Abstract
Imaging mass spectrometry (IMS) technologies are capable of mapping a wide array of biomolecules in diverse cellular and tissue environments. IMS has emerged as an essential tool for providing spatially targeted molecular information due to its high sensitivity, wide molecular coverage and chemical specificity. One of the major challenges for mapping the complex cellular milieu is the presence of many isomers and isobars present in these samples. This challenge is traditionally addressed using orthogonal LC-based analysis, though, common approaches such as chromatography and electrophoresis are not able to be performed at timescales that are compatible with most imaging applications. Ion mobility offers rapid, gas-phase separations that are readily integrated with IMS workflows in order to provide additional data dimensionality that can improve signal-to-noise, dynamic range, and specificity. Here, we highlight recent examples of ion mobility coupled to imaging mass spectrometry and highlight their importance to the field.
Keywords: Ion Mobility, Imaging Mass Spectrometry, Tissue Analysis, Metabolites, Lipids, Proteins, Trapped Ion Mobility Spectrometry, TIMS, Drift tube ion mobility spectrometry, DTIMS, Travelling Wave Ion Mobility Spectrometry, TWIMS, High-field Asymmetric Waveform Ion Mobility, FAIMS, IMS, MALDI, DESI, IR-MALDESI, LESA, liquid microjunction, LAESI
Introduction:
The biology of multicellular organisms is intricate, involving molecular interactions that are coordinated within cells, tissue microenvironments, and across organ systems.1 To fully characterize the biomolecular underpinnings of processes such as homeostasis and pathogenesis, technologies that can capture this molecular diversity while maintaining spatial context are essential.2 One such tool, imaging mass spectrometry (IMS), achieves this by combining the molecular sensitivity and specificity of mass spectrometry (MS) with cellular and tissue sampling technologies that enable rapid, high spatial resolution. IMS is a label-free technology that provides ion maps that are easily correlated to tissue histology for a diverse array of biological specimens.3–9 While alternative techniques such as fluorescence-based approaches can provide high resolution images, the number of molecular channels is limited because of excitation and emission overlap between fluorophores.10 In contrast, other techniques such as Raman or infrared spectroscopy are sensitive to many types of molecular features, but often at the expense of chemical specificity.11,12 IMS offers an untargeted mapping capability for hundreds to thousands of molecules in a single experiment. Moreover, IMS provides the analytical flexibility to investigate different classes of biomolecules including small metabolites,13,14 lipids,15,16 drugs,17,18 glycans,19,20 peptides,21,22 and proteins.23,24 However, IMS faces challenges of dynamic range, peak capacity, and the ability to structurally identify observed species. Traditionally, other mass spectrometry-based techniques rely on orthogonal separation methods such as chromatography or electrophoresis to separate complex mixtures and provide an additional dimension of information. However, practical considerations such as throughput and sampling limitations minimize their applicability to most imaging technology approaches.
Ion mobility offers effective separation within milliseconds25 as well as providing additional molecular information. While there are many different types of ion mobility devices, all achieve molecular separation by exposing analytes to opposing forces where a force is applied to analytes in one direction by collisions with an inert gas and in the opposite direction by a voltage gradient (Figure 1). Molecules are affected differentially based on their size, charge and mass where factors such as temperature and pressure can have dramatic effects as defined by the Mason-Schamp equation.26 Interactions between analytes and the inert gas are defined differently for each ion mobility technique,27 but in all cases discrepancies between molecular size-to-charge ratio determine mobility and cause molecules to exit from the ion mobility cell at differing times, referred to as the drift or arrival time.28,29 Similar to retention times in chromatography, these arrival times can often distinguish molecules of similar mass. Moreover, ion mobility can be used to calculate the collision cross section (CCS), the 3-dimensional surface area of an ion, that provides additional structural information.30–32 This has proven valuable in several ways, including estimation of a molecule’s binding affinity33 or for some applications in other areas such as the prediction of blood-brain barrier permeation.34 Ion mobility has effectively analyzed a wide range of analyte classes without the need for significant changes to the system such as switching mobile or stationary phases in chromatography. Not only is ion mobility capable of separating discrete analyte classes, but it is often used to distinguish both isobaric35,36,37 and isomeric38,39,40 species.
Figure 1:
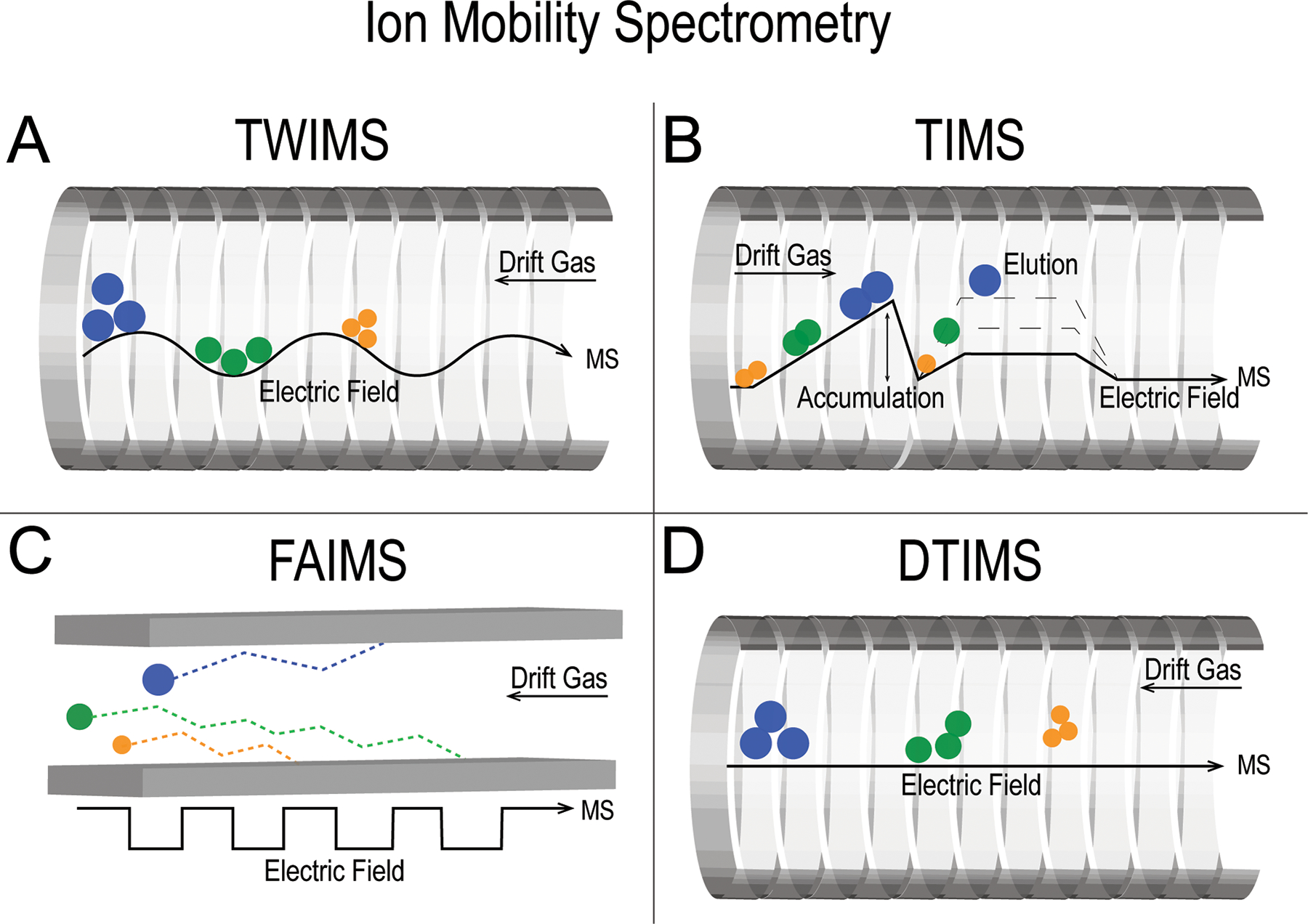
Overview of ion mobility techniques covered in this article. (A) Travelling wave ion mobility spectrometry (TWIMS), (B) trapped ion mobility spectrometry (TIMS), (C) high-field asymmetric waveform ion mobility (FAIMS) and (D) drift tube ion mobility spectrometry (DTIMS).
The ion separation capability of ion mobility has major advantages when coupled to imaging mass spectrometry to significantly simplify spectral complexity. Moreover, use of ion mobility for discrimination of ions with similar or equal m/z is invaluable for direct tissue analysis where MS instruments cannot distinguish structural isomers using mass resolving power alone.
Ion mobility has been integrated with several IMS techniques including matrix-assisted laser desorption/ionization (MALDI),41,42 desorption electrospray ionization (DESI),43,44 laser-assisted electrospray ionization (LAESI),45,46 liquid-extraction surface analysis (LESA),47,48 liquid-microjunction surface sampling probe (LMJ-SSP),49 and infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI)50 (Figure 2). Briefly, MALDI relies on laser desorption for introduction of analytes to the mass spectrometer, where LESA, DESI and LMJ-SSP are electrospray-based techniques which use solvent to desorb analytes for detection.8,51–53 LAESI and IR-MALDESI utilize both laser desorption and electrospray for analyte introduction and ionization.54,55 For detailed descriptions of these ionization sources, see these recent reviews.56–58 While secondary ion mass spectrometry (SIMS) is also a prevalent IMS ion source,56,57 it has not yet been coupled to ion mobility and will therefore not be covered in this perspective. Regardless of IMS ion source, ion mobility provides efficient ion separation prior to mass analysis59 and increased signal-to-noise (S/N).36,60,61 As the resolving power of ion mobility technologies continues to improve, applications of ion mobility-IMS to discriminate biomolecules closer in m/z without the need for ultra-high mass resolving power mass spectrometers will be of significant benefit. In this article, we briefly describe recent advances in the field of ion mobility-IMS. We also discuss how recent advances impact the current state of the ion mobility-IMS field as well as implications for the future. For further background, the reader is directed to the review from Sans, Feider et al.62
Figure 2:
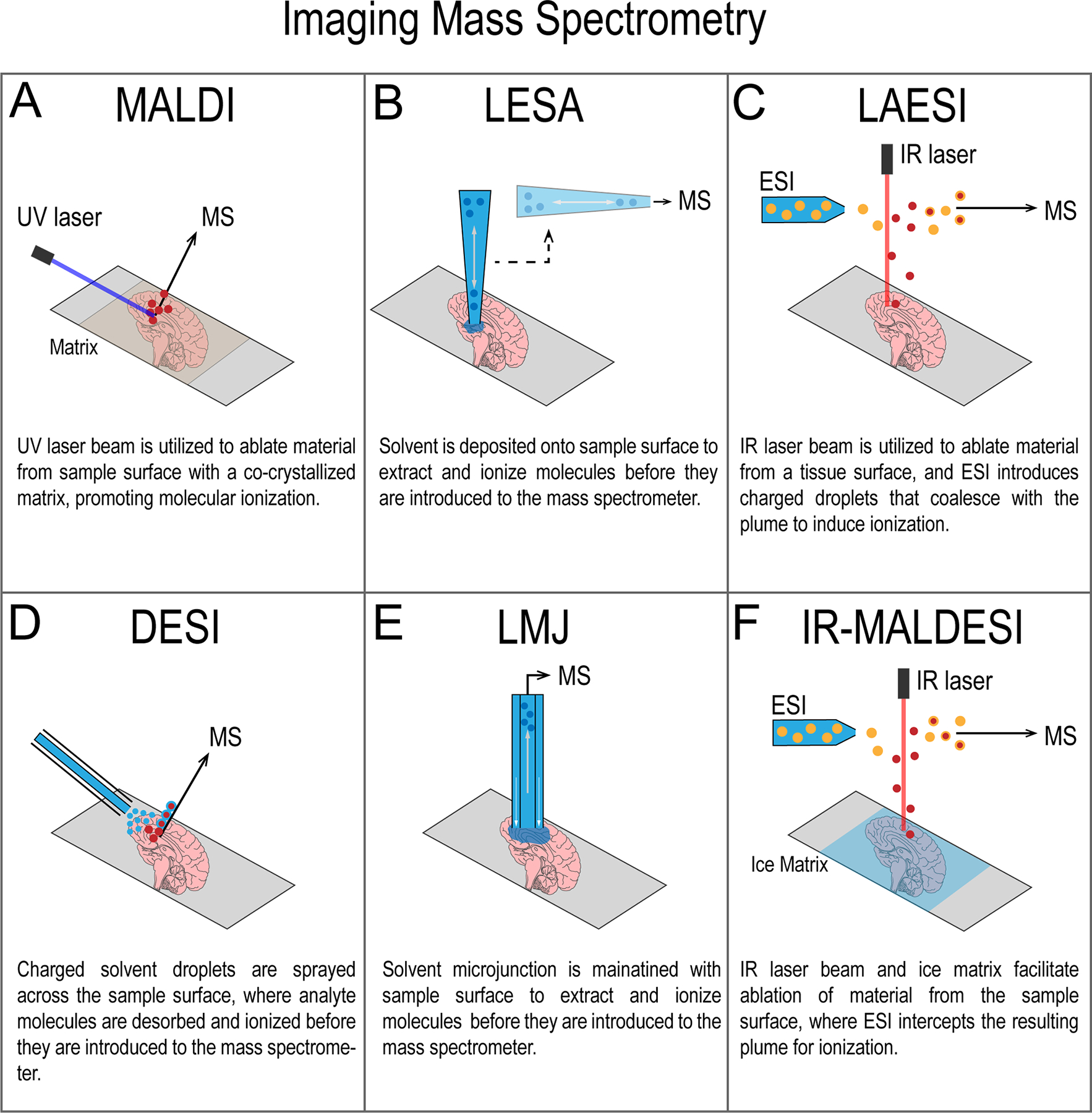
Overview of imaging mass spectrometry ion sources discussed. (A) Matrix-assisted laser desorption/ionization, (B) liquid extraction surface analysis (LESA), (C) laser ablation electrospray ionization (LAESI), (D) desorption electrospray ionization (DESI), (E) liquid microjunction (LMJ), and (F) infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI).
