Abstract
BACKGROUND: Patients with advanced cutaneous squamous cell carcinoma (cSCC) frequently have high tumor mutation burdens (TMBs) but cannot tolerate immunotherapy due to comorbid conditions or already immunosuppressed states. OBJECTIVE: We considered whether these patients might be good candidates for targeted therapy if unique genetic mutations are identified. METHODS: Biopsies of primary tumors or metastases of advanced cSCC from seven patients were sent for FoundationOne testing. Genomic alterations and TMBs were compiled from these samples and used to tailor therapy when possible. Patients were followed for changes in their disease burden. RESULTS: Eight biopsies taken from seven patients were sent for FoundationOne testing. Sixty-three genomic alterations were identified. Thirteen genes had mutations occur more than once, with mutations in TP53 being the most frequently identified (100% of patients). In one patient, an ERBB3 mutation was identified, and lapatinib was added to nivolumab for a six-month course of treatment, after which point the patient experienced stabilization of disease without progression for two years as of the most recent follow-up. CONCLUSION: More routine investigation of cSCC tumors with next-generation sequencing can help to identify unique mutations that respond favorably to targeted therapy in these notoriously difficult-to-treat malignancies.
Keywords: Cutaneous squamous cell carcinoma, FoundationOne, next-generation sequencing, targeted therapy
Cutaneous squamous cell carcinoma is a frequently encountered primary cutaneous malignancy, comprising approximately 20 percent of all skin cancers, with an increasing incidence predicted in the coming years.1 Systemic therapy options are limited for patients with advanced cSCC with distant metastases or locally advanced disease. Therapeutic options are limited not only by a lack of matched trial options, but also by the dearth of treatments that have been approved by the United States Food and Drug Administration. Further, advanced cSCC often has a high tumor mutation burden (TMB), which may be responsive to immunotherapy, but this treatment is often contraindicated as many patients are immunosuppressed or have a history of organ transplantation.2 As such, targeted therapy would be ideal for these challenging-to-treat cancers. The following case series adds to existing pooled genomic data of alterations identified with next-generation sequencing (NGS) in seven patients with locally advanced, recurrent or metastatic cSCC.
CASE PRESENTATIONS
Patient 1. A 69-year old woman with a history of cSCC of the left scalp was initially treated with Moh’s surgery with local recurrence and lung metastases two months later. Wide local excision and modified neck dissection were then performed, followed by radiation and cisplatin, which was stopped at Week 5 due to thrombocytopenia and acute kidney injury. Treatment with 5-fluorouracil/carboplatin and cetuximab was started but discontinued after liver metastases were found. Biopsy of a metastatic lesion was then sent for FoundationOne testing (Table 1). No targetable alterations were identified. Patient received 10 cycles of pembrolizumab before she had progressive disease. She completed one cycle of paclitaxel before transitioning to hospice care.
TABLE 1.
Genomic findings (TMB, microsatellite status, and mutations) for each biopsy
| Gene identification | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6, Biopsy 1 | Patient 6, Biopsy 2 | Patient 7 | Aberration Details | Targetable Aberration |
|---|---|---|---|---|---|---|---|---|---|---|
| TMB | High, 56 Muts/Mb | High, 115 Muts/Mb | High, 31 Muts/Mb | Intermediate, 11 Muts/Mb | High, 25 Muts/Mb | Intermediate, 7 Muts/Mb | Intermediate, 12 Muts/Mb | High, 75 Muts/Mb | -- | -- |
| Microsatellite Status | Stable | Stable | Stable | Stable | Stable | Stable | Stable | Not noted | -- | -- |
| TP53 | x | x | x | x | x | x | x | x | Table 2 | |
| BRCA2 | x† | x† | x† | x† | Table 2 | x (Patients 2, 4, and 6) | ||||
| RB1 | x | x | x | x | Table 2 | |||||
| FAT1 | x | x | x | Table 2 | ||||||
| CDKN2A | x | x | x† | Table 2 | x (Patient 7) | |||||
| KMT2C (MLL3) | x | x | x | Table 2 | ||||||
| MLL2 | x | x | x | Table 2 | ||||||
| FLT1 | x | x | Table 2 | |||||||
