Abstract
Cognitive dysfunction is among the hallmark symptoms of Neurofibromatosis 1, and accordingly, loss of the Drosophila melanogaster ortholog of Neurofibromin 1 (dNf1) precipitates associative learning deficits. However, the affected circuitry in the adult CNS remained unclear and the compromised mechanisms debatable. Although the main evolutionarily conserved function attributed to Nf1 is to inactivate Ras, decreased cAMP signaling on its loss has been thought to underlie impaired learning. Using mixed sex populations, we determine that dNf1 loss results in excess GABAergic signaling to the central for associative learning mushroom body (MB) neurons, apparently suppressing learning. dNf1 is necessary and sufficient for learning within these non-MB neurons, as a dAlk and Ras1-dependent, but PKA-independent modulator of GABAergic neurotransmission. Surprisingly, we also uncovered and discuss a postsynaptic Ras1-dependent, but dNf1-independnet signaling within the MBs that apparently responds to presynaptic GABA levels and contributes to the learning deficit of the mutants.
Keywords: cAMP, Drosophila, GABA, learning, neurofibromatosis 1, Ras1
Significance Statement
Two decades after initially reporting the learning deficit upon Neurofibromin loss, we reveal that it is consequent of excess GABAergic signaling to mushroom body (MB) neurons. Significantly, dNf1 activity within non-MB neurons is necessary and sufficient for normal associative learning. We demonstrate that unlike prior suggestions, GABAergic modulation in these neurons does not require cAMP/PKA, but rather regulated Ras signaling. However, a novel Ras1-dependent mechanism in the MBs may affect resident cAMP levels in response to elevated GABAergic stimulation providing a plausible explanation of the unsettled link between cAMP levels and Nf1 function.
Introduction
Neurofibromatosis Type 1 (NF1) is an autosomal dominant, multisymptomatic, and characteristically clinically variable disorder and the most prevalent of the neurocutaneous syndromes (Yap et al., 2014; Nix et al., 2020). NF1 is the result of mutations in Neurofibromin, a regulatory protein of multiple cellular processes, including Ras inactivation via its GTPase-activating domain and apparently activation of adenylyl cyclase by a yet unknown mechanism (Diggs-Andrews and Gutmann, 2013). Neurofibromin is a large ubiquitous cytoplasmic protein, but most abundant in the brain, the spinal cord, and the peripheral nervous system; 50%-80% of children with NF1 manifest impairments in executive functions, attention, language, visual perception, and learning (Shilyansky et al., 2010a; Diggs-Andrews and Gutmann, 2013). However, the molecular and cellular circuitries whose perturbation underlies these cognitive dysfunctions remain poorly understood, but results from animal models have yielded valuable mechanistic insights (Diggs-Andrews and Gutmann, 2013).
Loss of the conserved Drosophila dNf1 ortholog results in organismal size reduction, circadian and associative learning, and memory deficits (Walker et al., 2012) and behavioral inflexibility (King et al., 2016), in accord to human pathology. Intrinsic mushroom body (MB) neurons are central to olfactory learning and memory in Drosophila (Davis, 2005; Hige, 2018). The MBs are bilateral clusters of ∼2500 neurons in the dorsal posterior brain, with their dendrites situated ventrally to their somata and their fasciculated axons projecting anteriorly and bifurcating to form medial (β, β’ and γ) and dorsal (α and α’) lobes (Tanaka et al., 2008). Unexpectedly, transgenic restoration of dNf1 in the adult Drosophila CNS, with explicit exclusion of the MBs, appears sufficient to reverse the associative learning deficits of dNf1 null mutants (Gouzi et al., 2011). Given the cardinal role of MBs for associative learning (Hige, 2018) and prior reports implicating these neurons in the deficient learning of the mutants (Buchanan and Davis, 2010), it was rather surprising that dNf1 appeared required outside the MBs for the process. In accord with this observation, rescue of the learning deficit and reduced size of the mutants depended on attenuation of the receptor tyrosine kinase, anaplastic lymphoma kinase (dAlk) outside the MBs (Gouzi et al., 2011). However, the neurons housing this dAlk-dNf1 functional interaction remained elusive. Therefore, as neurons projecting to the MBs with roles in learning and memory have been described (Tanaka et al., 2008; Chen et al., 2012), we aimed to identify the neuronal circuit where dNf1 is necessary and sufficient for negatively reinforced associative learning in Drosophila.
The growth deficiency of dNf1 homozygotes depends on cAMP/PKA signaling within the neuroendocrine cells of the Drosophila larval ring gland (Walker et al., 2013). However, the implication of this signaling pathway in cognitive deficits in flies and mice (The et al., 1997; Guo et al., 2000; Tong et al., 2002; Brown et al., 2010; Diggs-Andrews and Gutmann, 2013), remained controversial as the learning deficits of Nf1+/− mice appear entirely attributable to excessive Ras activity (Costa and Silva, 2002; Li et al., 2005). We address this discordance by investigating whether cAMP signaling is implicated autonomously or nonautonomously in neurons where we define that dNf1 is required for associative learning.
Materials and Methods
Drosophila culture and strains
Drosophila crosses were set up in standard wheat-flour-sugar food supplemented with soy flour and CaCl2 and cultured at 25°C and 50%-70% humidity with a 12 h light/dark cycle. The dNf1E2 mutant has been described previously (Walker et al., 2006; Gouzi et al., 2011). Transgenic fly strains used in this work were as follows: UAS-dNf1 (Walker et al., 2006), UAS-Nf1RNAi (Bloomington Drosophila Stock Center #25845), UAS-mCD8::GFP (Lee and Luo, 1999), UAS-GadRNAi (Vienna Drosophila RNAi Center #32344), trans-Tango (UAS-myrGFP.QUAS-mtdTomato-3xHA; Bloomington Drosophila Stock Center #77124) (Talay et al., 2017), UAS-PKAC1 (Bloomington Drosophila Stock Center #35554), murine UAS-PKA* (Walker et al., 2013), UAS-AlkRNAiKK (Vienna Drosophila RNAi Center #KK107083), UAS-AlkDN (Bazigou et al., 2007), UAS-Ras1RNAi (Vienna Drosophila RNAi Center #28129), UAS-Ras2RNAi (Vienna Drosophila RNAi Center #6225), UAS-rlsem (Martin-Blanco, 1998), UAS-rlRNAi (Bloomington Drosophila Stock Center #43121), UAS-DsorRNAi(Vienna Drosophila RNAi Center #40026), UAS-Ras1V12 (Karim and Rubin, 1998), TubGal80ts (McGuire et al., 2004), and the MB-specific Gal80 (MB-Gal80), which suppresses expression in the MBs (Krashes et al., 2007). The Gal4-driver lines used in this study were as follows: Leo-Gal4 (Messaritou et al., 2009), OK72-Gal4 (Bloomington Drosophila Stock Center #6486) (Acebes and Ferrús, 2001), c739-Gal4 (Bloomington Drosophila Stock Center #7362), Gad-Gal4 (Bloomington Drosophila Stock Center #51630), Alk(38)-Gal4 (Gouzi et al., 2011), Cha-Gal4 (Bloomington Drosophila Stock Center #6798), NP5288-Gal4 (Kyoto Drosophila Genetic Resource Center #104937), c316-Gal4 (Bloomington Drosophila Stock Center #30830), G0431-Gal4 (Chen et al., 2012), Or83b-Gal4 (Bloomington Drosophila Stock Center #23909), Ras1-Gal4 (Gouzi et al., 2011), Elav[C155]-Gal4, and c772-Gal4 (Bloomington Drosophila Stock Center #6494). All strains were backcrossed into the Cantonised-w1118 isogenic background for six generations.
Behavioral analyses and conditioning
Conditioning and testing were performed under dim red light at 24°C-25°C and 73%-80% humidity. All animals were 2-6 d old, collected under light CO2 anesthesia 1 d before testing, and kept in food vials in groups of 50-70 at 25°C or 18°C as appropriate for strains with Gal80ts-dependent temporal restriction of transgene expression (TARGET). For experiments using the TARGET system, all animals were raised at 18°C and transgene expression was induced maximally by placing 3- to 5-d-old adults at 30°C for 48-72 h. All flies were transferred to fresh vials 1 h before testing and acclimated to the training temperature (25°C) and darkness for 30-60 min before training.
Olfactory learning in the negatively reinforced paradigm coupling aversive odors as conditioned stimuli (CS+ and CS–) with the electric shock unconditioned stimulus (US) was used to assess learning as described previously (Gouzi et al., 2011). For training, 50-70 flies of both sexes were loaded into a training tube, lined with an electrifiable grid, where they received the following sequence of stimuli: 30 s of an odor paired with 6 pulses of 90 V electric shock (CS+), 30 s of air, 30 s of a second odor with no electric shock pulses (CS–), and finally 30 s of air. We used benzaldehyde (Sigma Millipore) and 3-octanol (OCT) (ACROS Organics), as standard odorants. Two groups of animals of the same genotype were trained simultaneously, one to avoid benzaldehyde, the other 3-octanol, while the complementary odorant was used as the respective control. The animals were transferred to a T-maze apparatus immediately and allowed to choose between the two odors converging in the middle for 90 s. Because the time between testing and US/CS coupling is 3 min, the initial performance assessment is that of 3 min memory, which we refer to as learning. Performance was measured by calculating an index (PI), as the fraction of flies that avoided the shock-associated odor minus the fraction that avoided the control odor reflected learning because of one of the conditioning stimuli and represented half of the performance index. One performance index was calculated as the average of the half-learning indexes for each of the two groups of animals trained to complementary conditioning stimuli and ranges from 100 to 0, for perfect to no learning, respectively. All behavioral experiments were conducted in a balanced design, where all genotypes involved in an experiment were tested per day and the experimental set was replicated at least once with flies from different crosses and a different time period (biological replicates).
Drug administration
The phosphodiesterase-4 inhibitor rolipram (4-[3-(cyclopentyloxy)-4-methoxyphenyl]-2-pyrrolidinone) was from Cayman Chemicals and was dissolved in DMSO per the manufacturer’s instructions and mixed into brewer’s yeast paste to a final concentration of 1 mm; ∼60 flies of both sexes were placed in plastic vials containing 0.5 ml of this paste and allowed to feed ad libidum for 5 h before training (Kanellopoulos et al., 2012). Food dye was added to monitor homogeneity and to ensure that flies were actually ingesting the food; ∼1 h before conditioning, flies were transferred into normal fly-food vials. The no drug groups were fed with the same yeast paste, but containing DMSO instead of drug. No differences in learning performance have been observed in flies fed with DMSO, relative to no DMSO controls (Gouzi et al., 2011).
Immunohistochemical analysis and data processing
Whole-mount adult (2- to 6-d-old) brains were dissected in cold PBS, fixed in 4% paraformaldehyde for 20 min at room temperature and permeabilized using 0.3% Triton X-100 in PBS (PBST). Samples were blocked in 10% normal goat serum (diluted in 0.3% PBST) for 90 min at room temperature, and incubated with primary antibodies for one or two overnights at 4°C. Samples were washed (PBST) 3 times for 15 min each at room temperature and incubated with secondary antibodies for 2 h at room temperature or overnight at 4°C. The samples were then washed again at least 3 times for 15 min at room temperature and mounted on a slide using Dako fluorescent mounting medium.