Traveling Wave Ion Mobility Spectrometry
Traveling wave ion mobility spectrometry (TWIMS) is perhaps the most common form of ion mobility coupled with IMS. Briefly, TWIMS achieves gas-phase separation by transmitting ions into a cell lined with a series of ring electrodes where they are radially confined with a RF voltage (Figure 1A), and ion propulsion is achieved by applying an additional DC voltage to a pair of adjacent ring electrodes. This voltage is then pulsed down the length of the TWIMS cell, creating a travelling electric field wave that ions follow through an inert gas, such as nitrogen.63,64 Differential propulsion of ions through this gas leads to separation as a function of effective size, shape, and charge. Similar to MS, ion mobility performance is often measured in terms of resolving power, but it is important to note that these values are often calculated differently across ion mobility methods. As such, care should be taken when making inter-platform comparisons as elegantly discussed in a review by May and McLean65 and later addressed by Dodds et al.31 Within this perspective, we will use a CCS-based calculation (CCS/ΔCCS) that allows for cross-platform comparisons whenever possible. TWIMS commonly yields a resolving power of ~40 (CCS/ΔCCS),31 which is sufficient for resolving many isobaric ion species36,66 and some isomeric ions, often with the aid of chemical derivatization67 or metal adduction.68,69 CCS values for many analyte classes are also frequently calculated using TWIMS-based analyses, leading to the curation of several databases for species such as proteins and steroids.30,70 Despite being newer than some other ion mobility techniques, utilization of TWIMS already extends across many analytical disciplines. In particular, TWIMS has proven to lend itself to analysis of large proteins and protein complexes, even becoming popular for use in native mass spectrometry.71–73 Upon commercialization, TWIMS has also been incorporated into many imaging mass spectrometry workflows for increasing the chemical information obtained from a pixel and applied to various analyte classes in an imaging context.60,74,75 For example, TWIMS has been coupled to many ion sources including MALDI,36,76–80 DESI,60,74,80,81 LAESI,82,83 and LESA (Figure 2A–D).48,61,84,85
TWIMS coupled to a DESI ion source was reported by Towers et al. in 2018 for the investigation of intact proteins and peptides from mouse liver tissue sections.60 Ion mobility drift time data from TWIMS proved invaluable in this work by providing ‘spectral filtration’ that allowed for separation of endogenous species from chemical noise. Mass spectral data was partitioned into three regions of mobility, allowing for the removal of many mass interferences for proteins and increasing S/N. This work illustrates the power of coupling TWIMS to imaging mass spectrometry for enhanced visualization of proteins and peptides. This suggests that TWIMS may afford improved dynamic range for IMS enabling key insights into complex biological questions within more molecularly heterogenous organ systems in the future. In another case, the Cooper group demonstrated LESA-based imaging coupled to TWIMS for the analysis of even larger analytes, intact proteins and protein complexes.84 Here, the authors employed native-like solvents containing ammonium acetate to their LESA workflow to allow protein complexes to remain in a native-like state. The detection of low charge state proteins directly from tissue along with CCS values calculated using TWIMS suggested that proteins retained a folded state through the analysis. Uniquely, this approach provides a way to determine how protein structure might change in relation to their morphological distributions in tissue. It is important to note that analysis of intact proteins and protein complexes is a highly elusive endeavor and determining native structure is a point of much discussion in the scientific community.85–87 In another example, Hale and colleagues showed that mobility information provided by TWIMS enabled filtering of spectral noise that contributed to improved ion image quality.61 For example, the authors demonstrated that without mobility filtering, ion images of heme-bound hemoglobin tetramer collected on mouse kidney was homogenously distributed, but incorporating ion mobility information uncovered localization to blood vessels within the kidney renal pelvis. This observation touches on an important issue for imaging mass spectrometry where potentially confounding factors such as spectral complexity, dynamic range, and chemical specificity present challenges in accurately determining spatial distributions.88,89 While these difficulties have not yet been fully addressed in the field, this work provides an example of how ion mobility can enhance molecular imaging performance.
Small molecules and lipids are often difficult to analyze due to the presence of isobaric and isomeric species that impede mass spectral interpretation. Barre, Rocha and coworkers utilized a prototype “μMALDI” source with enhanced speed and spatial resolution combined with the separating capabilities of ion mobility for IMS of lipids (Figure 3).36 In an image collected from rat brain tissue, the authors highlighted the drift time separation of two isobaric species (Figure 3A–C), ([PC(18:1_18:0)+K]+,m/z 826.60) and unidentified m/z 826.47, demonstrating the ability of TWIMS to reduce spectral complexity and provide a clear map of their distinct spatial distributions (Figure 3D–E). These two ions were easily distinguished in drift time space (Figure 3A–C), demonstrating ion mobility separation for enhanced discrimination of analytes while increasing S/N in an IMS experiment. While challenges associated with direct tissue sampling limit molecular coverage, these studies illustrate how technological advancements such as ion mobility, lend to spatial investigation of an ever-expanding list of biomolecular classes.
Figure 3:
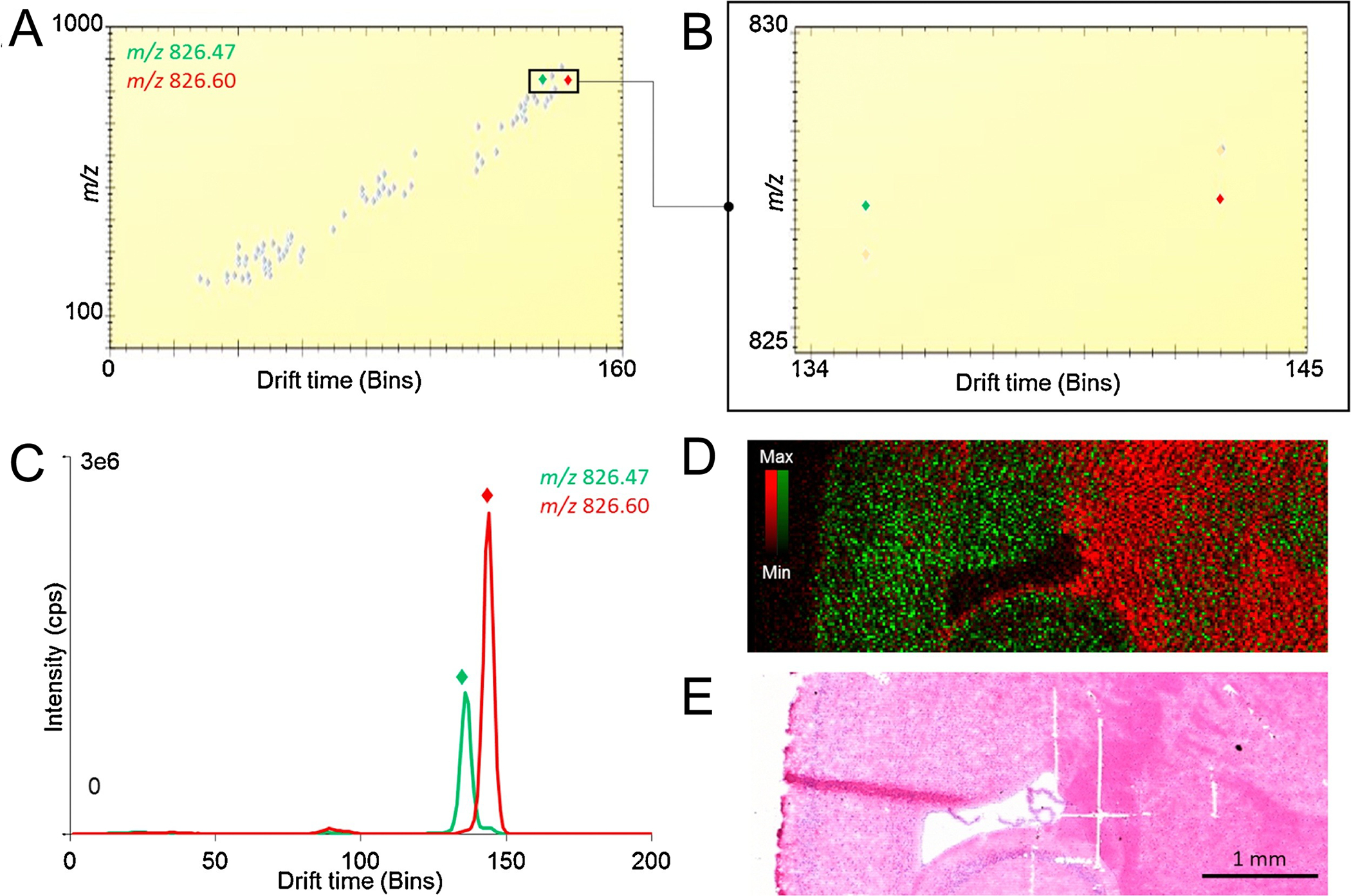
TWIMS provides an orthogonal level of separation for isobaric ions, aiding in the elucidation of distinct spatial distributions in murine brain tissue. Data was collected in continuous rastering mode at 20 μm spatial resolution on a Synapt G2-Si mass spectrometer equipped with a prototype “μMALDI” ion source. (A) Ion mobiligram averaged from the full m/z range acquired. (B) Enlargement of drift range 134–145 (bins) and m/z 825–830. (C) Extracted ion mobiligram for [PC(18:1_18:0)+K]+ (green) and unidentified m/z 826.60 (red). (D) Overlay image of both species demonstrating unique distributions. (E) H&E stained image of tissue post-IMS. This figure is adapted with permission from ref36, Barre, F. et al. International Journal of Mass Spectrometry (2020). Copyright 2019 Elsevier.
A recent advancement in high performance ion mobility is cyclic TWIMS (cTWIMS).90 Because TWIMS resolving power is, in part, a function of the length of the cell, this next generation of TWIMS allows for theoretically unlimited resolving power by designing the cell to be cyclical, or continuous. This cyclical design means that ions can cycle around the cell multiple times, increasing the effective ‘length’ of the mobility device and thereby significantly increasing the potential resolving power. Using cTWIMS, Giles et al. demonstrated resolving powers of up to 750 (CCS/ΔCCS).90 While the increase in ion mobility resolving power is infinite in theory, two major practical factors define its limitations. First, imperfect radial confinement of ions leads to increased signal loss as a function of path length.91 To provide resolving powers as high as those described above, many passes through the cTWIMS device are required, necessitating relatively high ion signal abundances to withstand radial ion loss. Second, the cyclical nature of the device leads to a phenomenon called “wrap around” where ions with different mobilities can lap one another as they cycle through the device, leading to ions with higher mobility overlapping ions with lower mobility. This inherently makes it difficult to determine the number of passes each ion has made through the cTWIMS cell and makes data difficult to interpret. To address this issue, researchers often use mass filtering prior to ion mobility to allow only a small m/z window into the cTWIMS device to limit the range of mobilities within the cell at any given time. Even with these limitations, cTWIMS offers one of the highest ion mobility resolving powers recorded. This enhancement has obvious implications for IMS where traditional separation techniques are not applicable. Sisley et al. has demonstrated the use of cTWIMS with a LESA-IMS workflow to study proteins within the murine brain.48 In sum, a single pass within the cTWIMS cell provided detection of 30 proteins, where traditional LESA-IMS only yielded 16 proteins. In this case, analysis of rat kidney was hampered by highly abundant interfering α– and β–globin chains of hemoglobin, but mobility separation from a single pass of cTWIMS allowed for detection of an additional 60 proteins. Excellent results were obtained from multiple passes through the cTWIMS cell where 1, 2, and 3 cycles were compared for the analysis of rat kidney tissue. Using heat-maps of arrival time vs m/z, the data could be dissected into discrete regions of interest that allowed for removal of interferences and improved sensitivity for protein detection (Figure 4A). The authors found that more passes led to higher degrees of separation and therefore greater distinction of proteins (Figure 4B). While only currently commercially available for integration with DESI and electrospray-based techniques like LESA, future cTWIMS utilization for IMS has great potential to introduce a new generation of high-resolution gas-phase separations to molecular imaging experiments.
