Abstract
After spinal cord injury (SCI), patients face many physical and psychological issues including intestinal dysfunction and comorbidities, strongly affecting quality of life. The gut microbiota has recently been suggested to influence the course of the disease in these patients. However, to date only two studies have profiled the gut microbiota in SCI patients, months after a traumatic injury. Here we characterized the gut microbiota in a large Italian SCI population, within a short time from a not only traumatic injury. Feces were collected within the first week at the rehabilitation center (no later than 60 days after SCI), and profiled by 16S rRNA gene-based next-generation sequencing. Microbial profiles were compared to those publicly available of healthy age- and gender-matched Italians, and correlated to patient metadata, including type of SCI, spinal unit location, nutrition and concomitant antibiotic therapies. The gut microbiota of SCI patients shows distinct dysbiotic signatures, i.e. increase in potentially pathogenic, pro-inflammatory and mucus-degrading bacteria, and depletion of short-chain fatty acid producers. While robust to most host variables, such dysbiosis varies by lesion level and completeness, with the most neurologically impaired patients showing an even more unbalanced microbial profile. The SCI-related gut microbiome dysbiosis is very likely secondary to injury and closely related to the degree of completeness and severity of the lesion, regardless of etiology and time interval. This microbial layout could variously contribute to increased gut permeability and inflammation, potentially predisposing patients to the onset of severe comorbidities.
Subject terms: Spinal cord diseases, Dysbiosis, Microbiome
Introduction
A spinal cord injury (SCI) is a devastating event resulting in motor and/or sensory deficit, as well as vegetative dysfunction with huge implications for all aspects of the patient’s life and environment, and a high cost on society1. The clinical picture and functioning outcome depend on the level of injury and its completeness. Intestinal dysfunction is an almost inevitable occurrence after SCI of any etiology, traumatic and non-traumatic2,3. Alterations in motor and secretory functions and in digestion and absorption mechanisms can affect all segments of the digestive tract, from pharynx to the anal canal, but, incomprehensibly, only a few studies have investigated intestinal pathophysiology after SCI: only five papers have studied the alterations underlying clinically relevant events in the SCI patient, such as pathological gastroesophageal reflux resulting in airway inflammation and possibly pneumonia4.
Colorectal dysfunction is the most studied issue probably because defecation management greatly affects lifestyle and Quality of Life (QoL) in persons with SCI. On the one hand, the transport of contents through the colon is delayed and, on the other hand, continence and evacuation efficiency are impaired. Consequently, intestinal emptying is no longer complete and does not involve the same colonic tract as healthy subjects5. Persons with SCI and reduced mobility, having bowel motion may not be able to reach a toilet in time and episodes of fecal and/or gas incontinence or continuous soiling and leakage may occur. Persons with SCI report unsatisfactory bowel management as the primary cause of QoL impairment, to an even greater extent than problems regarding walking, bladder management and sexuality6. This awareness arises a while after injury, in relation to acceptance of new life conditions concerning mobility and autonomy7,8.
Studies investigating the colonic motor pattern after SCI are few and show contrasting results9–11. More recent studies concluded that intact neural transmission between the spinal cord and higher centers is not essential for normal colorectal responses to feeding and distension12. The peristaltic motor pattern producing massive ab-oral transport of contents consists of particularly intense contractions called High Amplitude Propagating Contractions (HAPC) that overlap the normal segmentation contractions and create the conditions, in the presence of feces and/or gas in the lumen, for evacuation to occur when socially convenient13,14. This propulsive contractile activity is absent in SCI patients15, particularly upon awakening, which distinguishes them from patients with Slow Transit Constipation of functional origin, in which HAPCs are less frequent, less vigorous, but still present16,17. The pathophysiological mechanisms that correlate lesion level and degree of completeness with the type of motor behavior of the different colonic segments are not fully understood, given their different innervation. However, it is established that the transport of solid and gaseous contents through the colon in people with SCI is pathologically slowed as a consequence of reduced motor and peristaltic activity, reduced visceral perception, anal sphincter dysfunction, inadequate abdominal muscle tone and the person’s reduced immobility. This causes incomplete evacuation, formation of fecal residues, fecal impaction and unwanted defecation and/or overflow fecal incontinence18,19.
The gastrointestinal tract, especially the colon, hosts a complex community of bacteria, but also viruses, fungi and parasites, i.e., the gut microbiota, which performs multiple crucial functions for homeostasis20. In addition to allowing the extraction of energy from otherwise indigestible carbohydrates and synthesizing vitamins, the gut microbiota modulates the development and functionality of the immune, endocrine and nervous systems, through the production of bioactive molecules that can enter the bloodstream and reach extra-intestinal organs21. Recent population studies showed that the Bristol Stool Form Scale (BSFS) score was among the strongest explanatory factors for gut microbiota variation22,23: stool consistency and frequency are associated with specific microbiota components and growth rates, suggesting that particular ecological networks are established depending on the context24,25. This is very relevant as other studies have shown that bolus size and consistency affect propulsion speed, thus indicating that propulsion in the large bowel is not a simple reflex but a more complex process involving an adaptable neuromechanical loop26. This may imply that an empty colon has no motor activity and that an altered microbiota composition, i.e., dysbiosis, has a profound role in the pathophysiology of colon disorders, as already shown in several functional syndromes, such as chronic constipation, Irritable Bowel Syndrome and other colon motility disorders27–29. Investigating variations in the composition of the intestinal microbiota after SCI, in consideration of its deep relationship with Enteric Nervous System functionality and, consequently, colonic motor activity30,31 constitutes an important element to understand the pathophysiology of intestinal dysfunctions in SCI patients. Interestingly, following an experimental injury in mice, it was shown that the perilesional area is significantly larger in the presence of severe dysbiosis, which exacerbates neurological impairment and spinal cord pathology after SCI32. Not least, peculiar dysbiotic profiles may favor the onset of sepsis and organ infectious complications, so frequent in the clinical course after SCI. Dysbiosis in critically ill patients, such as after severe and sudden insults33, can lead to sepsis, from Systemic Inflammatory Response Syndrome to the almost always fatal Multi Organ Dysfunction Syndrome34–36. According to Wallace and colleagues, trauma in its various forms is likely to cause dysbiotic changes in the gut microbiota, which are strongly influenced by the variety of local and systemic phenomena, and in turn act pathologically, favoring the onset and severity of comorbidities. With specific regard to SCI, the authors point out that understanding how the gut microbiome changes following injury and whether it has a role in potentiating inflammation or protecting against secondary injury and infections, holds great promise for the development of microbiota-targeted intervention strategies to enhance patient care and potentially improve outcomes37.
To our knowledge, only Gungor et al.38 and Zhang et al.39 investigated the composition of the gut microbiota in SCI patients. The former assessed the microbiota in 30 patients ≥ 12 months after traumatic SCI (15 with complete lesion, above T6 level, and 15 with cauda equina syndrome), compared to 10 healthy subjects, all males of the same age. The latter evaluated 43 male patients at least 6 months after complete traumatic SCI (20 tetraplegics and 23 paraplegics), compared to 23 healthy male adults, aged 18–60 years. Both studies documented the presence of dysbiosis after SCI and suggested that it may play a role in neurological recovery.
