Abstract
Classification of clinical symptoms and diagnostic microbiology are essential to effectively employ antimicrobial therapy for lower respiratory tract infections (LRTIs) in a timely manner. Empirical antibiotic treatment without microbial identification hinders the selective use of narrow-spectrum antibiotics and effective patient treatment. Thus, the development of rapid and accurate diagnostic procedures that can be readily adopted by the clinic is necessary to minimize non-essential or excessive use of antibiotics and accelerate patient recovery from LRTI-induced damage. We developed and validated a multiplex real-time polymerase chain reaction (mRT-PCR) assay with good analytical performance and high specificity to simultaneously detect four bacterial pathogens causing pneumonia: Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, and Moraxella catarrhalis. The analytical performance of mRT-PCR against target pathogens was evaluated by the limit of detection (LOD), specificity, and repeatability. Two hundred and ten clinical specimens from pneumonia patients were processed using an automatic nucleic acid extraction system for the “respiratory bacteria four” (RB4) mRT-PCR assay, and the results were directly compared to references from bacterial culture and/or Sanger sequencing. The RB4 mRT-PCR assay detected all target pathogens from sputum specimens with a coefficient of variation ranging from 0.29 to 1.71 and conservative LOD of DNA corresponding to 5 × 102 copies/reaction. The concordance of the assay with reference-positive specimens was 100%, and additional bacterial infections were detected from reference-negative specimens. Overall, the RB4 mRT-PCR assay showed a more rapid turnaround time and higher performance that those of reference assays. The RB4 mRT-PCR assay is a high-throughput and reliable tool that assists decision-making assessment and outperforms other standard methods. This tool supports patient management by considerably reducing the inappropriate use of antibiotics.
Introduction
According to the World Health Organization (WHO), lower respiratory tract infections (LRTIs) were the main cause of morbidity and mortality leading to 3 million deaths worldwide in 2016 [1]. LRTIs, caused by various bacteria and viruses, are associated with different clinical symptoms and etiologies affected by age, sex, and season [2]. Owing to complicated clinical symptoms, efforts to reduce the global burden of LRTIs using preventive and treatment strategies require timely identification of pathogens [3].
LRTIs fall into two categories based on their origin: community-acquired pneumonia (CAP) and hospital-acquired pneumonia (HAP) [4]. In the Asia-Pacific region, 11 species of bacteria and eight species of viruses causing CAP are examined by serology, culture, immunofluorescence assay, or polymerase chain reaction (PCR) [5]. Notably, in LRTI patients with severe CAP, Staphylococcus aureus, Klebsiella pneumoniae, and Pseudomonas aeruginosa are the most common species, accounting for 28.6%, 28.6%, and 17.9%, respectively [6], of infections and are identified by diagnostic culture methods. In addition, despite low incidence, Moraxella catarrhalis has been recently recognized as an important emerging pathogen because it shows prevalent resistance to some antibiotics through beta-lactamase production [7].
Rapid identification of pathogens and determination of susceptibility profiles are critical for prompting appropriate therapy [8]. Nevertheless, the current standard diagnosis to detect respiratory bacteria is a culture-based procedure, which typically takes 24–72 h to process. Culture-based diagnosis of LRTI specimens has low sensitivity and negative predictive value (NPV); only approximately 30% of CAP patients receive correct microbiological identification [9]. Therefore, a combination of different tests, such as culture, antigen, and serology detection of pathogens, is commonly requested [10]. Thus, as the demand of rapid and highly precise methods for microbial identification increases, PCR-based assays would be an appropriate system to allow patient specimens to be examined on a large scale easily within quarter of a day [11, 12].
Current molecular diagnostic panels lack coverage of diverse respiratory pathogens. In the last few decades, conventional PCR assays have been performed for rapid identification of a wide range of pathogens; however, it requires the tube to be opened for post-detection analysis, leading to putative cross-contamination of the amplicon and consequent false-positive results [13]. Recently, the TaqMan method (Applied Biosystems, Inc., CA, USA) has been developed in the multiplex real-time PCR (mRT-PCR) platform, enabling quantitative applications [14]. The mRT-PCR assay is highly sensitive, specific, rapid, and less prone to false-positives [13].
Here, we developed a high-throughput mRT-PCR assay for the rapid and reliable identification of key respiratory bacterial pathogens causing pneumonia: S. aureus, M. catarrhalis, K. pneumoniae, and P. aeruginosa. Direct comparison to bacterial culture and/or a sequencing method (reference assays) in LRTI specimens was used to validate the effectiveness of our high-throughput respiratory bacteria four (RB4) mRT-PCR assay.
Materials and methods
Primer/PROBE design
Candidate assay targets for the four key bacteria were selected based on publications. National Center for Biotechnology Information basic local alignment search tool (NCBI-BLAST) for nucleotide sequences was used to compare candidate oligonucleotides to targets in GenBank, including large-scale sequence databases. Assays were redesigned or modified using GeneRunner version 6.0 and NCBI-BLAST to optimize for multiplex performance. The specificity of optimized oligonucleotide sequences was analyzed in silico by the multAlin interface. Sequences were also screened by alignments within sequence databases for species selectivity. Considering these assessments, four targets were selected for the pathogen assay and/or designed for a dual priming oligonucleotide-based multiplex PCR assay for specificity [15]. The sequences of primers and probes are described in Table 1.
