Systematic hospital-based surveillance of rotavirus strains has been conducted in Taiwan to track baseline strain prevalence before and during the introduction of vaccines and to document any strain changes that occur as vaccine use increases (1). Both Rotarix and RotaTeq have been available via Taiwan's private pharmaceutical market since September 2006. As a result of more rigorous surveillance and the exclusive use of gene sequencing for strain genotyping, a variety of unusual strains were detected between 2005 and 2010 (2,3). A strain with rare VP7 and VP4 genotypes, RVA/Human-wt/TWN/04-97s379/2008/G8P[14] (hereafter referred to as 04-97s379), was identified in a 23-month-old boy treated for fever, diarrhea, and vomiting at an outpatient clinic in Changhua, Taiwan. This was the only G8P[14] strain identified among the 1,273 human rotaviruses that were genotyped during this 6-year surveillance period in Taiwan. Because human G8P[14] strains have only been reported in a few countries, including only a single country in the World Health Organization Western Pacific Region, Australia (4), it was of interest to characterize the G8P[14] rotavirus strain detected in Taiwan.
The oligonucleotide primers used to amplify and sequence full-length or partial open reading frames of the VP7 (1,062 bp), VP4 (831 bp), VP6 (1,356 bp), and NSP4 (738 bp) genes (GenBank accession numbers, JX1543843, JX1542665, JX1543853, and JX1542003, respectively) have been described elsewhere (3). For phylogenetic analysis, nucleotide sequences of related strains were retrieved from GenBank and compared with the Taiwanese strain 04-97s379 using the MEGA4 software (5).
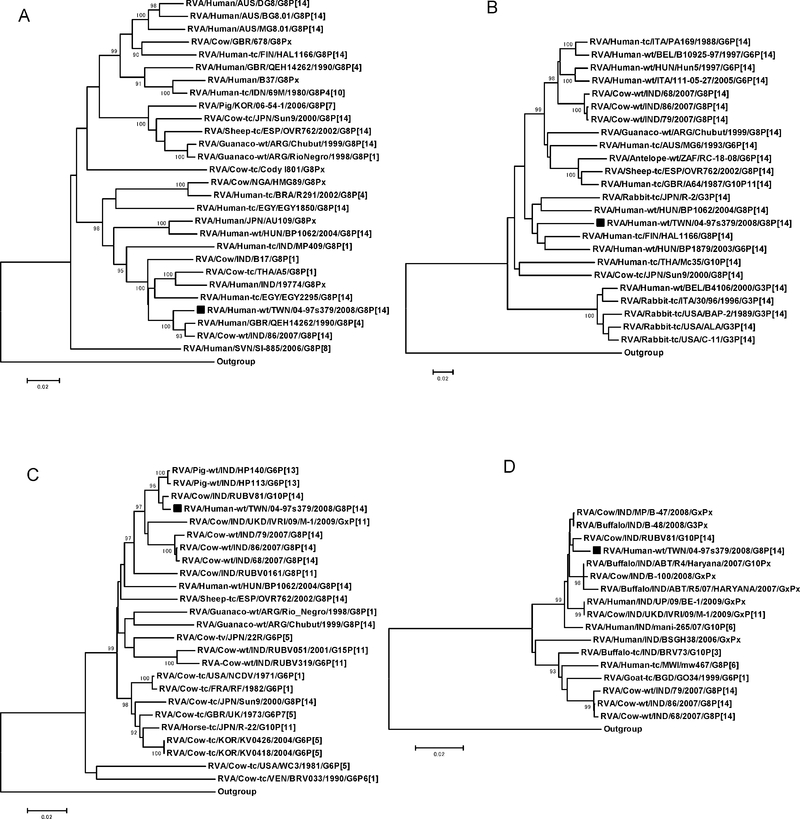
Our analysis showed that the G8 VP7 gene of 04-97s379 is most closely related to that of certain Indian bovine G8 strains (strains 68 and 86; ≥97.8% nucleotides [nt]) (Fig. 1A). The VP4 gene of 04-97s379 was found to share < 90% nt identity with the VP4 gene sequesnce in most P[14] strains, although the VP4 of the suspected zoonotic Finnish G8P[14] strain, HAL1166, had slightly higher nt similarity (90.3%) (Fig. 1B). The VP6 and NSP4 genes were also sequenced to confirm the presumed animal origin of the 04-97s379 strain. The I2 VP6 genotype was found to be most similar to that of an Indian bovine strain (RUBV81, 99.4%) (Fig. 1C), and E2 NSP4 genotype was also most closely related to that of RUBV81, along with a few additional Indian bovine strains (e.g., B-48; up to 98.9% nt identity) (Fig. 1D). Overall, three (VP7, VP6, and NSP4) of the four genes investigated in this study were found to be more closely related to homologues in select Asian bovine rotavirus strains, including some other G8P[14] rotaviruses (e.g., strain 86), than to homologues in human G8P[14] rotavirus strains that have been detected outside Taiwan. This finding suggests that the genome of 04-97s379 is at least partially of bovine origin.
