Abstract
O-GlcNAcylation is the modification of serine and threonine residues with β-N-acetylglucosamine (O-GlcNAc) on intracellular proteins. This dynamic modification is attached by O-GlcNAc transferase (OGT) and removed by O-GlcNAcase (OGA) and is a critical regulator of various cellular processes. Furthermore, O-GlcNAcylation is dysregulated in many diseases, such as diabetes, cancer, and Alzheimer’s disease. However, the precise role of this modification and its cycling enzymes (OGT and OGA) in normal and disease states remains elusive. This is partially due to the difficulty in studying O-GlcNAcylation with traditional genetic and biochemical techniques. In this review, we will summarize recent progress in chemical approaches to overcome these obstacles. We will cover new inhibitors of OGT and OGA, advances in metabolic labeling and cellular imaging, synthetic approaches to access homogeneous O-GlcNAcylated proteins, and cross-linking methods to identify O-GlcNAc-protein interactions. We will also discuss remaining gaps in our toolbox for studying O-GlcNAcylation and questions of high interest that are yet to be answered.
Graphical Abstract
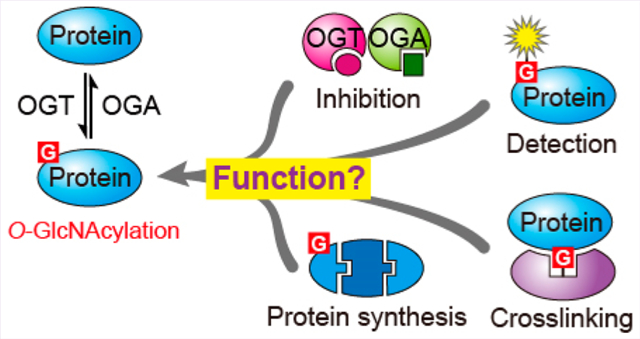
O-GlcNAcylation is the attachment of O-linked β-N-acetyl glucosamine (O-GlcNAc) onto serine or threonine residues of numerous intracellular proteins (Figure 1). This essential post-translational modification (PTM) was an unexpected discovery by the Hart lab in 19841 and was later found ubiquitously in all eukaryotes except yeast.2,3 Distinct from the cell surface glycans that mainly exist as oligosaccharides or polysaccharides, O-GlcNAcylation is a unique monosaccharide modification in the nucleus and cytoplasm.4 To date, over 4000 O-GlcNAcylated proteins have been reported.5 The glycosylation of these proteins is all catalyzed by a single pair of opposing enzymes: O-GlcNAc transferase (OGT) transfers O-GlcNAc from the sugar donor uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) to various protein substrates,6 while O-GlcNAcase (OGA) removes this modification7 (Figure 1). This stands in great contrast to phosphorylation, which is controlled by hundreds of kinases and phosphatases.8 Although the molecular mechanisms regulating the O-GlcNAc cycling enzymes remain a challenging ongoing question, emerging evidence points to modulation of OGT by PTMs9,10 and protein–protein interactions.11,12 OGT’s activity is also highly regulated by cellular concentrations of UDP-GlcNAc,13 which is derived from glucose and incorporates fatty acid, nucleotide, and amino acid metabolites via the hexosamine biosynthetic pathway (HBP; Figure 1).14 Consequently, O-GlcNAcylation level changes in rapid response to the fluctuation of the nutrient and metabolic status of the cell to modulate proteins’ stability, activity, and localization. Taken together, O-GlcNAcylation serves as a nutrient sensor that dynamically regulates protein functions.
Figure 1.
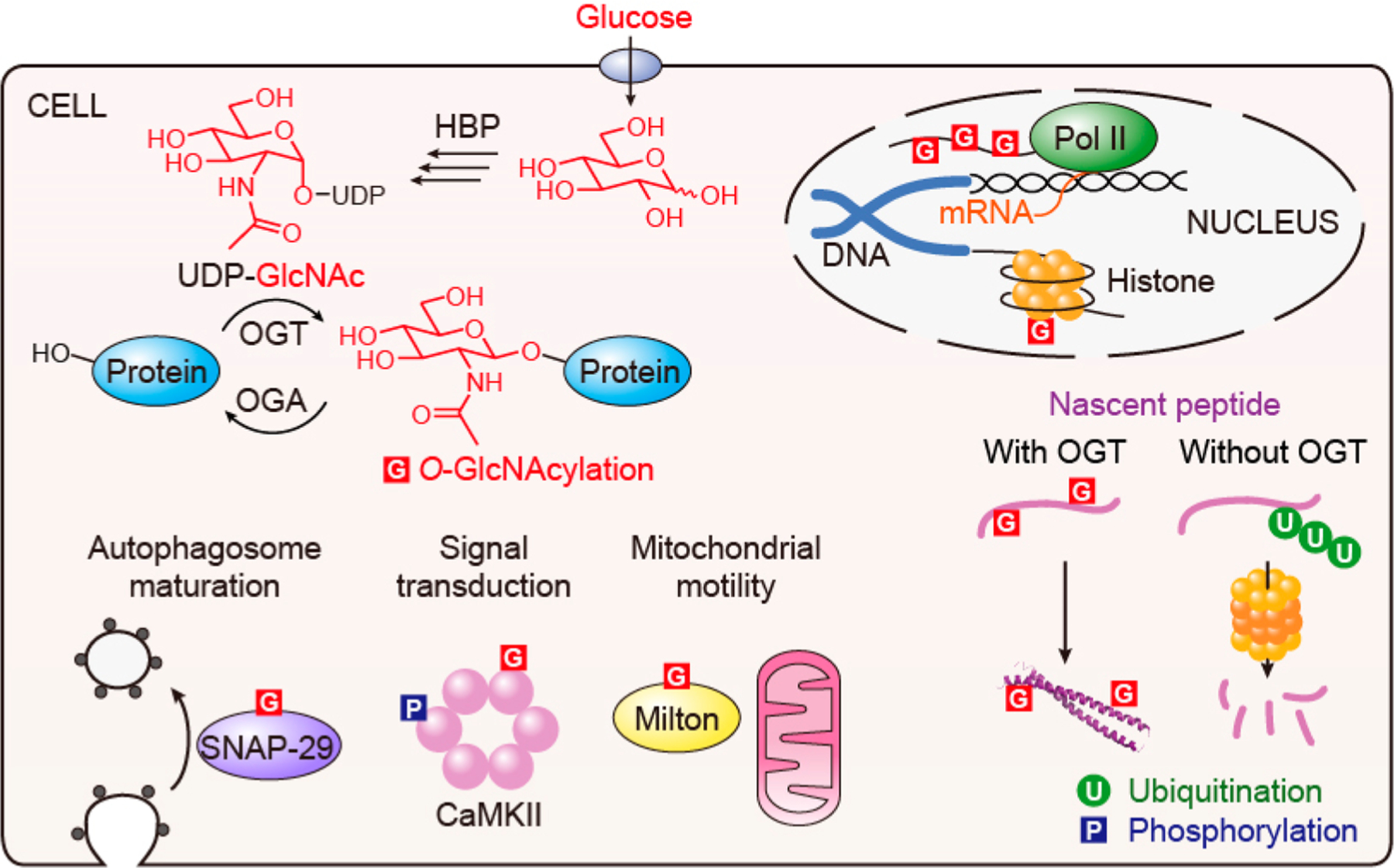
O-GlcNAcylation cycle and the biological processes regulated by dynamic O-GlcNAcylation in cells. HBP: hexosamine biosynthetic pathway. Pol II: RNA polymerase II. SNAP-29: synaptosomal-associated protein 29. CaMKII: Ca2+/calmodulin-dependent protein kinase II.