Table 1. Oligonucleotide primers and probes for the real time PCR to detect target pathogens and an internal control.
| Pathogen | Target gene | Type | Sequences (5’-3’) | Tm (°C) | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| K. pneumoniae | yphG | F. primer | GAGTTAGGGAAACGAACATTGTGIIIIIGGCAGTGCTC | 58 | 137 | [16] |
| R. primer | TCTCTATCGGACAGACGTCGGIIIIIAAAGAGGTTC | 59 | ||||
| probe | aFAM-TTCATTGGCATCATCACTTAGCGAC | 65.1 | ||||
| P. aeruginosa | regA | F. primer | GCTTCATCGACAGCATCGGIIIIIACTGACGCCA | 58.1 | 111 | [17] |
| R. primer | CGGCTTTTTCTCTGGTCTGTGIIIIIGTTCCGCTGT | 59.1 | ||||
| probe | bHEX-AACACAAACGCACTCGGAAAAATCG | 68.5 | ||||
| S. aureus | nuc | F. primer | GATGGCTATCAGTAATGTTTCGAIIIIICAATACRCAA | 56.3 | 213 | [18] |
| R. primer | GTCGCAGGTTCTTTATGTAATTTTIIIIITGAAGTTGCA | 57.4 | ||||
| probe | cTexas red-CAAGTCTAAGTAGCTCAGCARATGCATCA | 67.1 | ||||
| M. catarrhalis | copB | F. primer | ATTCGTGGCATGGGTCATAAT | 58.5 | 182 | [9] |
| R. primer | GTAACAATCGCACCRTTGGTT | 56.8~59.3 | ||||
| probe | dCy5.5-CACCAAGGTCGCTTTATGCTAGACCC | 68.7 | ||||
| Internal control | HBB | F. primer | GGCATAAAAGTCAGGGCAGAIIIIICTATTGCT | 56.9 | 158 | - |
| R. primer | CCAACTTCATCCACGTTCACCIIIIICCACAGGG | 59.0 | ||||
| probe | eCy5-CCTGAGGAGAAGTCTGCCGTTACTGC | 68.8 |
Probes were labeled with FAM, HEX, Texas red, Cy5.5, and Cy5 and detected at 520, 556, 616, 694, and 669 nm, respectively. All the probes were had BHQ as a quencher at 3’ end.
Abbreviations: Tm, melting temperature; F. primer, forward primer; R. primer, reverse primer; probe, fluorescently labeled primer; R: A or G.
Control isolates
The positive control for the four bacterial strains used in confirmation assays were as follows: S. aureus for American Type Culture Collection (ATCC) 29213, M. catarrhalis for Korean Culture Center of Microorganisms (KCCM) 42706, K. pneumoniae for ATCC 13883, and P. aeruginosa for ATCC 27853. Plasmids containing target genes were generated by cloning amplicons with the pLUG-prime TA-cloning vector system (iNtRON Biotechnology, Inc., Seongnam, Korea). Plasmid DNA was diluted in DNA-spin (iNtRON) to 1 copy/reaction via serial dilution for PCR optimization and quantification standards. S. auricularis and S. haemolyticus were the predominantly isolated respiratory specimens in Seegene Medical Foundation (SGMF) for use as a negative control. Most of these controls were commercially supplied as DNA extracts from ATCC, Korean Collection for Type Cultures (KCTC), KCCM, and Korea Bank for Pathogenic Viruses (KBPV).
Nucleic acid extraction
All patient samples were simultaneously processed using an automated nucleic acid extraction system, the MagNA PURE 96 (Roche, Inc., Basel, Switzerland), according to the manufacturer’s instructions. The specimens were pre-treated with 1 mL of phosphate-buffered saline solution (Biosesang, Co., Seongnam, Korea) and vigorously vortexed for 10 s. Each sample was divided into 200 μL for nucleic acid extraction. Nucleic acid samples were eluted with 100 μL elution buffer and stored at −20°C until use.
Multiplex real-time PCR
PCR procedures were carried out using the CFX96 instrument (Bio-Rad Laboratories, Inc., Irvine, CA, USA). RB4 mRT-PCR assays were performed in a total reaction volume of 20 μL, comprising μL oligonucleotide mixture, 5 μL 4X PCR enzyme, and 5 μL of nucleic acid extract. PCR was conducted with the following parameters: 95°C for 15 min in the first step, followed by 38 cycles of 95°C for 10 s for denaturation and 60°C for 60 s for annealing/extension. Test runs were validated when positive controls for each amplification target were positive and negative controls (no template) were negative. The RB4 mRT-PCR assay is designed to detect K. pneumoniae on FAM, P. aeruginosa on HEX, S. aureus on Texas red, and M. catarrhalis on the Cy5.5 channel. To prevent false-negative results, the human hemoglobin subunit beta (HBB) was simultaneously amplified and was the detected housekeeping gene used as an endogenous internal control.
