Abstract
The SH-SY5Y cell line is often used as a surrogate for neurons in cell-based studies. This cell line is frequently differentiated with all-trans retinoic acid (ATRA) over a 7-day period, which confers neuron-like properties to the cells. However, no analysis of proteome remodeling has followed the progress of this transition. Here, we quantitatively profiled over 9400 proteins across a 7-day treatment with retinoic acid using state-of-the-art mass spectrometry-based proteomics technologies, including FAIMS, real-time database searching, and TMTpro16 sample multiplexing. Gene ontology analysis revealed that categories with the highest increases in protein abundance were related to the plasma membrane/extracellular space. To showcase our data set, we surveyed the protein abundance profiles linked to neurofilament bundle assembly, neuron projections, and neuronal cell body formation. These proteins exhibited increases in abundance level, yet we observed multiple patterns among the queried proteins. The data presented represent a rich resource for investigating temporal protein abundance changes in SH-SY5Y cells differentiated with retinoic acid. Moreover, the sample preparation and data acquisition strategies used here can be readily applied to any analogous cell line differentiation analysis.
Keywords: SPS-MS3, multinotch, TMTpro, eclipse, ATRA, retinoic acid
Graphical Abstract
INTRODUCTION
Transformed neuron-like cell lines are often used to overcome the limitations of primary mammalian neurons, which can no longer be propagated once differentiated into mature neurons. As such, cell culture models are often used to study drug neurotoxicity and neurodegenerative diseases, for example, Parkinson’s or Alzheimer’s disease.1-4 The human-derived cell line SH-SY5Y is commonly used as an in vitro model for human neurons.5 This cell line, which originated from a bone marrow biopsy of a patient with neuroblastoma, can be induced to differentiate into cells with a neuron-like phenotype. As such, SH-SY5Y is generally used as a model of neuronal function and differentiation in neuroscience studies as the cell line can reproduce many biochemical and morphological properties of neurons.6 Specifically, SH-SY5Y cells have been used to investigate neurotoxicity, neuroprotection, neuroblastoma tumorigenesis, neuronal differentiation and metabolism, as well as other aspects of neuroadaptive and neurodegenerative processes.4,7
Retinoic acid (all-trans-retinoic acid, ATRA) is a well-established inducer of a neuronal phenotype in SH-SY5Y cells.8,9 Retinoic acid is a vitamin A derivative with wide-ranging cellular differentiation-promoting properties.10 This metabolite’s method of action involves binding to a DNA-bound receptor, thereby affecting transcription factors, which may induce or repress the expression of nearby genes.11 As such, retinoic acid induces profound changes in the cellular proteome. SH-SY5Y cells differentiate primarily to a cholinergic neuron phenotype in response to at least 7 days of retinoic acid treatment, typically at a concentration of 10 μM.12 Specifically, neuronal differentiation involves the formation and extension of neuronal processes, induction of neuron-specific enzymes, and production of neurotransmitters.13-15 Studies frequently use either undifferentiated16-19 and retinoic acid-differentiated20-23 SH-SY5Y cells. Some studies have compared undifferentiated and differentiated SH-SY5Y cells, but they highlight only specific proteins and pathways.8 Until now, the comprehensive analysis of proteome remodeling during differentiation of SH-SY5Y has not yet been assessed.
Sample multiplexing strategies, such as isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMT), have a wide range of benefits for profiling protein abundance at the proteome level.24,25 Isobaric tagging allows multiple samples to be analyzed simultaneously, thereby reducing costs and instrument time, permitting multiple comparisons in one experiment, and producing fewer missing values within a sample set. Sample multiplexing strategies are also valuable for time-course investigations.26,27 Here, we applied such an approach to investigate SH-SY5Y cell differentiation.
We used a TMTpro-based workflow to quantitatively profile the differentiation of SH-SY5Y cells following treatment with retinoic acid. First, we performed an “end-point” experiment comparing SH-SY5Y cells following 7 days of retinoic acid treatment versus mock treatment in a TMT6-plex format and observed hundreds of significant protein alterations. Next, we expanded our study using TMTpro16 reagents28 that enabled the temporal abundance profiling of over 9400 proteins in SH-SY5Y cells with single-day resolution during a 7-day retinoic acid treatment. We also highlighted proteins related to neurofilament bundle assembly, neuron projections, and neuronal cell body formation. These data, which were collected on a Orbitrap Eclipse mass spectrometer with FAIMS and real-time database searching (RTS), represent the largest temporal protein abundance profiling data set for retinoic acid differentiation in SH-SY5Y cells to date. With this data set, we provide a rich resource that is amenable to further data mining to study individual proteins, pathways, and interactions thereof related to retinoic acid-induced SH-SY5Y differentiation.
EXPERIMENTAL SECTION
Materials
Tandem mass tag (TMT) isobaric reagents were from Thermo Fisher Scientific (Waltham, MA). All-trans-retinoic acid (R2625) was purchased from Sigma (St. Louis, MO). Water and organic solvents were from J.T. Baker (Center Valley, PA). Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) was from Life Technologies (Waltham, MA). Trypsin was purchased from Pierce Biotechnology (Rockford, IL) and LysC from Wako Chemicals (Richmond, VA). Unless otherwise noted, all other chemicals were from Pierce Biotechnology (Rockford, IL). SH-SY5Y cell line (CRL-2266) was from the American Type Culture Collection (Manassas, VA).
Cell Growth and Harvesting
Methods of cell propagation followed techniques used previously.29,30 In brief, upon ~90% confluency, we aspirated the growth media and washed the cells three times with ice-cold phosphate-buffered saline. For our end-point experiment, a single dish of cells was propagated into six dishes at low density. Three dishes were treated with 10 μM retinoic acid and three without for 7 days. Cells were washed and fresh media was added daily. For our time-course experiment, two separate dishes of cells were each divided into eight dishes. Cells were either harvested on the appropriate day or spent media was aspirated and fresh media was added. All experiments with retinoic acid were conducted in subdued light conditions. We noted neurite outgrowth, as expected, as SH-SY5Y cells exhibited typical neuronal morphology in the presence of retinoic acid after several days.8 At the appropriate time point, the cells were dislodged with a nonenzymatic reagent, harvested by trituration following the addition of 10 mL of phosphate-buffered saline, pelleted by centrifugation at 3000g for 5 min at 4 °C, and the supernatant was removed. Five hundred microliters of 200 mM EPPS, 8 M urea, pH 8.5 supplemented with 1× Pierce Protease Inhibitors were added directly to each pellet.
Cell Lysis and Protein Digestion
Cells were homogenized by 12 passes through a 21-gauge (1.25 inches long) needle and incubated at 4 °C with gentle agitation for 30 min. The homogenate was sedimented at 21 000g for 5 min. Protein concentrations were determined from the supernatant using the bicinchoninic acid (BCA) assay. Proteins were subjected to disulfide bond reduction with 5 mM tris (2-carboxyethyl) phosphine (15 min, room temperature) and alkylation with 10 mM iodoacetamide (20 min in the dark, room temperature) followed by quenching with 10 mM dithiotreitol (15 min in the dark, room temperature). Trichloroacetic acid (TCA) precipitation was performed prior to protease digestion. Samples, resuspended in 200 mM EPPS, pH 8.5, were digested for 14 h at room temperature with LysC (100:1 protein-to-protease ratio). Trypsin was then added also at a 100:1 protein-to-protease ratio and the samples were incubated at 37 °C for 6 h.
Tandem Mass Tag Labeling
For the end-point experiment, 100 μg (in 5 μL of acetonitrile) of TMT reagent was added to the peptides (50 μg) along with 15 μL of acetonitrile for a final acetonitrile concentration of ~30% (v/v). After incubating for 1 h at room temperature, we quenched the reaction with hydroxylamine to a final concentration of 0.3%. The TMT-labeled samples were pooled at a 1:1 ratio across the samples. For the time-course experiment, 120 μg (in 6 μL of acetonitrile) of TMTpro reagent was added to the peptides (50 μg) with 14 μL of acetonitrile to achieve a final concentration of ~30% (v/v). Following incubation for 1 h at room temperature, the reaction was quenched with hydroxylamine to a final concentration of 0.3%. TMTpro-labeled samples were pooled at a 1:1 ratio across all 16 samples. For each experiment, the pooled sample was vacuum centrifuged and subjected to C18 solid-phase extraction (Sep-Pak, Waters).