The primary objective of the present study was to characterize the composition of the intestinal microbiota in a larger population of patients, of both genders, with traumatic and non-traumatic SCI, in the acute and early post-acute phase (< 60 days from SCI), as compared with a healthy Italian population of overlapping age, gender and geographical origin. Secondary objectives were to correlate the microbiota composition with patient characteristics (gender, age, intestine characteristics, Body Mass Index—BMI), type of SCI (traumatic or non-traumatic etiology, lesion level and completeness), spinal unit location, nutrition and concomitant antibiotic therapies.
Results
Study cohort description
One hundred SCI patients were enrolled from Spinal Units and Neuro-Rehabilitation Centers in nine different Italian cities: Bari, Brescia, Catania, Costa Masnaga, Milan, Imola, Pietra Ligure, Pavia and Perugia (Supplementary Fig. S1). The number of patients enrolled is evenly distributed among the North (31), Center (30) and South (39) of the country, so that dissimilarities due to different regional eating habits/lifestyles can possibly be identified.
Demographic, neurological and clinical characteristics of enrolled SCI patients are shown in Table 1. As expected, the majority of patients are male and etiology is traumatic. The mean age is very similar. The percentages of patients in the three groups according to lesion level and in the four related to AIS score, allow a meaningful comparison among them (please, see also “Methods”). In accordance with inclusion criteria, the average time interval between injury and fecal sampling is about one month with minimal variation among centers.
Table 1.
Demographic, neurological, anthropometric and clinical characteristics of the 100 enrolled Italian patients with SCI.
| Spinal unit | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imola | Pavia | Brescia | Pietra L | Baria | Catania | Perugia | Milan | Barib | Costa M | |||
| Pts (no.) | 10 | 8 | 8 | 10 | 12 | 16 | 10 | 7 | 11 | 8 | 100 | |
| Male (%) | 100 | 75 | 87.5 | 60 | 66.6 | 68.7 | 100 | 57.1 | 63.6 | 75 | 76% | |
| Age, years (mean ± SD) | 45 ± 14 | 51 ± 20 | 51 ± 14 | 53 ± 13 | 50 ± 19 | 54 ± 13 | 47 ± 15 | 60 ± 12 | 49 ± 14 | 43 ± 21 | 51 ± 15 | |
| Traumatic etiology (%) | 100 | 87.5 | 87.5 | 70 | 75 | 68.7 | 30 | 28.5 | 63.6 | 62.5 | 68% | |
| Neurologic lesion level | C1–C6 (%) | 20 | 25 | 12.5 | 30 | 58.3 | 31.2 | 40 | 28.5 | 18.8 | 12.5 | 29% |
| C7–T5 (%) | 40 | 25 | 25 | 60 | 0 | 6.3 | 20 | 43 | 18.8 | 37.5 | 25% | |
| T6–L5 (%) | 40 | 50 | 62.5 | 10 | 41.7 | 62.5 | 40 | 28.5 | 63.4 | 50 | 46% | |
| AIS score | A (%) | 70 | 50 | 37.5 | 30 | 50 | 12.5 | 10 | 43 | 18.2 | 62.5 | 36% |
| B (%) | 0 | 25 | 25 | 20 | 0 | 12.5 | 30 | 14.3 | 27.2 | 12.5 | 16% | |
| C (%) | 0 | 25 | 37.5 | 50 | 41.6 | 43.7 | 10 | 14.3 | 54.6 | 12.5 | 31% | |
| D (%) | 30 | 0 | 0 | 0 | 8.4 | 31.3 | 50 | 28.4 | 0 | 12.5 | 17% | |
| Time interval between lesion and fecal sampling | Days ± SD | 21 ± 8 | 30 ± 33 | 18 ± 10 | 34 ± 16 | 32 ± 16 | 29 ± 17 | 23 ± 18 | 36 ± 22 | 19 ± 4 | 28 ± 1 | 27 ± 5 |
| 1–15 (%) | 20 | 50 | 50 | 20 | 27.3 | 25 | 40 | 14.3 | 18.8 | 25 | 27% | |
| 16–30 (%) | 70 | 12.5 | 37.5 | 10 | 27.3 | 43.7 | 40 | 28.7 | 81.2 | 50 | 41% | |
| 31–60 (%) | 10 | 37.5 | 12.5 | 70 | 54.4 | 31.3 | 10 | 57 | 0 | 25 | 31% | |
| BMI | < 18.5 (%) | 0 | 12.5 | 0 | 0 | 0 | 6.3 | 0 | 14.3 | 0 | 12.5 | 4% |
|
≥ 18.5 ≤ 24.9 (%) |
50 | 62.5 | 62.5 | 100 | 66.7 | 37.5 | 70 | 71.4 | 91 | 62.5 | 66% | |
| ≥ 25 (%) | 50 | 25 | 37.5 | 0 | 33.3 | 56.2 | 30 | 14.3 | 9 | 25 | 30% | |
| Nutrition | OS (%) | 0 | 75 | 50 | 50 | 75 | 93.7 | 70 | 100 | 81.8 | 62.5 | 67% |
| EN and/or PN (%) | 100 | 25 | 50 | 50 | 25 | 6.3 | 30 | 0 | 18.2 | 37.5 | 33% | |
| BSFS | Type 1–3 (%) | 0 | 0 | 50 | 40 | 33.4 | 50 | 0 | 42.8 | 36.6 | 37.5 | 30% |
| Type 4–5 (%) | 80 | 37.5 | 50 | 60 | 66.6 | 50 | 100 | 57.2 | 54.4 | 37.5 | 60% | |
| Type 6–7 (%) | 20 | 62.5 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 25 | 10% | |
| Antibiotic intake | No (%) | 70 | 62.5 | 50 | 40 | 33.4 | 18.7 | 30 | 28.5 | 36.3 | 50 | 40% |
| Yes, 3 days (%) | 10 | 25 | 50 | 50 | 66.6 | 50 | 40 | 43 | 36.3 | 37.5 | 42% | |
| Yes, 10 days (%) | 20 | 12.5 | 0 | 10 | 0 | 31.3 | 30 | 28.5 | 27.4 | 12.5 | 18% | |
As for the time interval between lesion and fecal sampling, patients were grouped into the following categories: ≤ 15 days, between 16 and 30 days, and between 31 and 60 days. As for BMI, patients were divided into the following categories: underweight (BMI < 18.5 kg/m2), normal weight (BMI between 18.5 and 24.9 kg/m2) and overweight/obesity (BMI ≥ 25 kg/m2). OS per os nutrition, EN enteral nutrition, PN parenteral nutrition, BSFS Bristol Stool Form Scale. With regards to the antibiotic intake, “No” stands for untreated patients in the past 10 days, “Yes, 3 days” for patients who were given antibiotics in the 3 days before fecal sampling, “Yes, 10 days” for patients who were given antibiotics between 4 and 10 days before fecal sampling. In the last column the total of patients and the average in years/days and in percentages per each variable/group are reported.
aOspedale Policlinico.
bICS Maugeri.