The primary and secondary antibodies were diluted in blocking solution. The primary antibodies were used as follows: mouse anti-GFP (3E6, Invitrogen, 1:400), rabbit anti-GABA (A2052, Sigma Millipore, 1:50), rabbit anti-GFP (A-6455, Invitrogen, 1:400), mouse anti-HA (sc-7392, Santa Cruz Biotechnology, 1:400), rabbit anti-Leo (1:1000) (Skoulakis and Davis, 1996), and mouse anti-FasII (1:5) (Developmental Studies Hybridoma Bank). The secondary antibodies used were as follows: goat anti-mouse and anti-rabbit conjugated with Alexa-Fluor secondary antibodies (1:400, all from Invitrogen). Confocal laser microscopy was performed using the Leica Microsystems TCS SP8 Confocal Microscope. Serial optical sections of 0.75 μm thickness were obtained from the fixed whole-mount adult brain samples. For the representative images shown, each experiment has been successfully reproduced at least 3 times and was performed on multiple days. Images were formatted using Fiji software and Adobe Photoshop CS2.
Western blot analyses
Single female fly heads at 2-5 d after eclosion were homogenized in 1× Laemmli buffer (50 mm Tris, pH 6.8, 100 mm DTT, 5% 2-mercaptoethanol, 2% SDS, 10% glycerol, and 0,01% bromophenol blue) and loaded per lane. Proteins were transferred to PVDF membrane and probed with the following antibodies: mouse anti-β-tubulin E7 (Developmental Studies Hybridoma Bank) at 1:500, mouse anti-pERK (Sigma Millipore) at 1:2000, and rabbit anti-ERK (Cell Signaling Technology) at 1:2000. Appropriate HRP-conjugated secondary antibodies were applied at 1:5000. Proteins were visualized with chemiluminescence (ECL Plus, GE Healthcare).
Statistical analysis of behavioral data
Untransformed (raw) behavioral data were analyzed parametrically with the JMP 7.1 statistical software package (SAS Institute) as described previously (Moressis et al., 2009). Following initial ANOVA, least square means (LSM) contrast analyses were performed between the experimental group (in gray throughout all figures) and its genetic or treatment controls, using α = 0.01 unless mentioned otherwise (§). The level of significance was adjusted for the experiment-wise error rate. Detailed results of all planned comparisons (LSM contrast analyses) are reported in the text.
Results
dNf1 is required in OK72 neurons for normal olfactory associative learning
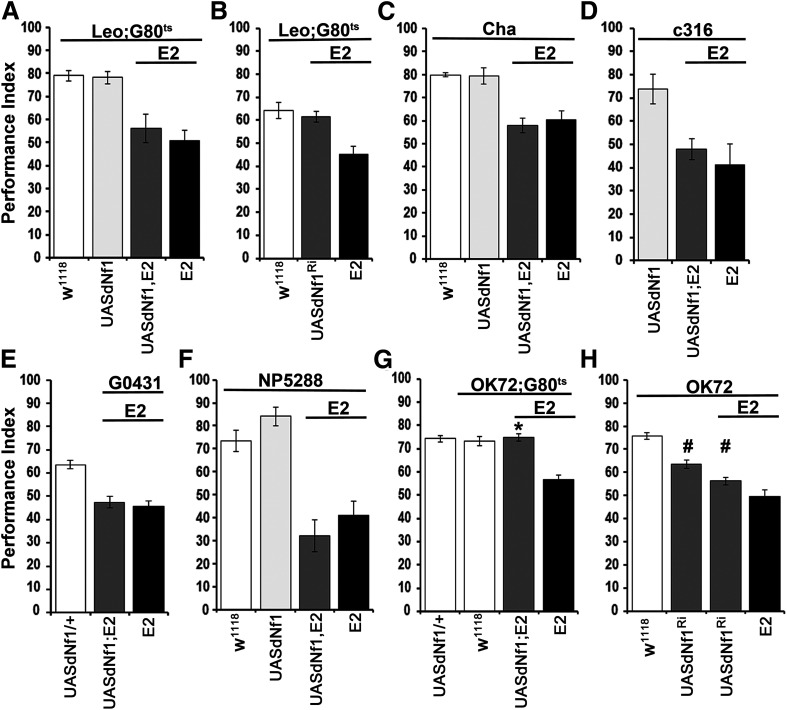
Initially, the finding that dNf1 function is required for negatively reinforced associative learning outside the MBs (Gouzi et al., 2011) was independently confirmed. Adult-specific expression of a UAS-dNf1 transgene (Walker et al., 2006) under the strong pan-MB driver Leo-Gal4 (Messaritou et al., 2010) failed to restore learning in dNf1E2 null mutants (Fig. 1A; ANOVA: F(3,33) = 9.7133, p < 0.0001; subsequent LSM: LeoGAL4/+;Ε2,Gal80ts/UASdNf1,E2 vs LeoGAL4/+;Ε2,Gal80ts/E2, p = 0.4050). Conversely, adult-specific RNAi-mediated dNf1 attenuation in the MBs did not impair associative learning (Fig. 1B; ANOVA: F(2,30) = 10.177, p = 0.0005; subsequent LSM: LeoGAL4/+;UASdNf1RNAi/E2,Gal80ts vs LeoGAL4/+;Gal80ts/+, p = 0.5653). Therefore, although present in their dendrites (Gouzi et al., 2011), clearly dNf1 presence in the MBs is not necessary for normal olfactory associative learning.
Figure 1.
Adult-specific requirement of dNf1 in OK72 neurons for olfactory associative learning. Performance assessment 3 min after conditioning in control (white bars), mutant (black bars), experimental (dark gray bars), and transgene (UAS-dNf1)-expressing (light gray bars) animals. The genotypes are indicated below each bar. Data are mean ± SEM. *Statistically significant difference of the experimental from the mutant flies. #Statistically significant difference of the experimental from the control flies. G80ts indicates the ubiquitously expressed temperature-sensitive Gal4 repressor Tub-Gal80ts. A, Adult-specific expression of an Nf1 transgene throughout the MBs under Leo-Gal4 does not restore learning in dNf1E2 homozygotes. B, RNAi-mediated (UASdNf1Ri) dNf1 attenuation in MBs (gray bar) does not affect learning. C, Restoration of dNf1 expression within cholinergic neurons under Cha-Gal4 (gray bar) does not reverse the learning deficit of dNf1E2 homozygotes (black bar), which remained significantly different from that of controls (white bar). The UAS-Nf1 transgene-expressing flies (light gray bar) presented the same learning performance with control flies. D, UAS-Nf1 expression in dorsal paired medial neurons under c316-Gal4 does not restore learning to dNf1E2 homozygotes. E, UAS-Nf1 expression in dorsal anterior lateral neurons under the G0431-Gal4 driver does not restore associative learning to mutant homozygotes. F, UAS-Nf1 expression in antennal lobe projection neurons, including the APL, marked with the NP5288-Gal4 driver does not restore associative learning to mutant homozygotes. G, Adult-specific UAS-Nf1 expression in neurons marked with the OK72-Gal4 driver (gray bar) reverses the associative learning deficit of dNf1E2 nulls. H, RNAi-mediated dNf1 abrogation (UASdNf1Ri) in OK72 neurons in control flies and dNf1E2 heterozygotes (gray bars) induces significant learning deficits.
To search for neurons within the adult CNS where dNf1 is necessary and sufficient for normal associative learning, we introduced Gal4 drivers marking neurons implicated in the process (Guven-Ozkan and Davis, 2014) to dNf1E2 null mutants and asked for learning deficit reversal on UASdNf1 expression therein. Initially, dNf1 was restored within cholinergic neurons, but this did not reverse the learning deficit of null homozygotes (Fig. 1C; ANOVA: F(3,29) = 15.7783, p < 0.0001; subsequent LSM: ChaGAL4;UASdNf1,E2/E2 vs ChaGAL4;E2/E2, p = 0.5369). Furthermore, dNf1 restoration within the dorsal paired medial MB intrinsic neurons (Waddell et al., 2000) under c316-Gal4 (Fig. 1D), or the dorsal anterior lateral MB extrinsic neurons (Fig. 1E) under G0431-Gal4 (Chen et al., 2012) did not rescue the learning deficit of dNf1 nulls (ANOVA: F(2,17) = 6.2708, p = 0.0105; subsequent LSM: c316GAL4/+;UASdNf1,E2/E2 vs c316GAL4/+;E2/E2, p = 0.5079 and ANOVA: F(2,57) = 20.2124, p < 0.0001; subsequent LSM: G0431GAL4/UASdNf1;E2/E2 vs G0431GAL4/+;E2/E2, p = 0.6117, respectively). Finally, we assayed neurons marked by NP5288-Gal4, which include antennal lobe projection neurons, presynaptic to the MBs and lateral horn but also the GABAergic anterior-paired-lateral (APL) neurons (Tanaka et al., 2008), known to be involved in associative learning (Liu and Davis, 2009). However, UASdNf1 expression therein failed again (ANOVA: F(3,22) = 17.0161, p < 0.0001; subsequent LSM: NP5288GAL4/+;UASdNf1,E2/E2 vs NP5288GAL4/+;E2/E2, p = 0.2886) to restore learning in dNf1E2 homozygotes (Fig. 1F).
Having excluded the MBs and main MB-associated neurons involved in dNf1-mediated learning, additional available Gal4 drivers were screened, including OK72-Gal4, where surprisingly, adult-specific dNf1 reinstatement fully restored associative learning to the mutants (Fig. 1G; ANOVA: F(3,46) = 18.9052, p < 0.0001; subsequent LSM: OK72GAL4/UASdNf1;E2,Gal80ts/E2 vs OK72GAL4/+;E2,Gal80ts/E2, p < 0.0001). This outcome demonstrates that dNf1 loss in the mutants does not result in developmental defects underlying their learning deficit. Corroborating this interpretation, RNAi-mediated dNf1 attenuation in OK72-marked neurons precipitated a strong learning deficit (Fig. 1H; ANOVA: F(3,65) = 37.0918, p < 0.0001; subsequent LSM: OK72GAL4/+ vs OK72GAL4/+;UASdNf1RNAi/+, p < 0.0001; OK72GAL4/+ vs OK72GAL4/+;UASdNf1RNAi/E2 p < 0.0001). The RNAi-mediating transgene appears to be inefficient, as on its own yielded a partial deficit (LSM following ANOVA above: OK72GAL4/+;UASdNf1RNAi/+ vs OK72GAL4/+;UASdNf1RNAi/E2 p = 0.0071 and vs OK72GAL4/+;E2/E2 p < 0.0001), while in combination with one dNf1E2 mutant allele phenocopied the learning deficit of the null homozygotes (LSM: OK72GAL4/+;UASdNf1RNAi/E2 vs OK72GAL4/+;E2/E2 p = 0.0187). Consequently, the RNAi-mediating transgene is used in null heterozygous background henceforth. Significantly, the collective results demonstrate that dNf1 is acutely necessary and sufficient within adult OK72-marked neurons to support normal olfactory learning.