Figure 4:
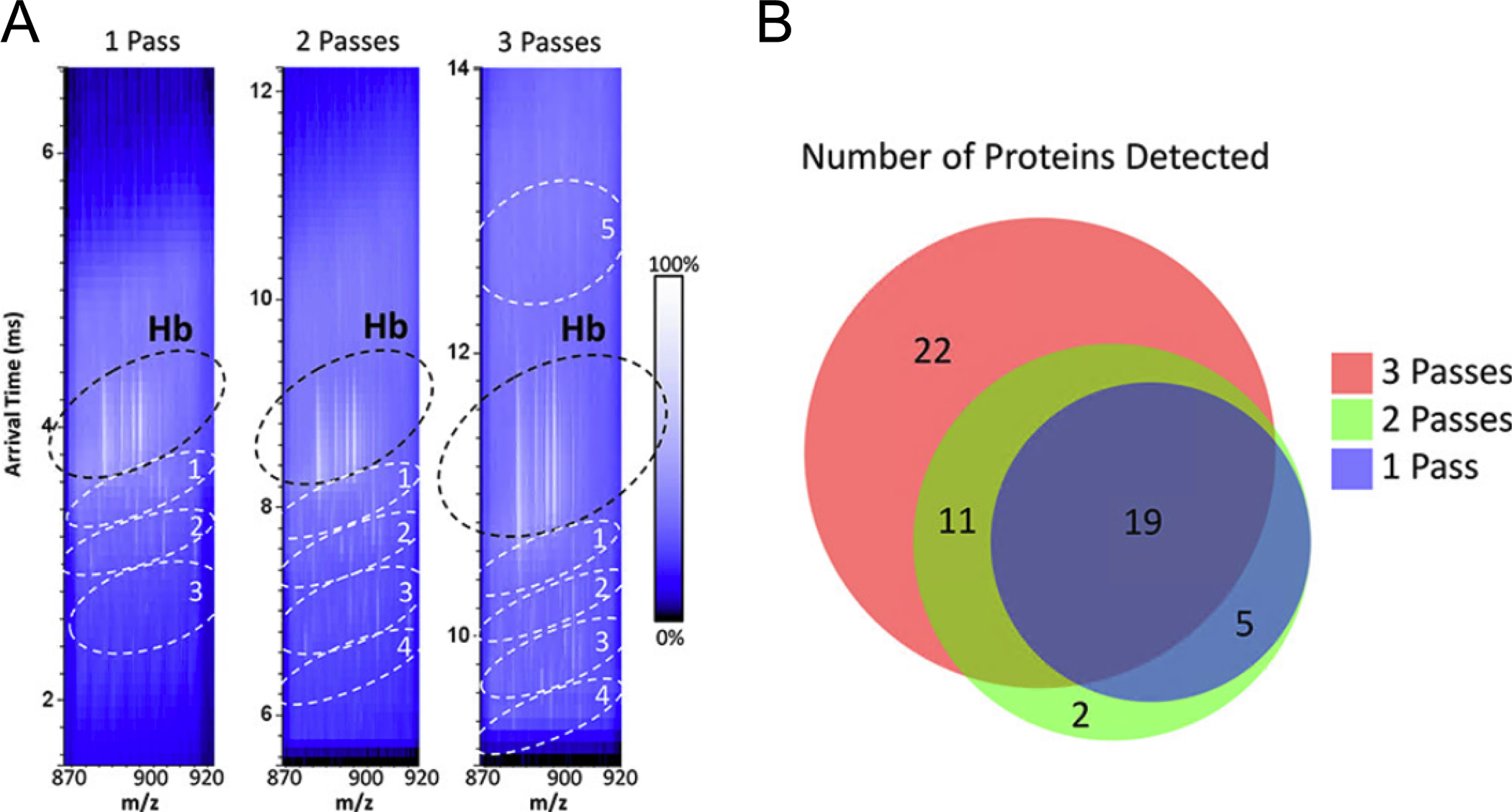
Increasing passes of ions through the cTWIMS device results in higher degrees of separation and more proteins detected from a LESA experiment of murine kidney tissue. LESA experiments were performed with a microjunction diameter of ~1.5 mm and data were collected using a prototype cTWIMS-MS mass spectrometer with quadrupole isolation of m/z 870 – 920. (A) 2D heatmaps plotted with arrival time vs m/z for 1, 2, and 3 passes through the device with regions of interest circled. (B) Venn diagram of protein numbers detected for each number of cTWIMS passes. This figure is adapted with permission from ref48, Sisley, E.K. et al. Analytical Chemistry (2020). Copyright 2020 American Chemical Society.
Another recent development in traveling wave ion mobility is stuctures for lossless ion manipulations (SLIM) first demonstrated by Smith and coworkers.92–94 SLIM utilizes RF ion confinement within a printed circuit board device and mobility separation is achieved by application of DC potentials for ion manipulation through an inert gas in a traveling wave-based approach. Some SLIM designs are capable of ultra-high resolving powers, with some reported CCS-based values of up to ~1860.95 Although examples of imaging with this technology are still limited, Nagy et al. demonstrated high resolution SLIM separations with LESA for imaging of disaccharides within a tripartite culture of peat moss, cyanobacteria, and fungi.96 Despite SLIM-IMS still being in its infancy, it has clear potential for substantial future contributions to spatial interrogation of complex biological systems.
Trapped Ion Mobility Spectrometry
Trapped ion mobility spectrometry (TIMS) is a high resolving power ion mobility technology introduced by Fernandez-Lima and coworkers in 2011.97 TIMS separations are carried out in the first vacuum stage of a mass spectrometer consisting of an augmented ion funnel with an entrance funnel, TIMS tunnel, and exit funnel (Figure 1B).97,98 In brief, ions are accumulated in the device, where analytes with differing mobilities are trapped and separated by opposing forces. Moving forward through the funnel, ions are propelled by a carrier gas. In the opposite direction, force is applied to the ions using an axial electric field gradient. To elute trapped ions, the electric field gradient is gradually reduced/scanned resulting in transmission of ions with ascending mobilities. Note, this is reverse from most other ion mobility techniques that transmit ions based on descending mobility, accomplished by applying a forward force using an electric field and collisions with the gas counteracts this forward motion. The scan rate of a TIMS experiment is defined by the scan time (ms) and voltage range, which correspond to user-defined mobility values. Scan rates dictate the separation resolution, with slower scan rates resulting in higher resolutions. To date, resolving power of >300 for singly charged species and >200 for doubly charged species have been reported.99
For IMS applications, TIMS was first introduced in a MALDI Q-TOF platform in 2019 by Spraggins and coworkers.37 MALDI TIMS IMS studies have successfully demonstrated enhanced data quality over images collected with TIMS disabled.37 Specifically, TIMS separations greatly increase the peak capacity of an IMS experiment, as reported by Neumann et al., who have detected >900 more spectral features by utilizing TIMS in comparison to Q-TOF-only mode when analyzing metabolites within human kidney tissue.100 In addition, TIMS successfully resolved matrix peaks from metabolite signal and demonstrated the separation of both isobaric and isomeric metabolites with distinct spatial localizations within human kidney tissue.100 TIMS has also been utilized in spatial interrogation of lipids. The Caprioli research group previously demonstrated the use of a MALDI timsTOF prototype platform for increased specificity of lipid analysis through in situ separation of closely isobaric lipid species.37 Phosphatidylcholines (PC) with unique adducts, [PC(34:3)+H]+ and [PC(32:0)+Na]+, have a mass difference of 3 mDa which would require a mass resolving power of over 300,000 to distinguish, and cannot be separated by the mass resolving power of the Q-TOF platform. However, with gas phase separations afforded by TIMS, the two species elute at different mobility values and can be resolved in mobility space, presenting an alternative to ultra-high mass resolution instruments. The two PCs have distinct spatial localizations within the analyzed mouse pup tissue, with the protonated species localizing to the entire tissue, with the exception of the brain and the spinal cord (Figure 5A), and the sodiated species localizing to the brain, spine, and intestines (Figure 5B). Similarly, Fu et. al. have demonstrated in situ mapping of isobaric and isomeric PCs in freshwater crustacean tissue.40 They report the separation of isobaric species: [PC (16:0/18:1)+Na]+ and [PC(18:1/18:3)+H]+ / [PC(16:0–20:4)+H]+ and distinguish their unique distributions in the muscle and oocytes of a female gammarid. In a follow up manuscript, Djambazova and coworkers have demonstrated the MALDI TIMS separation of isomeric lipid standards that vary in acyl chain composition, as well as those that vary in double bond position and geometry within acyl chains.101 Lastly, the authors also show in situ separation of lipid isomers that localize to different regions of a whole-body mouse pup.
Figure 5:
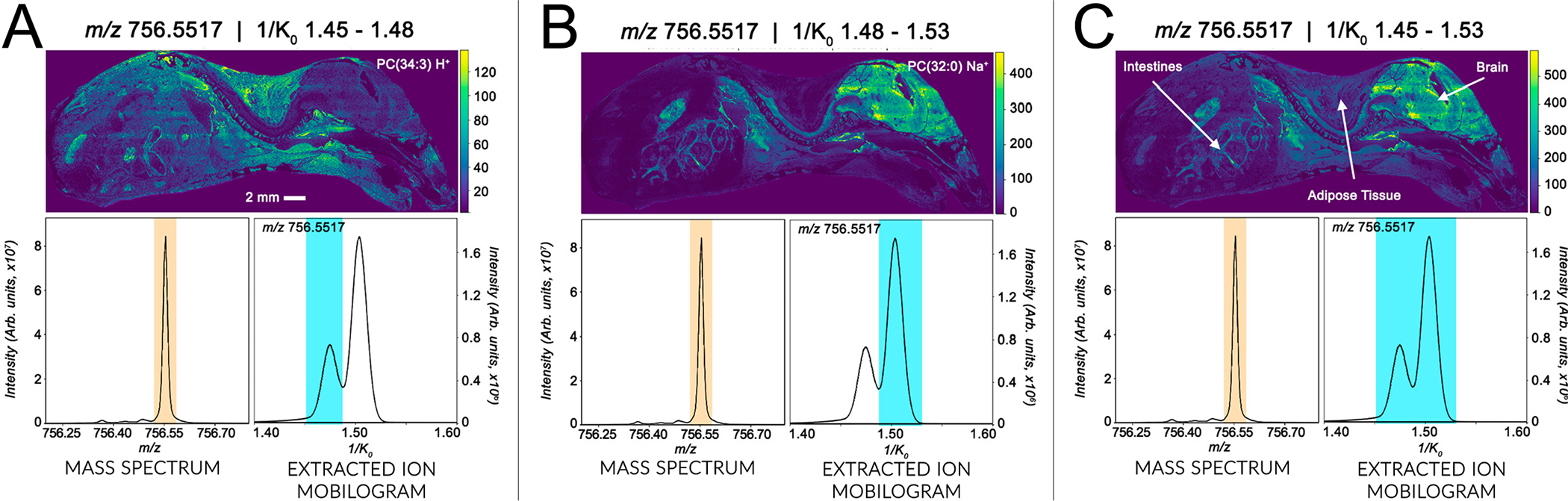
TIMS analysis of a whole-body mouse pup uncovers distinct spatial distributions of isobaric species indistinguishable by the mass spectrometer alone. Ion images were acquired at 50 μm spatial resolution with a 400 ms TIMS scan time on a prototype timsTOF fleX equipped with a MALDI ion source. (A) Ion image produced from a selected mobility region for [PC(34:3)+H]+. (B) Ion image produced from selected mobility region for [PC(32:0)+Na]+. (C) Ion image produced without mobility selection, demonstrating domination by the higher intensity [PC(32:0)+Na]+.This figure is adapted with permission from ref37, Spraggins, J.M. et al. Analytical Chemistry (2019). Copyright 2019 American Chemical Society.
Since TIMS is a relatively new introduction to the field of imaging mass spectrometry, it has not been integrated with other IMS techniques to the best of our knowledge, although we eagerly anticipate this. Furthermore, although TIMS has been used for metabolomic, and lipidomic analysis, it has not yet been used to study other molecular classes in an imaging context. Beyond imaging applications, TIMS has been utilized for in-depth lipidomic102–104 and proteomic104–106 analyses alike. Of note, Fernandez-Lima and coworkers have demonstrated LC-TIMS separations of isomeric lipid standards,103 including double bond position/geometry isomers, isomers that vary in acyl chain position (sn-position) and various combinations thereof. Although TIMS has only provided lipid isomer separation in imaging on a limited basis,40,101 these results clearly indicate the potential for further isomer distinction and mapping. Furthermore, the separation and tentative identification of different lipid classes, as well as the partial separation of sn-position isomers from a non-targeted analysis of human plasma, have also been shown. Vasilopoulou et al. carried out a comprehensive characterization of the lipidome in small volumes of human plasma using nanoLC and TIMS.107 Utilizing the parallel accumulation-serial fragmentation (PASEF) capabilities of the timsTOF platform, they enabled fragmentation of 15 precursors in each TIMS scan. With this tool, they report identifications of over three times as many lipids as with standard TIMS-MS/MS. This study speaks to the utility of PASEF in lipid analysis for enhanced sensitivity within a TIMS experiment. If applied to imaging, PASEF has enormous potential to make MS/MS a much more prevalent practice in the field, and greatly improve the information obtained from a single imaging experiment.