The gut microbiota of SCI patients segregates from that of healthy controls
The gut microbiota of SCI patients was profiled by 16S rRNA gene-based next-generation sequencing and compared with that of age- and sex-matched healthy Italians (aged 45.8 ± 14.1 years), whose data are publicly available40–43. A total of 1,171,676 high-quality reads (mean ± SD, 11,717 ± 6088) were generated and clustered into 12,442 OTUs at 97% identity.
No differences were observed in alpha diversity between SCI patients and healthy controls (p = 0.8, Wilcoxon rank sum test) (Fig. 1A). On the other hand, the Principal Coordinates Analysis (PCoA) of inter-individual variation, based on Bray–Curtis distances between the genus-level profiles, revealed a significant separation between the study groups (p = 0.001, R2 = 0.15; permutation test with pseudo-F ratios) (Fig. 1B). This data was confirmed by Random Forests (error rate at discriminating SCI patients vs. healthy controls, 1%).
Figure 1.
Diversity of the gut microbiota of SCI patients as compared to healthy controls. (A) Box plots showing the distribution of alpha diversity values, according to the inverse Simpson index in SCI patients as compared to age- and sex-matched healthy Italian controls. (B) Principal Coordinates Analysis based on Bray–Curtis distances between the genus-level microbial profiles of SCI patients and healthy controls. A significant separation was found (p = 0.001, permutation test with pseudo-F ratios).
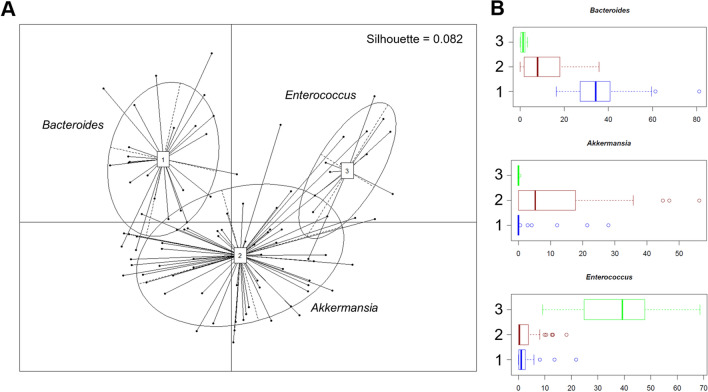
In line with the available literature44, the microbial profiles of both SCI patients and healthy controls are dominated by Firmicutes (mean relative abundance, SCI vs. controls: 50.9% vs. 54.8%) and Bacteroidetes (22.8% vs. 34.3%), with the families Ruminococcaceae (15.5% vs. 23.6%), Lachnospiraceae (9.4% vs. 18.5%) and Bacteroidaceae (16.1% vs. 16.1%) being the most represented regardless of health status (Fig. 2A,B). Nevertheless, many differences emerged, suggesting an altered composition of the gut bacterial community after SCI. In particular, SCI patients are enriched in several families variously involved in inflammation-based disorders45–48, such as Coriobacteriaceae (3.8% vs. 0.5%), Enterococcaceae (6.3% vs. 0.03%), Lactobacillaceae (2.5% vs. 0.2%), Streptococcaceae (5.4% vs. 0.6%), Methanobacteriaceae (0.3% vs. 0.002%) and Enterobacteriaceae (8.3% vs. 0.5%), as well as Verrucomicrobiaceae (7.2% vs 0.4%) (p < 0.001, Wilcoxon rank sum test). Moreover, they showed a decrease in Prevotellaceae (0.7% vs. 12.6%), Clostridiaceae (0.6% vs 1.0%) and the short-chain fatty acid (SCFA)-producing family, Ruminococcaceae (15.5% vs. 23.6%) (p < 0.001) (Fig. 2B). Consistent data were obtained at the genus level and confirmed by Random Forests, resulting in the identification of the following highly discriminatory taxa: Methanobrevibacter (0.3% vs. 0.002), an unclassified genus belonging to Coriobacteriaceae (0.8% vs. 0.1%), Streptococcus (5.2% vs. 0.6%), Enterococcus (6.3% vs. 0.03%), Klebsiella (3.0% vs. 0.05%), Akkermansia (7.2% vs. 0.4%), Faecalibacterium (1.1% vs. 5.2%), and Coprococcus (0.5% vs. 2.8%) (p < 0.001) (Fig. 2C). Interestingly, enterotype analysis revealed that SCI patients could be stratified into three distinct groups, based on the Jensen-Shannon distance metrics, which were dominated by Bacteroides (enterotype-like group 1), Akkermansia (enterotype-like group 2) and Enterococcus (enterotype-like group 3), respectively (p < 0.001, Kruskal–Wallis test) (Fig. 3). The vast majority (64%) of samples fell into group 2, 26% into group 1 and the remaining 10% into group 3.
Figure 2.
Compositional structure of the gut microbiota in SCI. Pie charts showing the average relative abundance of the most abundant phyla (A) and families (B) in SCI patients as compared to age- and sex-matched healthy Italian controls. Only taxa with relative abundance > 0.1% in at least 20 samples for phyla and > 0.01% in at least 30 samples for families are shown. (C) Box plots showing the distribution of the relative abundance values of discriminant genera between SCI patients and healthy controls, according to Random Forests. p < 0.001, Wilcoxon rank sum test.
Figure 3.
Enterotype-like groups in SCI patients. (A) Principal Coordinates Analysis of enterotype clusters in SCI patients. Three enterotypes were identified by using the Jensen-Shannon divergence distance metric. The ellipse covers 67% of the samples belonging to a cluster. The silhouette coefficient is shown at the top right. (B) Box plots showing the distribution of the relative abundance values of discriminant genera among the three enterotype-like groups. p < 0.001, Kruskal–Wallis test.
The gut microbiota profiles of SCI patients stratify by lesion level and AIS score
PCoA analysis based on Bray–Curtis distances between the genus-level profiles of SCI patients showed no segregation by time interval since the injury (≤ 15 days, between 16 and 30 days, over 30 days) (p > 0.05, permutation test with pseudo-F ratios) (Fig. 4), suggesting that microbiota dysbiosis is unrelated to the tremendous metabolic and neurological conditions that characterize spinal shock, which usually ends within the first 20–30 days after the lesion. Similarly, no separation was observed by well-known variables to be associated with the gut microbiota22,43,49,50, i.e. age, BMI (under vs. normal vs. overweight/obesity), type of nutrition (per os vs. enteral/parenteral) and BSFS (1–3, 4–5, 6–7), nor gender or etiology of the lesion (traumatic vs. non-traumatic) (p > 0.05) (Supplementary Figs. S2–S7). However, a few microbial signatures consistent with literature were highlighted, including lower representation of Bacteroidetes members and Faecalibacterium, and enrichment of enterobacteria in patients following enteral/parenteral nutrition vs. those fed per os (p ≤ 0.04, Wilcoxon rank sum test) (Supplementary Fig. S4)51. As for BMI, underweight SCI patients were characterized by increased proportions of Dialister and Fusobacterium, overweight ones by increased amounts of Enterococcus, while Prevotella was distinctive of normal-weight patients (p ≤ 0.03) (Supplementary Fig. S3).
