyl transferases. Chemistry & Biology, 3, 923–936. [DOI] [PubMed] [Google Scholar]
- Leão P. N., Pereira A. R., Liu W. T., Ng J., Pevzner P. A., Dorrestein P. C., König G. M., Vasconcelos V. M., Gerwick W. H. (2010). Synergistic allelochemicals from a freshwater cyanobacterium. Proceedings of the National Academy of Sciences of the USA, 107, 11183–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao T., Castelão G., Korobeynikov A., Monroe E. A., Podell S., Glukhov E., Allen E. E., Gerwick W. H., Gerwick L. (2017). Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proceedings of the National Academy of Sciences of the USA, 114, 3198–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. U., Kwak H. B., Pi S. H., You H. K., Byeon S. R., Ying Y., Luesch H., Hong J., Kim S. H. (2011). In vitro and in vivo osteogenic activity of largazole. ACS Medicinal Chemistry Letters, 2, 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikoski N., Fewer D. P., Jokela J., Wahlsten M., Rouhiainen L., Sivonen K. (2010). Highly diverse cyanobactins in strains of the genus Anabaena. Applied and Environmental Microbiology, 76, 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Luo D., Luesch H. (2019). Advances in exploring the therapeutic potential of marine natural products. Pharmacological Research, 147, 104373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. C., Pakrasi H. B. (2019). Engineering cyanobacteria for production of terpenoids. Planta, 249, 145–154. [DOI] [PubMed] [Google Scholar]
- Linington R. G., Clark B. R., Trimble E. E., Almanza A., Urena L. D., Kyle D. E., Gerwick W. H. (2009). Antimalarial peptides from marine cyanobacteria: Isolation and structural elucidation of gallinamide A. Journal of Natural Products, 72, 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Pakrasi H. B. (2018). Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microbial Cell Factories, 17, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Mazmouz R., Ongley S. E., Chau R., Pickford R., Woodhouse J. N., Neilan B. A. (2017). Directing the heterologous production of specific cyanobacterial toxin variants. ACS Chemical Biology, 12, 2021–2029. [DOI] [PubMed] [Google Scholar]
- Liu T., Mazmouz R., Pearson L. A., Neilan B. A. (2019). Mutagenesis of the microcystin tailoring and transport proteins in a heterologous cyanotoxin expression system. ACS Synthetic Biology, 8, 1187–1194. [DOI] [PubMed] [Google Scholar]
- Long P. F., Dunlap W. C., Battershill C. N., Jaspars M. (2005). Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem, 6, 1760–1765. [DOI] [PubMed] [Google Scholar]
- Ma A. T., Schmidt C. M., Golden J. W. (2014). Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Applied and Environmental Microbiology, 80, 6704–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado I. M., Atsumi S. (2012). Cyanobacterial biofuel production. Journal of Biotechnology, 162, 50–56. [DOI] [PubMed] [Google Scholar]
- Malla S., Sommer M. O. (2014). A sustainable route to produce the scytonemin precursor using Escherichia coli. Green Chemistry, 16, 3255–3265. [Google Scholar]
- Mason C. P., Edwards K. R., Carlson R. E., Pignatello J., Gleason F. K., Wood J. M. (1982). Isolation of chlorine-containing antibiotic from the freshwater cyanobacterium Scytonema hofmanni. Science, 215, 400–402. [DOI] [PubMed] [Google Scholar]
- Mazard S., Penesyan A., Ostrowski M., Paulsen I. T., Egan S. (2016). Tiny microbes with a big impact: The role of cyanobacteria and their metabolites in shaping our future. Marine Drugs, 14, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Dhakal D., Han J. M., Lim H. N., Jung H. J., Yamaguchi T., Sohng J. K. (2019). Production of a novel tetrahydroxynaphthalene (THN) derivative from Nocardia sp. CS682 by metabolic engineering and its bioactivities. Molecules (Basel, Switzerland), 24, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaser R., Paul V. J., Luesch H. (2013). Modular strategies for structure and function employed by marine cyanobacteria: Characterization and synthesis of pitinoic acids. Organic Letters, 15, 4050–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougiakos I., Bosma E. F., Ganguly J., van der Oost J., van Kranenburg R. (2018). Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: Advances and prospects. Current Opinion in Biotechnology, 50, 146–157. [DOI] [PubMed] [Google Scholar]
- Myronovskyi M., Luzhetskyy A. (2019). Heterologous production of small molecules in the optimized Streptomyces hosts. Natural Product Reports, 36, 1281–1294. [DOI] [PubMed] [Google Scholar]
