Abstract
Plasmodesmata (PD) are membrane-lined pores that connect adjacent cells to mediate symplastic communication in plants. These intercellular channels enable cell-to-cell trafficking of various molecules essential for plant development and stress responses, but they can also be utilized by pathogens to facilitate their infection of hosts. Some pathogens or their effectors are able to spread through the PD by modifying their permeability. Yet plants have developed various corresponding defense mechanisms, including the regulation of PD to impede the spread of invading pathogens. In this review, we aim to illuminate the various roles of PD in the interactions between pathogens and plants during the infection process. We summarize the pathogenic infections involving PD and how the PD could be modified by pathogens or hosts. Furthermore, we propose several hypothesized and promising strategies for enhancing the disease resistance of host plants by the appropriate modulation of callose deposition and plasmodesmal permeability based on current knowledge.
Keywords: plasmodesmata, plant pathogens, disease resistance, callose, callose synthase, cell-to-cell movement
Introduction
Throughout their life span, plants are constantly challenged by pathogens, namely fungi, bacteria, and viruses (Jones and Dangl, 2006). It is well-known that plant cells can respond to pathogens autonomously. Plants recognize microbe-associated molecular patterns (MAMPs) via pattern recognition receptors (PRRs) on the cell membrane, which initiates a series of signaling events and activates pattern-triggered immunity (PTI; Ranf, 2017; Saijo et al., 2018). Some pathogens, however, can produce effectors capable of inhibiting PTI to overcome the host immune system (Grant et al., 2006; Le Fevre et al., 2015; Toruno et al., 2016). A second defense response, which is activated by recognizing pathogenic effectors with corresponding nucleotide-binding leucine-rich repeat (NLR) proteins of hosts, is called effector-triggered immunity (ETI; Cui et al., 2015). Recently, the PTI and ETI systems were found to share common elements and to interact with each other (Ngou et al., 2021; Yuan et al., 2021). Besides the cell-autonomous immunity within infected regions, uninfected host cells could also establish immune responses in what is known as systemic acquired resistance (SAR; Klessig et al., 2018). To gain SAR, signaling molecules must move from infected cells to distal uninfected tissues (Wendehenne et al., 2014; Singh et al., 2017). Further, SAR confers an immune “memory” in hosts enabling them to activate defense responses more quickly and effectively when exposed to another pathogen attack (Conrath, 2006; Ramirez-Prado et al., 2018; Hake and Romeis, 2019; Guerra et al., 2020).
Plant cells have evolved unique cell-wall-spanning structures, termed PD, that link neighboring cells for their symplastic communication (Epel, 1994; Lucas et al., 2009). The typical PD are composed of plasma membrane (PM), cytoplasmic sleeve, and desmotubule derived from the endoplasmic reticulum (ER) (Zambryski and Crawford, 2000; Zambryski, 2008). Various key PD-localized proteins and lipids have been identified, including actin, receptor-like kinases, glycosylphosphatidylinositol-anchor proteins, remorins, sphingolipids, and sterols (Fernandez-Calvino et al., 2011). By controlling the intercellular exchange of both micromolecules and macromolecules, PD are functionally critical during the development of plants and in their responses to abiotic and biotic stresses (Maule, 2008; Lee and Lu, 2011; Lee et al., 2011; Han et al., 2014; Lee, 2014; Sager and Lee, 2014; Cui and Lee, 2016; Wu et al., 2016; Reagan et al., 2018; Miyashima et al., 2019; Yan and Liu, 2020). The aperture of the PD pore, which determines the size exclusion limit (SEL), is a major determinant of PD permeability (Lucas and Lee, 2004; Peters et al., 2021). This PD aperture is dynamically controlled by the deposition and degradation of callose within the cell walls near the neck of PD (Amsbury et al., 2017; Wu et al., 2018). Callose synthases (CalSs) and β-1,3 glucanases (BGs) govern the production and degradation of callose, respectively, fulfilling crucial roles in various developmental and physiological processes of plants (Chen and Kim, 2009; Zavaliev et al., 2011). PD-LOCALIZED PROTEINS (PDLPs) and PLASMODESMATA CALLOSE-BINDING PROTEINS (PDCBs) are two key protein families that positively regulate the dynamics of callose accumulation at PD (Simpson et al., 2009; Lee et al., 2011). Apart from callose, the architecture of PD also affects their conductivity and functioning. In this respect, PD may be classified as type I or II according to the status of the cytoplasmic sleeve between the PM and desmotubule. Compared with type II, the structure of type I PD lacks a visible cytoplasmic sleeve and internal tethers (Nicolas et al., 2017). Loss of function of the PHLOEM UNLOADING MODULATOR gene results in the lack of type II PD, whereas enhances PD permeability, fosters phloem unloading, and accelerates root elongation (Yan et al., 2019).
Beyond the key roles in plant development, the PD can participate in plant-pathogen interactions (Faulkner et al., 2013; Wang et al., 2013; Brunkard and Zambryski, 2017; Cheval and Faulkner, 2018). Specifically, PD facilitate the intercellular transport of mobile signal molecules, such as azelaic acid and glycerol-3-phosphate, needed for the establishment of SAR (Singh et al., 2017). Yet many pathogens and effectors can also spread in a cell-to-cell manner via PD to hasten the infection (Lent et al., 1991; Waigmann et al., 1994; Benitez-Alfonso et al., 2010; Cao et al., 2018). Currently, it remains an open question how plants modulate the timing of PD closure and the movement of SAR signals and pathogenic effectors to achieve the defense response. We speculate the apoplastic trafficking of immune molecules might provide an alternative way to impede the spread of pathogens or effectors in the case of blocked PD (Lim et al., 2016b; Singh et al., 2017). Further study of PD in the battle between plants and pathogens is gaining interest and becoming important. Here, we summarize the studied mobile pathogens and effectors, the recognition between pathogens and PD, and the antagonistic regulation of PD by plants and pathogens. Finally, we propose several hypothesized strategies to assist hosts in their battle against pathogens via the appropriate modulation of PD.
Pathogens Exploit Pd to Facilitate Host Infections
Through the PD, the cell-to-cell movement of a variety of molecules is possible. But based on this intercellular connection, phytopathogens have evolved mechanisms that take advantage of PD as gateways to facilitate their host infections (Figure 1). To do this, pathogens encode their own proteins and recruit or interact with the host proteins to target and modify the PD, either directly or indirectly (Table 1).
Figure 1.
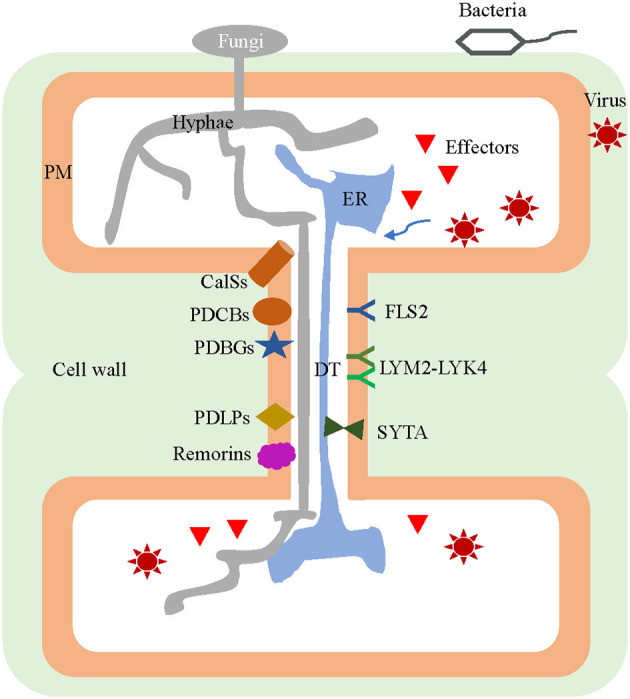
Plasmodesmal components involved in both pathogen infection of plants and host defense responses. Virions, effectors of fungi and bacteria, as well as the fungal hyphae are transported from infected cells to the neighboring healthy cells through the plasmodesmata (PD). Many PD-localized components are jointly exploited by both pathogens and host plants, for use in their interaction during an infection. The pathogens inhibit callose synthesis by inactivating CalSs, suppressing PDCBs, destabilizing PDLPs, and/or recruiting PDBGs to assist in their intercellular movement. Viruses can interact with SYTA by relying on MP to remodel the PD. Conversely, PD harbor specific plasmodesmal PM-located receptors of LYM2-LYK4 complex and FLS2 for perception of fungal elicitor chitin and bacterial flagellin, respectively. During infection, some CalS and PDLP genes can be induced so as to promote callose accumulation and PD closure. The remorins in the membrane microdomains of plasmodesmal PM interact with MP and impede the movement of the virus among host cells. PD, plasmodesmata; DT, desmotubule; CalS, callose synthase; PDCB, PLASMODESMATA CALLOSE-BINDING PROTEIN; PDLP, PD-LOCALIZED PROTEIN; PDBG, plasmodesmal-localized β-1,3 glucanase; SYTA, synaptotagmin A; MP, movement protein; PM, plasma membrane; LYM2, LYSM-CONTAINING GPI-ANCHORED PROTEIN 2; LYK4, LysM-CONTAINING RECEPTOR-LIKE KINASE 4; FLS2, FLAGELLIN SENSING.
Table 1.