Figure 4.
The gut microbiota dysbiosis in SCI patients is stable over time. Principal Coordinates Analysis based on Bray–Curtis distances between the genus-level microbial profiles of SCI patients according to the time interval between injury and fecal sampling (≤ 15 days, between 16 and 30 days, over 30 days). No significant segregation was found (p > 0.05, permutation test with pseudo-F ratios).
A slight separation in the ordination space was observed by recruitment center (p = 0.003, R2 = 0.13; permutation test with pseudo-F ratios) (Supplementary Fig. S8). In particular, the samples from the spinal unit of Pietra Ligure form a cluster slightly separated from the others (p ≤ 0.04, R2 = 0.03). A barely separable grouping was also observed for Catania, with respect to Bari (Ospedale Policlinico), Costa Masnaga, Milan and Imola (p ≤ 0.04, R2 ≤ 0.02). Random Forests-based analysis confirmed the data, i.e., that the rehabilitation center does not strongly affect the patients’ microbiota (error rate at discriminating between centers, 83%), with only the samples from Catania being quite accurately discriminated (i.e., 10/16). Such a discrimination is mainly related to a lower representation of Streptococcus and Faecalibacterium while greater proportions of Bacteroides and Rikenellaceae in the Catania-derived samples compared to the other spinal units (p ≤ 0.04, Kruskal–Wallis test) (Supplementary Fig. S8). According to the enterotype analysis, 7/16 patients from Catania fell in fact in the enterotype-like group 1 dominated by Bacteroides.
Interestingly, the genus-level layouts of SCI patients were found to stratify by AIS score, i.e., there was a significant separation between patients with AIS A and B vs. those with AIS C and D (p = 0.03, R2 = 0.03; permutation test with pseudo-F ratios) (Fig. 5A). Significant segregation was also found by injury severity: more severity for patients with AIS A or B and neurological level of lesion between C1 to C6 and C7 to T6, vs. less severity for patients with AIS A or B, but lesion level between T7 and L5 together with patients with AIS C or D and any neurological level of lesion (p = 0.04, R2 = 0.02) (Fig. 5B). At the taxonomic level, the genera Bacteroides and Faecalibacterium, and unclassified members of Lachnospiraceae were overrepresented in patients with grade C or D impairment, whereas Lactobacillus was more abundant in AIS grade A or B patients, especially in those with a C1–C6 cervical lesion (p ≤ 0.04, Wilcoxon rank sum test) (Fig. 5C). In the latter patient subset, Lactobacillus OTUs were variously assigned to the species L. rhamnosus, L. gasseri, L. helveticus, L. fermentum and L. mucosae. As for injury severity, the proportions of the mucus degrader Akkermansia, as well as those of Oscillospira, Dialister, and unclassified genera of Lachnospiraceae and [Mogibacteriaceae] were reduced in the group having more severe SCI (p ≤ 0.05) (Fig. 5D). When looking at the enterotype distribution, we found that although the enterotype-like group 2 (dominated by Akkermansia) was the most represented in both patients with more (59%) and less injury severity (66%), the enterotype-like group 3 (dominated by Enterococcus) accounted for a greater percentage of those having more severe SCI (18% vs 7% in patients with less severe injury). It should be noted that more than half of all patients had taken antibiotics but this percentage was higher in those with more severe SCI (76% vs 56% in those with less severe injury).
Figure 5.
The gut microbiota profiles of SCI patients stratify by AIS score and severity of injury. Principal Coordinates Analysis based on Bray–Curtis distances between the genus-level microbial profiles of SCI patients according to AIS score (A) and SCI severity (B). A significant separation by both covariates was found (p ≤ 0.04, permutation test with pseudo-F ratios). More severity, AIS score A or B and neurological level of lesion between C1 to C6 and C7 to T6; less severity, AIS score A or B and lesion level between T7 and L5, and AIS score C or D and every neurological level of lesion. Box plots showing the distribution of the relative abundance values of discriminatory genera by AIS score (C) and SCI severity (D). p ≤ 0.05, Wilcoxon rank sum test.
As expected, a significant segregation was found between the gut microbiota structures of SCI patients who had or not been given antibiotic therapy, with a further stratification by timing of administration (i.e., in the 3 days or between 4 and 10 days before fecal sampling) (p = 0.001, R2 = 0.07; permutation test with pseudo-F ratios) (Fig. 6A). In particular, the proportions of Enterococcus and Klebsiella were far greater in patients who had received antibiotics in the 3 days before sampling (p ≤ 0.03, Kruskal–Wallis test). Enterococcus and Klebsiella OTUs were mainly assigned to E. faecium and K. pneumoniae, respectively. On the other hand, typically health-associated microbiota components, such as Lachnospiraceae and Ruminococcaceae members, were greatly reduced by recent antibiotics administration, but their levels after a few days (i.e., 4–10) were overall comparable to those of patients who had not taken therapy (p ≤ 0.02). A similar trend was observed for Collinsella, Bacteroidetes members (i.e., Rikenellaceae and [Barnesiellaceae]) and Akkermansia (p ≤ 0.01) (Fig. 6B). As expected, the Enterococcus enterotype (group 3) was particularly represented in patients who had received antibiotics in the 3 days before sampling (24%) but its proportion dropped after 4–10 days (6%), up to 0% in those who had not received antibiotics.
Figure 6.
Impact of antibiotics administration on the gut microbiota of SCI patients. (A) Principal Coordinates Analysis based on Bray–Curtis distances between the genus-level microbial profiles of SCI patients according to antibiotics administration and timing (i.e., no antibiotic therapy, and therapy in the 3 days or between 4 and 10 days before fecal sampling). A significant separation among groups was found (p = 0.001, permutation test with pseudo-F ratios). (B) Box plots showing the distribution of the relative abundance values of discriminant genera. p ≤ 0.05, Kruskal–Wallis test.
