Abstract
The ability of mammalian taste cells to respond to fatty acids (FAs) has garnered significant attention of late and has been proposed to represent a sixth primary taste. With few exceptions, studies on FA taste have centered exclusively on polyunsaturated FAs, most notably on linoleic acid. In the current study, we have identified an additional FA receptor, GPR84, in the gustatory system that responds to the medium-chain saturated FAs (MCFAs) in male mice. GPR84 ligands activate both Type II and Type III taste cells in calcium imaging and patch-clamp recording assays. MCFAs depolarize and lead to a rise in intracellular free [Ca2+] in mouse taste cells in a concentration-dependent fashion, and the relative ligand specificity in taste cells is consistent with the response profile of GPR84 expressed in a heterologous system. A systemic Gpr84−/− mouse model reveals a specific deficit in both the neural (via chorda tympani recording) and behavioral responses to administration of oral MCFAs compared with WT mice. Together, we show that the peripheral taste system can respond to an additional class of FAs, the saturated FAs, and that the cognate receptor necessary for this ability is GPR84.
Keywords: fat taste, GPR84, medium-chain fatty acids, saturated fatty acids, taste cells
Significance Statement
Recognition of fatty acids by taste receptor cells in the oral cavity has been suggested to underlie the taste cue for dietary fat. In vitro and in vivo studies in both animal models and humans supports the notion fat may represent an additional taste primer. However, virtually all these studies have focused on the ability of polyunsaturated fatty acids, such as linoleic acid, to activate taste pathways. We have identified GPR84 in the taste system that underlies the ability of mice to recognize and respond to medium chain saturated fatty acids. The ability to sense the taste of saturated fats may be important for controlling the intake of this class of fats whose intake is a greater concern for human health.
Introduction
Historically, the peripheral gustatory system was believed to respond to only five basic taste qualities corresponding to the human perceptions of salty, sour, sweet, bitter, and umami. Recent studies have demonstrated that the taste system responds to diverse compounds reflecting the taste of carbonation (Chandrashekar et al., 2009), astringency (Schiffman et al., 1992), pungency (Hata et al., 2012), and fat (Gilbertson et al., 1997), which do not fit neatly within the five broad taste classes.
Of these nonclassical tastants, fat has received significant attention because dietary fat intake associates with the current epidemic of obesity (Bray et al., 2004). Accumulating evidence has supported the idea of a “taste of fat” (Running et al., 2015) and that the prototypical fat stimuli are free fatty acids (FAs) (Gilbertson et al., 1997; Mattes, 2009). Gilbertson et al. (1997) were the first to show that cis-polyunsaturated FAs (cis-PUFAs) were able to activate native taste cells by acting as open channel blockers of delayed rectifying potassium (DRK) channels, so it was assumed that there were additional receptors upstream of DRK channels to initiate their voltage-dependent opening. The Cluster of Differentiation 36 (CD36) (Laugerette et al., 2005; Passilly-Degrace et al., 2009) and FA-activated GPCRs (Matsumura et al., 2009; Cartoni et al., 2010; Galindo et al., 2012) were identified in the peripheral taste system, where they may have overlapping roles (Ozdener et al., 2014), and together have been hypothesized to operate upstream of the FA-responsive DRK channels (Gilbertson and Khan, 2014).
GPCRs initiate the transduction events underlying sweet, bitter, and umami taste (Chaudhari and Roper, 2010). Support for the involvement of GPCR-based FA reception in taste system has come on several fronts. Two bottle taste preference tests have shown that knocking out GPR120 (FFA4) or GPR40 (FFAR1) significantly reduced the preference for linoleic acid in mice (Cartoni et al., 2010). GPR120 has been implicated in FA taste in a variety of species, including mice, chicken (Sawamura et al., 2015; Kawabata et al., 2018), and humans (Galindo et al., 2012). The GPR120-mediated FA transduction pathway in the mouse taste system encompasses the same elements found in other GPCR-mediated taste pathways, including activation of PLCβ2, IP3R3-mediated Ca2+ release from the endoplasmic reticulum, and activation of transient receptor potential channel type M5 (TrpM5) (Liu et al., 2011), which initiates the opening of FA-sensitive DRK channels.
With few exceptions, the search for a taste of fat has focused exclusively on cis-PUFAs. Little research has been done on saturated fats, despite the fact that they pose a greater threat to human health (Hamulka et al., 2018). Mattes and colleagues have shown that humans had detectable oral thresholds for several medium-chain saturated FAs (MCFAs) (Mattes, 2009; Running and Mattes, 2014; Running et al., 2015), although the mechanism underlying this ability was not elucidated. GPR84 is a novel GPCR expressed predominantly in hematopoietic tissues and can be activated by MCFAs of C9 to C14 in length (Wang et al., 2006). While the role of GPR84 in the immune system is clear, an additional role in nutrient sensing has been suggested (Ichimura et al., 2009).
In this study, we have found that GPR84 is expressed in native mouse taste cells, and the ability of mice to taste MCFAs is consistent with the ligand specificity and concentration dependence of heterologously expressed GPR84. Using a systemic Gpr84−/− mouse model, we have shown that MCFAs induce membrane depolarization and release of calcium in taste cells in a GPR84-dependent fashion. Recordings from the chorda tympani (CT) nerve reveal that MCFAs elicit concentration-dependent increases in neural activity that is lost specifically in Gpr84-deficient mice. Finally, behavioral assays reveal MCFAs act as a conditioned taste stimulus in WT mice, but not in mice lacking GPR84. Together, we propose that GPR84 is a taste receptor for MCFAs and the taste system can recognize saturated fats, revealing the taste of fat is broader than previously recognized.
Materials and Methods
Cell culture
A HEK293 cell line was designed to express GPR84 in an inducible fashion under control by the tetracycline promoter. It was constructed by and a generous gift of International Flavor and Fragrances. The specific constructs being used was an inducible Gpr84 + Gqi9 (a chimeric G-protein) (Wang et al., 2006). This cell line was cloned, validated by PCR/qPCR, and verified for function in FLIPR-based calcium assays. All cells were cultured in the cell culture medium DMEM with GlutaMax (Invitrogen) supplemented with 10% tetracycline-free FBS (Thermo Fisher Scientific); 10 µg/ml Blasticidin S HCl (Invitrogen) and 100 µg/ml Hygromycin B (Invitrogen) were added to the medium to select the antibiotic-resistant cells. To induce the expression of transfected Gpr84 receptor genes, 0.5 µg/ml doxycycline (Sigma Millipore) was added to the culture medium 24 h before experiments. Noninduced cells served as controls. Cells were plated on glass coverslip 2-4 h before calcium imaging.
Animals
The GPR84 KO strain (Gpr84−/−) was created by Taconic on a pure C57BL/6N background. It was made by incorporating a KO gene, in which the single exon was replaced by a LacZ/Neo cassette by homologous recombination. Gpr84−/− mice used in the experiments were generated by mating Gpr84+/− and Gpr84−/−mice, and subsequently identified by genotyping. The production of GFP-PLCβ2 and GFP-GAD67 strains has been described in detail previously (Chattopadhyaya et al., 2004; Kim et al., 2006). All experiments were performed on adult (2-6 months) male mice, and they were raised with water and normal mouse chow provided ad libitum in a room with 12 h:12 h day/night cycle. All procedures involving animals were approved by the Utah State University and University of Central Florida Institutional Animal Care and Use Committees and were performed in accordance with American Veterinary Medical Association guidelines.
Solutions
Tyrode’s buffer contained the following (in mm): 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES, 10 glucose, and 10 Na pyruvate; pH was adjusted to 7.40 with NaOH and osmolarity to 300-330 mOsm/L with NaCl, if necessary. Calcium-magnesium free Tyrode’s (Ca2+-Mg2+ free Tyrode’s) solution contained the following (in mm): 140 NaCl, 5 KCl, 2 BAPTA, 10 HEPES, 10 glucose, and 10 Na pyruvate; adjusted pH to 7.40 with NaOH and, as needed, osmolarity to 300-330 mOsm/L with NaCl. Enzyme mixture used for taste cell isolation contained collagenase I (1 mg/ml), dispase II (2.45 mg/ml), and trypsin inhibitor (1 mg/ml) in Tyrode’s buffer. Solution used to dilute fura-2 AM for calcium imaging on taste cells (HHP with 2% FBS) contained the following: 98% 1× HBSS, 1% HEPES, 1% Na pyruvate, and 2% heat-inactivated FBS. Intracellular solution used in whole-cell patch-clamp recording contained the following (in mm): 140 K gluconate, 1 CaCl2, 2 MgCl2, 10 HEPES, 11 EGTA, 3 ATP, and 0.5 GTP; pH was adjusted to 7.20 with KOH and osmolarity was adjusted to 305-315 mOsm/L with K gluconate. All the fatty acids (FAs) (sodium salt) were purchased from Sigma Millipore and were dissolved in Tyrode’s solution (cellular assays) or water (behavioral assays) to make test solutions. Capric acid (Sigma Millipore) used in nerve recording was dissolved in water and homogenized by vortex for 10 min.