| NOTCH1 | x | x | Table 2 | |||||||
| LRP1B | x | x | Table 2 | |||||||
| NOTCH3 | x | x | Table 2 | |||||||
| BCORL1 | x | x | Table 2 | |||||||
| TERT-promoter | x | x | Table 2 | |||||||
| ASXL1 | x | W930* | ||||||||
| JUN | x | Amplification | ||||||||
| HRAS | x† | G13D | x | |||||||
| SMARCB1 | x† | Splice site 629-1G>A | x | |||||||
| CIC | x | R422* | ||||||||
| IKZF1 | x | G158S, Q366* | ||||||||
| PREX2 | x | E421K | ||||||||
| SPOP | x | E47K | ||||||||
| EZH2 | x† | Y646N | x | |||||||
| ARID2 | x | Q651* | ||||||||
| NOTCH2 | x | Q206* | ||||||||
| TNFRSF14 | x | G124fs*66 | ||||||||
| FANCA | x | E692* | ||||||||
| VHL | x | L188V | ||||||||
| CCNE1 | x | Equivocal | ||||||||
| NFKB1A | x | Equivocal | ||||||||
| NKX2-1 | x | Equivocal | ||||||||
| AKT1 | x† | Amplification | x | |||||||
| PIK3R2 | x | E296* | ||||||||
| ERBB3 | x† | R135C | x | |||||||
| NF2 | x† | Q400* | x | |||||||
| CARD11 | x | R888C | ||||||||
| RUNX1T1 | x | R394W |
Redundant mutations (seen in more than one biopsy) are shown in green rows
†Mutations identified as candidates for targeted therapy
Patient 2. A 56-year-old man with a history of cSCC of the right scalp with locally advanced disease had completed treatment with cetuximab and radiation, but had progression of disease within four months. Superficial parotidectomy and right modified lymph node dissection were performed for control of metastatic disease into the right parotid gland and sent for FoundationOne testing (Table 1). Three potentially targetable aberrations were identified on sequencing. He completed 20 cycles with pembrolizumab and has been clinically stable for one year.
Patient 3. A 96-year-old woman with a history of locally advanced malignant melanoma with spindle cell differentiation, Breslow depth of 1.8mm, of the nose and central face presented to the clinic. Surgery was not performed on the melanoma following initial biopsy due to the patient’s underlying Parkinson’s disease. One year later, a biopsy was taken on a cSCC on her thigh and sent for FoundationOne testing (Table 1). While awaiting FoundationOne results, the patient received one dose of talimogene laherparepvec for treatment of her melanoma. Radiotherapy was planned for management of her cSCC pending FoundationOne testing (with one targetable aberration identified); however, she quickly declined following her initial dose of talimogene laherparepvec and did not survive long enough to pursue further treatment.
Patient 4. A 65-year-old man with a history of scleroderma and nonsurgically resectable cSCC of the right parietal scalp presented. A second biopsy was taken six months after diagnosis and sent for FoundationOne testing (Table 1) and one targetable mutation was identified. He completed radiation and two cycles of pembrolizumab before he passed away from complications of pulmonary hypertension secondary to his underlying scleroderma.
Patient 5. A 62-year-old man with a history of cSCC of the right temple was treated with radiation followed by recurrent, locally advanced disease without metastases. FoundationOne testing was conducted on a locally recurrent lesion, with no targetable genetic alterations identified (Table 1). Wide local excision, parotidectomy, and neck dissection were performed with negative margins. At his most recent follow-up, cemiplimab was planned; however, the patient has not been seen by any providers in the six months since that visit.
Patient 6. A 63-year-old woman with a history of BRCA2-positive breast cancer (status post-lumpectomy and chemoradiation without hormone therapy) and cSCC of the scalp was initially treated with surgical excision. Fifteen years following her initial diagnosis, local recurrence of the cSCC was noted, and biopsy was performed, followed by wide local excision. She was treated with surgery followed by radiation, cetuximab, carboplatin, and olaparib. She had progression of her disease with local recurrence and metastatic disease to the neck and lymph nodes two years after chemotherapy, and a new biopsy was sent for FoundationOne testing (Table 1). Two targetable genomic alterations were identified, but the patient passed away from complications of osteomyelitis of the skull and subdural empyema before therapy could be further discussed.