Viral, fungal, and bacterial pathogens and the effectors that move cell-to-cell through the plasmodesmata of attacked plants.
| Pathogens | MPs/Effectors | Function | References | ||
|---|---|---|---|---|---|
| Virus | Tobacco mosaic virus (TMV) | MP30 | MPs bind viral RNAs and increase the SEL of PD in the form of ribonucleoprotein complexes. | Wolf et al., 1989; Brill et al., 2000; Peña and Heinlein, 2012; Pitzalis and Heinlein, 2017 | |
| Carnation mottled carmovirus (CarMV) | P7 and P9 | Vilar et al., 2002 | |||
| Turnip crinkle virus (TCV) | P8 and P9 | Hacker et al., 1992; Li et al., 1998 | |||
| Melon necrotic spot virus (MNSV) | P7A and P7B | MPs bind viral RNAs and transit through PD in the form of ribonucleoprotein complexes. TGBp1 of PVX and TGBp2 and TGBp3 of PVX and PMTV increase the PD SEL Tamai and Meshi, 2001; Howard et al., 2004; Haupt et al., 2005 | Genoves et al., 2006 | ||
| Pelargonium flower break virus (PFBV) | P7 and P12 | Martinez-Turino and Hernandez, 2011 | |||
| Potato virus X (PVX) |  |
TGB: TGBp1, TGBp2 and TGBp3 | Tilsner et al., 2013 | ||
| Bamboo mosaic virus (BaMV) | Chou et al., 2013 | ||||
| Barley stripe mosaic virus (BSMV) | Lim et al., 2008; | ||||
| Poasemi latent virus (PSLV) | Shemyakina et al., 2011 | ||||
| Potato mop-top virus (PMTV) | Zamyatnin et al., 2004 | ||||
| Beet yellows virus (BYV) | Hsp70h, 64kDa protein, two capsid proteins, and 6-kDa hydrophobic protein | Alzhanova et al., 2000; Dolja, 2003; Avisar et al., 2008 | |||
| Tobacco etch virus | Capsid protein (CP) | The CP is required for cell-to-cell and long-distance movement of virus. | Dolja et al., 1995 | ||
| Cowpea mosaic virus (CPMV) | MP 58K and 48K |  |
MPs form movement tubles to replace PD desmotubule. | Pouwels et al., 2004; Ritzenthaler and Hofmann, 2007 | |
| Grapevine fanleaf virus (GFLV) | MP 2B | Laporte et al., 2003; Amari et al., 2010 | |||
| Cauliflower mosaic virus (CaMV) | MP P1 | Thomas and Maule, 1995 | |||
| Broad bean wilt virus 2 | MP VP37 | Xie et al., 2016 | |||
| Turnip mosaic virus (TuMV) | 6K2 protein | 6K2 induces vesicle formation for intercellular movement through PD. | Grangeon et al., 2013 | ||
| Viroids | Potato spindle tuber viroid (PSTVd) | / | 11 RNA loop motifs are critical for cell-to-cell mvement. | Ding et al., 1997; Zhong et al., 2008 | |
| Fungi | Magnaporthe oryzae (M. oryzae) | / | IH seek for the pit fields, followed by crossing the PD channels into adjacent cells with constricted hyphae. | Kankanala et al., 2007 | |
| BSA3 | BSA3 locates near PD. | Mosquera et al., 2009 | |||
| PWL2 and BAS1 | PWL2 and BAS1were delivered into the cytoplasm of rice cells by biotrophic interfacial complex (BIC), and finally into neighboring cells via PD. | Khang et al., 2010 | |||
| Melampsora larici-populina | AvrL567 | AvrL567 accumulates at PD. | Germain et al., 2018 | ||
| Ustilago maydis | Cmu1 | Cmu1 could likely spread to the neighboring cells through PD and repress SA biosynthesis in host plants. | Djamei et al., 2011 | ||
| Phytophthora brassicae | RxLR3 | RxLR3 reduce the callose deposition around PD by interacting with and inhibiting CalS1, CalS2, and CalS3 | Tomczynska et al., 2020 | ||
| Fusarium graminearum | Fusaoctaxin A | Fusaoctaxin A inhibit expression of PDCB-like genes and CalS genes. | Jia et al., 2019 | ||
| Fusarium oxysporum | Avr2 and Six5 | Avr2-Six5 effector pair alters plasmodesmatal conductivity to facilitate intercellular movement of Avr2. | Cao et al., 2018 | ||
| Bacteria | Candidatus Liberibacter asiaticus (CLas) | / | CLas move through sieve pores, and cause callose overproduction and sieve-pore plugging in phloem. | Koh et al., 2012; Achor et al., 2020 | |
| Pseudomonas syringae pv. tomato (Pst) DC3000 | HopO1-1 | HopO1-1 targets to PD and destabilizes PDLP7 and PLDP5. | Aung et al., 2020 | ||
| Pseudomonas syringae pv. tomato (Pst) DC3000 | HopK1, HopY1, HopF2, HopU1, HopH1, HopC1, HopN1, HopAA1, HopAF1, HopP1, HopAB2, HopE1, HopAO1, HopA1, HopX1, HopB1 | Hop effectors move from infected cells to neighboring cells through PD. | Li et al., 2020 | ||
Viral Spread Through PD
Plant viruses are biotrophic pathogens that utilize the transcriptional machinery of hosts to replicate and propagate with them. To overcome the cell wall barrier, viruses may exploit the PD to engage in cell-to-cell movement and thereby systemically spread throughout the host plants. Viruses encode movement proteins (MPs) to target to and dilate PD (Heinlein and Epel, 2004; Waigmann et al., 2004; Lucas, 2006). For example, a plasmodesmal localization signal sequence in tobacco mosaic virus (TMV) and sugar canemosaic virus MPs was found necessary and sufficient for PD localization (Yuan et al., 2016; Cheng et al., 2017). Viruses harbor different transport strategies based on differing MP numbers (Epel, 2009). TMV encodes only a single MP, which binds to its RNA and increase the SEL of PD in the form of ribonucleoprotein complexes (Wolf et al., 1989; Brill et al., 2000; Asurmendi et al., 2004; Peña and Heinlein, 2012). In cowpea mosaic virus, grapevine fanleaf virus, and cauliflower mosaic virus, the MPs reorganize and expand the PD pores by forming a movement tubule (Thomas and Maule, 1995; Laporte et al., 2003; Pouwels et al., 2004). Carnation mottled virus encodes two small MPs (DGBp1 and DGBp2) by the double-gene block module (Epel, 2009; Hull, 2014). An early model was proposed, in which DGBp2 interacts with DGBp1:vRNA and drives the transportation of this ternary complex to PD via the endomembrane system. This model is not perfect, however, because of some inconsistencies and the mechanism of carmovirus movement is believed to be more complicated (Navarro et al., 2019). The triple gene block module encodes three MPs termed TGBp1, TGBp2, and TGBp3; the TGBp2 and TGBp3 are ER membrane-associated proteins and they form both homologous and heterologous complexes (Morozov and Solovyev, 2003; Lim et al., 2008). Binding of TGBp2/TGBp3 to TGBp1:vRNA allows for the targeting of vRNA to PD and its cell-to-cell spread in the host (Epel, 2009). Beet yellows virus (BYV) assembles five MPs to facilitate its intercellular movement, including four viral components-an Hsp70h, a 64kDa protein, and two capsid proteins-and a none-structural 6-kDa hydrophobic protein (Alzhanova et al., 2000). The Hsp70h autonomously targets to PD and its ATPase activity drives the intercellular translocation of BYV (Dolja, 2003; Avisar et al., 2008).
As mentioned before, some plant viruses reorganize the inner structure of PD to produce a movement tubule while passing through it (Thomas and Maule, 1995; Laporte et al., 2003; Pouwels et al., 2004). In this process, MPs are assembled into tubular structures by interacting with the host PDLPs, and this replaces the PD desmotubule to leave only a simple PM-lined tunnel remaining, which aids the viral transport (Amari et al., 2010). The pdlp1/2/3 triple mutant showed a significant reduction of tubule formation along with diminished local and systemic spread of infection, indicating the important roles of PDLPs (Amari et al., 2010). The cytoskeletons, which are involved in the physical formation and structural operation of PD, are also the modification targets of certain viruses (Liu et al., 2005; Prokhnevsky et al., 2005; Wright et al., 2007; Avisar et al., 2008). One study proved that the MPs of Cucumber mosaic virus and TMV are able to sever F-actin, weakening the integrity of PD, thereby allowing larger molecules to pass (Su et al., 2010).
Callose around the PD plays a critical role in regulating their permeability and symplastic communication (Amsbury et al., 2017; Wu et al., 2018). Decreasing this callose was shown to result in an enhanced viral infection (Bucher et al., 2001; Li et al., 2012), whereas increasing callose in the β-1,3-glucanase-deficient and atbg pap mutants slowed the spread of the virus (Iglesias and Meins, 2000; Zavaliev et al., 2013). Nevertheless, viruses can facilitate their intercellular movement in hosts by limiting the synthesis of callose and promoting its degradation at PD. For example, potato virus Y is capable of inducing the activity of a class I β-1,3-glucanase and suppressing callose accumulation in a strain-nonspecific manner, which may explain why some viruses are still able to spread in resistant-genotype hosts (Chowdhury et al., 2020).
PD is a compelling type of membrane contact site, perhaps best illustrated by the specialization of the ER and the PM at the sites of cell-to-cell junctions (Tilsner et al., 2016). The desmotubule and PM together provide a cytoplasmic conduit for intercellular transport (Roberts and Oparka, 2003; Brunkard et al., 2015). Plant synaptotagmin A (SYTA), a membrane protein, can be recruited to form ER-PM contact sites adjacent to the PD. But viral MPs can interact with SYTA to remodel these contact sites to alter PD and aid viral movement (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014; Levy et al., 2015; Pitzalis and Heinlein, 2017). Recently, the multiple C2 domains and transmembrane region protein family were reported to act as ER-PM tethers specifically at PD (Brault et al., 2019). Further studies need to clearly elucidate the role of the ER-PM membrane in PD functioning and identify more PD tethering machineries that participate in the interactions between pathogens and plants.