Analytical performance
The LOD was obtained from the positive control assessments for K. pneumoniae, P. aeruginosa, S. aureus, and M. catarrhalis. Each control was serially diluted into 103, 5 × 102, 102, 5 × 101, 101, and 1 copies/reaction. LOD tests were performed 50 times for these concentrations. Likewise, repeatability tests were determined 20 times for intra-assay coefficients of variation (CV): high for 103 copies/reaction, medium for 5 × 102 copies/reaction, low for 102 copies/reaction, and very low for 5 × 101 copies/reaction. Fifty strains of bacteria and 19 strains of viruses were selected for reactivity tests.
Clinical LRT specimens
A total of 210 anonymized residual sputum specimens from patients presenting symptoms of pneumonia were obtained and preserved for routine procedures between June and August 2018. All specimens were classified into two groups (positive or negative) with reference assays. Samples were confirmed as positive if more than one result from reference assays were positive.
Microbial identification
Well-mixed sputum specimens were cultured on blood and MacConkey agar plates. The plates were incubated for 24 h at 37°C. The colonies from culture plates were deposited on an assay plate, and 1 μL 70% formic acid and 1 μL matrix solution were added. Thereafter, the plate was analyzed using a Bruker Biotyper MALDI-TOF (Bruker, Bremen, Germany).
Sanger sequencing
In the event of a discrepancy between culture assays and PCR assays, Sanger sequencing was performed as an additional confirmatory test in a different institute (Cosmogenetech, Co., Seoul, Korea). The results matched, and over 95% were regarded as references. Sequencing data were analyzed using NCBI-BLAST.
Statistical analysis
All statistical analyses were performed using SPSS version 26.0 (IBM, Co., NY, USA) for Windows. The analytical Cohen’s kappa defined statistical significance only if P-values were ≤0.05. The CVs were determined for mRT-PCR platforms using measurements obtained 20 times from double runs and presented as means and standard deviations. The LOD of mRT-PCR assays, the concentration of the sample detected as positive with 95% confidence, was estimated to fit the probit regression model. For the diagnostic test, sensitivity, specificity, positive predictive values (PPVs), and NPVs were used to compare each RB4 mRT-PCR assay to the reference result and subsequently estimate the diagnostic accuracy of the pathogens.
Ethics statement
Ethical aspects for this study were reviewed and approved by the Seegene Medical Foundation Institutional Review Board (approval number, SMF-IRB-2021-001), provided that after conducting the original test, the remaining anonymous sputum specimens were used. All data were fully anonymized administrative data without patient identifiers, and patient consent was waived by the institutional review board.
Results
Design of the RB4 mRT-PCR assay
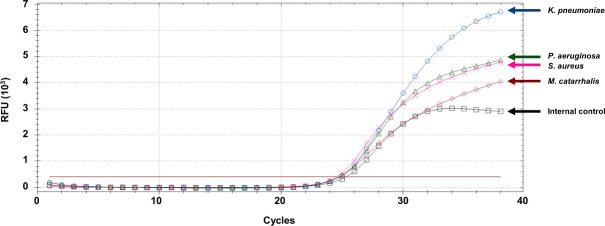
The four pathogens selected for the RB4 mRT-PCR assay, K. pneumoniae, P. aeruginosa, S. aureus, and M. catarrhalis, are species with clinical significance and were detected at 1.3–24.7%, 0–8%, 0.4–10.4%, 0.3–15%, respectively, from pneumonia patients [5]. yphG and regA were selected for the detection of K. pneumoniae and P. aeruginosa, respectively, because they have been evaluated with great sensitivity in the literature [16, 17]. A thermostable nuclease gene, nuc, was selected to detect S. aureus, owing to its greater specificity [18]. A highly conserved region of copB was selected for M. catarrhalis detection [9], and HBB was used as an internal control (Table 1). All target genes were efficiently amplified, without inhibiting one another, in the PCR (Fig 1).
Fig 1. RB4 mRT-PCR assay performed using positive controls corresponding to Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Moraxella catarrhalis, and the internal control, respectively.
The data were analyzed from cycle 1 to 38.
Performance of analytical specificity
To confirm the specificity of the RB4 mRT-PCR assay, other bacterial and viral strains were tested (Table 2). The coverage of the four strains was 100% for each assay target, and no cross-reactivity was observed, indicating great specificity to all targets. The specificity against the 62 negative controls was 100%.
Table 2. Sixty-nine isolates of global origin for analytical specificity of target pathogens.
