Abstract
Sylvatic populations of Triatoma infestans represent a challenge to Chagas disease control as they are not targeted by vector control activities and may play a key role in post-spraying house re-infestation. Understanding sylvatic foci distribution and gene flow between sylvatic and domestic populations is crucial to optimize vector control interventions and elucidate the development and spread of insecticide resistance. Herein, the genetic profiles of five Andean T. infestans populations from Bolivia with distinct insecticide susceptibility profiles were compared. Multilocus genotypes based on eight microsatellites and the DNA sequence of a fragment of the cytochrome B (cytB) gene were obtained for 92 individuals. CytB haplotypes were analyzed with previously reported Bolivian T. infestans haplotypes to evaluate putative historical gene flow among populations. Each specimen was also screened for two nucleotide mutations in the sodium channel gene (kdr), related to pyrethroid resistance (L1014 and L9251). Significant genetic differentiation was observed among all populations, although individuals of admixed origin were detected in four of them. Notably, the genetic profiles of adjacent domestic and sylvatic populations of Mataral, characterized by higher levels of insecticide resistance, support their common ancestry. Only one sylvatic individual from Mataral carried the kdr mutation L1014, suggesting that this mechanism is unlikely to cause the altered insecticide susceptibility observed in these populations. However, as the resistance mutation is present in the area, it has the potential to be selected under insecticidal pressure. Genetic comparisons of these populations suggest that insecticide resistance is likely conferred by ancient trait(s) in T. infestans sylvatic populations, which are capable of invading domiciles. These results emphasize the need for stronger entomological surveillance in the region, including early detection of house invasion, particularly post-spraying, monitoring for resistance to pyrethroids and the design of integrative control actions that consider sylvatic foci around domestic settings and their dispersion dynamics.
Keywords: Triatoma infestans, Bolivia, Insecticide resistance, Sylvatic populations, Genetic structure, Gene flow
1. Introduction
Chagas disease is caused by Trypanosoma cruzi, a protozoan parasite that infects humans as well as domestic and sylvatic mammals. This disease can be transmitted by approximately 150 species of hemipteran insect vectors of which Triatoma infestans (Klug 1834) (Hemiptera: Reduviidae, Triatominae) is the most important in the southern countries of Latin America (Gurtler et al., 2008; Schofield et al., 2006). The most recent reports estimate that Chagas disease affects approximately 6 million people worldwide; approximately 70 million people are at risk of infection in 21 Latin American countries (WHO, 2015).
Domiciliary populations of T. infestans have been successfully controlled by spraying with pyrethroid insecticides in parts of southern South America (Schofield et al., 2006; Zerba, 1999); this contributed to the reduction in the geographical distribution of T. infestans from an estimated 6.28 million km2 in the 1960s to less than 1 million km2 today (Gorla, 2002; Schofield et al., 2006). Moreover, several countries and regions in South America, have been certified as having interrupted domestic vector transmission (Coura et al., 2014).
T. infestans has been found in sylvatic foci in several areas in the Gran Chaco and Andean regions, some reportedly invading houses, and are associated with T. cruzi transmission to humans (Breniere et al., 2017; Buitrago et al., 2013; Buitrago et al., 2016; Buitrago et al., 2010; Ceballos et al., 2009; Ceballos et al., 2011; Cortez et al., 2007; Noireau et al., 2000; Noireau et al., 2005; Rolón et al., 2011; Waleckx et al., 2012; Waleckx et al., 2011). Sylvatic populations are not targeted by vector control interventions and could re-invade houses treated with insecticide, especially if the population has resistant variants that can be selected under insecticidal pressure. Certain sylvatic populations from Bolivia have shown low susceptibility to pyrethroid insecticides (Roca-Acevedo et al., 2011; Depickère et al., 2012), and some also exhibited a lower sensitivity to fenitrothion (Santo-Orihuela et al., 2013). Furthermore, in the Andean region, post-spraying household reinfestation has been attributed partially to the dispersal of individuals from sylvatic populations (Noireau et al., 2005; Ceballos et al., 2011; Breniere et al., 2013; Waleckx et al., 2015) and domestic pyrethroid resistance has also been reported (Gomez et al., 2014).
Pyrethroid resistance in T. infestans was initially described in Argentina and Bolivia in the late 1990s. Increasing reports indicate that multiple natural populations are resistant to insecticides, and significant variability in susceptibility levels has been observed among them (Germano et al., 2010a; Mougabure-Cueto and Picollo, 2015; Pessoa et al., 2015; Picollo et al., 2005), at both the macro-geographic scale and even between ecotopes within the same household and peridomicile (Echeverria et al., 2018). Based on identification of a hybrid zone composed of Andean and non-Andean T. infestans from the Argentinean-Bolivian border, it has been proposed that the elimination of T. infestans by pyrethroid insecticides in Brazil, Chile, Uruguay and parts of Argentina and Paraguay indicated that resistance was less common in non-Andean regions and that Andean populations may have a particular genetic background that enabled the development of resistance more rapidly than non-Andean populations (Panzera et al., 2014). However, other studies have demonstrated that some of the highest resistance levels ever reported in T. infestans were in non-Andean individuals in Argentina (Picollo et al., 2005; Fronza et al., 2016); the evolutionary origins of pyrethroid resistance in T. infestans remains debated (Panzera et al., 2014). In these latter populations with extremely high pyrethroid resistance, which required 1000 times the amount of insecticide to kill the same proportion of susceptible individuals (Fronza et al., 2016), resistance was implicated as the cause of local control program failure. Similarly, in other parts of the Gran Chaco, rapid house re-infestation post-spraying and insecticide resistance are growing causes for concern (Cavallo et al., 2016; Cecere et al., 2006; Depickère et al., 2012; Gurevitz et al., 2011; Gurevitz et al., 2013; Lardeux et al., 2010; Piccinali et al., 2018; Cecere et al., 2019; Perez-Cascales et al., 2020).
Resistance traits are considered pre-adaptive, implying that there are a few resistant insects in each natural population, which would survive under pressure of insecticide exposure, selecting for the resistant genetic variants that would be inherited and in turn, become more frequent in subsequent generations. The dispersal rate of resistant traits will depend on pre-existing genetic variation in natural populations, selection intensity, fitness cost associated with expressing the resistant variant, local adaptation and levels of gene flow among populations. Such resistance dynamics have been reported in populations of other medically important vector species, particularly Aedes aegypti (Aguirre-Obando et al., 2015; Marcombe et al., 2013).
Elucidating population genetic dynamics through high resolution genetic markers in the context of well characterized insecticide resistance profiles could test these hypotheses and provide invaluable information for an integrated vector control approach. Establishing the role of sylvatic populations of T. infestans as potential sources of re-infestation and understanding the impact of insecticide resistance on triatomine population genetic diversity is crucial to safeguard the continued effectiveness of current control efforts. Previous studies using high resolution molecular markers demonstrated that genetic differentiation is detectable up to the capture site level (local populations) if sufficient numbers of bugs are collected per site to constitute a representative population for genetic analysis (Breniere et al., 2013; Marcet et al., 2008; Pérez de Rosas et al., 2008; Perez de Rosas et al., 2013).
The aim of this work was to establish and characterize the genetic profiles of four sylvatic and one domestic Andean populations of T. infestans from Bolivia, and to interpret their genetic relationships in the context of their differing levels of insecticide susceptibility. Moreover, an evaluation of the mechanisms likely responsible for pyrethroid resistance observed in these populations, including esterase enzymes and kdr mutations is discussed.
2. Materials and methods
2.1. Insect origin
Collection methods were described in detail in (Roca-Acevedo et al., 2011). Briefly, T. infestans populations were sampled from domiciliary areas in Mataral, Cochabamba Department, Bolivia (hereby named Mat-D) and a sylvatic area located approximately 2 km away (Mat-S). Sylvatic bugs were also collected from additional sites in Cochabamba (Ilicuni and 20 de Octubre) and Potosí (Kirus Mayu) Departments. Sylvatic triatomines were sampled from rock-piles using mouse-baited sticky traps (Noireau et al., 1999). From each population a colony was established with a minimum of 10 founder individuals, and maintained at 28 ± 1 °C, 50% RH, with a photoperiod of 12:12 (L:D) h at the Centro de Investigaciones de Plagas e Insecticidas (CIPEIN, CITEFA-CONICET), Buenos Aires, Argentina. Rearing conditions are described in detail elsewhere (Núñez and Segura, 1987; Picollo et al., 1976). The insecticide resistance profile of F1 individuals from these populations was established by toxicological bioassays, performed according to World Health Organization (WHO, 1994) standard protocols (Roca-Acevedo et al., 2011), after which these samples were preserved for molecular analysis. Genomic DNA was obtained from three legs of each bug using the Wizard Genomic Purification Kit (Promega®) following the manufacturer recommendations.
2.2. Multilocus microsatellite genotyping
A multilocus genotype (MLG) was obtained for each individual based initially on nine microsatellite loci, developed and optimized for T. infestans, following the amplification conditions previously described (Marcet et al., 2006; Marcet et al., 2008). Five loci were amplified individually (Tinf_ms3, Tinf_ms19, Tinf_ms23, Tinf_ms56 and Tinf_ms64) and multiplex PCR reactions were carried out with 2 pairs of loci with the same annealing temperature and different dye colors (Tinf_ms5 with Tinf_ms42 and Tinf_ms22 with Tinf_ms27). DNA fragment detection with 1 bp resolution was performed with an automated DNA sequencer (ABI 3130, Applied Biosystems) and size determination was obtained with GeneMapper 4.1 (Applied Biosystems). Ninety-two colony-reared, second-third generation individuals were sampled for microsatellite genotyping (Table 1).