Chloroplasts are the organelle responsible for not only the generation of small molecules and secondary metabolites important for plant defense, but also the origination of signals in response to developmental and environmental cues (Ganusova and Burch-Smith, 2019). Nevertheless, particular plant viral proteins can interact with chloroplast proteins to impair the defense of hosts and facilitate the infection of virus (Zhao et al., 2016; Bhattacharyya and Chakraborty, 2018). During Potato virus X (PVX) infection, the viral p25 protein interacts with the chloroplast protein ferredoxin 1 (FD1) to reduce its mRNA and protein levels, resulting in a dramatic decrease of PD callose accumulation that is probably associated with the reduction in phytohormones abscisic acid (ABA) and salicylic acid (SA) (Yang et al., 2020). Arabidopsis INCREASED SIZE EXCLUSION LIMIT (ISE) 2 encodes a chloroplast DEAH RNA helicase, whose mutation increases the branched PD formation and intercellular trafficking (Kobayashi et al., 2007). The ISE2 expression can be induced by the infection of TMV or turnip mosaic virus in Nicotiana benthamiana. However, ISE2-overexpressing plants are more susceptible to viral infection, without any influence on callose deposition (Ganusova et al., 2017). These findings imply a still, as of yet unknown mechanism of ISE2-mediated chloroplast-nucleus signaling in the interactions between PD and viruses.
Viroids are the smallest known pathogenic agents, consisting only of circular single-stranded RNAs that replicate autonomously and traffic themselves systemically throughout their hosts via the vascular tissue phloem (Flores et al., 2005). Viroids differ from viruses in having unique structural, functional, and evolutionary properties (Flores et al., 2005). Work by Ding et al. (1997) demonstrated that potato spindle tuber viroid (PSTVd) can move rapidly from the initially injected mesophyll cells which are interconnected by PD into neighboring cells, whereas it was retained in mature guard cells lacking PD connections. The PSTVd consists of 27 RNA loop motifs flanked by short helices, of which 11 loops were identified as critical for its intercellular movement (Zhong et al., 2008). A small RNA from the virulence-modulating region of PSTVd can suppress the expression of tomato CalS11-like and CalS12-like genes, pointing to a hypothesized mechanism of viroid movement through PD (Adkar-Purushothama et al., 2015). More mechanisms underpinning the regulation of viroid intercellular trafficking by RNA motifs and cellular factors are reviewed by Takeda and Ding (2009).
Fungal Infection by Invasive Hyphae (IH) and Effectors
Perhaps the best example of how a fungal pathogen can spread through PD is the study of the hemibiotrophic rice blast fungus, Magnaporthe oryzae (M. oryzae; Kankanala et al., 2007; Sakulkoo et al., 2018). By means of the enormous turgor pressure generated by their appressoria, M. oryzae breaches the outer cell surface and produces special hyphae named the penetration peg (Howard and Valent, 1996). When entering the epidermal cell lumen, this penetration peg expands to form primary hyphae, which differentiate into bulbous invasive hyphae (IH; Heath et al., 1990). These IH are encased in a plant-derived extra-invasive hyphal membrane outside their cell wall. Then, the bulbous IH seek out pit fields composed of PD clusters in the cell wall, after which they crossing the PD channels into adjacent cells using constricted hyphae (Kankanala et al., 2007). Callose occlusions around the PD were found absent only during the early stages (24–27 h post-inoculation) of invasion in the first rice cell; hence, over this period the PD stayed open, indicating the fungus is able to suppress the callose deposition at pit fields in the host at a specific time before invading the neighboring cells (Sakulkoo et al., 2018). Consistent with this key role of PD, another investigation revealed the failure of IH to move into mature guard cells from neighboring cells due to the degeneration of PD (Kankanala et al., 2007).
Furthermore, the mobile effectors PATHOGENICITY TOWARD WEEPING LOVEGRASS (PWL2) and BIOTROPHY-ASSOCIATED SECRETED (BAS1) produced by M. oryzae can move in a cell-to-cell fashion to facilitate host infection (Khang et al., 2010). Both PWL2 and BAS1 are released by IH into the cytoplasm of rice cells by a biotrophic interfacial complex, and move into non-invaded neighboring cells via PD before the spread of IH, which was presumed to better prepare the host cells for the following invasion of IH (Khang et al., 2010). Pmk1, a single fungal mitogen-activated protein kinase, regulates the expression of secreted fungal effectors that inhibit ROS (reactive oxygen species) generation and callose deposition at the PD in rice (Sakulkoo et al., 2018). Accordingly, inhibiting Pmk1 prevents M. oryzae from infecting adjacent plant cells, leaving it trapped in the present cell, yet without affecting the biotrophic interfacial complex structure and hyphae morphology (Sakulkoo et al., 2018). These findings indicate the importance of PD for the cell-to-cell invasion of rice cells by M. oryzae during the infection process.
The RxLR3 effector produced by Phytophthora brassicae can interact with and inhibit CalS1, CalS2, and CalS3, to reduce the callose deposition around PD, so as to promote symplastic trafficking (Tomczynska et al., 2020). In wheat, three PDCB-like genes and seven CalS genes are suppressed by the virulence factor Fusaoctaxin A during Fusarium graminearum infection, which suggests this pathogen may interfere with normal callose accumulation and disrupt the PD status of host plants (Jia et al., 2019). The effectors Avr2 and Six5 secreted by F. oxysporum interact at the PD during its infection of tomato; however, Avr2 only moves cell-to-cell in the presence of Six5, while Six5 alone does not alter plasmodesmal conductivity (Cao et al., 2018). Generally, however, the consensus PD-targeting signal peptides of such pathogen effectors have yet to be identified.
Bacterial Infection With Symplastic Trafficking
Presently, the cell-to-cell spread of bacteria has been mostly reported to occur in the sieve tubes of phloem tissues. Candidatus Liberibacter asiaticus (CLas) is a phloem-inhabiting bacterium that causes a destructive disease of citrus trees called Huanglongbing (HLB), which is achieved by its spread via sap flow in the phloem throughout the host plants (Bove, 2006). The cells of CLas adhere to the plasma membrane of those phloem cells positioned specifically adjacent to the sieve pores, and the ensuing morphology changes there enable its movement (Achor et al., 2020). Although we know HLB-infected phloem cells undergo callose accumulation and sieve-pore plugging (Koh et al., 2012; Achor et al., 2020), there is still no evidence showing CLas passing through PD between cells in other plant tissues. The interaction between CLas and phloem cells evidently needs more careful investigation. Usually, bacterial pathogens do not cross the cell wall, probably because their suitable habitat is mostly limited to the apoplastic spaces between plant cells, unlike viruses and fungi which spread intercellularly during local and systemic infections (reviewed by Lee and Lu, 2011). Still, bacteria can release specific effector molecules into plant cells not unlike fungi do, which then move through the PD to spread intercellularly in the host (Li et al., 2020; Figure 1). Only a few effectors have been studied to date. A notable example is the effector protein HopO1-1 of Pseudomonas syringae pv. tomato (Pst) DC3000, a putative mono-ADP-ribosyltransferase. The amino acids in position 41 to 283 (C-terminal end residue) of HopO1-1 are required for its localization to PD (Aung et al., 2020). Once there, HopO1-1 enhances the PD-dependent intercellular molecular flux by destabilizing the PDLP7 and PLDP5 proteins of hosts without affecting their transcript levels (Aung et al., 2020). Further, Li et al. (2020) recently proved that the movement of 16 Hop effectors of Pst DC3000 move from transformed cells into neighboring cells through PD depends on their molecular weights. Among them, HopAF1 was characterized by the highest PD-dependent movement, which can nonetheless be inhibited by callose overproduction (Li et al., 2020). This study provided robust evidence that the effectors of bacteria, like fungi, may possess an intercellular mobile ability. It would seem those mobile effectors exploit different mechanisms when interacting with the host during its infection, a topic that warrants further investigation.
Utilization of Pd by Hosts for Defense
In plant-pathogen interactions, plants have evolved two protein families to recognize pathogens: PM-anchored PRR receptors for PAMPs and intracellular NLR receptors for pathogens effectors (reviewed by Dodds and Rathjen, 2010). The lysin motif (LysM) domain-containing protein CHITIN ELICTOR BINDING PROTEIN (CEBiP) and the receptor-like kinases FLAGELLIN SENSING (FLS2) respectively recognize chitin and flagellin (Kaku et al., 2006; Shimizu et al., 2010; Bücherl et al., 2017). The plasmodesmal PM that is enriched with particular proteins and lipids will integrate extracellular signals differently from the other remaining PM. Increasing numbers of receptors and kinases have been shown to be active in or recruited to plasmodesmal PM (Stahl et al., 2013; Grison et al., 2019; Hunter et al., 2019). A PD-located receptor, LYSM-CONTAINING GPI-ANCHORED PROTEIN 2 (LYM2)/ CEBiP, responds to chitin and signaling, thereby reducing the molecular flux through PD (Faulkner et al., 2013). A receptor complex called LYM2-LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE 4 (LYK 4) (Table 3) found localized at plasmodesmal PM is utilized for plant defense in response to fungal chitin (Cheval et al., 2020). Downstream chitin signaling triggers the phosphorylation of the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN D via a calcium-dependent protein kinase, leading to callose deposition and eventual PD closure. Intriguingly, FLS2 was observed in the vicinity of PD and mediates flg22-triggered changes of PD-mediated trafficking (Faulkner et al., 2013). This phenomenon suggests FLS2 may have an unconsidered role in recognizing flagellin at PD. More receptors at the plasmodesmal PM await discovery.
Table 3.
