Abstract
Background and study aims Large adenomas are sometimes misidentified as cancers during colonoscopy and are surgically removed. To address this overtreatment, we developed an artificial intelligence (AI) tool that identified cancerous pathology in vivo with high specificity. We evaluated our AI tool under the supervision of a government agency to obtain regulatory approval.
Patients and methods The AI tool outputted three pathological class predictions (cancer, adenoma, or non-neoplastic) for endocytoscopic images obtained at 520-fold magnification and previously trained on 68,082 images from six academic centers. A validation test was developed, employing 500 endocytoscopic images taken from various parts of randomly selected 50 large (≥ 20 mm) colorectal lesions (10 images per lesion). An expert board labelled each of the 500 images with a histopathological diagnosis, which was made using endoscopic and histopathological images. The validation test was performed using the AI tool under a controlled environment. The primary outcome measure was the specificity in identifying cancer.
Results The validation test consisted of 30 cancers, 15 adenomas, and five non-neoplastic lesions. The AI tool could analyze 83.6 % of the images (418/500): 231 cancers, 152 adenomas, and 35 non-neoplastic lesions. Among the analyzable images, the AI tool identified the three pathological classes with an overall accuracy of 91.9 % (384/418, 95 % confidence interval [CI]: 88.8 %–94.3 %). Its sensitivity and specificity for differentiating cancer was 91.8 % (212/231, 95 % CI: 87.5 %–95.0 %) and 97.3 % (182/187, 95 % CI: 93.9 %–99.1 %), respectively.
Conclusions The newly developed AI system designed for endocytoscopy showed excellent specificity in identifying colorectal cancer.
Introduction
Large colorectal polyps are sometimes surgically resected without undergoing endoscopic removal because endoscopists consider them unresectable or harboring invasive cancer. However, over 80 % of surgically resected polyps are actually adenomas rather than cancer 1 . This type of excessive overtreatment should be reduced, but pretreatment assessment of these large, complicated colorectal polyps is suboptimal. In a randomized controlled trial, endoscopic identification of invasive cancer was limited to around 70 % sensitivity and 70 % specificity, even with the aid of advanced endoscopic imaging modalities such as magnifying endoscopy or ultrasonography 2 . The low specificity in identifying deeply invasive submucosal cancer was also demonstrated in the recently published meta-analysis; the pooled sensitivity and specificity were 98 % and 65 %, respectively when magnifying narrow-band imaging was used for lesion assessment 3 . The relatively lower specificity for cancer recognition may contribute to the observed increase in unnecessary surgeries of large colorectal adenomas.
To reduce this overtreatment, we developed an artificial intelligence (AI)-driven decision support tool that could identify cancerous pathology during ongoing colonoscopy in real time with high accuracy. This system called EndoBRAIN-Plus (Cybernet System Corp., Tokyo, Japan), was designed to analyze images obtained using endocytoscopy (520-fold contact endomicroscopy, Olympus Corp., Tokyo, Japan) and we initially evaluated its efficacy in a pilot study 4 . However, clinical implementation of such an AI tool requires regulatory approval from a public entity. This report describes a clinical study that was performed to obtain regulatory approval from the Japan Pharmaceuticals and Medical Device Agency (PMDA). Based on the present study, EndoBRAIN-Plus was granted regulatory approval on July 16th 2020 (No. 30200BZX00235000).
Patients and methods
Patients and study design
This study was a benchmark test to evaluate the value of EndoBRAIN-Plus designed for endocytoscopy with regard to its discriminatory capability in identifying colorectal cancer. This was a multicenter clinical study in which we collected training data (i. e., endoscopic images) for constructing the AI model from a total of six academic centers, while the validation data (i. e., endoscopic images) were collected from one university hospital. The study protocol was designed under supervision of the PMDA and the study was conducted in a highly controlled environment.
Preparation of the validation test
First, pooled data for the validation test were produced according to the following inclusion/exclusion criteria to secure the variability of the test data and credibility of the gold standards. Inclusion criteria were as follows: 1. Lesions measuring ≥ 20 mm detected and removed between January 2016 and January 2019 at Showa University Northern Yokohama Hospital; 2. > 20 endocytoscopic images of sufficient quality available for each lesion; 3. Patients aged ≥ 20 years; and 4. Informed consent obtained. Exclusion criteria were as follows: 1. Lesions in which the targeted area for endomicroscopic observation was unclear; 2. No standard endoscopy image obtained; and 3. Sessile serrated lesions (excluded because there was still large inter-rater variance among pathologists regarding histological diagnosis of serrated polyps 5 ). These criteria were applied to the endoscopy database of a consecutive series of 5,605 colorectal lesions from 2,483 patients, which resulted in the extraction of data on 242 lesions.