O-GlcNAcylation plays significant roles in a broad range of cellular processes, and here we highlight a few of them from recent publications (Figure 1). Previous studies have uncovered the functional impacts of O-GlcNAcylation on a number of transcription factors including p53,15 FoxO1,16 and Sp1.17 In contrast, the investigation of O-GlcNAcylation on the central transcriptional machinery RNA polymerase II (Pol II) remains a big challenge, as numerous glycosylation sites are located on its highly repetitive C-terminal domain (CTD)18,19 and no specific O-GlcNAcylated CTD antibody is currently available. Despite this, in vitro evidence has suggested that O-GlcNAcylated Pol II facilitates the assembly of transcription preinitiation complex (PIC) onto DNA promoters.20 In addition, OGT was found in the protein complex with Pol II and enriched at promoter regions in cells. More recently, OGA was reported as an important component of the transcription elongation complex.21 O-GlcNAcylation also complements phosphorylation as a modulator of signal transduction. In an interesting example, high glucose conditions induced O-GlcNAcylation of Ca2+/calmodulin-dependent protein kinase II (CaMKII). This activates the kinase to regulate downstream signaling, indicating that O-GlcNAc directly links intracellular glucose level to signal transduction (Figure 1).22 Additional examples have been extensively reviewed.23,24 O-GlcNAcylation also influences cytoskeletal motor proteins such as mitochondrial motor-adaptor protein milton.25 This occurs in response to high glucose stimulation and tunes the motility of mitochondria in neurons (Figure 1). O-GlcNAcylation is likewise involved in autophagy, as O-GlcNAcylation of synaptosomal-associated protein 29 (SNAP-29) regulates the maturation of autophagosome in accordance with nutrient levels (Figure 1).26 Moreover, cellular feeding is in part governed by O-GlcNAcylation, particularly through insulin production and signaling.27,28 Surprisingly, OGT also appears to have proteolytic activity, as it was found to cleave host cell factor 1 (HCF-1) in the same active site that catalyzes O-GlcNAcylation.29 This activates HCF-1 to become a key regulator of the cell cycle.30 A more recent study reported that the proteolysis of HCF-1 involves unprecedented O-GlcNAcylation on the cleaving glutamate residue.31 Overall, the discoveries highlighted here represent only a handful of the cellular processes that O-GlcNAcylation and its cycling enzymes regulate, underscoring their broad impacts in cell physiology.
Growing evidence supports an essential role of O-GlcNAcylation and its cycling enzymes in human diseases. Increased OGT and O-GlcNAcylation levels occur in various types of cancer and have been found to promote cancer growth and progression.32 In diabetic cells, a number of insulin signaling proteins and mitochondrial proteins are also hyper O-GlcNAcylated.22,33 Alzheimer’s disease is thought to arise from tau protein oligomerization, which is promoted by its hyperphosphorylation.34,35 O-GlcNAcylation and phosphorylation of tau have been shown to be mostly reciprocal in vitro, and tau O-GlcNAcylation appears to reduce its oligomerization in vivo.36 Despite the significance of O-GlcNAcylation in normal and disease states, our mechanistic understanding on many of these processes remains elusive, such as the regulation of OGT substrate specificity and the functional impacts of its dysregulation in pathogenesis. Limiting our ability to answer these questions is the difficulty of studying O-GlcNAcylation in a specific manner using traditional genetic and biochemical approaches, such as gene deletion or routine profiling of the proteome. In this review, we will highlight recently developed chemical approaches to detect O-GlcNAcylation and investigate its functions, serving as new avenues to overcome the inherent challenges of this modification.
O-GLCNAC CYCLING ENZYMES
OGT and Its Inhibitors.
OGT is a multidomain protein belonging to the 41 family of glycosyltransferase in the CAZy database (Figure 2a). Kinetic and structural studies have established that OGT glycosylates substrates in an ordered bi-bi mechanism.37–39 OGT binds UDP-GlcNAc first, followed by the peptide substrate. After sugar transfer, glycosylated peptide leaves prior to UDP. The binding site for UDP-GlcNAc is primarily comprised of two lobes of the catalytic domain (N-Cat and C-Cat in Figure 2a), separated by the intervening domain (Int-D), which is conserved in metazoans with its function remaining to be characterized.37 A unique structural feature of OGT is the N-terminal tetratricopeptide repeat (TPR) domain, which forms an extended α-helical tunnel and has been suggested to participate in protein substrate binding.40 The length of the TPR domain distinguishes the three splicing isoforms of OGT (ncOGT, mOGT, and sOGT) found in humans (Figure 2a), although differences in their functions are mostly unclear. The substrate recognition of OGT remains an open question as no consensus sequence motif has been identified in substrate proteins. To address this, efforts have focused on the crystallization of OGT-substrate complexes. The first crystal structure of human OGT was obtained with a truncated construct containing 4.5 repeats of the 13.5 TPRs of ncOGT (called OGT4.5) in complex with UDP and CKII peptide substrate.37 Following that, additional complex structures of OGT4.5 with TAB1, HCF-1, RBL2, Ret, keratin-7, and lamin B1 peptides have been solved.29,39,41 These structures have revealed that short peptides are mainly anchored by UDP-GlcNAc bound in the OGT active site, as well as backbone interactions with adjacent TPR residues. Limited interactions with peptide side-chains may explain the tolerance of OGT for a broad range of substrates. The structures have also assisted in designing OGT inhibitors, which are discussed below. While OGT4.5 is fully capable of glycosylating a number of peptide substrates, it is incompetent in glycosylating protein substrates, indicating that the extended TPR domain is crucial for protein substrate binding. Because of the flexible nature of TPR, no crystal structure of full-length ncOGT is currently available. In addition, the substrate-binding mode of OGT beyond the immediate surroundings of the active site remains elusive.
Figure 2.
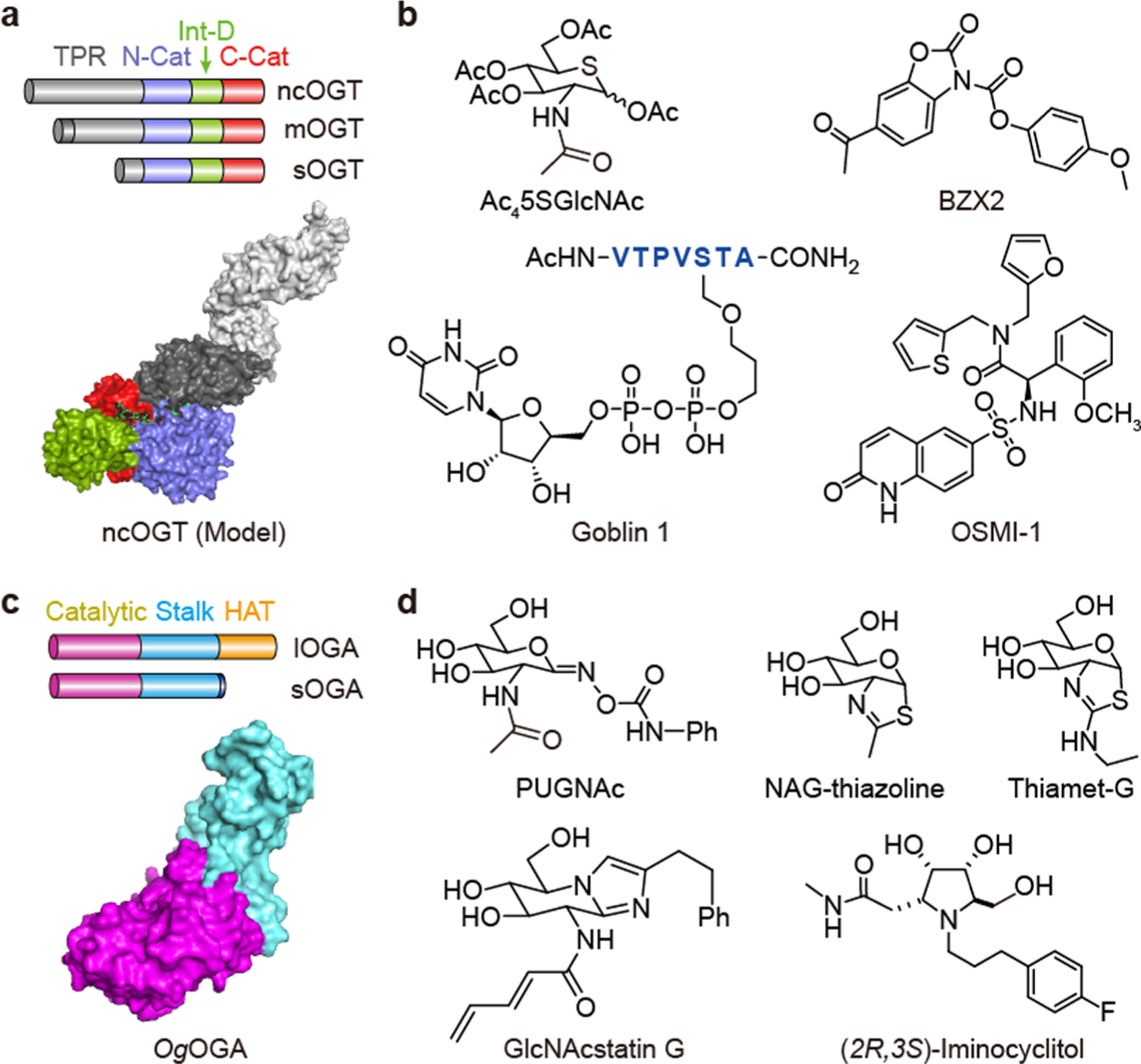
O-GlcNAc cycling enzymes and their inhibitors. (a) Schematic representation of the three isoforms of human OGT (ncOGT, mOGT, and sOGT) and the model of full-length ncOGT built from the crystal structures of OGT4.5 (PDB: 3PE4)37 and TPR domain (PDB: 1W3B).40 TPR: tetratricopeptide repeat domain. N-Cat: N-terminal catalytic domain. C-Cat: C-terminal catalytic domain. Int-D: intervening domain. (b) Inhibitors of OGT. (c) Schematic representation of the two isoforms of human OGA (lOGA and sOGA) and the crystal structure of a bacterial OGA, OgOGA (PDB: 2XSB).55 Catalytic: catalytic domain. Stalk: stalk domain. HAT: pseudo-histone acetyl transferase domain. (d) Inhibitors of OGA.