Previous reports have also shown TIMS to be a powerful tool for structural investigation of proteins and peptides. Recently, nESI-TIMS-MS has been used for high throughput screening of peptide topoisomers. The Fernandez-Lima group has demonstrated the separation and identification of lasso peptides from their branched-cyclic analogs, which is challenging as their structural stability is due to subtle differences in the tertiary structures.104 Furthermore, similar to previous literature, they report increased TIMS separations via metal ion adduction (Na, K, and Cs salt). All of these studies lay the groundwork for future IMS applications in which TIMS separations can be essential for addressing increasingly complex biological problems including clinical applications.
High-Field Asymmetric Waveform Ion Mobility
High-field asymmetric waveform ion mobility (FAIMS), or differential mobility spectrometry, is another form of ion mobility commonly exploited for use in IMS. FAIMS achieves gas-phase separation of ions at atmospheric pressure based on their mobility prior to entering the mass spectrometer.108–110 FAIMS functions by applying a dispersion field, a high frequency asymmetric waveform of alternating low and high fields, perpendicular to the ion trajectory through the device (Figure 1C). This alternating dispersion field results in ions oscillating through a buffer gas, usually nitrogen or helium, causing deviation from their initial trajectory as a function of their respective mobilities through bombarding gas. This deviation ultimately leads to neutralization upon collision with the electrode wall. Selective ion transmission is made possible by the superimposition of a secondary DC field known as the compensation voltage which acts to correct the deviation of ions within a select mobility window, allowing transmission of ions with specific mobilities. Unlike other ion mobility methods, FAIMS is not usually used to provide orthogonal information such as drift time or CCS but to increase signal-to-noise, sensitivity, and dynamic range.109 FAIMS operates at atmospheric pressure, and it has not yet been coupled with sub-atmospheric IMS ion sources, such as mid-low pressure MALDI sources. While modifications to recently developed atmospheric MALDI sources could make MALDI-FAIMS possible, it has not yet been reported. For this reason, all examples of FAIMS applied to IMS to date have been conducted with ambient ion sources such as DESI and LESA.
FAIMS has recently shown great promise for increasing imaging data quality when coupled to a LMJ-SSP (Figure 2E) for protein analysis in rat brain and human ovarian cancer tissue sections.49 Fieder et al. demonstrated use of LMJ-SSP that provides sensitive protein sampling at the expense of spatial resolution (~500–1000 μm). Selective transmission by FAIMS resulted in detection of many proteins that were not seen with LMJ-SSP alone (Figure 6A). Furthermore, implementation of FAIMS yielded dramatic noise reduction resulting in S/N increases of up to 7-fold for some proteins (Figure 6B and C). These dramatic data improvements, however, came at the expense of sensitivity outside the m/z range for which FAIMS was optimized and signal loss of several proteins was observed. In addition, colocalization of several proteins to the tumor region were uncovered when FAIMS was coupled to LMJ-SSP for imaging of healthy and high grade serous ovarian cancer tissues. In later work, Garza et al. went on to demonstrate the utility of FAIMS for IMS of intact proteins on tissue when coupled to DESI.111 When compared to data acquired without FAIMS, DESI-FAIMS-IMS of mouse brain demonstrated a significant increase in S/N of several high abundance proteins. Here, the authors acknowledged that only highly abundant proteins were detected, necessitating further optimization for less abundant species. However, this study does provide groundwork for further development of technologies for performing atmospheric pressure gas-phase separations.
Figure 6:
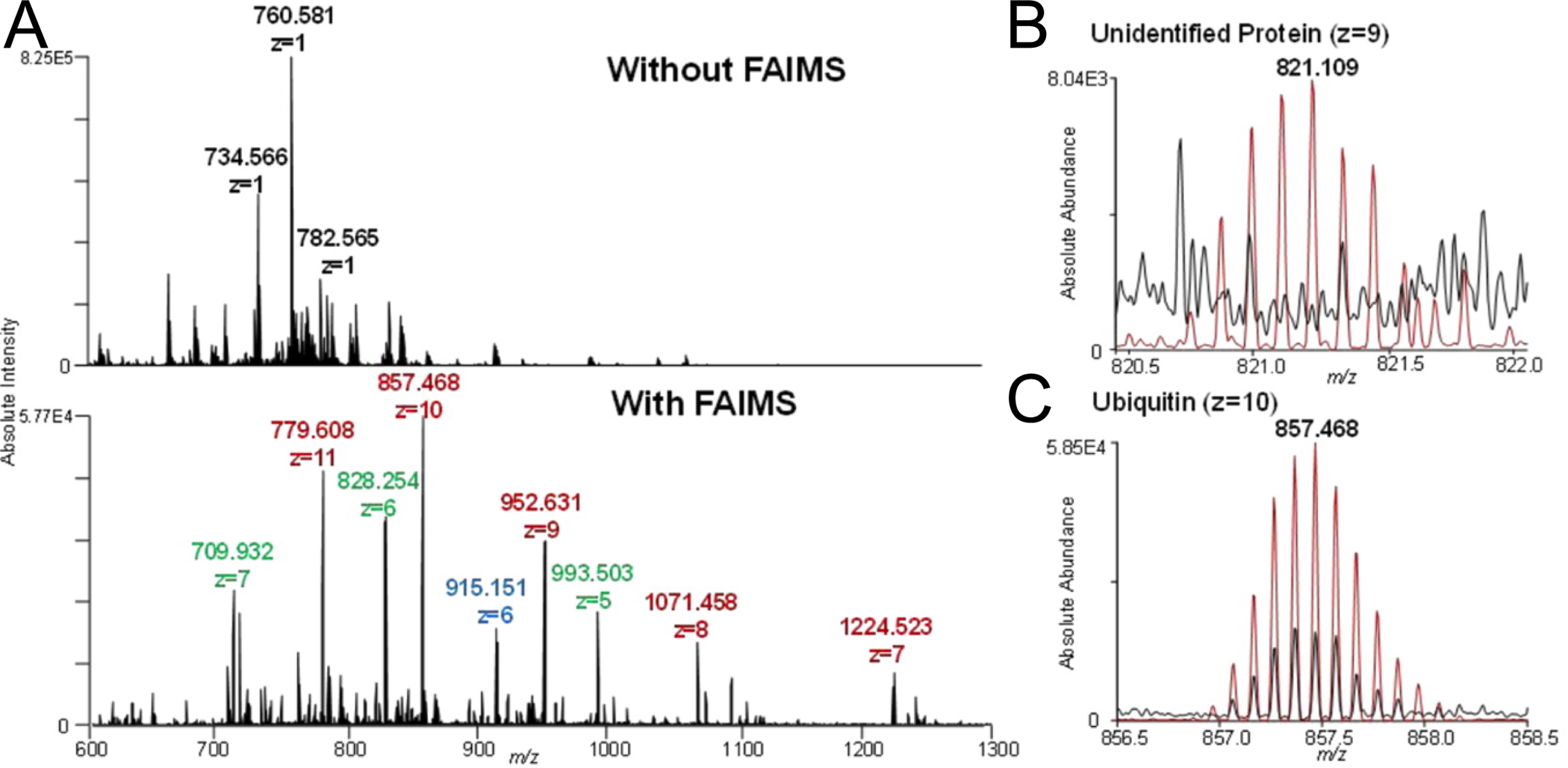
LMJ-SSP analysis of rat brain tissue increases in S/N and number of proteins observed when FAIMS is applied. Data were collected using a Q Exactive mass spectrometer equipped with an ultraFAIMS device using an ND chip and optimized for proteins between 4 – 12 kDa. (A) Spectral interferences are significantly reduced with FAIMS. (B) Signal detected of an unidentified protein (z=9) with, (red) and without, (black) FAIMS. (C) Signal detected of a ubiquitin (z=10) with, (red) and without, (black) FAIMS. This figure is adapted with permission from ref49, Feider, C.L. et al. Analytical Chemistry (2016). Copyright 2016 American Chemical Society.
While the examples described above were conducted with a planar FAIMS device, Griffiths et al. recently showed the benefits of applying a cylindrical FAIMS device for protein detection using LESA-IMS.112 Implementation of this cylindrical FAIMS device allowed for a dynamic multistep compensation workflow wherein several voltages were used per pixel that provided increased protein coverage. While advantageous to overall data quality, this enhancement was to the detriment of acquisition time, requiring three to four minutes per pixel. When LESA-FAIMS was used for imaging of rat testes, kidney and brain, up to a 22-fold increase in detected proteins were observed above data collected with FAIMS voltages turned off. Although performed on a more sensitive mass spectrometer, the use of cylindrical FAIMS produced a dramatic increase in proteins detected compared to previous work conducted with a planar FAIMS device.84 Advantages of implementing a cylindrical FAIMS device in an imaging platform are especially clear when compared to data acquired from bottom-up proteomics studies resulting in similar numbers of detected proteins.113 There is a trade-off, however, as this work provides spatial information of detected proteins, it comes at the expense of fragmentation-based identification afforded by bottom-up proteomics. Several other recent FAIMS advancements have been made with implications for use in IMS. Bowman et al. recently investigated the separating power of FAIMS for lipid isomers using electrospray ionization.114 Careful internal calibration with lipid standards allowed the authors to separate many types of lipids: seven out of nine sn and chain length, three out of four double bond position, and seven out of nine cis/trans lipid isomer pairs. In another recent application of FAIMS for isomer separation, Shliaha et al. coupled FAIMS to an LC-MS/MS workflow for the middle-down investigation of mouse embryonic stem cell histone post translation modifications (PTM).115 The authors found that FAIMS provided separation of many isomeric proteoforms with the same PTMs on different residues, resulting in discrimination of 25 out of 40 tested forms of histone H3. While these advancements dramatically increase duration of analysis, this work clearly establishes the power of FAIMS for the separation of peptide isomers that could prove highly beneficial for future use in IMS.
Drift Tube Ion Mobility Spectrometry
The design of drift tube ion mobility spectrometry (DTIMS) is rooted in the early days of ion mobility, making it perhaps the simplest modern form of these gas-phase separation technologies. DTIMS functions by transmitting ions through an inert gas within a drift tube comprised of stacked ring electrodes. Ions are driven through the cell by a uniform electric field where they collide with drift gas (Figure 1D). Ions with a larger CCS encounter more drift gas molecules compared to ions with a smaller CCS, allowing separation as a function of mobility.65 DTIMS has been demonstrated in varying pressures regimes, but reduced pressure (<5 Torr)116,117 provides efficient ion transfer from the ion mobility cell to the high vacuum mass spectrometer and is therefore advantageous for coupling to IMS.118 The simplistic design of DTIMS does not, however, equate to low performance as DTIMS can routinely achieve moderate-high resolving power (~60 (CCS/ΔCCS)65,119 and produces consistent CCS measurements.32 It also readily couples to MS and is commonly used to provide analyte separation for increased peak capacity and reduced spectral complexity.120–122 Moreover, DTIMS has proven invaluable for offering orthogonal information about analytes in a MS experiment, being a primary technology used to measure absolute CCS.65,123 Of note, DTIMS is the only form of ion mobility that can provide CCS values without calibration.124 This is an inherent benefit often leading to increased accuracy as improper calibration produces inaccurate CCS values.30,125 Stow and coworkers recently demonstrated the robustness of DTIMS for measuring CCS values in an interlaboratory study where they reported a relative standard deviation of 0.29%.32 This level of robustness has also been exploited by several groups to compile DTIMS CCS databases.124,126–129 Despite its clear utility and use in other MS fields, a limited number of examples of DTIMS-IMS experiments have been reported in recent years. This may be the result of early success of the TWIMS Q-TOF MS technology that was compatible with a variety of sources. Likewise, custom integration of DTIMS with modern IMS platforms poses many challenges such as instrument synchronization, and communication.50
The Woods laboratory has published numerous examples of successful utilization of MALDI-DTIMS.130–132 In addition, the McLean and Caprioli research groups reported DTIMS coupled to MALDI IMS using a custom MALDI source capable of 200 μm spatial resolution. They demonstrated the power of DTIMS to distinguish isobaric lipid and peptide species, allowing for their clear distinction that was not afforded by the mass spectrometer alone.42 Additionally, DTIMS can be coupled to other ion sources capable of IMS, such as DESI, LAESI and IR-MALDESI demonstrating the potential for its expanded use in the field.43,45,46,50,83,133,134
In 2014, Jackson and coworkers utilized MALDI-DTIMS for imaging various lipid classes where distinct mobility trends for each lipid subclass was shown to correlate with lipid head group size.132 Lipids with smaller head groups (e.g. glycerophosphates with only a proton at the sn-3 position) had the fastest drift times corresponding to the highest mobility. An important consideration of this study that extends to all examples of IMS datasets with ion mobility, is data manageability. Without a system for data reduction, IMS file sizes with added ion mobility dimensionality can reach enormous sizes. In this case, the authors addressed this problem by restricting mass and drift time ranges to attenuate file size. In spite of this challenge, DTIMS provided an orthogonal element to imaging data collected from mouse cerebrum permitting easier lipid assignment. This work serves an example of the data dimensionality that DTIMS provides to an imaging experiment.