- Myronovskyi M., Rosenkränzer B., Nadmid S., Pujic P., Normand P., Luzhetskyy A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metabolic Engineering, 49, 316–324. [DOI] [PubMed] [Google Scholar]
- Na D., Lee D. (2010). RBSDesigner: Software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics, 26, 2633–2634. [DOI] [PubMed] [Google Scholar]
- Nakahira Y., Ogawa A., Asano H., Oyama T., Tozawa Y. (2013). Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in cyanobacterium Synechococcus elongatus PCC 7942. Plant & Cell Physiology, 54, 1724–1735. [DOI] [PubMed] [Google Scholar]
- Navarro-Muñoz J. C., Selem-Mojica N., Mullowney M. W., Kautsar S. A., Tryon J. H., Parkinson E. I., De Los Santos E. L., Yeong M., Cruz-Morales P., Abubucker S., Roeters A., Lokhorst W., Fernandez-Guerra A., Dias Cappelini L. T., Goering A. W., Thomson R. J., Metcalf W. W., Kelleher N. L., Barona-Gomez F., Medema M. H. (2020) A computational framework to explore large-scale biosynthetic diversity. Nature Chemical Biology, 16, 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. H., Berla B. M., Pakrasi H. B. (2015). Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. PCC 6803. Applied and Environmental Microbiology, 81, 6857––6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. T., Dhakal D., Pham V. T. T., Nguyen H. T., Sohng J. K. (2020). Recent advances in strategies for activation and discovery/characterization of cryptic biosynthetic gene clusters in Streptomyces. Microorganisms, 8, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T. C., Lin G. M., Xie L. R., Wang Z. Q., Xing W. Y., Zhang J. Y., Zhang C. C. (2019). Expanding the potential of CRISPR-Cpf1 based genome editing technology in the cyanobacterium Anabaena PCC 7120. ACS Synthetic Biology, 8, 170–180. [DOI] [PubMed] [Google Scholar]
- Nudler E., Gottesman M. E. (2002). Transcription termination and anti-termination in E. coli. Genes to Cells, 7, 755–768. [DOI] [PubMed] [Google Scholar]
- Ohbayashi R., Akai H., Yoshikawa H., Hess W. R., Watanabe S. (2016). A tightly inducible riboswitch system in Synechocystis sp. PCC 6803. Journal of General and Applied Microbiology, 62, 154–159. [DOI] [PubMed] [Google Scholar]
- Oliver N. J., Rabinovitch-Deere C. A., Carroll A. L., Nozzi N. E., Case A. E., Atsumi S. (2016). Cyanobacterial metabolic engineering for biofuel and chemical production. Current Opinion in Chemical Biology, 35, 43–50. [DOI] [PubMed] [Google Scholar]
- Onai K., Morishita M., Kaneko T., Tabata S., Ishiura M. (2004). Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: A simple and efficient method for gene transfer. Molecular Genetics and Genomics, 271, 50–59. [DOI] [PubMed] [Google Scholar]
- Ongley S. E., Bian X., Zhang Y., Chau R., Gerwick W. H., Müller R., Neilan B. A. (2013). High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chemical Biology, 8, 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega M. A., van der Donk W. A. (2016). New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chemical Biology, 23, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H., Lee K., Jang J. W., Hahn J. S. (2019). Metabolic engineering of Saccharomyces cerevisiae for production of shinorine, a sunscreen material, from xylose. ACS Synthetic Biology, 8, 346–357. [DOI] [PubMed] [Google Scholar]
- Pereira A. R., Kale A. J., Fenley A. T., Byrum T., Debonsi H. M., Gilson M. K., Valeriote F. A., Moore B. S., Gerwick W. H. (2012). The carmaphycins: New proteasome inhibitors exhibiting an α, β-epoxyketone warhead from a marine cyanobacterium. Chembiochem, 13, 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B. A., Admiraal S. J., Gramajo H., Cane D. E., Khosla C. (2001). Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science, 291, 1790–1792. [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., Lim W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy A. V., Sorrels C. M., Gerwick W. H. (2007). Cloning and biochemical characterization of the hectochlorin biosynthetic gene cluster from the marine cyanobacterium Lyngbya majuscula. Journal of Natural Products, 70, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Rosano G. L., Ceccarelli E. A. (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Frontiers in Microbiology, 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulet J., Taton A., Golden J. W., Arabolaza A., Burkart M. D., Gramajo H. (2018). Development of a cyanobacterial heterologous polyketide production platform. Metabolic Engineering, 49, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Abe K., Nakashima S., Ellinger J. J., Ferri S., Sode K., Ikebukuro K. (2015). Scaffold-fused riboregulators for enhanced gene activation in Synechocystis sp. PCC 6803. MicrobiologyOpen, 4, 