Abbreviation list.
| Abbreviation | Full name |
|---|---|
| ABA | Abscisic acid |
| A. thaliana | Arabidopsis thaliana |
| BG | β-1,3 glucanase |
| PD | Plasmodesmata |
| CalS | Callose synthase |
| CEBiP | CHITIN ELICTOR BINDING PROTEIN |
| DGB | Double gene block |
| ER | Endoplasmic reticulum |
| ETI | Effector-triggered immunity |
| FLS2 | FLAGELLIN SENSING |
| HLB | Huanglongbing |
| IH | Invasive hyphae |
| ISE | INCREASED SIZE EXCLUSION LIMIT |
| LysM | Lysin motif |
| LYM2 | LYSM-CONTAINING GPI-ANCHORED PROTEIN 2 |
| LYK4 | LysM-CONTAINING RECEPTOR-LIKE KINASE 4 |
| MAMPs | Microbe-associated molecular patterns |
| MP | Movement protein |
| NLR | Nucleotide-binding leucine-rich repeat |
| PDBG | Plasmodesmal-lacalized β-1,3 glucanase |
| PDCB | PLASMODESMATA CALLOSE-BINDING PROTEIN |
| PDLP | PD-LOCALIZED PROTEIN |
| PM | Plasma membrane |
| PRRs | Pattern recognition receptors |
| PSTVd | Potato spindle tuber viroid |
| PTI | Pattern-triggered immunity |
| PVX | Potato virus X |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
| SEL | Size exclusion limit |
| SYTA | Synaptotagmin A |
| TGB | Triple gene block |
| TMV | Tobacco mosaic virus |
Being more than simply passive conduits for trafficking, PD also act as hubs capable of integrating multiple signals from the plant development and defense pathways. How do plants protect themselves from pathogens invasion relying on PD? The underlying molecular mechanisms have been elucidated by a few studies. Callose deposition at PD was proven able to restrict infection by pathogen (Cheval and Faulkner, 2018; Wu et al., 2018), suggesting one potential mechanism. The expression levels of CalS1, 5, 9, 10 and 12 genes were stimulated by Hyaloperonospora infection and a SA treatment, whereas the induction of CalS1 and CalS12 was significantly repressed in the npr1 mutant, thus implying a NPR1-dependent regulation (Dong et al., 2008). In the cals1 mutant, callose at the PD is not affected by either an SA treatment or Pseudomonas infection (Cui and Lee, 2016), which suggests CalS1 is essential for SA-mediated callose deposition. The pdlp1/2/3 triple mutant is more susceptible to the downy mildew pathogen Hyaloperonospora arabidopsidis, whereas PDLP1 overexpression increases callose deposition around the haustoria and enhances plant resistance (Caillaud et al., 2014). PDLP5, localized at the central region of PD, plays a positive role in conferring an enhanced innate immunity of host plants against bacterial pathogens in a SA-dependent manner, by modulating PD callose deposition (Lee et al., 2011; Wang et al., 2013). Enrichment of t18:0-based sphingolipids were found to facilitate the recruitment of PDLP5 proteins to PD, which consequently led to reduced PD conductivity and enhanced resistance to the fungal-wilt pathogen Verticillium dahlia and the bacterium Pst DC3000 (Liu et al., 2020). Remorins are plant-specific proteins found especially in PM microdomains (Raffaele et al., 2009). Applying SA to plants can trigger a remorin-dependent reorganization of lipid raft nanodomains at PD, thereby modifying the inner structure of PD to impede viral spreading in hosts (Huang et al., 2019). Further, remorins can physically interact with TGBp1, a MP of PVX, to impede the cell-to-cell spread of PVX in tomato leaves (Raffaele et al., 2009).
The number and architecture of PD vary among different cell types and plant developmental stages, which enables the dynamic changes of symplastic transport (Ormenese et al., 2000; Ehlers and Kollmann, 2001; Burch-Smith et al., 2011). During the floral transition of the shoot apical meristem in Sinapis alba, for example, the PD frequency increased substantially (Ormenese et al., 2000). While sink leaf cells may contain simple PD in excess of 90%, in stark contrast the source leaf cells mainly contain highly branched PD in Arabidopsis thaliana. Correspondingly, the PD in sink cells permit the transport of relatively large molecules, whereas tissues composed of source cells predominantly show a decline in their transport ability (Oparka et al., 1999). Tomato yellow leaf curl virus infection leads to an increased number of PD in susceptible tomato plants (Reuveni et al., 2015). Similarly, in Casuarina glauca nodules there are fewer PD, perhaps because of the cell enlargement combined with a failed secondary PD formation (Schubert et al., 2013). One study proved ABA negatively regulates PD permeability via callose induction, leading to restricted viral cell-to-cell spreading (Alazem and Lin, 2017). Another study showed treating plants with ABA can modify the number, width, and frequency of their PD (Kitagawa et al., 2019). Collectively, these findings indicate that host plants may reduce and modulate the density and architecture of PD to better defend against invading pathogens. Further investigation is arguably needed to explore in depth the functional PD regulators involved.
Strategies for Improving Disease Resistance of Hosts By Modulating PD
Overall, it is evident that PD can be employed as a weapon, by both pathogens and their hosts, who may compete for control of key PD sites. Although PD confer benefits to both pathogenic infections and their host defense responses (Tables 1, 2), we can try to impede the invasion of one or more pathogens by developing corresponding strategies capable of modifying the PD of the host accordingly. Due to the possible trade-off in functioning between the closure of PD and symplastic transmission of immune signals (Lim et al., 2016b), these strategies must feature quick and effective regulation of PD conductivity spatiotemporally. The prompt and timely induction of PD closure in hosts suffering pathogen attacks are thus speculated to block the trafficking of pathogens, effectors, and toxic molecules from the primary invaded cells into adjacent cells, as well as the needed nutrient import into invaded cells for pathogen growth (Lee et al., 2011; Zavaliev et al., 2011); this might weaken the necessity of systemic immune signal transport. Based on previous findings, we propose three promising hypothesized approaches to spatiotemporally induce callose overproduction and PD closure after pathogen invasion, which would be worth trying to improve plant resistance against enemies (Figure 2).
Table 2.
Experimentally studied Host proteins/lipids that can regulate the plasmodesmata for plant defense.
| Proteins/lipids | Function | References |
|---|---|---|
| LYM2-LYK4 | PD located LYM2-LYK4 recognize the chitin and trigger downstream signaling to reduce the molecular flux through PD. | Faulkner et al., 2013; Cheval et al., 2020 |
| FLS2 | FLS2 is observed in the vicinity of PD and mediates flg22-triggered changes of PD-mediated trafficking. | Faulkner et al., 2013 |
| RBOHD | RBOHD produce ROS that induces PD closure in the signaling cascade of LYM2-LYK4. | Cheval et al., 2020 |
| CalS1 | Callose deposition | Dong et al., 2008; Cui and Lee, 2016 |
| CalS12 | Callose deposition | Dong et al., 2008 |
| PDLP1 | Callose deposition | Caillaud et al., 2014 |
| PDLP5 | Callose deposition | Lee et al., 2011; Wang et al., 2013 |
| Calreticulin | Calreticulin interact directly with TMV MP and interferes with targeting of TMV MP to delay cell-to-cell movement of the virus. | Chen et al., 2005 |
| Remorins | Remorins interact with MP TGBp1 of PVX and impairs PVX movement. | Raffaele et al., 2009 |
| Remorins narrow the PD channels to impede virus spreading depended on SA signaling. | Huang et al., 2019 | |
| Sphingolipids | Sphingolipids recruited PDLP5 proteins to PD, which consequently results in the decreased PD conductivity. | Liu et al., 2020 |
Figure 2.
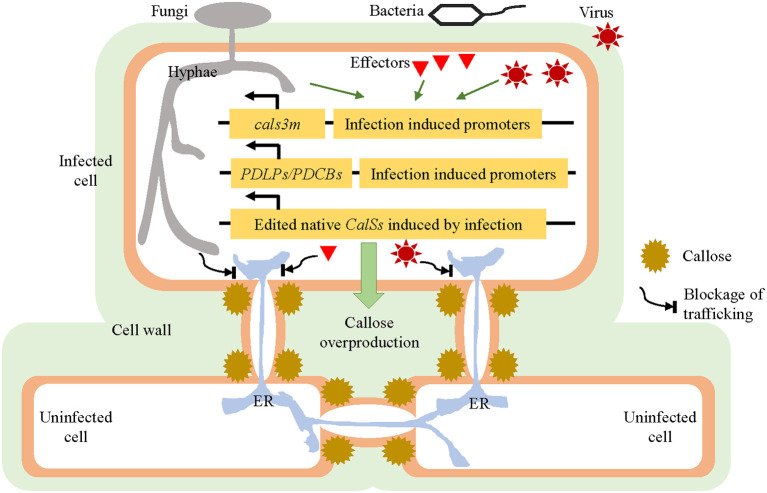
Strategies for improving the disease resistance of hosts by modulating their plasmodesmata (PD). Under normal plant growing conditions, the cals3m, PDLPs/PDCBs, and edited CalSs genes are expressed at a relatively low level, without affecting the plants' ordinary development. Once a pathogen attack is detected, the expression of cals3m, PDLPs/PDCBs, and edited CalSs could be induced quickly and strongly, resulting in prompt callose overproduction around the PD and PD closure; this is followed by imposing a blockage of connections between the primary invaded cells and neighboring uninfected cells, which should also stop the intercellular trafficking of pathogens or effectors. These hypothesized approaches are supposed to slow down the spread of pathogens and thus enhance the disease resistance of host plants. PD, plasmodesmata; CalS, callose synthase; PDLP, PD-Localized Protein; PDCB, Plasmodesmata Callose-Binding Protein.