Table 1.
Number of bugs per population evaluated per molecular marker:
| Collection site | msat | cytB | kdr | LD501 (95% CL) | RR1 (95% CL) |
|---|---|---|---|---|---|
| NFS* | - | - | - | 0.13 (0.11–0.15) | - |
| Mataral-D (Mat-D) | 19 | 19 | 13 | 2.25 (0.28–4.80) | 17.4 (11.88–25.43) |
| Mataral-S (Mat-S) | 11 | 11 | 6 | 1.53 (0.53–3.34) | 11.9 (9.43–14.93) |
| Kirus Mayu (KM) | 22 | 23 | 15 | 0.95 (0.49–1.66) | 7.4 (5.78–9.25) |
| flicuni (Ili) | 18 | 18 | 16 | 0.25 (0.14–0.39) | 1.9 (1.44–2.56) |
| 20 de Octubre (20oct) | 22 | 22 | 17 | 0.88 (0.08–1.84) | 6.8 (4.98–9.26) |
msat = microsatellites; cytB = cytochrome B; kdr: evaluated for 2 mutations in the sodium channel gene (L1014 and L9251). Toxicity of topically applied deltamethrin in first instars nymphs of T. infestans from Bolivia (1Data previously published (Roca-Acevedo et al., 2011)).
Susceptible strain reared at CIPEIN. LD50: Lethal dose to kill 50% of the sample measured in ng/insect; CL = confidence limit; RR: resistance ratio.
2.3. Microsatellite analysis
Allele number per locus and population were obtained directly after binning, and mean allele number among all loci were compared. The inbreeding coefficient Fis (Weir and Cockerham, 1984); allelic richness Ar (El Mousadik and Petit, 1996), gene diversity or expected heterozygosity (He) and observed heterozygosity (Ho) (Nei, 1987) were obtained with the Excel add-in MS_tools.xla for Microsoft Excel (Office 365) or with FSTAT2.9.3.2 (Goudet, 1995). Bonferroni correction was applied to determine p-values for multiple comparisons (Rice, 1989). The fit to Hardy-Weinberg (HW) equilibrium expectations was evaluated using the U score test available in Genepop 4.2 (Raymond and Rousset, 1995), under the null hypothesis of random union of gametes. Sample-size-corrected private (population-specific) allele frequency per locus (PA/L) was calculated in HP-RARE (Kalinowski, 2005). Multilocus linkage disequilibrium, estimated by the index of association (IA), was calculated in MULTILOCUS 1.3b (Agapow and Burt, 2001) and statistical significance was evaluated by comparison with a null distribution of 1000 randomizations.
Pairwise FST values and significance levels were calculated with ARLEQUIN 3.01 (Excoffier et al., 2005), as in Weir and Cockerham (1984). A Fisher exact test for population differentiation comparing genic and genotypic frequencies was implemented in Genepop 4.2 (Raymond and Rousset, 1995). Population clustering was explored using a neighbor-joining (NJ) tree based on pairwise distances (DAS: 1 proportion of shared alleles at all loci/n). A Mantel test for the effect of isolation by distance within populations (pairwise genetic vs. geographic distance) was performed in GenAlEx 6.5 using 10,000 random permutations (Peakall and Smouse, 2012). A Hierarchical Analysis of Molecular Variance (AMOVA) (Excoffier et al., 1992) was performed at three levels of population structure: among communities, among populations and among individuals in a population, in ARLEQUIN 3.01 (Excoffier et al., 2005).
2.4. Population structure
Population genetic structure was examined using the Bayesian model-based approach (Falush et al., 2007; Hubisz et al., 2009; Pritchard et al., 2000) implemented in STRUCTURE 2.3.4. Each individual MLG in the sample is probabilistically assigned to one of K populations, or jointly to two or more populations if their genotypes might have an admixed origin. Simultaneously, the method determines the number of significant K genetic clusters within the total sample. The number of clusters evaluated (=K) ranged from 1 to 10. The analysis was performed using 35 replicate runs per K value, a burn-in period length of 50,000 and a run length of 50,000. The analysis model used was the admixed and correlated allele frequencies, with no prior information on the origin of the individuals. The final selection of the sample K value was based on the log-probability of the data between successive K values (Evanno et al., 2005) implemented in the online version of STRUCTURE HARVESTER (Earl and vonHoldt, 2012).
2.5. Mitochondrial (mtDNA) analysis
The cytB gene fragment was amplified as previously described (Monteiro et al., 2003) for a total of 74 specimens from the five populations (Table 1). SeqManPro Software 14.1 (DNAstar) was used to assemble and align forward (F) and reverse (R) chromatograms and obtain the consensus sequence for each individual. MEGA 5 software (Tamura et al., 2011) was used to align multiple sequences and to examine the phylogenetic relationships among sequences. Standard genetic variability (haplotype diversity: Hd; and nucleotide diversity: π) and differentiation among sequences were evaluated using DnaSP version 5.1 software (Librado and Rozas, 2009). A Nexus matrix was constructed for haplotype network analysis in PopART using a median-joining model based on 1000 iterations with default parameters (Bandelt et al., 1999; Leigh and Bryant, 2015).
To evaluate the possible directionality and history of gene flow among populations, we compared the mitochondrial sequences obtained in this work with previously reported Andean T. infestans haplotypes. Sequences were trimmed from 666 bp to 388 bp and a second haplotype network was built using the same software and criteria including 25 sequences from sylvatic Bolivian T. infestans previously deposited in GenBank: HQ333215-HQ333239 (Quisberth et al., 2011; Waleckx et al., 2011).
2.6. Screening for mutations in the kdr gene
A fragment of the DNA sequence of the sodium channel gene was amplified and screened for two nucleotide mutations previously reported in T. infestans associated with pyrethroid knockdown resistance (kdr) (Capriotti et al., 2014; Fabro et al., 2012). The protocol has been previously described (Santo-Orihuela et al., 2017) and was used to screen 67 individuals from the five populations (Table 1).
3. Results
3.1. Population diversity analysis
Ninety-two individual T. infestans samples were grouped a priori into five populations based on their geographical origin (Table 1): Mataral domestic (Mat-D), Mataral sylvatic (Mat-S), Ilicuni (Ili), 20 de Octubre (20oct) and Kirus Mayu (KM) and were genotyped across eight polymorphic microsatellites. In contrast with genotyping data from T. infestans in Argentina and Paraguay, the locus Tims_64 did not amplify consistently across samples. In addition, locus Tims_65 did not amplify in either domestic or sylvatic Mat populations, indicative of null alleles (and a common trait between these two adjacent populations, discrepant from the others); therefore, neither loci were utilized in the final population analysis.
A total of 84 unique individual MLGs were identified among populations (Table 2). Genetic diversity levels were high and consistent across most populations as evidenced by similar frequencies of unique MLGs, levels of allelic richness (3.1–5.4); distance between shared alleles (0.38–0.67); private alleles per locus (0.42–1.26) and average gene diversity (0.44–0.74) (Table 2). Only minor differences were observed between populations with larger numbers of founding individuals (e.g. 20oct and KM), indicating minimal genetic diversity was lost by rearing population members prior to genetic analysis.
Table 2.
Population genetic parameters for Bolivian T. infestans based on eight microsatellite loci.
| Population | #MLG8/N | PA/L ± SE | av# alleles | Fis ± SE | av PIC | av Ar | He ± SE | Ho | %PL Hd | DAS ± SE | IA | IA P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mataral-D | 18/19 | 0.55 ± 0.09 | 5 | 0.052 ± 0.03 | 0.57 | 4.51 ± 1.91 | 0.63 ± 0.07 | 0.6 | 50 | 0.55 ± 0.12 | 0.40 | 0.007 |
| Mataral-S | 10/11 | 1.26 ± 0.22 | 5.5 | 0.22 ± 0.04** | 0.65 | 5.37 ± 2.07 | 0.74 ± 0.05 | 0.57 | 100 | 0.67 ± 0.19 | 1.08 | <0.001 |
| Kirus Mayu | 20/22 | 0.45 ± 0.12 | 4.3 | 0.108 ± 0.03** | 0.47 | 3.69 ± 1.83 | 0.52 ± 0.09 | 0.47 | 85.7 | 0.45 ± 0.15 | 0.65 | <0.001 |
| Ilicuni | 18/18 | 0.75 ± 0.11 | 4.9 | 0.047 ± 0.05 | 0.58 | 4.34 ± 1.50 | 0.64 ± 0.07 | 0.61 | 62.5 | 0.54 ± 0.14 | 0.45 | 0.012 |
| 20 de Octubre | 18/22 | 0.42 ± 0.13 | 3.6 | −0.047 ± 0.07** | 0.37 | 3.06 ± 0.96 | 0.44 ± 0.06 | 0.46 | 50 | 0.34 ± 0.17 | 1.44 | <0.001 |
#MGL8/N: number of complete multilocus genotype for 8 loci / number of individuals genotyped per population; PA/L: private alleles per locus; av# alleles: average number of alleles per locus Fis: inbreeding coefficient (Weir and Cockerham, 1984)
Significant Heterozygote deficit (p < 0.05).; av PIC: averaged Polymorphism Information Content (Botstein et al., 1980). av Ar: averaged allele richness (El Mousadik and Petit, 1996); He: expected heterozygosity = average gene diversity (Nei, 1987); Ho: observed heterozygosity; % PL Hd: percentage of polymorphic loci displaying a deficit in heterozygosity; DAS = Pairwise distance between alleles. IA: index of association that suggests multilocus linkage disequilibrium if p < 0.05.
