Abstract
Yield enhancement is a top priority for soybean (Glycine max Merr.) breeding. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) is a major integrator in flowering pathway, and it is anticipated to be capable of regulating soybean reproductive stages through its interactions with other MADS-box genes. Thus, we produced transgenic soybean for a constitutive expression of a maize SOC1 (ZmSOC1). T1 transgenic plants, in comparison with the nontransgenic plants, showed early flowering, reduced height of mature plants, and no significant impact on grain quality. The transgenic plants also had a 13.5–23.2% of higher grain weight per plant than the nontransgenic plants in two experiments. Transcriptome analysis in the leaves of 34-day old plants revealed 58 differentially expressed genes (DEGs) responding to the expression of the ZmSOC1, of which the upregulated FRUITFULL MADS-box gene, as well as the transcription factor VASCULAR PLANT ONE-ZINC FINGER1, contributed to the promoted flowering. The downregulated gibberellin receptor GID1B could play a major role in reducing the plant height. The remaining DEGs suggested broader effects on the other unmeasured traits (e.g., photosynthesis efficiency and abiotic tolerance), which could contribute to yield increase. Overall, modulating expression of SOC1 in soybean provides a novel and promising approach to regulate plant growth and reproductive development and thus has a potential either to enhance grain yield or to change plant adaptability.
Subject terms: Biotechnology, Plant sciences
Introduction
Soybean (Glycine max (L.) Merr.) is the world’s most important food legume to fight hunger and to feed a future with the growing population due to its high quality and high content of both protein and oil1. From 1994 to 2018, the world’s total hectare for soybean production increased at an average annual rate of 3.0%, the total production increased annually at 4.2%, and the yield per hectare increased at 1.2% (http://www.fao.org/faostat/en/#data/QC/visualize). Apparently, two main agronomic factors, including expansion of cropped land areas and yield increases, contributed the sustainable increase in soybean production in the past decades. Further expansion of cropped land areas becomes limited due to the decreasing availability of arable land (http://www.fao.org/3/y4252e/y4252e06.htm)2–4. Therefore, yield increases will be critical to meet the increasing demand of soybean over a wider range of environmental conditions for increasing populations worldwide4.
Crop yield is often determined by genotype and environment. Accordingly, mitigating of the negative environmental impact of climate change on crop yield can be achieved either by developing new agricultural practice to optimize plant growth conditions through or by breeding new genotypes for high yields4. For soybean, historical gains of new cultivars in grain yield over the past 84 years of breeding had been driven by linear increases in light interception and energy conversion and partitioning efficiencies5. The evidence of greater chlorophyll content and greater sink capacity late in the growing season in more recently released soybean varieties suggested that energy conversion/partitioning efficiencies were of importance for seed yield increase, although no consistent changes in photosynthetic capacity with 80 years of soybean breeding were observed6. Photosynthesis enhancement through breeding was considered a viable strategy to increase soybean yield due to their high positive genetic correlation7. Overall, enhancing photosynthetic efficiency of leaves, improving root traits for high nitrogen fixation and water usage efficiency, and increasing flower initiation and reducing flower abortion are considered to be the major genetic solutions to increase soybean yield7–9. In addition, increase in adaptability of soybean cultivars is desirable for expending soybean production area. Alternatively, similar to other legume crops, enhancing plant resilience to abiotic and biotic stresses is anticipated to reduce yield losses10–12.
Transgenic soybean is a major commercialized biotech crop. In the Approval Database of genetically modified (GM) crops, 41 GM soybean events are listed, including 11 events with a GM trait of modified oil/fatty acid, one event “Verdeca HB4 Soybean” containing a sunflower (Helianthus annuus) HAHB-4 gene for drought tolerance13,14, and one event containing Arabidopsis thaliana B-BOX32 domain gene (BBX32) for enhanced photosynthesis/yield15,16 (http://www.isaaa.org). Herbicide tolerance (HT) and insect resistance (IR) are the major GM traits. In 2018, 123.5 million hectares soybeans were planted in the world, of which 95.9 million hectares (78%) were transgenics with either HT (69.3 million hectares) or HT/IR (26.6 million hectares), total accounting for 50% of the global area of biotech crops (http://www.isaaa.org). Currently, neither of the HB4 soybean for abiotic tolerance nor the BBX32 soybean for enhanced photosynthesis/yield has been made available for commercialization (http://www.isaaa.org). Recent field tests suggested that the HB4 soybeans were able to increase seed yield in warm and dry environment by changing plant architecture and improving drought tolerance14.