Taste cell isolation
Taste cell isolation has been previously described in detail (Liu et al., 2011). Briefly, tongues were removed, rinsed, and then injected with enzyme mixture under the lingual epithelium. They were then incubated at room temperature in Tyrode’s solution with O2 bubbled for 45 min. The lingual epithelium was separated from the underlying muscle layer and pinned flat in a Sylgard-lined Petri dish, followed by incubation in Ca2+-Mg2+ free Tyrode’s solution for 10 min, and subsequently in the same enzyme mixture for another 2-7 min. Taste cells were isolated by gentle suction with a fire-polished pipette and placed on a glass coverslip or slide coated with Corning Cell-Tak Cell and Tissue Adhesive for calcium imaging or patch-clamp recording. The same protocol was used for RNA isolation, with the exception that tongues were bubbled in O2 with Ca2+-Mg2+ free Tyrode’s solution and taste buds and cells were expelled into a 0.5 microcentrifuge tube containing RNALater (Thermo Fisher Scientific) for each taste papillae before RNA isolation.
Real-time PCR
RNA extraction and analysis were performed by RNeasy Plus Micro Kit (QIAGEN) and RNA integrity was assessed using Experion RNA HighSens Analysis Kit (Bio-Rad), and first-strand cDNA was synthesized by iScript cDNA Synthesis Kit (Bio-Rad). All the procedures above were performed according to the manufacturers’ protocols. Commercially available TaqMan Gene Expression Assays were then used to detect the expression of GPR84. Real-time PCR analyses were performed on the Applied Biosystems QuantStudio three Real-Time PCR Systems (Thermo Fisher Scientific) according to manufacturer’s instructions using TaqMan protocols. Final reaction cocktail (25 µl) contains the following: 1× TaqMan Fast MasterMix, 0.2 mm dNTPs, 2.5 mm Mg2+, 0.2 µm forward and reverse primers for GAPDH, 0.2 µm GAPDH probe, 1× TaqMan gene expression mix for the genes tested (mouse GPR84: assay ID Mm02620530_s1; mouse Gustducine: assay IDMm011653313_m1; mouse Trpm5: assay ID Mm01129032_m1; Thermo Fisher Scientific), and 1.5 µl template cDNA (Thermo Fisher Scientific). The GAPDH probe was VIC-labeled, and the GPR84 probe was FAM-labeled. The expression levels of GPR84 from fungiform and circumvallate were normalized to the expression level of the housekeeping gene GAPDH within the same sample by the equation: ΔCT(GPR84) = CT(GPR84)– CT(GAPDH). CT was defined as the number of cycles required for the fluorescence signal to cross 30 fluorescent units. Relative gene expression of Gpr84 was normalized to Trpm5 expression in fungiform papillae by the equation ΔΔCT = ΔCT(Gpr84) – ΔCT(Trpm5). Finally, the ΔΔCT was exponentially transformed to represent the relative expression (R) to the calibrator: .
Calcium imaging
The procedure of calcium imaging has been described in detail in previous studies (Liu et al., 2011; Shah et al., 2012). Cell lines were loaded with 4 µm of the ratiometric intracellular calcium indicator fura-2 AM (Invitrogen) in Tyrode’s with 0.25% pluronic acid (Thermo Fisher Scientific) solution, and incubated at 37°C for 45 min in the cell incubator. Isolated taste cells on coverslips were loaded with the fura-2 AM in HHP buffer with 2% FBS for 1 h at room temperature in the dark. The coverslips were then mounted onto an imaging chamber (RC-25F or RC-26Z, Warner Instruments) and visualized on an inverted microscope (Nikon TE-100). The excitation wavelengths of 340 and 380 nm were generated by a fast-switching monochromator (Bentham FSM150, Intracellular Imaging) to excite fura-2. Each cell was outlined, and the fluorescence in the outlined area was recorded by an imaging software IncytIm2 (Intracellular Imaging) through a CCD camera linked to the microscope. A standard curve of the ratio of fluorescence (340 nm/380 nm) versus calcium concentration was calibrated beforehand using a Calcium Calibration Buffer Kit (Invitrogen) with pentapotassium salt of fura-2 as the indicator. During calcium imaging, the calcium concentration was directly calculated by the calibration curve from the ratio of fluorescence. MCFA solutions were applied by a bath perfusion system at a flow rate of 4 ml/min for 2.5 min, followed by 1 min of 0.1% FA free BSA in Tyrode’s solution and then regular Tyrode’s solution to wash away the FAs until the calcium signal returned to the baseline. The criterion for responsive cells is their calcium peak amplitude was at least 8 times the SD of baseline prestimulus interval. The calcium response was quantified by the area under the curve (AUC), which was calculated with Gaussian multipeak function in Origin 7/8 (OriginLab). The calcium response between the taste cells from WT and Gpr84−/− mice were compared by an unpaired two-tailed Student’s t test, and significance was set at α = 0.05. To generate the dose–response curves for lauric and capric acids on HEK293 cells with induced GPR84 receptors, a series of lauric or capric acid at concentrations of 1, 3, 10, 30, and 100 µm were applied to the cells, and AUC was calculated as the calcium response. Dose–response curves were fitted using Prism software (GraphPad Software) to the following equation:
where Ymax and Ymin are plateaued in the units of the y axis.
Whole-cell patch-clamp recording
An Axopatch-200B amplifier (Molecular Devices) in current-clamp mode was used to record the capric acid-induced membrane potential change in taste cells. Borosilicate patch pipettes were pulled by a Sutter P-97 puller (Sutter Instruments) and fire polished by a microforge (model MF-9; Narishige). The resistances of the pipettes were 4-11 mΩ. pCLAMP software (version 10, Molecular Devices) was used to deliver commands and collect data, and it was interfaced to the amplifier with a Digidata 1344 A/D digitizer (Molecular Devices). Data were collected at 2-5 kHz and filtered online at 1 kHz. To record the membrane potential, a seal with the resistance higher than 1 GΩ was needed between the patch pipette and the cell membrane, and then more suction was applied to rupture the membrane patch to reach a whole-cell mode. The membrane potentials were recorded while holding the cell at its zero-current level, and capric acid-induced currents were recorded at a holding potential of −100 mV. Focal application of capric acid was delivered by a pipette positioned near the cell and controlled by PicoSpritzer III (Parker Hannifin). The membrane potentials and currents induced by capric acid recorded in WT and Gpr84−/− taste cells were compared by an unpaired, two-tailed Student’s t test, and significance was set at α = 0.05.
Nerve recording
Mice were anesthetized with urethane (∼2 g/kg, i.p.) and tracheostomized to facilitate breathing. A ventral approach was used to expose the CT, which was cut near the tympanic bulla and placed on a platinum-iridium electrode (A-M Systems) for recording. A reference electrode was placed in the nearby tissue. The anterior tongue was stimulated with different tastants, including NaCl (30 mm), monosodium glutamate (10 mm), denatonium benzoate (1 mm), HCl (1 mm), Steviose (25 mm), and capric acid (0.04, 0.1, 1, and 5 mm). The tastants, with the exception of capric acid, were applied for 15 s and rinsed with water for 30-45 s, or until the signal returned to baseline. Capric acid was applied for 30 s because of the lower magnitude nerve response and its slightly slower onset compared with other tastants. Nerve responses were amplified by model 3000 AC/DC differential amplifier (A-M Systems) and collected with DataWave Technologies SciWorks Experimenter software (DataWave Technologies). NH4Cl (100 mm) was applied at the start and the end of each experiment, and the nerve response of each tastant (15 s) was normalized to the average NH4Cl response (15 s) within each session.
Conditioned taste aversion (CTA)
Details of the CTA assay have been described in previous studies (Pittman et al., 2008; Liu et al., 2011). Briefly, mice were placed on a 23.5 h water restriction schedule for the duration of the CTA assay starting at 24 h before water training, where they were given 30 min access to water each day. This level of water deprivation caused maximal lick rates (∼5 licks/s) to water and other preferred (nonaversive) taste stimuli on testing days. Water training and testing were both performed in an MS-160 Davis Rig gustatory behavioral apparatus (Med Associates). First, mice were trained with water for 3-4 d until they exhibit stable numbers of licks for each of the presentations. Taste aversions were then initiated by pairing the conditioned stimulus (CS) and the unconditioned stimulus (US) over the next 3 consecutive days. Five minutes of free access of capric acid (600 µm) were given as the CS, and then the same CS was given orally with syringes right before 150 mm of LiCl (or 150 mm NaCl for the control group) was injected intraperitoneally (20 ml/kg body weight dosage) as the US, which could induce gastric distress, with the signs including lethargy, lying in an extended prone position, and stopping exploration in the cage. On testing days, a fan was placed near the animal chamber to provide constant airflow along the longitudinal axis of the stimulus delivery tray to reduce the olfactory effect of the FAs. In addition to water, the test solutions included capric acid (10, 30, 100, 300, and 600 µm), linoleic acid (100 µm), undecanoic acid (300 µm), lauric acid (300 µm), sucrose (100 mm), denatonium benzoate (2 mm), and NaCl (100 mm). The sequences of the tastants were arranged randomly, and the test session included 2 blocks of 12 presentations with shutter opening for 5 s, and the wait times for the first lick were 150 s. The total number of licks per stimulus was averaged between the 2 trials and normalized relative to the lick numbers to water presentations. Trials with zero licks, while infrequent, were removed from the analysis. The differences between LiCl and NaCl-injected groups within each tastant were compared by an unpaired, two-tailed Student’s t test and significance was set at α = 0.05.