Although it had the lowest number of individuals available for testing, Mat-S displayed the highest parameters related to genetic diversity (average number of alleles = 5.5, Ar = 5.37, He = 0.74, av PIC = 0.65). Moreover, individuals from this area demonstrated the highest number of private alleles per locus (1.26) and Fis value (0.22) indicative that this may be an ancestral population with restricted intra-population gene flow (i.e. sub-structure) (Table 2). The populations 20oct, KM and Mat-S presented significant deviations from HW equilibrium, and all due to significant heterozygote deficits.
3.2. Inter-population subdivision and gene flow
Both genic and genotypic Fisher tests for pairwise population differentiation were highly significant for all pairs compared. A hierarchical AMOVA, which evaluated the distribution of genetic diversity, demonstrated that the majority of variation (71.5%) was within individuals, compared to 24.1% and 4.4% among populations and among individuals within populations, respectively.
A population neighbor joining (NJ) clustering analysis based on pair-wise genetic distances DAS (Nei et al., 1983) is shown in Fig. 1. The NJ topology was consistent with geographical origin and FST distances. The more proximate populations, both Mat populations, were grouped together and closer to Ili (separated by approximately 60 km), while the more distant populations of 20oct and KM were more closely related.
Fig. 1.
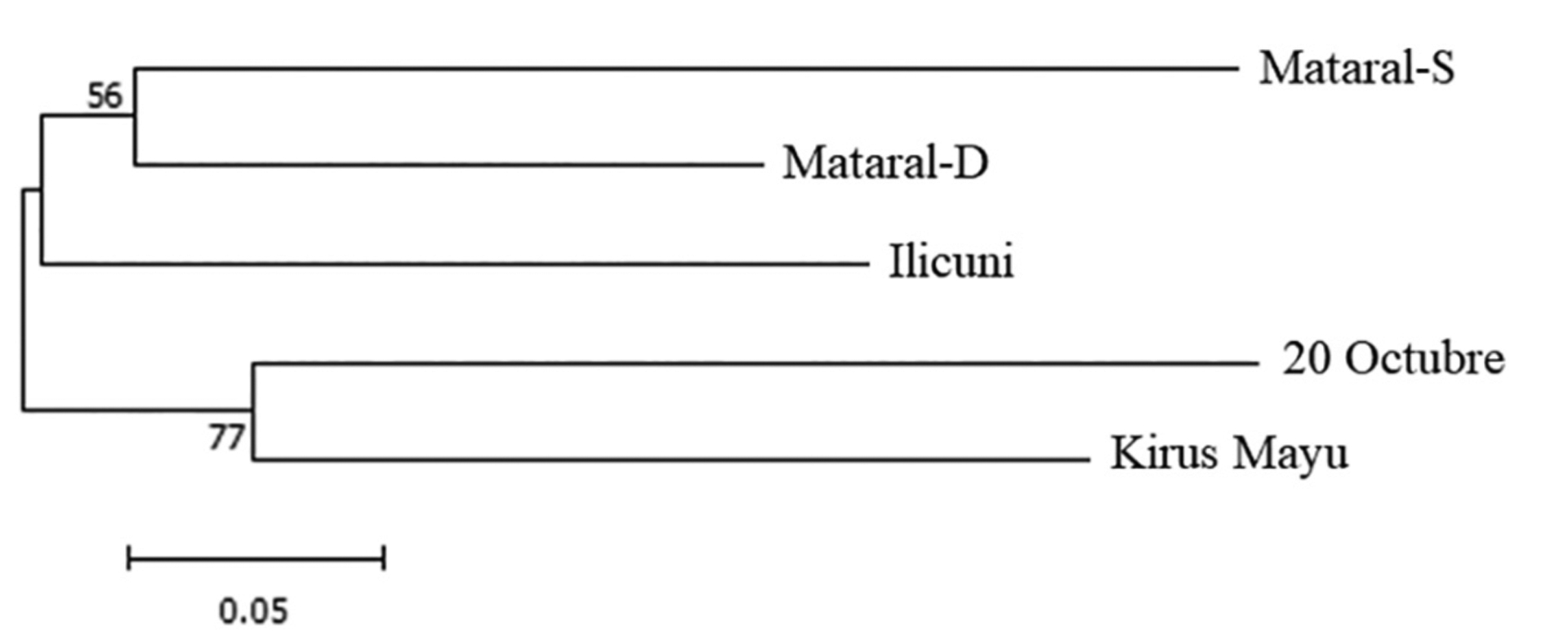
Neighbor joining (NJ) tree based on population pairwise DAS distances (Nei et al., 1983) estimated with the MLGs of 8 loci across 1000 pseudoreplicates.
Estimates of population subdivision based on allele frequency and identity (FST) indicated significant genetic differentiation between all populations, with the lowest FST values found between both populations of Mat and Ili (Table 3). FST values among sylvatic populations spanning a greater geographical distance (e.g. FST = 0.27 between Mat-S and 20oct; 162.5 km) were largely equivalent to those observed between domestic and sylvatic triatomines sampled from a more restricted area (FST = 0.22 between Mat-D and KM; 57 km) and between closer sylvatic populations (FST = 0.22 between Mat-S and KM 102.06 km), suggesting little gene flow occurred among populations throughout the geographic range considered. The Mantel test conducted to evaluate the significance of the correlation between the geographical and genetic distances between population pairs, showed a slight but significant adjustment to an isolation by distance model (RXY = 0.413; p< 0.01).
Table 3.
FST values (geographic distances) in a pair-wise comparisons between populations.
| 20oct | Ili | KM | Mat-S | Mat-D | |
|---|---|---|---|---|---|
| 20oct | - | ** | ** | ** | ** |
| Ili | 0.272 (151.76 km) | - | ** | ** | ** |
| KM | 0.323 (62.92 km) | 0.282 (104.63 km) | - | ** | ** |
| Mat-S | 0.272 (162.50 km) | 0.144 (56.02 km) | 0.223 (102.06 km) | - | ** |
| Mat-D | 0.278 (159.74 km) | 0.163 (56.94 km) | 0.216 (99.07 km) | 0.145 (3.38 km) | - |
(p < 0.005)
A Bayesian assignment of individual genotypes determined an optimal number of five genetic clusters to describe the dataset (Figs. 2 and 3), corresponding broadly to the five populations assigned by capture site. Most individual MLGs clustered significantly with other members from the same geographic location. However, in all sylvatic populations, individuals of admixed origin or first generation migrants were detected (Fig. 3).
Fig. 2.
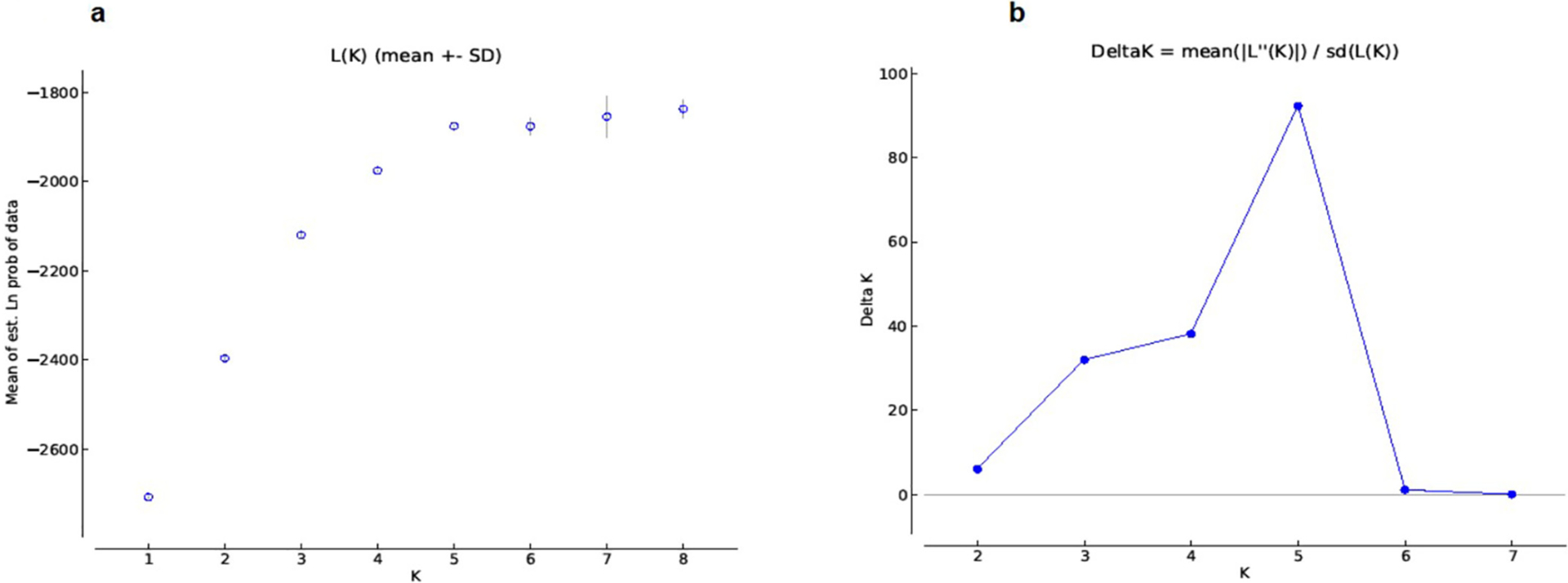
Bayesian cluster analysis results of T. infestans populations from sylvatic and domestic areas in Bolivia. a) Mean of estimated Ln likelihood of the data (Ln(P)) versus K number of genetic clusters considered (Pritchard et al., 2000). b) Graphic representation of Delta K = mean ((|L′′(K)|)/SD(L(K)) (Evanno et al., 2005) to evaluate the rate of change in the Ln(P) versus the number of genetic clusters considered (K = 1–10).