dNf1 is acutely required in adult non-MB OK72-marked neurons to support learning
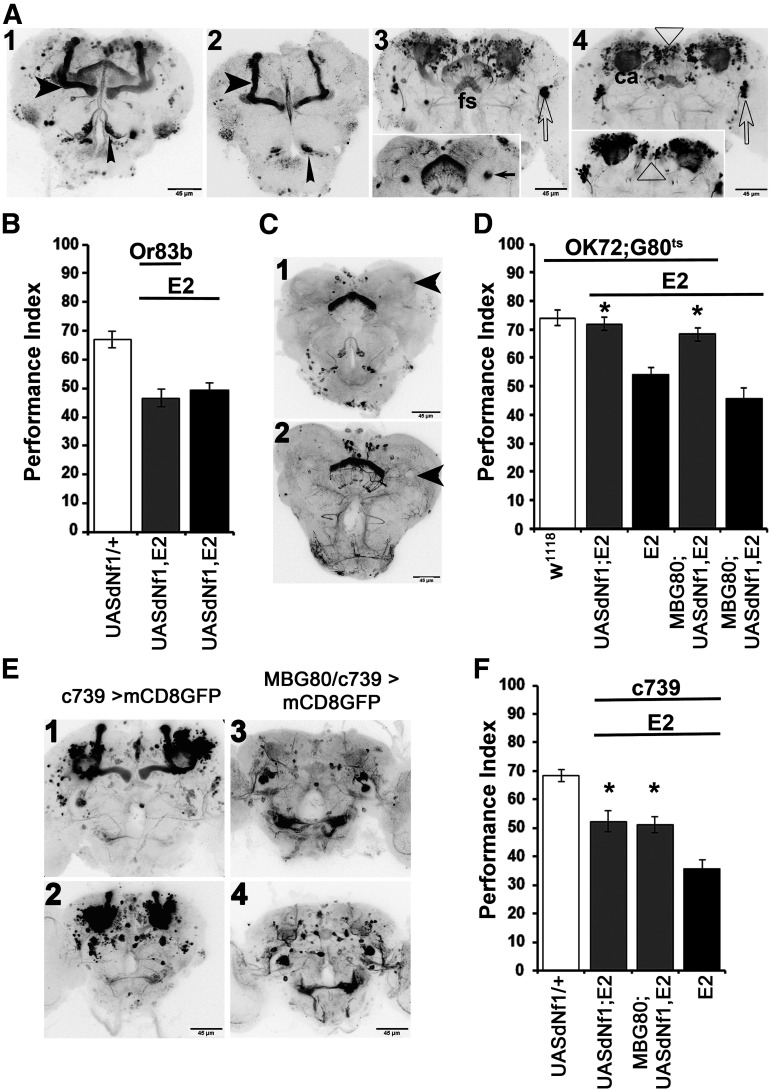
OK72-Gal4 is expressed, although in a restricted pattern, in embryonic, larval, and adult MBs (de Haro et al., 2010), and we observed significant levels of GFP immunoreactivity primarily in α/β MB neurons (Fig. 2A1,A2), in the fan-shaped body (Fig. 2A3) and two large clusters of lateral neurons with axons appearing to project to MB dendrites (Fig. 2A3,A4). Other prominent GFP-positive structures included the MB dendrites (Fig. 2A4) and a constellation of neurons in the superior medial protocerebrum (SMP; Fig. 2A3,A4).
Figure 2.
MB-independent requirement of dNf1 in OK72 neurons for olfactory associative learning. A, Grayscale inverted confocal images of adult brains stained with anti-GFP driven by the OK72-Gal4 driver, arranged from anterior to posterior (A1–A4): A1, AL glomeruli (narrow arrowhead) and MB lobes (wide arrowhead). Note the constellation of positive neurons between the vertical MB lobes in the center of the brain. A2, A single confocal section at the anterior of the brain clearly indicating expression in the α/β MB lobes (wide arrowhead) and AL glomeruli (narrow arrowhead). A3, A mid-frontal view of a brain revealing the constellation of positive neurons at the SMP, as well as symmetrical clusters of lateral neurons (open arrow pointing to the right cluster). The fan-shaped body (fs) is indicated with the staining pattern flanked by the MB pedunculi, better revealed in the single confocal section in the insert (arrows). A4, A posterior view of the brain reveals the MB dendrites/calyces (ca), the posterior SMP neurons (open triangle), and the lateral clusters (open arrow). The single confocal plain insert focuses on the posterior SMP neurons (open triangle). B, UAS-Nf1 expression in olfactory receptor neurons and antennal lobe glomeruli marked with the Or83b-Gal4 driver do not restore associative learning to dNf1E2 homozygotes. C, Anti-GFP stained grayscale inverted confocal images of adult brains of OK72-Gal4/MBG80 driving UAS-mcD8GFP in an anterior (C1) and mid-posterior (C2) view. Although staining remains in the fs, antennal lobe, and SMP neurons, the area occupied by the MBs does not express GFP (large arrowheads). D, Adult-specific dNf1 expression within the OK72 marked neurons, excluding the MBs, restores associative learning in dNf1E2homozygotes. MBG80 encodes a Gal80 repressor expressed constitutively within MB neurons. The performance of OK72-Gal4/MBG80; UAS-Nf1,dNf1E2/TubG80ts,dNf1E2 (second gray bar) was significantly different from that of animals not expressing the transgene (black bars), but not from controls (white bar). *Statistically significant difference of the experimental from the mutant flies. E, Confocal images of adult brains stained with anti-GFP driven by the c739-Gal4 driver in anterior (E1) and posterior (E2) views, illustrating strong MB expression as well as a constellation of non-MB GFP-positive neurons, especially in the posterior of the brain (E2). These non-MB neurons appear prominently in the anterior (E3) and mostly in the posterior (E4) of c739-Gal4/MBG80 brains where GFP expression in the MBs is eliminated. F, c739-Gal4;UAS-Nf1,dNf1E2/dNf1E2 animals (first gray bar) express transgenic dNf1 in c739 neurons, and this partially restores learning performance in dNf1E2nulls equally well as in animals expressing the transgene under c739 at the exclusion of the MBs (second gray bar). *Statistically significant difference of the experimental from the mutant flies.
The OK72 driver is also expressed in a group of 50-70 cells in the third antennal segment and antennal lobe glomeruli VM1, VM4, and DL1 (Devaud et al., 2003), as indicated in Figure 2A1, A2. However, UASdNf1 expression under Or83b-GAL4, which marks ∼80% of olfactory neurons and the antennal glomeruli (Wang et al., 2003), did not rescue (ANOVA: F(2,35) = 15.5027, p < 0.0001 subsequent LSM: Or83bGAL4/UASdNf1;E2/E2 vs UASdNf1/+;E2/E2 p = 0.5235) the dNf1E2learning deficit (Fig. 2B). Hence, rescue under OK72-Gal4 was not consequent of protein restoration in olfactory neurons and glomeruli, in accord with the lack of olfactory, or footshock sensory deficits in the mutants (Gouzi et al., 2011).
Because OK72 marks α/β MB neurons, dNf1 may still be necessary therein, although not sufficient for learning. To address this possibility, the UASdNf1 transgene was expressed under OK72-Gal4 but coexpressing the constitutive Gal4 suppressor MB-Gal80 (Krashes et al., 2007), which eliminated expression in MBs (Fig. 2C). Significantly, dNf1 restoration in adult non-MB OK72-marked neurons, fully rescued the learning deficit (ANOVA: F(4,49) = 17.6434, p < 0.0001 subsequent LSM: OK72GAL4/MBG80;UASdNf1,E2/E2,Gal80ts vs MBG80/+;UASdNf1,E2/E2, p < 0.0001), just as it did when expressed in all neurons marked with this driver (LSM: OK72GAL4/UASdNf1;E2,Gal80ts/E2 vs OK72GAL4/+;E2,Gal80ts/E2, p < 0.0001, but note that p = 0.3148 compared with OK72GAL4/MBG80;UASdNf1,E2/E2,Gal80ts), as shown in Figure 2D. Therefore, dNf1 expression within the MBs is neither necessary nor sufficient for olfactory associative learning, but rather it is required within other types of OK72 marked neurons for the process.
This conclusion was independently verified with c739-GAL4, which is preferentially expressed in the α/β MB neurons, but also in a number of unidentified neurons distributed throughout, but especially in the posterior and dorsomedial CNS (Fig. 2E1,E2). Use of this driver was reported to improve learning in dNf1 mutants (Buchanan and Davis, 2010) and consistently, it partially rescued the learning deficit of dNf1E2 homozygotes (Fig. 2F; ANOVA: F(3,46) = 21.5259, p < 0.0001 subsequent LSM: c739GAL4/UASdNf1;E2/E2 vs c739GAL4/+;E2/E2, p = 0.0006). However, dNf1E2 homozygotes expressing UASdNf1 under c739, but not in the MBs (Fig. 2E3,E4), presented the same level of partial rescue (Fig. 2F; LSM after ANOVA above: c739GAL4/MBG80;UASdNf1,E2/E2 vs c739GAL4/+;E2/E2, p = 0.0004), as mutant flies expressing the transgene under the full c739 pattern. Notably, c739 expression outside the MBs includes cells in the vicinity of OK72-marked neurons, including the SMP cluster (Fig. 2E2,E4), which likely account for the learning improvement, rather than expression in the MBs. Collectively then, dNf1 expression in a subset of OK72-Gal4 marked neurons outside the MBs is necessary and sufficient to support olfactory learning.
GABAergic dysregulation within OK72 neurons underlies the learning deficits on dNf1 loss
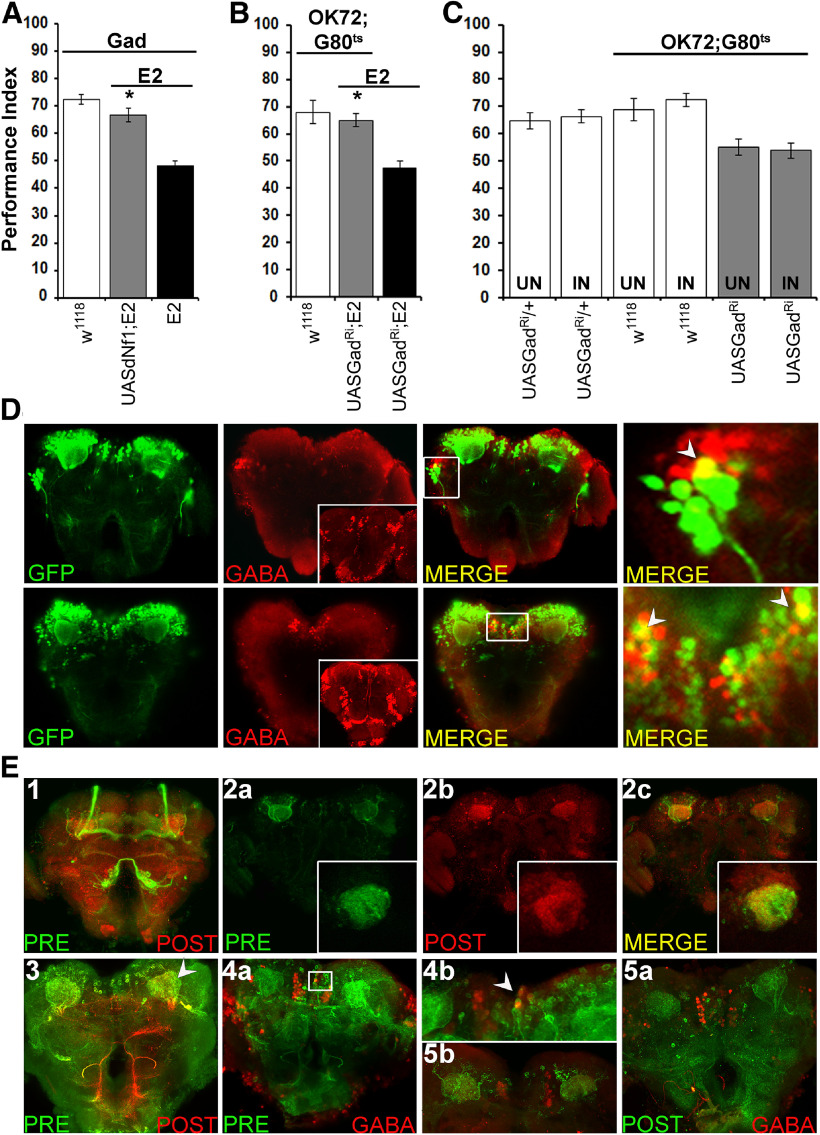
Given the complex OK72 expression pattern, lack of a functional requirement for dNf1 in cholinergic neurons for learning (Fig. 1C) and reports from Nf1+/− mice and humans (Cui et al., 2008; Shilyansky et al., 2010b), we asked whether any of the OK72-marked neurons are inhibitory, expressing glutamic acid decarboxylase (GAD), the enzyme requisite for GABA production (Kulkarni et al., 1994). Importantly, restoring dNf1 expression in GABAergic neurons of null homozygotes fully reversed their deficient learning (Fig. 3A; ANOVA: F(2,39) = 42.4689, p < 0.0001 subsequent LSM: GadGAL4/UASdNf1;E2/E2 vs GadGAL4/+;E2/E2, p < 0.0001). This strongly suggests that dNf1 loss from OK72-marked GABAergic neurons underlies the learning deficit of the null animals. To validate this notion, GAD was abrogated in OK72 neurons of adult dNf1E2 homozygotes, which in accord with the hypothesis fully restored their deficient learning (Fig. 3B; ANOVA: F(2,32) = 15.6612, p < 0.0001 subsequent LSM: OK72GAL4/+;UASGadRNAi/Gal80ts vs UASGadRNAi,E2/E2, p < 0.0001). Therefore, at least a subset of the OK72-marked neurons must be GABAergic.