Experimental design and statistical analysis
Calcium imaging data analyses, based on the AUC and analyzed in Origin 9.6 (OriginLab) and GraphPad Prism, version 8.0 (GraphPad Software), were used to perform statistical analyses. All data were obtained from experiments run at least 3 times from multiple samples and are shown as mean ± SEM. Generally, statistical analysis was performed using Student’s t test and two-way ANOVA. Asterisks represent significant differences. All error bars indicate SE measurement. In our experience, false positive responses caused by artifacts related to switching solutions are uncommon with calcium imaging, patch-clamp recording, and nerve recording. If any unusual responses related to solutions changes are seen, they would be evident on changing both to and from stimuli in a similar fashion and these data would not be used for subsequent analyses.
Results
GPR84 is expressed in taste cells
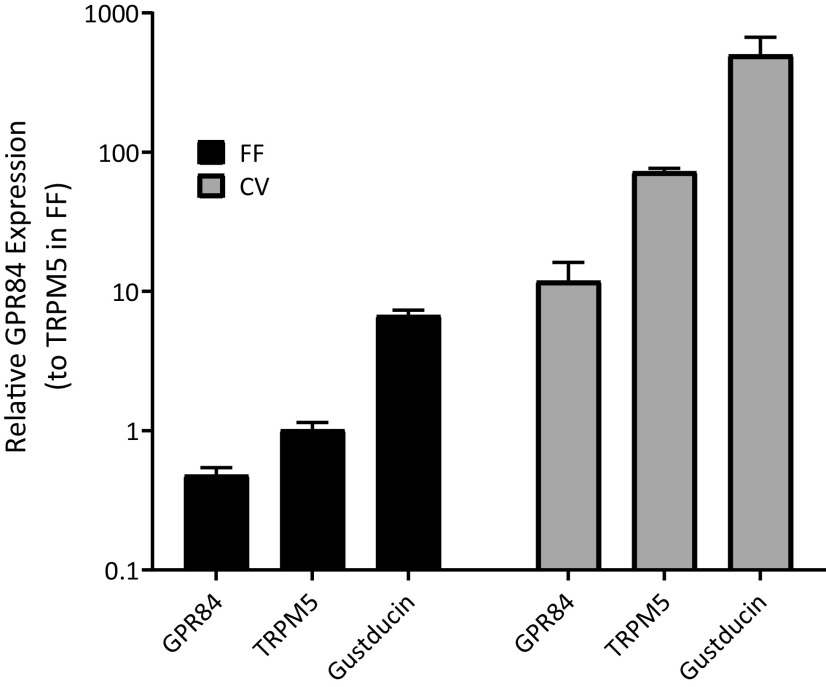
Previous studies have shown that GPR84 is the receptor for MCFAs (Venkataraman and Kuo, 2005; Wang et al., 2006). To determine whether GPR84 is the MCFA receptor in taste cells, the mRNA expression of Gpr84 in taste cells isolated from fungiform and circumvallate papillae were examined separately by qPCR. Trpm5 and gustducin (Gnat3), genes with high abundance in taste cells (Kaske et al., 2007; Liu et al., 2011), were used to compare the expression of Gpr84 in taste cells (Fig. 1). The mRNA of Gpr84 expressed significantly in both fungiform and circumvallate taste papillae, with the higher expression in circumvallate than fungiform papillae.
The relative mRNA expression of Gpr84 in the taste cells from fungiform and circumvallate papillae (n = 3 replicates, where n = 4 or 5 mice for each sample). Data are expressed relative to the expression of Trpm5 in the fungiform papilla (calibrator) as described in the text.
Ligand specificity of GPR84
As a means to compare the specificity and concentration-response functions for FAs in the taste system with GPR84, calcium imaging was performed to characterize GPR84 in a cell line that has been designed to express this receptor in an inducible fashion under control by the tetracycline promoter. By using functional calcium imaging, the intracellular calcium rise stimulated by a variety of FAs was recorded in HEK293 cells with induced GPR84 receptors and Gqi9. The results showed that all MCFAs, caproic (C6:0, Fig. 2A), caprylic (C8:0, Fig. 2B), capric (C10:0, Fig. 2C), undecanoic (C11:0, Fig. 2D), and lauric (C12:0, Fig. 2E), elicited a robust and reversible calcium increase in the cells induced to express GPR84 receptors as opposed to the noninduced control cells where there were no calcium responses. All MCFAs elicitied a similar response profile with a rapid rising phase followed by adaptation despite the continued presence of the MCFA. Interestingly, one additional long chain saturated FA (SFA), arachidic acid (C20:0), was also able to elicit a calcium response from the engineered HEK cell line (Fig. 2F).
Figure 2.
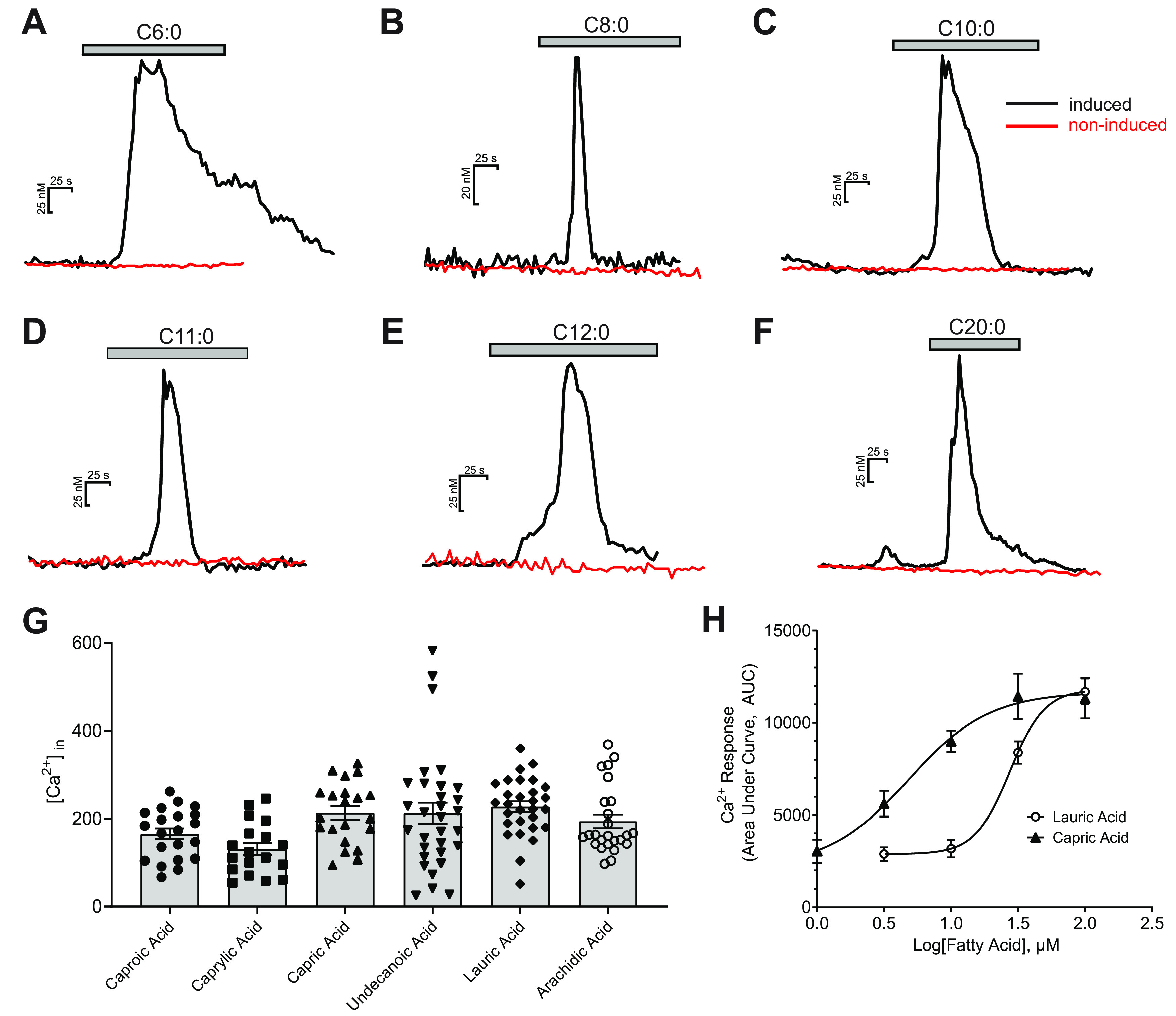
Ligand specificity of GPR84 receptor. Ligand specificity was investigated by functional calcium imaging in HEK293 cell line with inducible GPR84 receptors. Bath application of caproic (C6:0, A), caprylic acid (C8:0, B), capric acid (C10:0, C), undecanoic acid (C11:0, D), lauric acid (C12:0, E), and arachidic acid (C20:0, F) all elicited a robust and reversible calcium response in the cells with induced GPR84 receptors, while they did not elicit any calcium increase in the noninduced control cells (red lines). G, The individual and mean ± SEM calcium responses of GPR84-HEK293 cells to the FAs above (caproic 165.5 ± 12.3 nm, caprylic acid 130.8 ± 14.0 nm, capric acid 213.0 ± 15.2 nm, undecanoic acid 212.5 ± 24.1 nm, lauric acid 227.5 ± 12.2 nm, and arachidic acid 193.6 ± 15.4 nm). Noninduced cells showed no measurable changes in intracellular calcium. H, Dose–response curves of lauric and capric acid. The EC50 values for lauric and capric acids were 27.4 ± 1.1 μm (n = 53-81 cells per point) and 4.4 ± 1.3 μm (n = 17-55 cells per point), respectively.