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto I., Abe K., Kawai S., Tsukakoshi K., Sakai Y., Sode K., Ikebukuro K. (2018). Improving the induction fold of riboregulators for cyanobacteria. RNA Biology, 15, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H. M., Mirsky E. A., Voigt C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotechnology, 27, 946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Reyes L.A., Luesch H. (2015). Biological targets and mechanisms of action of natural products from marine cyanobacteria. Natural Product Reports, 32, 478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia L., Martín L., Mangues R., Ferrer-Miralles N., Vázquez E., Villaverde A. (2016). Recombinant pharmaceuticals from microbial cells: A 2015 update. Microbial Cell Factories, 15, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Merino M., Singh A. K., Ducat D. C. (2019). New applications of synthetic biology tools for cyanobacterial metabolic engineering. Frontiers Bioengineering and Biotechnology, 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Bagdasarian M. M., Scholz P., Lurz R., Rückert B., Bagdasarian M. (1984). Replication of the broad host range plasmid RSF1010: Requirement for three plasmid-encoded proteins. Proceedings of the National Academy of Sciences of the USA, 81, 654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. W., Nelson J. T., Rasko D. A., Sudek S., Eisen J. A., Haygood M. G., Ravel J. (2005). Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proceedings of the National Academy of Sciences of the USA, 102, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta J., Peebles C. A. (2020). Improving heterologous protein expression in Synechocystis sp. PCC 6803 for alpha-bisabolene production. Metabolic Engineering Communications, 10, e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S. W., Yang J. S., Cho H. S., Yang J., Kim S. C., Park J. M., Kim S., Jung G. Y. (2015). Predictive combinatorial design of mRNA translation initiation regions for systematic optimization of gene expression levels. Scientific Reports, 4, 4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J. L., Zhang Z. C., Yin X. Y., Chen M., Hao F. H., Wang K., Feng J. L., Xu H. F., Yin Y. C., Tang H. R., Qiu B. S. (2018). UV-B induced biosynthesis of a novel sunscreen compound in solar radiation and desiccation tolerant cyanobacteria. Environmental Microbiology, 20, 200–213. [DOI] [PubMed] [Google Scholar]
- Shi S., Liang Y., Zhang M. M., Ang E. L., Zhao H. (2016). A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metabolic Engineering, 33, 19–27. [DOI] [PubMed] [Google Scholar]
- Shishido T. K., Humisto A., Jokela J., Liu L., Wahlsten M., Tamrakar A., Fewer D. P., Permi P., Andreote A. P., Fiore M. F., Sivonen K. (2015). Antifungal compounds from cyanobacteria. Marine Drugs, 13, 2124–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S. A., Marahiel M. A. (2005). Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chemical Reviews, 105, 715–738. [DOI] [PubMed] [Google Scholar]
- Siewers V., Chen X., Huang L., Zhang J., Nielsen J. (2009). Heterologous production of non-ribosomal peptide LLD-ACV in Saccharomyces cerevisiae. Metabolic Engineering, 11, 391–397. [DOI] [PubMed] [Google Scholar]
- Skinnider M. A., Dejong C. A., Rees P. N., Johnston C. W., Li H., Webster A. L., Wyatt M. A., Magarvey N. A. (2015). Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Research, 43, 9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski M. J., Zhou H., Claesen J., Shen B., Fischbach M. A., Voigt C. A. (2016). Synthetic biology to access and expand nature's chemical diversity. Nature Reviews Microbiology, 14, 135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Tsai S. C. (2007). The type I fatty acid and polyketide synthases: A tale of two megasynthases. Natural Product Reports, 24, 1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Summers M. L., Sobotka R. (2020). Cyanobacterial sigma factors: Current and future applications for biotechnological advances. Biotechnology Advances, 40, 107517. [DOI] [PubMed] [Google Scholar]