| Group | organism | Source | FAM | HEX | Cal Red 610 | Quasar 705 |
|---|---|---|---|---|---|---|
| Bacteria | Klebsiella pneumoniae | ATCC 13883 | + | - | - | - |
| Klebsiella pneumoniae | KCCM 42750 | + | - | - | - | |
| Pseudomonas aeruginosa | ATCC 27853 | - | + | - | - | |
| Pseudomonas aeruginosa | KCCM 11266 | - | + | - | - | |
| Staphylococcus aureus | ATCC 29213 | - | - | + | - | |
| Staphylococcus aureus | KCCM 32395 | - | - | + | - | |
| Moraxella catarrhalis | KCCM 42706 | - | - | - | + | |
| Corynebacterium glucuronolyticum | ATCC 51860 | - | - | - | - | |
| Enterobacter aerogenes | ATCC 13048 | - | - | - | - | |
| Enterobacter aerogenes | KCCM 12177 | - | - | - | - | |
| Enterobacter cloacae | ATCC 13047 | - | - | - | - | |
| Enterococcus faecalis | KCTC 5290 | - | - | - | - | |
| Enterococcus faecalis | KCCM 12117 | - | - | - | - | |
| Enterococcus faecium | KCTC 13225 | - | - | - | - | |
| Haemophilus influenzae | ATCC 9007 | - | - | - | - | |
| Klebsiella ornithinolytica | KCCM 41044 | - | - | - | - | |
| Klebsiella oxytoca | ATCC 13182 | - | - | - | - | |
| Klebsiella planticola | KCCM 11649 | - | - | - | - | |
| Klebsiella quasipneumoniae sub | ATCC 700603 | - | - | - | - | |
| Lactobasillus crispatus | ATCC 33820 | - | - | - | - | |
| Lactobasillus gasseri | ATCC 33323 | - | - | - | - | |
| Lactobasillus jensenii | ATCC 25258 | - | - | - | - | |
| Moraxella atlantae | KCCM 11242 | - | - | - | - | |
| Moraxella osloensis | KCTC 52865 | - | - | - | - | |
| Plasmodium falciparum | ATCC 30932 | - | - | - | - | |
| Proteus mirabilis | ATCC 29906 | - | - | - | - | |
| Pseudomonas alcaligenes | KCTC 12029 | - | - | - | - | |
| Pseudomonas basaltis | KCTC 22136 | - | - | - | - | |
| Pseudomonas fluorescens | KCTC 1767 | - | - | - | - | |
| Pseudomonas putida | KCTC 1642 | - | - | - | - | |
| Pseudomonas taetrolens | KCTC 12501 | - | - | - | - | |
| Staphylococcus auricularis | ATCC 33753 | - | - | - | - | |
| Staphylococcus caprae | KCTC 3583 | - | - | - | - | |
| Staphylococcus epidermidis | Clinical isolate | - | - | - | - | |
| Staphylococcus gallinarum | KCTC 3585 | - | - | - | - | |
| Staphylococcus haemolyticus | Clinical isolate | - | - | - | - | |
| Staphylococcus pasteuri | KCTC 13167 | - | - | - | - | |
| Staphylococcus Saprophyticus | KCTC 3345 | - | - | - | - | |
| Streptococcus agalactiae | ATCC 29213 | - | - | - | - | |
| Streptococcus downei | KCTC 5808 | - | - | - | - | |
| Streptococcus gordonii | KCTC 3671 | - | - | - | - | |
| Streptococcus mitis | ATCC 29213 | - | - | - | - | |
| Streptococcus mutans | KCTC 5365 | - | - | - | - | |
| Streptococcus oralis | KCTC 5672 | - | - | - | - | |
| Streptococcus parasanguinis | ATCC15912 | - | - | - | - | |
| Streptococcus pneumoniae | ATCC 49619 | - | - | - | - | |
| Streptococcus pyogenes | KCTC 19615 | - | - | - | - | |
| Streptococcus sanguinis | KCTC 3284 | - | - | - | - | |
| Streptococcus pneumoniae | KCCM 40410 | - | - | - | - | |
| Haemophilus influenzae | KCCM 42099 | - | - | - | - | |
| Virus | BK virus | ATCC VR837 | - | - | - | - |
| Coxasackie virus B3 | ATCC VR30 | - | - | - | - | |
| Dengue virus type 2 | ATCC VR1584 | - | - | - | - | |
| Dengue virus type 4 | ATCC VR1254CAF | - | - | - | - | |
| Herpes virus type 2 | ACTT VR734 | - | - | - | - | |
| Human Adenovirus type 18 | ATCC VR1095 | - | - | - | - | |
| Human Adenovirus type 40 | ATCC VR931 | - | - | - | - | |
| Human Adenovirus type 8 | ATCC VR1368 | - | - | - | - | |
| Human coronavirus OC43 | ATCC VR1558 | - | - | - | - | |
| Human Parainfluenzavirus | ATCC VR1380 | - | - | - | - | |
| Human Respiratory syncytial virus A | ATCC VR41 | - | - | - | - | |
| Influenza A H1N1 | KBPV VR33 | - | - | - | - | |
| Influenza A H1N1 | ATCC VR219 | - | - | - | - | |
| Influenza A H1N1 | ATCC VR897 | - | - | - | - | |
| Influenza A H1N1 | ATCC VR1683 | - | - | - | - | |
| Influenza A H3N2 | ATCC VR547 | - | - | - | - | |
| Influenza A H3N2 | ATCC VR822 | - | - | - | - | |
| Influenza A H3N2 | ATCC VR810 | - | - | - | - | |
| Influenza A H3N2 | ATCC VR544 | - | - | - | - |
RB4 mRT-PCR assays were performed using 69 strains from public centers corresponding to target species. The clinical isolates were identified by MALDI-TOF at the Department of Laboratory medicine in SGMF
Abbreviations: ATCC, American Type Culture Collection; KCTC, Korean Collection for Type Cultures; KCCM, Korean Culture Center of Microorganisms; KBPV, Korea Bank for Pathogenic Viruses; SGMF, Seegene Medical Foundation.