Patient 7. An 84-year-old man with a history of unresectable cSCC of the left zygoma was treated with palliative surgical debulking followed by cryosurgery of the base, radiation, and nivolumab with progression of disease found after eight cycles of therapy. Repeat debulking was done, with biopsy sent for FoundationOne testing and three targetable mutations reported (Table 1). Lapatinib was added to nivolumab based on the identification of an ERBB3 mutation via FoundationOne. He completed six months with this regimen before discontinuation and has had stable disease without local recurrence for two years, continuing routine follow-up in the clinic.
RESULTS
Advanced cSCC poses a significant challenge in management, from recognizing what constitutes high-risk disease to identifying treatment options. While there is not one unified definition of high-risk cSCC (HRcSCC), multiple staging systems for the identification of tumors with high-risk features have been published. Common themes among the different systems include a tumor size of 2cm or greater, perineural invasion, and poor histologic differentiation.3 Certain patient characteristics are generally recognized as increasing the risk for poor outcomes, with chronic ultraviolet radiation exposure, advanced age, and immunosuppression (from medication, autoimmune disease, or solid organ transplant) frequently mentioned.1,3,4 All patients in this case series were older than 55 years of age, with varying degrees of historical ultraviolet exposure. One patient had underlying autoimmune disease (Patient 4, scleroderma), and two had other active or prior malignancies (Patient 3, melanoma, and Patient 6, breast cancer).
For patients not definitively managed with initial surgical excision—due to tumor size/invasiveness or patient inability to tolerate surgery, radiation therapy has been a mainstay of management. In those patients with extensive local spread and/or metastases that are not managed by radiotherapy, systemic therapy is the next line of management.2,3 All patients in our case series underwent radiation treatments except for one (Patient 3), who was unable to tolerate either surgical excision or radiation secondary to advanced age (96 years) and generalized frailty precluding aggressive intervention—that same patient was also unable to tolerate any systemic therapy prior to their death. Of the remaining six patients, one (patient 1) received platinum-based chemotherapy prior to NGS sampling, one (Patient 2) received treatment with epidermal growth factor receptor (EGFR) inhibition with cetuximab prior to NGS testing, and one (Patient 6) was on a multiagent regimen with cetuximab, carboplatin, and olaparib (targeting BRCA2 given the history of breast cancer) prior to testing. All three of these patients exhibited disease progression despite their respective treatments.