Discussion
To the best of our knowledge, this is the first study on the intestinal microbiota in acute and early post-acute SCI patients, some of them still in a phase of spinal shock, being in the first 10–12 weeks after injury. Consistently with Gungor et al.38 and Zhang et al.39, their gut microbiota appears dysbiotic and shows distinct signatures compared to age- and sex-matched healthy controls from the same geographical location. In addition to a reduction in beneficial SCFA producers (e.g. Faecalibacterium and Coprococcus), which probably represents a non-specific, shared response to diseases52, SCI patients show increased proportions of potentially pathogenic and pro-inflammatory bacteria, such as Enterococcus, Streptococcus and Klebsiella. These include well-known nosocomial pathogens, associated with urinary tract infections, bacteremia, and elevated rates of mortality in critically ill patients36,53,54. As expected, Enterococcus and Klebsiella were more abundant in patients who had received antibiotic therapy in the 3 days prior to fecal sampling. According to previous findings, their bloom could be closely related to the drop in oxidative stress-sensitive SCFA-producing commensals, with increased epithelial oxygenation and thus oxygen availability for facultative anaerobic pathogens55,56. Moreover, SCI patients were enriched in Methanobacteriaceae, Coriobacteriaceae and Verrucomicrobiaceae, specifically Akkermansia. The archaeal species Methanobrevibacter smithii was found to increase in critically ill patients, possibly due to its intrinsic resistance to antibiotics54. However, it should be noted that the primer combination we used was shown to be not optimal for Archaea detection57, which may have biased our results. On the other hand, Coriobacteriaceae members have been shown to reduce the expression of tight-junction proteins, possibly leading to gut leakage and metabolic endotoxemia58. The role of the mucus degrader Akkermansia is instead more controversial, since it has been proposed as next-generation probiotics for metabolic disorders59, but also shown to induce pro-inflammatory T lymphocyte responses and potentially exacerbate the symptoms of neurodegenerative disorders, such as Multiple Sclerosis and Parkinson’s disease60,61. It is plausible that in a compromised clinical state, high amounts of Akkermansia contribute to the erosion of the mucus layer, compromising the barrier function and facilitating access to epithelia by enteropathogens, with consequently increased inflammatory tone.
Apart from the decrease in SCFA producers, the discriminatory taxa herein identified do not overlap with the results by Gungor et al.38 and Zhang et al.39. The reasons may include technical issues related to library preparation and bioinformatics, exiguity of their patient population, ethnicity and geographic location (including lifestyle, environmental exposure, socio-economic status, etc.)23,62 and, not least, the chronicity of SCI in their population. Furthermore, patients who had taken antibiotics and/or probiotics in the previous 3 weeks38 and month39 were excluded, so it was not possible to make between-study comparisons on the impact of the antibiotic intake on the intestinal microbiota of SCI patients.
The gut microbiota dysbiosis in SCI is robust to several variables known to be associated with the microbiota variation, i.e., age, gender, BMI, type of nutrition and BSFS23,43,49,50,63, thus corroborating the hypothesis that the onset of dysbiosis is secondary to SCI. On the other hand, our data showed an impact, albeit limited, of the spinal unit, with the Catania-derived samples being more easily discriminated against by others. It is not unusual to find inter-hospital differences, possibly related to the hospital environment itself, not only food related, as previously discussed64. While caution must be taken in associating discriminatory taxa with other variables, in the light of the data commented above we believe that the center-related variation should not have biased our results.
No differences were found in relation to time since injury as well as etiology, be it traumatic or non-traumatic. The previous studies and obviously also the animal studies with experimental lesions, concerned only traumatic injuries so this is the first demonstration that dysbiosis is associated with SCI of any origin. In contrast, we found that the gut microbiota is significantly different between AIS A or B patients, i.e., with motor complete lesion, and patients with motor incomplete injury, i.e., AIS C or D. The previous studies38,39 had not explored this correlation, because only complete patients were included, however the need to verify these data had already been reported65. In particular, the proportions of Bacteroides, Faecalibacterium and Lachnospiraceae are reduced in complete patients, suggesting further loss of health-promoting microorganisms and reduced bioavailability of SCFAs. On the contrary, in these patients Lactobacillus is more present. Lactobacillus spp. are low-virulence subdominant commensals, widely used as probiotics; however, this genus and/or certain species have also been found to be overabundant in some inflammatory disorders, including obesity, coronary heart disease and heart failure66–68. Lactobacilli are also frequently resistant to vancomycin, which could explain their persistence under critical health conditions69.
The division of patients into a group with severe SCI, i.e., AIS A or B and C1-T6 lesion level, and a less neurologically compromised group, i.e., AIS A or B with T6-L5 lesion level and all AIS C or D, confirmed that the severity of intestinal dysbiosis is related to the severity of the neurological damage. In particular, the under-representation of health-associated taxa, such as Lachnospiraceae, Mogibacteriaceae and Oscillospira52,70,71, in more severe SCI further stresses the presence of an unbalanced bacterial ecosystem with reduced SCFA production capacity, potentially contributing to increased intestinal permeability and inflammation. Oscillospira is able to utilize host glycans as well as Akkermansia, whose relative abundance was also found to be reduced in more severe vs. less severe SCI. In addition, the most neurologically impaired patients showed reduced proportions of Dialister, a lactate-utilizing genus inversely related to inflammation and recently proposed as a potential lead for psychobiotics72. Interestingly, Dialister was also found to be lower in complete SCI in both Turkish38 and Chinese patients39. On the other hand, the enterotype analysis showed that the enterotype-like group 3 (Enterococcus dominated) was far more represented in patients with more severe SCI, suggesting a possible involvement of this pathobiont. As discussed above, this could be closely related to antibiotic intake, as a high percentage of patients with more severe injury had taken antibiotics, particularly in the 3 days prior to fecal sampling. The reasons for the variations in these taxa remain unclear and will require further investigation. Given the recent acquisition of knowledge about the important role of the intestinal microbiome in neuropathic and visceral pain73,74, it would have been interesting to stratify the microbiota data also in relation to pain symptoms, but the sample size for every different neurologic damage was too small to achieve meaningful results. Moreover, this study regards the very early post-acute phase after SCI and for the study of neuropathic pain it would be too early, as neuropathic pain usually develops up to 6 months after onset.
In summary, patients in the acute phase after SCI are characterized by distinctive alterations of the intestinal microbiota, which appear to be stable at least during the first 60 days of injury. Dysbiosis does not vary by gender, age, BMI, type of hospital nutrition, location of the spinal unit where hospitalized, BSFS or etiology. In contrast, it varies according to the degree of completeness and severity of the lesion: patients with tetraplegia and motor complete lesions show a more compromised gut microbiota profile. However, these data may be partially biased by antibiotic intake and need to be validated in larger independent cohorts.
Once confirmed, this knowledge may help to better understand and hopefully predict the onset and severity of common post-injury comorbidities, such as sepsis, organ infection, immunological break-down, catabolic condition, intestinal and bladder complications, by clarifying the reasons why the recovery of some patients is more impaired than others.
Methods
Patients and fecal sampling
We scheduled to enroll around 100 consecutive patients from 10 Italian Spinal Units and Centers for Neurologic Rehabilitation, non-competitively, according to the following inclusion criteria:
age ≥ 18 and ≤ 70 years, both male and female
traumatic or non-traumatic SCI following vascular myelopathy or transverse myelitis
onset within 60 days
SCI of any neurological level ≥ L5, complete or incomplete, classifiable as grade AIS A, B, C, D. The International standards for neurological classification of SCI represent the gold standard assessment for documentation of the level and severity of SCI75. The lesion completeness is defined according to the American Spinal Injury Association Impairment Scale (AIS) with the following grades: AIS A: complete, AIS B: sensory incomplete, AIS C and D: motor incomplete and AIS E: normal function76.
no more than 7 days since first admission to Spinal Unit.