Second, 50 lesions were randomly extracted from the 242 pooled lesions as validation test material. For this trial, the PMDA requested that test sets represent clinical practice in terms of variety of lesions. Therefore, we performed a stratified data extraction so that the ratio of invasive cancer, adenomas, and non-neoplastic lesions was 6:3:1, which was roughly in line with the prevalence of lesions that were 20 mm or larger in clinical practice (unpublished data from clinical records at Showa University Northern Yokohama Hospital between 2016 and 2018). After applying this data extraction, the final test set of 50 lesions included 30 invasive cancers, 15 adenomas, and five non-neoplastic lesions.
Third, 10 different endoscopic images were selected from each of the 50 lesions. In this extraction, an expert panel, including two board-certified endoscopists, one board-certified pathologist, and three external reviewers who had board certification in gastrointestinal endoscopy, was organized to ensure the quality of the validation data. This committee was monitored independently by the external contract research organization, EPS Corp. (Tokyo, Japan), to ensure its credibility. The two expert endoscopists together with the expert pathologist went through all endoscopic images, videos (if available), and pathological specimens of each subject’s 50 lesions to identify which part of the pathological image corresponded to the obtained endocytoscopic image, namely a 1:1 match between pathological and endoscopic images. In this process, the pathologist divided the pathological specimens into the three classes by marking either invasive cancer, adenoma, or non-neoplastic on the pathology slides, considering the possible heterogeneity in a pathological specimen ( Fig. 1 ). After 10 fully-labelled endoscopic images were prepared, the three external reviewers thoroughly reviewed the extracted images. In this process, the reviewers assessed all 500 images independently and voted “qualified” or “non-qualified” according to their image qualities. The voting was based on majority rule and was strictly monitored by EPS Corp. As a result, no images were identified as “non-qualified”.
Fig. 1.
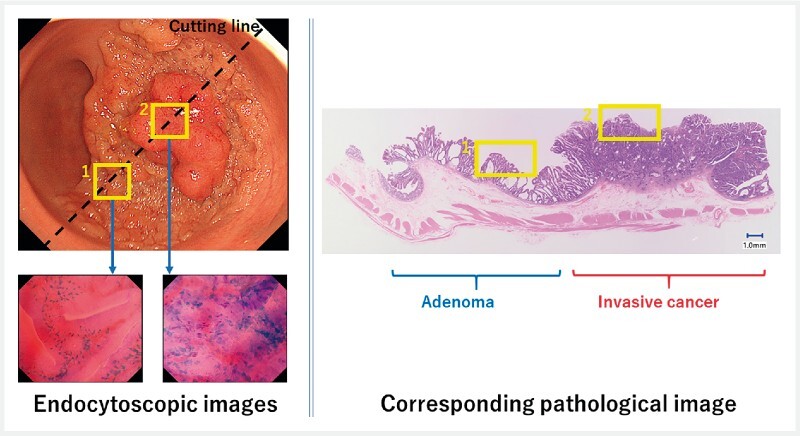
Because some lesions have pathological heterogeneity, all endoscopic images and corresponding pathological images were rigorously reviewed by multiple experts. An endocytoscopic picture taken from area “1” corresponded with an adenomatous component of the pathological specimen, while that taken from area “2” corresponded with a cancerous component.
Finally, the two board-certified endoscopists double-checked the following: 1. Patient consent was secured for the use of selected images; 2. The image satisfied the inclusion/exclusion criteria; 3. 1:1 matching between endoscopic and pathological images was correct; and 4. No overlap was observed in the test images. After this, the personal identifiable information in the test data was anonymized. The test data were stored in the same place as the data that were isolated from the study institutions.
Endocytoscopy and the AI system
Video 1 A large colorectal lesion was predicted to be an invasive cancer using the EndoBRAIN-Plus during an ongoing colonoscopy.