Inducible Callose Overproduction by icals3m System
Vatén et al. (2011) developed a system, named icals3m, which blocks PD-mediated trafficking by inducing the overproduction of callose surrounding PD in a cell-specific manner. The icals3m system has been widely applied to the studies of intercellular trafficking of proteins and small RNAs in biological processes. For example, the symplastic movements of the transcription factor SHORT-ROOT and microRNA165 between the stele and the endodermis were confirmed by the study in plants expressing pCRE1::icals3m and p6xUAS::icals3m (Vatén et al., 2011). The cals3m was also used to investigate cell-cell connectivity between pericycle cells, founder cells, and the neighboring tissues during lateral root formation and patterning in Arabidopsis thaliana (Benitez-Alfonso et al., 2013). In the shoot apical meristem, cals3m expression could lead to abnormal development and differentiation due to limited movement of WUSCHEL (Daum et al., 2014). Inducible blocking of symplastic signaling going in and out of endodermis by cals3m disrupts the coordinated growth and development of roots, which includes an increase of cell layers and the misspecification of stele cells (Wu et al., 2016).
The icals3m system provides a wonderful tool for spatially and temporally modulating the aperture of PD. The strategy is to introduce the icals3m under the control of pathogen infection-induced promoters into the hosts. Therefore, PD trafficking should get blocked, due to ectopic callose synthesis, once pathogens invade the host cells. The best situation would be that where the attacking pathogens are trapped in primary infected cells without any further spread. In such a case, the usual trafficking of immune signals might not be required even they are also affected. The following three important points likely merit consideration as prerequisites for this approach. First, the promoters must be induced only by pathogen invasion, so they remain inactive or active at very low levels under normal conditions. Otherwise, the callose produced by cals3m might interfere with the usual growth and development of host plants. Second, the promoters must respond to the pathogen invasion as soon as possible, preferably prior to the start of its spread into the second plant cell. Third, the induced activities of the promoters must be high enough to yield sufficient callose to constrict the PD. It is known that plant defense responses vary within the same host and among differing ones against different pathogens, so the screening, analysis, and testing for appropriate promoters are crucial steps.
Inducible Callose Overproduction Utilizing the PDLPs/PDCBs
PDLPs and PDCBs are well known for being positive regulators of callose production. Compared to wild-type plants, overexpression of PDLP5 restricts the movement of the symplastic tracers CFDA and GFP and some MPs, and conversely the reduction of PDLP5 leads to increased intercellular trafficking (Lee et al., 2011). These findings indicate that changes in PDLP5 expression were sufficient to regulate both basal PD permeability and MP movement. Similarly, overexpression of PDLP1 decreased the efficiency of protein diffusion through PD (Thomas et al., 2008). Furthermore, the overexpression of both PDLP1 and PDLP5 enhanced plant resistance against pathogens revealing a positive relationship between the levels of PDLPs and plant resistance (Lee et al., 2011; Caillaud et al., 2014). The PDCBs are located at the outer neck region of PD, and greater expression of PDCB1 can lead to increased callose deposition and reduced cell-to-cell trafficking (Simpson et al., 2009). Therefore, we speculate that a timely increase in the expression of PDLPs or PDCBs, or both, could make same contribution to plant defense as cals3m. The same selective promoters mentioned above in icals3m system may be applied to drive the expression of PDLPs and PDCBs to increase the callose deposition at the initially infected cells during the onset of infection, thereby preventing pathogens from continuing to invade uninfected tissues. However, a study showed that PDLP5-overexpressing plants are still susceptibility to turnip crinkle virus (Lim et al., 2016a), probably due to the ability of virus to alter the aperture of PD (Singh et al., 2017). It is hoped that our approach will prove useful for helping to augment plant resistance to some pathogens to a certain extent. It cannot be expected to inhibit all possible pathogen infections facing host plants due to their different and unknown pathogenic mechanisms.
Gene Editing of Native Callose Synthases in Hosts
Vatén et al. (2011) identified three allelic semidominant A. thaliana mutants called cals3-1d, -2d, and -3d, which showed aberrant unloading patterns due to the blockage of PD. The cals3-1d, cals3-2d, and cals3-3d mutations lead to non-synonymous amino acid changes of R84K, R1926K, and P189L, respectively. By combining the two mutations of R84K and R1926K together, the enzymatic activity of encoded callose synthase (cals3m) is increased by 10 to 50% (Vatén et al., 2011). In brief, such mutations in CALS3 can foster the increased production of callose and reduced aperture of PD that together impair cell-to-cell trafficking activity. This raises an intriguing hypothesis: the introduction of same-site mutations of cals3m into other native CalS genes that are quickly and dramatically induced by pathogen attacks, may function similarly as cals3m, precluding the introduction of an exogenous gene resource. CRISPR technology is a suitable choice for gene editing (Zaynab et al., 2020). For instance, it was reported that the expression levels of CalS1 and CalS12 were highly induced in response to biotic stresses (Dong et al., 2008; Cui and Lee, 2016). Through sequence alignments, we found that both 84R and 1926R of CalS3 are conserved in CalS1 and CalS12 (Supplementary Figure 1), suggesting the feasibility of generating cals1m and cals12m similarly. However, a pre-test for screening those modifications that do not interfere with the normal functioning of plants in the absence of pathogens is still necessary. When pathogens attack, these improved CalS proteins are then functioning at high efficiency.
Future Perspectives
Over the last few decades, findings have increasingly emerged which are helpful for addressing how pathogens modify the PD structure and permeability to facilitate their intercellular movement and how plants manipulate PD to impede pathogenic infections. It is known that various PD-localized components are involved in the interactions between pathogens and plants, but many questions about mechanistic differences in how PD are regulated remain unanswered. For example, is there any conserved molecular mechanism conferring symplastic mobility to various pathogens? How do some pathogens or effectors overcome the blockage of PD by callose, and why do others fail to? How do plants manage themselves to gain control over the modification of PD when competing for this with pathogens during an infection? Previously, high-resolution electron microscopy and genetic approaches have greatly advanced our understanding of PD structure and function. Methodological improvements in the isolation and purification of PD may be helpful for identifying new PD components and examining their modifications that occur during interactions between pathogens and plants. PD regulation by pathogens and plants could provide us with a new perspective for the genetic improvement of plant disease resistance.
Author Contributions
JL, LZ, and DY wrote the manuscript. DY and JL designed the figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (32070192), the Program of Introducing Talents of Discipline to Universities (111 Project, D16014), and the Scientific Research Foundation for Advanced Talents of Henan University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.644870/full#supplementary-material
Sequence alignments of callose synthase proteins in A. thaliana. Protein sequences from AtCalS1 (NP_563743.2), AtCalS2 (Q9SL03.3), AtCalS3 (Q9LXT9.3), AtCalS4 (Q9LTG5.2), AtCalS5 (Q3B724.1), AtCalS6 (Q9LYS6.2), AtCalS7 (NP_172136.2), AtCalS8 (Q9LUD7.2), AtCalS9 (Q9SFU6.2), AtCalS10 (ACV04899.1), AtCalS11 (Q9S9U0.1), and AtCalS12 (Q9ZT82.1). The red boxes and asterisks denote those amino acids mutated in cals3m which are conserved in CalS1 and CalS12.