Off-Line Basic pH Reversed-Phase (BPRP) Fractionation
We fractionated the pooled, labeled peptide sample using BPRP HPLC.31 We used an Agilent 1200 pump equipped with a degasser and 220 and 280 nm detectors. Peptides were subjected to a 50 min linear gradient from 5 to 35% acetonitrile in 10 mM ammonium bicarbonate pH 8 at a flow rate of 0.6 mL/min over an Agilent 300Extend C18 column (3.5 μm particles, 4.6 mm ID, and 250 mm in length). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated into 24 superfractions.32 Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each consolidated fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
Liquid Chromatography and Tandem Mass Spectrometry
Mass spectrometric data were collected on an Orbitrap Lumos (end-point experiment) or Eclipse (time-course experiment) mass spectrometer coupled to a Proxeon NanoLC-1200 UHPLC. The 100 μm capillary column was packed with 35 cm of Accucore 150 resin (2.6 μm, 150 Å; Thermo Fisher Scientific). The scan sequence began with an MS1 spectrum (Orbitrap analysis, resolution 120 000, 350–1400 Th, automatic gain control (AGC) target 5 × 105, maximum injection time 100 ms). Data were acquired for 90 min per fraction. SPS-MS3 analysis was used to reduce ion interference.33,34 The MS2 stage analysis consisted of collision-induced dissociation (CID), quadrupole ion trap analysis, automatic gain control (AGC) 2 × 104, normalized collision energy (NCE) 35, q-value 0.25, maximum injection time 120 ms, and isolation window at 0.7 Th. Following the acquisition of each MS2 spectrum, we collected an MS3 spectrum in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches. MS3 precursors were fragmented by HCD and analyzed using the Orbitrap (NCE 65, AGC 1.5 × 105, maximum injection time 150 ms, resolution was 50 000). The NCE at the MS3 stage for TMTpro samples was set at 45, as per previous recommendation.28 For TMTpro16 data, we used the real-time search option using a human database and we limited MS3 scans to two peptides per protein per fraction.35 For data acquisition including FAIMS, the dispersion voltage (DV) was set at 5000 V, the compensation voltages (CVs) were set at −40, −60, and −80 V, and TopSpeed parameter was set at 1 s per CV.
Data Analysis
Spectra were converted to mzXML via MSconvert.36 Database searching included all entries from the Human UniProt Database (downloaded: August 2019) that was concatenated with that database with all protein sequences in the reversed order. Searches were performed using a 50 ppm precursor ion tolerance and a 0.9 Da product ion tolerance in conjunction with Comet searches and linear discriminant analysis.37,38 TMT tags on lysine residues and peptide N-termini (+229.163 Da for TMT and + 304.207 Da for TMTpro) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as variable. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR).39,40 PSM filtering was performed using a linear discriminant analysis (LDA), as described previously38 and then assembled further to a protein-level FDR of 1%.40 Proteins were quantified by summing reporter ion counts across all PSMs, also as described previously.41 Reporter ion intensities were adjusted to correct for the isotopic impurities of the different TMT reagents according to manufacturer specifications. The signal-to-noise (S/N) measurements of peptides assigned to each protein were summed and normalized such that the sum of the signal for all proteins in each channel was equivalent and so we accounted for equal protein loading. Finally, each abundance measurement was scaled, such that the summed signal-to-noise for that protein across all channels equaled 100, generating a relative abundance (RA) measurement. Data analysis and visualization were performed in R or Microsoft Excel. We used Bioconductor42 and the time-course package,43 which applies a multivariate empirical Bayes model to rank temporally changing proteins and calculate the Hotelling T2 statistic.
Data Access
RAW files will be made available upon request. The data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the data set identifier PXD020807. In the Supporting Information, we have included tables listing protein names, gene symbols, and TMT quantitation values for both the end-point (Supplemental Table 1) and time-course (Supplemental Table 2) data sets. We have also included lists of peptides, associated protein names, gene symbols, TMT quantification values, retention time, and isolation purity for the end-point (Supplemental Table 3) and time-course (Supplemental Table 4) data sets.
RESULTS AND DISCUSSION
Isobaric Tag-Based Proteomics Permitted the Relative Protein Abundance Profiling of SH-SY5Y Cell Line Differentiation Induced by Retinoic Acid Treatment
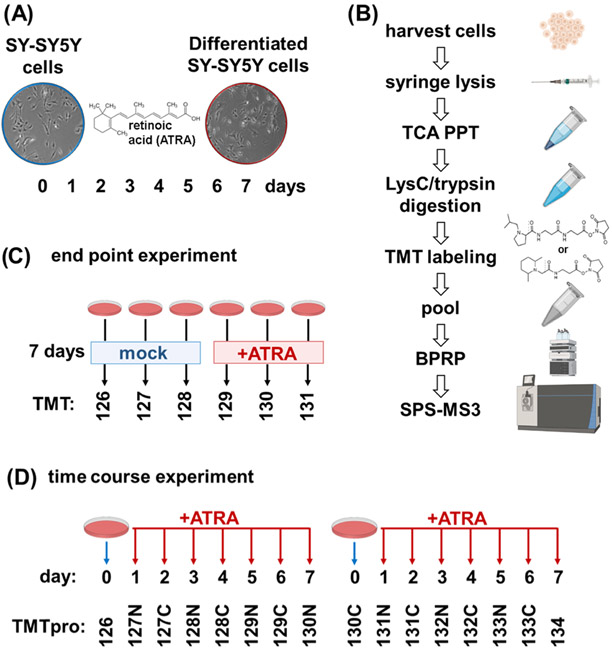
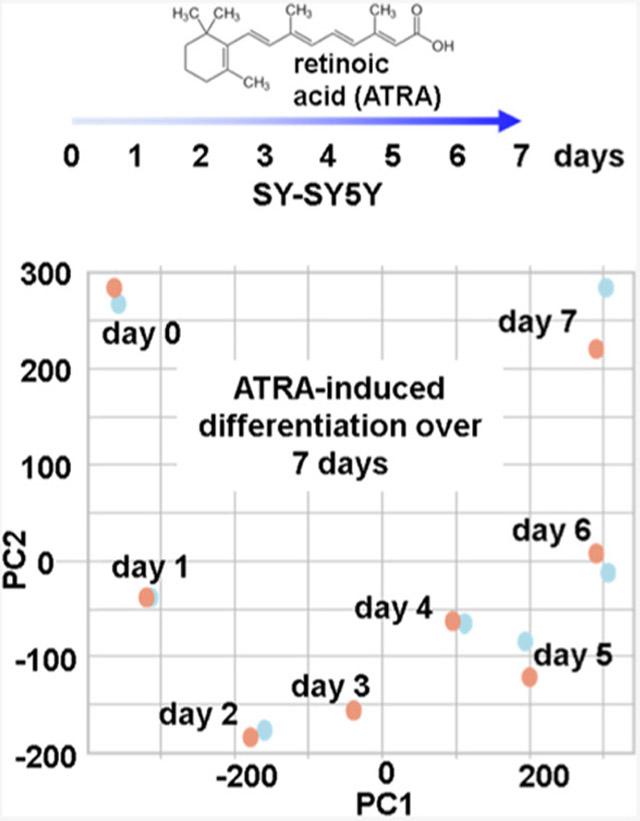
We investigated proteome remodeling in the SH-SY5Y cell line resulting from retinoic acid treatment using an isobaric tagging strategy. Our experimental design consisted of treating SH-SY5Y cells with 10 μM of retinoic acid for a 7-day period (Figure 1A). The samples were processed for mass spectrometry analysis using a modified SL-TMT protocol45 (Figure 1B). Cells were harvested and syringe-lysed in 8 M urea. Cysteines were reduced and alkylated, and proteins were TCA precipitated and digested with LysC followed by trypsin. We used TCA for precipitation as protein concentration was <1 mg/mL, which limits the use of chloroform–methanol precipitation, as is the typical precipitation strategy in the SL-TMT protocol.46 Peptides for each condition were labeled with TMT, pooled, fractionated by basic pH reversed-phase chromatography, and analyzed by SPS-MS3. In the initial experiment (the “end-point” experiment), we compared three biological replicates each of SH-SY5Y cells treated for 7 days with retinoic acid and mock-treated cells as a TMT6-plex experiment (Figure 1C). After observing considerable changes in the proteome, we designed a time-course experiment using TMTpro16 reagents.28 TMTpro16 facilitated the arrangement of eight time points (Days 0–7) in biological duplicate.
