ABSTRACT
Leishmania (Mundinia) martiniquensis is a kinetoplastid parasite that was first isolated in 1995 on Martinique. We report the first complete genome for Leishmania martiniquensis from Asia, isolate LSCM1, strain LV760, which was sequenced using combined short-read and long-read technologies. This will facilitate greater understanding of the evolution of the geographically dispersed subgenus Mundinia.
ANNOUNCEMENT
Parasites of the genus Leishmania infect a wide range of hosts, including humans. After malaria, leishmaniasis is the second most common cause of death due to parasitic infection (1), and it can be manifested in three main forms, i.e., visceral, cutaneous, and mucocutaneous. Around 100 countries have reported at least one of these three forms (2). The subgenus Mundinia (3) has been reported in dispersed locations worldwide, namely, Thailand (4), Martinique (5), Switzerland (6), Australia (7), and Ghana (8), but the only complete Mundinia genome, prior to the present work, is that of Leishmania (Mundinia) martiniquensis MAR LEM2494 (GenBank accession number GCA_000409445.2). We now report the complete genome of L. (M.) martiniquensis isolate Chiang Mai 1, strain LV760 (WHO code MHOM/TH/2012/LSCM1), which was originally obtained by bone marrow aspiration from a 52-year-old male patient from northern Thailand (9).
Genomic DNA isolation, cultivation, and extraction were performed using an in vitro culture system that had been developed previously for Leishmania (Mundinia) orientalis axenic amastigotes (10). Parasites were grown in Schneider's insect medium at 26°C as promastigotes and then in M199 medium supplemented with 10% fetal calf serum (FCS), 2% stable human urine, 1% basal medium Eagle vitamins, and 25 μg/ml gentamicin sulfate, with subpassage to fresh medium every 4 days to sustain parasite growth and viability. The extracted DNA concentration was assessed using a Qubit fluorometer, microplate reader, and agarose gel electrophoresis. All sequencing libraries were based on the same extracted DNA sample to avoid any inconsistency.
Short-read library construction and sequencing were contracted to BGI (Shenzhen, China) (DNBseq libraries producing paired-end reads [270 bp and 500 bp] using the Illumina HiSeq platform) and Aberystwyth University (Aberystwyth, UK) (TruSeq Nano DNA libraries producing paired-end reads [300 bp] using the Illumina MiSeq platform). We performed long-read library preparation and sequencing according to the Nanopore protocol (SQK-LSK109) on R9 flow cells (FLO-MIN106). Read quality was assessed using MultiQC (11).
We assembled the long reads with Flye (12), using default parameters, to generate chromosome-scale scaffolds. Then, using Minimap2 (13) and SAMtools (14), we mapped the short reads onto the assembled scaffolds to compensate for erroneous bases within the long reads and to create consensus sequences. After polishing of the assembly with Pilon (15), another round of consensus short-read mapping was performed. Then, we removed duplicated contigs and sorted the remainder according to length using Funannotate (16). Finally, we separated the chimeric sequences and performed scaffolding using RaGOO (17) with the Leishmania major Friedlin strain genome (GCA_000002725.2) (18) as a reference guide, aligning all 36 chromosomes for our assembly with the exception of six small contigs totaling 70,152 bp.
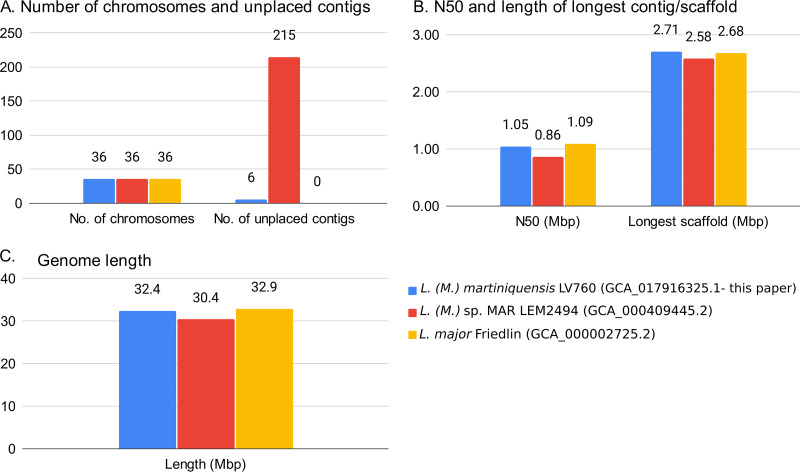
The analysis workflow for assembly and annotation, including software versions and parameters used, was performed using Snakemake (19) and is available online for reproducibility purposes (https://github.com/hatimalmutairi/LGAAP). Figure 1 compares our assembly with other complete genomes.
FIG 1.
Assembly comparison of L. (M.) martiniquensis LV760 (this paper) with L. martiniquensis MAR LEM2494 and L. major Friedlin.
We assessed assembly completeness with BUSCO (20) using the lineage data set for the phylum Euglenozoa, containing 130 single-copy orthologues from 31 species, and we found 128 of the orthologues to be present (98.5% completeness). We carried out functional annotation and prediction using the MAKER2 (21) annotation pipeline in combination with AUGUSTUS (22) gene prediction software. Table 1 shows additional summary metrics for sequencing, assembly, and annotation.
TABLE 1.