Figure 3.
Olfactory associative learning requires dNf1 in GABAergic OK72-marked neurons. A–C, Performance was assessed within 3 min after conditioning in control (white bars), mutant (black bars), and experimental (gray bars) animals. The genotypes are indicated below each bar, representing the mean ± SEM. *Statistically significant difference of the experimental from mutant flies. G80ts indicates the ubiquitously expressed temperature-sensitive Gal4 repressor Tub-Gal80ts. A, Expression of the UAS-Nf1 transgene within GABAergic neurons restores normal learning to dNf1E2 nulls. B, Adult-specific Gad abrogation (UASGadRi) within OK72 neurons reverses the learning deficit of dNf1E2 homozygotes to levels not significantly different from that of controls. C, RNAi-mediated abrogation of Gad (UASGadRi) in OK72 neurons of adult WT flies does not alter learning at the restrictive (18°C, UN) or the permissive temperature (30°C, IN), respectively (gray bars). White bars represent the control genotypes, raised at both temperatures. D, All images are at 40× magnification. Size bar indicates relative size. Anti-GFP staining (green), anti-GABA staining (red), and merging of the two (yellow) in representative optical sections from fly brains where GFP expression is driven by OK72-Gal4. The insets in the GABA panels are maximum projections to demonstrate ample penetration of the anti-GABA antibody not obvious in the optical sections used for colocalization. Colocalization of GFP and GABA immunofluorescence is detected laterally (top) and the SMP (bottom). Right panels (both top and bottom), Magnifications of the respective marked boxes, showing colocalization in single cells. E, In flies bearing the trans-Tango components, driving ligand and myrGFP expression under OK72-Gal4 (green) results in HA-mtdTomato expression in postsynaptic MB dendrites (red). Anterior (E1) and posterior (E3) view of maximum projections of presynaptic and postsynaptic neurons under OK72Gal4-driven TANGO. E2a–E2c, Representative optical sections of presynaptic (OK72; green) and postsynaptic (red) neurons at the dendritic level, where yellow represents their interaction. Insets, Higher magnification of confocal images from different brains at the level of MB dendrites. E4a, Posterior view maximum projection of OK72 presynaptic neurons (green) and GABA-positive neurons (red). White box represents the area magnified (80×) in E4b with the white arrow indicating colocalization. E5a, Posterior view maximum projection of postsynaptic to OK72-marked neurons (green) and GABAergic ones (red), clearly showing no colocalization. E5b, Area in the posterior of a different brain than in E5a verifying independently the lack of GABA (red) colocalization with postsynaptic to OK72-marked neurons (green).
Although somewhat depressed, learning was not significantly different in adult WT flies expressing the UAS-GadRNAi transgene under OK72-Gal4;Gal80ts compared with nonexpressing siblings (Fig. 3C; ANOVA: F(5,69) = 6.5681, p < 0.0001 subsequent LSM: OK72GAL4/+;UASGadRNAi/Gal80ts Uninduced vs Induced, p = 0.7104). Hence, reduction or loss of GABA from GABAergic OK72-marked neurons does not underlie the learning deficit of the null Nf1 mutant homozygotes. Collectively, the results indicate that GABA production or release is elevated in OK72 neurons upon dNf1 loss. However, GABAergic activity from OK72-marked neurons does not limit learning in control animals, as activity of the also GABAergic APL neurons does (Liu et al., 2009). It is unlikely that dNf1 regulates APL-mediated GABAergic activity since its expression therein in the mutants did not restore learning (Fig. 1F). Therefore, it is likely that the OK72-marked GABAergic neurons define a novel circuit, which in addition to the APL is involved in Drosophila associative learning.
Immunohistochemical experiments revealed GABA and OK72-Gal4-driven GFP colocalization in a small subset of cells within the lateral horn and a distinct group in the SMP (Fig. 3D). This is not consequent of reduced anti-GABA penetration as demonstrated in the respective panel inserts (Fig. 3D; anti-GABA). Therefore, these specific GABAergic OK72 neurons represent the minimal number of cells where dNf1 is required for normal olfactory associative learning. Unfortunately, we have been unable thus far to identify a Gal4 driver restricted to these SMP neurons to formally test this assertion. However, we traced the synaptic connections of OK72-marked neurons, taking advantage of the trans-TANGO system of trans-synaptic circuit analysis (Talay et al., 2017). Presynaptic neurons are marked by mcD8GFP expression and express a trans-synaptic ligand that activates mtdTomato expression in post-synaptic neurons.
The distributed expression of OK72-Gal4 under TANGO resulted in a complex expression pattern (Fig. 3E1,E3). It is interesting that neurons of the MB core appeared to be presynaptic (Fig. 3E1, green), most likely to other MB neurons via lateral connections. Since dNf1 expression or abrogation within MB neurons did not rescue the learning deficit of null mutants, or precipitate any learning deficiency respectively, such potential synaptic connections do not contribute to the phenotype. It should be noted that expression levels were not as high as reported (Talay et al., 2017), with some neurons marked with OK72Gal4 directly driving mcD8GFP expression (Fig. 2A), but not visible under TANGO. This was most notable with the somata of lateral neurons projecting to the MBs (Fig. 2A3,A4). This may result from weak expression of the driver within these neurons, or a resolution limitation of the TANGO system, but some of the driver-marked neurons could be presynaptic to (Fig. 3E2a) and in contact with MB dendrites (Fig. 3E2b). This agrees with the maximum projection image in the posterior of the brain, where presynaptic OK72-marked neurons appear to synapse with the postsynaptic MB dendrites (calyces) (Fig. 3E2c). Therefore, at least one set of OK72-marked neurons appear to impinge on the MBs, and we posit that these are likely GABAergic. This is strongly suggested by the importance of the MBs for associative learning, the role of GABAergic neurons in the process (Liu et al., 2007; Liu and Davis, 2009), and the apparent functional upregulation of GABA in a subset of OK72 neurons on dNf1 loss (Fig. 3A,B).
To address this possibility, brains expressing TANGO under OK72-Gal4 were stained with anti-GABA. Although the antibody does not stain GABAergic projections, but rather mostly cell bodies (Fig. 3D, anti-GABA stained inserts), limited colocalization was observed in the SMP area (Fig. 3E4a; and magnified in Fig. 3E4b), consistent with the notion that some OK72-expressing presynaptic neurons in that area are GABAergic. This was confirmed with staining for the TANGO postsynaptic component of the driver with anti-HA (Fig. 3E5a,5b). The punctate pattern of neurons postsynaptic to OK72 is evident, and GABAergic neurons clearly do not colocalize with them. Collectively, the results suggest that GABAergic OK72-marked neurons are likely presynaptic to the MBs.
cAMP signaling is not required in OK72 neurons for Nf1-mediated learning
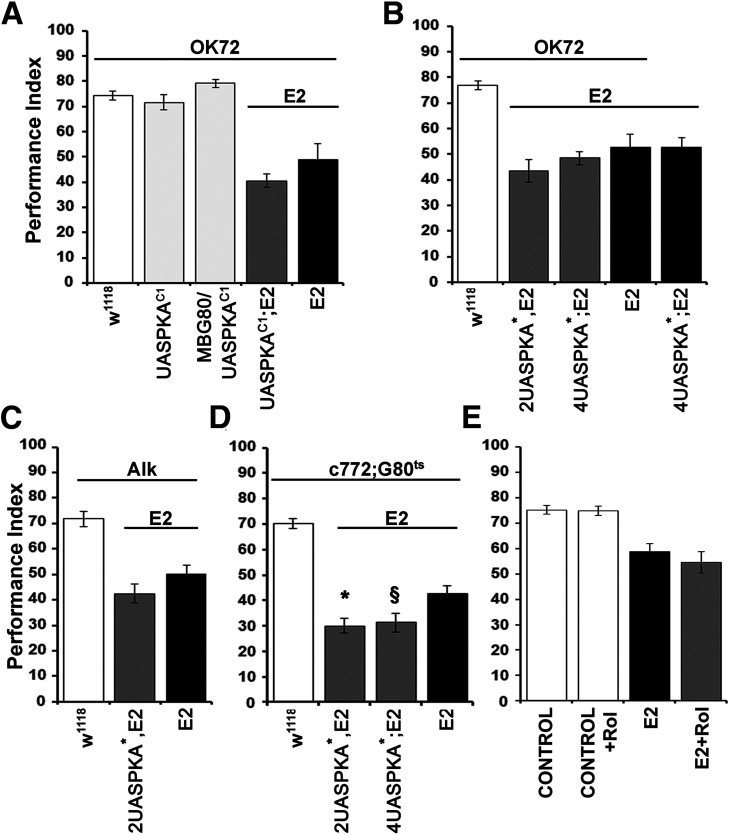
To explore potential mechanisms used within OK72-marked neurons for Nf1-mediated learning, we manipulated signaling pathways therein in the context of Nf1 null mutants. Prior studies have suggested that Nf1 regulates cAMP levels both in Drosophila and mouse (Guo et al., 1997; The et al., 1997; Tong et al., 2002; Hannan et al., 2006). Indeed, the learning deficits of Drosophilapartial Nf1 partial null mutants have been attributed to disrupted adenylyl cyclase function and compromised cAMP-mediated signaling (Guo et al., 2000). Disrupted cAMP signaling was also reported causative of the reduced size of dNf1 null homozygotes (Walker et al., 2013).
To address whether compromised cAMP signaling within OK72 neurons underlies the learning deficits of dNf1 nulls, we expressed therein constitutively active Drosophila (Fig. 4A; PKAC1) and murine (Fig. 4B; PKA*) PKA catalytic subunit transgenes, reported to rescue the small size of mutant homozygotes (Walker et al., 2013). However, neither fly (Fig. 4A; ANOVA: F(4,58) = 34.7329, p < 0.0001 subsequent LSM: OK72GAL4/UASPKAC1;E2/E2 vs OK72GAL4/+;E2/E2, p = 0.073), nor murine transgenes reversed the dNf1E2 deficient learning, even when expressed at variable levels (2UAS vs 4UAS) for the murine transgenes (Fig. 4B; ANOVA: F(4,45) = 28.6341, p < 0.0001 subsequent LSM: OK72GAL4/4UASPKA*;E2/E2 vs 4UASPKA*/+;E2/E2, p = 0.3272 and OK72GAL4/+;2UASPKA*,E2/E2 vs 4UASPKA*/+;E2/E2, p = 0.0845). Failure of these constitutively active PKA catalytic subunit transgenes to rescue the learning deficit of dNf1 nulls strongly suggests that signaling via PKA is not required within OK72-marked neurons for dNf1-mediated learning.