Fig. 3.
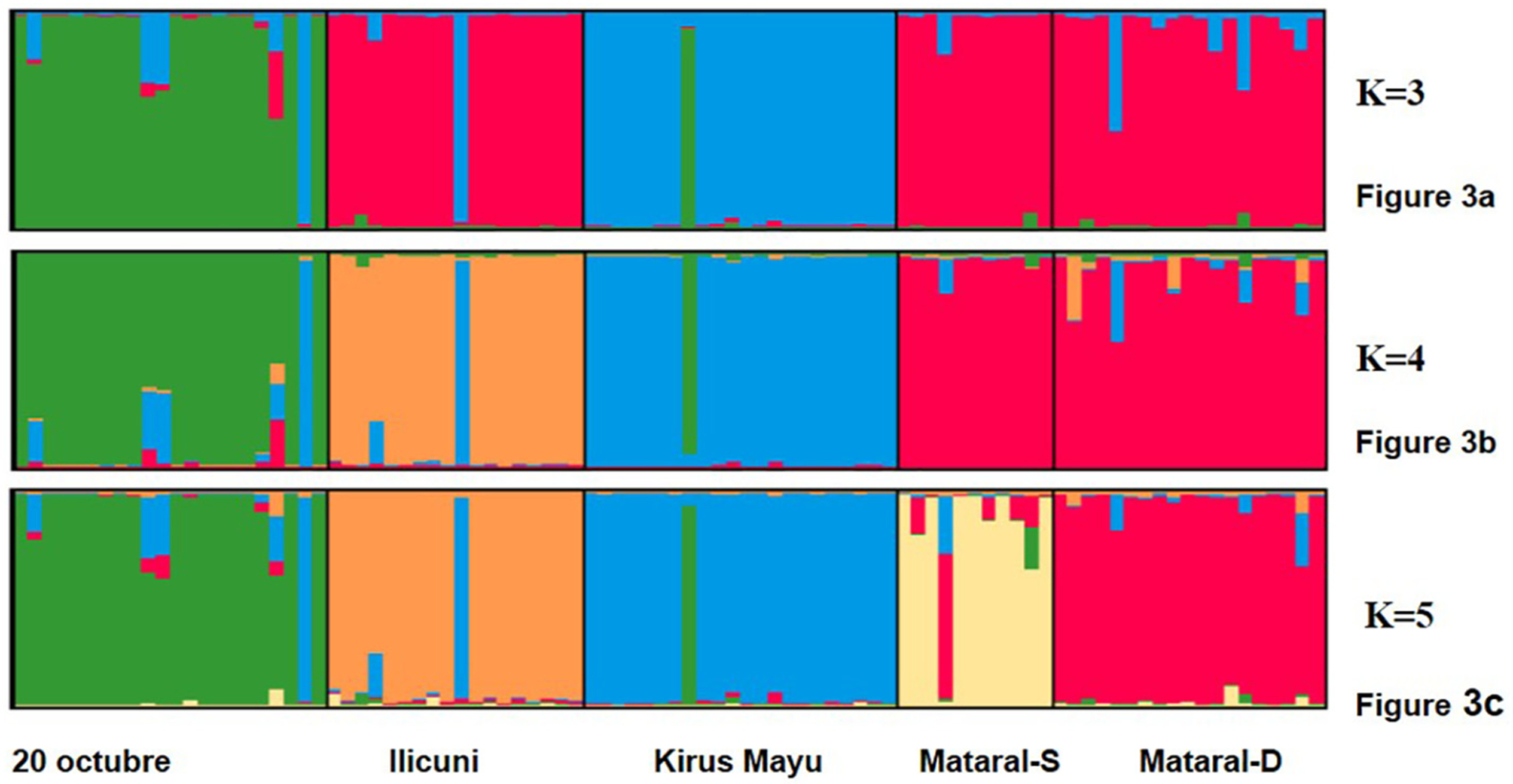
Bar plot representing the individual assignment probability of each MLG to a specific genetic cluster. Each vertical bar represents an individual and the different colors are the probability that each individual was assigned to a particular genetic cluster (K). The hierarchical approach allows for comparison of individual MLG assignment results when considering K = 3, K = 4 and K = 5 genetic clusters.
A hierarchical comparison of alternative clustering models reflects the relative genetic distance among MLGs. This analysis shows the resulting clustering pattern of MLGs from the whole sample when using different K values (i.e. considering alternative numbers of genetic clusters). The results are presented in Fig. 3, which showed the closer genetic relationship between both Mat populations and Ili, as the individuals from those populations clustered together when considering four or three possible genetic clusters to assign the whole dataset.
3.3. Mitochondrial genetic clustering among populations and individuals
A fragment of the cytB gene was amplified and sequenced from 74 individuals (20oct = 14, Ili = 15, KM = 15, Mat-S = 11 and Mat-D = 19) and assembled into a 666 bp alignment. A total of eight haplotypes were observed, including an ambiguous (i.e. heteroplasmid) sequence detected in two individuals from KM (ht; haplotypes in Table 4). All haplotypes are accessible from GenBank (MH763648 to MH763654). Haplotypes were determined by 15 variable sites that were mostly synonymous except for sites 28 and 625, carried by haplotypes XVI and XLIX (Table 4).
Table 4.
Variable sites among the cytochrome B (cytB) haplotypes detected in sylvatic populations of Andean T. infestans in Bolivia.
| Variable sites (nucleotide positions) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotypes | 28 | 162 | 279 | 402 | 450 | 480 | 526 | 549 | 564 | 576 | 588 | 600 | 625 | 646 | 636 |
| I | G | A | A | A | A | A | C | G | T | A | T | C | G | T | T |
| XXIV | . | . | . | G | G | . | T | . | . | T | . | . | . | . | . |
| XVI | A | G | . | . | . | . | . | . | . | T | . | . | . | . | C |
| ht | A | G | R | . | . | R | . | . | . | Y | . | . | . | Y | C |
| XLIX | A | G | G | . | . | G | . | . | . | C | . | . | A | C | C |
| XXVII | . | G | . | . | . | . | . | . | . | T | . | . | . | . | C |
| XLV | . | G | . | . | . | . | . | . | . | T | . | . | . | . | C |
| XLVI | . | . | . | . | . | . | . | A | . | T | C | . | . | . | . |
| XLVII | . | . | . | . | . | . | . | A | C | T | C | . | . | Y | C |
Bold font indicates non-synonymous substitutions. R and Y are the IUPAC nucleotide code indicating ambiguous base position (A/G) and (C/T) respectively. ht.: heteroplasmid sequence (i.e. individuals carrying more than one mitochondrial haplotype). Haplotype I was among the first one described for T. infestans in Bolivia (Monteiro et al., 1999) and found widely distributed in several populations since then. Italic font indicates the variable sites shared between haplotypes sequenced in this study and those previously deposited in GenBank for Bolivian Andean T.infestans populations.
The most common mitochondrial genotype was XVI, identified in 21 isolates from 20oct, Ili and KM, but absent in both Mat populations (Table 5 and Fig. 4). Mat-D and Ili displayed the highest haplotype diversity (Hd = 0.65 and 0.59, respectively), and were the populations that also presented the most diverse haplotypes (π = 0.0056 and 0.0044, respectively) (Table 5). Both Mat populations presented private unique haplotypes (XLV in Mat-D and XLVI and XLVII in Mat-S, respectively). Genetic divergence of Mat-S was also evidenced as only one haplotype (XXIV) was shared with other populations (Mat-D and Ili). All haplotypes found in the domestic population were shared or closely related to sylvatic ones (Fig. 4).
Table 5.
Cytochrome B (cytB) haplotype distribution per population and diversity parameters.
| Population | XXVII | XXIV | XVI | XLV | XLVI | XLVII | XLIX | ht | # cytB seqs | Hd ± SD | π ± SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mat-D | 7 | 9 | 3 | 19 | 0.649 ± 0.061 | 0.00437 ± 0.00033 | |||||
| Mat-S | 1 | 9 | 1 | 11 | 0.455 ± 0.17 | 0.00241 ± 0.0011 | |||||
| KM | 1 | 2 | 10 | 2 | 15 | 0.41 ± 0.154 | 0.00312 ± 0.0011 | ||||
| Ili | 6 | 8 | 1 | 15 | 0.59 ± 0.077 | 0.00563 ± 0.00097 | |||||
| 20oct | 11 | 3 | 14 | 0.363 ± 0.13 | 0.00272 ± 0.00098 | ||||||
| Total | 8 | 16 | 21 | 3 | 9 | 1 | 14 | 2 | 74 |
Hd = Haplotype diversity, π = nucleotide diversity, SD = standard deviation. ht = heteroplasmid.
Fig. 4.