Many recent advancements in DTIMS resolving power135–139 may provide the impetus for investigators to develop further strategies for integrating DTIMS with IMS. Ekelof et al. described the first inclusion of DTIMS to IR-MALDESI (Figure 2F) for effective imaging.50 Validation of the modified instrument demonstrated calculated CCS differences of less than 3.1% of previously reported values. Of note in this study, both proteins and small molecules were efficiently ionized by IR-MALDESI, allowing for accurate calculation of CCS for analytes in low and high mass ranges by this custom IR-MALDESI-DTIMS system. The authors demonstrated effective analysis of caffeine, and mobility separation of glucose and fructose when applied to analysis of Coca-Cola®. This separation presents obvious advantages for use in imaging of small molecules where MS/MS is often insufficient for isomer distinction, a situation that severely hampers analysis.140 In contrast to previous work coupling IR-MALDESI with trap-based instruments,141–144 this system provided the necessary dynamic range to detect intact proteins, lipids and small molecules in an image of an oak leaf. Data acquired from this proof-of-concept experiment has implications for future utilization of IR-MALDESI coupled to DTIMS-MS for structurally informative spatial interrogation of biomolecules.
Conclusions and Future Directions
The examples above clearly demonstrate the advantages of integrating ion mobility with IMS and the need to continue developing these technologies for biological imaging experiments. Throughout history, new technologies have continuously driven scientific advancement. Modern instrumentation and molecular imaging capabilities enable researchers to make connections between molecular, spatial and temporal domains with the goal of fully describing the interplay of biomolecular species in cellular environments. The aspiration of this work is to uncover how the molecular components of cells, tissues and organ systems interact to drive disease; ultimately enabling more effective precision treatments and therapies. IMS is ideally positioned to bridge the gap between the many -omics (e.g. lipidomics, proteomics, and metabolomics) that are foundational to modern biomedical research. This is made evident in the prominent role IMS is playing in recently established, large-scale research initiatives such as the NIH human biomolecular atlas program (HuBMAP, hubmapconsortium.org),145 NIH kidney precision medicine program (KPMP, kpmp.org), and the Cancer Research Grand Challenge in the United Kingdom (cancerresearchuk.org). As the field moves beyond demonstrating the capabilities of IMS towards more impactful biological applications, removing the ambiguity of peak annotation is critical. To fully describe the molecular drivers of biological processes, structural identification is paramount in understanding their mechanistic underpinnings. The extra data dimension afforded by ion mobility-IMS (x,y,z positions, m/z, and mobility), provides a path forward for improving sensitivity, dynamic range, and specificity in a way that is fully compatible with the imaging experiment.
It is important to note that this added data dimension presents significant computational challenges. As instruments become more sensitive and spatial resolution is improved, the number of pixels (e.g. spectra) collected in a single image increases dramatically which can produce terabyte sized datasets. This challenge is only compounded by the addition of ion mobility, which adds an extra dimension to the data matrix and exponentially increases its resulting footprint. While this has obvious ramifications for data analysis and processing, it also significantly impacts raw data storage, access, and sharing. There are relatively few databanks willing to store such datasets, especially without significant cost to the authors or journal, creating a considerable barrier for data sharing. As such, this can have a negative impact on scientific reproducibility and collaboration. Some ways of dealing with large data sizes include binning or removing noise profiles to reduce the number of data points across the dataset. While effective at minimizing the data footprint, it removes noise and low intensity peaks, making it difficult to accurately calculate critical data features such as absolute S/N, mass resolution, CCS, and limits of detection/quantitation. As such, we anticipate the development of more advanced compression algorithms and efficient file types that facilitate data sharing and processing while maintaining an accurate representation of the data.
Few commercial software packages support visualization of ion mobility-IMS data and even fewer support statistical analysis. Because of this sparsity, many labs resort to custom software to visualize, analyze, and annotate ion mobility-IMS data. This limits the number of scientific groups without dedicated computational members or collaborators who can readily perform these experiments and utilize the performance of ion mobility-IMS for biological applications. Ultimately, we foresee an increase in commercial software capable of handling ion mobility-IMS data in the near future, in addition to the publication and distribution of lab-built, stand-alone software. For example, the Muddiman group is currently updating MSiReader to include ion mobility support50,146,147 while the Van de Plas37,100,101 and Laskin148 laboratories are each presently developing software capable of handling ion mobility-IMS data. Readily available and robust data analysis software that is reasonably easy to use will be more quickly incorporated within academic labs and enable higher quality analysis and interpretation of these complex data.
The future of ion mobility-IMS certainly includes instrumental developments that improve sensitivity, transmittance, speed, and resolving power. Significant technical improvements to these attributes will unquestionably enhance visualization and analysis of all chemical classes. In particular, ion mobility-IMS is well suited to clarify the biological relevance of metabolic isomers, lipid double bond position, and native protein conformers, all of which are difficult to study with other analytical approaches. Not only will ion mobility information improve our understanding of normal and disease states within many biological systems, but it will allow us to generate testable hypotheses in order to deeply interrogate molecularly complex systems. When considering the field in entirety, MALDI and DESI compose a majority of ion mobility-IMS publications, despite the high spatial resolution and orthogonal chemical coverage afforded by SIMS. This can partially be attributed to fewer number SIMS instruments used for R&D compared to other systems, low source pressure, and high energy of secondary ions entering the mass spectrometer. Despite these engineering difficulties, SIMS analysis would benefit from ion mobility separations because there is an abundance of isomers and isobars present within the m/z range most accessible to SIMS analysis (<2 kDa).
This article has summarized recent advances within the field of integrated ion mobility and IMS and the potential of this technology to help unravel the molecular complexity of living systems. With maturation of these technologies and the future development of hardware and software that will usher in higher performance capabilities, the outlook for ion mobility-IMS for imaging experimentation and discovery is exciting.
Acknowledgements
Support was provided by the NIH Common Fund and National Institute of Diabetes and Digestive and Kidney Diseases (U54DK120058 awarded to J.M.S. and R.M.C.), NIH National Institute of Allergy and Infectious Disease (R01AI138581 awarded to J.M.S.), the National Science Foundation Major Research Instrument Program (CBET - 1828299 awarded to J.M.S. and R.M.C.)., and by the NIH National Institute of General Medical Sciences (2P41GM103391 awarded to R.M.C.). E.K.N. is supported by a National Institute of Environmental Health Sciences training grant (T32ES007028).
References:
- (1).Bryant DM; Mostov KE From Cells to Organs: Building Polarized Tissue. Nat. Rev. Mol. Cell Biol Nature Publishing Group; November 2008, pp 887–901. 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Norris JL; Caprioli RM Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/ Ionization Imaging Mass Spectrometry in Biological and Clinical Research. 10.1021/cr3004295. [DOI] [PMC free article] [PubMed]
- (3).Caprioli RM; Farmer TB; Gile J Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem 1997, 69 (23), 4751–4760. 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- (4).Wiseman JM; Puolitaival SM; Takáts Z; Cooks RG; Caprioli RM Mass Spectrometric Profiling of Intact Biological Tissue by Using Desorption Electrospray Ionization. Angew. Chemie Int. Ed 2005, 44 (43), 7094–7097. 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- (5).Ifa DR; Wiseman JM; Song Q; Cooks RG Development of Capabilities for Imaging Mass Spectrometry under Ambient Conditions with Desorption Electrospray Ionization (DESI). Int. J. Mass Spectrom 2007, 259 (1–3), 8–15. 10.1016/j.ijms.2006.08.003. [DOI] [Google Scholar]
- (6).Robichaud G; Barry JA; Garrard KP; Muddiman DC Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. J. Am. Soc. Mass Spectrom 2013, 24, 92–100. 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nemes P; Barton AA; Li Y; Vertes A Ambient Molecular Imaging and Depth Profiling of Live Tissue by Infrared Laser Ablation Electrospray Ionization Mass Spectrometry. Anal. Chem 2008, 80 (12), 4575–4582. 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- (8).Eikel D; Vavrek M; Smith S; Bason C; Yeh S; Korfmacher WA; Henion JD Liquid Extraction Surface Analysis Mass Spectrometry (LESA-MS) as a Novel Profiling Tool for Drug Distribution and Metabolism Analysis: The Terfenadine Example. Rapid Commun. Mass Spectrom 2011, 25 (23), 3587–3596. 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- (9).Laskin J; Heath BS; Roach PJ; Cazares L; Semmes OJ Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Anal. Chem 2012, 84 (1), 141–148. 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Du Z; Lin J-R; Rashid R; Maliga Z; Wang S; Aster JC; Izar B; Sorger PK; Santagata S Qualifying Antibodies for Image-Based Immune Profiling and Multiplexed Tissue Imaging. Nat. Protoc 10.1038/s41596-019-0206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Neumann EK; Comi TJ; Spegazzini N; Mitchell JW; Rubakhin SS; Gillette MU; Bhargava R; Sweedler JV Multimodal Chemical Analysis of the Brain by High Mass Resolution Mass Spectrometry and Infrared Spectroscopic Imaging. Anal. Chem 2018, 90 (19), 11572–11580. 10.1021/acs.analchem.8b02913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Baker MJ; Trevisan J; Bassan P; Bhargava R; Butler HJ; Dorling KM; Fielden PR; Fogarty SW; Fullwood NJ; Heys KA; et al. Using Fourier Transform IR Spectroscopy to Analyze Biological Materials. Nat. Protoc 2014, 9 (8), 1771–1791. 10.1038/nprot.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mulder IA; Esteve C; Wermer JH; Hoehn M; Tolner EA; Van Den Maagdenberg AMJM; Mcdonnell LA; Mcdonnell L Funnel-Freezing versus Heat-Stabilization for the Visualization of Metabolites by Mass Spectrometry Imaging in a Mouse Stroke Model. Proteomics 2016, 16, 1652–1659. 10.1002/pmic.201500402. [DOI] [PubMed] [Google Scholar]