Potent and specific inhibitors would be powerful tools to investigate the biological functions of OGT. Several promising inhibitors have been developed recently employing either substrate analogues or new chemical scaffolds identified from high-throughput screens (HTS; Figure 2b). UDP-5SGlcNAc, which mimics the sugar donor UDP-GlcNAc, is one of the most potent OGT inhibitors in vitro (Ki of 8 μM).42 Its cell permeable precursor Ac45SGlcNAc can be metabolically converted to UDP-5SGlcNAc through the GlcNAc salvage pathway to inhibit endogenous OGT. Another example is UDP-peptide conjugate Goblin 1, which is a bisubstrate-linked inhibitor with an IC50 of 8 μM in vitro.43 However, this conjugate is not cell permeable due to the negatively charged diphosphate group. Meanwhile, HTS led to the discovery of a quinolinone-6-sulfonamide scaffold.44 Structure optimization has identified OSMI-1, which demonstrated improved potency (IC50 of 2.7 μM) and selectivity over substrate analogues to block OGT’s activity in vitro, although its specificity in cells requires further elucidation.45 Intriguingly, OSMI-1 has been applied to demonstrate that inhibition of OGT’s activity attenuates the replication of herpes simplex virus (HSV), similarly as genetic siRNA silencing of OGT.46 Another interesting scaffold discovered from this HTS is benzoxazoli-none (BZX).44 Optimization of this scaffold gave BZX2, which covalently inhibits OGT with a novel double displacement mechanism and cross-links two important residues (Lys842 and Cys917) in the enzyme active site.47 While this inhibitor is highly effective in vitro, its specificity is low and thus not suitable for cellular studies. These new inhibitors possess significantly improved properties; however, none of them is potent or specific enough for in vivo applications. Furthermore, new strategies to develop substrate-specific inhibitors that allow targeting of O-GlcNAcylation on an individual or small family of proteins are a desired but unmet need.
OGA and Its Inhibitors.
The enzyme that specifically cleaves O-GlcNAcylation is OGA, belonging to the CAZy glycoside hydrolase family 84.7 There are two splicing isoforms of OGA in humans: long OGA (lOGA) and short OGA (sOGA) (Figure 2c). lOGA consists of an N-terminal catalytic domain and a C-terminal pseudo-histone acetyl transferase (HAT) domain, linked by a stalk domain between them. sOGA lacks the pseudo-HAT domain and displays lower catalytic activity.48,49 Since human OGA (hOGA) has not been amenable for crystallization, the structures of four bacterial OGAs have been solved: Bacteroides thetaiotaomicron hexosaminidase (BtGH84),36,50,51 Clostridium perfringens NagJ (CpOGA),52–54 Oceanicola granulosus glycosidase (OgOGA),55 and Thermobaculum terrenum glycoside hydrolase (TtOGA).56 These bacterial homologues share significant sequence similarity with hOGA in the catalytic domain, but not in the stalk domain or the pseudo-HAT domain, which have been suggested to contribute to hOGA substrate recognition. Nevertheless, these crystal structures of bacterial homologues and the biochemical characterization of hOGA have provided significant insights into the catalytic mechanism of O-GlcNAc hydrolysis and guided the rational design of OGA inhibitors.57,58
Several highly potent and selective OGA inhibitors have been reported, most of which are transition state mimics (Figure 2d). PUGNAc is one of the first potent OGA inhibitors with a Ki of 46 nM, but it also inhibits other glycoside hydrolases.59,60 GlcNAcstatin G improved upon this with a Ki of 4.1 nM for hOGA.61 Although it possesses a remarkable 900000-fold selectivity over β-hexosaminidase (the most structurally related enzyme to hOGA) in vitro, it is difficult to synthesize and has low water solubility, thus limiting its application in vivo. NAG-thiazoline represents another OGA inhibitor scaffold.62 A slight modification of this compound produced the potent inhibitor thiamet-G (Ki = 21 nM against hOGA) with excellent selectivity (37000-fold) over β-hexosaminidase.36 Moreover, the synthesis of this inhibitor is straightforward, and the compound can cross the blood—brain barrier for potential applications in neurodegenerative diseases. For instance, thiamet-G has been employed to raise O-GlcNAcylation level in mouse models of Alzheimer’s disease, which reduced protein aggregation and alleviated Alzheimer’s symptoms.36,63 More recently, stereoisomeric iminocyclitols have also been reported to inhibit hOGA.64 Different isomers demonstrated distinct inhibition effects, with several exhibiting low nanomolar activity against hOGA in vitro. One of the isomers (Figure 2d) showed good bioavailability in mice and could also pass the blood—brain barrier.64 For more detailed reviews on OGA inhibitors, please refer to refs 65 and 66.
DETECTION OF O-GLCNACYLATION
Lectins and Small Molecule Probes.
One of the main challenges in the field is detecting O-GlcNAcylation with high sensitivity and specificity. This modification has been traditionally detected using lectins (e.g., WGA)67 and antibodies (e.g., CTD110.6).68 Despite their widespread use, they potentially recognize other types of sugars,69 and very few site-specific O-GlcNAc antibodies are currently available. Recently, a “synthetic lectin” was reported with high selectivity and 25-fold improved affinity for β-O-GlcNAc versus WGA.70 In another study, the recombinant fungal lectin rPVL was also shown to be more specific than WGA and about 10-fold more potent.71 Additionally, rPVL contains multiple O-GlcNAc binding sites, which can bind to multivalent O-GlcNAc with an affinity 1900-fold greater than to monovalent O-GlcNAc. A different approach has exploited the ability of bacterial CpOGA to recognize the O-GlcNAc proteome.72 Even though CpOGA can hydrolyze O-GlcNAc from human cell lysates, its physiological role and recognition specificity require further investigation as no dynamic O-GlcNAcylation was detected in bacteria. Besides detecting global O-GlcNAcylation, development of site-specific antibodies and lectins will be invaluable for future O-GlcNAc detection.
Metabolic labeling is another prevailing strategy to detect O-GlcNAcylation. Similar to Ac45SGlcNAc, other GlcNAc analogues with bioorthogonal handles (e.g., Ac4GlcNAz,73 Ac36AzGlcNAc,74 and Ac4GlcNAlk75 in Figure 3a) can be metabolically converted to UDP sugars in cells for O-GlcNAc enrichment and detection. These probes have been reviewed extensively.5,76 By exploiting the endogenous epimerases, GalNAc sugars can also be converted to UDP-GlcNAc analogues in cells to monitor O-GlcNAcylation. A recent application of Ac4GalNAz (Figure 3a) has discovered that O-GlcNAcylation could be a cotranslational process and protects nascent peptides from ubiquitination and degradation (Figure 1).77,78 A new GlcNAc analogue (Ac34dGlcNAz, Figure 3a) lacking the 4′-OH group was reported to reduce nonspecific incorporation into extracellular glycans and increase resistance to OGA hydrolysis, thus leading to elevated specificity and sensitivity for O-GlcNAc detection.79
Figure 3.