Determination of analytical sensitivity
Analytical sensitivity and LOD were estimated with 50 replicates of positive control bacteria at six different concentrations, from 1 to 103 copies/reaction (Table 3). The levels of 95% LOD were obtained using probit analysis: 143.88 copies/reactions for K. pneumoniae, 212.32 copies/reactions for P. aeruginosa, 166.72 copies/reactions for S. aureus, and 73.11 copies/reactions for M. catarrhalis. The RB4 mRT-PCR assay exhibited 100% reproducibility for all target bacteria as low as 5 × 102 copies/reaction, except for M. catarrhalis, where the assays were approximately five times more sensitive. The LOD was approximately 10–102 copies/reaction, depending on the targets.
Table 3. Evaluation of detection limit of target pathogens.
| Pathogen | DNA Conc. (Copies/rxn) | Reactions | Positive | Positive rate (%) | LOD 95% level (Copies/rxn) |
|---|---|---|---|---|---|
| K. pneumoniae | 1 × 103 | 50 | 50 | 100 | 143.88 |
| 5 × 102 | 50 | 50 | 100 | ||
| 1 × 102 | 50 | 45 | 90 | ||
| 5 × 101 | 50 | 16 | 32 | ||
| 1 × 101 | 50 | 0 | 0 | ||
| 1 | 50 | 0 | 0 | ||
| P. aeruginosa | 1 × 103 | 50 | 50 | 100 | 212.32 |
| 5 × 102 | 50 | 50 | 100 | ||
| 1 × 102 | 50 | 25 | 50 | ||
| 5 × 101 | 50 | 0 | 0 | ||
| 1 × 101 | 50 | 0 | 0 | ||
| 1 | 50 | 0 | 0 | ||
| S. aureus | 1 × 103 | 50 | 50 | 100 | 166.72 |
| 5 × 102 | 50 | 50 | 100 | ||
| 1 × 102 | 50 | 34 | 68 | ||
| 5 × 101 | 50 | 3 | 6 | ||
| 1 × 101 | 50 | 0 | 0 | ||
| 1 | 50 | 0 | 0 | ||
| M. catarrhalis | 1 × 103 | 50 | 50 | 100 | 73.11 |
| 5 × 102 | 50 | 50 | 100 | ||
| 1 × 102 | 50 | 50 | 100 | ||
| 5 × 101 | 50 | 42 | 84 | ||
| 1 × 101 | 50 | 11 | 22 | ||
| 1 | 50 | 0 | 0 |
RB4 mRT-PCR reactions performed using serially diluted positive controls. The LOD 95% data were calculated using the probit model.
Abbreviations: Conc., concentration; rxn, reaction; LOD, limit of detection.
Determination of analytical repeatability
Repeatability was measured by comparing bacterial loads of 100 replicates using the RB4 mRT-PCR assay: 20 positive controls at four different concentrations, high (103 copies/reaction), medium (5 × 102 copies/reaction), low (102 copies/reaction), and very low (5 × 101 copies/reaction), and 20 negative controls (Table 4). The number of positive tests for four target pathogens, out of total 80 replicates, were 80 (100%) at high, 80 (100%) at medium, 61 (76.3%) at low, and 25 (31.3%) at very low concentrations. The best repeatability was obtained from M. catarrhalis detection; 77 replicates were quantified as positive in all tested concentrations, resulting in a 96.3% repeatability. The repeatability values for K. pneumoniae, P. aeruginosa, and S. aureus were 81.3% (65/80), 62.5% (50/80), and 67.5% (54/80), respectively. Overall, the results indicated that the RB4 mRT-PCR assay accurately quantifies the target bacterial load from positive controls at a concentration as low as 5 × 102 copies/reaction.
Table 4. Evaluation of detection repeatability of pathogens using mRT-PCR.
| Pathogen | Conc. | Reactions | Positive | Mean ± SD | CV (%) |
|---|---|---|---|---|---|
| K. pneumoniae | High | 20 | 20 | 32.78 ± 0.22 | 0.67 |
| Medium | 20 | 20 | 33.77 ± 0.32 | 0.95 | |
| Low | 20 | 18 | 36.35 ± 0.69 | 0.66 | |
| Very low | 20 | 7 | 37.48 ± 0.30 | 0.29 | |
| negative | 20 | 0 | - | - | |
| P. aeruginosa | High | 20 | 20 | 34.00 ± 0.19 | 0.56 |
| Medium | 20 | 20 | 35.48 ± 0.48 | 1.36 | |
| Low | 20 | 10 | 37.68 ± 0.21 | 0.57 | |
| Very low | 20 | 0 | - | - | |
| negative | 20 | 0 | - | - | |
| S. aureus | High | 20 | 20 | 34.05 ± 0.20 | 0.59 |
| Medium | 20 | 20 | 35.01 ± 0.21 | 0.60 | |
| Low | 20 | 13 | 37.37 ± 0.33 | 0.87 | |
| Very low | 20 | 1 | 37.56 | - | |
| negative | 20 | 0 | - | - | |
| M. catarrhalis | High | 20 | 20 | 32.04 ± 0.16 | 0.50 |
| Medium | 20 | 20 | 32.99 ± 0.23 | 0.70 | |
| Low | 20 | 20 | 35.22 ± 0.43 | 1.22 | |
| Very low | 20 | 17 | 36.74 ± 0.63 | 1.71 | |
| negative | 20 | 0 | - | - |
RB4 mRT-PCR reactions performed using positive controls in the amount of 103, 5 × 102, 102, and 5 × 101 copies/reaction corresponding to high, medium, low, and very low concentrations, respectively.