We developed the AI software tool, EndoBRAIN-Plus, for endocytoscopy (CF-290EC, Olympus Corp.) using translational engineering research 4 . Endocytoscopy is performed using a contact light microscopy system (520-fold magnification; focusing depth, 35 μm; field of view, 570 × 500 μm) integrated into the distal tip of a colonoscope, enabling in vivo cellular visualization after staining with 1.0 % methylene blue simply by pulling a hand-operated lever 6 . EndoBRAIN-Plus was designed to pick up a total of 312 variables from one endocytoscopic image that reflects textures characterized according to the contrast difference between each adjacent pixel. In this feature extraction, we did not develop any hand-crafted feature extraction. Instead, the AI system automatically picked up overall texture of each image which may characterized the lesion pathologies. Subsequently, the support vector machine, one of the most common machine learning methods, classified the image as either invasive cancer, adenoma, or non-neoplastic based on the extracted features obtained in real time during the colonoscopy ( Video 1 ) 4 6 . The probability of these predictions was also shown as 0 % to 100 %. Specific pathological predictions would not be displayed when the prediction probability was below 70 %, or the image was not of sufficient quality to be analyzed. The classification performance of the support vector machine was optimized using 68,082 images as training material which were fully labelled with corresponding three-class pathologies. These images were collected from the six academic centers in Japan.
EndoBRAIN-Plus testing
The test was performed using an original computer program that sequentially analyzed test images under the independent supervision of EPS Corp ( Fig. 2 ). EndoBRAIN-Plusʼs diagnostic ability to identify three class subcategories was evaluated with the pathology which was labelled in each image as the gold standard. Images that were non-analyzable owing to their low prediction probability or low quality were excluded from the analysis.
Fig. 2.
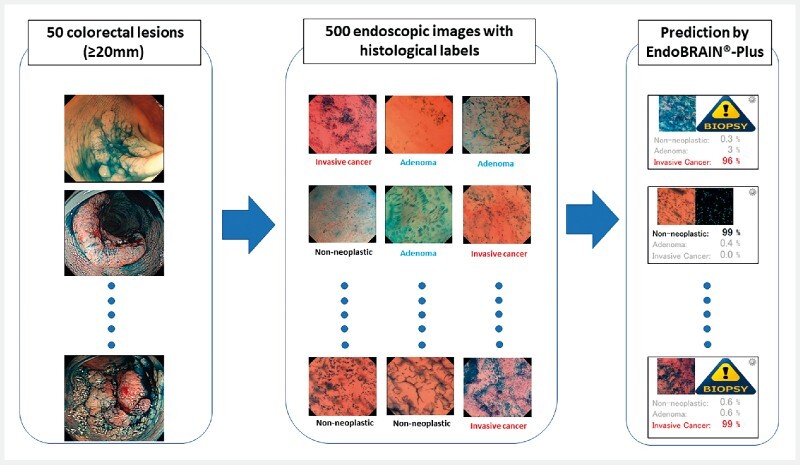
Overview of the validation study. Five hundred endocytoscopic images selected from 50 large colorectal lesions (≥ 20 mm) were analyzed using EndoBRAIN-Plus. The agreement between the diagnostic prediction of EndoBRAIN-Plus and the corresponding histopathological diagnosis of each endocytoscopic image was calculated.
The evaluation was performed on September 19, 2019 under the approval of the Ethics Committee of Showa University Northern Yokohama Hospital (No. 18H085; granted on July 18, 2019). Informed consent was obtained from all involved patients. The study was registered in the UMIN clinical trial registry (UMIN000037977).
Outcome measurements
We primarily investigated if the lower boundary of the 95 % confidence interval (CI) of specificity in identifying invasive cancer exceeded 90 % with EndoBRAIN-Plus.
Secondary outcomes were: 1) sensitivity of EndoBRAIN-Plus to differentiate invasive cancer; 2) sensitivity/specificity of EndoBRAIN-Plus to differentiate between neoplastic and non-neoplastic lesions; and 3) rate at which EndoBRAIN-Plus could analyze images.
As a post-hoc sub-analysis, lesion-based accuracy was also assessed. In this assessment, the majority rule was adopted; the most dominant prediction out of 10 predictions per each lesion was considered the final diagnosis of EndoBRAIN-Plus. Lesions with heterogenous pathologies (e. g., invasive cancer in adenoma) were excluded from this subanalysis.