Figure 1.
Experimental overview. (A) SH-SY5Y has been shown to differentiate when treated with all-trans-retinoic acid (ATRA) over a 7-day period and to acquire neuron-like properties. (B) Experimental workflow. Samples were processed using the SL-TMT protocol (45). (C) Layout of TMT6-plex end-point experiment. Triplicates of untreated and 7-day retinoic acid-treated SH-SY5Y cells were arranged in a 3 × 2 TMT6-plex experiment. (D) Layout of TMTpro16-plex time-course. SH-SY5Y cells treated with retinoic acid were harvested over the course of 7 days. Biological replicate samples from Day 0 to Day 7 were arranged in a TMTpro16-plex. ATRA, all-trans retinoic acid; BPRP, basic pH reversed-phase; TCA PPT, trichloroacetic acid precipitation.
We quantified a total of 8575 proteins for the TMT6-plex end-point experiment, representing one of the largest isobaric-tagged SH-SY5Y proteomes.16 These proteins were inferred from 73 827 peptides of which 60 135 were unique in sequence. These data were collected on a FAIMSpro-equipped47,48 Orbitrap Lumos mass spectrometer using SPS-MS3. We acquired the time-course TMTpro16 experiment on the latest-generation Orbitrap Eclipse mass spectrometer equipped with a FAIMSpro interface and using real-time searching (RTS) to decide on-the-fly whether or not to collect an MS3 scan for a given MS2 scan.35,49 This analysis enabled the quantification of 9418 proteins. These proteins were inferred from 93,380 peptides of which 64 578 were unique in sequence. This data set marked the largest number of SH-SY5Y-derived proteins quantified to date. These data are tabulated in Table 1.
Table 1.
Data Set Overview
| dysregulateda | |||||
|---|---|---|---|---|---|
| total peptides | unique peptides | proteins | up | down | |
| end point | 73 827 | 60 135 | 8575 | 399 | 191 |
| time course | 93 380 | 64 578 | 9418 | n/a | n/a |
Fold change > ∣2∣ and Benjamini–Hochberg corrected p-value < 0.01, not applicable.
Several Hundred Proteins Were Differentially Expressed When SH-SY5Y Cells Were Mock-Treated or Treated with Retinoic Acid for 7 Days
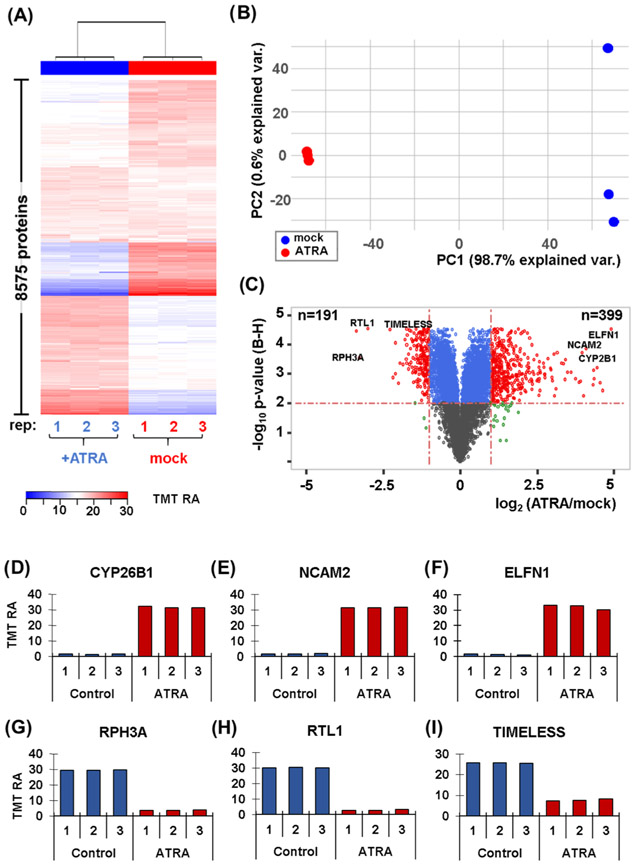
We investigated further the abundance profiles of the 8575 proteins quantitated in our end-point experiment. First, we performed hierarchical clustering incorporating Euclidean distance and Ward linkage using the TMT relative abundance values (Figure 2A). We noted very tight clustering among biological replicates, with a clear split between treated and untreated cells. We next performed principal components analysis (PCA) in efforts to determine how much variation could be explained by a given variable. Agreeing with the tight hierarchical clustering, PCA showed a distinct split between treated and untreated cells (Figure 2B). We determined that 98.7% of the variance in the experiment can be explained by the first principal component (PC1), which divides the samples by treatment. These data showed that retinoic acid clearly has a significant impact on proteome remodeling of SH-SY5Y cells. We illustrated these data in two dimensions using a volcano plot. Here, we plotted the Benjamini–Hochberg-corrected p-value versus the log 2 ratio of the average TMT reporter ion signal-to-noise value of the retinoic acid-treated sample (ATRA) to that of the mock-treated control (mock) (Figure 2C). We set stringent significance thresholds consisting of a Benjamini–Hochberg-corrected p-value < 0.01 and an absolute fold change of 2 or greater. These thresholds determined 191 and 399 proteins to be statistically significant in terms of being of lower or higher abundance, respectively, when the cells were treated with retinoic acid.
Figure 2.
Proteome profiling of SH-SY5Y cells with and without a 7-day retinoic acid treatment. (A) Heat map illustrates the relative abundance of the 8,575 proteins quantified in this experiment. Each protein abundance value sums to 100 across the six channels. (B) Principal components analysis (PCA) of the six samples depicts the variance explained by the first two principal components (PC). (C) Volcano plot illustrating the Benjamini–Hochberg (BH)-corrected p-value of retinoic acid (ATRA) vs. mock control and the fold change for these values. Highlighted on the volcano plot are six of the most altered proteins resulting from retinoic acid treatment. These proteins include: (D) CYP26B1, cytochrome P450 26B1; (E) NCAM2, neural cell adhesion molecule 2; (F) ELFN1, extracellular leucine-rich repeat and fibronectin type III.(G) RPH3A, rabphilin-3A; (H) RTL1, retrotransposon-like protein 1; and (I) TIMELESS.
We proceeded to subject these short lists of dysregulated proteins to Gene Ontology (GO) enrichment analysis using the DAVID interface50 with the entire list of quantified proteins as background. We required 10 proteins per category and an FDR < 0.05. Proteins having higher relative abundance when cells were treated with retinoic acid were categorized mainly as cell membrane, cell surface, cell migration, or related proteins (Table 2 top). On the contrary, proteins with significantly lower abundance when cells were treated with retinoic acid were categorized mainly as cell division, cell proliferation, or DNA-interaction-related proteins (Table 2 bottom). The names of proteins in each category listed in Table 2 are included in Supplemental Table 5. We noted that these classifications did not discriminate whether the function of the protein was permissive or inhibitory, only that a given protein had some role in that gene ontology classification. Nonetheless, a wide range of biological processes and molecular functions were affected by retinoic acid treatment.
Table 2.