References
- Achor D., Welker S., Ben-Mahmoud S., Wang C., Folimonova S. T., Dutt M., et al. (2020). Dynamics of candidatus liberibacter asiaticus movement and sieve-pore plugging in citrus sink cells. Plant Physiol. 182, 882–891. 10.1104/pp.19.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkar-Purushothama C. R., Brosseau C., Giguere T., Sano T., Moffett P., Perreault J. P. (2015). Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 27, 2178–2194. 10.1105/tpc.15.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M., Lin N. S. (2017). Antiviral roles of abscisic acid in plants. Front. Plant Sci. 8:1760. 10.3389/fpls.2017.01760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzhanova D. V., Hagiwara Y., Peremyslov V. V., Dolja V. V. (2000). Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268, 192–200. 10.1006/viro.1999.0155 [DOI] [PubMed] [Google Scholar]
- Amari K., Boutant E., Hofmann C., Schmitt-Keichinger C., Fernandez-Calvino L., Didier P., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 6:e1001119. 10.1371/journal.ppat.1001119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsbury S., Kirk P., Benitez-Alfonso Y. (2017). Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 69, 105–115. 10.1093/jxb/erx337 [DOI] [PubMed] [Google Scholar]
- Asurmendi S. R. H., Berg J. C. K., Beachy R.N. (2004). Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 101, 1415–1420. 10.1073/pnas.0307778101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Kim P., Li Z., Joe A., Kvitko B., Alfano J. R., et al. (2020). Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues. Plant Cell 32, 595–611. 10.1105/tpc.19.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D., Prokhnevsky A. I., Dolja V. V. (2008). Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J. Virol. 82, 2836–2843. 10.1128/JVI.02246-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Pendle A., Miyashima S., Helariutta Y., Maule A. (2013). Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 26, 136–147. 10.1016/j.devcel.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A. J. (2010). Plasmodesmata: gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23, 1403–1412. 10.1094/MPMI-05-10-0116 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D., Chakraborty S. (2018). Chloroplast: the Trojan in plant-virus interaction. Mol. Plant Pathol. 19, 504–518. 10.1111/mpp.12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J. M. (2006). Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. 10.4454/jpp.v88i1.82825372135 [DOI] [Google Scholar]
- Brault M. L., Petit J. D., Immel F., Nicolas W. J., Glavier M., Brocard L., et al. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep. 20:e47182. 10.15252/embr.201847182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill L. M., Nunn R. S., Kahn T. W., Yeager M., Beachy R. N. (2000). Recombinant tobacco mosaic virus movement protein is an RNA-binding, α-helical membrane protein. Proc. Natl. Acad. Sci. U.S.A. 87, 7112–7117. 10.1073/pnas.130187897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard J. O., Runkel A. M., Zambryski P. C. (2015). The cytosol must flow: intercellular transport through plasmodesmata. Curr. Opin. Cell Biol. 35, 13–20. 10.1016/j.ceb.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Brunkard J. O., Zambryski P. C. (2017). Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 35, 76–83. 10.1016/j.pbi.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Bucher G. L., Tarina C., Heinlein M., Serio F. D., Fred Meins J., Iglesias V. A. (2001). Local expression of enzymatically active class I β-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28, 361–369. 10.1046/j.1365-313X.2001.01181.x [DOI] [PubMed] [Google Scholar]
- Bücherl C. A., Jarsch I. K., Schudoma C., Segonzac C., Mbengue M., Robatzek S., et al. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6:e25114. 10.7554/eLife.25114.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T. M., Stonebloom S., Xu M., Zambryski P. C. (2011). Plasmodesmata during development: re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma 248, 61–74. 10.1007/s00709-010-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M. C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S. J., Jones A. M., et al. (2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10:e1004496. 10.1371/journal.ppat.1004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Blekemolen M. C., Tintor N., Cornelissen B. J. C., Takken F. L. W. (2018). The fusarium oxysporum avr2-six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of avr2. Mol. Plant 11, 691–705. 10.1016/j.molp.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Chen M. H., Tian G. W., Gafni Y., Citovsky V. (2005) Effects of Calreticulin on Viral Cell-to-Cell Movement. Plant Physiol. 183, 1866–1876. 10.1104/pp.105.064386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Y., Kim J. Y. (2009). Callose synthesis in higher plants. Plant Signal Behav. 4, 489–492. 10.4161/psb.4.6.8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Dong M., Xu Q., Peng L., Yang Z., Wei T., et al. (2017). Dissecting the molecular mechanism of the subcellular localization and cell-to-cell movement of the sugarcane mosaic virus P3N-PIPO. Sci. Rep. 7:9868. 10.1038/s41598-017-10497-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheval C., Faulkner C. (2018). Plasmodesmal regulation during plant-pathogen interactions. New Phytol. 217, 62–67. 10.1111/nph.14857 [DOI] [PubMed] [Google Scholar]
- Cheval C., Samwald S., Johnston M. G., De Keijzer J., Breakspear A., Liu X., et al. (2020). Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc. Natl. Acad. Sci. U.S.A. 117, 9621–9629. 10.1073/pnas.1907799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y. L., Hung Y. J., Tseng Y. H., Hsu H. T., Yang J. Y., Wung C. H., et al. (2013). The stable association of virion with the triple-gene-block protein 3-based complex of Bamboo mosaic virus. PLoS Pathog. 9:e1003405. 10.1371/journal.ppat.1003405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R. N., Lasky D., Karki H., Zhang Z., Goyer A., Halterman D., et al. (2020). HCPro suppression of callose deposition contributes to strain-specific resistance against potato virus Y. Phytopathology 110, 164–173. 10.1094/PHYTO-07-19-0229-FI [DOI] [PubMed] [Google Scholar]
- Conrath U. (2006). Systemic acquired resistance. Plant Signal Behav. 1, 179–184. 10.4161/psb.1.4.3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- Cui W., Lee J.-Y. (2016). Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2:16034. 10.1038/nplants.2016.34 [DOI] [PubMed] [Google Scholar]
- Daum G., Medzihradszky A., Suzaki T., Lohmann J. U. (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14619–14624. 10.1073/pnas.1406446111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Kwon M.-O., Hammond R., Owens R. (1997). Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12, 931–936. 10.1046/j.1365-313X.1997.12040931.x [DOI] [PubMed] [Google Scholar]
- Djamei A., Schipper K., Rabe F., Ghosh A., Vincon V., Kahnt J., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478, 395–398. 10.1038/nature10454 [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Dolja V. V. (2003). Beet yellows virus: the importance of being different. Mol. Plant Pathol. 4, 91–98. 10.1046/j.1364-3703.2003.00154.x [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Haldeman-Cahill R., Montgomery A. E., Vandenbosch K. A., Carrington J. C. (1995). Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 206, 1007–1016. 10.1006/viro.1995.1023 [DOI] [PubMed] [Google Scholar]
- Dong X., Hong Z., Chatterjee J., Kim S., Verma D. P. S. (2008). Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229, 87–98. 10.1007/s00425-008-0812-3 [DOI] [PubMed] [Google Scholar]
- Ehlers K., Kollmann R. (2001). Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma 216, 1–30. 10.1007/BF02680127 [DOI] [PubMed] [Google Scholar]
- Epel B. L. (1994). Plasmodesmata: composition, structure and trafficking. Plant Mol. Biol. 26, 1343–1356. 10.1007/BF00016479 [DOI] [PubMed] [Google Scholar]
- Epel B. L. (2009). Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1,3-glucanases. Semin. Cell Dev. Biol. 20, 1074–1081. 10.1016/j.semcdb.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Faulkner C., Petutschnig E., Benitez-Alfonso Y., Beck M., Robatzek S., Lipka V., et al. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 110, 9166–9170. 10.1073/pnas.1203458110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., et al. (2011). Arabidopsis plasmodesmal proteome. PLoS ONE 6:e18880. 10.1371/journal.pone.0018880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Hernández C., Martínez de Alba A. E., Daròs J.-A., Di Serio F. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43, 117–139. 10.1146/annurev.phyto.43.040204.140243 [DOI] [PubMed] [Google Scholar]
- Ganusova E. E., Burch-Smith T. M. (2019). Review: Plant-pathogen interactions through the plasmodesma prism. Plant Sci. 279, 70–80. 10.1016/j.plantsci.2018.05.017 [DOI] [PubMed] [Google Scholar]
- Ganusova E. E., Rice J. H., Carlew T. S., Patei A., Perrodin-Njoku E., et al. (2017). Altered expression of chloroplast protein affects the outcome of virus and nematode infection. Mol. Plant Microbe In 30, 478–488. 10.1094/MPMI-02-17-0031-R [DOI] [PubMed] [Google Scholar]
- Genoves A., Navarro J. A., Pallas V. (2006). Functional analysis of the five melon necrotic spot virus genome-encoded proteins. J. Gen. Virol. 87, 2371–2380. 10.1099/vir.0.81793-0 [DOI] [PubMed] [Google Scholar]
- Germain H., Joly D. L., Mireault C., Plourde M. B., Letanneur C., Stewart D., et al. (2018). Infection assays in Arabidopsis reveal candidate effectors from the poplar rust fungus that promote susceptibility to bacteria and oomycete pathogens. Mol. Plant Pathol. 18, 191–200. 10.1111/mpp.12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R., Jiang J., Wan J., Agbeci M., Zheng H., Lalibert,é J. F. (2013). 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 4:351. 10.3389/fmicb.2013.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. R., Fisher E. J., Chang J. H., Mole B. M., Dangl J. L. (2006). Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60, 425–449. 10.1146/annurev.micro.60.080805.142251 [DOI] [PubMed] [Google Scholar]
- Grison M. S., Kirk P., Brault M. L., Wu X. N., Schulze W. X., Benitez-Alfonso Y., et al. (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol. 181, 142–160. 10.1104/pp.19.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra T., Schilling S., Hake K., Gorzolka K., Sylvester F.-P., Conrads B., et al. (2020). Calcium-dependent protein kinase 5 links calcium signaling with N-hydroxy-L-pipecolic acid- and SARD1-dependent immune memory in systemic acquired resistance. N. Phytol. 225, 310–325. 10.1111/nph.16147 [DOI] [PubMed] [Google Scholar]
- Hacker D. L., Petty I. T., Wei N., Morris T. J. (1992). Turnipcrinkle virus genes required for RNA replication and virus movement. Virology 186, 1–8. 10.1016/0042-6822(92)90055-T [DOI] [PubMed] [Google Scholar]