Exclusion criteria were: non-traumatic etiology different from vascular myelopathy or transverse myelitis (others causes for SCI (neoplasms, discitis, etc.) do not guarantee acute onset and precise lesion level), presence of pre-existing intestinal pathologies such as Inflammatory Bowel Disease, chronic hepatitis, Celiac Disease, neoplasm, extensive intestinal resection, ongoing diarrhea (more than 6 watery evacuations per day and/or fecal volume > 250 ml over 24 h), intake of probiotics after injury, ongoing severe sepsis, pregnancy, brain injury and/or coma status associated with SCI, and inability to give informed consent.
A single fecal sample was collected for each patient during a spontaneous or scheduled evacuation, within the first week of hospitalization at the rehabilitation center. Samples were stored at − 20 °C and sent under temperature-controlled conditions from all Centers to the same Microbiology Laboratory, where they were kept at − 80 °C until analysis.
Endpoints
For the Primary Objective: deviation of the gut microbiota profile of patients with SCI from that of the healthy population living in the same geographical area, matched by gender and age.
For the Secondary Objective: correlation between the gut microbiota profile of SCI patients and sex, age, traumatic and non-traumatic etiology, lesion level, AIS score, interval between injury and fecal sampling, BMI at admission, type of nutrition, stool type at the time of sampling and any previous or ongoing antibiotic therapy. As for lesion level, patients were arbitrarily divided into 3 groups according to the severity of sensory-motor impairment and integrity of the Sympathetic Nervous System: C1–C6 group includes high tetraplegia; C7–T6 includes tetraplegia with residual upper limb function and paraplegia with impairment of the sympathetic pathways and involvement of abdominal muscles; T7–L5 injuries result in paraplegia without sympathetic involvement of a part for the large bowel. Regarding the time interval from the injury, in order to evaluate the potential impact of the spinal shock, which usually ends within the first 20–30 days after the lesion, we selected the following cut-off values: until 15 days, between 16 and 30 days, and over 30 days.
Microbial DNA extraction and 16S rRNA gene sequencing
Microbial DNA was extracted from approximately 250 mg of fecal sample using the repeated bead-beating plus column method as previously reported77. Briefly, the samples were suspended in 1 mL of lysis buffer (500 mM NaCl, 50 mM Tris–HCl pH 8, 50 mM EDTA, 4% SDS) and processed three times in a FastPrep instrument (MP Biomedicals, Irvine, CA) at 5.5 movements/s for 1 min, in the presence of four 3-mm glass beads and 0.5 g of 0.1-mm zirconia beads (BioSpec Products, Bartlesville, OK). Stool particles were pelleted after 15 min of incubation at 95 °C, and nucleic acids were precipitated by adding 10 M ammonium acetate and one volume of isopropanol. The pellets were then washed with 70% ethanol and resuspended in 10 mM Tris–HCl, 1 mM EDTA pH 8.0 (TE) buffer. After treatment with 10 mg/mL DNase-free RNase at 37 °C for 15 min, the samples were subjected to protein removal and column-based DNA purification following the manufacturer’s instructions (DNeasy Blood & Tissue kit; QIAGEN, Hilden, Germany).
The V3–V4 hypervariable region of 16S rDNA was amplified by using the 341F and 785R primers with added Illumina adapter overhang sequences as previously described48. After amplicon purification with a magnetic bead-based clean-up system (Agencourt AMPure XP; Beckman Coulter, Brea, CA), indexed libraries were prepared by limited-cycle PCR using Nextera technology, further cleaned as above, and pooled at equimolar concentration. The final library was denatured with 0.2 N NaOH and diluted to 6 pM with a 20% PhiX control. Paired-end sequencing was performed on an Illumina MiSeq platform (Illumina, San Diego, CA), as per manufacturer’s guidelines, at Wellmicro Srl (Bologna, Italy).
Bioinformatics and statistics
Raw sequences were processed using a pipeline combining PANDAseq78 and QIIME79. Length- and quality-filtered reads were clustered into Operational Taxonomic Units (OTUs) at 97% similarity using UCLUST80. All singleton OTUs and chimeras were discarded. Taxonomy assignment was conducted using the RDP classifier against the Greengenes database (May 2013 release). For species-level identification, OTUs of interest were subjected to BLAST analysis.
Publicly available sequences of the gut microbiota from age- and sex-matched healthy Italian subjects were downloaded and processed as above for comparative purposes. Specifically, we recovered sequences from De Filippis et al.40 (76 Italians; NCBI SRA SRP042234), Turroni et al.41 (1 Italian; NCBI SRA PRJNA340060), Schnorr et al.42 (8 Italians; MG-RAST mgp12183) and Biagi et al.43 (15 elderly Italians; MG-RAST mgp17761). Alpha diversity was calculated using the inverse Simpson index. Principal Coordinates Analysis (PCoA) was performed on Bray–Curtis distances between the genus-level profiles.
Statistical analysis was performed in R 3.3.2 (https://www.r-project.org/) using the packages ‘made4’81, ‘vegan’82 and ade483. A permutation test with pseudo-F ratios (function ‘adonis’ in vegan) was used to assess the significance of separation in PCoA. The contribution of covariates to the ordination space was determined using the function ‘envfit’ of vegan. Wilcoxon rank sum test was used to assess significant differences in diversity and taxon relative abundance between groups while Kruskal–Wallis test was used for multiple comparisons. The impact of clinical variables on microbiota structure was also evaluated by Random Forests84, using the packages RandomForest and rfPermute function. Stratification into enterotypes was achieved following the enterotyping tutorial provided in the R environment by EMBL (http://enterotype.embl.de/enterotypes.html). Briefly, samples were clustered based on genus-level relative abundances using the Jensen-Shannon distance (JSD) and the Partitioning Around Medoids (PAM) clustering algorithm. The optimal number of clusters was chosen by maximizing the Calinski–Harabasz (CH) index. Cluster validation was performed using the silhouette coefficient. The results of clustering were visualized on a PCoA plot by the ade4 package. When appropriate, p values were adjusted for multiple comparisons using the Benjamini–Hochberg correction. A false discovery rate (FDR) < 0.05 was considered as statistically significant.
Ethical declarations
The study was approved by the institutional ethical board (Comitato Etico Interaziendale Bologna-Imola) of the coordinating center (Montecatone Rehabilitation Institute), and subsequently by all participating centers (University of Bologna, ASST Gaetano Pini CTO, ICS Maugeri Bari, Ospedale Valduce, ICS Maugeri Pavia, Ospedale Santa Corona, Ospedale Cannizzaro, Azienda Ospedaliera di Perugia, Fondazione Teresa Camplani Domus Salutis Brescia, Ospedale Policlinico Bari), and conducted in accordance with the Declaration of Helsinki. All study participants completed a written consent form prior to their inclusion in the study.
Supplementary Information
Acknowledgements
The Authors are indebted to Dr. Cecilia Baroncini, Scientific Office of the Montecatone Rehabilitation Institute, for the secretarial assistance and all of them wish to thank Dr. Roberto Nardulli, MD, the past-Director of the Spinal Unit in Cassano Murge, for his indorsement to this work.