Abbreviations: Conc., concentration; SD, standard deviation; CV, coefficient of variation
Performance evaluation in clinical specimens
In the clinical test of 210 specimens, 73 and 137 sputum specimens obtained from pneumonia patients were negative and positive in reference assays, respectively. The sensitivity and NPVs of the RB4 mRT-PCR assay, compared to reference assays, were 100% for all target pathogens, and the specificity was 92.36% to K. pneumoniae, 85.71% to P. aeruginosa, 96.13% to S. aureus, and 98.96% to M. catarrhalis. PPVs were 85.71% for K. pneumoniae, 77.78% for P. aeruginosa, 90.16% for S. aureus, and 90.00% for M. catarrhalis (Table 5). Together, the RB4 mRT-PCR assay, compared to other standard methods, showed a better performance in clinical samples.
Table 5. Comparison of two platforms for the clinical qualification of target pathogens.
| Pathogen | RB4 mRT-PCR | Reference assays | Kappa Value | p | Sen | Spe | PPV | NPV | DA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | |||||||||
| K. pneumoniae | Positive | 66 | 11 | 77 | 0.884 | <0.001 | 100 | 92.36 | 85.71 | 100 | 94.76 |
| Negative | 0 | 133 | 133 | ||||||||
| Total | 66 | 144 | 210 | ||||||||
| P. aeruginosa | Positive | 70 | 20 | 90 | 0.800 | <0.001 | 100 | 85.71 | 77.78 | 100 | 90.48 |
| Negative | 0 | 120 | 120 | ||||||||
| Total | 70 | 140 | 210 | ||||||||
| S. aureus | Positive | 55 | 6 | 61 | 0.929 | <0.001 | 100 | 96.13 | 90.16 | 100 | 97.14 |
| Negative | 0 | 149 | 149 | ||||||||
| Total | 55 | 155 | 210 | ||||||||
| M. catarrhalis | Positive | 18 | 2 | 20 | 0.942 | <0.001 | 100 | 98.96 | 90.00 | 100 | 99.05 |
| Negative | 0 | 190 | 190 | ||||||||
| Total | 18 | 192 | 210 | ||||||||
Analysis of Kappa value, p-value, sensitivity, specificity, positive predictive value, negative predictive value, diagnostic accuracy assessed using the RB4 mRT-PCR assay and reference assay in different target pathogens.
Abbreviations: Sen, sensitivity; Spe, specificity; PPV, positive predictive value; NPV, negative predictive value; DA, diagnostic accuracy.
Discussion
Clinical specimens from pneumonia patients frequently contain coinfections, and mRT-PCR has an advantage to simultaneously diagnose pathogens in a rapid and accurate manner [19, 20]. In this study, we developed and evaluated the RB4 mRT-PCR assay for simultaneous detection of four different bacterial pathogens causing pneumonia, including K. pneumonia, P. aeruginosa, S. aureus, and M. catarrhalis. The performance of the RB4 mRT-PCR assay had analytical specificity for the four pathogens. The consistency of reference assays was >0.80 (kappa value, P < 0.001), and the clinical performance presented 100% reliability for all target pathogens (Table 5).
Sputum specimen retrieval is better accepted by patients for the diagnosis of LRTIs because sputum can be obtained easily and noninvasively [21]. Therefore, the RB4 mRT-PCR assay has been validated for use on sputum specimens rather than nasopharyngeal swabs or aspirates. Moreover, sputum specimens contain housekeeping genes that are stably preserved in human cells and can serve as internal controls [22]. The housekeeping gene HBB was used to ensure proper sampling and adequate target amplification for internal quality [23]. Although microscopy can determine sputum quality based on the number of polymorphonuclear leukocytes (≥10) and squamous epithelial cells (<25) [24], the use of HBB as an internal control correctly determines the quality of the specimen, procedure of nucleic acid extraction, and presence of PCR inhibition [25, 26].
The advantage of the RB4 mRT-PCR assay is its effectiveness on both reference materials and clinical specimens and its correct determination of all positive and negative strains (Table 2). Moreover, 10 (six single and four mixed infections) of the 73 (13.7%) negative specimens and 118 of the 137 (86.1%) positive specimens were matched, whereas RB4 mRT-PCR assays detected an additional 19 bacterial infections from reference-positive samples: 15 (78.9%) single infections and four (21.1%) mixed infections (S1 Table). The RB4 mRT-PCR assay procedure includes automated sample processing and internal specimen quality check, and it can report results within 4 h of specimen receipt on a standard mRT-PCR platform.
There are several noteworthy points in this study. (1) This assay lacks quantitative molecular levels and correlation between Ct values and colony-forming units of bacterial pathogens for LRTI diagnosis; however, it showed stable Ct values with 0.29–1.71% CV (Table 4) [9, 27]. These results indicated that the RB4 mRT-PCR platform is more sensitive than reference assays, even at low copy numbers (Table 3). (2) The assay results were validated with sputum specimens and not with other LRTI specimens, such as endotracheal aspirate and bronchoalveolar lavage [28]. There is a need to improve the efficiency of the mRT-PCR assay and develop test kits compatible with a broad range of LRTI specimens.