Although not significant (Fig. 2G), MCFAs between 10 and 12 carbons showed higher average calcium responses. Capric and lauric acids are more common in food products, so they were used as representative MCFAs throughout the majority of this study. To characterize the ligand sensitivity of GPR84, the concentration-response functions for activation of GPR84 by lauric and capric acids were investigated using ratiometric calcium imaging. Figure 2H shows the concentration-response plots measured from the cells with induced GPR84 receptors in response to a series of lauric (C12:0) or capric acid (C10:0) at concentrations ranging from 1 to 100 µm. The EC50 values for lauric and capric acid were 27.4 ± 1.1 µm (n = 53-81) and 4.4 ± 1.3 µm (n = 17-55), respectively.
MCFAs activated mouse taste cells
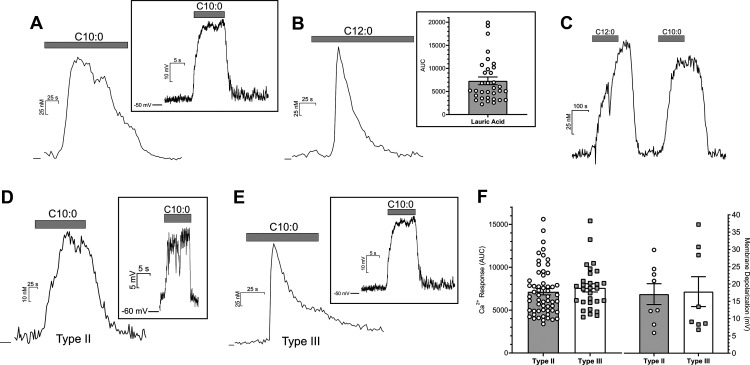
To explore the ability of mouse taste cells to respond to MCFAs, two different cellular assays were used with capric acid (C10:0) and lauric acid (C12:0) as representative MCFA stimuli. Intracellular calcium concentration change is a reliable indicator of cell activity, so functional ratiometric calcium imaging using fura-2 AM was conducted on single taste cells from WT mice, which were stimulated with capric acid or lauric acid. Bath application of capric and lauric acid (100 µm) both induced a robust intracellular Ca2+ rise in single taste cells (Fig. 3A and Fig. 3B, respectively), and they were able to stimulate the same taste cell in a similar fashion (Fig. 3C); ∼8.3% of all taste cells responded to capric acid in the calcium imaging experiments (n = 314 cells). Figure 3B (inset) shows the range of measurable calcium responses in response to lauric acid (C12:0) in isolated taste cells. To verify the calcium imaging results, whole-cell patch-clamp recording was performed on mouse taste cells. Capric acid (200 µm) was applied locally with a stimulus pipette positioned near the isolated taste cells, where it elicited reversible cell membrane depolarization (Fig. 3A, inset). Both calcium imaging and patch-clamp recording experiments were consistent with the interpretation that MCFAs were able to activate native mouse taste cells.
Figure 3.
Taste cell responses induced by MCFAs. Bath application of capric acid (C10:0; 100 μm, A) and lauric acid (C12:0; 100 μm, B) elicited intracellular calcium rises in mouse taste cells. B, Inset, The range of calcium responses seen in taste cells to lauric acid. Focal application of capric acid (200 μm) induced a membrane depolarization in taste cells (A, inset). Lauric acid and capric acid induced similar calcium responses in the same taste cells (C). Cells were held at zero current level to measure membrane potential in current-clamp mode. MCFAs also induced robust calcium increases and membrane depolarization (insets) in GFP-positive taste cells from both GFP-PLCβ2 (Type II cells; D) and GFP-GAD67 mice (Type III cells; E). F, both Type II and Type III cells show similar calcium response magnitudes and degree of membrane depolarization in response to a mixture of capric acid (100 μm) and lauric acid (100 μm). Data presented show only those cells that met the criterion for a responsive cell.
Additional experiments were conducted to determine the subtypes of taste cells that respond to MCFAs. Both functional calcium imaging and patch-clamp recording were performed using the isolated GFP-labeled taste cells from mice expressing a Type II (Receptor) cell marker (PLCβ2) and/or a Type III (Presynaptic) cell marker (GAD67). In calcium imaging, the criterion used to determine a calcium response was its reversibility and with a peak amplitude that was at least 10 times the variance of the prestimulus interval. MCFAs (100 µm of lauric and capric acids mixture) elicited calcium responses in 37.3% (62 of 166) GFP-positive taste cells from GFP-PLCβ2 mice (Fig. 3D,F), and in 55.8% (29 of 52) GFP-positive taste cells from GFP-GAD67 mice (Fig. 3E,F), which suggested that MCFAs could activate both Type II and Type III taste cells in mice, and the elicited calcium responses were similar in magnitude in both types (Fig. 3F). As illustrated in Figure 3D, E, MCFA responses in Type II cells appeared consistently slower in onset and in adaptation time course than those from Type III cells. The latter were qualitatively more similar to the calcium responses in HEK cells expressing GPR84 (Fig. 2). We have not characterized systematically these differences in the present study.
To verify the ability of MCFAs to activate both cell types, patch-clamp recording was also performed on the taste cells from both GFP-labeled strains. Consistent with the calcium imaging data, capric acid induced reversible cell membrane depolarization in the GFP-labeled taste cells from both strains of mice (Fig. 3D,E, insets). On average, 200 µm capric acid induced a 17.1 ± 3.0 mV (n = 8) depolarization from rest in Type II cells and a 17.8 ± 4.3 mV (n = 8) depolarization in Type III cells (Fig. 3F). Unlike timing differences in the MCFA-induced rise in calcium, no timing differences in membrane potential changes were noted between Type II and Type III cells. Together, the cell-based assays were consistent with both Type II and Type III taste cells being MCFA-sensitive.
MCFA-induced calcium responses and membrane depolarization were reduced in Gpr84−/− taste cells
To study the functional role of GPR84 in MCFA taste transduction, we used a transgenic mouse model that is Gpr84-deficient (Gpr84−/−). This homozygous global KO model was viable and appeared qualitatively normal in terms of weight gain and percent body fat under normal mouse chow over a 20 week period. A similar phenotype was observed in previous studies using similar KO models (Venkataraman and Kuo, 2005; Du Toit et al., 2018). Taste cells were isolated from Gpr84−/− mice, and both functional calcium imaging and current-clamp recording were used to characterize the taste cell responses to a prototypical MCFA, capric acid. In current-clamp recording, capric acid was rapidly applied via a stimulus pipette positioned near the cells, and it induced a rapid membrane depolarization in WT taste cells (Fig. 4A; 18.6 ± 5.4 mV, n = 12). This depolarization was significantly reduced (t(14) = 5.32, p = 0.0001, unpaired t test) in Gpr84−/− taste cells (Fig. 4A,C; 2.6 ± 0.9 mV, n = 8). The resting membrane potential for Gpr84−/− mice was −54.7 ± (−19.3) mV (n = 8), which was not significantly different from WT mice (−45.8 ± (−13.2) mV, n = 12, t(18) = 1.77, p = 0.093, unpaired t test). In calcium imaging, the percentage of the responsive taste cells was calculated for each mouse type (Fig. 4D), and Gpr84−/− mice showed a significantly lower percentage of taste cells (1.0 ± 0.6%, n = 14) that responded to capric acid than WT mice (8.5 ± 1.2%, t(22) = 6.318, n = 10, p = 0.0001, unpaired t test). And consistent with the electrophysiology data, calcium imaging showed a significantly smaller intracellular calcium increase in Gpr84−/− taste cells elicited by capric acid (AUC: 3060 ± 542, t(33) = 2.92, p = 0.006, unpaired t test) compared with the responses in WT taste cells (Fig. 4B,C; AUC: 16,777 ± 2728).
Figure 4.