Author contributions
All underlined authors, listed in alphabetical order, as well as M.B. and M.C.B. contributed equally to the study for Investigation. Conceptualization: G.B., M.C. and S.T. Data curation: M.C.B. and F.D.A. Formal analysis: S.T., F.D.A. Funding acquisition: G.B. Project administration: G.B., S.T. and T.G. Resources: P.B., S.T. and F.D.A. Supervision: G.B. Validation: S.T., P.B. and C.K. Visualization: S.T., F.D.A. and G.B. Writing—original draft: G.B., S.T. and F.D.A. Writing—review & editing: S.T., C.K. and G.B. All authors had reviewed and approved the final manuscript.
Funding
Non-profit study funded by each clinical centre with its own financial resources, also to cover the costs of the molecular tests, with a financial grant by SIMS, the Italian Society for Spinal Cord.
Data availability
Sequence reads were deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA; BioProject ID PRJNA724686).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92027-z.
References
- 1.World Health Organization. International perspectives on spinal cord injury. https://www.who.int/publications-detail/international-perspectives-on-spinal-cord-injury. Accessed 26 May 2020.
- 2.Krogh K, Christensen P. Neurogenic colorectal and pelvic floor dysfunction. Best Pract. Res. Clin. Gastroenterol. 2009;23:531–543. doi: 10.1016/j.bpg.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Awad RA. Neurogenic bowel dysfunction in patients with spinal cord injury, myelomeningocele, multiple sclerosis and Parkinson’s disease. World J. Gastroenterol. 2011;17:5035–5048. doi: 10.3748/wjg.v17.i46.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radulovic M, et al. Greatly increased prevalence of esophageal dysmotility observed in persons with spinal cord injury. Dis. Esophagus. 2015;28:699–704. doi: 10.1111/dote.12272. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen MM, et al. Colorectal transport during defecation in subjects with supraconal spinal cord injury. Spinal Cord. 2013;51:683–687. doi: 10.1038/sc.2013.58. [DOI] [PubMed] [Google Scholar]
- 6.Glickman S, Kamm MA. Bowel dysfunction in spinal-cord-injury patients. Lancet. 1996;347:1651–1653. doi: 10.1016/S0140-6736(96)91487-7. [DOI] [PubMed] [Google Scholar]
- 7.Pardee C, Bricker D, Rundquist J, MacRae C, Tebben C. Characteristics of neurogenic bowel in spinal cord injury and perceived quality of life. Rehabil. Nurse. 2012;37:128–135. doi: 10.1002/RNJ.00024. [DOI] [PubMed] [Google Scholar]
- 8.Burns AS, et al. Phenomenological study of neurogenic bowel from the perspective of individuals living with spinal cord injury. Arch. Phys. Med. Rehabil. 2015;96:49–55. doi: 10.1016/j.apmr.2014.07.417. [DOI] [PubMed] [Google Scholar]
- 9.Glick ME, et al. Colonic dysfunction in patients with thoracic spinal cord injury. Gastroenterology. 1984;86:287–294. doi: 10.1016/0016-5085(84)90413-X. [DOI] [PubMed] [Google Scholar]
- 10.Bruninga K, Camilleri M. Colonic motility and tone after spinal cord and cauda equine injury. Am. J. Gastroenterol. 1997;92:891–894. [PubMed] [Google Scholar]
- 11.Fajardo NR, et al. Decreased colonic motility in persons with chronic spinal cord injury. Am. J. Gastroenterol. 2003;98:128–134. doi: 10.1111/j.1572-0241.2003.07157.x. [DOI] [PubMed] [Google Scholar]
- 12.Suttor VP, et al. Colorectal responses to distension and feeding in patients with spinal cord injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1344–1349. doi: 10.1152/ajpgi.90408.2008. [DOI] [PubMed] [Google Scholar]
- 13.Moreno-Osset E, et al. Association between postprandial changes in colonic intraluminal pressure and transit. Gastroenterology. 1989;96:1265–1273. doi: 10.1016/S0016-5085(89)80013-7. [DOI] [PubMed] [Google Scholar]
- 14.Herbst F, et al. Gastrointestinal transit and prolonged ambulatory colonic motility in health and faecal incontinence. Gut. 1997;41:381–389. doi: 10.1136/gut.41.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ancha HR, et al. Absence of high amplitude propagating contractions in subjects with chronic spinal cord injury. World J. Gastroenterol. 2010;16:5435–5439. doi: 10.3748/wjg.v16.i43.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazzocchi G, et al. Postprandial colonic transit and motor activity in chronic constipation. Gastroenterology. 1990;98:686–693. doi: 10.1016/0016-5085(90)90289-D. [DOI] [PubMed] [Google Scholar]
- 17.Dinning PG, et al. Pancolonic spatiotemporal mapping reveals regional deficiencies in, and disorganization of colonic propagating pressure waves in severe constipation. Neurogastroenterol. Motil. 2010;22:e340–349. doi: 10.1111/j.1365-2982.2010.01597.x. [DOI] [PubMed] [Google Scholar]
- 18.Krogh K, Mosdal C, Laurberg S. Gastrointestinal and segmental colonic transit times in patients with acute and chronic spinal cord lesions. Spinal Cord. 2000;38:615–621. doi: 10.1038/sj.sc.3101066. [DOI] [PubMed] [Google Scholar]
- 19.Qi Z, Middleton JW, Malcolm A. Bowel dysfunction in Spinal Cord Injury. Curr. Gastroenterol. Rep. 2018;20:47. doi: 10.1007/s11894-018-0655-4. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert J, et al. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turroni S, Brigidi P, Cavalli A, Candela M. Microbiota-host transgenomic metabolism, bioactive molecules from the inside. J. Med. Chem. 2018;61:47–61. doi: 10.1021/acs.jmedchem.7b00244. [DOI] [PubMed] [Google Scholar]
- 22.Zhernakova A, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018;24:1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 24.Vandeputte D, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon HJ, et al. Is stool frequency associated with the richness and community composition of gut microbiota? Intest. Res. 2019;17:419–426. doi: 10.5217/ir.2018.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer NJ, Dinning PG, Brookes SJ, Costa M. Insights into the mechanisms underlying colon motor patterns. J. Physiol. 2016;594:4099–4116. doi: 10.1113/JP271919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley EMM, Spiller RC. Constipation and the microbiome: Lumen versus mucosa. Gastroenterology. 2016;150:300–303. doi: 10.1053/j.gastro.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Shin A, Preidis GA, Shulman R, Kashyap PC. The gut microbiome in adult and pediatric functional gastrointestinal disorders. Clin. Gastroenterol. Hepatol. 2019;17:256–274. doi: 10.1016/j.cgh.2018.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casen C, et al. Deviations in human gut microbiota: A novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther. 2015;42:71–83. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Giorgio R, Blandizzi C. Targeting enteric neuroplasticity: Diets and bugs as new key factors. Gastroenterology. 2010;138:1663–1666. doi: 10.1053/j.gastro.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 31.Osadchiy V, Martin CR, Mayer EA. The gut–brain axis and the microbiome: Mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019;17:322–332. doi: 10.1016/j.cgh.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kigerl KA, et al. Gut dysbiosis impairs recovery after spinal cord injury. J. Exp. Med. 2016;213:2603–2620. doi: 10.1084/jem.20151345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayakawa M, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig. Dis. Sci. 2011;56:2361–2365. doi: 10.1007/s10620-011-1649-3. [DOI] [PubMed] [Google Scholar]
- 34.McDonald D, et al. Extreme dysbiosis of the microbiome in critical illness. mSphere. 2016;31:1–4. doi: 10.1128/mSphere.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assimakopoulos SF, et al. Gut-origin sepsis in the critically ill patients: pathophysiology and treatment. Infection. 2018;46:751–760. doi: 10.1007/s15010-018-1178-5. [DOI] [PubMed] [Google Scholar]