Figure 4.
The cAMP/PKA pathway is not implicated in Nf1-related learning deficit within OK72 neurons. Performance assessment after conditioning of control (white bars), mutant (black bars), experimental (dark gray bars), and transgene-overexpressing (light gray bars) animals. The genotypes of all animals are indicated below each bar. Data are mean ± SEM. G80ts indicates the ubiquitously expressed temperature-sensitive Gal4 repressor Tub-Gal80ts. A, Expression of a constitutively active Drosophila PKA transgene in OK72 neurons (dark gray bar) does not rescue the dNf1E2 learning deficit (black bar) relative to controls (white bar). The second and third bars (light gray) represent learning of WT animals expressing the UAS-PKAC1 transgene in OK72 neurons, including or excluding the MBs, respectively, and are indistinguishable from the control. B, Expression of a constitutively active murine PKA* transgene with 2 (first gray bar) or 4 (second gray bar) UAS elements in OK72 neurons does not reverse the learning deficit of dNf1E2 mutant homozygotes (black bars). The performance of both null mutants (black bars) and PKA* transgene-expressing mutant animals (gray bars) is significantly different from that of control flies (white bar). C, Pan-neuronal expression, under the Alk-Gal4 driver, of a constitutively active murine PKA* transgene with 2 UAS elements (gray bar) fails to reverse the learning deficiency of dNf1E2 homozygotes (black bar). D, Expression of a constitutively active murine PKA* transgene with 2 (first gray bar) or 4 (second gray bar) UAS elements in adult MBs does not rescue the learning deficit of dNf1E2 homozygotes. *Statistically significant difference of the experimental from mutant flies. §Statistical difference of the respective experimental group from the mutant flies (p = 0.0277). E, Rolipram administration (1 mm) to dNf1E2 null homozygotes (gray bar) does not restore learning, which remains indistinguishable from that of untreated mutant flies (black bar). Treated and untreated control flies (white bars) perform significantly different from the mutant groups. Control flies are OK72-Gal4 homozygotes, whereas the genotype of E2 mutants is OK72-Gal4;E2 homozygotes.
Alternatively, PKA activity may be compromised in dNf1E2 homozygotes in neurons not marked by OK72, such as the MBs, where it is known to be required for normal learning (Skoulakis et al., 1993). Such MB-specific kinase activity could in principle explain the reported reversal of the dNf1 learning deficit with acute global PKA* expression (Guo et al., 2000). We addressed this possibility using the strong Alk-GAL4 driver, which highly overlaps the dNf1 endogenous pattern in the adult CNS (Gouzi et al., 2011), to drive the constitutively active murine 2UASPKA* transgene, but again failed to rescue the dNf1E2learning deficit (Fig. 4C; ANOVA: F(2,29) = 21.2205, p < 0.0001 subsequent LSM: AlkGAL4/+; 2UASPKA*,E2/E2 vs AlkGAL4/+;E2/E2, p = 0.1286). Moreover, acute constitutive murine PKA activity within adult MBs under c772Gal4; Gal80ts also failed to rescue the dNf1E2learning deficit and in fact depressed learning further (Fig. 4D; ANOVA: F(3,31) = 23.2169, p < 0.0001 subsequent LSM: c772-GAL4/+;2UASPKA*,E2/E2,Gal80ts vs c772GAL4/+;E2,Gal80ts/E2, p = 0.005 and c772GAL4/4UASPKA*;E2,Gal80ts/E2 vs c772GAL4/+;E2,Gal80ts/E2, p = 0.0277). This strongly argues against a model of PKA activation within the MBs in response to dNf1-mediated signals from OK72 neurons.
Finally, we aimed to investigate the possibility that learning in dNf1E2flies may require cAMP elevation independent of PKA activation, or require regulated activation of the endogenous PKA. To that end, we raised global cAMP levels using the phosphodiesterase inhibitor rolipram, shown to be effective in elevating cAMP levels and restoring learning in dFmr1 deficient flies (Kanellopoulos et al., 2012). However, systemic cAMP elevation did not alter the learning deficit of the null mutants (Fig. 4E; ANOVA: F(3,46) = 15.0271, p < 0.0001 subsequent LSM: E2+rolipram vs E2, p = 0.2819). Collectively, these results do not support the notion that compromised cAMP levels, or PKA signaling consequent to dNf1 loss, are causal of the associated learning deficits.
dAlk-mediated signaling via Nf1/Ras1 within OK72-GABAergic neurons is essential for associative learning
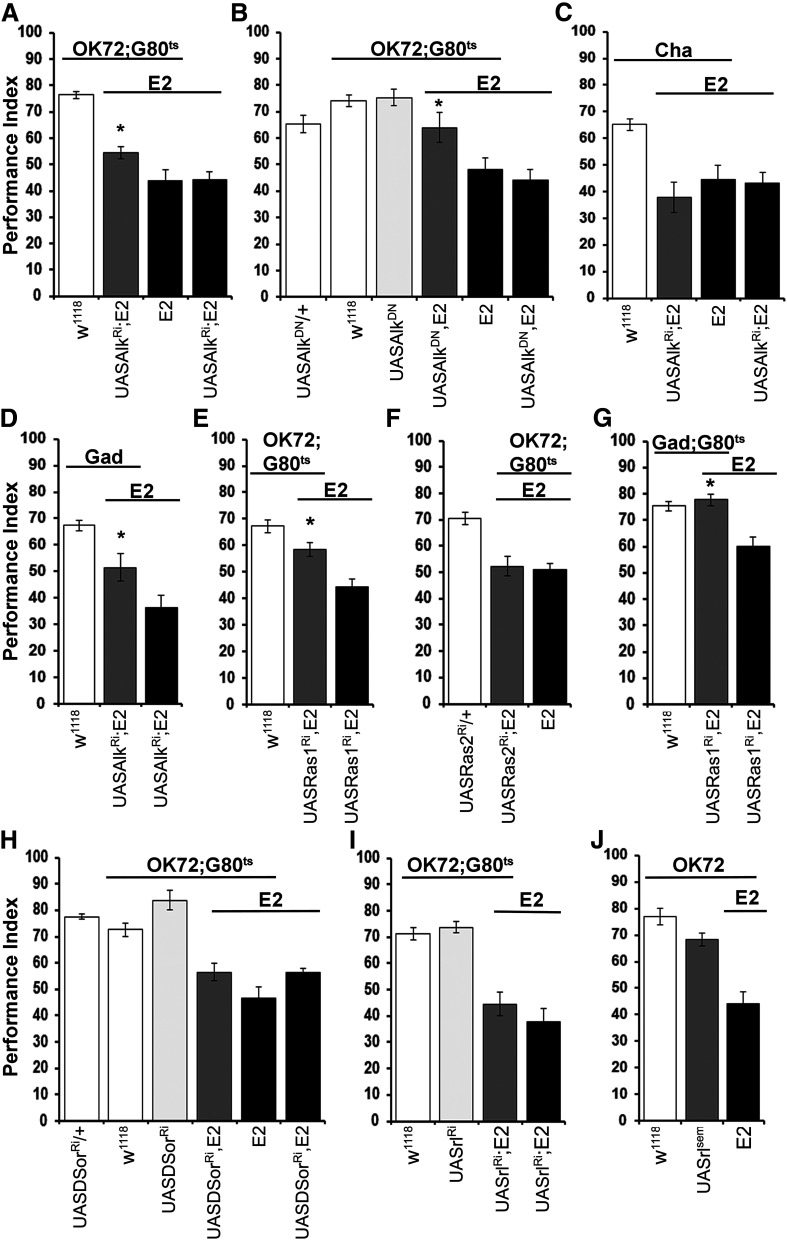
Previous work has shown that abrogation of the receptor tyrosine kinase dAlk in the adult CNS at the exclusion of the MBs proved sufficient to reverse the learning deficit of dNf1 null mutants (Gouzi et al., 2011). Given the role of GABAergic OK72-marked neurons in the dNf1E2 learning deficit, we investigated whether these are the neurons where dAlk abrogation reverses the deficient learning of the nulls. We used two different transgenes to attenuate dAlk activity within adult OK72 neurons: a UASAlkRNAi (Fig. 5A) and the dominant negative form UASAlkDN (Fig. 5B) (Bazigou et al., 2007). Indeed, reduction of dAlk expression in these neurons largely (Fig. 5A; ANOVA: F(3,50) = 45.2064, p < 0.0001 subsequent LSM: OK72GAL4/UASAlkRNAi;E2,Gal80ts/E2 vs UASAlkRNAi/+;E2/E2, p = 0.0031), albeit not completely, restored olfactory learning in dNf1 nulls. However, complete rescue of the learning deficit was achieved by adult-specific expression of the much stronger (Gouzi et al., 2011) dominant negative transgenic dAlkDN protein in OK72 neurons (Fig. 5B; ANOVA: F(5,58) = 10.1953, p < 0.0001 subsequent LSM: OK72GAL4/+; UASAlkDN,E2/E2,Gal80ts vs UASAlkDN,E2/E2, p = 0.0005). Moreover, suppressing dAlk levels specifically within cholinergic (Fig. 5C) neurons did not rescue the learning deficit (ANOVA: F(3,28) = 12.6667, p < 0.0001 subsequent LSM: ChaGAL4/UASAlkRNAi;E2/E2 vs UASAlkRNAi/+;E2/E2, p = 0.3847). In contrast, dAlk attenuation in GABAergic neurons improved learning in dNf1E2 flies (Fig. 5D; ANOVA: F(2,46) = 16.7035, p < 0.0001 subsequent LSM: GadGAL4/UASAlkRNAi;E2/E2 vs UASAlkRNAi/+;E2/E2, p = 0.0104). These results support the notion that dAlk-dependent compensation of dNf1 loss is specific to OK72-marked GABAergic neurons.