Sample size calculation and statistical analysis
We hypothesized that the lower limit of the 95 % CI for the specificity of EndoBRAIN-Plus in recognizing invasive cancer was 90 % or greater. A total of 159 non-cancer images were required with an assumption of a specificity of 98.9 % 4 , with an alpha error of 0.5, and a power of 0.9. Because the ratio of cancer to non-cancer images was considered to be 4:6, a total of 400 images would be required. We decided to secure 500 images in consideration of the presence of non-analyzable images. To secure the independence and variety of the 500 images, we decided to take 10 images from each of the 50 different lesions. Because multiple images were taken from one lesion, intraclass correlation might have been considered. To avoid this, we intentionally selected images taken from different parts of the lesion. This could be achieved because the scanning area of endocytoscopy is very small; thus, basically the area from where each image was taken was completely different among the ten images.
Measures of accuracy were expressed as 95 % CIs using an exact binominal method. Parametric continuous variables were expressed as means with standard deviation and nonparametric variables as median with interquartile ranges (IQRs). All statistical analyses were performed using SPSS Statistics, ver.24 (IBM Corp.) by an external statistician (Nishimura Chiaki, CN Medical Data Research, Tokyo, Japan). All authors had access to the study data and reviewed and approved the final manuscript.
Results
Patient and lesion characteristics
The 50 lesions used for the validation comprised 30 invasive cancers, 15 adenomas, and five non-neoplastic lesions, which were collected from 47 patients (25 male and 22 female, median age 65.5 ± 12.2 years). The detailed data are presented in Table 1 . The median size of the lesions was 31.5 mm (IQR, 24 to 40 mm). Regarding location, 26 were located in the right colon, 19 in the left colon, and 10 in the rectum. A total of 28 lesions were superficial type tumors including seven polypoid polyps and 21 slightly elevated lesions, according to the Paris classification 7 . Histopathological evaluation confirmed that there were 8 T1 cancers and 22 T2–4 cancers in the invasive cancer group, 13 high-grade adenomas and two low-grade adenomas in the adenoma group, and three juvenile polyps, one Peutz-Jeghers type polyp, and one inflammatory polyp in the non-neoplastic lesion group.
Table 1. Characteristics of lesions used in the validation test.
| Number | 50 |
| Size (interquartile range), mm | 31.5 (24–40) |
| Location | |
| Right colon | 26 |
| Left colon | 19 |
| Rectum | 5 |
| Morphology | |
| Advanced type | 22 |
| Superficial type | |
| Polypoid (Is, Ip) | 7 |
| Slightly elevate (IIa) | 21 |
| Histopathology | |
| Invasive cancer | |
| T1 | 8 |
| T2–4 | 22 |
| Adenoma | |
| High-grade 1 | 13 |
| Low-grade | 2 |
| Non-neoplastic | |
| Juvenile polyp | 3 |
| Peutz-Jeghers type polyp | 1 |
| Inflammatory polyp | 1 |
An intramucosal cancer was classified as a high-grade adenoma.
Diagnostic performance of EndoBRAIN-Plus
Of the images, 84 % (418/500) were recognized as analyzable by EndoBRAIN-Plus, which included 231 cancer, 152 adenoma, and 35 non-neoplastic images. Table 2 shows the agreement between the prediction using EndoBRAIN-Plus and the corresponding histopathology for these analyzable images. Among the analyzable images, EndoBRAIN-Plus identified the three pathology classes with an overall accuracy of 91.9 % (384/418, 95 % CI: 88.8 %–94.3 %). Its sensitivity and specificity for differentiating cancer was 91.8 % (212/231, 95 % CI: 87.5 %–95.0 %) and 97.3 % (182/187, 95 % CI: 93.9 %–99.1 %), respectively. Regarding the differentiation of neoplastic polyps ( = the sum of invasive cancers and adenomas) from non-neoplastic polyps, EndoBRAIN-Plus showed a sensitivity of 98.7 % (378/383, 95 % CI: 97.0 %–99.6 %) and a specificity of 77.1 % (27/35, 95 % CI: 59.9 %–89.6 %). The results are summarized in Table 3 .