Modulation of plant reproductive develop through genetic manipulation is a powerful approach to impact soybean yield17. As the only GM soybean for enhanced photosynthesis/yield in the GM Approval Database, BBX32 soybean contained an ectopically expressed BBX32, which could, at least, affect photoperiodic regulation of flowering through the interaction of B-box family proteins [e.g., CONSTANS-LIKE 3 (COL3) in Arabidopsis]15,16,18,19. In Arabidopsis, COL3 is a protein-binding partner of CONSTANS (CO)18,20, which interacts with two major downstream flowering pathway integrators, including FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1). The CO-FT-SOC1 module plays an essential role in plant reproductive development and flowering time control21–23. In soybean, ten FT homologs have been identified, of which functional analysis of GmFT2a and GmFT5a has been conducted in Arabidopsis and soybean24,25. As a member of MADS-box gene family, homologs of SOC1 in soybean play significant roles in floral development and flowering time control26–28. Ectopic expression of either a soybean SOC1 homolog GmGAL1/GmSOC1 in Arabidopsis or a GmSOC1-LIKE in Lotus corniculatus promoted flowering29,30; however, functional analysis of these two genes in soybean has not been reported. Arabidopsis Agamous-like MADS-box protein AGL1 played an essential role in ovule development, seed coat development, and endosperm formation31. To our knowledge, soybean AGL1 (GmAGL1) is the only MADS-box gene that has been studied in soybean through overexpression, which resulted in early flowering32,33.
Modulating expression of MADS-box genes have a potential to increase crop yield34–36. For example, increasing the expression of a homolog of the Arabidopsis FUL/AGL8 gene of maize (ZMM28) enhanced maize grain yield in the field37. The SOC1 protein is a MIKC protein. In Arabidopsis, the SOC1 gene is a positive regulator of the downstream FUL/AGL8 gene21. Accordingly, enhancing expression of the SOC1 gene may have a potential to increase crop yield at least through the FUL/AGL8 gene. In literatures, constitutive expression of a maize (Zea mays) SOC1 gene (ZmSOC1 or ZmMADS1) resulted in accelerated flowering time and reduced overall plant height in both Arabidopsis and maize38. In this study, we transformed the ZmSOC1 into soybean to examine the possible impact of a major MADS-box gene on soybean plant growth and flowering. Meanwhile, we conducted transcriptome analysis of transgenic and nontransgenic plants to reveal the overall impact of ectopic expression of ZmSOC1 on expression of other associated genes in soybean. The results demonstrate that ectopic expression of a monocot (maize) derived SOC1 is efficient to regulate plant reproductive development in a dicot (soybean) and thus has a potential either to enhance grain yield or to change plant adaptability.
Materials and methods
Vector construction
The sequence of the maize SOC1 gene (ZmSOC1 or ZmMADS1) has been published in GenBank (accession numbers: NM_001111682.1 and HQ858775.1)38. A 696-bp ZmSOC1 derived from the cDNA of the maize inbred line B104 was cloned into the pKANNIBAL plasmid to form a ZmSOC1 expression cassette CaMV 35S-ZmSOC1-Ocs, which was successfully inserted into the binary vector pTF101.1 to produce get pTF101.1-CaMV35S-ZmSOC1-Ocs (herein pTF101.1-ZmSOC: see the details in Fig. 1A). Sanger sequencing data confirmed the ZmSOC1 sequence. The pTF101.1-ZmSOC1 was transformed into Agrobacterium tumefaciens strain EHA105. All recombinant DNA works were conducted in compliance with relevant institutional, national, and international guidelines and legislation.