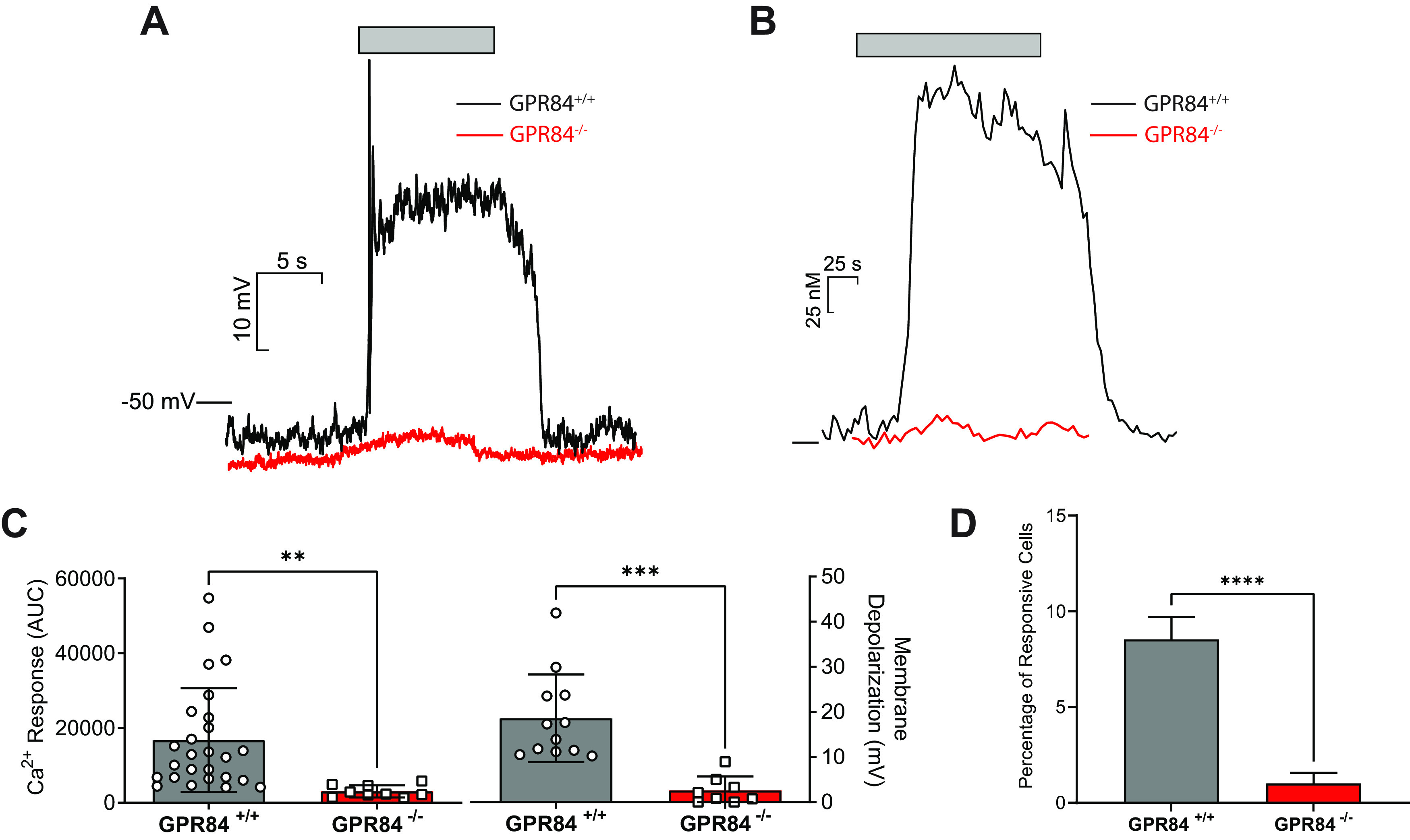
The membrane depolarization and intracellular calcium responses induced by MCFAs in Gpr84−/− taste cells were significantly lower than WT taste cells. Focal application of capric acid (CA, 200 μm) induced a membrane depolarization in WT taste cells, and this depolarization was significantly reduced in Gpr84−/− taste cells (A). The intracellular calcium rises elicited by MCFAs in Gpr84−/− taste cells were significantly lower than WT taste cells. Capric acid (100 μm) induced an intracellular calcium increase in WT taste cells, and it was reduced in Gpr84−/− taste cells (B). The calcium response as measured by AUC (C, left) and membrane depolarization (C, right) of these two groups were calculated (mean ± SEM) and compared via an unpaired two-tailed Student’s t test. The percentage of the responsive taste cells in calcium imaging was calculated for each mouse in WT and Gpr84−/− groups and compared via an unpaired two-tailed Student’s t test (D). Imaging data presented show only those cells that met the criterion for a responsive cell. **p < 0.01. ***p < 0.001. ****p < 0.0001.
Gpr84−/− mice showed significantly reduced taste nerve response to MCFAs
To test whether GPR84 contributes specifically to the MCFA taste responses, whole-nerve recordings were performed on CT nerves isolated from WT and GPR84−/− mice while stimulating the tongue with various tastants. CT nerve recordings from WT mice showed dose-dependent responses to different concentrations of capric acid (0.04, 0.1, 1, and 5 mm; Fig. 5A), suggesting that the capric acid taste signal could be transmitted from taste cells to taste nerve fibers. Two-way ANOVA showed that genotype had a significant effect on nerve response (F(3,39) = 10.92, p < 0.0001). In GPR84−/− mice, the CT nerve response was significantly reduced when stimulated with capric acid with concentrations higher than 1 mm compared with WT mice (Fig. 5C,E; 1 mm CA: p = 0.030; 5 mm CA: p = 0.002, unpaired t test), while the nerve responses to sweet, salty, bitter, umami, and sour were similar in the absence or presence of GPR84 (Fig. 5B,D,F).
Figure 5.
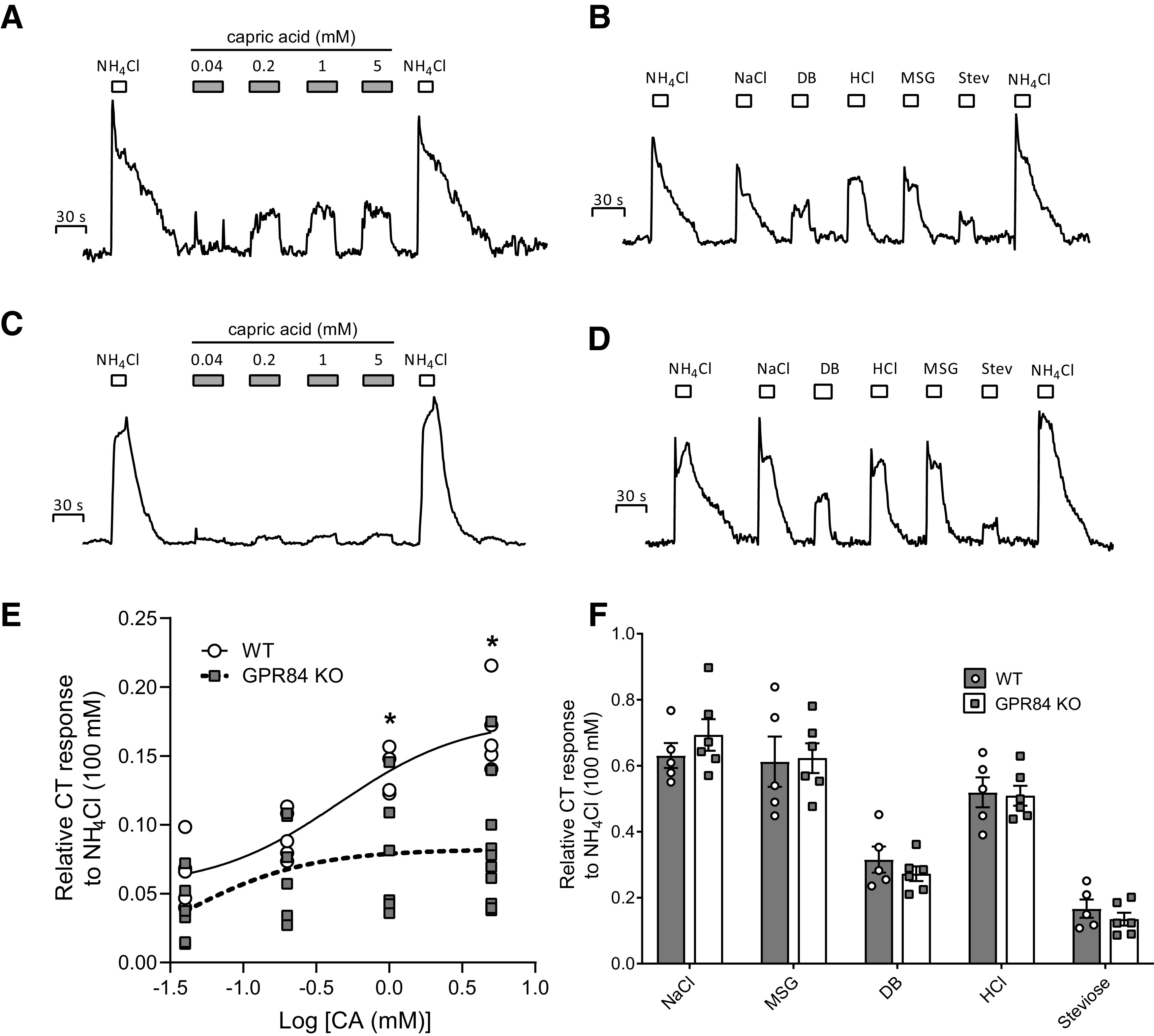
Integrated CT nerve responses to different concentrations of capric acid (30 s) (A,C) and various tastants (15 s) in WT mice and Gpr84−/− mice (B and D, respectively). NH4Cl (100 mm) was applied (15 s) before and after the session as a reference. Bars represent the duration of the stimulus. AUC of the integrated response for each tastant in WT and Gpr84−/− mice was calculated and compared via unpaired two-tailed Student’s t tests. Responses were normalized to the response to 100 mm NH4Cl. Responses show the mean ± SEM (n = 5-10 mice). No significant differences were observed between the WT (black bars) and KO (open bars) mice (F) when stimulated with NaCl (30 mm), MSG (10 mm), DB (1 mm), HCl (1 mm), and Steviose (25 mm); however, the nerve response to capric acid was significantly decreased at concentrations of 1 mm (t(8) = 2.64, p = 0.030, unpaired t test) and 5 mm (t(13) = 3.79, p = 0.002, unpaired t test) in GPR84−/− mice (E). *p < 0.05, Significant differences between two groups.