Table 2. Agreement between prediction using EndoBRAIN-Plus and corresponding histopathology of endocytoscopic images.
| Pathology | ||||
| Invasive cancer | Adenoma | Non-neoplastic | ||
| Prediction of EndoBRAIN-Plus | Invasive Cancer | 212 | 3 | 2 |
| Adenoma | 18 | 145 | 6 | |
| Non-neoplastic | 1 | 4 | 27 | |
Table 3. Summary of the validation test using EndoBRAIN-Plus.
| Point estimate (%) | 95 % confidence interval (%) | |||
| Lower boundary | Upper boundary | |||
| Differentiation of invasive cancer | Sensitivity | 91.8 | 87.5 | 95.0 |
| Specificity | 97.3 | 93.9 | 99.1 | |
| Differentiation of neoplastic lesions | Sensitivity | 98.7 | 97.0 | 99.6 |
| Specificity | 77.1 | 59.9 | 89.6 | |
As a subanalysis, lesion-based accuracy based on the majority rule was assessed. In this assessment, 44 lesions with homogenous histology were assessed (24 invasive cancer, 15 adenomas, and 5 non-neoplastic). Table 4 illustrates the results of this lesion-based analysis. All but one prediction were correct based on the majority rule (43/44; accuracy was 97.7 % [95 % CI, 88.0 %–99.9 %]).
Table 4. Lesion-based agreement between the prediction using EndoBRAIN-Plus and the corresponding histopathology of endocytoscopic images.
| Pathology | ||||
| Invasive cancer | Adenoma | Non-neoplastic | ||
| Prediction of EndoBRAIN-Plus | Invasive Cancer | 23 | ||
| Adenoma | 1 | 15 | ||
| Non-neoplastic | 5 | |||
Prediction of EndoBRAIN-Plus was determined based on the majority rule. Lesions with heterogenous pathologies were excluded from the analysis.
With regard to non-analyzable images to EndoBRAIN-Plus, its rate differs according to pathology of the lesions: The rates of non-analyzable images were 11.8 % (95 % CI, 8.2 %–16.4 %: 31/262) in invasive cancers, 19.1 % (13.8 %–25.5 %: 36/188) in adenomas, and 30.0 % (17.9 %–44.6 %: 15/50) in non-neoplastic lesions, respectively.
Images misdiagnosed by EndoBRAIN-Plus
According to Table 2 , an image of invasive cancer which was predicted as non-neoplastic by EndoBRAIN-Plus was an exposed area of desmoplastic reaction in an invasive cancer where aggregation of inflammatory cells (not cancer cells) was observed. This might be why EndoBRAIN-Plus showed “non-neoplastic” instead of outputting “invasive cancer”. Two images of non-neoplastic polyps which were predicted as invasive cancer came from inflammatory polyps. Surface of the inflammatory polyps were covered with lots of inflammatory cells some of which were enlarged due to active inflammation, which mimicked appearance of cancer cells. Therefore, these two images might be misdiagnosed as invasive cancer.
Discussion
This is the first benchmark study to clarify the value of an AI-assisted colonoscopy system in identifying colorectal cancer under the strictly controlled environment proposed by the regulatory body. EndoBRAIN-Plus provided 97.3 % (95 % CI: 93.9 % to 99.1 %) specificity in differentiating invasive cancer, which met the predefined primary endpoint. This high degree of specificity can potentially contribute to a reduction in overtreatment (i. e., surgery) of large adenomas. Moreover, its high sensitivity of 91.8 % (95 %CI: 87.5 %–95.0 %) can also contribute to the strength of the system because it may also reduce the need for “two-step” therapy for invasive cancer (i. e., endoscopic resection followed by subsequent surgery).
Accurate identification of cancer invasion before treatment is required to optimize the management of patients with large colorectal lesions who are sometimes directly sent to surgery because of the lack of a correct diagnosis. Patients with these polyps, then, are subject to risk of complications and mortality caused by surgery which was reported as about 2 % 8 . To reduce overtreatment, various measures have been taken including the use of advanced endoscopic techniques; however, their benefits were limited 2 . In this study, we tried to overcome this unfavorable situation by applying our AI technology to improve optical diagnosis of invasive cancer, especially with a strong focus on obtaining regulatory approval. Apparently, identification of the histologically worst area in a lesion is a prerequisite of using EndoBRAIN-Plus in an appropriate way. We did not assess how endoscopists identify these areas in the study, which will be an important research topic in the future.