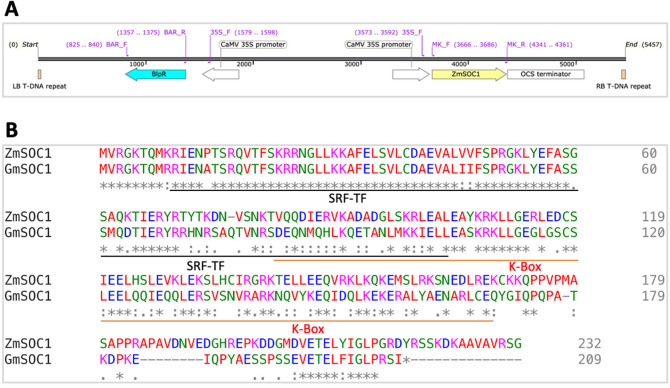
Figure 1.
(A) Diagram of the T-DNA region of the pTF101.1-ZmSOC1 vector. The primer positions are included in the diagram. LB left border, RB right border. (B) Protein sequence alignment of the ZmSOC1 and soybean’s GmSOC1 (iTAK: Glyma.18G224500.2_CDS). The protein sequence of the cloned ZmSOC1 is identical to that derived from either the HQ858775.1 or a part of the NM_001111682.1. GmSOC1 is the same as the sequence detected in the soybean transcriptome reference (cultivar Jack) (DN17539_c0_g1).
Plant transformation
Mature seeds of soybean cultivar Jack were used to produce cotyledonary node explants for transformation. Transformation and regeneration were conducted essentially by following the protocol by Pat and Wang39. About 400 explants were used in each transformation. The co-cultivation time was five days. After co-cultivation, the total number of shoots produced in 4 weeks and the total number of shoots elongated in 4–14 weeks were counted. After transferred to rooting medium, the number of elongated shoots produced roots was counted. All T0 plants were transferred to soil and grown in the greenhouse to produce T1 seeds for phenotyping T1 plants. Nontransgenic (NT) seedlings were used as a control. The plants were grown under a 16-h photoperiod at 21–30 °C. Phenotypic variations (e.g., date of the appearance of the first flower, the node number where the first flower appeared, the date of the appearance of the first pod, flower shape and structure, leaf color and leaf shape, plant architecture, and plant height) among the T0 plants were recorded. Young leaves, about 0.2 g for each greenhouse-growing plant, were harvested for DNA isolation and polymerase chain reaction (PCR) analysis of the transgenes. The transformation frequency was calculated as the percentage of the inoculated explants which produced the PCR positive T0 plants. The transformation experiment was repeated twice.
Phenotypic evaluation of T1 and T2 plants
Early- and late-planted T1 plants were evaluated in two experiments. In experiment #1, the seeds were sowed on May 04, 2020, and the plants were growing in the greenhouse and moved to a secured field under natural environment conditions at East Lansing, Michigan on June 01, 2020. In experiment #2, the seeds were sowed on June 04, 2020 and the plants were growing in the field shared with the plants of experiment #1. Depending on the availability of the seeds, 1–40 seeds per line for all the T0 line/plant that produced seeds were sowed to evaluate T1 transgenic plants and to produce T2 seeds. 150 mg L−1 glufosinate ammonium (GS) was initially used to screen herbicide resistant transgenic plants by painting a half of a leaf along the midvein for each of the T1 seedlings. GS-resistant plants were transferred to one-gallon pots (20 cm diameter × 16 cm height). DNA from the GS-resistant plants was isolated for PCR analysis of the transgenes.
For phenotypic evaluation, the data collected for each plant included: (1) Date of seed germination; (2) Date of the appearance of first flower; (3) Node position where the first flower appeared; (4) Date of the first pod set; (5) Plant height measured and branch number and pod number counted at 141 for experiment #1 and 110 day for experiment #2 after sowing; (6) Pictures taken at different developmental stages; and (7) Seed protein, oil, and fiber contents were measured using a Grain Analyser (Infratec™ 1241, FOSS Analytical AB, Denmark). Fatty acids were extracted as described by Bubeck et al. from two seeds of each plant40. Fatty acid composition was determined by gas chromatography.