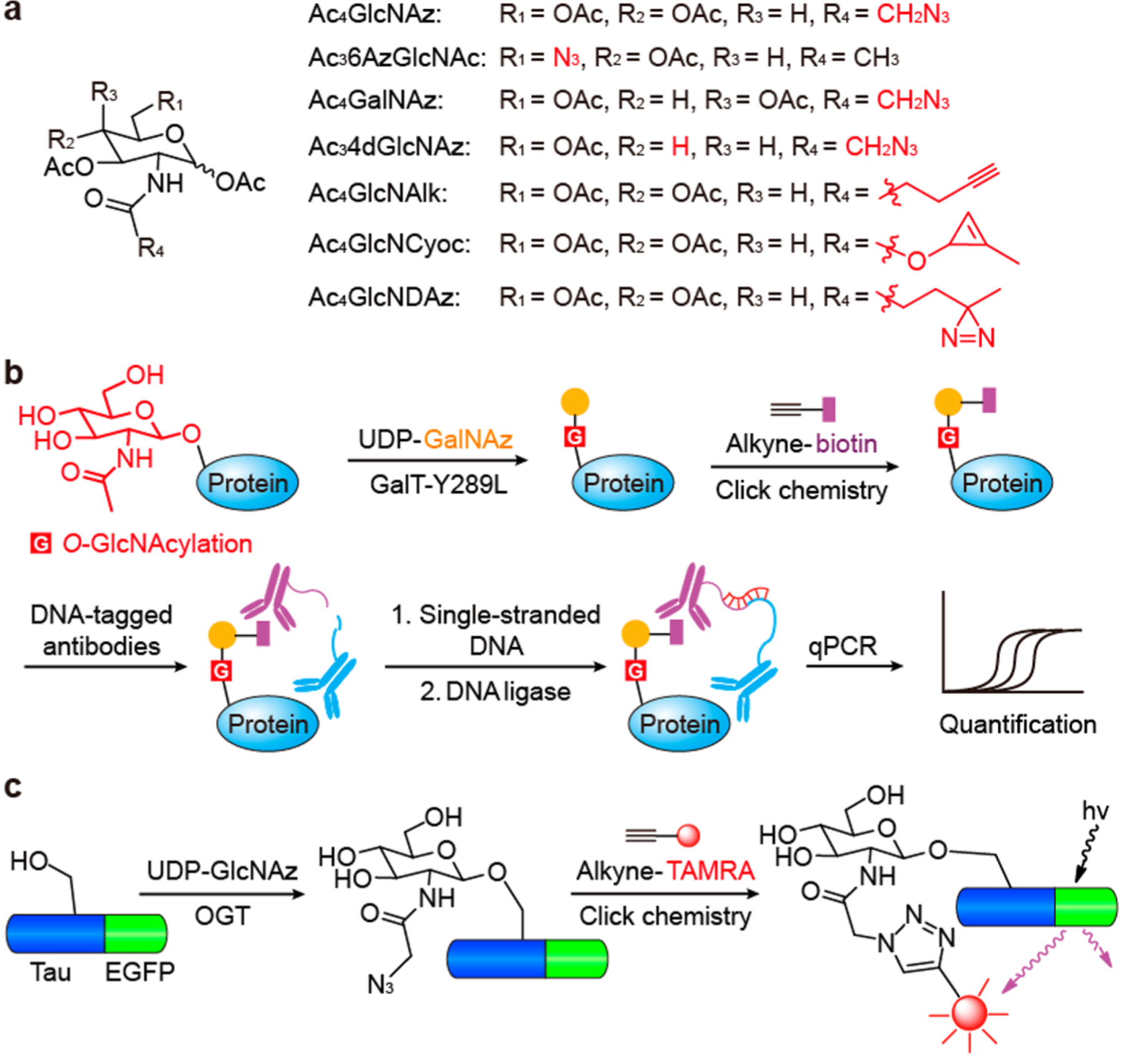
Chemical methods to detect O-GlcNAcylation. (a) GlcNAc and GalNAc analogues containing different functional groups that have been applied in metabolic labeling of O-GlcNAcylation. (b) The principle of Glyco-seek for sensitive quantification of O-GlcNAcylation. (c) O-GlcNAc imaging is enabled by O-GlcNAc metabolic labeling and EGFP fusion. EGFP: enhanced green fluorescent protein. TAMRA: carboxytetramethylrhodamine.
Copper-catalyzed or copper-free (e.g., cyclooctyne) click chemistry has been widely employed to detect O-GlcNAcylation in combination with the above metabolic probes. Owing to the toxicity of copper to cells and nonspecific reactivity of strained alkynes, other metal free, bioorthogonal “click” reactions have been developed, such as the inverse electron demand Diels–Alder (DAinv) reaction between 1,2,4,5-tetrazines and an alkene dienophile. One such study demonstrated its potential by employing methylcyclopropene GlcNAc analogue Ac4GlcNCyoc (Figure 3a) for intracellular protein labeling.80 Notably, few extracellular glycans were detected. DAinv could potentially allow dual labeling of different targets in cells along with azide–alkyne click chemistry. Continued improvement in the specificity and incorporation efficiency of metabolic probes will advance their applications in O-GlcNAc biology.
Sensitivity is another huge challenge in O-GlcNAc detection since many O-GlcNAcylated proteins are present in low abundance and low O-GlcNAc stoichiometry. A method termed Glyco-seek was recently shown to increase the sensitivity of O-GlcNAc detection by several orders of magnitude (Figure 3b).81 In this study, O-GlcNAc was first labeled by a mutant galactosyltransferase (GalT-Y289L) with GalNAz using an established chemoenzymatic method.82 Following click chemistry, O-GlcNAc residues were conjugated to biotin epitopes. DNA-tagged antibodies specific for biotin and the target protein bring the attached DNA strands into proximity if the protein is O-GlcNAcylated, and this can be quantified by DNA ligation and quantitative PCR. This new technique can detect unprecedented attomole levels of glycosylated proteins, although it may not be that effective for discovering new OGT substrates. Further progress toward more sensitive and specific methods for O-GlcNAc detection will aid research into the functions of this dynamic modification.
Mass Spectrometry.
Mass spectrometry (MS) has become the primary technique for discovering new OGT substrates. As discussed above, sensitivity is often an issue in O-GlcNAc detection. The problem is even more pronounced in MS methods due to the profound ion suppression effect of O-GlcNAcylated peptides. Consequently, enrichment of O-GlcNAcylated proteins/peptides is often essential for adequate detection. Normally this involves either enrichment with WGA or metabolic labeling followed by conjugation to biotin with click chemistry as discussed above. The difficulty of eluting biotinylated proteins/peptides from streptavidin beads promoted the development of cleavable biotin linkers. A photocleavable linker has been successfully applied to enrich O-GlcNAc; however, it is difficult to cleave quantitatively.83,84 Diazobenzene linkers that can be cleaved under mild conditions with dithionite have also been employed in O-GlcNAc studies74 but suffer from a similar problem.84,85 A silane group was later shown to be efficiently cleaved with dilute acid for the enrichment of O-GlcNAcylated proteins and exhibited greatly improved selectivity.85,86 Recently, a linker containing the 1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)ethyl (Dde) group was shown to be quantitatively cleaved by hydrazine and, importantly, gave a positively charged amine for better MS ionization.87 Using immunoblot, this probe displayed better efficiency of global labeling and cleavage than the commonly used photocleavable alkyne-biotin in a test of cell lysates. Additionally, it could easily detect α-crystallin added to the cell lysates using MS. However, it has yet to be employed in MS detection of O-GlcNAcylation on endogenous proteins.