Mice lacking Gpr84−/− showed reduced taste responsiveness to MCFAs
To determine the role of GPR84 in MCFA taste transduction in vivo, CTA assays were performed on both WT and Gpr84−/− mice, and then taste responsiveness was compared between these two groups. Mice were injected with either LiCl or NaCl (controls) immediately following a single exposure to 600 µm of lauric acid. The results of the CTA assays showed that LiCl-injected WT mice significantly avoided lauric acid at concentrations higher than 30 µm (Fig. 6A), which suggested that lauric acid was an effective taste stimulus in vivo (30 µm: t(13) = 2.51, p = 0.026; 100 µm: t(13) = 2.97, p = 0.011; 300 µm: t(13) = 9.19, p < 0.001; 600 µm: t(13) = 4.26, p < 0.001; unpaired t test for all comparisons). The taste aversion to lauric acid had generalized to both capric and undecanoic acid (Fig. 6B) as expected since they all were known ligands for GPR84 (compare Fig. 2). There was no cross generalization to other GPCR-mediated taste stimuli, such as sweet, bitter, or linoleic acid (Fig. 6B). Gpr84−/− mice, however, did not show any significant aversions for lauric acid with concentrations up to 300 µm (Fig. 6C), and there were no significant aversions to other tastants tested, including capric and undecanoic acids (Fig. 6D). The results of the behavior studies suggested that the lack of GPR84 receptors significantly reduced the taste responsiveness of mice to MCFAs.
Figure 6.
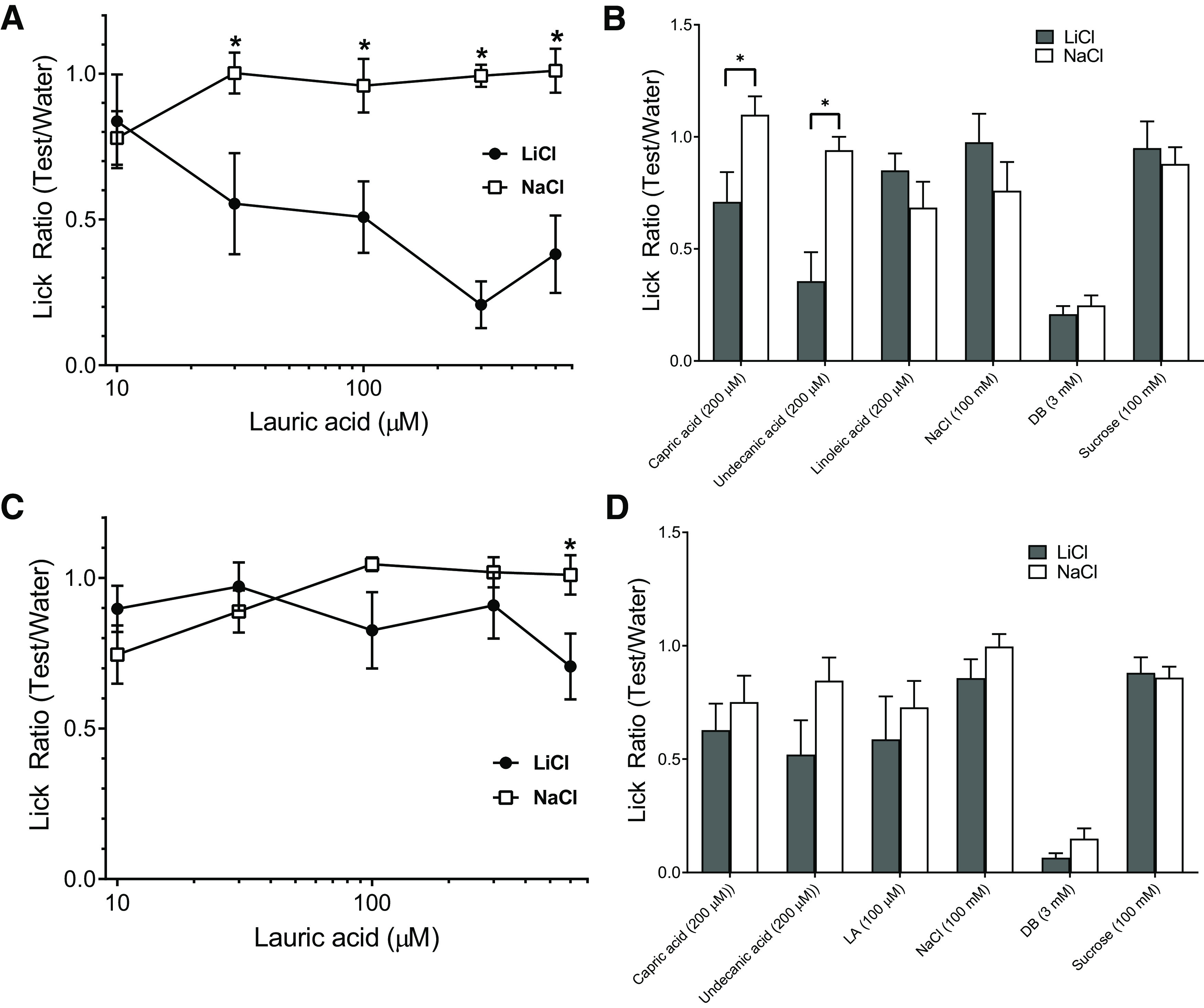
WT mice were able to detect MCFAs, and Gpr84−/− mice showed lower taste sensitivities to MCFAs than WT mice. A, Lick ratios (mean ± SEM) for different concentrations of lauric acid (10, 30, 100, 300, 600 μm) in WT mice on day 2 after generation of a CTA to lauric acid (n = 7 for LiCl group, and n = 8 for NaCl group). C, Gpr84−/− male mice (n = 7 for LiCl group, and n = 9 for NaCl group) on day 2 after CTA. *p < 0.05, significant differences between two groups (unpaired two-tailed Student’s t test). B, D, Lick ratios (mean ± SEM) for other MCFAs and GPCR-mediated taste stimuli in WT mice and Gpr84−/− mice, respectively.
Discussion
FA signaling has been of considerable interest of late because of its important role in normal physiology (Calder, 2015a,b; Miyamoto et al., 2016) and its aberrant signaling related to a number of diseases (Bazinet and Laye, 2014; Ghosh et al., 2017; Yazici and Sezer, 2017; Bessone et al., 2019). As such, there has been effort made toward the identification of functional receptors and transport proteins that underlie the physiological actions of free FAs. One of the main classes of FA-sensitive proteins are the GPCRs that exist in a wide variety of fat sensitive tissues. These GPCRs respond to a variety of different FA classes, including members of the polyunsaturated and monounsaturated FAs (GPR120; GPR40), the MCFAs (GPR84), and the short-chain saturated FAs (GPR41, GPR43) (Yonezawa et al., 2013).
It has been almost two decades since the ability of free FAs to activate taste cells was initially identified (Gilbertson et al., 1997); and with few exceptions, subsequent research has focused on the long-chain PUFAs, especially linoleic acid, in studies related to the taste properties of free FAs. However, except for the few reports have shown that MCFAs are detectable in humans (Mattes, 2009; Running and Mattes, 2014; Mouillot et al., 2019), no other studies have reported data on the effects of MCFAs in the mammalian taste system, despite the general consensus that the Western diet includes other forms of FAs, such as MCFAs and short-chain saturated FAs, and it is particularly important to limit saturated fat intake (German and Dillard, 2004). To help fill this void, we attempted to determine whether, like the PUFAs, SFAs could activate the mammalian taste system and elicit behavioral responses consistent with these compounds having their own taste quality.
The current study has shown that MCFAs are able to activate mouse taste cells in vitro and elicit concentration-dependent taste nerve responses in vivo. MCFAs are also effective taste stimuli in vivo as they generated CTAs in mice that did not significantly cross generalize to other tastants, including PUFAs. Moreover, mRNA of GPR84 is expressed in mouse taste cells, and GPR84 functioned as a receptor for SFAs from C6 to C12. A Gpr84−/− mouse model was used, and cellular, neural, and behavioral assays were all consistent with the interpretation that GPR84 was critical in gustatory recognition of MCFAs.