- 36.Agudelo-Ochoa GM, et al. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes. 2020;10:1–16. doi: 10.1080/19490976.2019.1707610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace DJ, et al. Spinal cord injury and the human microbiome: Beyond the brain–gut-axis. Neurosurg. Focus. 2019;46:E11. doi: 10.3171/2018.12.FOCUS18206. [DOI] [PubMed] [Google Scholar]
- 38.Gungor B, Adiguzel E, Gursel I, Yilmaz B, Gursel M. Intestinal microbiota in patients with spinal cord injury. PLoS One. 2016;11:1–10. doi: 10.1371/journal.pone.0145878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, et al. Gut microbiota dysbiosis in male patients with chronic traumatic complete spinal cord injury. J. Transl. Med. 2018;16:353. doi: 10.1186/s12967-018-1735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Filippis F, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 41.Turroni S, et al. Fecal metabolome of the Hadza hunter-gatherers: A host-microbiome integrative view. Sci. Rep. 2016;6:32826. doi: 10.1038/srep32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnorr SL, et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biagi E, et al. Gut microbiota and extreme longevity. Curr. Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project. Nature. 569, 641–648 (2019). [DOI] [PMC free article] [PubMed]
- 45.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsilingiri K, et al. Probiotic and postbiotic activity in health and disease: Comparison on a novel polarised ex-vivo organ culture model. Gut. 2012;61:1007–1015. doi: 10.1136/gutjnl-2011-300971. [DOI] [PubMed] [Google Scholar]
- 47.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancello R, et al. Effect of short-term dietary intervention and probiotic mix supplementation on the gut microbiota of elderly obese women. Nutrients. 2019;11:3011. doi: 10.3390/nu11123011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 50.Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 51.D'Amico F, et al. Enteral nutrition in pediatric patients undergoing hematopoietic SCT promotes the recovery of gut microbiome homeostasis. Nutrients. 2019;11:2958. doi: 10.3390/nu11122958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8:1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bajaj JS, et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2019;17:756–765. doi: 10.1016/j.cgh.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 54.Ravi A, et al. Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb. Genom. 2019;5:e000293. doi: 10.1099/mgen.0.000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livanos AE, et al. Rapid gastrointestinal loss of Clostridial Clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS ONE. 2018;13:e0200322. doi: 10.1371/journal.pone.0200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byndloss MX, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–575. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer MA, Güllert S, Neulinger SC, Streit WR, Schmitz RA. Evaluation of 16S rRNA gene primer pairs for monitoring microbial community structures showed high reproducibility within and low comparability between datasets generated with multiple archaeal and bacterial primer pairs. Front. Microbiol. 2016;7:1297. doi: 10.3389/fmicb.2016.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Depommier C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cekanaviciute E, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerhardt S, Mohajeri MH. Changes of colonic bacterial composition in Parkinson's disease and other neurodegenerative diseases. Nutrients. 2018;10:708. doi: 10.3390/nu10060708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deschasaux M, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018;24:1526–1531. doi: 10.1038/s41591-018-0160-1. [DOI] [PubMed] [Google Scholar]
- 63.Santos-Marcos JA, et al. Sex differences in the gut microbiota as potential determinants of gender predisposition to disease. Mol. Nutr. Food Res. 2019;63:e1800870. doi: 10.1002/mnfr.201800870. [DOI] [PubMed] [Google Scholar]
- 64.Biagi E, et al. Early gut microbiota signature of aGvHD in children given allogeneic hematopoietic cell transplantation for hematological disorders. BMC Med. Genom. 2019;12:49. doi: 10.1186/s12920-019-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kigerl KA, Mostacada K, Popovich PG. Gut microbiota are disease-modifying factors after traumatic spinal cord injury. Neurotherapeutics. 2018;15:60–67. doi: 10.1007/s13311-017-0583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Million M, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Kamo T, et al. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS ONE. 2017;12:e0174099. doi: 10.1371/journal.pone.0174099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamashita T. Intestinal immunity and gut microbiota in atherogenesis. J. Atheroscler. Thromb. 2017;24:110–119. doi: 10.5551/jat.38265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halpin AL, et al. Intestinal microbiome disruption in patients in a long-term acute care hospital: A case for development of microbiome disruption indices to improve infection prevention. Am. J. Infect. Control. 2016;44:830–836. doi: 10.1016/j.ajic.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konikoff T, Gophna U. Oscillospira: A central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 71.Oki K, et al. Comprehensive analysis of the fecal microbiota of healthy Japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol. 2016;16:284. doi: 10.1186/s12866-016-0898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valles-Colomer M, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 73.Bazzocchi, G., Balloni, M. & Turroni, S. Microbiome and chronic pelvic pain. In Chronic Pelvic Pain and Pelvic Dysfunction. Assessment and Multidisciplinary Dysfunctions (eds. Giammò, A., Biroli, A.) 71–80 (Springer Nature Switzerland AG, 2021).
- 74.Lagomarsino VN, Kostic AD, Chiu IM. Mechanisms of microbial–neuronal interactions in pain and nociception. Neurobiol. Pain. 2020;9:100056. doi: 10.1016/j.ynpai.2020.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.American Spinal Injury Association. International standards for neurological classification of spinal cord injury. (American Spinal Injury Association, 2019). https://asia-spinalinjury.org/. Accessed 26 May 2020.
- 76.Kirschblum S, Waring W. Updates for the international standards for neurological classification of spinal cord injury. Phys. Med. Rehabil. Clin. N. Am. 2014;25:505–517. doi: 10.1016/j.pmr.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Barone M, et al. Gut microbiome response to a modern Paleolithic diet in a Western lifestyle context. PLoS ONE. 2019;14:e0220619. doi: 10.1371/journal.pone.0220619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Masella AP, et al. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2011;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 81.Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21:2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- 82.Oksanen, J., et al. vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan (2019). Accessed 26 April 2021.
- 83.Dray, S. & Dufour, A. The ade4 Package: Implementing the duality diagram for ecologists. J. Stat. Softw.22. http://www.jstatsoft.org/v22/i04 (2007). Accessed 26 April 2021.
- 84.Breiman L. Random forests. Mach. Learn. 2011;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads were deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA; BioProject ID PRJNA724686).