A powerful strategy to identify low abundance O-GlcNAcylation is quantitative proteomics on isotopically recoded glycopeptides. In a recent example, a biotin-dibromide cleavable tag was applied to detect O-GlcNAcylated proteins of significantly lower abundance than was previously achievable.86 Another study fed 13C labeled glucose to living cells to monitor O-GlcNAc turnover and compare to other types of PTMs.88 O-GlcNAcylation turnover rate was found to spread over a wide range (0.02 h−1 to 1.6 h−1) but was generally slower than phosphorylation and acetylation. Other isotopic labeling strategies such as stable isotope labeling with amino acids in cell culture (SILAC) and β-elimination Michael addition (BEMAD) with deuterated DTT (d6) have also been used to quantitatively investigate O-GlcNAc and have been covered in previous reviews.5,89
Another challenge of MS detection of O-GlcNAcylation is its lability under collision-induced fragmentation methods (e.g., collision-induced dissociation (CID), higher-energy collisional dissociation (HCD)). This problem was partially addressed with the development of electron transfer dissociation (ETD)90 and electron capture dissociation (ECD),91 which enable the fragmentation of glycopeptides exclusively along the backbone while leaving the glycosidic bond intact. These fragmentation methods are capable of site mapping for PTMs such as O-GlcNAcylation, though both suffer from lower sequence coverage. Several studies have overcome these problems by utilizing more than one type of fragmentation.5 An improved technique, called electron transfer and higher-energy collision dissociation (EThcD), offers ETD fragmentation prior to HCD for more complete fragmentation and more confident site mapping of O-GlcNAcylation.92 One study applied this method to human leukocyte antigen (HLA) class 1 peptides, which are recognized on the cell surface by the immune system to distinguish healthy versus infected or cancerous cells.93 They discovered O-GlcNAcylation on HLA peptides and found it extended with up to four other monosaccharides. The authors proposed this as the first example of intracellular O-GlcNAc being used as the base of an extracellular glycan. This study demonstrates the value of improved fragmentation techniques toward studying O-GlcNAc biology. Further progress will allow more efficient O-GlcNAc detection with higher sequence coverage.
Determining the stoichiometry of O-GlcNAcylation is another area of interest in the field. The O-GlcNAc residue is neutral and too small (203 Da) to produce a noticeable shift in SDS-PAGE gel, making it difficult to monitor the O-GlcNAcylation level of a particular protein. A mass-tagging approach has been devised in which a large polyethylene glycol (PEG) tag can be ligated to an O-GlcNAc residue, which allows bands of O-GlcNAcylated proteins to be discerned on SDS-PAGE gel based on the number of sugars.94 More recently, intact protein mass spectrometry has also been employed for this purpose. A recent study detected around 20 residues of O-GlcNAc modified on the CTD of Pol II.19 It also demonstrated that the addition and removal of O-GlcNAc from the CTD occurs via distributive mechanisms by OGT and OGA, respectively. Intact protein MS was particularly essential in this case due to the repetitive nature of the CTD and the highly heterogeneous O-GlcNAcylation states, which make O-GlcNAc site assignment extremely difficult. Intact protein MS also gave better resolution for quantifying O-GlcNAc stoichiometry than mass tags. This was the first application of mass spectrometry to study the mechanism of dynamic O-GlcNAcylation and should prove useful for other O-GlcNAcylated proteins.
Fluorescence Imaging.
Visualization of specific O-GlcNAcylated proteins in living cells will enable studying the spatial and temporal distribution of O-GlcNAcylated proteins in physiological and disease states. A Förster resonance energy transfer (FRET) method was introduced a decade ago for this purpose.95 However, it requires the substrate to be genetically fused to two fluorescent proteins and a lectin, potentially affecting its endogenous O-GlcNAcylation and localization. A newer study has fused only enhanced green fluorescent protein (EGFP) to tau (Figure 3c),96 a known OGT substrate and a critical protein in Alzheimer’s disease. First, the authors metabolically labeled O-GlcNAcylated proteins with Ac4GalNAz. Following click chemistry with alkyne-TAMRA and imaging with fluorescence lifetime imaging microscopy (FLIM-FRET), they were able to monitor O-GlcNAcylated tau and β-catenin in living cells. Since FLIM-FRET detects donor fluorescence lifetime rather than acceptor fluorescence intensity, it reduced the background and increased the imaging sensitivity and resolution. A similar strategy utilizing methylcyclopropene-tetrazine click chemistry has imaged several O-GlcNAcylated proteins including OGT and p53 in cells.97 This may prove to be useful considering the previously mentioned drawbacks of alkyne–azide click chemistry. Improved imaging techniques will facilitate characterizing the localization of O-GlcNAcylation in diseases and identifying any cellular heterogeneity among them.
FUNCTIONAL CHARACTERIZATION
Synthetic Proteins.
A lack of access to homogeneous samples of O-GlcNAcylated proteins has significantly hampered our understanding on the properties and functions of this modification in a site-specific manner. This challenge is being overcome with two general synthetic strategies. One employs native chemical ligation (NCL) and expressed protein ligation (EPL) to accomplish total protein synthesis. The first report on semisynthesis of a GlcNAcylated protein was accomplished with the preparation of S-GlcNAcylated CK2α by EPL. This synthetic glycoprotein was successfully applied to establish that S-GlcNAcylation on CK2α modulates its phosphorylation and substrate selectivity.98 More recently, NCL and EPL were applied in combination to generate O-GlcNAcylated-Thr72 of α-synuclein (Figure 4a), a protein implicated in neurodegenerative diseases such as Parkinson’s disease.99 They were able to demonstrate that O-GlcNAcylation on Thr72 reduces aggregation and toxicity of α-synuclein. Another group used a similar approach to produce O-GlcNAcylated-Ser400 of the tau protein for the first time, which will be valuable considering the lack of knowledge on the role of site-specific tau O-GlcNAcylation.100
Figure 4.
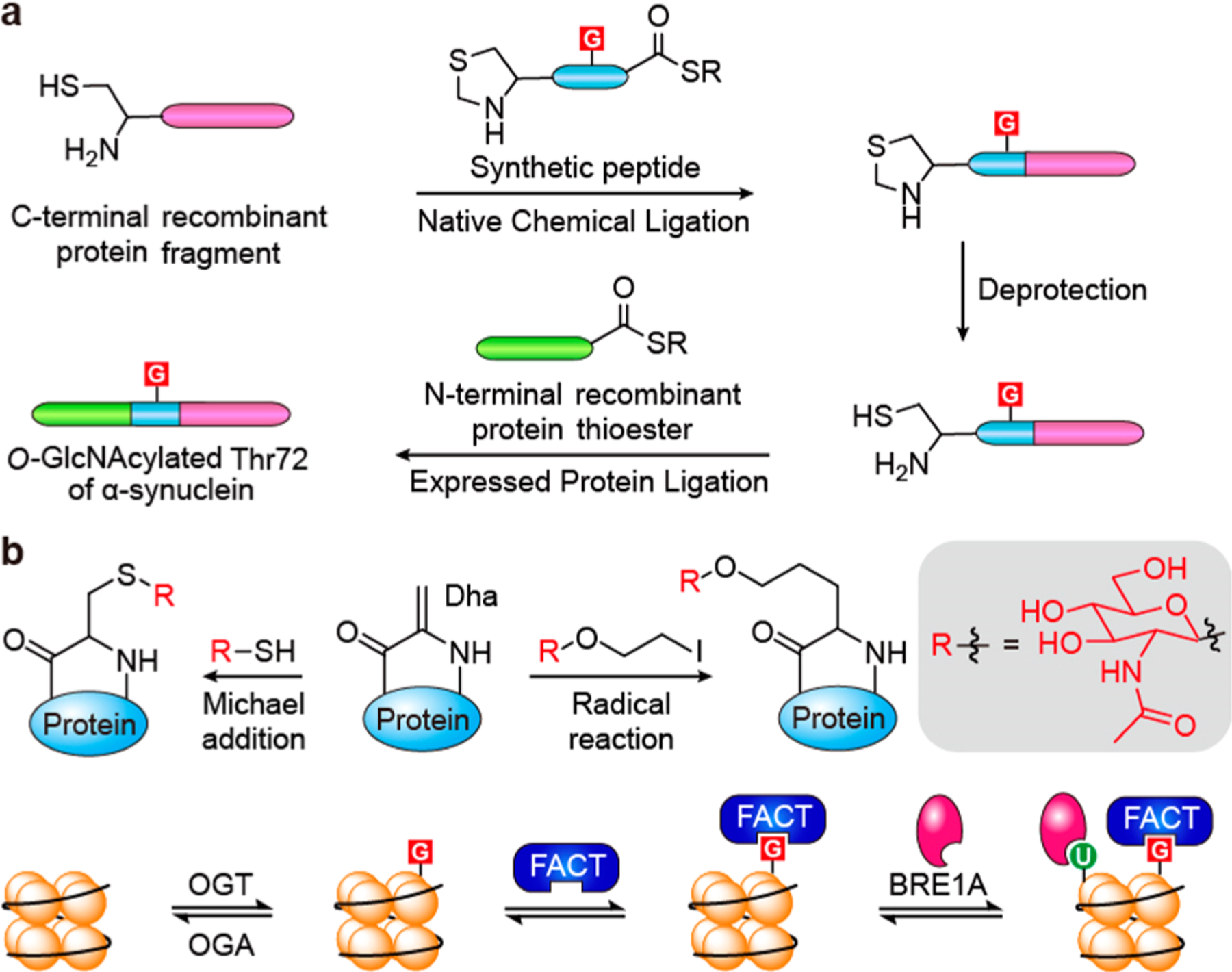
Synthetic protein methods facilitating the functional investigation on O-GlcNAcylated proteins. (a) Native chemical ligation and expressed protein ligation methods for the synthesis of O-GlcNAcylated α-synuclein protein. (b) In a different strategy, GlcNAc-thiol or halogenated O-GlcNAc can be attached to dehydroalanine (Dha) through Michael addition or radical reaction, respectively. The S-GlcNAc on Cys112 of histone H2B promotes binding with the FACT protein complex, as well as histone ubiquitination by BRE1A enzyme. U: ubiquitination. FACT: facilitates chromatin transcript.