Gene Ontology Classifications for Differentially Abundant Proteins
| abundance | category | term | count | % | FDR |
|---|---|---|---|---|---|
| up | BP | cell adhesion | 26 | 6.6 | 4.46 × 10−2 |
| up | BP | extracellular matrix organization | 22 | 5.6 | 4.46 × 10−6 |
| up | BP | cell migration | 15 | 3.8 | 4.22 × 10−2 |
| up | BP | interferon-γ-mediated signaling | 10 | 2.5 | 3.86 × 10−2 |
| up | CC | extracellular exosome | 142 | 36.0 | 4.37 × 10−22 |
| up | CC | plasma membrane | 134 | 34.1 | 5.39 × 10−5 |
| up | CC | extracellular space | 70 | 17.8 | 5.36 × 10−9 |
| up | CC | focal adhesion | 50 | 12.7 | 5.15 × 10−21 |
| up | CC | Cell surface | 47 | 11.9 | 9.10 × 10−13 |
| up | CC | Cell–cell junction | 21 | 5.3 | 2.52 × 10−2 |
| up | CC | extracellular matrix | 20 | 5.1 | 2.47 × 10−2 |
| up | CC | actin cytoskeleton | 19 | 4.8 | 1.24 × 10−3 |
| up | CC | membrane raft | 16 | 4.1 | 4.48 × 10−2 |
| down | BP | Cell division | 39 | 20.9 | 7.07 × 10−25 |
| down | BP | DNA replication | 31 | 16.7 | 1.09 × 10−26 |
| down | BP | DNA repair | 23 | 12.4 | 6.41 × 10−12 |
| down | BP | Cell proliferation | 17 | 9.1 | 2.18 × 10−3 |
| down | BP | DNA replication initiation | 15 | 8.1 | 6.14 × 10−17 |
| down | CC | nucleus | 118 | 63.4 | 4.65 × 10−20 |
| down | CC | Cytoplasm | 78 | 41.9 | 3.38 × 10−2 |
| down | CC | nuclear chromatin | 14 | 7.5 | 7.70 × 10−5 |
| down | MF | protein binding | 133 | 71.5 | 2.30 × 10−8 |
| down | MF | DNA binding | 41 | 22.0 | 2.97 × 10−4 |
| down | MF | ATP binding | 37 | 19.9 | 1.16 × 10−3 |
| down | MF | Chromatin binding | 23 | 12.4 | 1.09 × 10−7 |
Threshold for including a given GO category: >10 proteins per category and a false discovery rate (FDR) ≤ 0.05. BP, biological pathway; CC, cellular component; MF, molecular function.
We also subjected the lists to Kyoto Encyclopedia of Genes and Genomes (KEGG)51 pathway analysis using the STRING52 interface. These results were in line with those determined from the GO enrichment analyses. Specifically, proteins that increased in abundance when cells were treated with retinoic acid were observed in several pathways, including membrane protein-related extracellular matrix (ECM)– receptor interaction and cholesterol metabolism, as well as retinol metabolism (Table 3 top). Meanwhile, proteins with significantly lower abundance when cells were treated with retinoic acid were in pathways reflecting the GO enrichment analysis, specifically, DNA-related pathways and the cell cycle (Table 3 bottom). The names of proteins in each pathway in Table 3 are included in Supplemental Table 6. These data reinforced that a wide variety of cellular pathways were affected by retinoic acid treatment.
Table 3.
KEGG Pathway Classifications for Differentially Abundant Proteins
| abundance | category | pathway | count | % | FDR |
|---|---|---|---|---|---|
| up | hsa04512 | ECM–receptor interaction | 11 | 13.6 | 1.60 × 10−4 |
| up | hsa04979 | cholesterol metabolism | 5 | 10.4 | 3.63 × 10−2 |
| up | hsa05410 | hypertrophic cardiomyopathy | 8 | 10.1 | 6.10 × 10−3 |
| up | hsa00830 | retinol metabolism | 6 | 10.0 | 2.49 × 10−2 |
| down | hsa03030 | DNA replication | 16 | 44.4 | 9.75 × 10−19 |
| down | hsa03430 | mismatch repair | 5 | 21.7 | 1.20 × 10−4 |
| down | hsa03410 | base excision repair | 5 | 15.2 | 4.80 × 10−4 |
| down | hsa04110 | cell cycle | 16 | 13.0 | 9.05 × 10−12 |
| down | hsa00240 | pyrimidine metabolism | 11 | 11.0 | 2.20 × 10−7 |
| down | hsa03420 | nucleotide excision repair | 5 | 10.9 | 1.80 × 10−3 |
| down | hsa03440 | homologous recombination | 4 | 10.0 | 8.50 × 10−3 |
Threshold for including a given KEGG pathway category: Number of proteins were ≥10% of the background gene count per category and a false discovery rate (FDR) ≤ 0.05.
We highlighted several proteins that showed elevated changes in abundance in the TMT6-plex data set. We noted three examples of upregulated proteins that were involved in the metabolism of retinoic acid or have well-studied functions in neurons. First, CYP26B1 (cytochrome P450 26B1) is involved in the metabolism of retinoic acid (RA), rendering it inactive through oxidation53 (Figure 2D). Next, NCAM2 (neural cell adhesion molecule 2) plays a prominent role in the projection of axons54 (Figure 2E). Finally, ELFN1 (extracellular leucine-rich repeat and fibronectin type III domain containing 1) is a postsynaptic protein that modulates the temporal dynamics of interneuron recruitment55 (Figure 2F). Moreover, we also highlighted three examples of downregulated proteins that have roles in protein transport and DNA processing. First, RPH3A (rabphilin-3A) plays a crucial role in docking and fusion steps of regulated exocytosis and is implicated in intracellular protein transport56 (Figure 2G). Next, RTL1 (retrotransposon-like protein 1) is a retrotransposon that reverts RNA into DNA through the process of reverse transcription using an RNA transposition intermediate57 (Figure 2H). Finally, TIMELESS (a human homolog of the identically named Drosophila protein) has a role in controlling DNA replication, DNA repair, maintenance of genome stability, and in the regulation of the circadian clock58 (Figure 2I). We will interrogate these proteins again in the quantitative time-course data set.
Differences in the Relative Protein Abundance Profiles of SH-SY5Y Cells Were Traced Over a 7-Day Treatment of Retinoic Acid
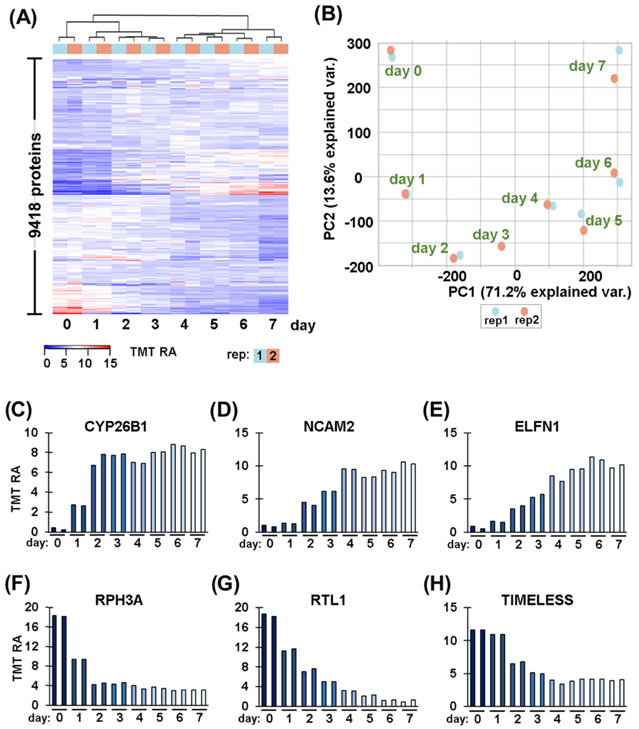
After noting striking proteome remodeling in the end-point assay, we examined the protein profile alterations over the course of retinoic acid treatment. We measured the quantitative temporal proteome profile of 9418 proteins across eight time points in duplicate. As with the end-point assay, we performed hierarchical clustering with Euclidean distance and Ward linkage using TMT relative abundance values (Figure 3A). We noted very tight clustering among biological duplicates with adjacent time points grouped closely together, as expected. We also observed a major split between Day 3 and 4 time points, potentially indicating that major proteome remodeling occurred between these 2 days. We again performed principal components analysis (PCA) to explore how much variation can be explained by a certain variable (Figure 3B). PCA showed tight clustering between duplicates, as in the hierarchical clustering heat map. PC1 explained 71.2% of the variance in the experiment, which was associated with length of treatment. More specifically, we can observe that the pattern along the PC1 axis was chronological, that is, the leftmost data points were Day 0 and the days were sequential from left to right until Day 7, which was the rightmost point on the plot. These data supported that retinoic acid clearly had a significant impact on proteome remodeling of SH-SY5Y cells and that we can track the increase or decrease in protein abundance over time.