Of the six patients who were able to tolerate some form of systemic therapy, five were treated with some form of immunotherapy, either EGFR inhibition with cetuximab (Patients 2 and 6) or programmed cell death protein 1 (PD-1) inhibition with pembrolizumab (Patients 1, 2, and 4) or nivolumab (Patient 7). Patients 1, 2, and 7 all had high TMBs on FoundationOne testing, but only Patient 2 had a sustained clinical response to immune checkpoint inhibition with pembrolizumab and did not require further treatment. Patient 1 experienced disease progression despite immunotherapy and, with no genetic targets identified on NGS, transitioned to hospice care. Patient 7 also experienced disease progression on nivolumab, but demonstrated clinical response to treatment with lapatanib, targeted to an ERBB3 mutation reported on NGS. Patient 5 also returned a high TMB but no targetable aberrations on testing and, at the most recent follow-up, had been chosen for initiation of cemiplimab (anti-PD-1), the only systemic therapy for locally advanced/metastatic cSCC in patients who are not candidates for surgical resection or curative radiation approved by the Food and Drug Administration.5
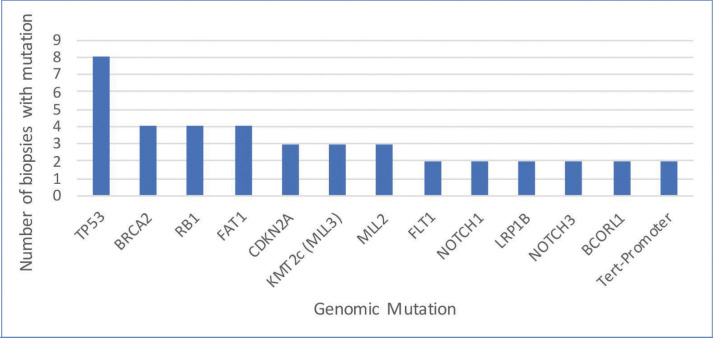
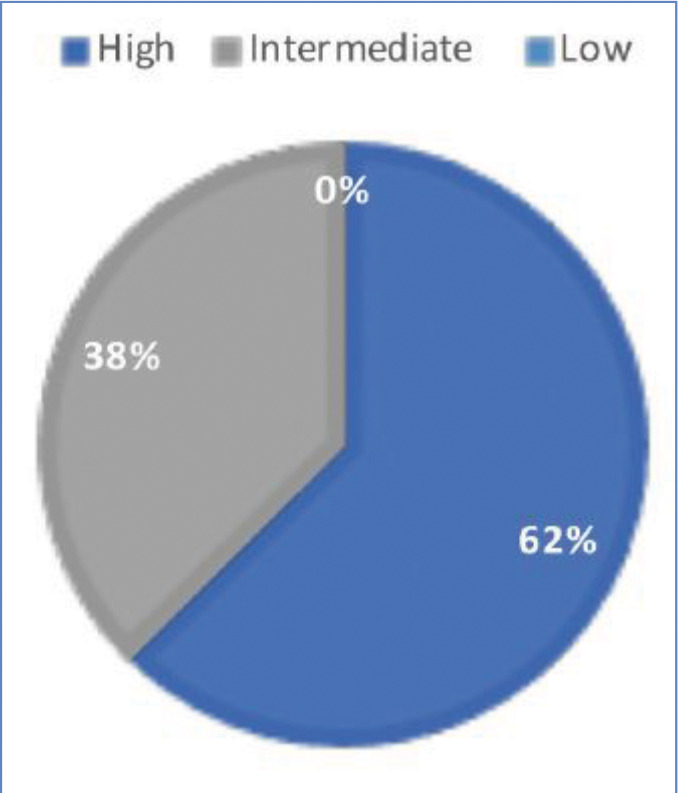
In total, 63 genomic alterations were identified in eight biopsies from seven patients by FoundationOne testing (Table 1). These 63 alterations occurred across a total of 36 different genes. Thirteen genes had mutations occur more than one time (Figure 1). Mutations of TP53 occurred most often and were present in each biopsy sent. Four mutations occurred each in FAT1, BRCA2, and RB1. Three mutations occurred in CDKN2a, KMT2C (MML3), and MML2. Two mutations occurred in BCORL2, FLT1, NOTCH3, LRP1B, NOTCH1, and Tert-promoter. Mutations in the remaining genes reported occurred only once. TMB was reported as high in 62 percent of patients (5/8 biopsies) (Figure 2). Microsatellite status was reported as stable in 7 of 8 biopsies, and was not reported in the remaining biopsy.
FIGURE 1.
Mutation frequency of the most common genomic alterations in advanced cutaneous squamous cell carcinoma (mutations that occurred in more than one biopsy)
FIGURE 2.
Tumor mutation burden in eight biopsies of advanced cSCC, where low TMB is five or less Muts/Mb, intermediate TMB is six to 19 Muts/Mb, and high TMB is 20 or greater Muts/Mb12
Two patients (Patients 1 and 5) returned no targetable genomic findings on FoundationOne testing. Five patients (Patients 2, 3, 4, 6, and 7) all returned between one and three targetable alterations on testing (Tables 1 and 2). Patients 3, 4, and 6 died during or shortly after the period between the identification of genomic aberrations and potential initiation of targeted therapy. Patient 2, who is still living, completed and responded well to 20 cycles of pembrolizumab.
TABLE 2.