An alternative strategy exploits the unnatural amino acid dehydroalanine (Dha), which can be incorporated into a protein with several methods.101,102 The Michael acceptor of Dha can react with a thiol nucleophile such as GlcNAc-thiol (Figure 4b) to generate specifically S-GlcNAcylated protein without the need for ligating fragments. This approach has been implemented in a couple of studies.103,104 In particular, production of histone H2B S-GlcNAcylation on Cys112 using this method has found that S-GlcNAcylation increased binding with several proteins including the facilitates chromatin transcript (FACT) complex, which led to a new proposed mechanism for FACT mediated H2B ubiquitination by BRE1A (Figure 4b).104 S-GlcNAcylated proteins are resistant to OGA hydrolysis, which is advantageous for O-GlcNAc detection but may impede experiments aimed at O-GlcNAc dynamics. A novel approach applying free radical chemistry to attach PTMs to Dha through a carbon–carbon bond was recently published by two independent groups (Figure 4b).105,106 One of the studies found that a protein O-GlcNAcylated with this method was still a substrate for OGA, even though it contains extra carbon atoms between the α-carbon and the sugar.105 Although protein (re)folding may limit the utility of these synthetic approaches for larger proteins, they present vast opportunities for understanding the properties and functions of O-GlcNAcylation in a site-specific manner.
O-GlcNAc-Protein Interactions.
Modulation of protein–protein interactions is believed to be an important function of O-GlcNAcylation. The interacting complex between O-GlcNAc monosaccharide modified protein and the binding protein, however, is generally too weak to be pulled down. To address this issue, an N-acyl diazirine functional group was introduced to GlcNAc (called Ac4GlcNDAz, Figure 3a) to cross-link interacting proteins upon UV irradiation.107 However, OGT prefers UDP-GlcNAc over UDP-GlcNDAz, which is problematic due to already low cross-linking efficiencies of diazirine. This issue was alleviated by the discovery of mutant OGT(C917A) that favors UDP-GlcNDAz over UDP-GlcNAc, leading to significantly improved O-GlcNDAz incorporation.108 The ability to efficiently cross-link O-GlcNAc with interacting proteins will be highly valuable for mapping the interactome of O-GlcNAcylated proteins for functional characterization. Isotopic labeling methods such as SILAC can also help overcome sensitivity issues.
CONCLUSION AND FUTURE PERSPECTIVES
A number of new strategies for studying O-GlcNAcylation have been published in recent years. Improved OGT inhibitors have been introduced to complement OGA inhibitors already in use. However, off-target effects still plague these inhibitors; thus more specific ones are still needed. Metabolic labeling with novel GlcNAc analogues and advances in mass spectrometry have aided its detection on both known and new proteins. Methods to obtain specifically O-GlcNAcylated proteins are also proving effective for investigating the function of O-GlcNAcylation in a much more targeted way. Further application of these synthetically modified proteins will undoubtedly lead to new discoveries on the essential roles of this unique modification. One function of O-GlcNAcylation of particular interest is its ability to modulate protein–protein interactions, for which photoactivatable GlcNAc analogues will provide considerable insights. Strategies that afford more specific and higher yields of photo-cross-linking will enable the study of interactions that are weaker and involve less abundant proteins.
Many goals remain to be reached, including structures of OGT and OGA bound to protein substrates, improved MS detection, and site-specific O-GlcNAc antibodies. Ultimately, these advances will allow us to answer important questions such as the role of O-GlcNAcylation in transcriptional and epigenetic regulation. In addition, its function in neurobiology is of particular importance owing to its potential role in Alzheimer’s disease and other neurodegenerative diseases but has not yet reached a full understanding. Answers to these questions will lead to better knowledge of the pathogenesis of diseases and the development of new therapeutics.
ACKNOWLEDGMENTS
We thank members of the Jiang laboratory for critical reading of the manuscript and providing helpful suggestions. This research was supported by University of Wisconsin—Madison startup funds.
KEYWORDS:
- post-translational modification (PTM)
the covalent modification of a protein during or after translation, normally accomplished by enzymes
- glycosylation
a post-translational modification where a carbohydrate is attached to a protein
- O-GlcNAcylation
the modification of serine or threonine residues of intracellular proteins with O-linked β-N-acetylglucosamine
- lectin
a highly specific carbohydrate-binding protein or macromolecule
- click chemistry
ligation reactions that are modular, high yielding, wide in scope and highly exothermic so that they give only one reaction product; includes azide–alkyne cycloadditions as discussed here
- EThcD
a mass spectrometry fragmentation technique that produces fragments from electron transfer dissociation followed by higher-energy collisional dissociation in a single MS/MS spectrum to provide more sequence information and to improve sequence coverage
- Förster resonance energy transfer
distance-dependent nonradiative transfer of energy between two molecules
- native chemical ligation
a conjugation reaction between a peptide-alpha-thioester and a peptide bearing an N-terminal cysteine used to construct a longer polypeptide chain while retaining the native peptide backbone
- expressed protein ligation
a protein semisynthesis method that permits the in vitro ligation of a recombinant polypeptide C-terminal thioester with a synthetic peptide bearing an N-terminal cysteine; the C-terminal thioester is obtained using a mutant intein splicing protein
- diazirine
a class of organic molecules consisting of a carbon bound to two nitrogen atoms, which are double-bonded to each other, forming a three-membered ring; upon UV irradiation, diazirines generate reactive carbenes that can cross-link with nearby molecules
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Torres CR, and Hart GW (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem 259, 3308–3317. [PubMed] [Google Scholar]
- (2).Hart GW (1997) Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem 66, 315–335. [DOI] [PubMed] [Google Scholar]
- (3).Halim A, Bohse Larsen IS, Neubert P, Joshi HJ, Petersen BL, Vakhrushev SY, Strahl S, and Clausen H (2015) Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc. Natl. Acad. Sci. U. S. A 112, 15648–15653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Holt GD, and Hart GW (1986) The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J. Biol. Chem 261, 8049–8057. [PubMed] [Google Scholar]
- (5).Ma J, and Hart GW (2014) O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Haltiwanger RS, Blomberg MA, and Hart GW (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem 267, 9005–9013. [PubMed] [Google Scholar]
- (7).Gao Y, Wells L, Comer FI, Parker GJ, and Hart GW (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem 276, 9838–9845. [DOI] [PubMed] [Google Scholar]
- (8).Kramer I (2016) Signal Transduction, 3rd ed., Elvesier, London. [Google Scholar]
- (9).Song M, Kim HS, Park JM, Kim SH, Kim IH, Ryu SH, and Suh PG (2008) O-GlcNAc transferase is activated by CaMKIV-dependent phosphorylation under potassium chloride-induced depolarization in NG-108–15 cells. Cell. Signalling 20, 94–104. [DOI] [PubMed] [Google Scholar]
- (10).Seo HG, Kim HB, Kang MJ, Ryum JH, Yi EC, and Cho JW (2016) Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci. Rep 6, 34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Iyer SPN, and Hart GW (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem 278, 24608–24616. [DOI] [PubMed] [Google Scholar]
- (12).Cheung WD, Sakabe K, Housley MP, Dias WB, and Hart GW (2008) O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem 283, 33935–33941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kreppel LK, and Hart GW (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem 274, 32015–32022. [DOI] [PubMed] [Google Scholar]
- (14).Peterson SB, and Hart GW (2016) New insights: a role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol 51, 150–161. [DOI] [PubMed] [Google Scholar]
- (15).Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, and Cho JW (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol 8, 1074–1083. [DOI] [PubMed] [Google Scholar]