Figure 3.
Temporal differences in protein expression. (A) Heat map illustrates the relative abundance of the 9418 proteins quantified in this time-course experiment. The abundance values for each protein sum to 100 across the 16 channels. (B) Principal components analysis (PCA) of the 16 samples depicts the variance explained by the first two principal components (PC). Temporal expression patterns of proteins highlighted in Figure 2: (C) CYP26B1, cytochrome P450 26B1; (D) NCAM2, neural cell adhesion molecule 2; (E) ELFN1, extracellular leucine-rich repeat and fibronectin type III, (F) RPH3A, rabphilin-3A; (G) RTL1, retrotransposon-like protein 1; and (H) TIMELESS.
We revisited the proteins highlighted in Figure 2D-I to determine the pattern of protein abundance alteration over the 7-day retinoic acid treatment. We focused on three proteins that exhibited lower abundance with retinoic acid treatment. First, CYP26B1 showed a sharp increase over 2 days of treatment and then leveled off (Figure 3C). Similarly, NCAM2 exhibited a gradual increase after Day 2 that plateaued at Day 4 (Figure 3D). In contrast, ELFN1 showed a gradual increase over the first 6 days of treatment and dropped slightly on the final day of treatment (Figure 3E). Likewise, we again highlighted three examples of downregulated proteins. First, RPH3A displayed a decrease in abundance of about 2-fold over the first 2 days of retinoic acid treatment before leveling off (Figure 3F). RTL1 exhibited a gradual decrease over the first 6 days of treatment (Figure 3G). Finally, the abundance of TIMELESS began to decrease on Day 2 and then decreased more than 50% by Day 4 (Figure 3H). We noted that in this limited sampling, different patterns of protein abundance alterations emerged.
Next, we assessed the statistical significance of our temporally changing proteins. We applied the time-course package in R Bioconductor, which used a multivariate empirical Bayes model to rank temporally altered proteins.43 More precisely, we employed the mb.long function to calculate a Hotelling T2 statistic and rank proteins according to their temporal abundance alterations.59 The mb.long function is used to compute multivariate empirical Bayes Statistics for longitudinal replicated time-course data. Specifically, the mb.long function computes the T2 statistic of differential expression for longitudinal replicated time-course data allowing for the ranking of genes or proteins.60 We have included the Hotelling T2 statistic for each protein in this data set as a column in Supplemental Table 3. We next compared the GO classifications determined from the end-point assay to that of the time course. We graphically illustrated the significant temporally altered proteins by plotting the log 10 of the Hotelling T2 statistic versus the ratio of the Day 7 to the Day 0 time point (Supplemental Figure 1A). We set stringent significance thresholds consisting of an absolute fold change of 2 or greater and a Hotelling T2 statistic of 322.836, which encompasses about 30% of the data.27 As such, the number of significantly changing proteins approached the number in the end-point data set. Here, 202 and 345 proteins decreased or increased temporally, respectively, during the 7-day retinoic acid treatment.
We again subjected these short lists of proteins to GO enrichment analysis using the DAVID interface,50 with the entire list of quantified proteins as background. As with the previous analysis, we required 10 proteins per category and an FDR < 0.05. As expected, the analysis revealed categories on par with those highlighted in the end-point data set (Table 2). As in the previous data set, proteins of higher abundance in treated cells were categorized as cell migration, cell surface, cell junction, or related proteins (Supplemental Figure 1B). On the contrary, proteins with significantly lower abundance in treated cells were categorized mainly as cell proliferation, cell division, or DNA-related proteins (Supplemental Figure 1C). Although both data sets produced similar results, the time-course data set permitted us to determine when the largest shift in abundance for a given protein change occurred to single-day resolution.
Neuron-Related Proteins Demonstrated a Time-Dependent Increase in Their Relative Abundance Following Retinoic Acid Treatment
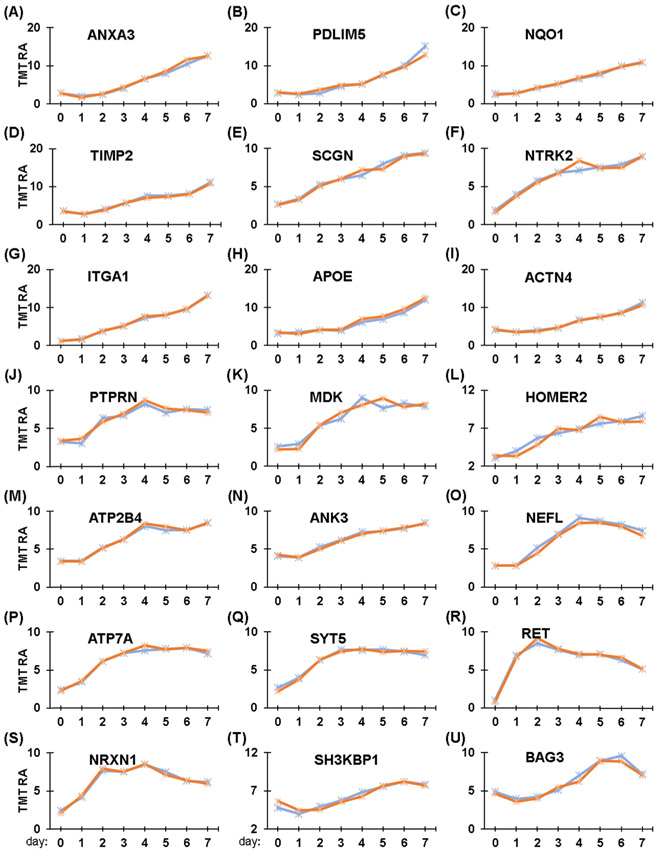
We showcased our time-course data set further to examine alterations in proteins with roles in neuronal development. As we mentioned previously, retinoic acid-differentiated SH-SY5Y cells are commonly used in neuroscience research, as they acquire more neuron-like properties after treatment. We used the time-course data to extract proteins with functions in neurofilament bundle assembly, neuron projections, and neuronal cell body formation. The temporal abundance profiles of these 21 proteins are illustrated in Figure 4A-U.
Figure 4.
Temporal protein profiles of neuron-related proteins. (A–U) Proteins associated with neurons, particularly neurite outgrowth, are highlighted. These proteins include: (A) Annexin A3, ANXA3; (B) PDZ and LIM domain protein 5, PDLIM5; (C) NAD(P)H dehydrogenase [quinone] 1, NQO1; (D) Metalloproteinase inhibitor 2, TIMP2; (E) Secretagogin, SCGN; (F) BDNF/NT-3 growth factors receptor, NTRK2; (G) Integrin alpha1, ITGA1; (H) Apolipoprotein E, APOE; (I) α-actinin-4, ACTN4; (J) Receptor-type tyrosine-protein phosphatase-like N, PTPRN; (K) Midkine, MDK; (L) Homer protein, HOMER2; (M) Plasma membrane calcium-transporting ATPase 4, ATP2B4; (N) Ankyrin-3, ANK3; (O) Neurofilament light polypeptide, NEFL; (P) Copper-transporting ATPase 1, ATP7A; (Q) Synaptotagmin-5, SYT5; (R) Proto-oncogene tyrosine-protein kinase receptor, RET; (S) Neurexin-1, NRXN1; (T) SH3 domain-containing kinase-binding protein 1, SH3KBP1; and (U) BAG family molecular chaperone regulator 3, BAG3.