Summary of genome sequencing, assembly, and annotation metrics for L. (M.) martiniquensis LV760
| Feature | Value |
|---|---|
| Total no. of reads | 24,128,044 |
| No. of MiSeq reads | 4,462,344 |
| No. of HiSeq reads | 18,716,360 |
| No. of MinION reads (read N50 [bp]) | 949,340 (15,090) |
| No. of bases (Gb) | 19.24 |
| Genome coverage (×) | 277.9 |
| Total no. of scaffolds | 42 |
| Genome size (bp) | 32,413,670 |
| N50 (bp) | 1,046,741 |
| GC content (%) | 59.90 |
| No. of Ns (% of genome) | 50 (0.0002) |
| No. of genes | 7,967 |
| Gene density (no. of genes/Mb) | 245.8 |
| No. of exons | 7,969 |
| Mean gene length (bp) | 1,857 |
| Total length of coding sequences (Mb [% of genome]) | 14.80 (45.66) |
Data availability.
The assembly and annotations are available under GenBank assembly accession number GCA_017916325.1. The master record for the whole-genome sequencing project is available under accession number JAFEUZ000000000.1. Raw sequence reads are available under accession number PRJNA691531.
ACKNOWLEDGMENT
This work is funded by a Ph.D. studentship grant to H.A. from the Ministry of Health and Public Health Authority of Saudi Arabia.
Contributor Information
Derek Gatherer, Email: d.gatherer@lancaster.ac.uk.
Antonis Rokas, Vanderbilt University.
REFERENCES
- 1.Ghorbani M, Farhoudi R. 2018. Leishmaniasis in humans: drug or vaccine therapy? Drug Des Devel Ther 12:25–40. doi: 10.2147/DDDT.S146521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burza S, Croft SL, Boelaert M. 2018. Leishmaniasis. Lancet 392:951–970. doi: 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- 3.Espinosa OA, Serrano MG, Camargo EP, Teixeira MMG, Shaw JJ. 2018. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145:430–442. doi: 10.1017/S0031182016002092. [DOI] [PubMed] [Google Scholar]
- 4.Jariyapan N, Daroontum T, Jaiwong K, Chanmol W, Intakhan N, Sor-Suwan S, Siriyasatien P, Somboon P, Bates MD, Bates PA. 2018. Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasit Vectors 11:351. doi: 10.1186/s13071-018-2908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desbois N, Pratlong F, Quist D, Dedet JP. 2014. Leishmania (Leishmania) martiniquensis n. sp. (Kinetoplastida: Trypanosomatidae), description of the parasite responsible for cutaneous leishmaniasis in Martinique Island (French West Indies). Parasite 21:12. doi: 10.1051/parasite/2014011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobsiger L, Muller N, Schweizer T, Frey CF, Wiederkehr D, Zumkehr B, Gottstein B. 2010. An autochthonous case of cutaneous bovine leishmaniasis in Switzerland. Vet Parasitol 169:408–414. doi: 10.1016/j.vetpar.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Rose K, Curtis J, Baldwin T, Mathis A, Kumar B, Sakthianandeswaren A, Spurck T, Low Choy J, Handman E. 2004. Cutaneous leishmaniasis in red kangaroos: isolation and characterisation of the causative organisms. Int J Parasitol 34:655–664. doi: 10.1016/j.ijpara.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Kwakye-Nuako G, Mosore MT, Duplessis C, Bates MD, Puplampu N, Mensah-Attipoe I, Desewu K, Afegbe G, Asmah RH, Jamjoom MB, Ayeh-Kumi PF, Boakye DA, Bates PA. 2015. First isolation of a new species of Leishmania responsible for human cutaneous leishmaniasis in Ghana and classification in the Leishmania enriettii complex. Int J Parasitol 45:679–684. doi: 10.1016/j.ijpara.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Pothirat T, Tantiworawit A, Chaiwarith R, Jariyapan N, Wannasan A, Siriyasatien P, Supparatpinyo K, Bates MD, Kwakye-Nuako G, Bates PA. 2014. First isolation of Leishmania from northern Thailand: case report, identification as Leishmania martiniquensis and phylogenetic position within the Leishmania enriettii complex. PLoS Negl Trop Dis 8:e3339. doi: 10.1371/journal.pntd.0003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanmol W, Jariyapan N, Somboon P, Bates MD, Bates PA. 2019. Axenic amastigote cultivation and in vitro development of Leishmania orientalis. Parasitol Res 118:1885–1897. doi: 10.1007/s00436-019-06311-z. [DOI] [PubMed] [Google Scholar]
- 11.Ewels P, Magnusson M, Lundin S, Kaller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 13.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li WC, Wang TF. 2021. PacBio long-read sequencing, assembly, and Funannotate reannotation of the complete genome of Trichoderma reesei QM6a. Methods Mol Biol 2234:311–329. doi: 10.1007/978-1-0716-1048-0_21. [DOI] [PubMed] [Google Scholar]
- 17.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Müller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schäfer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mölder F, Jablonski KP, Letcher B, Hall MB, Tomkins-Tinch CH, Sochat V, Forster J, Lee S, Twardziok SO, Kanitz A, Wilm A, Holtgrewe M, Rahmann S, Nahnsen S, Köster J. 2021. Sustainable data analysis with Snakemake. F1000Res 10:33. doi: 10.12688/f1000research.29032.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 21.Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembly and annotations are available under GenBank assembly accession number GCA_017916325.1. The master record for the whole-genome sequencing project is available under accession number JAFEUZ000000000.1. Raw sequence reads are available under accession number PRJNA691531.