Figure 5.
dNf1 engages Alk and Ras1 in OK72-GABAergic neurons to mediate learning. Performance assessment after conditioning of control (white bars), mutant (black bars), experimental (dark gray bars), and transgene-overexpressing (light gray bars) animals. The genotypes of all animals are indicated below each bar. Data are mean ± SEM. *Statistically significant difference in the performance of the experimental from mutant flies. G80ts indicates the ubiquitously expressed temperature-sensitive Gal4 repressor Tub-Gal80ts. A, Adult-induced dAlk abrogation in OK72 neurons ameliorates the Nf1E2 learning deficit. The performance of UASAlkRi-expressing mutants (gray bar) is significantly different from that of the nulls (black bars). B, Adult-induced dAlk inhibition in OK72 neurons rescues Nf1E2 learning defects. ANOVA indicated significant effects on UASAlkDN expression (dark gray bar), compared with Nf1E2 homozygotes (black bars), but not compared with controls (white bars). UASAlkDN expressing WT flies (light gray bar) perform equally to control animals. C, dAlk suppression specifically in cholinergic neurons (gray bar) does not alter the learning deficit of dNf1E2homozygotes (black bars). The performance of UASAlkRi-expressing mutant flies (gray bar) is significantly different from that of control animals (white bar). D, RNAi-mediated dAlk abrogation in GABAergic neurons (gray bar) improves the learning deficiency of Nf1E2 null flies. E, Adult-specific dRas1 abrogation in OK72 neurons of null mutants ameliorates their learning deficiency. F, Adult-specific abrogation of dRas2 within OK72 neurons does not reverse the learning deficit of dNf1E2 homozygotes. G, RNAi-mediated adult-specific dRas1 attenuation in GABAergic neurons fully rescues the dNf1E2 learning deficiency. H, Adult-specific RNAi-mediated abrogation of MEK (UASDsorRi) in OK72 neurons fails to rescue the learning deficit of dNf1E2 homozygotes (dark gray bar). I, Adult specific RNAi-mediated abrogation of MAPK (UASrlRi) in OK72 neurons does not restore the learning deficits of dNf1E2 homozygotes. J, Overexpression of a constitutively active MAPK transgene (UASrlsem) in OK72 neurons does not precipitate learning deficits in WT flies.
dAlk is a typical receptor tyrosine kinase, transducing signals through the Ras/ERK pathway (Gouzi et al., 2005; Palmer et al., 2009). The main function ascribed to Neurofibromin is negative regulation of Ras signaling (Xu et al., 1990; Costa et al., 2002), potentially intercepting activated dAlk signals (Gouzi et al., 2011). Therefore, we investigated the possibility that hyperactivation of one of the Drosophila Ras proteins within OK72 GABAergic neurons underlies the deficient learning of dNf1 nulls. Indeed, RNAi-mediated adult-specific attenuation of Ras1 within OK72 neurons partially rescued the learning deficit of null homozygotes (Fig. 5E; ANOVA: F(2,49) = 19.2928, p < 0.0001 subsequent LSM: OK72GAL4/+;UASRas1RNAi,E2/E2,Gal80ts vs UASRas1RNAi,E2/E2, p = 0.0007, but vs OK72GAL4/+;Gal80ts/+, p = 0.0257). In contrast, attenuation of Ras2 did not alter the dNf1E2 learning deficit (Fig. 5F; ANOVA: F(2,31) = 14.9055, p < 0.0001 subsequent LSM: OK72GAL4/UASRas2RNAi;E2,Gal80ts/E2 vs OK72GAL4/+;E2,Gal80ts/E2, p = 0.7144). Furthermore, complete rescue of the dNf1E2 learning deficit was attained on adult-specific Ras1 attenuation in GABAergic neurons (Fig. 5G; ANOVA: F(2,59) = 14.5783, p < 0.0001 subsequent LSM: GadGAL4/+;UASRas1RNAi,E2/E2,Gal80ts vs UASRas1RNAi,E2/E2, p < 0.0001). Therefore, Ras1 overactivation on dNf1 loss within OK72-marked GABAergic neurons underlies the resultant associative learning deficit, most likely by increased GABA production and/or release to MB dendrites.
Loss of dNf1 results in neuronal ERK overactivation underlying the reduced body size of mutant homozygotes (Walker et al., 2006), while pan-neuronal expression of constitutively active ERK (UAS-rlsem) (Martin-Blanco, 1998) yielded smaller animals (Gouzi et al., 2011). Conversely, pan-neuronal dAlk overexpression in adult Drosophila elevated phospho-ERK and RNAi-mediated dAlk abrogation reduced the activated kinase in head lysates (Gouzi et al., 2011). Hence, it is possible that dNf1 loss leads to hyperactivation of the Ras/ERK cascade within OK72 neurons precipitating GABA elevation and deficient learning. However, RNAi-mediated attenuation of the fly MEK ortholog Dsor (Fig. 5H), or of the MAPK Rolled (rl) (Fig. 5I), did not reverse the learning deficit of null homozygotes (Fig. 5H; ANOVA: F(5,41) = 21.7710, p < 0.0001 subsequent LSM: OK72Gal4/+;UASDsorRNAi,E2/E2,Gal80ts vs UASDsorRNAi,E2/E2, p = 0.9482 and Fig. 5I; ANOVA: F(3,45) = 30.2013, p < 0.0001 subsequent LSM: OK72GAL4/UASrlRNAi;E2,Gal80ts/E2 vs UAS-rlRNAi/+;E2/E2, p = 0.1971, respectively). In agreement with these results, expression of the constitutively active ERK rlsem within OK72 neurons did not yield deficient leaning in otherwise WT animals (Fig. 5J; ANOVA: F(2,40) = 26.2560, p < 0.0001 subsequent LSM: OK72GAL4/UASrlsem vs OK72GAL4/+;E2/E2, p < 0.0001). Therefore, unless the RNAis have very little efficacy, hyperactivation of MEK and ERK within OK72 neurons does not appear to account for the learning deficits of dNf1 nulls.
Collectively, the results suggest that signals, originated by dAlk and transduced to Ras1 to trigger downstream effectors, are intercepted and regulated by dNf1 within OK72 neurons. Moreover, Ras1 effectors other than the canonical MEK and ERK are implicated either in regulation of GAD activity or GABA release within these neurons. Experiments to define these effectors and their mode of action on GABA production or release are currently ongoing.
Ras1 hyperactivation within the MBs of dNf1 nulls attenuates learning
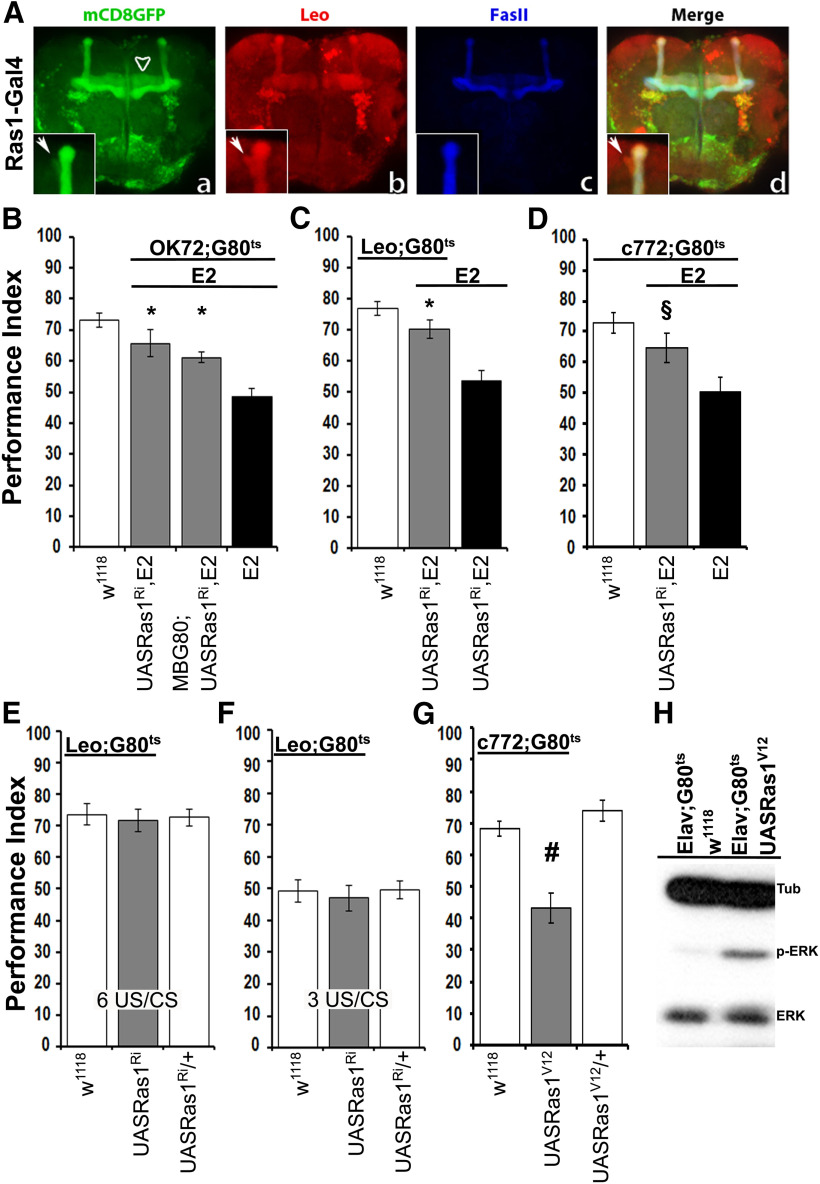
Because Ras1 abrogation under OK72 improved learning of dNf1 homozygotes (Fig. 5E), we investigated whether its effects were solely because of its activity in GABAergic neurons (Fig. 5G), or the MBs where Ras1 appears preferentially expressed (Fig. 6A). Importantly, Ras1 attenuation in OK72-marked neurons with exclusion of the MBs resulted in significant rescue of the dNf1E2 learning deficit (Fig. 6B; ANOVA: F(3,37) = 18.7081, p < 0.0001, subsequent LSM: OK72GAL4/MBG80; UASRas1RNAi,E2/E2,Gal80ts vs OK72GAL4/+;E2,Gal80ts/E2, p = 0.0007), similar to that when Ras1 was also abrogated in the MBs (Fig. 6B; LSM following initial ANOVA above: OK72GAL4/+; UASRas1RNAi,E2/E2,Gal80ts vs OK72GAL4/+;E2,Gal80ts/E2, p < 0.0001). However, reversal of the learning deficit appeared more efficient when the OK72 pattern included the MBs (Fig. 6B; LSM after ANOVA above: OK72GAL4/MBG80; UASRas1RNAi,E2/E2,Gal80ts vs OK72GAL4/+; Gal80ts/+, p = 0.0005 and OK72GAL4/+; UASRas1RNAi,E2/E2,Gal80ts vs OK72GAL4/+; Gal80ts/+, p = 0.0542). Therefore, we investigated whether Ras1 activity is required within the MBs in addition to GABAergic neurons. Remarkably, acute abrogation of Ras1 in adult MBs under the broad MB driver Leo-GAL4 ameliorated the deficient learning of dNf1E2nulls (Fig. 6C; ANOVA: F(2,34) = 16.9274, p < 0.0001, LSM: LeoG4/+;UASRas1RNAi,E2/E2,Gal80ts vs UASRas1RNAi,E2/E2, p = 0.0003), which was recapitulated under an independent more restricted MB driver (Fig. 6D; ANOVA: F(2,33) = 6.6027, p < 0.0041, LSM: c772G4/+;UASRas1RNAi,E2/E2,Gal80ts vs c772GAL4/+;E2,Gal80ts/E2, p = 0.0312). Interestingly, Ras1 attenuation in the MBs of control animals did not affect learning (Fig. 6E; ANOVA F(2,42) = 0.0883, p = 0.9156) , even after reduced training conditions (Fig. 6F; ANOVA F(2,47) = 0.1663, p = 0.8473). This indicates that Ras1 is hyperactivated specifically within the MBs of the dNf1 nulls and this attenuates learning even when excessive GABA from OK72 neurons is ameliorated. However, Ras1 in the MBs does not appear to play a role in learning of control animals. In agreement with this notion, adult-specific expression of an ERK hyperactivating (Fig. 6H), constitutively active RasV12 protein in the MBs precipitated a significant learning deficit in otherwise WT flies (Fig. 6G; ANOVA: F(2,22) = 21.7134, p < 0.0001, LSM: c772GAL4/+;UASRas1V12/Gal80ts vs UASRas1V12/+, p < 0.0001 and vs c772GAL4/+;Gal80ts/+, p = 0.0002).
Figure 6.