We observed several different patterns of increasing abundance of certain proteins over the 7-day treatment with retinoic acid. Several proteins exhibited a gradual increase in abundance, specifically annexin A3 (Figure 4A), PDZ and LIM domain protein 5 (Figure 4B), NAD(P)H dehydrogenase [quinone] 1 (Figure 4C), α-actinin-4 (Figure 4D), Metalloproteinase inhibitor 2 (Figure 4E), Secretagogin (Figure 4F), BDNF/NT-3 growth factors receptor (Figure 4G), integrin alpha1 (Figure 4H), and apolipoprotein E (Figure 4I). In contrast, the abundance of some proteins increased on the second day of retinoic acid treatment, including receptor-type tyrosine-protein phosphatase-like N (Figure 4J), midkine (Figure 4K), Homer protein (Figure 4L), plasma membrane calcium-transporting ATPase 4 (Figure 4M), ankyrin-3 (Figure 4N), and neurofilament light polypeptide (Figure 4O). In addition, two proteins increased in abundance gradually and plateaued after 4 days, specifically, copper transporting ATPase 1 (Figure 4P) and synaptotagmin-5 (Figure 4Q). Two other proteins increased rapidly in abundance and reached their maximum after approximately 2 days, specifically, proto-oncogene tyrosine-protein kinase receptor (Figure 4R) and neurexin-1 (Figure 4S). Finally, SH3 domain-containing kinase-binding protein 1 (Figure 4T) and BAG family molecular chaperone regulator 3 (Figure 4U) decreased slightly in abundance during the first day of treatment with retinoic acid but peaked at Day 6. Although most of these proteins showed a similar pattern, no consensus increase was apparent, which supports that complex time-dependent mechanisms were regulating the retinoic acid-mediated differentiation of SH-SY5Y cells.
Several transcriptomic profiling studies have also investigated the differentiation of SH-SY5Y cells with retinoic acid. Here, we compare our data to two such reports. First, Pezzini, et al. performed RNA-seq analysis on retinoic acid-treated SH-SY5Y cells differentiation with subsequent growth in neurobasal medium.61 Although the differentiation conditions differed from our workflow, similar conclusions were drawn as dysregulated proteins were implicated in remodeling of the plasma membrane and the cytoskeleton, also had implications in neuritogenesis, which agreed with our gene ontology data (Table 2) and Figure 4. In fact, 146 of the 399 proteins that we deemed as significantly higher in abundance upon retinoic acid treatment were also significantly upregulated in Pezzini, et al. Likewise, 85 of the 191 proteins that we determined to be significantly lower in abundance upon retinoic acid treatment were significantly downregulated in that publication as well. Second, Korecka, et al. investigated retinoic acid differentiation of SH-SY5Y cells via a genome-wide transcriptional analysis.62 Cell growth conditions were slightly different from ours as cells were propagated on Matrigel for that study. Nonetheless, like Pezzini, et al. and our own work, dysregulated proteins included those associated with proliferation, neuronal differentiation, and development. Moreover, 91 of our 399 proteins with significantly higher abundance upon retinoic acid treatment were also significantly upregulated and 24 of our 191 proteins with significantly lower in abundance were significantly downregulated in Korecka, et al. These researchers highlighted the neural cell adhesion molecule, NCAM2, which is highly involved in neuronal compartmentalization, which was 12-fold upregulated by Day 8. We similarly observed an almost 10-fold increase in the abundance of NCAM2 after 7 days of retinoic acid treatment (Figure 3D). Although discrepancies among studies are expected due to differences in treatment lengths, media, and growth conditions, our findings specifically those related to plasma membrane proteome remodeling, neuron projections, and neuronal cell body formation were in agreement.
CONCLUSIONS
We quantitatively profiled over 9400 proteins across the 7-day retinoic acid treatment using TMTpro16 reagents and a FAIMS-equipped Orbitrap Eclipse mass spectrometer with real-time database searching. This data set represents a rich resource for the study of temporal protein abundance, which can be used for further data mining for deeper studies into retinoic acid differentiation in SH-SY5Y cells. We highlighted proteins involved in neuron projections; however, many other biological insights can be extracted from these data. Moreover, the sample preparation and data acquisition strategies used herein can be applied to virtually any biological system with a sequenced genome. Although retinoic acid is the most common method to induce differentiation in SH-SY5Y cells, several other compounds are also known to differentiate this cell line. For instance, SH-SY5Y cells may be driven toward a mature dopaminergic phenotype by retinoic acid treatment with coadministration of phorbol esters, such as 12-O-tetradecanoyl-phorbol-13 acetate (TPA).7,63 In addition, exposure of SH-SY5Y cells to dibutyryl cyclic AMP (dbcAMP) results in neurite extension.64 Staurosporine, a pan-kinase inhibitor, can trigger neurite formation and cell cycle arrest in SH-SY5Y cells.65 Similarly, culturing SH-SY5Y cells in neurobasal medium with the addition of B27 has resulted in enhanced differentiation.66 Other compounds including treatment with cholesterol,67 vitamin D,68 and insulin69 have been shown to promote neuronal differentiation in SH-SY5Y cells. In summary, the quantitative proteomics techniques outlined herein can be used to temporally profile the alterations in protein abundance for any mechanism of cellular differentiation, including, but not limited to, those mentioned above.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the members of the Gygi Lab at Harvard Medical School, in particular Ramin Rad. This work was funded in part by NIH/NIGMS grant GM67945 (S.P.G.) and R01 GM132129 (J.A.P.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00614.
Protein differences in the time-course data set (Supplemental Figure 1) (PDF)
Proteins quantified in the end-point TMT6 data set (Supplemental Table 1) (XLSX)
Proteins quantified in the time-course TMTpro16 data set (Supplemental Table 2) (XLSX)
Peptides quantified in the end-point TMT6-plex data set (Supplemental Table 3) (XLSX)
Peptides quantified in the time-course TMT16pro data set (Supplemental Table 4) (XLSX)
Expanded gene ontology classifications for differentially abundant proteins (Supplemental Table 5) (XLSX)
Expanded KEGG pathway classifications for differentially abundant proteins (Supplemental Table 6) (XLSX)
The authors declare no competing financial interest.