The RB4 mRT-PCR assay can potentially serve as an improved decision-making tool during LRTI treatment. Faster and more accurate diagnosis of pathogens would promote the use of narrow- over broad-spectrum antibiotics and substantially reduce the inappropriate use of antibiotics.
Supporting information
The RB4 mRT-PCR assays detected an additional 19 bacterial infections from reference-positive assays.
(PDF)
Acknowledgments
We would like to thank Editage (www.editage.co.kr) for English language editing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Lee SH, Ruan SY, Pan SC, Lee TF, Chien JY, Hsueh PR. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect. 2019;52(6):920–8. Epub 2019/12/07. doi: 10.1016/j.jmii.2019.10.009 ; PubMed Central PMCID: PMC7185395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchatchouang S, Nzouankeu A, Kenmoe S, Ngando L, Penlap V, Fonkoua MC, et al. Bacterial Aetiologies of Lower Respiratory Tract Infections among Adults in Yaounde, Cameroon. Biomed Res Int. 2019;2019:4834396. Epub 2019/05/24. doi: 10.1155/2019/4834396 ; PubMed Central PMCID: PMC6500673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creer DD, Dilworth JP, Gillespie SH, Johnston AR, Johnston SL, Ling C, et al. Aetiological role of viral and bacterial infections in acute adult lower respiratory tract infection (LRTI) in primary care. Thorax. 2006;61(1):75–9. Epub 2005/10/18. doi: 10.1136/thx.2004.027441 ; PubMed Central PMCID: PMC2080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kara S, Akcay MS, Ekici Unsal Z, Bozkurt Yilmaz HE, Habesoglu MA. Comparative analysis of the patients with community-acquired pneumonia (CAP) and health care-associated pneumonia (HCAP) requiring hospitalization. Tuberk Toraks. 2019;67(2):108–15. Epub 2019/08/16. doi: 10.5578/tt.68421 . [DOI] [PubMed] [Google Scholar]
- 5.Song JH, Huh K, Chung DR. Community-Acquired Pneumonia in the Asia-Pacific Region. Semin Respir Crit Care Med. 2016;37(6):839–54. Epub 2016/12/14. doi: 10.1055/s-0036-1592075 ; PubMed Central PMCID: PMC7171710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wongsurakiat P, Chitwarakorn N. Severe community-acquired pneumonia in general medical wards: outcomes and impact of initial antibiotic selection. BMC Pulm Med. 2019;19(1):179. Epub 2019/10/18. doi: 10.1186/s12890-019-0944-1 ; PubMed Central PMCID: PMC6794881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu SF, Lin YT, Chen TL, Siu LK, Hsueh PR, Huang ST, et al. Antimicrobial resistance of Moraxella catarrhalis isolates in Taiwan. J Microbiol Immunol Infect. 2012;45(2):134–40. Epub 2011/12/14. doi: 10.1016/j.jmii.2011.09.004 . [DOI] [PubMed] [Google Scholar]
- 8.Yoo IY, Huh K, Shim HJ, Yun SA, Chung YN, Kang OK, et al. Evaluation of the BioFire FilmArray Pneumonia Panel for rapid detection of respiratory bacterial pathogens and antibiotic resistance genes in sputum and endotracheal aspirate specimens. Int J Infect Dis. 2020;95:326–31. Epub 2020/03/18. doi: 10.1016/j.ijid.2020.03.024 . [DOI] [PubMed] [Google Scholar]
- 9.Gadsby NJ, McHugh MP, Russell CD, Mark H, Conway Morris A, Laurenson IF, et al. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect. 2015;21(8):788 e1–e13. Epub 2015/05/20. doi: 10.1016/j.cmi.2015.05.004 ; PubMed Central PMCID: PMC4509705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50(2):202–9. Epub 2009/12/18. doi: 10.1086/648678 ; PubMed Central PMCID: PMC7107844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh HJ, Kim JY, Kwon HJ, Yun SA, Lee MK, Lee NY, et al. Performance Evaluation of Allplex Respiratory Panels 1, 2, and 3 for Detection of Respiratory Viruses and Influenza A Virus Subtypes. J Clin Microbiol. 2017;55(2):479–84. Epub 2016/12/03. doi: 10.1128/JCM.02045-16 ; PubMed Central PMCID: PMC5277517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YJ, Shin KS, Lee KH, Kim YR, Choi JH. Clinical Characteristics of Macrolide-Resistant Mycoplasma pneumoniae from Children in Jeju. J Korean Med Sci. 2017;32(10):1642–6. Epub 2017/09/07. doi: 10.3346/jkms.2017.32.10.1642 ; PubMed Central PMCID: PMC5592178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HY, Kim S, Kim J, Park SD, Uh Y, Lee H. Multiplex real-time PCR assay for rapid detection of methicillin-resistant staphylococci directly from positive blood cultures. J Clin Microbiol. 