Our metrics for taste cell activation in the present study were twofold. We used both ratiometric calcium imaging and patch-clamp recording to determine whether MCFAs could lead to intracellular calcium rise and cell depolarization, respectively. The calcium responses induced by MCFAs were similar with those produced by linoleic acid in respect to amplitude and were found in a subset of Type II and Type III cells (Liu et al., 2011). The EC50 values for lauric acid and capric acid were also comparable to earlier work (Wang et al., 2006) in the GPR84-expressing HEK cells. We confirmed that the GPR84 ligands include MCFAs from C6 to C12 and included C20, the latter of which is unexpected based on previous results. This may represent the different nature of the assays used to determine ligand specificity of GPR84. The time course of calcium changes induced by MCFAs was qualitatively different from those elicited by PUFAs in chemosensory cells (Liu et al., 2011; Shah et al., 2012). Responses to MCFAs in the present study show a more rapid onset and a significant degree of adaptation (compare Fig. 3), a feature shared by calcium responses in HEK cells expressing GPR84 (compare Fig. 2). PUFA-elicited calcium responses in mouse taste cells by contrast showed a much slower onset (rising phase) and generally did not show adaptation. The membrane depolarization elicited by MCFAs were generally smaller than those attributed to linoleic acid in taste cells (Liu et al., 2011) and in enteroendocrine cells (Shah et al., 2012). These differences may reflect the fact that MCFAs activate taste cells via a different pathway than PUFAs or activate a unique subset of taste cells entirely. There were obvious differences between Type II and Type III cells in the timing of the MCFA-induced intracellular calcium rise (compare Fig. 3D,E). Broadly speaking, Type II cells had a slower onset and longer adaptation time course than Type III cells. Presently, we do not understand the nature of these differences, but it may reflect differences in the MCFA transduction pathway(s) between the two cell types. In the current study, it was beyond the scope to elucidate the GPR84-MCFA transduction pathway or to determine the cell specificity to different FA classes. As mentioned, GPR84 in other systems operates via a pertussis toxin-sensitive Gi/o pathway, and net reduction in cAMP (Wei et al., 2017). Additional experiments will be required to determine whether this is the case in the taste system as well. Further, PUFAs, but not MCFAs, have been shown to directly inhibit DRK channels in taste cells (Gilbertson et al., 1997), leading to a prolonged and enhanced depolarization.
Both indicators of cell activity, the MCFA-induced rise in intracellular calcium and membrane depolarization, were dependent on the expression of GPR84. Loss of GPR84 in our global KO model resulted in a greatly reduced ability of taste cells to respond to MCFAs. As expected, knocking out of GPR84 significantly diminished the taste cell responses activated by capric acid. However, there were still small residual responses in both intracellular calcium rise (Fig. 4B) and membrane depolarization (Fig. 4A), suggesting that an additional unidentified receptor(s) may also contribute to MCFA taste transduction in the gustatory system. One possible receptor, GPR40, has been shown to be activated by some MCFAs (Briscoe et al., 2003); however, it has not been studied extensively in taste cells within this context.
We used whole CT nerve recording to assess the neural signal generated by MCFAs in mice. Interestingly, for PUFA taste signaling, comparatively few studies have conclusively demonstrated that linoleic acid alone reliably produced activity in the CT nerves (Stratford et al., 2008; Cartoni et al., 2010; Yasumatsu et al., 2019) despite its role in carrying the FA taste signal (Pittman and Contreras, 1998; Stratford et al., 2006). We have shown in the present study reliable, concentration-dependent responses to MCFAs from the mouse CT. Similar to GPR120 (Cartoni et al., 2010; Yasumatsu et al., 2019) in linoleic acid-induced CT activity, we show that loss of GPR84 greatly abrogates activity in the CT nerve during stimulation with MCFAs. Again, as with the case of the MCFA-stimulated cell activity, loss of GPR84 did not completely inhibit neural capric acid responses, and there were still modest increases in CT activity. A recent single fiber CT study by Ninomiya and colleagues (Yasumatsu et al., 2019) identified a so-called “F-fiber” that seems to be largely responsible for carrying the PUFA signal. As we did not perform single-fiber recording in the present study, it is unclear whether the neural response to MCFAs acting via GPR84 is carried in the same subset of CT fibers as PUFAs via GPR120.
The identification of an additional functional gustatory FA receptor, GPR84, whose ligands include the SFAs, has implication for the control of dietary intake for this important class of compounds. The PUFA receptors, GPR120, GPR40, and CD36 have all been shown to varying degrees to contribute to oral and postoral fat (i.e., FA) preference (Abumrad, 2005; Sclafani et al., 2007; Cartoni et al., 2010; Keller et al., 2012; Sclafani et al., 2013; Ozdener et al., 2014; Douglas Braymer et al., 2017) and eventually in dietary fat intake. A similar role could be hypothesized for GPR84 for the preference for lipids high in saturated fats. While largely believed to be expressed predominantly in hematopoietic cells related to immune system function, GPR84 had been identified in the gut where it has been suggested to play a regulatory role in metabolism and gut homeostasis (Tan et al., 2017). A recent paper has found GPR84 expressed in the fungiform taste papillae in humans (Costanzo et al., 2019), and this may represent the receptor underlying earlier work demonstrating that MCFAs were perceived by humans (Mattes, 2009). Thus, as is the case for the other FA receptors, there is expression of GPR84 throughout the oral cavity and enteric nervous system where this receptor may contribute to the chemosensory recognition of dietary saturated fat.
In conclusion, we have identified a new role for GPR84 in the oral cavity in mammals: that of a gustatory receptor for MCFAs. Support for this role has come from its expression in chemosensory taste cells, its involvement in cell-based and neural responses to MCFAs, and its critical function in the behavioral recognition of MCFAs in mice. Together, the data strongly suggest that there is a transduction pathway initiated by GPR84 in the taste system for SFAs. Given that virtually all research into fat taste has focused on PUFAs, such as linoleic acid, this finding should open up important new avenues for research, in the same manner as our original research on effects of PUFAs on DRK channels in rodent taste cells (Gilbertson et al., 1997) helped raise awareness concerning fat taste.
Footnotes
This work was supported by National Institute of Health Award R01DC013318.
The authors declare no competing financial interests.
References
- Abumrad NA (2005) CD36 may determine our desire for dietary fats. J Clin Invest 115:2965–2967. 10.1172/JCI26955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet RP, Laye S (2014) Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat Rev Neurosci 15:771–785. 10.1038/nrn3820 [DOI] [PubMed] [Google Scholar]
- Bessone F, Razori MV, Roma MG (2019) Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell Mol Life Sci 76:99–128. 10.1007/s00018-018-2947-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray GA, Paeratakul S, Popkin BM (2004) Dietary fat and obesity: a review of animal, clinical and epidemiological studies. Physiol Behav 83:549–555. 10.1016/j.physbeh.2004.08.039 [DOI] [PubMed] [Google Scholar]
- Briscoe CP, Tadayyon M, Andrews JL, Benson WG, Chambers JK, Eilert MM, Ellis C, Elshourbagy NA, Goetz AS, Minnick DT, Murdock PR, Sauls HR, Shabon U, Spinage LD, Strum JC, Szekeres PG, Tan KB, Way JM, Ignar DM, Wilson S, et al. (2003) The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J Biol Chem 278:11303–11311. 10.1074/jbc.M211495200 [DOI] [PubMed] [Google Scholar]
- Calder PC (2015a) Marine omega-3 fatty acids and inflammatory processes: effects, mechanisms and clinical relevance. Biochim Biophys Acta 1851:469–484. 10.1016/j.bbalip.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Calder PC (2015b) Functional roles of fatty acids and their effects on human health. JPEN J Parenter Enteral Nutr 39:18S–32S. 10.1177/0148607115595980 [DOI] [PubMed] [Google Scholar]
- Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N, le Coutre J, Ninomiya Y, Damak S (2010) Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci 30:8376–8382. 10.1523/JNEUROSCI.0496-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Yarmolinsky D, von Buchholtz L, Oka Y, Sly W, Ryba NJ, Zuker CS (2009) The taste of carbonation. Science 326:443–445. 10.1126/science.1174601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ (2004) Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci 24:9598–9611. 10.1523/JNEUROSCI.1851-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N, Roper SD (2010) The cell biology of taste. J Cell Biol 190:285–296. 10.1083/jcb.201003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Liu D, Nowson C, Duesing K, Archer N, Bowe S, Keast R (2019) A low-fat diet up-regulates expression of fatty acid taste receptor gene FFAR4 in fungiform papillae in humans: a co-twin randomised controlled trial. Br J Nutr 122:1212–1220. 10.1017/S0007114519002368 [DOI] [PubMed] [Google Scholar]