- Hake K., Romeis T. (2019). Protein kinase-mediated signalling in priming: Immune signal initiation, propagation, and establishment of long-term pathogen resistance in plants. Plant Cell Environ. 42, 904–917. 10.1111/pce.13429 [DOI] [PubMed] [Google Scholar]
- Han X., Kumar D., Chen H., Wu S., Kim J. Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65, 1737–1749. 10.1093/jxb/ert422 [DOI] [PubMed] [Google Scholar]
- Haupt S., Cowan G. H., Ziegler A., Roberts A. G., Oparka K. J., Torrance L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17, 164–181. 10.1105/tpc.104.027821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M. C., Valent B., Howard R. J., Chumley F. G. (1990). Interactions of two strains of Magnaporthe grisea with rice, goosegrass, and weeping lovegrass. Can. J. Bot. 68, 1627–1637. 10.1139/b90-209 [DOI] [Google Scholar]
- Heinlein M., Epel B. L. (2004). Macromolecular transport and signaling through plasmodesmata. Int. Rev. Cytol. 235, 93–164. 10.1016/S0074-7696(04)35003-5 [DOI] [PubMed] [Google Scholar]
- Howard A. R., Heppler M. L., Ju H. J., Krishnamurthy K., Payton M. E., Verchot-Lubicz J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328, 185–197. 10.1016/j.virol.2004.06.039 [DOI] [PubMed] [Google Scholar]
- Howard R. J., Valent B. (1996). Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. 10.1146/annurev.micro.50.1.491 [DOI] [PubMed] [Google Scholar]
- Huang D., Sun Y., Ma Z., Ke M., Cui Y., Chen Z., et al. (2019). Salicylic acid-mediated plasmodesmal closure via remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U.S.A. 1176, 21274–21284. 10.1073/pnas.1911892116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. (2014). Plant to plant movement, in Plant Virology, fifth edition, ed. Hull R. (Boston, Academic Press), 669–751. 10.1016/B978-0-12-384871-0.00012-1 [DOI] [Google Scholar]
- Hunter K., Kimura S., Rokka A., Tran H. C., Toyota M., Kukkonen J. P., et al. (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180, 2004–2021. 10.1104/pp.19.00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias V. A., Meins M., Jr. (2000). Movement of plant viruses is delayed in β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal sizeexclusion limit and enhanced callose deposition. Plant J. 21, 157–166. 10.1046/j.1365-313x.2000.00658.x [DOI] [PubMed] [Google Scholar]
- Jia L. J., Tang H. Y., Wang W. Q., Yuan T. L., Wei W. Q., Pang B., et al. (2019). A linear nonribosomal octapeptide from Fusarium graminearum facilitates cell-to-cell invasion of wheat. Nat. Commun. 10:922. 10.1038/s41467-019-08726-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. J., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. 10.1105/tpc.106.046300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C. H., Berruyer R., Giraldo M. C., Kankanala P., Park S.-Y., Czymmek K., et al. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22, 1388–1403. 10.1105/tpc.109.069666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Tomoi T., Fukushima T., Sakata Y., Sato M., Toyooka K., et al. (2019). Abscisic acid acts as a regulator of molecular trafficking through plasmodesmata in the moss Physcomitrella patens. Plant Cell Physiol. 60, 738–751. 10.1093/pcp/pcy249 [DOI] [PubMed] [Google Scholar]
- Klessig D. F., Choi H. W., Dempsey D. A. (2018). Systemic acquired resistance and salicylic acid: past, present, and future. Mol. Plant Microbe Interact. 31, 871–888. 10.1094/MPMI-03-18-0067-CR [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Otegui M. S., Krishnakumar S., Mindrinos M., Zambryski P. (2007). INCREASED SIZE EXCLUSION LIMIT2 Encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19, 1885–1897. 10.1105/tpc.106.045666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh E.-J., Zhou L., Williams D. S., Park J., Ding N., Duan Y.-P., et al. (2012). Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacterasiaticus”. Protoplasma 249, 687–697. 10.1007/s00709-011-0312-3 [DOI] [PubMed] [Google Scholar]
- Laporte C., Vetter G., Loudes A. M., Robinson D. G., Hillmer S., Stussi-Garaud C., et al. (2003). Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15, 2058–2075. 10.1105/tpc.013896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre R., Evangelisti E., Rey T., Schornack S. (2015). Modulation of host cell biology by plant pathogenic microbes. Annu. Rev. Cell Dev. Biol. 31, 201–229. 10.1146/annurev-cellbio-102314-112502 [DOI] [PubMed] [Google Scholar]
- Lee J. Y. (2014). New and old roles of plasmodesmata in immunity and parallels to tunneling nanotubes. Plant Sci. 221-222, 13–20. 10.1016/j.plantsci.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Lu H. (2011). Plasmodesmata: the battleground against intruders. Trends Plant Sci. 16, 201–210. 10.1016/j.tplants.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., et al. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23, 3353–3373. 10.1105/tpc.111.087742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lent J. V., Storms M., van der Meer F., Weilink J., Goldbach R. (1991). Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protoplasts. J. General Virol. 72, 2615–2623. 10.1099/0022-1317-72-11-2615 [DOI] [PubMed] [Google Scholar]
- Levy A., Zheng J. Y., Lazarowitz S. G. (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 25, 2018–2025. 10.1016/j.cub.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Lazarowitz S. G. (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. U.S.A. 107, 2491–2496. 10.1073/pnas.0909080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhao Y., Liu C., Yao G., Wu S., Hou C., et al. (2012). Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant ell Rep. 31, 905–916. 10.1007/s00299-011-1211-y [DOI] [PubMed] [Google Scholar]
- Li W. Z., Qu F., Morris T. J. (1998). Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244, 405–416. 10.1006/viro.1998.9125 [DOI] [PubMed] [Google Scholar]
- Li Z., Variz H., Chen Y., Liu S., Aung K. (2020). Plasmodesmata-dependent intercellular movement of bacterial effectors. BioRxiv. 10.1101/2020.12.10.420240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G., Kachroo A., Kachroo P. (2016a). Role of plasmodesmata and plasmodesmata localizing proteins in systemic immunity. Plant Signal Behav. 11:e1219829. 10.1080/15592324.2016.1219829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G., Shine M. B., de Lorenzo L., Yu K., Cui W., Navarre D., et al. (2016b). Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19, 541–549. 10.1016/j.chom.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Lim H. S., Bragg J. N., Ganesan U., Lawrence D. M., Yu J., Isogai M., et al. (2008). Triple gene block protein interactions involved in movement of Barley stripe mosaic virus. J. Virol. 82, 4991–5006. 10.1128/JVI.02586-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Z., Blancaflor E. B., Nelson R. S. (2005). The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 138, 1853–1865. 10.1104/pp.105.065722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhang T., Liu Z., Chen X., Guo H., Ju B., et al. (2020). Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5-stimulated callose accumulation in Arabidopsis. Mol. Plant 13, 128–143. 10.1016/j.molp.2019.10.013 [DOI] [PubMed] [Google Scholar]
- Lucas W. J. (2006). Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344, 169–184. 10.1016/j.virol.2005.09.026 [DOI] [PubMed] [Google Scholar]
- Lucas W. J., Ham B. K., Kim J. Y. (2009). Plasmodesmata-bridging the gap between neighboring plant cells. Trends Cell Biol. 19, 495–503. 10.1016/j.tcb.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Lucas W. J., Lee J. Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5, 712–726. 10.1038/nrm1470 [DOI] [PubMed] [Google Scholar]
- Martinez-Turino S., Hernandez C. (2011). A membrane-associated movement protein of Pelargonium flower break virus shows RNA-binding activity and contains a biologically relevant leucine zipper-like motif. Virology 413, 310–319. 10.1016/j.virol.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Maule A. J. (2008). Plasmodesmata: structure, function and biogenesis. Curr. Opin. Plant Biol. 11, 680–686. 10.1016/j.pbi.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Miyashima S., Roszak P., Sevilem I., Toyokura K., Blob B., Heo J. O., et al. (2019). Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565, 490–494. 10.1038/s41586-018-0839-y [DOI] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G. (2003). Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84, 1351–1366. 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M. C., Khang C. H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21, 1273–1290. 10.1105/tpc.107.055228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J. A., Sanchez-Navarro J. A., Pallas V. (2019). Key checkpoints in the movement of plant viruses through the host. Adv. Virus Res. 104, 1–64. 10.1016/bs.aivir.2019.05.001 [DOI] [PubMed] [Google Scholar]
- Ngou B. P. M., Ahn H., Ding P., Jones J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 592, 110–115. 10.1038/s41586-021-03315-7 [DOI] [PubMed] [Google Scholar]
- Nicolas W. J., Grison M. S., Trépout S., Gaston A., Fouch,é M., Cordelières F. P., et al. (2017). Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 3:17082. 10.1038/nplants.2017.82 [DOI] [PubMed] [Google Scholar]
- Oparka K. J., Roberts A. G., Boevink P., Cruz S. S., Roberts I., Pradel K. S., et al. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. 10.1016/S0092-8674(00)80786-2 [DOI] [PubMed] [Google Scholar]
- Ormenese S., Havelange A., Deltour R., Bernier G. (2000). The frequency of plasmodesmata increases early in the whole shoot apical meristem of Sinapis alba L. during foral transition. Planta 211, 370–375. 10.1007/s004250000294 [DOI] [PubMed] [Google Scholar]
- Peña E. J., Heinlein M. (2012). RNA transport during TMV cell-to-cell movement. Front. Plant Sci. 3:193. 10.3389/fpls.2012.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W. S., Jensen K. H., Stone H. A., Knoblauch M. (2021). Plasmodesmata and the problems with size: interpreting the confusion. J. Plant Physiol. 257:153341. 10.1016/j.jplph.2020.153341 [DOI] [PubMed] [Google Scholar]
- Pitzalis N., Heinlein M. (2017). The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J. Exp. Bot. 69, 117–132. 10.1093/jxb/erx334 [DOI] [PubMed] [Google Scholar]
- Pouwels J., Van Der Velden T., Willemse J., Borst J. W., Van Lent J., Bisseling T., et al. (2004). Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. J. Gen. Virol. 85, 3787–3796. 10.1099/vir.0.80497-0 [DOI] [PubMed] [Google Scholar]