Differentiation between T1a and T1b cancers is the ultimate goal of optical diagnosis. However, most of the endoscopists do not seem to reach this level; according to the latest, population-based, prospective trial conducted in Dutch, more than 60 % of T1 cancers were misdiagnosed as adenomas by the endoscopists 9 . Therefore, there is a large room for improvement of identifying T1 cancer before diving into subclassification problem of T1 cancer. Thus, we prioritized a clinically more relevant topic in the present study, namely identification of T1 cancer.
Thus far, there have been several AI software tools on the market with regulatory approval (e. g., GI-Genius, Medtronic Corp. Dublin, Ireland; EndoBRAIN, Cybernet Corp. Tokyo, Japan; CAD-EYE, Fujifilm Corp., Tokyo, Japan; and DISCOVERY, Hoya Corp., Tokyo, Japan). However, no AI tool that can identify cancer was on the market. Therefore, to the best of our knowledge, this was the first official benchmark test that has been monitored by a regulatory body to evaluate the feasibility of using an AI software tool designed to differentiate invasive cancer in real time during a colonoscopy. Increased availability of variable AI tools may accelerate their implementation according to the clinical guideline; in 2019, the European Society of Gastrointestinal Endoscopy published a practice guideline in which implementation of AI technology in colonoscopy practice was first recommended (weak recommendation) 10 .
The regulatory process related to the development of an AI software tool such as EndoBRAIN-Plus required several special steps 11 . Before starting this study, we consulted several times with the Japanese regulatory body, the PMDA. During these consultations, we discussed how to develop the study protocol and how to obtain official permission for the protocol. Finally, the PMDA authorized an ex vivo study because EndoBRAIN-Plus is a second-reader type AI system and not a concurrent type system; second-reader systems are considered to have less risk, and thus, a more robust assessment such as a randomized controlled trial was not considered necessary. Actually, EndoBRAIN-Plus is never activated unless an endoscopist intentionally pushes the endoscope’s image capture button. Therefore, inspection of the target lesion by an endoscopist always precedes output from the EndoBRAIN-Plus. We were asked by the PMDA to conduct our study under very strict control measures to ensure the quality of the ex vivo study, in which all procedures related to the production of the validation set were thoroughly monitored by an external independent entity. The advantage of the present study is that we disclosed all the details of the methodologies that were strictly supervised by Japan’s regulatory body. Dissemination of this information may possibly accelerate the regulatory process of other research groups and industries, and ultimately result in early implementation of AI in endoscopy practice.
This study had several limitations. First, it was a retrospective study whose results were likely to be overestimated. The proportion of non-analyzable, 16 % in the present study, is expected to be higher in clinical practice. This might hamper the widespread application of this AI tool in a clinical setting. Second, the specificity in discrimination of neoplastic change was limited to 77.1 %, which appears to be much lower than previously reported for similar systems 6 . This may be because the large non-neoplastic polyps included in the study were those that were not easily identified as non-neoplastic, even by experts; these were giant juvenile polyps, Peutz–Jeghers polyps, and inflammatory polyps, all of which were superficially similar to neoplastic lesions. No hyperplastic polyps were included in the study, which are usually typical, easy-to-identify non-neoplastic polyps. However, the lesion-based accuracy based on the majority rule was as high as 97.7 % which may mitigate this weakness of the system. Third, EndoBRAIN-Plus could not predict the specific invasive depth of the lesion in the submucosal layer, which is used as a predictor for the risk of metastasis in certain clinical guidelines 12 . Per the study result, endoscopist would still need to be proficient enough to assess if there is possibility of superficial invasion only and resection is technically possible or not, which should be addressed in the following studies. Fourth, an endocytoscope is not widely available in the West though it has been already on the market in Europe. Requirement of expertise and experience in it is another limiting factor. Fifth, we prepared the test data that consisted of endoscopic images from only one institution, which may hinder generalizability of the study results. Sixth, because we excluded sessile serrated lesions from the study, the efficacy of EndoBRAIN-Plus for these lesions were not established.
Conclusions
In conclusion, EndoBRAIN-Plus showed sufficient diagnostic capability in the discrimination of invasive cancer. Its high specificity may contribute to a reduction in unnecessary surgeries for large adenomas. Future prospective trials will help validate the results of the present study.