2014;52(6):1911–20. Epub 2014/03/22. doi: 10.1128/JCM.00389-14 ; PubMed Central PMCID: PMC4042743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Hwang KA, Ahn JH, Nam JH. Evaluation of EZplex MTBC/NTM Real-Time PCR kit: diagnostic accuracy and efficacy in vaccination. Clin Exp Vaccine Res. 2018;7(2):111–8. Epub 2018/08/17. doi: 10.7774/cevr.2018.7.2.111 ; PubMed Central PMCID: PMC6082673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, et al. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007;35(6):e40. Epub 2007/02/09. doi: 10.1093/nar/gkm051 ; PubMed Central PMCID: PMC1874606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garza-Ramos U, Silva-Sanchez J, Martinez-Romero E, Tinoco P, Pina-Gonzales M, Barrios H, et al. Development of a multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol. 2015;15:64. Epub 2015/04/18. doi: 10.1186/s12866-015-0396-6 ; PubMed Central PMCID: PMC4361152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon KE, Lee DY, Trevors JT, Beaudette LA. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci Total Environ. 2007;382(1):121–9. Epub 2007/04/28. doi: 10.1016/j.scitotenv.2007.02.039 . [DOI] [PubMed] [Google Scholar]
- 18.Montazeri EA, Khosravi AD, Jolodar A, Ghaderpanah M, Azarpira S. Identification of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from burn patients by multiplex PCR. Burns. 2015;41(3):590–4. Epub 2014/12/03. doi: 10.1016/j.burns.2014.08.018 . [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Li X, Wang W, Jia Y, Lin F, Xu J. The prevalence of respiratory pathogens in adults with community-acquired pneumonia in an outpatient cohort. Infect Drug Resist. 2019;12:2335–41. Epub 2019/08/24. doi: 10.2147/IDR.S213296 ; PubMed Central PMCID: PMC6679678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu XM, Xu JX, Jiang LX, Deng LR, Gu ZM, Xie XY, et al. Design and Evaluation of a Novel Multiplex Real-Time PCR Melting Curve Assay for the Simultaneous Detection of Nine Sexually Transmitted Disease Pathogens in Genitourinary Secretions. Front Cell Infect Microbiol. 2019;9:382. Epub 2019/11/30. doi: 10.3389/fcimb.2019.00382 ; PubMed Central PMCID: PMC6861374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyu J, Zhang J, Ren X. Detection and identification of bacterial pathogens directly from sputum samples by pyrosequencing. J Med Microbiol. 2019;68(3):368–73. Epub 2019/01/12. doi: 10.1099/jmm.0.000917 . [DOI] [PubMed] [Google Scholar]
- 22.Etoh K. Evaluation of a real-time PCR assay for the diagnosis of Pneumocystis pneumonia. Kurume Med J. 2008;55(3–4):55–62. Epub 2008/01/01. doi: 10.2739/kurumemedj.55.55 . [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Chen JX, Yang S, Fu XP, Zhang Z, Chen KH, et al. Selection of reliable reference genes for gene expression study in nasopharyngeal carcinoma. Acta Pharmacol Sin. 2010;31(11):1487–94. Epub 2010/11/06. doi: 10.1038/aps.2010.115 ; PubMed Central PMCID: PMC4003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YJ, Shin S, Roh EY, Yoon JH, Kim DK, Chung HS, et al. Acceptability of sputum specimens for diagnosing pulmonary tuberculosis. J Korean Med Sci. 2015;30(6):733–6. Epub 2015/06/02. doi: 10.3346/jkms.2015.30.6.733 ; PubMed Central PMCID: PMC4444473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruhanya V, Jacobs GB, Glashoff RH, Engelbrecht S. Clinical Relevance of Total HIV DNA in Peripheral Blood Mononuclear Cell Compartments as a Biomarker of HIV-Associated Neurocognitive Disorders (HAND). Viruses. 2017;9(11). Epub 2017/11/01. doi: 10.3390/v9110324 ; PubMed Central PMCID: PMC5707531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez LF, Torres IP, Jimenez AM, McEwen JG, de Bedout C, Pelaez CA, et al. Detection of Histoplasma capsulatum in Organic Fertilizers by Hc100 Nested Polymerase Chain Reaction and Its Correlation with the Physicochemical and Microbiological Characteristics of the Samples. Am J Trop Med Hyg. 2018;98(5):1303–12. Epub 2018/03/14. doi: 10.4269/ajtmh.17-0214 ; PubMed Central PMCID: PMC5953352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi H, Saito R, Miya S, Tanaka Y, Miyamura N, Kuda T, et al. Development of quantitative real-time PCR for detection and enumeration of Enterobacteriaceae. Int J Food Microbiol. 2017;246:92–7. Epub 2017/03/09. doi: 10.1016/j.ijfoodmicro.2016.12.015 . [DOI] [PubMed] [Google Scholar]
- 28.Baldassarri RJ, Kumar D, Baldassarri S, Cai G. Diagnosis of Infectious Diseases in the Lower Respiratory Tract: A Cytopathologist’s Perspective. Arch Pathol Lab Med. 2019;143(6):683–94. Epub 2018/09/12. doi: 10.5858/arpa.2017-0573-RA . [DOI] [PubMed] [Google Scholar]