- Stovicek V., Holkenbrink C., Borodina I. (2017). CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Research, 17 (5), fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L.T., Phyo M.Y. (2020). Marine cyanobacteria: A source of lead compounds and their clinically-relevant molecular targets. Molecules (Basel, Switzerland), 25, 2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wu Y., Deng J., Chen N., Zheng Z., Wei Y., Luo X., Keasling J. D. (2020). Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites, 10, E320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., van der Donk W. A. (2012). Structural characterization of four prochlorosins: A novel class of lantipeptides produced by planktonic marine cyanobacteria. Biochemistry, 51, 4271–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A., Ecker A., Diaz B., Moss N. A., Anderson B., Reher R., Reher R, Leão T. F., Simkovsky R., Dorrestein P. C., Gerwick L., Gerwick W. H. (2020). Heterologous expression of cryptomaldamide in a cyanobacterial host. ACS Synthetic Biology, 9, 3364–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A., Unglaub F., Wright N. E., Zeng W. Y., Paz-Yepes J., Brahamsha B., Palenik B., Peterson T. C., Haerizadeh F., Golden S. S., Golden J. W. (2014). Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Research, 42, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teece B.L., George S.C., Djokic T., Campbell K.A., Ruff S.W., Van Kranendonk M.J. (2020). Biomolecules from fossilized hot spring sinters: Implications for the search for life on Mars. Astrobiology, 20, 537–551. [DOI] [PubMed] [Google Scholar]
- Temme K., Hill R., Segall-Shapiro T. H., Moser F., Voigt C. A. (2012). Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Research, 40, 8773–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuan N. H., An T. T., Canh N. X., Shrestha A., Sohng J. K., Dhakal D. (2019). Recent advances in exploration and biotechnological production of bioactive compounds in three cyanobacterial genera: Nostoc, Lyngbya, and Microcystis. Frontiers in Chemistry, 7, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Wang J., Shen X., Rey J. F., Yuan Q., Yan Y. (2017). Fundamental CRISPR-Cas9 tools and current applications in microbial systems. Synthetic and Systems Biotechnology, 2, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz J. I., Schwalen C. J., Patel P. S., Maxson T., Blair P. M., Tai H. C., Zakai U. I., Mitchell D. A. (2017). A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nature Chemical Biology, 13, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada K., Shinki S., Kaneko A., Murakami K., Irie K., Murai M., Miyoshi H., Dan S., Kawaji K., Hayashi H., Kodama E.N. (2020). Synthetic biology based construction of biological activity-related library of fungal decalin-containing diterpenoid pyrones. Nature Communications, 11, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K., Sakai Y., Shono C., Sakamoto I., Tsukakoshi K., Hihara Y., Sode K., Ikebukuro K. (2017). Applying a riboregulator as a new chromosomal gene regulation tool for higher glycogen production in Synechocystis sp. PCC 6803. Applied Microbiology and Biotechnology, 101, 8465–8474. [DOI] [PubMed] [Google Scholar]
- Ungerer J., Pakrasi H. B. (2016). Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Scientific Reports, 6, 39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Santen J. A., Jacob G., Singh A. L., Aniebok V., Balunas M. J., Bunsko D., Neto F. C., Castaño-Espriu L., Chang C., Clark T. N., Cleary Little J. L., (2019). The natural products atlas: An open access knowledge base for microbial natural products discovery. ACS Central Science, 5, 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan R., Gale G. A., Schiavon A. A., Puzorjov A., Malm J., Gillespie M. D., Vasudevan R, Gale GA, Schiavon AA, Puzorjov A, Malin J, Gillespie MD, Vavitsas K., Zulkower V., Wang B., Howe C. J., Lea-Smith D. (2019). CyanoGate: A Golden Gate modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. Plant Physiology, 180, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videau P., Wells K. N., Singh A. J., Eiting J., Proteau P. J., Philmus B. (2020). Expanding the natural products heterologous expression repertoire in the model cyanobacterium Anabaena sp. strain PCC 7120: Production of Pendolmycin and Teleocidin B-4. ACS Synthetic Biology, 9, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videau P., Wells K. N., Singh A. J., Gerwick W. H., Philmus B. (2016). Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: Production of lyngbyatoxin A. ACS Synthetic Biology, 5, 978–988. [DOI] [PubMed] [Google Scholar]
- Vogel A. I. M., Lale R., Hohmann-Marriott M. F. (2017). Streamlining recombination-mediated genetic engineering by validating three neutral integration sites in Synechococcus sp. PCC 7002. Journal of Biological Engineering, 11, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Eckert C., Maness P. C., Yu J. (2018). A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synthetic Biology, 7, 276–286. [DOI] [PubMed] [Google Scholar]