Most commonly found gene mutations identified by specific aberrations in each biopsy
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6, Biopsy 1 | Patient 6, Biopsy 2 | Patient 7 | |
|---|---|---|---|---|---|---|---|---|
| TP53 | P278S, R248W | R342*, V143G | H179Y, Q317*, Q331*-subclonal | P250L | C277F, G279fs*65 | M160fs*10 | M160fs*10 | R196*, R282W |
| BRCA2 | W2619*† | P2771fs*6† | 12315fs*12,† N2288fs*41 | 1315fs*12† | ||||
| RB1 | Q217*, splice site 1960 +1G>A | Q217* | P591fs*8 | P591fs*8 | ||||
| FAT1 | Q1483*, R1070* | E2351*, N2147fs*28 | C3738fs*12 | |||||
| CDKN2A | p16INK4a P48L | p16INK4a loss and p14ARF loss exons 2–3 | p16INK4a D84Y,† p14ARF R98L† | |||||
| KMT2C (MLL3) | R3405fs*15 | R4549C | splice site 14344-1G>A | |||||
| MLL2 | D1203* | Q773*, R2915* | E1682fs*40 | |||||
| FLT1 | S287F | M66I-subclonal | - | |||||
| NOTCH1 | H2033fs*6 | W745* | ||||||
| LRP1B | Q3834* | G4199E | ||||||
| NOTCH3 | Amplification | R113* | ||||||
| BCORL1 | R1090* | Q438* | ||||||
| TERT-promoter | 146C>T | 139_-138CC>TT |
*Aberrations identified as candidates for targeted therapy
For one patient (Patient 7) in this series, results of FoundationOne testing were applied with significant clinical benefit. Patient 7, who initially presented with metastatic cSCC with primary tumor of the left zygoma, had undergone four surgical excisions and radiation, and was previously on nivolumab and experienced disease progression. A unique mutation in ERBB3 was identified on testing, lapatinib was added to nivolumab, and the patient had marked improvement; they have been stable without local recurrence off of therapy for two years.
DISCUSSION
FoundationOne testing assays all genes known to be somatically altered in human solid tumors that have been validated as therapeutic targets; currently, this assay includes 315 genes and introns of 28 genes involved in rearrangements, as well as TMB and microsatellite status, where high, medium, and low TMBs are defined as 20 or more Muts/Mb, six to 19 Muts/Mb, and five or less Muts/Mb, respectively.6 In our case series of eight biopsies taken from seven patients (one patient had testing repeated on a separate lesion more than one year after initial biopsy), FoundationOne analysis identified a total of 63 unique genetic aberrations across 36 different genes (Tables 1 and 2). Thirteen genes had mutations in more than one patient, with TP53 being the most frequently mutated (100% of biopsies). Other genes with redundant mutations, in descending order of frequency, were BRCA2 and RB1 (4 biopsies each); FAT1, CDKN2A, KMT2C (MLL3), and MLL2 (3 biopsies each); and FLT1, NOTCH1, LRP1B, NOTCH3, BCORL1, and TERT-promoter (2 biopsies each). Of those genes, targetable alterations were reported in BRCA2 (specific mutations listed in Table 2) and CDKN2A. The CDKN2A mutation was only reported to be targetable in Patient 7 (p16INK4a D84Y and p14ARF R98L), based on preclinical data suggesting that loss-of-function mutations in p16INK4a may be responsive to the addition of CDK4/6.7 Other targetable mutations that occurred once each were found in HRAS, SMARCB1, EZH2, AKT1, ERBB3, and NF2 (Table 2). Zilberg et al8 reported a series of NGS results for 10 high-risk cSCC samples—in their study, frequently observed mutations also included TP53 (10/10 samples), AKT1 (5/10 samples), and RB1 (5/10 samples). Other redundant mutations that were not similarly reported in our series were ATM, APC, ERBB4, GNAQ, and ABL1 (all 6/10) and KIT and PIK3CA (both 5/10). The NGS panel used in that study identified targetable mutations in eight different genes; the only one also identified in our series was that in HRAS.8 Overall, Zilberg et al reported targetable mutations in 60 percent of their cases (comparable to targetable mutations found in 5/7 of our patients; 71%). Notably, all of the samples in the Zilberg et al publication were taken from non-metastatic cases of cSCC, while our series included patients with metastatic disease and, while targetable mutations were identified, no patients were treated as part of the study by Zilberg et al; therefore, patient outcomes are outside the scope of the report.
Many of the commonly identified aberrations in our case series are not limited to cSCC. Other studies have identified mutations in TP53, CDKN2A, RB1, FAT1, and KMT2C (MLL3) as driver genes for the development and progression of malignancy.9–13 TP53 has widely been recognized for its tumor-suppression function; however, despite being widely mutated in a number of cancers, it has remained an elusive target for therapy.13 RB1 has been long-established as a gatekeeper of the cell cycle, inactivation of FAT1 is theorized to have a role in tumorigenesis via upregulation of Wnt signaling, and KMT2C (MLL3) has shown involvement in transcription regulation.12–14 Loss of CDKN2A function has been thought to contribute to immunotherapy resistance in melanoma and other solid organ tumors via the concomitant loss of JAK2 and subsequent resistance to interferon-γ15—all three patients in our case series who demonstrated CDKN2A alterations either had partial response to immunotherapy or progression of disease while on immunotherapy.