REFERENCES
- (1).Zhang M; Zhang YQ; Wei XZ; Lee C; Huo DS; Wang H; Zhao ZY Differentially expressed long-chain noncoding RNAs in human neuroblastoma cell line (SH-SY5Y): Alzheimer’s disease cell model. J. Toxicol. Environ. Health, Part A 2019, 82, 1052–1060. [DOI] [PubMed] [Google Scholar]
- (2).de Medeiros LM; De Bastiani MA; Rico EP; Schonhofen P; Pfaffenseller B; Wollenhaupt-Aguiar B; Grun L; Barbe-Tuana F; Zimmer ER; Castro MAA; Parsons RB; Klamt F Cholinergic Differentiation of Human Neuroblastoma SH-SY5Y Cell Line and Its Potential Use as an In vitro Model for Alzheimer’s Disease Studies. Mol. Neurobiol 2019, 56, 7355–7367. [DOI] [PubMed] [Google Scholar]
- (3).Azzolin VF; Barbisan F; Lenz LS; Teixeira CF; Fortuna M; Duarte T; Duarte M; da Cruz IBM Effects of Pyridostigmine bromide on SH-SY5Y cells: An in vitro neuroblastoma neurotoxicity model. Mutat. Res., Genet. Toxicol. Environ. Mutagen 2017, 823, 1–10. [DOI] [PubMed] [Google Scholar]
- (4).Xie HR; Hu LS; Li GY SH-SY5Y human neuroblastoma cell line: in vitro cell model of dopaminergic neurons in Parkinson’s disease. Chin. Med. J 2010, 123, 1086–1092. [PubMed] [Google Scholar]
- (5).Kovalevich J; Langford D Considerations for the Use of SH-SY5Y Neuroblastoma Cells in Neurobiology. Methods in Molecular Biology; Humana Press: Totowa, NJ, 2013; Vol. 1078, pp 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Biedler JL; Roffler-Tarlov S; Schachner M; Freedman LS Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978, 38, 3751–3757. [PubMed] [Google Scholar]
- (7).Presgraves SP; Ahmed T; Borwege S; Joyce JN Terminally differentiated SH-SY5Y cells provide a model system for studying neuroprotective effects of dopamine agonists. Neurotoxic. Res 2003, 5, 579–98. [DOI] [PubMed] [Google Scholar]
- (8).Cheung YT; Lau WK; Yu MS; Lai CS; Yeung SC; So KF; Chang RC Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology 2009, 30, 127–35. [DOI] [PubMed] [Google Scholar]
- (9).Nicolini G; Miloso M; Zoia C; Di Silvestro A; Cavaletti G; Tredici G Retinoic acid differentiated SH-SY5Y human neuroblastoma cells: an in vitro model to assess drug neurotoxicity. Anticancer Res. 1998, 18, 2477–2481. [PubMed] [Google Scholar]
- (10).Melino G; Thiele CJ; Knight RA; Piacentini M Retinoids and the control of growth/death decisions in human neuroblastoma cell lines. J. Neuro-Oncol 1997, 31, 65–83. [DOI] [PubMed] [Google Scholar]
- (11).Pogenberg V; Guichou JF; Vivat-Hannah V; Kammerer S; Perez E; Germain P; de Lera AR; Gronemeyer H; Royer CA; Bourguet W Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. J. Biol. Chem 2005, 280, 1625–1633. [DOI] [PubMed] [Google Scholar]
- (12).Lopes FM; Schroder R; da Frota ML Jr; Zanotto-Filho A; Muller CB; Pires AS; Meurer RT; Colpo GD; Gelain DP; Kapczinski F; Moreira JC; Fernandes Mda C; Klamt F Comparison between proliferative and neuron-like SH-SY5Y cells as an in vitro model for Parkinson disease studies. Brain Res. 2010, 1337, 85–94. [DOI] [PubMed] [Google Scholar]
- (13).Teppola H; Sarkanen JR; Jalonen TO; Linne ML Morphological Differentiation Towards Neuronal Phenotype of SH-SY5Y Neuroblastoma Cells by Estradiol, Retinoic Acid and Cholesterol. Neurochem. Res 2016, 41, 731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sallmon H; Hoene V; Weber SC; Dame C Differentiation of human SH-SY5Y neuroblastoma cells by all-trans retinoic acid activates the interleukin-18 system. J. Interferon Cytokine Res 2010, 30, 55–58. [DOI] [PubMed] [Google Scholar]
- (15).Fowler CJ; O’Neill C; Almqvist P; Nilsson S; Wiehager B; Winblad B Muscarinic receptors coupled to inositol phospholipid breakdown in human SH-SY5Y neuroblastoma cells: Effect of retinoic acid-induced differentiation. Neurochem. Int 1989, 15, 73–79. [DOI] [PubMed] [Google Scholar]
- (16).Paulo JA; Gygi SP Nicotine-induced protein expression profiling reveals mutually altered proteins across four human cell lines. Proteomics 2017, 17, No. 1600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Xue S; Jia L; Jia J Hypoxia and reoxygenation increased BACE1 mRNA and protein levels in human neuroblastoma SH-SY5Y cells. Neurosci. Lett 2006, 405, 231–235. [DOI] [PubMed] [Google Scholar]
- (18).Levites Y; Youdim MB; Maor G; Mandel S Attenuation of 6-hydroxydopamine (6-OHDA)-induced nuclear factor-kappaB (NF-kappaB) activation and cell death by tea extracts in neuronal cultures. Biochem. Pharmacol 2002, 63, 21–29. [DOI] [PubMed] [Google Scholar]
- (19).Levites Y; Amit T; Youdim MB; Mandel S Involvement of protein kinase C activation and cell survival/ cell cycle genes in green tea polyphenol (−)-epigallocatechin 3-gallate neuroprotective action. J. Biol. Chem 2002, 277, 30574–80. [DOI] [PubMed] [Google Scholar]
- (20).Waetzig V; Haeusgen W; Andres C; Frehse S; Reinecke K; Bruckmueller H; Boehm R; Herdegen T; Cascorbi I Retinoic acid-induced survival effects in SH-SY5Y neuroblastoma cells. J. Cell. Biochem 2019, 120, 5974–5986. [DOI] [PubMed] [Google Scholar]
- (21).Murillo JR; Goto-Silva L; Sanchez A; Nogueira FCS; Domont GB; Junqueira M Quantitative proteomic analysis identifies proteins and pathways related to neuronal development in differentiated SH-SY5Y neuroblastoma cells. EuPA Open Proteomics 2017, 16, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jahn K; Wieltsch C; Blumer N; Mehlich M; Pathak H; Khan AQ; Hildebrandt H; Frieling H A cell culture model for investigation of synapse influenceability: epigenetics, expression and function of gene targets important for synapse formation and preservation in SH-SY5Y neuroblastoma cells differentiated by retinoic acid. J. Neural Transm 2017, 124, 1341–1367. [DOI] [PubMed] [Google Scholar]
- (23).Wang K; Zhou F; Zhu X; Zhang K; Huang B; Zhu L; Zhu L Neuroprotective properties of ciliary neurotrophic factor on retinoic acid (RA)-predifferentiated SH-SY5Y neuroblastoma cells. Folia Neuropathol. 2014, 2, 121–127. [DOI] [PubMed] [Google Scholar]
- (24).Ross PL; Huang YN; Marchese JN; Williamson B; Parker K; Hattan S; Khainovski N; Pillai S; Dey S; Daniels S; Purkayastha S; Juhasz P; Martin S; Bartlet-Jones M; He F; Jacobson A; Pappin DJ Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 2004, 3, 1154–69. [DOI] [PubMed] [Google Scholar]
- (25).Thompson A; Schafer J; Kuhn K; Kienle S; Schwarz J; Schmidt G; Neumann T; Hamon C Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem 2003, 75, 1895–904. [DOI] [PubMed] [Google Scholar]
- (26).Clements DR; Murphy JP; Sterea A; Kennedy BE; Kim Y; Helson E; Almasi S; Holay N; Konda P; Paulo JA; Sharif T; Lee PW; Weekes MP; Gygi SP; Gujar S Quantitative Temporal in Vivo Proteomics Deciphers the Transition of Virus-Driven Myeloid Cells into M2 Macrophages. J. Proteome Res 2017, 16, 3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Murphy JP; Stepanova E; Everley RA; Paulo JA; Gygi SP Comprehensive Temporal Protein Dynamics during the Diauxic Shift in Saccharomyces cerevisiae. Mol. Cell Proteomics 2015, 14, 2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li J; Van Vranken JG; Pontano Vaites L; Schweppe DK; Huttlin EL; Etienne C; Nandhikonda P; Viner R; Robitaille AM; Thompson AH; Kuhn K; Pike I; Bomgarden RD; Rogers JC; Gygi SP; Paulo JA TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 2020, 17, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Paulo JA; Urrutia R; Banks PA; Conwell DL; Steen H Proteomic analysis of a rat pancreatic stellate cell line using liquid chromatography tandem mass spectrometry (LC-MS/MS). J. Proteomics 2011, 75, 708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Paulo JA; Urrutia R; Banks PA; Conwell DL; Steen H Proteomic analysis of an immortalized mouse pancreatic stellate cell line identifies differentially-expressed proteins in activated vs non-proliferating cell states. J. Proteome Res 2011, 10, 4835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang Y; Yang F; Gritsenko MA; Wang Y; Clauss T; Liu T; Shen Y; Monroe ME; Lopez-Ferrer D; Reno T; Moore RJ; Klemke RL; Camp DG 2nd; Smith RD Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 2011, 11, 2019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Paulo JA; O’Connell JD; Everley RA; O’Brien J; Gygi MA; Gygi SP Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteomics 2016, 148, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gygi JP; Yu Q; Navarrete-Perea J; Rad R; Gygi SP; Paulo JA Web-Based Search Tool for Visualizing Instrument Performance Using the Triple Knockout (TKO) Proteome Standard. J. Proteome Res 2019, 18, 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Paulo JA; O’Connell JD; Gygi SPA Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass Spectrom 2016, 27, 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schweppe DK; Eng JK; Yu Q; Bailey D; Rad R; Navarrete-Perea J; Huttlin EL; Erickson BK; Paulo JA; Gygi SP Full-Featured, Real-Time Database Searching Platform Enables Fast and Accurate Multiplexed Quantitative Proteomics. J. Proteome Res 2020, 19, 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chambers MC; Maclean B; Burke R; Amodei D; Ruderman DL; Neumann S; Gatto L; Fischer B; Pratt B; Egertson J; Hoff K; Kessner D; Tasman N; Shulman N; Frewen B; Baker TA; Brusniak MY; Paulse C; Creasy D; Flashner L; Kani K; Moulding C; Seymour SL; Nuwaysir LM; Lefebvre B; Kuhlmann F; Roark J; Rainer P; Detlev S; Hemenway T; Huhmer A; Langridge J; Connolly B; Chadick T; Holly K; Eckels J; Deutsch EW; Moritz RL; Katz JE; Agus DB; MacCoss M; Tabb DL; Mallick P A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol 2012, 30, 918–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Beausoleil SA; Villen J; Gerber SA; Rush J; Gygi SP A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 2006, 24, 1285–92. [DOI] [PubMed] [Google Scholar]
- (38).Huttlin EL; Jedrychowski MP; Elias JE; Goswami T; Rad R; Beausoleil SA; Villen J; Haas W; Sowa ME; Gygi SP A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Elias JE; Gygi SP Target-decoy search strategy for mass spectrometry-based proteomics. In Methods in Molecular Biology; Humana Press, 2010; Vol 604, pp 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–14. [DOI] [PubMed] [Google Scholar]
- (41).McAlister GC; Huttlin EL; Haas W; Ting L; Jedrychowski MP; Rogers JC; Kuhn K; Pike I; Grothe RA; Blethrow JD; Gygi SP Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Anal. Chem 2012, 84, 7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Reimers M; Carey VJ Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006, 411, 119–134. [DOI] [PubMed] [Google Scholar]
- (43).Tai YCS; Terence P A multivariate empirical Bayes statistic for replicated microarray time course data. Ann. Stat 2006, 34, 2387–2412. [Google Scholar]
- (44).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Navarrete-Perea J; Yu Q; Gygi SP; Paulo JA Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J. Proteome Res 2018, 17, 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Winter D; Dehghani A; Steen H Optimization of cell lysis and protein digestion protocols for protein analysis by LC-MS/MS. In Methods in Molecular Biology; Humana Press: New York, NY, 2015; Vol. 1295, pp 259–273. [DOI] [PubMed] [Google Scholar]
- (47).Schweppe DK; Rusin SF; Gygi SP; Paulo JA Optimized Workflow for Multiplexed Phosphorylation Analysis of TMT-Labeled Peptides Using High-Field Asymmetric Waveform Ion Mobility Spectrometry. J. Proteome Res 2020, 19, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Schweppe DK; Prasad S; Belford MW; Navarrete-Perea J; Bailey DJ; Huguet R; Jedrychowski MP; Rad R; McAlister G; Abbatiello SE; Woulters ER; Zabrouskov V; Dunyach JJ; Paulo JA; Gygi SP Characterization and Optimization of Multiplexed Quantitative Analyses Using High-Field Asymmetric-Waveform Ion Mobility Mass Spectrometry. Anal. Chem 2019, 91, 4010–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Erickson BK; Mintseris J; Schweppe DK; Navarrete-Perea J; Erickson AR; Nusinow DP; Paulo JA; Gygi SP Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. J. Proteome Res 2019, 18, 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Huang DW; Sherman BT; Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- (51).Kanehisa M; Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Szklarczyk D; Franceschini A; Wyder S; Forslund K; Heller D; Huerta-Cepas J; Simonovic M; Roth A; Santos A; Tsafou KP; Kuhn M; Bork P; Jensen LJ; von Mering C STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).MacLean G; Abu-Abed S; Dolle P; Tahayato A; Chambon P; Petkovich M Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech. Dev 2001, 107, 195–201. [DOI] [PubMed] [Google Scholar]
- (54).Winther M; Berezin V; Walmod PS NCAM2/OCAM/RNCAM: cell adhesion molecule with a role in neuronal compartmentalization. Int. J. Biochem. Cell Biol 2012, 44, 441–446. [DOI] [PubMed] [Google Scholar]
- (55).Sylwestrak EL; Ghosh A Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science 2012, 338, 536–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Yuan Q; Ren C; Xu W; Petri B; Zhang J; Zhang Y; Kubes P; Wu D; Tang W PKN1 Directs Polarized RAB21 Vesicle Trafficking via RPH3A and Is Important for Neutrophil Adhesion and Ischemia-Reperfusion Injury. Cell Rep. 2017, 19, 2586–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Riordan JD; Keng VW; Tschida BR; Scheetz TE; Bell JB; Podetz-Pedersen KM; Moser CD; Copeland NG; Jenkins NA; Roberts LR; Largaespada DA; Dupuy AJ Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet. 2013, 9, No. e1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Engelen E; Janssens RC; Yagita K; Smits VA; van der Horst GT; Tamanini F Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS One 2013, 8, No. e56623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hotelling H The Generalization of Student’s Ratio. Ann. Math. Stat 1931, 2, 360–378. [Google Scholar]
- (60).Tai YC; Speed TP A multivariate empirical Bayes statistic for replicated microarray time course data. Ann. Stat 2006, 34, 2387–2412. [Google Scholar]
- (61).Pezzini F; Bettinetti L; Di Leva F; Bianchi M; Zoratti E; Carrozzo R; Santorelli FM; Delledonne M; Lalowski M; Simonati A Transcriptomic Profiling Discloses Molecular and Cellular Events Related to Neuronal Differentiation in SH-SY5Y Neuroblastoma Cells. Cell Mol. Neurobiol 2017, 37, 665–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Korecka JA; van Kesteren RE; Blaas E; Spitzer SO; Kamstra JH; Smit AB; Swaab DF; Verhaagen J; Bossers K Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One 2013, 8, No. e63862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Påhlman S; Odelstad L; Larsson E; Grotte G; Nilsson K Phenotypic changes of human neuroblastoma cells in culture induced by 12-O-tetradecanoyl-phorbol-13-acetate. Int. J. Cancer 1981, 28, 583–589. [DOI] [PubMed] [Google Scholar]
- (64).Kume T; Kawato Y; Osakada F; Izumi Y; Katsuki H; Nakagawa T; Kaneko S; Niidome T; Takada-Takatori Y; Akaike A Dibutyryl cyclic AMP induces differentiation of human neuroblastoma SH-SY5Y cells into a noradrenergic phenotype. Neurosci. Lett 2008, 443, 199–203. [DOI] [PubMed] [Google Scholar]
- (65).Leli U; Shea TB; Cataldo A; Hauser G; Grynspan F; Beermann ML; Liepkalns VA; Nixon RA; Parker PJ Differential expression and subcellular localization of protein kinase C alpha, beta, gamma, delta, and epsilon isoforms in SH-SY5Y neuroblastoma cells: modifications during differentiation. J. Neurochem 1993, 60, 289–98. [DOI] [PubMed] [Google Scholar]
- (66).Sayas CL; Moreno-Flores MT; Avila J; Wandosell F The neurite retraction induced by lysophosphatidic acid increases Alzheimer’s disease-like Tau phosphorylation. J. Biol. Chem 1999, 274, 37046–52. [DOI] [PubMed] [Google Scholar]
- (67).Sarkanen JR; Nykky J; Siikanen J; Selinummi J; Ylikomi T; Jalonen TO Cholesterol supports the retinoic acid-induced synaptic vesicle formation in differentiating human SH-SY5Y neuroblastoma cells. J. Neurochem 2007, 102, 1941–1952. [DOI] [PubMed] [Google Scholar]
- (68).Reddy CD; Patti R; Guttapalli A; Maris JM; Yanamandra N; Rachamallu A; Sutton LN; Phillips PC; Posner GH Anticancer effects of the novel 1alpha, 25-dihydroxyvitamin D3 hybrid analog QW1624F2-2 in human neuroblastoma. J. Cell. Biochem 2006, 97, 198–206. [DOI] [PubMed] [Google Scholar]
- (69).Recio-Pinto E; Ishii DN Effects of insulin, insulin-like growth factor-II and nerve growth factor on neurite outgrowth in cultured human neuroblastoma cells. Brain Res. 1984, 302, 323–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.