- (14).Manier ML; Spraggins JM; Reyzer ML; Norris JL; Caprioli RM A Derivatization and Validation Strategy for Determining the Spatial Localization of Endogenous Amine Metabolites in Tissues Using MALDI Imaging Mass Spectrometry. J. Mass Spectrom 2014, 49 (8), 665–673. 10.1002/jms.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Thomas A; Charbonneau JL; Fournaise E; Chaurand P Sublimation of New Matrix Candidates for High Spatial Resolution Imaging Mass Spectrometry of Lipids: Enhanced Information in Both Positive and Negative Polarities after 1,5-Diaminonapthalene Deposition. Anal. Chem 2012, 84 (4), 2048–2054. 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- (16).Sparvero LJ; Amoscato AA; Kochanek PM; Pitt BR; Kagan VE; Bayär H Mass-Spectrometry Based Oxidative Lipidomics and Lipid Imaging: Applications in Traumatic Brain Injury. Journal of Neurochemistry. NIH Public Access December 2010, pp 1322–1336. 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cornett DS; Frappier SL; Caprioli RM MALDI-FTICR Imaging Mass Spectrometry of Drugs and Metabolites in Tissue. Anal. Chem 2008, 80 (14), 5648–5653. 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Heikkinen EM; Auriola S; Ranta VP; Demarais NJ; Grey AC; Del Amo EM; Toropainen E; Vellonen KS; Urtti A; Ruponen M Distribution of Small Molecular Weight Drugs into the Porcine Lens: Studies on Imaging Mass Spectrometry, Partition Coefficients, and Implications in Ocular Pharmacokinetics. Mol. Pharm 2019, 16 (9), 3968–3976. 10.1021/acs.molpharmaceut.9b00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Everest-Dass AV; Briggs MT; Kaur G; Oehler MK; Hoffmann P; Packer NH N-Glycan MALDI Imaging Mass Spectrometry on Formalin-Fixed Paraffin-Embedded Tissue Enables the Delineation of Ovarian Cancer Tissues. Mol. Cell. Proteomics 2016, 15 (9), 3003–3016. 10.1074/mcp.M116.059816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Drake RR; Powers TW; Norris-Caneda K; Mehta AS; Angel PM In Situ Imaging of N-Glycans by MALDI Imaging Mass Spectrometry of Fresh or Formalin-Fixed Paraffin-Embedded Tissue. Curr. Protoc. Protein Sci 2018, 94 (1). 10.1002/cpps.68. [DOI] [PubMed] [Google Scholar]
- (21).Hanrieder J; Zetterberg H; Blennow K MALDI Imaging Mass Spectrometry: Neurochemical Imaging of Proteins and Peptides. In Neuromethods; Humana Press Inc., 2019; Vol. 146, pp 179–197. 10.1007/978-1-4939-9662-9_15. [DOI] [Google Scholar]
- (22).Judd AM; Gutierrez DB; Moore JL; Patterson NH; Yang J; Romer CE; Norris JL; Caprioli RM A Recommended and Verified Procedure for in Situ Tryptic Digestion of Formalin-Fixed Paraffin-Embedded Tissues for Analysis by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry. 2019. 10.1002/jms.4384. [DOI] [PMC free article] [PubMed]
- (23).Dilillo M; Ait-Belkacem R; Esteve C; Pellegrini D; Nicolardi S; Costa M; Vannini E; De Graaf EL; Caleo M; McDonnell LA Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Sci. Rep 2017, 7 (1), 1–11. 10.1038/s41598-017-00703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Spraggins JM; Rizzo DG; Moore JL; Noto MJ; Skaar EP; Caprioli RM Next-Generation Technologies for Spatial Proteomics: Integrating Ultra-High Speed MALDI-TOF and High Mass Resolution MALDI FTICR Imaging Mass Spectrometry for Protein Analysis. Proteomics 2016, 16 (11–12), 1678–1689. 10.1002/pmic.201600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ruotolo BT; Gillig KJ; Stone EG; Russell DH Peak Capacity of Ion Mobility Mass Spectrometry: Separation of Peptides in Helium Buffer Gas. J. Chromatogr. B Anal. Technol. Biomed. Life Sci 2002, 782 (1–2), 385–392. 10.1016/S1570-0232(02)00566-4. [DOI] [PubMed] [Google Scholar]
- (26).Revercomb HE; Mason EA Theory of Plasma Chromatography/Gaseous Electrophoresis. A Review. Anal. Chem 1975, 47 (7), 970–983. 10.1021/ac60357a043. [DOI] [Google Scholar]
- (27).Gabelica V; Shvartsburg AA; Afonso C; Barran P; Benesch JLP; Bleiholder C; Bowers MT; Bilbao A; Bush MF; Campbell JL; et al. Recommendations for Reporting Ion Mobility Mass Spectrometry Measurements. Mass Spectrom. Rev 2019, 38 (3), 291–320. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wyttenbach T; Kemper PR; Bowers MT Design of a New Electrospray Ion Mobility Mass Spectrometer. Int. J. Mass Spectrom 2001, 212 (1–3), 13–23. 10.1016/S1387-3806(01)00517-6. [DOI] [Google Scholar]
- (29).Hofmann J; Hahm HS; Seeberger PH; Pagel K Identification of Carbohydrate Anomers Using Ion Mobility-Mass Spectrometry. Nature 2015, 526 (7572), 241–244. 10.1038/nature15388. [DOI] [PubMed] [Google Scholar]
- (30).Bush MF; Hall Z; Giles K; Hoyes J; Robinson CV; Ruotolo BT Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem 2010, 82 (22), 9557–9565. 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- (31).Dodds JN; May JC; McLean JA Correlating Resolving Power, Resolution, and Collision Cross Section: Unifying Cross-Platform Assessment of Separation Efficiency in Ion Mobility Spectrometry. Anal. Chem 2017, 89 (22), 12176–12184. 10.1021/acs.analchem.7b02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Anal. Chem 2017, 89 (17), 9048–9055. 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhao Y; Singh A; Li L; Linhardt RJ; Xu Y; Liu J; Woods RJ; Amster IJ Investigating Changes in the Gas-Phase Conformation of Antithrombin III upon Binding of Arixtra Using Traveling Wave Ion Mobility Spectrometry (TWIMS). Analyst 2015, 140 (20), 6980–6989. 10.1039/c5an00908a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Guntner AS; Thalhamer B; Klampfl C; Buchberger W Collision Cross Sections Obtained with Ion Mobility Mass Spectrometry as New Descriptor to Predict Blood-Brain Barrier Permeation by Drugs. Sci. Rep 2019, 9 (1), 1–10. 10.1038/s41598-019-55856-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gomez JD; Ridgeway ME; Park MA; Fritz KS Utilizing Ion Mobility to Identify Isobaric Post-Translational Modifications: Resolving Acrolein and Propionyl Lysine Adducts by TIMS Mass Spectrometry. Int. J. Ion Mobil. Spectrom 2018, 21 (3), 65–69. 10.1007/s12127-018-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Barré F; Rocha B; Dewez F; Towers M; Murray P; Claude E; Cillero-Pastor B; Heeren R; Porta Siegel T Faster Raster Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging of Lipids at High Lateral Resolution. Int. J. Mass Spectrom 2019, 437, 38–48. 10.1016/j.ijms.2018.09.015. [DOI] [Google Scholar]
- (37).Spraggins JM; Djambazova KV; Rivera ES; Migas LG; Neumann EK; Fuetterer A; Suetering J; Goedecke N; Ly A; Van de Plas R; et al. High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (TimsTOF) Mass Spectrometry. Anal. Chem 2019, 91 (22), 14552–14560. 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Williams JP; Bugarcic T; Habtemariam A; Giles K; Campuzano I; Rodger PM; Sadler PJ Isomer Separation and Gas-Phase Configurations of Organoruthenium Anticancer Complexes: Ion Mobility Mass Spectrometry and Modeling. J. Am. Soc. Mass Spectrom 2009, 20 (6), 1119–1122. 10.1016/j.jasms.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Adams KJ; Ramirez CE; Smith NF; Muñoz-Muñoz AC; Andrade L; Fernandez-Lima F Analysis of Isomeric Opioids in Urine Using LC-TIMS-TOF MS. Talanta 2018, 183, 177–183. 10.1016/j.talanta.2018.02.077. [DOI] [PubMed] [Google Scholar]
- 40.Fu T; Oetjen J; Chapelle M; Verdu A; Szesny M; Chaumot A; Degli-Esposti D; Geffard O; Clément Y; Salvador A; et al. In Situ Isobaric Lipid Mapping by MALDI-Ion Mobility Separation-Mass Spectrometry Imaging. J. Mass Spectrom 2020, e4531. 10.1002/jms.4531. [DOI] [PubMed] [Google Scholar]
- (41).Woods AS; Ugarov M; Egan T; Koomen J; Gillig KJ; Fuhrer K; Gonin M; Schultz JA Lipid/Peptide/Nucleotide Separation with MALDI-Ion Mobility-TOF MS. Anal. Chem 2004, 76 (8), 2187–2195. 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- (42).McLean JA; Ridenour WB; Caprioli RM Profiling and Imaging of Tissues by Imaging Ion Mobility-Mass Spectrometry. J. Mass Spectrom 2007, 42 (8), 1099–1105. 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- (43).Weston DJ; Bateman R; Wilson ID; Wood TR; Creaser C Direct Analysis of Pharmaceutical Drug Formulations Using Ion Mobility Spectrometry/Quadrupole-Time-of-Flight Mass Spectrometry Combined with Desorption Electrospray Ionization. Anal. Chem 2005, 77 (23), 7572–7580. 10.1021/ac051277q. [DOI] [PubMed] [Google Scholar]
- (44).Myung S; Wiseman JM; Valentine SJ; Takáts Z; Cooks RG; Clemmer DE Coupling Desorption Electrospray Ionization with Ion Mobility/Mass Spectrometry for Analysis of Protein Structure: Evidence for Desorption of Folded and Denatured States. J. Phys. Chem. B 2006, 110 (10), 5045–5051. 10.1021/jp052663e. [DOI] [PubMed] [Google Scholar]
- (45).Shrestha B; Vertes A High-Throughput Cell and Tissue Analysis with Enhanced Molecular Coverage by Laser Ablation Electrospray Ionization Mass Spectrometry Using Ion Mobility Separation. Anal. Chem 2014, 86 (9), 4308–4315. 10.1021/ac500007t. [DOI] [PubMed] [Google Scholar]
- (46).Stopka SA; Shrestha B; Maréchal É; Falconet D; Vertes A Metabolic Transformation of Microalgae Due to Light Acclimation and Genetic Modifications Followed by Laser Ablation Electrospray Ionization Mass Spectrometry with Ion Mobility Separation. Analyst 2014, 139 (22), 5945–5953. 10.1039/c4an01368a. [DOI] [PubMed] [Google Scholar]
- (47).Griffiths RL; Dexter A; Creese AJ; Cooper HJ Liquid Extraction Surface Analysis Field Asymmetric Waveform Ion Mobility Spectrometry Mass Spectrometry for the Analysis of Dried Blood Spots. Analyst 2015, 140 (20), 6879–6885. 10.1039/c5an00933b. [DOI] [PubMed] [Google Scholar]
- (48).Sisley EK; Ujma J; Palmer M; Giles K; Fernandez-Lima FA; Cooper HJ LESA Cyclic Ion Mobility Mass Spectrometry of Intact Proteins from Thin Tissue Sections. 2020. 10.1021/acs.analchem.9b05169. [DOI] [PMC free article] [PubMed]
- (49).Feider CL; Elizondo N; Eberlin LS Ambient Ionization and FAIMS Mass Spectrometry for Enhanced Imaging of Multiply Charged Molecular Ions in Biological Tissues. Anal. Chem 2016, 88 (23), 11533–11541. 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ekelöf M; Dodds J; Khodjaniyazova S; Garrard KP; Baker ES; Muddiman DC Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications. J. Am. Soc. Mass Spectrom 2020, 31 (3), 642–650. 10.1021/jasms.9b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hillenkamp F; Karas M; Beavis RC; Chait BT Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Biopolymers. Anal. Chem 1991, 63 (24), 1193A–1203A. 10.1021/ac00024a716. [DOI] [PubMed] [Google Scholar]
- (52).Takáts Z; Wiseman JM; Gologan B; Cooks RG Mass Spectrometry Sampling under Ambient Conditions with Desorption Electrospray Ionization. Science (80-. ). 2004, 306 (5695), 471–473. 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- (53).Van Berkel GJ; Kertesz V; Koeplinger KA; Vavrek M; Kong ANT Liquid Microjunction Surface Sampling Probe Electrospray Mass Spectrometry for Detection of Drugs and Metabolites in Thin Tissue Sections. J. Mass Spectrom 2008, 43 (4), 500–508. 10.1002/jms.1340. [DOI] [PubMed] [Google Scholar]
- (54).Nemes P; Vertes A Laser Ablation Electrospray Ionization for Atmospheric Pressure, in Vivo, and Imaging Mass Spectrometry. Anal. Chem 2007, 79 (21), 8098–8106. 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- (55).Sampson JS; Murray KK; Muddiman DC Intact and Top-Down Characterization of Biomolecules and Direct Analysis Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Coupled to FT-ICR Mass Spectrometry. J. Am. Soc. Mass Spectrom 2009, 20 (4), 667–673. 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bodzon-Kulakowska A; Suder P Imaging Mass Spectrometry: Instrumentation, Applications, and Combination with Other Visualization Techniques. Mass Spectrom. Rev 2016, 35 (1), 147–169. 10.1002/mas.21468. [DOI] [PubMed] [Google Scholar]
- (57).Spraker JE; Luu GT; Sanchez LM Imaging Mass Spectrometry for Natural Products Discovery: A Review of Ionization Methods. Natural Product Reports. Royal Society of Chemistry; February 1, 2020, pp 150–162. 10.1039/c9np00038k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Perez CJ; Bagga AK; Prova SS; Yousefi Taemeh M; Ifa DR Review and Perspectives on the Applications of Mass Spectrometry Imaging under Ambient Conditions. Rapid Commun. Mass Spectrom 2019, 33 (S3), 27–53. 10.1002/rcm.8145. [DOI] [PubMed] [Google Scholar]
- (59).Snel MF Ion Mobility Separation Mass Spectrometry Imaging. In Comprehensive Analytical Chemistry; Elsevier B.V., 2019; Vol. 83, pp 237–257. 10.1016/bs.coac.2018.09.001. [DOI] [Google Scholar]