Detection of transgenes
DNA was isolated from leaf tissues, 50–200 mg for each sample, using the cetyltrimethylammonium bromide (CTAB) method41. Three pairs of primers, bar-F and bar-R for the bar gene, and forward primers 35S-F (3′ portion of the CaMV 35S promoter) or MK-F and reverse primer MK_R for the ZmSOC1 gene, were used to detect the presence of transgenes in each sample (Figure S1A), and GmAct11_F and GmAct11_R primers were used as a DNA quality control to detect soybean’s actin gene42 (Table S1). PCR reaction conditions for all primer pairs started with an initial denaturation for two min at 94 °C, 30 cycles of 45 s at 94 °C, 60 s at 60 °C and 90 s at 72 °C, and a final extension for 10 min at 72 °C. All amplified PCR products were separated on 1.0% agarose gel containing ethidium bromide and visualized and photographed under UV light.
RNA sequencing and transcriptome analysis
The 5th young leaves near the shoot tips and exposed to sunshine at noon, 3–5 per plants, were harvested from 34-day old plants of three nontransgenic and six transgenic plants (i.e., 3 plants for each of the two transgenic lines) in experiment #2. The leaf samples were frozen immediately in liquid nitrogen, brought to lab, and stored at − 80 °C in a freezer for RNA isolation. Total RNA of each sample was isolated from about 500 mg leaf tissues using a CTAB method43 and was purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). On-Column DNase digestion with the RNase-free DNase Set was used to remove DNA in the RNA samples (Qiagen, Valencia, CA, USA). RNA quality was determined using the High Sensitivity RNA ScreenTape system (Agilent technologies, Santa Clara, CA). High quality RNA with an RNA integrity number (RIN) equivalent greater than 5.0 was used for sequencing and reverse transcription (RT) PCR analysis. Reverse-transcription of 3–5 μg RNA to cDNA was performed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Both regular RT-PCR and quantitative RT-PCR (qRT-PCR) using SYBR Green system (LifeTechnologies, Carlsbad, CA) were conducted to check the selected transcripts. The primers were designed according to the sequences of the RNA-seq (Table S1). QRT-PCR was performed on a Roche LightCycler® 480 Instrument II (Roche). RT-PCR products were separated and visualized on 1.0% agarose gel containing ethidium bromide. The reaction conditions for RT-PCR were 94 °C for 2 min, 35 cycles of 45 s at 94 °C, 60 s at 62 °C and 60 s at 72 °C, with a final 10 min extension at 72 °C. The reaction conditions for qRT-PCR were 95 °C for 5 min, 45 cycles of 30 s at 95 °C, 45 s at 62 °C and 30 s at 72 °C. Transcript levels within samples were normalized to Actin. Foldchanges were calculated using 2−∆∆Ct, where ∆∆Ct = (CtTARGET – CtNOM)transgenic – (CtTARGET – CtNOM)nontransgenic. Three biological samples and three technical replicates for each sample were used for the analysis of each transgenic and nontransgenic line.
The RNA samples were sequenced using the Illumina HiSeq4000 to generate 32–42 million 150 bp-paired end reads per sample. FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of sequencing reads for the per base quality scores. The reads with average scores greater than 38 were obtained and used for transcriptome analysis. A total of 72 million reads (MR) combined from a portion of the reads of the nine samples, 6.6–9.8 MR/sample, were assembled to develop a transcriptome reference using Trinity/2.8.544. The paired reads were aligned to the transcriptome reference to estimate and the abundance for each of a single read. The differentially expressed transcripts (DETs) with the false discovery rate (FDR) value below 0.05 were identified using the Trinity command “run_DE_analysis.pl --method edgeR”44. Pfam proteins were used to annotate the transcriptome reference.
Cytoscape 3.8.2 was used to construct gene networks of overrepresented gene ontology (GO) terms for the selected DETs under BiNGO’s default parameters with selected ontology file ‘GO_Full’ and selected organism ‘A. thaliana’45,46.
Statistical analysis
Statistical analysis of the phenotypic data was conducted using ANOVA and TukeyHSD in RStudio (Version 1.0.136).