Ras1 hyperactivation in the MBs contributes to the learning deficiency of dNf1 nulls. A, Confocal images illustrate the distribution of Ras1 in the MBs of adult flies. Ras1-Gal4 drives the expression of UAS-mCD8GFP, especially in the α/β MB lobes (Aa–Ad), the γ lobe (Aa, arrowhead), and at lower levels in the α/′β′ lobes, where it colocalizes with Leo that marks all MB lobes (Aa,Ab,Ad, arrow in the marked boxes). Marked boxes represent higher magnification of the α/α′ lobes. B–G, Performance assessment after conditioning in control (white bars), mutant (black bars), and experimental (gray bars) animals. The genotypes of all animals are indicated below each bar. Data are mean ± SEM. G80ts indicates the ubiquitously expressed temperature-sensitive Gal4 repressor Tub-Gal80ts. B, Adult-specific dRas1 abrogation within the OK72 marked neurons at the exclusion of MB expression (second gray bar) partially restores learning in dNf1E2 homozygotes. *Statistically significant difference of the experimental from mutant flies. C, dRas1 abrogation in MB neurons (Leo-Gal4 driver) during adulthood rescues the dNf1E2 learning deficits. *Statistically significant difference of the experimental from mutant flies. D, dRas1 abrogation in MB neurons (c772-Gal4 driver) during adulthood improves the dNf1E2 learning deficit. §Statistical difference of the experimental from the mutant flies (p = 0.0312). E, F, Adult-specific abrogation of Ras1 (UASRas1Ri) in MB neurons does not yield learning deficits in control flies. The performance of animals with reduced Ras1 within MBs was not significantly different from that of controls both after 6 (E) and 3 (F) CS-US pairings. G, Overexpression of the constitutively active Ras1 (UASRas1v12) in the MBs of WT adult flies yields learning deficits. #Statistically significant difference of the experimental from the control groups. H, Representative Western blot of head lysates from adult flies expressing pan-neuronally a constitutively active form of Ras1 (UASRas1v12) under Elav-Gal4;Gal80ts using an antibody against phosphorylated ERK (p-ERK), revealing elevated active kinase compared with control animals. The amount of total-ERK protein is not affected. The genotypes of animals used are indicated above the immunoblot. Tubulin levels serve as loading control.
Together, the data suggest Ras1 hyperactivation in the MBs of dNf1 nulls, possibly in response to GABA signals from presynaptic OK72 neurons, which ostensibly inhibits learning. Interestingly, an atypical Ras-mediated pathway regulating cAMP levels by suppressing Gαs activity has been reported in vertebrate neurons (Anastasaki and Gutmann, 2014) and a similar activity in Drosophila head lysates (Hannan et al., 2006). Therefore, hyperactivated Ras1 may lower cAMP levels and its attenuation in neurons using this pathway, as MBs appear to be, would elevate intracellular cAMP. Collectively then, regulated Ras1 activity is required within OK72-marked GABAergic neurons to mediate synthesis or release of GABA. In addition, GABA levels cell autonomously within MB neurons may be linked to, or acting in parallel to reported (Liu and Davis, 2009), or novel signaling pathways suppressing olfactory learning.
Discussion
We reveal GABAergic neurons apparently presynaptic to the MBs (Fig. 3D,E), where dNf1 is acutely required to support normal associative learning (Figs. 1G,H, 2D). Collectively, our results indicate that excess GABA, either because of elevated GAD activity (Fig. 3A,B) or GABA release on dNf1 loss, inhibits MB-mediated associative learning. This broadly agrees with results from Nf1+/− mice that also present increased GABAergic neurotransmission to the hippocampus, PFC, striatum, and amygdala and apparently underlies the deficits in visual-spatial, working memory, social learning, and attention they present (Costa and Silva, 2002; Cui et al., 2008; Shilyansky et al., 2010b; Diggs-Andrews and Gutmann, 2013; Molosh et al., 2014; Omrani et al., 2015).
The increase in GABAergic neurotransmission in dNf1 nulls depends on dAlk activation (Fig. 5A,B,D) and Ras1 engagement (Fig. 5E,G) within these OK72-marked neurons, but it is MEK- and ERK-independent (Fig. 5H–J). This suggests that a variant to the canonical Ras/ERK cascade is operant within these neurons modulating GABA levels. Similarly, an Alk (Weiss et al., 2017a,b) and Ras-dependent cascade results in ERK-mediated increase in GABAergic neurotransmission to the hippocampus of Nf1+/− mice (Costa et al., 2002; Cui et al., 2008). The molecular mechanism(s) underlying the elevated GABAergic neurotransmission to Drosophila MBs is under investigation, but identification of the OK72-marked inhibitory neurons housing them is a significant step toward their elucidation. These neurons are distinct from the also GABAergic APL neuron (Fig. 1F), whose activity normally suppresses the CS+ and is inhibited by learning (Liu et al., 2009; Liu and Davis, 2009).
Although as shown herein, these GABAergic OK72-marked neurons are cardinal to the dNf1 loss-mediated learning deficit, their normal function is less clear. GABAergic neurotransmission from the APL is thought to contribute to the fidelity and effectiveness of the CS+ (Liu and Davis, 2009). Consistent with this limiting inhibitory role, GAD reduction in APL neurons enhanced olfactory associative learning (Liu and Davis, 2009). However, GAD abrogation within GABAergic OK72-marked neurons did not affect learning in control flies (Fig. 3C), suggesting that normally they do not limit the process. Rather, activation of dAlk, a negative regulator of learning and memory (Gouzi et al., 2011, 2018) and Ras1 within these OK72-marked neurons may regulate GABA release modulating, but not suppressing, MB activation. We propose that hyperactivation of dAlk/Ras1-mediated signaling on dNf1 loss results in dysregulated, excessive, or persistent GABA levels, altering the excitatory to inhibitory balance of inputs to the MBs, thus impairing learning. Ongoing experiments address this hypothesis.
Significantly, our results demonstrate that constitutive activity of murine, or Drosophila PKA does not reverse the GABAergic excess of OK72 neurons (Fig. 4A,B) to rescue leaning in dNf1 null homozygotes. This strongly suggests that dNf1 loss does not result in decreased cAMP levels and consequently of endogenous PKA activity, at least in OK72 neurons. This conclusion is further supported by elevation of cAMP levels via rolipram-dependent Dnc phosphodiesterase inhibition (Kanellopoulos et al., 2012), which also did not ameliorate the deficient learning of dNf1E2 homozygotes (Fig. 3E). In genetic corroboration of the pharmacological results, broad expression in the fly CNS of constitutively active PKA also did not suppress the learning deficit of dNf1 nulls (Fig. 4C). Furthermore, acute activated PKA expression within adult MBs did not improve the deficient learning of the null mutants (Fig. 4D). These results argue against the notion that dNf1 loss precipitates a cAMP/PKA deficit within the fly CNS underlying the compromised associative leaning, in discord with suggestions from prior reports (Guo et al., 1997, 2000).
The collective evidence strongly supports the model whereby dNf1 is required within GABAergic OK72-marked neurons to directly or indirectly regulate the levels of inhibitory neurotransmission to the MBs. Loss of dNf1 therein presents excess GABA to the MBs, resulting in suppression of olfactory associative learning. However, Ras1 appears not to be hyperactivated only within GABAergic OK72-marked neurons of dNf1E2 nulls (Figs. 5E,G, 6B), but also in their MBs (Fig. 6C,D) and contributes to their deficient learning. This notion is supported by the constitutively active RasV12 transgene, which suppressed learning when acutely expressed in the MBs of control animals (Fig. 6G). Ras1 hyperactivation has been shown to result via PKCζ activation in suppression of Gαs and therefore reduced cAMP in vertebrate cultured neurons (Anastasaki and Gutmann, 2014), suggesting that this may also occur in the MBs of dNf1E2 homozygotes. Interestingly, an inhibitory role for Ras/Raf signaling for protein synthesis-dependent memory formation in MBs has been described recently (Noyes et al., 2020).
Importantly however, Ras1 activity in the MBs does not appear regulated by dNf1 because its abrogation therein did not phenocopy the learning deficits (Fig. 1B). In accord, dNf1 restoration within the MBs does not rescue the learning deficit of dNf1 nulls (Fig. 1A). It is then possible that dNf1-independent Ras1 hyperactivation within the MBs may result from excessive GABAergic activity, or the resultant imbalance of inhibitory to excitatory inputs into these neurons. Together, dNf1 loss appears to result in Ras1 hyperactivation within the MBs, which is not regulated by dNf1 within these neurons and could result in reduced cAMP levels. However, constitutively active PKA expression in the MBs (Fig. 4D), or rolipram treatment (Fig. 4E), did not reverse the deficient learning of the mutants. Indeed, strong activated catalytic PKA expression in dNf1E2 MBs appeared to reduce learning below that of null homozygotes (Fig. 4D). Collectively, these results indicate that GABAergic inhibition to the MBs on dNf1 loss must be relieved preceding, or concurrently with this putative MB-specific PKA elevation. Predictions and mechanistic aspects of the proposed model are currently under investigation.
Importantly, the learning defects of homozygotes for another null allele, dNf1P2, were reported reversed by acute expression of a constitutively active murine PKA subunit (hs-PKA*) expressed throughout the fly (Guo et al., 2000) and suggested to overcome the low cAMP levels in this mutant (Guo et al., 2000; Tong et al., 2002). The model proposed above offers reconciliation of our data and these reports noting the following. Reversal of the learning deficits of null dNf1 mutants with the constitutively active PKA has been obtained only under the heat shock promoter, which results in massive transgene expression throughout the fly, even without induction (Guo et al., 2000), probably because of position effects. The massive expression of the hs-PKA* transgene likely suffices to bypass the increased GABAergic inhibitory effects and elevate neuronal PKA activity within the MBs above a threshold that yields significant learning. Such levels of transgene induction are not likely in a tissue-limited manner and under Gal80ts, which appears to decrease UAS-driven transgene expression by ≥50% (Papanikolopoulou et al., 2018), under the conditions used herein. Although additional experiments are required to explore this hypothesis, it provides a putative resolution of a long-standing controversy, probably pertinent to vertebrate models where dopaminergic elevation of cAMP appears altered on Nf1 loss (Diggs-Andrews et al., 2013).
Footnotes
This work was supported in part by March of Dimes Grant 12FY012-189, the Strategic Development of the Biomedical Research Institute “Alexander Fleming” MIS 5002562 implemented under the Action for the Strategic Development on the Research and Technological Sector, funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and cofinanced by Greece and the European Union (European Regional Development Fund) and InfrafrontierGR (cofunded by Greece and the European Union, ERDF and NSRF 2014-2020) and DoD WS00321918. We would like to thank the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center and the Kyoto Drosophila Genetic Resource Center for stocks and Maro Loizou for expert technical help.
A. Moressis’ present address: Pharma Research and Early Development, Roche Innovation Center Basel, Switzerland.
The authors declare no competing financial interests.