NGS may also be useful in identifying patients who are less likely to respond to traditional chemotherapy. Reports of patients with NOTCH3 amplification in high-grade serous ovarian carcinoma have been attributed to carboplatin resistance; while only one patient in our series was found to have NOTCH3 amplification, that patient did not respond well to either carboplatin or cisplatin.16 Where cSCC patients have historically been treated with platinum-based chemotherapies that do not regularly result in sustained remission and have significant side effect profiles, immunotherapy has been identified as a more well-tolerated option with promising results coming from case series and ongoing clinical trials.17 Though the lack of prospective randomized phase III trials comparing chemotherapeutics and immunotherapies limits what final recommendations can be made regarding agent selection, early data from clinical studies include a PD-1 treatment response rate of up to 50 percent in locally recurrent/advanced cSCC, with even greater benefit seen when given in combination with an EGFR inhibitor.2 High TMB, routinely seen in cSCC and reflective of environmental DNA damage sustained from ultraviolet radiation, has been associated with long-term clinical benefit from immunotherapy in patients with melanoma, non-small-cell lung cancer, and urothelial cancer.18 However, caution should be used when considering these treatments in organ transplant recipients, as immune checkpoint inhibitors have been linked to rejection reactions in this patient population.2,19 The risks of transplant rejection with immunotherapy are especially significant when considering cSCC treatment. In organ transplant patients receiving immunosuppressive treatment, the risk of developing cSCC has been reported to be 65 to 250 times the risk in the general population.5 These patients represent a convergence of high-risk factors for the development of aggressive disease and poor tolerance of the most common systemic therapies. NGS offers the potential to identify unique genetic alterations that can be specifically targeted for tailored tumor treatment, circumventing the risks associated with immune checkpoint inhibitors. In nontransplanted patients, NGS also has utility for those who have progressed or who are intolerant of platinum-based chemotherapies and/or immunotherapy. As previously mentioned, some genetic alterations, such as in CDKN2A, have been reported to contribute to immunotherapy resistance—given the frequently high TMB seen in cSCC patients, there may be many more mutations that decrease efficacy of standard treatments, which would otherwise go unrecognized if not for NGS.15 This was demonstrated in our case series in patient 7, who had undergone multiple surgical resections and experienced disease progression while on anti-PD-1 therapy with nivolumab; in this patient, FoundationOne testing revealed a unique ERBB3 mutation that responded favorably to the addition of lapatinib. ERBB3 is a low-activity kinase that encodes a member of the EGFR family and requires the activity of other ERBB family members for full signaling function.20 Targeting ERBB2 has been proposed for those with ERBB3 mutations, and, as seen in our patient, treatment with ERBB2 inhibition via lapatinib resulted in clinical improvement and stable disease for two years as of the most recent follow-up.
CONCLUSION
The genomic profile of sampled cSCC is heterogeneous and highly variable amongst patients. Further, the majority of tumors harbored a high mutational burden, which has also been established as characteristic of this tumor type given the carcinogenic effect of ultraviolet light exposure.9,10 Genomic alterations in this series have predominantly affected well-known cancer-associated genes, including TP53, CDKN2A, RB1, FAT1, and KMT2C (MLL3). Similarly, previous research has identified these as potential driver mutations.9–11 The majority of these tumors are treated with some combination of EGFR inhibitors and anti-PD-1 antibodies. While results with anti-PD-1 antibody therapy are promising, with reported response rates of up to 50 percent in locally advanced and metastatic cSCC and more significant responses in combination with an EGFR inhibitor, a significant portion of these patients still experiences a suboptimal response to these therapies.2 Furthermore, the analysis of NGS results has been reported to reveal mutations that may predict resistance to immunotherapies.15 In many institutions, additional mutational analysis of progressive or recurrent tumors is not routinely performed as there are currently no established practice guidelines. Basket trials using NGS have not altered management practices of patients with advanced cSCC despite identifying molecular alterations.10 However, in our case series, we had one patient who was successfully treated based on the identification of an unusual mutation that responded well to lapatinib. While our case series was too limited in a sample size to draw any larger conclusions regarding the widespread applicability of NGS, the positive results suggest that, in the era of rapid genetic and molecular tumor analysis, personalized cancer care is possible if we are willing to investigate each patient and to act on the data obtained.