- Prokhnevsky A. I., Peremyslov V. V., Dolja V. V. (2005). Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 79, 14421–14428. 10.1128/JVI.79.22.14421-14428.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Bayer E., Lafarge D., Cluzet S., Retana S. G., Boubekeur T., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555. 10.1105/tpc.108.064279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Prado J. S., Abulfaraj A. A., Rayapuram N., Benhamed M., Hirt H. (2018). Plant immunity: from signalings to epigenetic control of defense. Trends Plant Sci. 23, 833–844. 10.1016/j.tplants.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Ranf S. (2017). Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 38, 68–77. 10.1016/j.pbi.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Reagan B. C., Ganusova E. E., Fernandez J. C., Mccray T. N., Burch-Smith T. M. (2018). RNA on the move: the plasmodesmata perspective. Plant Sci. 275, 1–10. 10.1016/j.plantsci.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Reuveni M., Debbi A., Kutsher Y., Gelbart D., Zemach H., Belausov E., et al. (2015). Tomato yellow leaf curl virus effects on chloroplast biogenesis and cellular structure. Physiol. Mol. Plant Pathol. 92, 51–58. 10.1016/j.pmpp.2015.08.001 [DOI] [Google Scholar]
- Ritzenthaler C., Hofmann C. (2007). Tubule-guided movement of plant viruses, in Viral Transport in Plants. Berlin: Springer Berlin Heidelberg; 63–83. 10.1007/7089_2006_105 [DOI] [Google Scholar]
- Roberts A. G., Oparka K. J. (2003). Plasmodesmata and the control of symplastic transport. Plant Cell Environment 26, 103–124. 10.1046/j.1365-3040.2003.00950.x [DOI] [Google Scholar]
- Sager R., Lee J.-Y. (2014). Plasmodesmata in integrated cell signalling insights from development and environmental signals and stresses. J. Exp. Bot. 65, 6337–6358. 10.1093/jxb/eru365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Loo E. P., Yasuda S. (2018). Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 93, 592–613. 10.1111/tpj.13808 [DOI] [PubMed] [Google Scholar]
- Sakulkoo W., Osés-Ruiz M., Garcia E. O., Soanes D. M., Littlejohn G. R., Hacker C., et al. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359, 1399–1403. 10.1126/science.aaq0892 [DOI] [PubMed] [Google Scholar]
- Schubert M., Koteyeva N. K., Zdyb A., Santos P., Voitsekhovskaja O. V., Demchenko K. N., et al. (2013). Lignification of cell walls of infected cells in Casuarina glauca nodules that depend on symplastic sugar supply is accompanied by reduction of plasmodesmata number and narrowing of plasmodesmata. Physiol. Plant 147, 524–540. 10.1111/j.1399-3054.2012.01685.x [DOI] [PubMed] [Google Scholar]
- Shemyakina E. A., Erokhina T. N., Gorshkova E. N., Schiemann J., Solovyev A. G., Morozov S. Y. (2011). Formation of protein complexes containing plant virus movement protein TGBp3 is necessary for its intracellular trafficking. Biochimie 93, 742–748. 10.1016/j.biochi.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A. J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21, 581–594. 10.1105/tpc.108.060145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Lim G.-H., Kachroo P. (2017). Transport of chemical signals in systemic acquired resistance. J. Integr. Plant Biol. 59, 336–344. 10.1111/jipb.12537 [DOI] [PubMed] [Google Scholar]
- Stahl Y., Grabowski S., Bleckmann A., Kuhnemuth R., Weidtkamp-Peters S., Pinto K. G., et al. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23, 362–371. 10.1016/j.cub.2013.01.045 [DOI] [PubMed] [Google Scholar]
- Su S., Liu Z., Chen C., Zhang Y., Wang X., Zhu L., et al. (2010). Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22, 1373–1387. 10.1105/tpc.108.064212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda R., Ding B. (2009). Viroid intercellular trafficking: RNA motifs, cellular factors and broad impacts. Viruses 1, 210–221. 10.3390/v1020210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai A., Meshi T. (2001). Cell-to-cell movement of potato virus X: the role of p12 and p8 encoded by the second and third open reading frames of the triple gene block. Mol. Plant-Microbe Interact. 14, 1158–1167. 10.1094/MPMI.2001.14.10.1158 [DOI] [PubMed] [Google Scholar]
- Thomas C. L., Bayer E. M., Ritzenthaler C., Fernandez-Calvino L., Maule A. J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6:e7. 10.1371/journal.pbio.0060007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. L., Maule A. J. (1995). Identification of structural domains within the cauliflower mosaic virus movement protein by scanning deletion mutagenesis and epitope tagging. Plant Cell 7, 561–572. 10.1105/tpc.7.5.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I. M., Chapman S. N., Oparka K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. 10.1083/jcb.201304003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Nicolas W., Rosado A., Bayer E. M. (2016). Staying tight: plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annu. Rev. Plant Biol. 67, 337–364. 10.1146/annurev-arplant-043015-111840 [DOI] [PubMed] [Google Scholar]
- Tomczynska I., Stumpe M., Doan T. G., Mauch F. (2020). A Phytophthora effector protein promotes symplastic cell-to-cell trafficking by physical interaction with plasmodesmata-localised callose synthases. New Phytol. 227, 1467–1478. 10.1111/nph.16653 [DOI] [PubMed] [Google Scholar]
- Toruno T. Y., Stergiopoulos I., Coaker G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. 10.1146/annurev-phyto-080615-100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A., Shimada-Beltran H., Levy A., Zheng J. Y., Javia P. A., Lazarowitz S. G. (2014). The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 5:584. 10.3389/fpls.2014.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A., Dettmer J., Wu S., Stierhof Y. D., Miyashima S., Yadav S. R., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21, 1144–1155. 10.1016/j.devcel.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Vilar M., Sauri A., Monne M., Marcos J. F., von Heijne G., Perez-Paya E., et al. (2002). Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J. Biol. Chem. 277, 23447–23452. 10.1074/jbc.M202935200 [DOI] [PubMed] [Google Scholar]
- Waigmann E., Lucas W. J., Citovsky V., Zambryski P. (1994). Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc. Nati. Acad. Sci. U.S.A. 91, 1433–1437. 10.1073/pnas.91.4.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Ueki S., Trutnyeva K., Citovsky V. (2004). The ins and outs of nondestructive cell-to-cell and systemic movement of plant Viruses. Critical Rev. Plant Sci. 23, 195–250. 10.1080/07352680490452807 [DOI] [Google Scholar]
- Wang X., Sager R., Cui W., Zhang C., Lu H., Lee J.-Y. (2013). Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25, 2315–2329. 10.1105/tpc.113.110676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D., Gao Q., Kachroo A., Kachroo P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20 127–134. 10.1016/j.pbi.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Wolf H., Deom C. M., Beachy R. N., Lucas W. J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379. 10.1126/science.246.4928.377 [DOI] [PubMed] [Google Scholar]
- Wright K. M., Wood N. T., Roberts A. G., Chapman S., Boevink P., Mackenzie K. M., et al. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic 8, 21–31. 10.1111/j.1600-0854.2006.00510.x [DOI] [PubMed] [Google Scholar]
- Wu S., O'lexy R., Xu M., Sang Y., Chen X., Yu Q., et al. (2016). Symplastic signaling instructs cell division, cell expansion, and cell polarity in the ground tissue of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. U.S.A. 113, 11621–11626. 10.1073/pnas.1610358113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-W., Kumar R., Iswanto A. B. B., Kim J.-Y. (2018). Callose balancing at plasmodesmata. J. Exp. Bot. 69, 5325–5339. 10.1093/jxb/ery317 [DOI] [PubMed] [Google Scholar]
- Xie L., Shang W., Liu C., Zhang Q., Sunter G., Hong J., et al. (2016). Mutual association of broad bean wilt virus 2 VP37-derived tubules and plasmodesmata obtained from cytological observation. Sci. Rep. 6:21552. 10.1038/srep21552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Liu Y. (2020). Diverse regulation of plasmodesmal architecture facilitates adaptation to phloem translocation. J. Exp. Bot. 71, 2505–2512. 10.1093/jxb/erz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Yadav S. R., Paterlini A., Nicolas W. J., Petit J. D., Brocard L., et al. (2019). Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat. Plants 5, 604–615. 10.1038/s41477-019-0429-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Lu Y., Wang F., Chen Y., Tian Y., Jiang L., et al. (2020). Involvement of the chloroplast gene ferredoxin 1 in multiple responses of Nicotiana benthamiana to Potato virus X infection. J. Exp. Bot. 71, 2142–2156. 10.1093/jxb/erz565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Lazarowitz S. G., Citovsky V. (2016). Identification of a functional plasmodesmal localization signal in a plant viral cell-to-cell-movement protein. mBio 7, e02052–e02015. 10.1128/mBio.02052-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 592, 105–109. 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P. (2008). Plasmodesmata. Curr. Biol. 18, R324–325. 10.1016/j.cub.2008.01.046 [DOI] [PubMed] [Google Scholar]
- Zambryski P., Crawford K. (2000). PLASMODESMATA: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16, 393–421. 10.1146/annurev.cellbio.16.1.393 [DOI] [PubMed] [Google Scholar]
- Zamyatnin A. A., Jr., Solovyev A. G., Savenkov E. I., Germundsson A., Sandgren M., et al. (2004). Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking. Mol. Plant Microbe Interact. 17, 921–930. 10.1094/MPMI.2004.17.8.921 [DOI] [PubMed] [Google Scholar]
- Zavaliev R., Levy A., Gera A., Epel B. L. (2013). Subcellular dynamics and role of Arabidopsis beta-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol. Plant Microbe Interact. 26, 1016–1030. 10.1094/MPMI-03-13-0062-R [DOI] [PubMed] [Google Scholar]
- Zavaliev R., Ueki S., Epel B. L., Citovsky V. (2011). Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma 248, 117–130. 10.1007/s00709-010-0247-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaynab M., Sharif Y., Fatima M., Afzal M. Z., Aslam M. M., Raza M. F., et al. (2020). CRISPR/Cas9 to generate plant immunity against pathogen. Microb. Pathog. 141:103996. 10.1016/j.micpath.2020.103996 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang X., Hong Y., Liu Y. (2016). Chloroplast in plant-virus interaction. Front. Microbiol. 7:1566. 10.3389/fmicb.2016.01565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Archual A. J., Amin A. A., Ding B. (2008). A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20, 35–47. 10.1105/tpc.107.056606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments of callose synthase proteins in A. thaliana. Protein sequences from AtCalS1 (NP_563743.2), AtCalS2 (Q9SL03.3), AtCalS3 (Q9LXT9.3), AtCalS4 (Q9LTG5.2), AtCalS5 (Q3B724.1), AtCalS6 (Q9LYS6.2), AtCalS7 (NP_172136.2), AtCalS8 (Q9LUD7.2), AtCalS9 (Q9SFU6.2), AtCalS10 (ACV04899.1), AtCalS11 (Q9S9U0.1), and AtCalS12 (Q9ZT82.1). The red boxes and asterisks denote those amino acids mutated in cals3m which are conserved in CalS1 and CalS12.