References
- Acebes A, Ferrús A (2001) Increasing the number of synapses modifies olfactory perception in Drosophila. J Neurosci 21:6264–6273. 10.1523/JNEUROSCI.21-16-06264.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, Gutmann DH (2014) Neuronal NF1/RAS regulation of cyclic AMP requires atypical PKC activation. Hum Mol Genet 23:6712–6721. 10.1093/hmg/ddu389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EM, Chen PL, Palmer RH, Salecker I (2007) Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell 128:961–975. 10.1016/j.cell.2007.02.024 [DOI] [PubMed] [Google Scholar]
- Brown JA, Gianino SM, Gutmann DH (2010) Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J Neurosci 30:5579–5589. 10.1523/JNEUROSCI.3994-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan ME, Davis RL (2010) A distinct set of Drosophila brain neurons required for neurofibromatosis type 1-dependent learning and memory. J Neurosci 30:10135–10143. 10.1523/JNEUROSCI.0283-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, Chiang AS (2012) Visualizing long-term memory formation in two neurons of the Drosophila brain. Science 335:678–685. 10.1126/science.1212735 [DOI] [PubMed] [Google Scholar]
- Costa RM, Silva AJ (2002) Molecular and cellular mechanisms underlying the cognitive deficits associated with neurofibromatosis 1. J Child Neurol 17:622–626; discussion 627-629, 646-651. 10.1177/088307380201700813 [DOI] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ (2002) Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415:526–530. 10.1038/nature711 [DOI] [PubMed] [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ (2008) Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135:549–560. 10.1016/j.cell.2008.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (2005) Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci 28:275–302. 10.1146/annurev.neuro.28.061604.135651 [DOI] [PubMed] [Google Scholar]
- de Haro M, Al-Ramahi I, Benito-Sipos J, López-Arias B, Dorado B, Veenstra JA, Herrero P (2010) Detailed analysis of leucokinin-expressing neurons and their candidate functions in the Drosophila nervous system. Cell Tissue Res 339:321–336. 10.1007/s00441-009-0890-y [DOI] [PubMed] [Google Scholar]
- Devaud JM, Keane J, Ferrus A (2003) Blocking sensory inputs to identified antennal glomeruli selectively modifies odorant perception in Drosophila. J Neurobiol 56:1–12. 10.1002/neu.10216 [DOI] [PubMed] [Google Scholar]
- Diggs-Andrews KA, Gutmann DH (2013) Modeling cognitive dysfunction in neurofibromatosis-1. Trends Neurosci 36:237–247. 10.1016/j.tins.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggs-Andrews KA, Tokuda K, Izumi Y, Zorumski CF, Wozniak DF, Gutmann DH (2013) Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann Neurol 73:309–315. 10.1002/ana.23793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzi JY, Moog-Lutz C, Vigny M, Brunet-de Carvalho N (2005) Role of the subcellular localization of ALK tyrosine kinase domain in neuronal differentiation of PC12 cells. J Cell Sci 118:5811–5823. 10.1242/jcs.02695 [DOI] [PubMed] [Google Scholar]
- Gouzi JY, Moressis A, Walker JA, Apostolopoulou AA, Palmer RH, Bernards A, Skoulakis EM (2011) The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet 7:e1002281. 10.1371/journal.pgen.1002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzi JY, Bouraimi M, Roussou IG, Moressis A, Skoulakis EM (2018) The Drosophila receptor tyrosine kinase Alk constrains long-term memory formation. J Neurosci 38:7701–7712. 10.1523/JNEUROSCI.0784-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HF, The I, Hannan F, Bernards A, Zhong Y (1997) Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science 276:795–798. 10.1126/science.276.5313.795 [DOI] [PubMed] [Google Scholar]
- Guo HF, Tong J, Hannan F, Luo L, Zhong Y (2000) A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 403:895–898. 10.1038/35002593 [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL (2014) Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem 21:519–526. 10.1101/lm.034363.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan F, Ho I, Tong JJ, Zhu Y, Nurnberg P, Zhong Y (2006) Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum Mol Genet 15:1087–1098. 10.1093/hmg/ddl023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T (2018) What can tiny mushrooms in fruit flies tell us about learning and memory? Neurosci Res 129:8–16. 10.1016/j.neures.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Kanellopoulos AK, Semelidou O, Kotini AG, Anezaki M, Skoulakis EM (2012) Learning and memory deficits consequent to reduction of the fragile X mental retardation protein result from metabotropic glutamate receptor-mediated inhibition of cAMP signaling in Drosophila. J Neurosci 32:13111–13124. 10.1523/JNEUROSCI.1347-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Rubin GM (1998) Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125:1–9. 10.1242/dev.125.1.1 [DOI] [PubMed] [Google Scholar]
- King LB, Koch M, Murphy KR, Velazquez Y, Ja WW, Tomchik SM (2016) Neurofibromin loss of function drives excessive grooming in Drosophila. 6:1083–1093. 10.1534/g3.115.026484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S (2007) Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron 53:103–115. 10.1016/j.neuron.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SJ, Newby LM, Jackson FR (1994) Drosophila GABAergic systems: II. Mutational analysis of chromosomal segment 64AB, a region containing the glutamic acid decarboxylase gene. Mol Gen Genet 243:555–564. 10.1007/BF00284204 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22:451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Li W, Cui Y, Kushner SA, Brown RA, Jentsch JD, Frankland PW, Cannon TD, Silva AJ (2005) The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol 15:1961–1967. 10.1016/j.cub.2005.09.043 [DOI] [PubMed] [Google Scholar]
- Liu X, Davis RL (2009) The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat Neurosci 12:53–59. 10.1038/nn.2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Krause WC, Davis RL (2007) GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron 56:1090–1102. 10.1016/j.neuron.2007.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Buchanan ME, Han KA, Davis RL (2009) The GABAA receptor RDL suppresses the conditioned stimulus pathway for olfactory learning. J Neurosci 29:1573–1579. 10.1523/JNEUROSCI.4763-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E (1998) Regulatory control of signal transduction during morphogenesis in Drosophila. Int J Dev Biol 42:363–368. [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004:pl6. 10.1126/stke.2202004pl6 [DOI] [PubMed] [Google Scholar]
- Messaritou G, Grammenoudi S, Skoulakis EM (2010) Dimerization is essential for 14-3-3zeta stability and function in vivo. J Biol Chem 285:1692–1700. 10.1074/jbc.M109.045989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaritou G, Leptourgidou F, Franco M, Skoulakis EM (2009) A third functional isoform enriched in mushroom body neurons is encoded by the Drosophila 14-3-3zeta gene. FEBS Lett 583:2934–2938. 10.1016/j.febslet.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Molosh AI, Johnson PL, Spence JP, Arendt D, Federici LM, Bernabe C, Janasik SP, Segu ZM, Khanna R, Goswami C, Zhu W, Park SJ, Li L, Mechref YS, Clapp DW, Shekhar A (2014) Social learning and amygdala disruptions in Nf1 mice are rescued by blocking p21-activated kinase. Nat Neurosci 17:1583–1590. 10.1038/nn.3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moressis A, Friedrich AR, Pavlopoulos E, Davis RL, Skoulakis EM (2009) A dual role for the adaptor protein DRK in Drosophila olfactory learning and memory. J Neurosci 29:2611–2625. 10.1523/JNEUROSCI.3670-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix JS, Blakeley J, Rodriguez FJ (2020) An update on the central nervous system manifestations of neurofibromatosis type 1. Acta Neuropathol 139:625–641. 10.1007/s00401-019-02002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes N, Walkinshaw E, Davis R (2020) Ras acts as a molecular switch between two forms of consolidated memory in Drosophila. Proc Natl Acad Sci USA 117:2133–2139. 10.1073/pnas.1819925117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A, van der Vaart T, Mientjes E, van Woerden GM, Hojjati MR, Li KW, Gutmann DH, Levelt CN, Smit AB, Silva AJ, Kushner SA, Elgersma Y (2015) HCN channels are a novel therapeutic target for cognitive dysfunction in Neurofibromatosis type 1. Mol Psychiatry 20:1311–1321. 10.1038/mp.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C, Hallberg B (2009) Anaplastic lymphoma kinase: signalling in development and disease. Biochem J 420:345–361. 10.1042/BJ20090387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolopoulou K, Grammenoudi S, Samiotaki M, Skoulakis E (2018) Differential effects of 14-3-3 dimers on Tau phosphorylation, stability and toxicity in vivo. Hum Mol Genet 27:2244–2261. 10.1093/hmg/ddy129 [DOI] [PubMed] [Google Scholar]
- Shilyansky C, Lee YS, Silva AJ (2010a) Molecular and cellular mechanisms of learning disabilities: a focus on NF1. Annu Rev Neurosci 33:221–243. 10.1146/annurev-neuro-060909-153215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilyansky C, Karlsgodt KH, Cummings DM, Sidiropoulou K, Hardt M, James AS, Ehninger D, Bearden CE, Poirazi P, Jentsch JD, Cannon TD, Levine MS, Silva AJ (2010b) Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc Natl Acad Sci USA 107:13141–13146. 10.1073/pnas.1004829107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis EM, Davis RL (1996) Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron 17:931–944. 10.1016/S0896-6273(00)80224-X [DOI] [PubMed] [Google Scholar]
- Skoulakis EM, Kalderon D, Davis RL (1993) Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron 11:197–208. 10.1016/0896-6273(93)90178-T [DOI] [PubMed] [Google Scholar]
- Talay M, Richman EB, Snell NJ, Hartmann GG, Fisher JD, Sorkac A, Santoyo JF, Chou-Freed C, Nair N, Johnson M, Szymanski JR, Barnea G (2017) Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96:783–795.e784. 10.1016/j.neuron.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K (2008) Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol 508:711–755. 10.1002/cne.21692 [DOI] [PubMed] [Google Scholar]
- The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, Gusella JF, Hariharan IK, Bernards A (1997) Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science 276:791–794. 10.1126/science.276.5313.791 [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y (2002) Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci 5:95–96. 10.1038/nn792 [DOI] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG (2000) The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell 103:805–813. 10.1016/S0092-8674(00)00183-5 [DOI] [PubMed] [Google Scholar]
- Walker JA, Tchoudakova AV, McKenney PT, Brill S, Wu D, Cowley GS, Hariharan IK, Bernards A (2006) Reduced growth of Drosophila neurofibromatosis 1 mutants reflects a non-cell-autonomous requirement for GTPase-activating protein activity in larval neurons. Genes Dev 20:3311–3323. 10.1101/gad.1466806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Gouzi JY, Bernards A (2012) Drosophila: an invertebrate model of NF1. In: Neurofibromatosis type 1 molecular and cellular biology (Upadhyaya M, Cooper DN, eds), pp 523–534. New York: Springer. [Google Scholar]
- Walker JA, Gouzi JY, Long JB, Huang S, Maher RC, Xia H, Khalil K, Ray A, Van Vactor D, Bernards R, Bernards A (2013) Genetic and functional studies implicate synaptic overgrowth and ring gland cAMP/PKA signaling defects in the Drosophila melanogaster neurofibromatosis-1 growth deficiency. PLoS Genet 9:e1003958. 10.1371/journal.pgen.1003958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R (2003) Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112:271–282. 10.1016/S0092-8674(03)00004-7 [DOI] [PubMed] [Google Scholar]
- Weiss JB, Weber S, Marzulla T, Raber J (2017a) Pharmacological inhibition of Anaplastic Lymphoma Kinase rescues spatial memory impairments in Neurofibromatosis 1 mutant mice. Behav Brain Res 332:337–342. 10.1016/j.bbr.2017.06.024 [DOI] [PubMed] [Google Scholar]
- Weiss JB, Weber SJ, Torres ER, Marzulla T, Raber J (2017b) Genetic inhibition of Anaplastic Lymphoma Kinase rescues cognitive impairments in Neurofibromatosis 1 mutant mice. Behav Brain Res 321:148–156. 10.1016/j.bbr.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F (1990) The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63:835–841. 10.1016/0092-8674(90)90149-9 [DOI] [PubMed] [Google Scholar]
- Yap YS, McPherson JR, Ong CK, Rozen SG, Teh BT, Lee AS, Callen DF (2014) The NF1 gene revisited: from bench to bedside. Oncotarget 5:5873–5892. 10.18632/oncotarget.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]