- (16).Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, and Hart GW (2008) O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem 283, 16283–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yang X, Su K, Roos MD, Chang Q, Paterson AJ, and Kudlow JE (2001) O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. U. S. A 98, 6611–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kelly WG, Dahmus ME, and Hart GW (1993) RNA polymerase II is a glycoprotein. Modification of the carboxy-terminal domain by O-GlcNAc. J. Biol. Chem 268, 10416–10424. [PubMed] [Google Scholar]
- (19).Lu L, Fan D, Hu CW, Worth M, Ma ZX, and Jiang J (2016) Distributive O-GlcNAcylation on the highly repetitive C-terminal domain of RNA polymerase II. Biochemistry 55, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, and Lewis BA (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem 287, 23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Resto M, Kim BH, Fernandez AG, Abraham BJ, Zhao K, and Lewis BA (2016) O-GlcNAcase is an RNA pol II elongation factor coupled to pausing factors SPT5 and TIF1β. J. Biol. Chem 291, 22703–22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, Copeland RJ, Despa F, Hart GW, Ripplinger CM, and Bers DM (2013) Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature 502, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hardiville S, and Hart GW (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bond MR, and Hanover JA (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol 208, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pekkurnaz G, Trinidad JC, Wang X, Kong D, and Schwarz TL (2014) Glucose regulates mitochondrial motility via milton modification by O-GlcNAc transferase. Cell 158, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Guo B, Liang Q, Li L, Hu Z, Wu F, Zhang P, Ma Y, Zhao B, Kovacs AL, Zhang Z, Feng D, Chen S, and Zhang H (2014) O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol 16, 1215–1226. [DOI] [PubMed] [Google Scholar]
- (27).Issad T, Masson E, and Pagesy P (2010) O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 36, 423–435. [DOI] [PubMed] [Google Scholar]
- (28).Lagerloef O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, and Huganir RL (2016) The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, and Walker S (2013) HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wysocka J, and Herr W (2003) The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci 28, 294–304. [DOI] [PubMed] [Google Scholar]
- (31).Janetzko J, Trauger SA, Lazarus MB, and Walker S (2016) How the glycosyltransferase OGT catalyzes amide bond cleavage. Nat. Chem. Biol 12, 899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Slawson C, and Hart GW (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Banerjee PS, Ma J, and Hart GW (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U. S. A 112, 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Johnson GVW, and Stoothoff WH (2004) Tau phosphorylation in neuronal cell function and dysfunction. J. Cell Sci 117, 5721–5729. [DOI] [PubMed] [Google Scholar]
- (35).Gong CX, Liu F, and Iqbal K (2016) O-GlcNAcylation: A regulator of tau pathology and neurodegeneration. Alzheimer’s Dementia 12, 1078–1089. [DOI] [PubMed] [Google Scholar]
- (36).Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, and Vocadlo DJ (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol 4, 483–490. [DOI] [PubMed] [Google Scholar]
- (37).Lazarus MB, Nam YS, Jiang J, Sliz P, and Walker S (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lazarus MB, Jiang J, Gloster TM, Zandberg WF, Whitworth GE, Vocadlo DJ, and Walker S (2012) Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol 8, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Schimpl M, Zheng X, Borodkin VS, Blair DE, Ferenbach AT, Schuettelkopf AW, Navratilova I, Aristotelous T, Albarbarawi O, Robinson DA, MacNaughtan MA, and van Aalten DMF (2012) O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol 8, 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, and Conti E (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol 11, 1001–1007. [DOI] [PubMed] [Google Scholar]
- (41).Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, Schuttelkopf AW, Albarbarawi O, and van Aalten DMF (2015) The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol 22, 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng LH, and Vocadlo DJ (2011) Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol 7, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Borodkin VS, Schimpl M, Gundogdu M, Rafie K, Dorfmueller HC, Robinson DA, and van Aalten DMF (2014) Bisubstrate UDP-peptide conjugates as human O-GlcNAc transferase inhibitors. Biochem. J 457, 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gross BJ, Kraybill BC, and Walker S (2005) Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc 127, 14588–14589. [DOI] [PubMed] [Google Scholar]
- (45).Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, Duveau DY, Tan ZW, Thomas CJ, and Walker S (2015) A small molecule that inhibits OGT activity in cells. ACS Chem. Biol 10, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Angelova M, Ortiz-Meoz RF, Walker S, and Knipe DM (2015) Inhibition of O-linked N-acetylglucosamine transferase reduces replication of herpes simplex virus and human cytomegalovirus. J. Virol 89, 8474–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Jiang J, Lazarus MB, Pasquina L, Sliz P, and Walker S (2012) A neutral diphosphate mimic crosslinks the active site of human O-GlcNAc transferase. Nat. Chem. Biol 8, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kim EJ, Kang DO, Love DC, and Hanover JA (2006) Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res 341, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li J, Huang CL, Zhang LW, Lin L, Li ZH, Zhang FW, and Wang P (2010) Isoforms of human O-GlcNAcase show distinct catalytic efficiencies. Biochemistry 75, 938–943. [DOI] [PubMed] [Google Scholar]
- (50).Dennis RJ, Taylor EJ, Macauley MS, Stubbs KA, Turkenburg JP, Hart SJ, Black GN, Vocadlo DJ, and Davies GJ (2006) Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol 13, 365–371. [DOI] [PubMed] [Google Scholar]
- (51).He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ (2010) Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J. Am. Chem. Soc 132, 1807–1809. [DOI] [PubMed] [Google Scholar]
- (52).Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, and Van Aalten DMF (2006) Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pathak S, Dorfmueller HC, Borodkin VS, and van Aalten DMF (2008) Chemical dissection of the link between Streptozotocin, O-GlcNAc, and pancreatic cell death. Chem. Biol 15, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Schimpl M, Borodkin VS, Gray LJ, and van Aalten DMF (2012) Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem. Biol 19, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Schimpl M, Schuettelkopf AW, Borodkin VS, and van Aalten DMF (2010) Human OGA binds substrates in a conserved peptide recognition groove. Biochem. J 432, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA, and van Aalten DMF (2015) Evidence for a functional O-linked N-acetylglucosamine (O-GlcNAc) system in the thermophilic bacterium Thermobaculum terrenum. J. Biol. Chem 290, 30291–30305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Macauley MS, Whitworth GE, Debowski AW, Chin D, and Vocadlo DJ (2005) O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem 280, 25313–25322. [DOI] [PubMed] [Google Scholar]
- (58).Cetinbas N, Macauley MS, Stubbs KA, Drapala R, and Vocadlo DJ (2006) Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844. [DOI] [PubMed] [Google Scholar]
- (59).Dong DL, and Hart GW (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem 269, 19321–19330. [PubMed] [Google Scholar]
- (60).Horsch M, Hoesch L, Vasella A, and Rast DM (1991) N-acetylglucosaminono-1,5-lactone oxime and the corresponding (phenylcarbamoyl)oxime. Novel and potent inhibitors of β-N-acetylglucosaminidase. Eur. J. Biochem 197, 815–818. [DOI] [PubMed] [Google Scholar]
- (61).Dorfmueller HC, Borodkin VS, Schimpl M, Zheng X, Kime R, Read KD, and van Aalten DMF (2010) Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem. Biol 17, 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Knapp S, Vocadlo D, Gao Z, Kirk B, Lou J, and Withers SG (1996) NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc 118, 6804–6805. [Google Scholar]
- (63).Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, and Vocadlo DJ (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol 8, 393–399. [DOI] [PubMed] [Google Scholar]
- (64).Bergeron-Brlek M, Goodwin-Tindall J, Cekic N, Roth C, Zandberg WF, Shan X, Varghese V, Chan S, Davies GJ, Vocadlo DJ, and Britton R (2015) A convenient approach to stereoisomeric iminocyclitols: generation of potent brain-permeable OGA inhibitors. Angew. Chem., Int. Ed 54, 15429–15433. [DOI] [PubMed] [Google Scholar]
- (65).Macauley MS, and Vocadlo DJ (2010) Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta, Gen. Subj 1800, 107–121. [DOI] [PubMed] [Google Scholar]