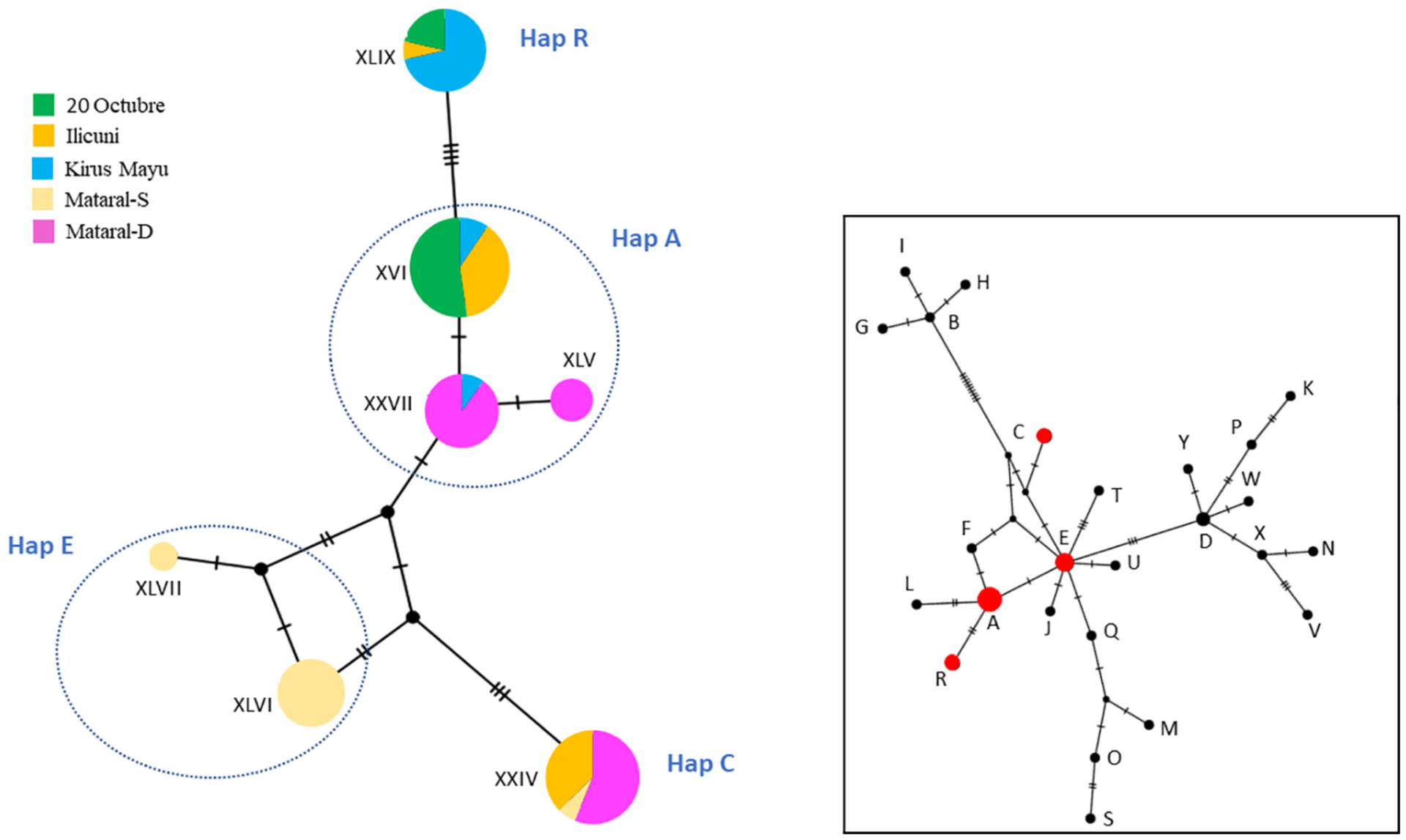
Left: haplotype network based on a median-joining model, constructed from sequences identified in this study. The size of the nodes is proportional to the abundance of each haplotype. The colors indicate the population of origin. The distance among haplotypes is represented by the number of mutational steps between them. Right: haplotype network including 25 additional Bolivian Andean T. infestans haplotypes (labelled A–Y) (Quisberth et al., 2011; Waleckx et al., 2011). Trimmed sequences from this study corresponded to four haplotypes previously reported: A, C, E and R (shown in red).
The network analysis, clustered haplotypes into two groups based on the substitution A/G in nucleotide position 162 (Table 4 and Fig. 4). One cluster contained haplotypes XLVI and XLVII (only detected in Mat-S) with XXIV, found in both Mat populations and Ili. The remaining haplotypes were found in all but the Mat-S population (Fig. 4).
In order to interpret the results of this study in the context of known phylogeographic patterns of T. infestans populations in Bolivia, we built a haplotype network with previously reported haplotypes for Bolivian T. infestans (Quisberth et al., 2011; Waleckx et al., 2011). As the sequences previously reported were only 388 bp long, the haplotypes we characterized were trimmed to that length for comparison. Consequently, some of our trimmed haplotypes were indistinguishable from each other within the fragment length considered, and were pooled into four haplotypes previously reported in Andean Bolivian populations: A, C, E and R (Waleckx et al., 2011), which are shown in red in Fig. 4.
Remarkably, the haplotype network built with sequences from previous studies of Bolivian T. infestans (Fig. 4) showed that the haplotypes found in Mat-S corresponded to a central node (i.e. ancient), haplotype E, found in populations from the “Central Andean area” collected in a sylvatic location in Mataral, Cochabamba, (Waleckx et al., 2011) and in domestic populations in Chuquisaca (Giordano et al., 2005; Waleckx et al., 2011). Another haplotype found in Mat-S, as well as in Mat-D and Ili (haplotype XXIV), corresponded to the previously reported haplotype C, a divergent haplotype considered a derivate from haplotype E, previously reported in domestic populations from Cochabamba (Waleckx et al., 2011; Waleckx et al., 2015).
In this comparison, the three haplotypes found in 20oct, KM, Ili and Mat-D populations, corresponded to that described as the most abundant haplotype A, previously detected in specimens from “Northern and Central Andean areas” (Waleckx et al., 2011). Finally the haplotype XLIX found in 20oct, KM and Ili populations, corresponded to haplotype R, considered a derivate of haplotype A (Waleckx et al., 2011). Haplotypes A, R and C had also been previously reported in domestic populations (Giordano et al., 2005).
3.4. Kdr screening
Among the sixty-seven samples screened for two mutations in the kdr gene, related to insecticide resistance in T. infestans, only one individual from Mat-S presented the resistant allele variant for the site L1014. No individual carrying the mutation L9251 was detected.
4. Discussion
Through high resolution genotyping of five Andean T. infestans populations, we provide insights into natural population genetic structure, the relationship between genetic diversity of sylvatic versus domestic triatomine populations and their respective insecticide resistance profiles. All populations were genetically distinct, with no evidence of current gene flow among them, which correlates with previously described heterogeneity in insecticide susceptibility profiles (Roca-Acevedo et al., 2011). The pattern of inter-population differentiation was consistent with a significant isolation-by-distance (IBD) model, with geographically closer populations being more genetically-related, specifically adjacent sylvatic-domestic populations of Mataral. This observation was supported by both mitochondrial and nuclear microsatellite markers. The genetic pattern of allelic similarity reflected possible shared ancestry and/or secondary contact between domestic and sylvatic Mat populations. This observation further supports previous reports of dynamic gene flow between sylvatic and domestic populations in the Bolivian Andes (Breniere et al., 2013; Quisberth et al., 2011; Waleckx et al., 2011).
Both Mat populations were characterized by considerably higher genetic diversity compared to the other sylvatic populations evaluated in this work; and between the Mat populations, genetic diversity and substructure were higher for the sylvatic population. The combination of high levels of homozygosity, number of private alleles per locus, number of private mitochondrial haplotypes and divergence of mitochondrial sequences, suggest that Mat-S may be ancestral to the other sylvatic populations evaluated here. This observation is supported by the occurrence of two mitochondrial haplotypes within this population that correspond to haplotype E (Fig. 4), which has been previously reported as an ancient haplotype among other Andean T. infestans populations (Waleckx et al., 2011).
The populations 20oct, KM and Mat-S presented significant deviations from HW equilibrium, and all due to significant heterozygote deficits. As the samples were arbitrarily grouped per community, the significant Fis values and hierarchical AMOVA, revealed the majority of variation was present within individuals, which was likely attributable to these population being comprised of admixed individuals from multiple, independent non-panmictic founding populations (Wahlund effect). This would also account for the higher linkage disequilibrium detected in these populations. High standard deviations associated with DAS values further supports the existence of intra-population sub-structuring. Alternatively, this genetic pattern could indicate that some members have undergone long-term inbreeding, following historical isolation. Both of these hypotheses would also be affected by our use of colony specimens (which may have undergone inbreeding during several generations of laboratory rearing) and not first-generation field captured individuals, a limitation of this study which must be acknowledged.