- Wang M., Carver J. J., Phelan V.V., Sanchez L. M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C. A., Luzzatto-Knaan T., Porto C. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology, 34, 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Jarmusch A. K., Vargas F., Aksenov A. A., Gauglitz J. M., Weldon K., Petras D., da Silva R., Quinn R., Melnik A.V., van der Hooft J. J. (2020). Mass spectrometry searches using MASST. Nature Biotechnology, 38, 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Ma X., Du G., Chen J. (2012). Overview of regulatory strategies and molecular elements in metabolic engineering of bacteria. Molecular Biotechnology, 52, 300–308. [DOI] [PubMed] [Google Scholar]
- Wang X., Liu W., Xin C., Zheng Y., Cheng Y., Sun S., Li R., Zhu X. G., Dai S. Y., Rentzepis P. M., Yuan J. S. (2016). Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proceedings of the National Academy of Sciences of the USA, 113, 14225–14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Hotta K., Praseuth A. P., Koketsu K., Migita A., Boddy C. N., Wang C. C., Oguri H., Oikawa H. (2006) Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nature Chemical Biology, 2, 423–428. [DOI] [PubMed] [Google Scholar]
- Weiz A. R., Ishida K., Makower K., Ziemert N., Hertweck C., Dittmann E. (2011). Leader peptide and a membrane protein scaffold guide the biosynthesis of the tricyclic peptide microviridin. Chemistry & Biology, 18, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Wells K. N., Videau P., Nelson D., Eiting J. E., Philmus B. (2018). The influence of sigma factors and ribosomal recognition elements on heterologous expression of cyanobacterial gene clusters in Escherichia coli. FEMS Microbiology Letters, 365, fny164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K. E., Ungerer J., Cobb R. E., Zhao H., Pakrasi H. B. (2016). CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microbial Cell Factories, 15, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchos J., Ascaso C., McKay C.P. (2006). Endolithic cyanobacteria in halite rocks from the hyperarid core of the Atacama Desert. Astrobiology, 6, 415–422. [DOI] [PubMed] [Google Scholar]
- Wijffels R. H., Kruse O., Hellingwerf K. J. (2013). Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Current Opinion in Biotechnology, 24, 405–413. [DOI] [PubMed] [Google Scholar]
- Wright A. V., Nuñez J. K., Doudna J. A. (2016). Biology and applications of CRISPR systems: Harnessing nature's toolbox for genome engineering. Cell, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Xiong W., Cano M., Wang B., Douchi D., Yu J. (2017). The plasticity of cyanobacterial carbon metabolism. Current Opinion in Chemical Biology, 41, 12–19. [DOI] [PubMed] [Google Scholar]
- Xu W., Klumbys E., Ang E. L., Zhao H. (2020). Emerging molecular biology tools and strategies for engineering natural product biosynthesis. Metabolic Engineering Communications, 10, e00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Burgard C., Popoff A., Zaburannyi N., Zipf G., Maier J., Bernauer H. S., Wenzel S. C., Müller R. (2018). Synthetic biology approaches and combinatorial biosynthesis towards heterologous lipopeptide production. Chemical Science, 9, 7510–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Cozad M. A., Holland D. A., Zhang Y., Luesch H., Ding Y. (2018). Photosynthetic production of sunscreen shinorine using an engineered cyanobacterium. ACS Synthetic Biology, 7, 664–671. [DOI] [PubMed] [Google Scholar]
- Yang G., Zhang Y., Lee N. K., Cozad M. A., Kearney S. E., Luesch H., Ding Y. (2017). Cyanobacterial Sfp-type phosphopantetheinyl transferases functionalize carrier proteins of diverse biosynthetic pathways. Scientific Reports, 7, 11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Zhang P., He W., Qin C., Chen S., Tao L., Wang Y., Tan Y., Gao D., Wang B., Chen Z. (2018). NPASS: Natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Research, 46, D1217–D1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. J., Tang X., Moore B. S. (2019). Genetic platforms for heterologous expression of microbial natural products. Natural Product Reports, 36 (9), 1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen M., Bruner S. D., Ding Y. (2018). Heterologous production of microbial ribosomally synthesized and post-translationally modified peptides. Frontiers in Microbiology, 9, 1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E (2008) Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie, International Edition, 47, 7756–7759. [DOI] [PubMed] [Google Scholar]
- Ziemert N., Ishida K., Weiz A., Hertweck C., Dittmann E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and Environmental Microbiology, 76, 3568–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]