- Douglas Braymer H, Zachary H, Schreiber AL, Primeaux SD (2017) Lingual CD36 and nutritional status differentially regulate fat preference in obesity-prone and obesity-resistant rats. Physiol Behav 174:120–127. 10.1016/j.physbeh.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Toit E, Browne L, Irving-Rodgers H, Massa HM, Fozzard N, Jennings MP, Peak IR (2018) Effect of GPR84 deletion on obesity and diabetes development in mice fed long chain or medium chain fatty acid rich diets. Eur J Nutr 57:1737–1746. 10.1007/s00394-017-1456-5 [DOI] [PubMed] [Google Scholar]
- Galindo MM, Voigt N, Stein J, van Lengerich J, Raguse JD, Hofmann T, Meyerhof W, Behrens M (2012) G protein-coupled receptors in human fat taste perception. Chem Senses 37:123–139. 10.1093/chemse/bjr069 [DOI] [PubMed] [Google Scholar]
- German JB, Dillard CJ (2004) Saturated fats: what dietary intake? Am J Clin Nutr 80:550–559. 10.1093/ajcn/80.3.550 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Gao L, Thakur A, Siu PM, Lai CW (2017) Role of free fatty acids in endothelial dysfunction. J Biomed Sci 24:50. 10.1186/s12929-017-0357-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson TA, Khan NA (2014) Cell signaling mechanisms of oro-gustatory detection of dietary fat: advances and challenges. Prog Lipid Res 53:82–92. 10.1016/j.plipres.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Gilbertson TA, Fontenot DT, Liu L, Zhang H, Monroe WT (1997) Fatty acid modulation of K+ channels in taste receptor cells: gustatory cues for dietary fat. Am J Physiol 272:C1203–C1210. 10.1152/ajpcell.1997.272.4.C1203 [DOI] [PubMed] [Google Scholar]
- Hamulka J, Glabska D, Guzek D, Bialkowska A, Sulich A (2018) Intake of saturated fatty acids affects atherogenic blood properties in young, Caucasian, overweight women even without influencing blood cholesterol. Int J Environ Res Public Health 15:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata T, Tazawa S, Ohta S, Rhyu MR, Misaka T, Ichihara K (2012) Artepillin C, a major ingredient of Brazilian propolis, induces a pungent taste by activating TRPA1 channels. PLoS One 7:e48072. 10.1371/journal.pone.0048072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura A, Hirasawa A, Hara T, Tsujimoto G (2009) Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat 89:82–88. 10.1016/j.prostaglandins.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Kaske S, Krasteva G, König P, Kummer W, Hofmann T, Gudermann T, Chubanov V (2007) TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci 8:49. 10.1186/1471-2202-8-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata Y, Kawabata F, Nishimura S, Tabata S (2018) Oral lipase activities and fat-taste receptors for fat-taste sensing in chickens. Biochem Biophys Res Commun 495:131–135. 10.1016/j.bbrc.2017.10.125 [DOI] [PubMed] [Google Scholar]
- Keller KL, Liang LC, Sakimura J, May D, van Belle C, Breen C, Driggin E, Tepper BJ, Lanzano PC, Deng L, Chung WK (2012) Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity (Silver Spring) 20:1066–1073. 10.1038/oby.2011.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Roberts C, Maruyama Y, Berg S, Roper S, Chaudhari N (2006) Faithful expression of GFP from the PLCbeta2 promoter in a functional class of taste receptor cells. Chem Senses 31:213–219. 10.1093/chemse/bjj021 [DOI] [PubMed] [Google Scholar]
- Laugerette F, Passilly-Degrace P, Patris B, Niot I, Febbraio M, Montmayeur JP, Besnard P (2005) CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest 115:3177–3184. 10.1172/JCI25299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Shah BP, Croasdell S, Gilbertson TA (2011) Transient receptor potential channel type M5 is essential for fat taste. J Neurosci 31:8634–8642. 10.1523/JNEUROSCI.6273-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S, Eguchi A, Mizushige T, Kitabayashi N, Tsuzuki S, Inoue K, Fushiki T (2009) Colocalization of GPR120 with phospholipase-Cbeta2 and alpha-gustducin in the taste bud cells in mice. Neurosci Lett 450:186–190. 10.1016/j.neulet.2008.11.056 [DOI] [PubMed] [Google Scholar]
- Mattes RD (2009) Oral detection of short-, medium-, and long-chain free fatty acids in humans. Chem Senses 34:145–150. 10.1093/chemse/bjn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto J, Hasegawa S, Kasubuchi M, Ichimura A, Nakajima A, Kimura I (2016) Nutritional signaling via free fatty acid receptors. Int J Mol Sci 17:450. 10.3390/ijms17040450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillot T, Szleper E, Vagne G, Barthet S, Litime D, Brindisi MC, Leloup C, Penicaud L, Nicklaus S, Brondel L, Jacquin-Piques A (2019) Cerebral gustatory activation in response to free fatty acids using gustatory evoked potentials in humans. J Lipid Res 60:661–670. 10.1194/jlr.M086587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdener MH, Subramaniam S, Sundaresan S, Sery O, Hashimoto T, Asakawa Y, Besnard P, Abumrad NA, Khan NA (2014) CD36- and GPR120-mediated Ca(2)(+) signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology 146:995–1005. 10.1053/j.gastro.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passilly-Degrace P, Gaillard D, Besnard P (2009) Orosensory perception of dietary lipids in mammals. Results Probl Cell Differ 47:221–238. 10.1007/400_2008_7 [DOI] [PubMed] [Google Scholar]
- Pittman DW, Contreras RJ (1998) Responses of single lingual nerve fibers to thermal and chemical stimulation. Brain Res 790:224–235. 10.1016/S0006-8993(98)00059-6 [DOI] [PubMed] [Google Scholar]
- Pittman DW, Smith KR, Crawley ME, Corbin CH, Hansen DR, Watson KJ, Gilbertson TA (2008) Orosensory detection of fatty acids by obesity-prone and obesity-resistant rats: strain and sex differences. Chem Senses 33:449–460. 10.1093/chemse/bjn012 [DOI] [PubMed] [Google Scholar]
- Running CA, Mattes RD (2014) Different oral sensitivities to and sensations of short-, medium-, and long-chain fatty acids in humans. Am J Physiol Gastrointest Liver Physiol 307:G381–G389. 10.1152/ajpgi.00181.2014 [DOI] [PubMed] [Google Scholar]
- Running CA, Craig BA, Mattes RD (2015) Oleogustus: the unique taste of fat. Chem Senses 40:507–516. 10.1093/chemse/bjv036 [DOI] [PubMed] [Google Scholar]
- Sawamura R, Kawabata Y, Kawabata F, Nishimura S, Tabata S (2015) The role of G-protein-coupled receptor 120 in fatty acids sensing in chicken oral tissues. Biochem Biophys Res Commun 458:387–391. 10.1016/j.bbrc.2015.01.125 [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Suggs MS, Sostman AL, Simon SA (1992) Chorda tympani and lingual nerve responses to astringent compounds in rodents. Physiol Behav 51:55–63. 10.1016/0031-9384(92)90203-e [DOI] [PubMed] [Google Scholar]
- Sclafani A, Ackroff K, Abumrad NA (2007) CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am J Physiol Regul Integr Comp Physiol 293:R1823–R1832. 10.1152/ajpregu.00211.2007 [DOI] [PubMed] [Google Scholar]
- Sclafani A, Zukerman S, Ackroff K (2013) GPR40 and GPR120 fatty acid sensors are critical for postoral but not oral mediation of fat preferences in the mouse. Am J Physiol Regul Integr Comp Physiol 305:R1490–R1497. 10.1152/ajpregu.00440.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah BP, Liu P, Yu T, Hansen DR, Gilbertson TA (2012) TRPM5 is critical for linoleic acid-induced CCK secretion from the enteroendocrine cell line, STC-1. Am J Physiol Cell Physiol 302:C210–C219. 10.1152/ajpcell.00209.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford JM, Curtis KS, Contreras RJ (2006) Chorda tympani nerve transection alters linoleic acid taste discrimination by male and female rats. Physiol Behav 89:311–319. 10.1016/j.physbeh.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Stratford JM, Curtis KS, Contreras RJ (2008) Linoleic acid increases chorda tympani nerve responses to and behavioral preferences for monosodium glutamate by male and female rats. Am J Physiol Regul Integr Comp Physiol 295:R764–R772. 10.1152/ajpregu.00916.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JK, McKenzie C, Marino E, Macia L, Mackay CR (2017) Metabolite-sensing G protein-coupled receptors-facilitators of diet-related immune regulation. Annu Rev Immunol 35:371–402. 10.1146/annurev-immunol-051116-052235 [DOI] [PubMed] [Google Scholar]
- Venkataraman C, Kuo F (2005) The G-protein coupled receptor, GPR84 regulates IL-4 production by T lymphocytes in response to CD3 crosslinking. Immunol Lett 101:144–153. 10.1016/j.imlet.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu X, Simonavicius N, Tian H, Ling L (2006) Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J Biol Chem 281:34457–34464. 10.1074/jbc.M608019200 [DOI] [PubMed] [Google Scholar]
- Wei L, Tokizane K, Konishi H, Yu HR, Kiyama H (2017) Agonists for G-protein-coupled receptor 84 (GPR84) alter cellular morphology and motility but do not induce pro-inflammatory responses in microglia. J Neuroinflammation 14:198. 10.1186/s12974-017-0970-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumatsu K, Iwata S, Inoue M, Ninomiya Y (2019) Fatty acid taste quality information via GPR120 in the anterior tongue of mice. Acta Physiol (Oxf) 226:e13215. 10.1111/apha.13215 [DOI] [PubMed] [Google Scholar]
- Yazici D, Sezer H (2017) Insulin resistance, obesity and lipotoxicity. Adv Exp Med Biol 960:277–304. 10.1007/978-3-319-48382-5_12 [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Kurata R, Yoshida K, Murayama MA, Cui X, Hasegawa A (2013) Free fatty acids-sensing G protein-coupled receptors in drug targeting and therapeutics. Curr Med Chem 20:3855–3871. 10.2174/09298673113209990168 [DOI] [PubMed] [Google Scholar]