The bioassays performed in these populations demonstrated three different levels of altered susceptibility (Roca-Acevedo et al., 2011): high levels of decreased susceptibility in Mat-D and Mat-S, moderate levels of decreased susceptibility in KM and 20oct and limited altered susceptibility in Ili (Table 1). The resistance ratios (RRs) obtained for these populations are not as high as values from other areas that have presented difficulties for vector control (i.e. RR > 2000 in El Juramento, Guemes, Chaco, Argentina (Fronza et al., 2016), RR50 = 541.6 in Tierras Nuevas, Tarija, Bolivia (Germano et al., 2010b) or RR = 818 in Taiguati in the Bolivian Chaco (Depickère et al., 2012)). However, in all but Ili, the RR values are above the level classified as resistant by PAHO guidelines, RR > 5 (fivefold higher than reference populations) (PAHO, 2005; Pessoa et al., 2015). Those guidelines also attribute altered susceptibility in field populations presenting RR < 5 to individual variability, which is not enough to cause resistance at the population level. As previously discussed (Depickère et al., 2012; Santo-Orihuela et al., 2013), elevated RR in a sylvatic population might result from at least three non-mutually exclusive hypotheses:
existence of naturally altered (lower) susceptibility to insecticides
development of resistance resulting from exposure to insecticides used in agriculture and/or vector control campaigns
contact and exchange of genetic variants between sylvatic and domestic resistant populations
In the context of this study, the decreased susceptibility to deltamethrin observed in all populations may represent a shared ancestral trait, which has undergone further intensification in Mataral following insecticidal treatment, particularly for the domestic population, with possible gene flow between the two Mat populations. In this scenario, the intermediate RRs of 20oct and KM would have been maintained by moderate selection pressure, likely the indirect effect of agricultural spraying; the difference between them could be explained by intrinsic intra-species variability, consistent with the distinct genetic patterns observed. However, the minimal altered susceptibility observed in Ili would imply that this trait had been almost lost in this population in the absence of selection. This pattern would be consistent with a natural or ancestral tolerance trait of the species that evolved independently of insecticide application (Mougabure-Cueto and Picollo, 2015) but could have been selected subsequently under pressure.
Alternatively, insecticide resistance could have been acquired independently in all populations following different histories of insecticide exposure, with individual population RR directly corresponding to the intensity of selection each population has undergone. Recent modelling studies have also demonstrated the impact of key environmental factors, which may drive the evolution of insecticide resistance in T. infestans, particularly, temperature (proposed to influence triatomine physiological stress as well as insecticide toxicity levels), precipitation (found to be negatively correlated with residual insecticide efficacy) and village size (reflecting smaller effective population sizes that are more prone to stochastic effects, e.g. genetic drift or quicker fixation/loss of resistant alleles) (Fronza et al., 2019). Further characterization of the ecological context of the populations in these areas is necessary to evaluate these hypothesis or alternative explanations for the observed genetic patterns.
Insecticide-based interventions are expected to impact vector population structure and levels of genetic diversity. Selective bottlenecks imposed by insecticidal pressure are likely to result in both reductions in genetic variability and population size, such that a small proportion of survivors, possessing the requisite advantageous resistance genes, survive and reproduce (i.e. diversification that would be reflected in a genetic pattern as multiple independent “island effects”). In general, insecticide resistant populations are expected to be less genetically diverse than a wild-type population, as they have likely undergone a period of selective pressure (i.e. insecticide exposure). And within this context, domestic populations are assumed to have been the subject of greater selection pressures, compared to their sylvatic counterparts and therefore be less diverse. Recent studies of T. infestans populations in the Argentinean Chaco support some of these assumptions, with pyrethroid resistant individuals characterized by lower levels of genetic diversity than to their resistant counterparts (Piccinali et al., 2020).
However, our study results challenge these traditional paradigms and may instead be explained by an alternative hypothesis for genetic diversification. In a given location, a population may undergo a selection process from insecticide exposure independent of neighboring triatomine populations (Pessoa et al., 2014). A previous work comparing the genetic profiles of populations under different vector control pressure (Marcet et al., 2008) described populations within a community recovering from a recent massive spraying as much more genetically structured and diverse than populations that were not under systematic vector control pressure. The authors proposed that the distinct observed genetic pattern reflected the cumulative effect of migration among adjacent populations for several generations, which generated a pattern of genetic “homogenization” even among populations located geographically far apart. In contrast, populations recovering from a pool of survivors of insecticide spraying were observed as discrete genetic entities at the individual capture site (i.e. one household). Pooling together such individuals from these “genetically discrete” populations with bugs from other sites, for a random sampling of a given area, (i.e. at the community level, pooling individuals from different houses in the same sample set), would result in a highly genetically diverse meta-population, that is actually composed of several subpopulations, with genetic substructure and gene flow restrictions within. In this study, the elevated genetic diversity of both Mat populations could represent the assemblage of individuals from distinct genetic units, as they have been under higher selective pressure than the sylvatic populations from 20oct, KM and Ili, located further away from the domestic area subjected to vector control. High levels of genetic diversity have also been reported in several domestic non-Andean T. infestans populations from the Gran-Chaco area (Perez de Rosas et al., 2013; Piccinali et al., 2009), of which many had been subject to extensive vector control pressure.
More importantly, the existence of genetically-diverse populations displaying high levels of altered insecticide resistance has potential implications for longer-term efficacy of control interventions, such that the likelihood of individuals surviving future selection events is greater when the original population is composed of more unique genetic variants. It is noteworthy that three haplotypes found in the sylvatic populations analyzed here (A, C and E) were also identified in domestic populations in Cochabamba Department (Giordano et al., 2005), suggesting household colonization by sylvatic populations in the Bolivian Andes is a pervasive and contemporary process.
Regarding the resistance mechanism(s) underlying the altered susceptibility patterns in Mat, analyses of P450 monoxygenases and 7-CP esterases (Roca-Acevedo et al., 2011; Roca-Acevedo and Picollo, 2017) showed a lack of association between enzymatic activities and susceptibility ratios, indicating that these enzymes were not responsible for modified susceptibility in these sylvatic populations. In this work, we evaluated the presence of two kdr mutations and found one individual from Mat-S carrying the mutation L1014 (Fabro et al., 2012). Although most likely not responsible for the mechanism(s) altering insecticide susceptibility in these populations, the presence of this resistance mutation in this area represents a risk for selection under pressure and potential development of higher resistance intensities (Mougabure-Cueto and Picollo, 2015; Sierra et al., 2016). The absence of kdr target site mutations in our Bolivian populations contrasts with recent reports from the Argentinean Gran Chaco, where high frequencies of the L925I substitution were implicated in pyrethroid resistance (Fronza et al., 2020). Future work to characterize the underlying metabolic mechanisms of resistance in these populations is warranted, including screening for novel kdr mutations A934V and K964R, recently identified in Triatoma mazzottii and Triatoma longipennis, respectively (Davila-Barboza et al., 2018), as well as investigating a possible role for integumental detoxification in T. infestans pyrethroid resistance (Dulbecco et al., 2018).
5. Conclusions
Sylvatic populations of T. infestans represent a challenge for vector control. These populations are not targeted by control activities and could play a key role in post-spraying house re-infestation. The results presented here suggest that there might be an ancient trait(s) that confers resistance in T. infestans sylvatic populations that are capable of invading domiciles. These observations emphasize the need for stronger entomological surveillance in the region, including early detection of house invasion, particularly post-spraying, monitoring for resistance to pyrethroids and the design of integrative control actions that consider both sylvatic foci around domestic settings as well as bug dispersion dynamics.
Acknowledgements
We thank Drs. François Noireau (1955-2011) and Mirko Rojas Cortez (Fundación SANIT) and the technicians of the Coordinación Nacional de Control de Vectores del Ministerio de Salud de Argentina for coordinating the field sampling.
Funding
This study received financial support from CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas. Argentina PIP 411/2012) and ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica-Argentina). DNA primers and probes were synthetized by the CDC’s core facilities and molecular laboratory reagents were provided by the CDC. LAM was supported by an American Society for Microbiology/Centers for Disease Control and Prevention Fellowship.
Footnotes
Availability of data and material
DNA sequences obtained in this study are available in GenBank.
Declaration of Competing Interest
The authors declare that they have no competing interests.
The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC).