- (60).Towers MW; Karancsi T; Jones EA; Pringle SD; Claude E Optimised Desorption Electrospray Ionisation Mass Spectrometry Imaging (DESI-MSI) for the Analysis of Proteins/Peptides Directly from Tissue Sections on a Travelling Wave Ion Mobility Q-ToF. J. Am. Soc. Mass Spectrom 2018, 29 (12), 2456–2466. 10.1007/s13361-018-2049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hale OJ; Sisley EK; Griffiths RL; Styles IB; Cooper HJ Native LESA TWIMS-MSI: Spatial, Conformational, and Mass Analysis of Proteins and Protein Complexes. J. Am. Soc. Mass Spectrom 2020, 31 (4), 873–879. 10.1021/jasms.9b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Sans M; Feider CL; Eberlin LS Advances in Mass Spectrometry Imaging Coupled to Ion Mobility Spectrometry for Enhanced Imaging of Biological Tissues. Current Opinion in Chemical Biology. Elsevier Ltd; February 1, 2018, pp 138–146. 10.1016/j.cbpa.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Giles K; Pringle SD; Worthington KR; Little D; Wildgoose JL; Bateman RH Applications of a Travelling Wave-Based Radio-Frequency-Only Stacked Ring Ion Guide. Rapid Commun. Mass Spectrom 2004, 18 (20), 2401–2414. 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- (64).Pringle SD; Giles K; Wildgoose JL; Williams JP; Slade SE; Thalassinos K; Bateman RH; Bowers MT; Scrivens JH An Investigation of the Mobility Separation of Some Peptide and Protein Ions Using a New Hybrid Quadrupole/Travelling Wave IMS/Oa-ToF Instrument. Int. J. Mass Spectrom 2007, 261 (1), 1–12. 10.1016/j.ijms.2006.07.021. [DOI] [Google Scholar]
- (65).May JC; McLean JA Ion Mobility-Mass Spectrometry: Time-Dispersive Instrumentation. Analytical Chemistry. American Chemical Society; February 3, 2015, pp 1422–1436. 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Stauber J; MacAleese L; Franck J; Claude E; Snel M; Kaletas BK; Wiel IMVD; Wisztorski M; Fournier I; Heeren RMA On-Tissue Protein Identification and Imaging by MALDI-Ion Mobility Mass Spectrometry. J. Am. Soc. Mass Spectrom 2010, 21 (3), 338–347. 10.1016/j.jasms.2009.09.016. [DOI] [PubMed] [Google Scholar]
- (67).Ahonen L; Fasciotti M; Gennäs G. B. af; Kotiaho T; Daroda RJ; Eberlin M; Kostiainen R Separation of Steroid Isomers by Ion Mobility Mass Spectrometry. J. Chromatogr. A 2013, 1310, 133–137. 10.1016/j.chroma.2013.08.056. [DOI] [PubMed] [Google Scholar]
- (68).Groessl M; Graf S; Knochenmuss R High Resolution Ion Mobility-Mass Spectrometry for Separation and Identification of Isomeric Lipids. Analyst 2015, 140 (20), 6904–6911. 10.1039/c5an00838g. [DOI] [PubMed] [Google Scholar]
- (69).Rister AL; Martin TL; Dodds ED Formation of Multimeric Steroid Metal Adducts and Implications for Isomer Mixture Separation by Traveling Wave Ion Mobility Spectrometry. J. Mass Spectrom 2019, 54 (5), 429–436. 10.1002/jms.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Hernández-Mesa M; Le Bizec B; Monteau F; García-Campaña AM; Dervilly-Pinel G Collision Cross Section (CCS) Database: An Additional Measure to Characterize Steroids. Anal. Chem 2018, 90 (7), 4616–4625. 10.1021/acs.analchem.7b05117. [DOI] [PubMed] [Google Scholar]
- (71).Ruotolo BT; Giles K; Campuzano I; Sandercock AM; Bateman RH; Robinson CV Biochemistry: Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science (80-. ). 2005, 310 (5754), 1658–1661. 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- (72).Chen SH; Chen L; Russell DH Metal-Induced Conformational Changes of Human Metallothionein-2A: A Combined Theoretical and Experimental Study of Metal-Free and Partially Metalated Intermediates. J. Am. Chem. Soc 2014, 136 (26), 9499–9508. 10.1021/ja5047878. [DOI] [PubMed] [Google Scholar]
- (73).Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson CV Membrane Proteins Bind Lipids Selectively to Modulate Their Structure and Function. Nature 2014, 510 (7503), 172–175. 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Ellis BM; Fischer CN; Martin LB; Bachmann BO; McLean JA Spatiochemically Profiling Microbial Interactions with Membrane Scaffolded Desorption Electrospray Ionization-Ion Mobility-Imaging Mass Spectrometry and Unsupervised Segmentation. Anal. Chem 2019, 91 (21), 13703–13711. 10.1021/acs.analchem.9b02992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Trim PJ; Henson CM; Avery JL; McEwen A; Snel MF; Claude E; Marshall PS; West A; Princivalle AP; Clench MR Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. Anal. Chem 2008, 80 (22), 8628–8634. 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- (76).Xu L; Kliman M; Forsythe JG; Korade Z; Hmelo AB; Porter NA; Mclean JA Profiling and Imaging Ion Mobility-Mass Spectrometry Analysis of Cholesterol and 7-Dehydrocholesterol in Cells Via Sputtered Silver MALDI. 10.1007/s13361-015-1131-0. [DOI] [PMC free article] [PubMed]
- (77).Trimpin S; Herath TN; Inutan ED; Wager-Miller J; Kowalski P; Claude E; Michael Walker J; Mackie K Automated Solvent-Free Matrix Deposition for Tissue Imaging by Mass Spectrometry. Anal. Chem 2010, 82 (1), 359–367. 10.1021/ac902065u. [DOI] [PubMed] [Google Scholar]
- (78).Harvey DJ; Scarff CA; Crispin M; Scanlan CN; Bonomelli C; Scrivens JH MALDI-MS/MS with Traveling Wave Ion Mobility for the Structural Analysis of N-Linked Glycans. J. Am. Soc. Mass Spectrom 2012, 23 (11), 1955–1966. 10.1007/s13361-012-0425-8. [DOI] [PubMed] [Google Scholar]
- (79).Ridenour WB; Kliman M; McLean JA; Caprioli RM Structural Characterization of Phospholipids and Peptides Directly from Tissue Sections by MALDI Traveling-Wave Ion Mobility-Mass Spectrometry. Anal. Chem 2010, 82 (5), 1881–1889. 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Škrášková K; Claude E; Jones EA; Towers M; Ellis SR; Heeren RMA Enhanced Capabilities for Imaging Gangliosides in Murine Brain with Matrix-Assisted Laser Desorption/Ionization and Desorption Electrospray Ionization Mass Spectrometry Coupled to Ion Mobility Separation. Methods 2016, 104, 69–78. 10.1016/j.ymeth.2016.02.014. [DOI] [PubMed] [Google Scholar]
- (81).Langridge JI; Claude E Matrix-Assisted Laser Desorption and Desorption Electrospray Ionization Mass Spectrometry Coupled to Ion Mobility. In Methods in Molecular Biology; Humana Press Inc., 2020; Vol. 2084, pp 245–265. 10.1007/978-1-0716-0030-6_16. [DOI] [PubMed] [Google Scholar]
- (82).Li H; Smith BK; Márk L; Nemes P; Nazarian J; Vertes A Ambient Molecular Imaging by Laser Ablation Electrospray Ionization Mass Spectrometry with Ion Mobility Separation. Int. J. Mass Spectrom 2015, 377 (1), 681–689. 10.1016/j.ijms.2014.06.025. [DOI] [Google Scholar]
- (83).Stopka SA; Vertes A Metabolomic Profiling of Adherent Mammalian Cells In Situ by LAESI-MS with Ion Mobility Separation. In Methods in Molecular Biology; Humana Press Inc., 2020; Vol. 2084, pp 235–244. 10.1007/978-1-0716-0030-6_15. [DOI] [PubMed] [Google Scholar]
- (84).Griffiths RL; Sisley EK; Lopez-Clavijo AF; Simmonds AL; Styles IB; Cooper HJ Native Mass Spectrometry Imaging of Intact Proteins and Protein Complexes in Thin Tissue Sections. Int. J. Mass Spectrom 2019, 437, 23–29. 10.1016/j.ijms.2017.10.009. [DOI] [Google Scholar]
- (85).Illes-Toth E; Cooper HJ Probing the Fundamentals of Native Liquid Extraction Surface Analysis Mass Spectrometry of Proteins: Can Proteins Refold during Extraction? Anal. Chem 2019, 91 (19), 12246–12254. 10.1021/acs.analchem.9b02075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Chandler SA; Benesch JL Mass Spectrometry beyond the Native State. Current Opinion in Chemical Biology. Elsevier Ltd; February 1, 2018, pp 130–137. 10.1016/j.cbpa.2017.11.019. [DOI] [PubMed] [Google Scholar]
- (87).Clemmer DE; Russell DH; Williams ER Characterizing the Conformationome: Toward a Structural Understanding of the Proteome. Accounts of Chemical Research. American Chemical Society; 2017, pp 556–560. 10.1021/acs.accounts.6b00548. [DOI] [PubMed] [Google Scholar]
- (88).Han J; Permentier H; Bischoff R; Groothuis G; Casini A; Horvatovich P Imaging of Protein Distribution in Tissues Using Mass Spectrometry: An Interdisciplinary Challenge. TrAC - Trends in Analytical Chemistry. Elsevier B.V. March 1, 2019, pp 13–28. 10.1016/j.trac.2018.12.016. [DOI] [Google Scholar]
- (89).Bouschen W; Spengler B Artifacts of MALDI Sample Preparation Investigated by High-Resolution Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization (SMALDI) Imaging Mass Spectrometry. Int. J. Mass Spectrom 2007, 266 (1–3), 129–137. 10.1016/j.ijms.2007.07.017. [DOI] [Google Scholar]
- (90).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem 2019, 91 (13), 8564–8573. 10.1021/acs.analchem.9b01838. [DOI] [PubMed] [Google Scholar]
- (91).Tang K; Shvarisburg AA; Lee HN; Prior DC; Buschbach MA; Li F; Tolmachev AV; Anderson GA; Smith RD High-Sensitivity Ion Mobility Spectrometry/Mass Spectrometry Using Electrodynamic Ion Funnel Interfaces. Anal. Chem 2005, 77 (10), 3330–3339. 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Webb IK; Garimella SVB; Tolmachev AV; Chen TC; Zhang X; Norheim RV; Prost SA; LaMarche B; Anderson GA; Ibrahim YM; et al. Experimental Evaluation and Optimization of Structures for Lossless Ion Manipulations for Ion Mobility Spectrometry with Time-of-Flight Mass Spectrometry. Anal. Chem 2014, 86 (18), 9169–9176. 10.1021/ac502055e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Garimella SVB; Ibrahim YM; Webb IK; Tolmachev AV; Zhang X; Prost SA; Anderson GA; Smith RD Simulation of Electric Potentials and Ion Motion in Planar Electrode Structures for Lossless Ion Manipulations (SLIM). J. Am. Soc. Mass Spectrom 2014, 25 (11), 1890–1896. 10.1007/s13361-014-0976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Tolmachev AV; Webb IK; Ibrahim YM; Garimella SVB; Zhang X; Anderson GA; Smith RD Characterization of Ion Dynamics in Structures for Lossless Ion Manipulations. Anal. Chem 2014, 86 (18), 9162–9168. 10.1021/ac502054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Deng L; Webb IK; Garimella SVB; Hamid AM; Zheng X; Norheim RV; Prost SA; Anderson GA; Sandoval JA; Baker ES; et al. Serpentine Ultralong Path with Extended Routing (SUPER) High Resolution Traveling Wave Ion Mobility-MS Using Structures for Lossless Ion Manipulations. Anal. Chem 2017, 89 (8), 4628–4634. 10.1021/acs.analchem.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Nagy G; Veličković D; Chu RK; Carrell AA; Weston DJ; Ibrahim YM; Anderton CR; Smith RD Towards Resolving the Spatial Metabolome with Unambiguous Molecular Annotations in Complex Biological Systems by Coupling Mass Spectrometry Imaging with Structures for Lossless Ion Manipulations. Chem. Commun 2019, 55 (3), 306–309. 10.1039/c8cc07482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Fernandez-Lima F; Kaplan DA; Suetering J; Park MA Gas-Phase Separation Using a Trapped Ion Mobility Spectrometer. Int. J. Ion Mobil. Spectrom 2011, 14 (2), 93–98. 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Fernandez-Lima FA; Kaplan DA; Park MA Note: Integration of Trapped Ion Mobility Spectrometry with Mass Spectrometry. Rev. Sci. Instrum 2011, 82 (12), 126106. 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Michelmann K; Silveira JA; Ridgeway ME; Park MA Fundamentals of Trapped Ion Mobility Spectrometry. J. Am. Soc. Mass Spectrom 2014, 26 (1), 14–24. 10.1007/s13361-014-0999-4. [DOI] [PubMed] [Google Scholar]
- (100).Neumann E; Migas L; Allen JL; Caprioli R; Van de Plas R; Spraggins J Spatial Metabolomics of the Human Kidney Using MALDI Trapped Ion Mobility Imaging Mass Spectrometry. 2020. 10.26434/CHEMRXIV.12118644.V2. [DOI] [PubMed]
- (101).Djambazova K; Klein DR; Migas L; Neumann E; Rivera E; Van de Plas R; Caprioli RM; Spraggins J Resolving the Complexity of Spatial Lipidomics with MALDI Trapped Ion Mobility Spectrometry. 2020. 10.26434/CHEMRXIV.12331652.V1. [DOI] [PubMed]
- (102).Baglai A; Gargano AFG; Jordens J; Mengerink Y; Honing M; van der Wal S; Schoenmakers PJ Comprehensive Lipidomic Analysis of Human Plasma Using Multidimensional Liquid- and Gas-Phase Separations: Two-Dimensional Liquid Chromatography–Mass Spectrometry vs. Liquid Chromatography–Trapped-Ion-Mobility–Mass Spectrometry. J. Chromatogr. A 2017, 1530, 90–103. 10.1016/j.chroma.2017.11.014. [DOI] [PubMed] [Google Scholar]