Fig. 1.
Phylogenetic trees of rotavirus VP7 (A), VP4 (B), VP6 (C), and NSP4 (D) genes. The black square indicates the RVA/Human-wt/TWN/04-97s379/2008/G8P[14] strain identified in Taiwan. Bootstrap values > 90 are indicated. The human strain WA was used as the outgroup.
In summary, to our knowledge, this is the first report of a human G8P[14] strain in Taiwan. The genetic analysis of 04-97s379 showed that it is divergent from other reported human isolates that carry the same G and P antigen types (e.g., isolates from Finland, Italy, Australia, Hungary, or Egypt) (6–9) and also that it differs from animal G8P[14] strains identified in Japan, India (bovine), Spain (ovine), or Argentina (guanaco) (7,10–12). Although the Taiwanese human G8P[14] strain was found to be somewhat similar to the Indian bovine G8P[14] strains when comparing the respective VP7 and VP6 genes, the VP4 and NSP4 genes were clearly divergent (11). This suggests an independent origin for 04-97s379. The source of the infection remained elusive because the patient had had no known contact with a diarrheic family member or an animal, which meant that the possible zoonotic origin could not be confirmed. The acquisition of this and other G8P[14] strains by children via environmental exposure or from symptomless carriers cannot be ruled out. To improve our understanding of the ecological and epidemiological features of sporadic putative zoonotic infections in infants and young children, the simultaneous surveillance of animal strains, environmental monitoring, and screening of asymptomatic individuals may be required.
Acknowledgments
This study was financially supported in part by research grant of "National Research Program for Genome Medicine" (supported grant: 94-0324-19-F-01-00-00, 95-0324-19-F01-00-00-00-35, 96-0324-01-F-01) and DOH98-DC-1005 from Centers for Disease Control, Department of Health, Execute Yuan, Taiwan. K.B. was supported by the Hungarian Academy of Sciences (Janos Bolyai scholarship; ’endule’ initiative; OTKA, PD76364, K100727).
Footnotes
Conflict of interest None to declare.
Disclaimer The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the funding agency or the Centers for Disease Control and Prevention (CDC). This article did receive clearance through the appropriate channels at the CDC prior to submission.
REFERENCES
- 1.Wu FT, Liang SY, Tsao KC, et al. (2009): Hospital-based surveillance and molecular epidemiology of rotavirus infection in Taiwan, 2005–2007. Vaccine, 27 (Suppl 5), F50–F54. [DOI] [PubMed] [Google Scholar]
- 2.Wu FT, Báanyai K, Huang JC, et al. (2011): Human infection with novel G3P[25] rotavirus strain in Taiwan. Clin. Microbiol. Infect, 17, 1570–1573. [DOI] [PubMed] [Google Scholar]
- 3.Wu FT, Báanyai K, Huang JC, et al. (2011): Diverse origin of P[19] rotaviruses in children with acute diarrhea in Taiwan: detection of novel lineages of the G3, G5, and G9 VP7 genes. J. Med. Virol, 83, 1279–1287. [DOI] [PubMed] [Google Scholar]
- 4.Swiatek DL, Palombo EA, Lee A, et al. (2010): Characterisation of G8 human rotaviruses in Australian children with gastroenteritis. Virus Res, 148, 1–7. [DOI] [PubMed] [Google Scholar]
- 5.Tamura K, Dudley J, Nei M, et al. (2007): MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol, 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- 6.Báanyai K, Bogdáan ÁA., Domonkos G, et al. (2009): Genetic diversity and zoonotic potential of human rotavirus strains, 2003–2006, Hungary. J. Med. Virol, 81, 362–370. [DOI] [PubMed] [Google Scholar]
- 7.Matthijnssens J, Potgieter CA, Ciarlet M, et al. (2009): Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol, 83, 2917–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medici MC, Abelli LA, Martinelli M, et al. (2008): Molecular characterization of VP4, VP6 and VP7 genes of a rare G8P[14] rotavirus strain detected in an infant with gastroenteritis in Italy. Virus Res, 137, 163–167. [DOI] [PubMed] [Google Scholar]
- 9.Holmes JL, Kirkwood CD, Gerna G, et al. (1999): Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch. Virol, 144, 1381–1396. [DOI] [PubMed] [Google Scholar]
- 10.Fukai K, Saito T, Inoue K, et al. (2004): Molecular characterization of novel P[14],G8 bovine group A rotavirus, Sun9, isolated in Japan. Virus Res, 105, 101–106. [DOI] [PubMed] [Google Scholar]
- 11.Chitambar SD, Arora R, Kolpe AB, et al. (2011): Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet. Microbiol, 148, 384–388. [DOI] [PubMed] [Google Scholar]
- 12.Ciarlet M, Hoffmann C, Lorusso E, et al. (2008): Genomic characterization of a novel group A lamb rotavirus isolated in Zaragoza, Spain. Virus Genes, 37, 250–265. [DOI] [PubMed] [Google Scholar]