References
- Agapow P-M, Burt A, 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1, 101–102. [Google Scholar]
- Aguirre-Obando OA, Bona ACD, Duque L,J,E, Navarro-Silva MA, 2015. Insecticide resistance and genetic variability in natural populations of Aedes (Stegomyia) aegypti (Diptera: Culicidae) from Colombia. Zoologia (Curitiba) 32, 14–22. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A, 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW, 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet 32, 314–331. [PMC free article] [PubMed] [Google Scholar]
- Breniere SF, Salas R, Buitrago R, Bremond P, Sosa V, Bosseno MF, Waleckx E, Depickere S, Barnabe C, 2013. Wild populations of Triatoma infestans are highly connected to intra-peridomestic conspecific populations in the Bolivian Andes. PLoS One 8, e80786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breniere SF, Buitrago R, Waleckx E, Depickere S, Sosa V, Barnabe C, Gorla D, 2017. Wild populations of Triatoma infestans: compilation of positive sites and comparison of their ecological niche with domestic population niche. Acta Trop. 176, 228–235. [DOI] [PubMed] [Google Scholar]
- Buitrago R, Waleckx E, Bosseno MF, Zoveda F, Vidaurre P, Salas R, Mamani E, Noireau F, Breniere SF, 2010. First report of widespread wild populations of Triatoma infestans (Reduviidae, Triatominae) in the valleys of La Paz, Bolivia. Am J Trop Med Hyg 82, 574–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago NLR, Bosseno MF, Waleckx E, Bŕemond P, Vidaurre P, Zoveda F, Brenìere SF, 2013. Risk of transmission of Trypanosoma cruzi by wild Triatoma infestans (Hemiptera: Reduviidae) in Bolivia supported by the detection of human blood meals. Infect. Genet. Evol 19, 141–144. [DOI] [PubMed] [Google Scholar]
- Buitrago R, Bosseno MF, Depickere S, Waleckx E, Salas R, Aliaga C, Barnabe C, Breniere SF, 2016. Blood meal sources of wild and domestic Triatoma infestans (Hemiptera: Reduviidae) in Bolivia: connectivity between cycles of transmission of Trypanosoma cruzi. Parasit. Vectors 9, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti N, Mougabure-Cueto G, Rivera-Pomar R, Ons S, 2014. L925I mutation in the Para-type Sodium Channel is associated with Pyrethroid resistance in Triatoma infestans from the Gran Chaco region. PLoS Negl. Trop. Dis 8, e2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo MJ, Amelotti I, Gorla DE, 2016. Invasion of rural houses by wild Triatominae in the arid Chaco. J. Vector Ecol 41, 97–102. [DOI] [PubMed] [Google Scholar]
- Ceballos LA, Piccinali RV, Berkunsky I, Kitron U, Gürtler RE, 2009. First finding of Melanic sylvatic Triatoma infestans (Hemiptera: Reduviidae) colonies in the argentine Chaco. J. Med. Entomol 46, 1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos LA, Piccinali RV, Marcet PL, Vazquez-Prokopec GM, Cardinal MV, Schachter-Broide J, Dujardin J-P, Dotson EM, Kitron U, Gürtler RE, 2011. Hidden sylvatic foci of the Main vector of Chagas disease Triatoma infestans: threats to the vector elimination campaign? PLoS Negl. Trop. Dis 5, e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U, 2006. Reinfestation sources for Chagas disease vector, Triatoma infestans, Argentina. Emerg. Infect. Dis 12, 1096–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere MC, Rodriguez-Planes LI, Vazquez-Prokopec GM, Kitron U, Gurtler RE, 2019. Community-based surveillance and control of chagas disease vectors in remote rural areas of the argentine Chaco: a five-year follow-up. Acta Trop. 191, 108–115. [DOI] [PubMed] [Google Scholar]
- Cortez MR, Emperaire L, Piccinali RV, Gurtler RE, Torrico F, Jansen AM, Noireau F, 2007. Sylvatic Triatoma infestans (Reduviidae, Triatominae) in the Andean valleys of Bolivia. Acta Trop. 102, 47–54. [DOI] [PubMed] [Google Scholar]
- Coura JR, Viñas PA, Junqueira ACV, 2014. Ecoepidemiology, short history and control of Chagas disease in the endemic countries and the new challenge for non-endemic countries. Mem. Inst. Oswaldo Cruz 109, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila-Barboza J, Villanueva-Segura OK, Lopez-Monroy B, Ponce-Garcia G, Bobadilla-Utrera C, Montes-Rincon M, Molina-Garza ZJ, Arredondo-Jimenez JI, Rodriguez-Sanchez IP, Manrique-Saide PC, Flores AE, 2018. Novel kdr mutations (K964R and A943V) in pyrethroid-resistant populations of Triatoma mazzottii and Triatoma longipennis from Mexico and detoxifying enzymes. Insect Sci. 00, 1–12. [DOI] [PubMed] [Google Scholar]
- Depickère S, Buitrago R, Siñani E, Baune M, Monje M, Lopez R, Waleckx E, Chavez T, Brenìere SF, 2012. Susceptibility and resistance to deltamethrin of wild and domestic populations of Triatoma infestans (Reduviidae: Triatominae) in Bolivia: new discoveries. In: Mem Inst Oswaldo Cruz, Rio de Janeiro, 107, pp. 1042–1047. [DOI] [PubMed] [Google Scholar]
- Dulbecco AB, Moriconi DE, Calderon-Fernandez GM, Lynn S, McCarthy A, Roca-Acevedo G, Salamanca-Moreno JA, Juarez MP, Pedrini N, 2018. Integument CYP genes of the largest genome-wide cytochrome P450 expansions in triatomines participate in detoxification in deltamethrin-resistant Triatoma infestans. Sci. Rep 8, 10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D, vonHoldt B, 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour 4, 359–361. [Google Scholar]
- Echeverria JE, Bustamante Gomez MB, D’Avila Pessoa GC, Cortez MR, Rodriguez AN, Diotaiuti LG, 2018. Resistance to deltamethrin by domestic and wild Triatoma infestans populations in the municipality of Toro Toro, Potosi, Bolivia. Parasit. Vectors 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mousadik A, Petit R, 1996. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor. Appl. Genet 92, 832–839. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier LG, Laval G, Schneider S, 2005. ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinformatics Online 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fabro J, Sterkel M, Capriotti N, Mougabure-Cueto G, Germano M, Rivera-Pomar R, Ons S, 2012. Identification of a point mutation associated with pyrethroid resistance in the Para-type sodium channel of Triatoma infestans, a vector of Chagas’ disease. Infect. Genet. Evol 12, 487–491. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK, 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronza G, Toloza AC, Picollo MI, Spillmann C, Mougabure-Cueto GA, 2016. Geographical variation of Deltamethrin susceptibility of Triatoma infestans (Hemiptera: Reduviidae) in Argentina with emphasis on a resistant focus in the Gran Chaco. J. Med. Entomol 53, 880–887. [DOI] [PubMed] [Google Scholar]
- Fronza G, Toloza AC, Picollo MI, Carbajo AE, Rodriguez S, Mougabure-Cueto GA, 2019. Modelling the association between deltamethrin resistance in Triatoma infestans populations of the Argentinian Gran Chaco region with environmental factors. Acta Trop. 194, 53–61. [DOI] [PubMed] [Google Scholar]
- Fronza G, Roca-Acevedo G, Mougabure-Cueto GA, Sierra I, Capriotti N, Toloza AC, 2020. Insecticide resistance mechanisms in Triatoma infestans (Reduviidae: Triatominae): the putative role of enhanced detoxification and knockdown resistance (kdr) allele in a resistant hotspot from the argentine Chaco. J. Med. Entomol 57, 837–844. [DOI] [PubMed] [Google Scholar]
- Germano MD, Roca Acevedo G, Mougabure Cueto GA, Toloza AC, Vassena CV, Picollo MI, 2010a. New findings of insecticide resistance in Triatoma infestans (Heteroptera: Reduviidae) from the Gran Chaco. J. Med. Entomol 47, 1077–1081. [DOI] [PubMed] [Google Scholar]
- Germano MD, Vassena CV, Picollo MI, 2010b. Autosomal inheritance of deltamethrin resistance in field populations of Triatoma infestans (Heteroptera: Reduviidae) from Argentina. Pest Manag. Sci 66, 705–708. [DOI] [PubMed] [Google Scholar]
- Giordano R, Cortez JC, Paulk S, Stevens L, 2005. Genetic diversity of Triatoma infestans (Hemiptera: Reduviidae) in Chuquisaca, Bolivia based on the mitochondrial cytochrome b gene. Mem. Inst. Oswaldo Cruz 100, 753–760. [DOI] [PubMed] [Google Scholar]
- Gomez MB, D’Avila GCP, Orellana ALG, Cortez MR, Rosa ACL, Noireau F, Diotaiuti LG, 2014. Susceptibility to deltamethrin of wild and domestic populations of Triatoma infestans of the Gran Chaco and the inter-Andean valleys of Bolivia. Parasit. Vectors 7, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorla D, 2002. Variables ambientales registradas por sensores remotos como indicadores de la distribución geográfica de Triatoma infestans (Heteroptera: Reduciidae) Ecología Austral, 12, pp. 117–127. [Google Scholar]
- Goudet J, 1995. FSTAT version 1.2: a computer program to calculate F-statistics. J. Hered 86, 485–486. [Google Scholar]
- Gurevitz JM, Ceballos LA, Gaspe MS, Alvarado-Otegui JA, Enriquez GF, Kitron U, Gurtler RE, 2011. Factors affecting infestation by Triatoma infestans in a rural area of the humid Chaco in Argentina: a multi-model inference approach. PLoS Negl. Trop. Dis 5, e1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz JM, Gaspe MS, Enriquez GF, Provecho YM, Kitron U, Gürtler RE, 2013. Intensified surveillance and insecticide-based control of the Chagas disease vector Triatoma infestans in the Argentinean Chaco. PLoS Negl. Trop. Dis 7, e2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler RE, Diotaiuti L, Kitron U, 2008. Commentary: Chagas disease: 100 years since discovery and lessons for the future. Int. J. Epidemiol 37, 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK, 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour 9, 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, 2005. HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol. Ecol. Notes 5, 187–189. [Google Scholar]