- (103).Fouque KJD; Ramirez CE; Lewis RL; Koelmel JP; Garrett TJ; Yost RA; Fernandez-Lima F Effective Liquid Chromatography-Trapped Ion Mobility Spectrometry-Mass Spectrometry Separation of Isomeric Lipid Species. Anal. Chem 2019, 91 (8), 5021–5027. 10.1021/acs.analchem.8b04979. [DOI] [PubMed] [Google Scholar]
- (104).Jeanne Dit Fouque K; Moreno J; Hegemann JD; Zirah verine; Rebuffat S; Fernandez-Lima F Identification of Lasso Peptide Topologies Using Native Nanoelectrospray Ionization-Trapped Ion Mobility Spectrometryо– Mass Spectrometry. Anal. Chem 2018, 90, 5139–5146. 10.1021/acs.analchem.7b05230. [DOI] [PubMed] [Google Scholar]
- (105).Jeanne Dit Fouque K; Garabedian A; Porter J; Baird M; Pang X; Williams TD; Li L; Shvartsburg A; Fernandez-Lima F Fast and Effective Ion Mobility-Mass Spectrometry Separation of d -Amino-Acid-Containing Peptides. Anal. Chem 2017, 89 (21), 11787–11794. 10.1021/acs.analchem.7b03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Garabedian A; Baird MA; Porter J; Jeanne Dit Fouque K; Shliaha PV; Jensen ON; Williams TD; Fernandez-Lima F; Shvartsburg AA Linear and Differential Ion Mobility Separations of Middle-Down Proteoforms. Anal. Chem 2018, 90 (4), 2918–2925. 10.1021/acs.analchem.7b05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Vasilopoulou CG; Sulek K; Brunner AD; Meitei NS; Schweiger-Hufnagel U; Meyer SW; Barsch A; Mann M; Meier F Trapped Ion Mobility Spectrometry and PASEF Enable In-Depth Lipidomics from Minimal Sample Amounts. Nat. Commun 2020, 11 (1), 1–11. 10.1038/s41467-019-14044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Kolakowski BM; Mester Z Review of Applications of High-Field Asymmetric Waveform Ion Mobility Spectrometry (FAIMS) and Differential Mobility Spectrometry (DMS). Analyst. Royal Society of Chemistry; August 20, 2007, pp 842–864. 10.1039/b706039d. [DOI] [PubMed] [Google Scholar]
- (109).Guevremont R High-Field Asymmetric Waveform Ion Mobility Spectrometry: A New Tool for Mass Spectrometry. Journal of Chromatography A. Elsevier November 26, 2004, pp 3–19. 10.1016/j.chroma.2004.08.119. [DOI] [PubMed] [Google Scholar]
- (110).Purves RW; Guevremont R Electrospray Ionization High-Field Asymmetric Waveform Ion Mobility Spectrometry-Mass Spectrometry. Anal. Chem 1999, 71 (13), 2346–2357. 10.1021/ac981380y. [DOI] [PubMed] [Google Scholar]
- (111).Garza KY; Feider CL; Klein DR; Rosenberg JA; Brodbelt JS; Eberlin LS Desorption Electrospray Ionization Mass Spectrometry Imaging of Proteins Directly from Biological Tissue Sections. Anal. Chem 2018, 90 (13), 7785–7789. 10.1021/acs.analchem.8b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Griffiths RL; Hughes JW; Abbatiello SE; Belford MW; Styles IB; Cooper HJ Comprehensive LESA Mass Spectrometry Imaging of Intact Proteins by Integration of Cylindrical FAIMS. Anal. Chem 2020, 92 (4), 2885–2890. 10.1021/acs.analchem.9b05124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Malkawi AK; Masood A; Shinwari Z; Jacob M; Benabdelkamel H; Matic G; Almuhanna F; Dasouki M; Alaiya AA; Rahman AMA Proteomic Analysis of Morphologically Changed Tissues after Prolonged Dexamethasone Treatment. Int. J. Mol. Sci 2019, Vol. 20, Page 3122 2019, 20 (13), 3122. 10.3390/IJMS20133122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Bowman AP; Abzalimov RR; Shvartsburg AA Broad Separation of Isomeric Lipids by High-Resolution Differential Ion Mobility Spectrometry with Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom 2017, 28 (8), 1552–1561. 10.1007/s13361-017-1675-2. [DOI] [PubMed] [Google Scholar]
- (115).Shliaha PV; Gorshkov V; Kovalchuk SI; Schwämmle V; Baird MA; Shvartsburg AA; Jensen ON Middle-Down Proteomic Analyses with Ion Mobility Separations of Endogenous Isomeric Proteoforms. Anal. Chem 2020, 92 (3), 2364–2368. 10.1021/acs.analchem.9b05011. [DOI] [PubMed] [Google Scholar]
- (116).McDaniel EW; Martin DW; Barnes WS Drift Tube-Mass Spectrometer for Studies of Low-Energy Ion-Molecule Reactions. Rev. Sci. Instrum 1962, 33 (1), 2–7. 10.1063/1.1717656. [DOI] [Google Scholar]
- (117).Moseley JT; Snuggs RM; Martin DW; McDaniel EW Mobilities, Diffusion Coefficients, and Reaction Rates of Mass-Indentified Nitrogen Ions in Nitrogen. Phys. Rev 1969, 178 (1), 240–248. 10.1103/PhysRev.178.240. [DOI] [Google Scholar]
- (118).Kanu AB; Dwivedi P; Tam M; Matz L; Hill HH Ion Mobility-Mass Spectrometry. Journal of Mass Spectrometry. January 2008, pp 1–22. 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- (119).Ibrahim YM; Baker ES; Danielson WF; Norheim RV; Prior DC; Anderson GA; Belov ME; Smith RD Development of a New Ion Mobility (Quadrupole) Time-of-Flight Mass Spectrometer. Int. J. Mass Spectrom 2015, 377 (1), 655–662. 10.1016/j.ijms.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Baker ES; Livesay EA; Orton DJ; Moore RJ; Danielson WF; Prior DC; Ibrahim YM; LaMarche BL; Mayampurath AM; Schepmoes AA; et al. An LC-IMS-MS Platform Providing Increased Dynamic Range for High-Throughput Proteomic Studies. J. Proteome Res 2010, 9 (2), 997–1006. 10.1021/pr900888b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Morris CB; May JC; Leaptrot KL; McLean JA Evaluating Separation Selectivity and Collision Cross Section Measurement Reproducibility in Helium, Nitrogen, Argon, and Carbon Dioxide Drift Gases for Drift Tube Ion Mobility–Mass Spectrometry. J. Am. Soc. Mass Spectrom 2019, 30 (6), 1059–1068. 10.1021/jasms.8b06014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Hinz C; Liggi S; Griffin JL The Potential of Ion Mobility Mass Spectrometry for High-Throughput and High-Resolution Lipidomics. Current Opinion in Chemical Biology. Elsevier Ltd; February 1, 2018, pp 42–50. 10.1016/j.cbpa.2017.10.018. [DOI] [PubMed] [Google Scholar]
- (123).Lanucara F; Holman SW; Gray CJ; Eyers CE The Power of Ion Mobility-Mass Spectrometry for Structural Characterization and the Study of Conformational Dynamics. Nature Chemistry. Nature Publishing Group; March 21, 2014, pp 281–294. 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- (124).Zheng X; Aly NA; Zhou Y; Dupuis KT; Bilbao A; Paurus VL; Orton DJ; Wilson R; Payne SH; Smith RD; et al. A Structural Examination and Collision Cross Section Database for over 500 Metabolites and Xenobiotics Using Drift Tube Ion Mobility Spectrometry. Chem. Sci 2017, 8 (11), 7724–7736. 10.1039/c7sc03464d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Bush MF; Campuzano IDG; Robinson CV Ion Mobility Mass Spectrometry of Peptide Ions: Effects of Drift Gas and Calibration Strategies. Anal. Chem 2012, 84 (16), 7124–7130. 10.1021/ac3014498. [DOI] [PubMed] [Google Scholar]
- (126).Dilger JM; Valentine SJ; Glover MS; Clemmer DE A Database of Alkaline-Earth-Coordinated Peptide Cross Sections: Insight into General Aspects of Structure. J. Am. Soc. Mass Spectrom 2013. 10.1007/s13361-013-0579-z. [DOI] [PubMed] [Google Scholar]
- (127).Dilger JM; Glover MS; Clemmer DE A Database of Transition-Metal-Coordinated Peptide Cross-Sections: Selective Interaction with Specific Amino Acid Residues. J. Am. Soc. Mass Spectrom 2017, 28 (7), 1293–1303. 10.1007/s13361-016-1592-9. [DOI] [PubMed] [Google Scholar]
- (128).Valentine SJ; Counterman AE; Clemmer DE A Database of 660 Peptide Ion Cross Sections: Use of Intrinsic Size Parameters for Bona Fide Predictions of Cross Sections. J. Am. Soc. Mass Spectrom 1999, 10 (11), 1188–1211. 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- (129).Picache JA; Rose BS; Balinski A; Leaptrot KL; Sherrod SD; May JC; McLean JA Collision Cross Section Compendium to Annotate and Predict Multi-Omic Compound Identities. Chem. Sci 2019, 10 (4), 983–993. 10.1039/c8sc04396e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Jackson SN; Wang HYJ; Woods AS; Ugarov M; Egan T; Schultz JA Direct Tissue Analysis of Phospholipids in Rat Brain Using MALDI-TOFMS and MALDI-Ion Mobility-TOFMS. J. Am. Soc. Mass Spectrom 2005, 16 (2), 133–138. 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- (131).Jackson SN; Ugarov M; Egan T; Post JD; Langlais D; Schultz JA; Woods AS MALDI-Ion Mohility-TOFMS Imaging of Lipids in Rat Brain Tissue. J. Mass Spectrom 2007, 42 (8), 1093–1098. 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Jackson SN; Barbacci D; Egan T; Lewis EK; Albert Schultz J; Woods AS MALDI-Ion Mobility Mass Spectrometry of Lipids in Negative Ion Mode. Anal. Methods 2014, 6 (14), 5001–5007. 10.1039/c4ay00320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Roscioli KM; Tufariello JA; Zhang X; Li SX; Goetz GH; Cheng G; Siems WF; Hill HH Desorption Electrospray Ionization (DESI) with Atmospheric Pressure Ion Mobility Spectrometry for Drug Detection. Analyst 2014, 139 (7), 1740–1750. 10.1039/c3an02113k. [DOI] [PubMed] [Google Scholar]
- (134).Stopka SA; Agtuca BJ; Koppenaal DW; Paša-Tolić L; Stacey G; Vertes A; Anderton CR Laser-Ablation Electrospray Ionization Mass Spectrometry with Ion Mobility Separation Reveals Metabolites in the Symbiotic Interactions of Soybean Roots and Rhizobia. Plant J. 2017, 91 (2), 340–354. 10.1111/tpj.13569. [DOI] [PubMed] [Google Scholar]
- (135).Ujma J; Giles K; Morris M; Barran PE New High Resolution Ion Mobility Mass Spectrometer Capable of Measurements of Collision Cross Sections from 150 to 520 K. Anal. Chem 2016, 88 (19), 9469–9478. 10.1021/acs.analchem.6b01812. [DOI] [PubMed] [Google Scholar]
- (136).Koeniger SL; Merenbloom SI; Clemmer DE Evidence for Many Resolvable Structures within Conformation Types of Electrosprayed Ubiquitin Ions. J. Phys. Chem. B 2006, 110 (13), 7017–7021. 10.1021/jp056165h. [DOI] [PubMed] [Google Scholar]
- (137).May JC; Russell DH A Mass-Selective Variable-Temperature Drift Tube Ion Mobility-Mass Spectrometer for Temperature Dependent Ion Mobility Studies. J. Am. Soc. Mass Spectrom 2011, 22, 1134–1145. 10.1007/s13361-011-0148-2. [DOI] [PubMed] [Google Scholar]
- (138).Merenbloom SI; Glaskin RS; Henson ZB; Clemmer DE High-Resolution Ion Cyclotron Mobility Spectrometry. Anal. Chem 2009, 81 (4), 1482–1487. 10.1021/ac801880a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Davis AL; Liu W; Siems WF; Clowers BH Correlation Ion Mobility Spectrometry. Analyst 2017, 142 (2), 292–301. 10.1039/c6an02249a. [DOI] [PubMed] [Google Scholar]
- (140).Porta Siegel T; Ekroos K; Ellis SR Reshaping Lipid Biochemistry by Pushing Barriers in Structural Lipidomics. Angew. Chemie Int. Ed 2019, 58 (20), 6492–6501. 10.1002/anie.201812698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Bokhart MT; Rosen E; Thompson C; Sykes C; Kashuba ADM; Muddiman DC Quantitative Mass Spectrometry Imaging of Emtricitabine in Cervical Tissue Model Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization. Anal. Bioanal. Chem 2015, 407 (8), 2073–2084. 10.1007/s00216-014-8220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Robichaud G; Barry JA; Muddiman DC IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. J. Am. Soc. Mass Spectrom 2014, 25, 319–328. 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Ekelöf M; Muddiman DC IR-MALDESI Method Optimization Based on Time-Resolved Measurement of Ion Yields. Anal. Bioanal. Chem 2018, 410 (3), 963–970. 10.1007/s00216-017-0585-2. [DOI] [PubMed] [Google Scholar]
- (144).Bokhart MT; Manni J; Garrard KP; Ekelöf M; Nazari M; Muddiman DC IR-MALDESI Mass Spectrometry Imaging at 50 Micron Spatial Resolution. J. Am. Soc. Mass Spectrom 2017, 28 (10), 2099–2107. 10.1007/s13361-017-1740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).The Human Body at Cellular Resolution: The NIH Human Biomolecular Atlas Program. Nature. Nature Publishing Group; October 10, 2019, pp 187–192. 10.1038/s41586-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Bokhart MT; Nazari M; Garrard KP; Muddiman DC MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J. Am. Soc. Mass Spectrom 2018, 29 (1), 8–16. 10.1007/s13361-017-1809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Robichaud G; Garrard KP; Barry JA; Muddiman DC MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. J. Am. Soc. Mass Spectrom 2013, 24 (5), 718–721. 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Sanchez DM; Creger S; Singla V; Kurulugama RT; Fjeldsted J; Laskin J An Ion Mobility-Mass Spectrometry Imaging Workflow. 2020. 10.26434/CHEMRXIV.12408083.V1. [DOI] [PubMed]