REFERENCES
- Di Nardo L, Pellegrini C, Di Stefani A et al. Molecular genetics of cutaneous squamous cell carcinoma: perspective for treatment strategies. J Eur Acad Dermatol Venereol. 2020;34(5):932–941. doi: 10.1111/jdv.16098. [DOI] [PubMed] [Google Scholar]
- Gellrich FF, Huning S, Beissert S et al. Medical treatment of advanced cutaneous squamous cell carcinoma. J Eur Acad Dermatol Venereol. 2019;33(Suppl 8):38–43. doi: 10.1111/jdv.16024. [DOI] [PubMed] [Google Scholar]
- Fu T, Aasi SZ, Hollmig ST. Management of high-risk squamous cell carcinoma of the skin. Curr Treat Options Oncol. 2016;17(7):34. doi: 10.1007/s11864-016-0408-2. [DOI] [PubMed] [Google Scholar]
- Roper E, Lum T, Palme CE et al. PD-L1 expression predicts longer disease free survival in high risk head and neck cutaneous squamous cell carcinoma. Pathology. 2017;49(5):499–505. doi: 10.1016/j.pathol.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Migden MR, Rischin D, Schmults CD et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous cell carcinoma. N Engl J Med. 2018;379(4):341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- Goodman AM, Kato S, Bazhenova L et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Sheng X, Wu X et al. Frequent genetic aberrations in the CDK4 pathway in acral melanoma indicate the potential for CDK4 inhibitors in targeted therapy. Clin Cancer Res. 2017;23(22):6946–6957. doi: 10.1158/1078-0432.CCR-17-0070. [DOI] [PubMed] [Google Scholar]
- Zilberg C, Lee MW, Yu B et al. Analysis of clinically relevant somatic mutations in high-risk head and neck cutaneous squamous cell carcinoma. Mod Pathol. 2018;31(2):275–287. doi: 10.1038/modpathol.2017.128. [DOI] [PubMed] [Google Scholar]
- Pickering CR, Zhou JH, Lee JJ et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20(24):6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LGT, Chandramohan R, West L et al. The molecular landscape of recurrent and metastatic head and neck cancers: insights from a precision oncology sequencing platform. JAMA Oncol. 2017;3(2):244–255. doi: 10.1001/jamaoncol.2016.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz AS, Hatice OG, Gillespie J et al. Differential mutation frequencies in metastatic cutaneous squamous cell carcinomas versus primary tumors. Cancer. 2017;123(7):1184–1193. doi: 10.1002/cncr.30459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Hanna GJ, Laga AC et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21(6):1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering CR, Zhou JH, Lee JJ et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20(24):6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LG, Kaufman AM, Gong Y et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45(3):253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Leonardelli S, Sucker A et al. Tumor CDKN2A-associated JAK2 loss and susceptibility to immunotherapy resistance. J Natl Cancer Inst. 2018;110(6):677–681. doi: 10.1093/jnci/djx271. [DOI] [PubMed] [Google Scholar]
- Park JT, Chen X, Tropè CG et al. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am J Pathol. 2010;177(3):1087–1094. doi: 10.2353/ajpath.2010.100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que ST, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: management of advanced and high-stage tumors. J Am Acad Dermatol. 2018;78(2):249–261. doi: 10.1016/j.jaad.2017.08.058. [DOI] [PubMed] [Google Scholar]
- Fancello L, Gandini S, Giuseppe Pelicci P et al. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer. 2019;7(1):183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittai AS, Oldham H, Cetnar J et al. Immune checkpoint inhibitors in organ transplant patients. J Immunother. 2017;40(7):277–281. doi: 10.1097/CJI.0000000000000180. [DOI] [PubMed] [Google Scholar]
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9(7):463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]