- Lardeux F, Depickere S, Duchon S, Chavez T, 2010. Insecticide resistance of Triatoma infestans (Hemiptera, Reduviidae) vector of Chagas disease in Bolivia. Tropical Med. Int. Health 15, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D, 2015. Popart: full-feature software for haplotype network construction. Methods Ecol. Evol 6, 1110–1116. [Google Scholar]
- Librado P, Rozas J, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Marcet PL, Lehmann T, Groner G, Gürtler RE, Kitron U, Dotson EM, 2006. Identification and characterization of microsatellite markers in the Chagas disease vector Triatoma infestans (Heteroptera: Reduviidae). Infect. Genet. Evol 6, 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet PL, Mora MS, Cutrera AP, Jones L, Gürtler RE, Kitron U, Dotson EM, 2008. Genetic structure of Triatoma infestans populations in rural communities of Santiago Del Estero, northern Argentina. Infect. Genet. Evol 8, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcombe S, Paris M, Paupy C, Bringuier C, Yebakima A, Chandre F, David J-P, Corbel V, Despres L, 2013. Insecticide-driven patterns of genetic variation in the dengue vector Aedes aegypti in Martinique Island. PLoS One 8, e77857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro FA, Perez R, Panzera F, Dujardin JP, Galvao C, Rocha D, Noireau F, Schofield C, Beard CB, 1999. Mitochondrial DNA variation of Triatoma infestans populations and its implication on the specific status of T. melanosoma. Mem. Inst. Oswaldo Cruz 94 (Suppl. 1), 229–238. [DOI] [PubMed] [Google Scholar]
- Monteiro FA, Barrett TV, Fitzpatrick S, Cordon-Rosales C, Feliciangeli D, Beard CB, 2003. Molecular phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus. Mol. Ecol 12, 997–1006. [DOI] [PubMed] [Google Scholar]
- Mougabure-Cueto G, Picollo MI, 2015. Insecticide resistance in vector Chagas disease: evolution, mechanisms and management. Acta Trop. 149, 70–85. [DOI] [PubMed] [Google Scholar]
- Nei M, 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [Google Scholar]
- Nei M, Tajima F, Tateno Y, 1983. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol 19, 153–170. [DOI] [PubMed] [Google Scholar]
- Noireau F, Flores R, Vargas F, 1999. Trapping sylvatic Triatominae (Reduviidae) in hollow trees. Trans. R. Soc. Trop. Med. Hyg 93, 13–14. [DOI] [PubMed] [Google Scholar]
- Noireau F, Bastrenta B, Catala S, Dujardin JP, Panzera F, Torres M, Perez R, Galvao C, Jurberg J, 2000. Sylvatic population of Triatoma infestans from the Bolivian Chaco: from field collection to characterization. Mem. Inst. Oswaldo Cruz 95 (Suppl. 1), 119–122. [DOI] [PubMed] [Google Scholar]
- Noireau F, Cortez MG, Monteiro FA, Jansen AM, Torrico F, 2005. Can wild Triatoma infestans foci in Bolivia jeopardize Chagas disease control efforts? Trends Parasitol. 21, 7–10. [DOI] [PubMed] [Google Scholar]
- Núñez JA, Segura EL, 1987. Rearing of Triatominae. In: Chagas’ Disease Vectors, 2, pp. 31–40. [Google Scholar]
- PAHO, 2005. Technical report: II Reunión técnica latinoamericana de monitoreo de resistencia a insecticidas en triatominos vectores de Chagas. Panamerican Health Organization, Panamá, 11–13 April. [Google Scholar]
- Panzera F, Ferreiro MJ, Pita S, Calleros L, Perez R, Basmadjian Y, Guevara Y, Breniere SF, Panzera Y, 2014. Evolutionary and dispersal history of Triatoma infestans, main vector of Chagas disease, by chromosomal markers. Infect. Genet. Evol 27, 105–113. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE, 2012. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Rosas AR, Segura EL, Fichera L, García BA, 2008. Macrogeographic and microgeographic genetic structure of the Chagas’ disease vector Triatoma infestans (Hemiptera: Reduviidae) from Catamarca, Argentina. Genetica 133, 247–260. [DOI] [PubMed] [Google Scholar]
- Perez de Rosas AR, Segura EL, Fusco O, Guinazu AL, Garcia BA, 2013. Fine-scale genetic structure in populations of the Chagas’ disease vector Triatoma infestans (Hemiptera, Reduvidae). Genetica 141, 107–117. [DOI] [PubMed] [Google Scholar]
- Perez-Cascales E, Sossa-Soruco VM, Breniere SF, Depickere S, 2020. Reinfestation with Triatoma infestans despite vigilance efforts in the municipality of Saipina, Santa Cruz, Bolivia: situational description two months after fumigation. Acta Trop. 203, 105292. [DOI] [PubMed] [Google Scholar]
- Pessoa GC, Dias LS, Diotaiuti L, 2014. Deltamethrin pyrethroid susceptibility characterization of Triatoma sordida Stal, 1859 (Hemiptera: Reduviidae) populations in the northern region of Minas Gerais, Brazil. Rev. Soc. Bras. Med. Trop 47, 426–429. [DOI] [PubMed] [Google Scholar]
- Pessoa GC, Vinas PA, Rosa AC, Diotaiuti L, 2015. History of insecticide resistance of Triatominae vectors. Rev. Soc. Bras. Med. Trop 48, 380–389. [DOI] [PubMed] [Google Scholar]
- Piccinali RV, Marcet PL, Noireau F, Kitron U, Gürtler RE, Dotson EM, 2009. Molecular population genetics and phylogeography of the Chagas disease vector Triatoma infestans in South America. J. Med. Entomol 46, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinali RV, Gaunt MW, Gurtler RE, 2018. A microsatellite-based analysis of house infestation with Triatoma infestans (Hemiptera: Reduviidae) after insecticide spraying in the argentine Chaco. J. Med. Entomol 55, 609–619. [DOI] [PubMed] [Google Scholar]
- Piccinali RV, Fronza G, Mougabure-Cueto GA, Toloza AC, 2020. Genetic structure of deltamethrin-resistant populations of Triatoma infestans (Hemiptera: Reduviidae) in the Gran Chaco. Parasitol. Res 10.1007/s00436-020-06789-y. [DOI] [PubMed] [Google Scholar]
- Picollo M, Wood E, Zerba E, Licastro S, Rúveda M, 1976. Laboratory test for measuring toxicity of insecticides in Triatoma infestans, Klug. Acta Bioquim 10, 67–70. [Google Scholar]
- Picollo MI, Vassena C, Santo Orihuela P, Barrios S, Zaidemberg M, Zerba E, 2005. High resistance to pyrethroid insecticides associated with ineffective field treatments in Triatoma infestans (Hemiptera: Reduviidae) from northern Argentina. J. Med. Entomol 42, 637–642. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P, 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisberth S, Waleckx E, Monje M, Chang B, Noireau F, Brenìere SF, 2011. “Andean” and “non-Andean” ITS-2 and mtCytB haplotypes of Triatoma infestans are observed in the Gran Chaco (Bolivia): population genetics and the origin of reinfestation. Infect. Genet. Evol 11, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F, 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86, 248–249. [Google Scholar]
- Rice W, 1989. Analyzing tables of statistical tests. Evolution 43, 223–225. [DOI] [PubMed] [Google Scholar]
- Roca-Acevedo G, Picollo MI, 2017. Identifying current and missing knowledge in the control of pyrethroid-resistant Triatoma infestans, vectors of Chagas disease. Global J. Health Sci 9, 47–56. [Google Scholar]
- Roca-Acevedo G, Cueto GM, Germano M, Orihuela PS, Cortez MR, Noireau F, Picollo MI, Vassena C, 2011. Susceptibility of sylvatic Triatoma infestans from Andeans valleys of Bolivia to deltamethrin and fipronil. J. Med. Entomol 48, 828–835. [DOI] [PubMed] [Google Scholar]
- Rolón M, Vega MC, Román F, Gómez A, Rojas de Arias A, 2011. First report of colonies of sylvatic Triatoma infestans (Hemiptera: Reduviidae) in the Paraguayan Chaco, using a trained dog. PLoS Negl. Trop. Dis 5, e1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo-Orihuela PL, Carvajal G, Picollo MI, Vassena CV, 2013. Toxicological and biochemical analysis of the susceptibility of sylvatic Triatoma infestans from the Andean valley of Bolivia to organophosphate insecticide. Mem. Inst. Oswaldo Cruz 108, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo-Orihuela PL, Vassena CV, Carvajal G, Clark E, Menacho S, Bozo R, Gilman RH, Bern C, Marcet PL, 2017. Toxicological, enzymatic, and molecular assessment of the insecticide susceptibility profile of Triatoma infestans (Hemiptera: Reduviidae, Triatominae) populations from rural communities of Santa Cruz, Bolivia. J. Med. Entomol 54, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ, Jannin J, Salvatella R, 2006. The future of Chagas disease control. Trends Parasitol. 22, 583–588. [DOI] [PubMed] [Google Scholar]
- Sierra I, Capriotti N, Fronza G, Mougabure-Cueto G, Ons S, 2016. Kdr mutations in Triatoma infestans from the Gran Chaco are distributed in two differentiated foci: implications for pyrethroid resistance management. Acta Trop. 158, 208–213. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waleckx E, Salas R, Huamán N, Buitrago R, Bosseno M-F, Aliaga C, Barnabé C, Rodriguez R, Zoveda F, Monje M, Baune M, Quisberth S, Villena E, Kengne P, Noireau F, Brenìere SF, 2011. New insights on the Chagas disease main vector Triatoma infestans (Reduviidae, Triatominae) brought by the genetic analysis of Bolivian sylvatic populations. Infect. Genet. Evol 11, 1045–1057. [DOI] [PubMed] [Google Scholar]
- Waleckx E, Depickere S, Salas R, Aliaga C, Monje M, Calle H, Buitrago R, Noireau F, Breniere SF, 2012. New discoveries of sylvatic Triatoma infestans (Hemiptera: Reduviidae) throughout the Bolivian Chaco. Am J Trop Med Hyg 86, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waleckx E, Gourbiere S, Dumonteil E, 2015. Intrusive versus domiciliated triatomines and the challenge of adapting vector control practices against Chagas disease. Mem. Inst. Oswaldo Cruz 110, 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- WHO, 1994. (World Health Organization). Protocolo de evaluación de efecto insecticida sobre triatominos. Acta Toxicológica Argentina 2, 29–32. [Google Scholar]
- WHO, 2015. Chagas disease in Latin America: An epidemiological update based on 2010 estimates. In: Organization WHEd.), Weekly Epidemiological Record (WER). WHO, Geneva, Switzerland, pp. 33–44. [PubMed] [Google Scholar]
- Zerba EN, 1999. Susceptibility and resistance to insecticides of Chagas disease vectors. Medicina (B Aires) 59 (Suppl. 2), 41–46. [PubMed] [Google Scholar]