- (66).Ostrowski A, and van Aalten DMF (2013) Chemical tools to probe cellular O-GlcNAc signalling. Biochem. J 456, 1–12. [DOI] [PubMed] [Google Scholar]
- (67).Hanover JA, Cohen CK, Willingham MC, and Park MK (1987) O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem 262, 9887–9894. [PubMed] [Google Scholar]
- (68).Comer FI, Vosseller K, Wells L, Accavitti MA, and Hart GW (2001) Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem 293, 169–177. [DOI] [PubMed] [Google Scholar]
- (69).Reeves RA, Lee A, Henry R, and Zachara NE (2014) Characterization of the specificity of O-GlcNAc reactive antibodies under conditions of starvation and stress. Anal. Biochem 457, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Rios P, Carter TS, Mooibroek TJ, Crump MP, Lisbjerg M, Pittelkow M, Supekar NT, Boons GJ, and Davis AP (2016) Synthetic receptors for the high-affinity recognition of O-GlcNAc derivatives. Angew. Chem., Int. Ed 55, 3387–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Machon O, Baldini S, Ribeiro J, Steenackers A, Varrot A, Lefebvre T, and Imberty A (2017) Recombinant fungal lectin as a new tool to investigate O-GlcNAcylation processes. Glycobiology 27, 123. [DOI] [PubMed] [Google Scholar]
- (72).Mariappa D, Selvan N, Borodkin VS, Alonso J, Ferenbach AT, Shepherd C, Navratilova IH, and van Aalten DMF (2015) A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development. Biochem. J 470, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, and Bertozzi CR (2003) A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A 100, 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Chuh KN, Zaro BW, Piller F, Piller V, and Pratt MR (2014) Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J. Am. Chem. Soc 136, 12283–12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Zaro BW, Yang YY, Hang HC, and Pratt MR (2011) Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1. Proc. Natl. Acad. Sci. U. S. A 108, 8146–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Kim EJ (2015) The utilities of chemical reactions and molecular tools for O-GlcNAc proteomic studies. ChemBioChem 16, 1397–1409. [DOI] [PubMed] [Google Scholar]
- (77).Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, and Bertozzi CR (2011) Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. U. S. A 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Zhu Y, Liu TW, Cecioni S, Eskandari R, Zandberg WF, and Vocadlo DJ (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol 11, 319–325. [DOI] [PubMed] [Google Scholar]
- (79).Li J, Wang J, Wen L, Zhu H, Li S, Huang K, Jiang K, Li X, Ma C, Qu J, Parameswaran A, Song J, Zhao W, and Wang PG (2016) An OGA-resistant probe allows specific visualization and accurate identification of O-GlcNAc-modified proteins in cells. ACS Chem. Biol 11, 3002–3006. [DOI] [PubMed] [Google Scholar]
- (80).Spaete AK, Schart VF, Haefner J, Niederwieser A, Mayer TU, and Wittmann V (2014) Expanding the scope of cyclopropene reporters for the detection of metabolically engineered glycoproteins by Diels-Alder reactions. Beilstein J. Org. Chem 10, 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Robinson PV, Tsai C, de Groot AE, McKechnie JL, and Bertozzi CR (2016) Glyco-seek: ultrasensitive detection of protein-specific glycosylation by proximity ligation PCR. J. Am. Chem. Soc 138, 10722–10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, and Hsieh-Wilson LC (2008) Direct ingel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc 130, 11576–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, and Hart GW (2010) Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 9, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Verhelst SHL, Fonovic M, and Bogyo M (2007) A mild chemically cleavable linker system for functional proteomic applications. Angew. Chem., Int. Ed 46, 1284–1286. [DOI] [PubMed] [Google Scholar]
- (85).Szychowski J, Mahdavi A, Hodas JJL, Bagert JD, Ngo JT, Landgraf P, Dieterich DC, Schuman EM, and Tirrell DA (2010) Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J. Am. Chem. Soc 132, 18351–18360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Woo CM, Spiciarich DR, Palaniappan KK, Iavarone AT, and Bertozzi CR (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Griffin ME, Jensen EH, Mason DE, Jenkins CL, Stone SE, Peters EC, and Hsieh-Wilson LC (2016) Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag. Mol. BioSyst 12, 1756–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Wang X, Yuan ZF, Fan J, Karch KR, Ball LE, Denu JM, and Garcia BA (2016) A novel quantitative mass spectrometry platform for determining protein O-GlcNAcylation dynamics. Mol. Cell. Proteomics 15, 2462–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kim EJ (2011) Chemical arsenal for the study of O-GlcNAc. Molecules 16, 1987–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, and Hunt DF (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 101, 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Zubarev RA, Kelleher NL, and McLafferty FW (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc 120, 3265–3266. [DOI] [PubMed] [Google Scholar]
- (92).Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, and Mohammed S (2012) Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem 84, 9668–9673. [DOI] [PubMed] [Google Scholar]
- (93).Marino F, Bern M, Mommen GPM, Leney AC, van Gaans-van den Brink JAM, Bonvin AMJJ, Becker C, van Els CACM, and Heck AJR (2015) Extended O-GlcNAc on HLA class-I-bound peptides. J. Am. Chem. Soc 137, 10922–10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, and Hsieh-Wilson LC (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol 6, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Carrillo LD, Krishnamoorthy L, and Mahal LK (2006) A cellular FRET-based sensor for β-O-GlcNAc, a dynamic carbohydrate modification involved in signaling. J. Am. Chem. Soc 128, 14768–14769. [DOI] [PubMed] [Google Scholar]
- (96).Lin W, Gao L, and Chen X (2015) Protein-specific imaging of O-GlcNAcylation in single cells. ChemBioChem 16, 2571–2575. [DOI] [PubMed] [Google Scholar]
- (97).Doll F, Buntz A, Spaete AK, Schart VF, Timper A, Schrimpf W, Hauck CR, Zumbusch A, and Wittmann V (2016) Visualization of protein-specific glycosylation inside living cells. Angew. Chem., Int. Ed 55, 2262–2266. [DOI] [PubMed] [Google Scholar]
- (98).Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, Zachara N, Etzkorn FA, Hart GW, Jeong JS, Blackshaw S, Zhu H, and Cole PA (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol 8, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, and Pratt MR (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson’s disease. Nat. Chem 7, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Schwagerus S, Reimann O, Despres C, Smet-Nocca C, and Hackenberger CPR (2016) Semi-synthesis of a tag-free O-GlcNAcylated tau protein by sequential chemoselective ligation. J. Pept. Sci 22, 327–333. [DOI] [PubMed] [Google Scholar]
- (101).Wang J, Schiller SM, and Schultz PG (2007) A biosynthetic route to dehydroalanine-containing proteins. Angew. Chem., Int. Ed 46, 6849–6851. [DOI] [PubMed] [Google Scholar]
- (102).Chalker JM, Gunnoo SB, Boutureira O, Gerstberger SC, Fernandez-Gonzalez M, Bernardes GJL, Griffin L, Hailu H, Schofield CJ, and Davis BG (2011) Methods for converting cysteine to dehydroalanine on peptides and proteins. Chem. Sci 2, 1666–1676. [Google Scholar]
- (103).Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, Schofield CJ, and Davis BG (2015) Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun 6, 7978–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Raj R, Lercher L, Mohammed S, and Davis BG (2016) Synthetic nucleosomes reveal that GlcNAcylation modulates direct interaction with the FACT complex. Angew. Chem., Int. Ed 55, 8918–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Wright TH, Bower BJ, Chalker JM, Bernardes GJL, Wiewiora R, Ng WL, Raj R, Faulkner S, Vallee MRJ, Phanumartwiwath A, Coleman OD, Thezenas ML, Khan M, Galan SRG, Lercher L, Schombs MW, Gerstberger S, Palm-Espling ME, Baldwin AJ, Kessler BM, Claridge TDW, Mohammed S, and Davis BG (2016) Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 354, 597. [DOI] [PubMed] [Google Scholar]
- (106).Yang A, Ha S, Ahn J, Kim R, Kim S, Lee Y, Kim J, Soell D, Lee HY, and Park HS (2016) A chemical biology route to site-specific authentic protein modifications. Science 354, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, and Kohler JJ (2012) Metabolic labeling enables selective photocrosslinking of O-GlcNac-modified proteins to their binding partners. Proc. Natl. Acad. Sci. U. S. A 109, 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Rodriguez AC, Yu SH, Li B, Zegzouti H, and Kohler J (2015) Enhanced transfer of a photocross-linking N-acetylglucosamine (GlcNAc) analog by an O-GlcNAc transferase mutant with converted substrate specificity. J. Biol. Chem 290, 22638–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]