Acknowledgements
This study was supported by Grants-in-Aid for Scientific Research (No. 19K08403) from the Japan Society for the Promotion of Science and the Research on Development of New Medical Devices and Project on Utilizing High-Definition Medical Imaging Data up to 8K Quality from the Japan Agency for Medical Research and Development, AMED. The funding source did not play a role in the design, conduct, or reporting of the study. The authors thank Olympus and Cybernet Systems for their equipment support in our study. The authors express gratitude to the following endoscopists for collecting colonoscopy data: Dr. Masayoshi Fukuda, Dr. Kenichiro Imai, Dr. Sayo Ito, Dr. Yoshihiro Kishida, Dr. Akiko Chino, Dr. Taishi Ide, Dr. Masahiro Igarashi, Dr. Hiroyuki Takamaru, Dr. Mai Ego, Dr. Masayoshi Yamada, Dr. Taku Sakamoto, Dr. Keiko Nakamura, Dr. Masau Sekiguchi, Dr. Yasuo Kakugawa, and Dr. Kensuke Shimura. They express gratitude to Dr. Nobunao Ikehara, Dr. Yoshiki Wada, and Dr. Hideyuki Miyachi for their contributions in the external committee. We thank Mark Abramovitz, PhD, from Edanz Group ( https://en-author-services.edanzgroup.com/ ) for editing a draft of this manuscript.
Footnotes
Competing interests Dr. Yuichi Mori and Dr. Misawa have received speaking honoraria from Olympus Corporation and ownership interest from Cybernet System Corporation. Dr. Kensaku Mori has received research funding from Cybernet System Corporation.
References
- 1.Bertelson N L, Kalkbrenner K A, Merchea A et al. Colectomy for endoscopically unresectable polyps: how often is it cancer? Dis Colon Rectum. 2012;55:1111–1116. doi: 10.1097/DCR.0b013e3182695115. [DOI] [PubMed] [Google Scholar]
- 2.Shimura T, Ebi M, Yamada T et al. Magnifying chromoendoscopy and endoscopic ultrasonography measure invasion depth of early stage colorectal cancer with equal accuracy on the basis of a prospective trial. Clin Gastroenterol Hepatol. 2014;12:662–668. doi: 10.1016/j.cgh.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 3.Chao G, Ye F, Li T et al. Estimation of invasion depth of early colorectal cancer using EUS and NBI-ME: a meta-analysis. Techniq Coloproctol. 2019;23:821–830. doi: 10.1007/s10151-019-02076-w. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K, Kudo S, Mori Y et al. Accuracy of diagnosing invasie colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798–802. doi: 10.1055/s-0043-105486. [DOI] [PubMed] [Google Scholar]
- 5.Hetzel J T, Huang C S, Coukos J A et al. Variation in the detection of serrated polyps in an average risk colorectal cancer screening cohort. Am J Gastroenterol. 2010;105:2656–2664. doi: 10.1038/ajg.2010.315. [DOI] [PubMed] [Google Scholar]
- 6.Mori Y, Kudo S E, Misawa M et al. Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 7.Anonymous Anonymous. The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3–S43. doi: 10.1016/s0016-5107(03)02159-x. [DOI] [PubMed] [Google Scholar]
- 8.Fox J P, Tyler J A, Vashi A A et al. A variation in the value of colectomy for cancer across hospitals: mortality, readmissions, and costs. Surgery. 2014;156:849–856. doi: 10.1016/j.surg.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Vleugels J LA, Koens L, Dijkgraaf M GW et al. Suboptimal endoscopic cancer recognition in colorectal lesions in a national bowel screening programme. Gut. 2020;69:977–980. doi: 10.1136/gutjnl-2018-316882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisschops R, East J E, Hassan C et al. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline – Update 2019. Endoscopy. 2019;51:1155–1179. doi: 10.1055/a-1031-7657. [DOI] [PubMed] [Google Scholar]
- 11.Kudo S E, Misawa M, Mori Y et al. Artificial intelligence-assisted system improves endoscopic identification of colorectal neoplasms. Clin Gastroenterol Hepatol. 2020;18:1874–1881. doi: 10.1016/j.cgh.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka S, Kashida H, Saito Y et al. JGES guidelines for colorectal endoscopic submucosal dissection/endoscopic mucosal resection. Dig Endosc. 2015;27:417–434. doi: 10.1111/den.12456. [DOI] [PubMed] [Google Scholar]