Results
Cloning of the ZmSOC1
The pTF101.1-ZmSOC1 contains a streptomycin/spectinomycin aminoglycoside adenylyltransferase gene (aadA) for bacterium selection and a bialaphos resistance (bar) gene under the CaMV 35S promoter conferring resistance to herbicide glufosinate (GS) for selection of transformed plant cells. In the 696-bp ZmSOC1 sequence, 694 bp are identical to the published 696-bp reference deposited in the GenBank (accession numbers HQ858775.1 and NM_001111682.1). The protein sequence of the cloned ZmSOC1 is identical to that derived from either the HQ858775.1 or a part of the NM_001111682.1. It has a 54.8% identity to the soybean’s SOC1 gene (GmSOC1) (iTAK: Glyma.18G224500.2_CDS), which is the sequence detected in the soybean transcriptome reference (cultivar Jack) assembled in this study (DN17539_c0_g1) (Fig. 1B).
Phenotypic variations in T0 transgenic plants
‘Jack’ was used to produce transgenic soybean plants because it was a transformable soybean cultivar using the bialaphos resistance (bar) gene as a selectable marker39,47. Herbicide-resistant plants were induced for half-seed explants after 2–4-week selection, 6–12-week elongation, and 2–4-week rooting (Figure S1). Of the total of 770 explants inoculated, 38 T0 transgenic lines/plants were produced in two experiments with transformation frequencies of 3.9% and 5.9%, respectively (Table S2). Remarkably, three of the 38 lines flowered and further formed seed pods during in vitro cultures (Fig. 2A,B).
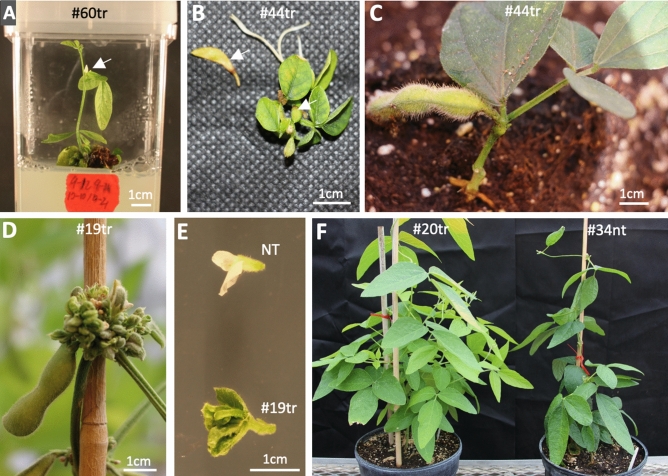
Figure 2.
Phenotypic variations of T0 transgenic ZmSOC1-OX (TR) and nontransgenic (NT) soybean plants cv. Jack grown under in vitro and greenhouse conditions. (A) Flowering of a TR line #60tr growing on the elongation medium. (B) Seed pod production from a TR line #34tr growing on the rooting medium. (C) The #34tr plant showed dwarfing, no branches, and produced a seed pod in 27 days after transferred to soil. (D,E) A representative TR line (#19tr) showing abnormal flowers. (F) A TR line #20tr produced more branches than a NT line #34nt. Arrows show flowers or seed pods.
Of the 38 T0 transgenic lines growing in the greenhouse under a 16-h photoperiod, 11 lines had no visible difference from the nontransgenic control and the rest 27 lines showed various phenotypic changes, including abnormal flowers or early flowering in seven lines, chimeric or increased numbers of branches in eight lines, and dead plants of two lines (Fig. 2C–I, Table S3; Figure S2). The lines with abnormal flowers or early flowering, which were rarely observed in our previous soybean transformations using other genes, were likely induced by the expression of the ZmSOC1, although the effect of tissue culture could also contribute to these phenotypic changes. Overall, 33 T0 transgenic lines produced seeds, and the lines with abnormal flowers produced no or a few seeds.
Phenotypic evaluation of T1 transgenic plants
Seedlings from 21 lines were screened in two experiments to identify transgenic plants for phenotyping. Herbicide-painting was effective in detecting the bar-expressing transgenic plants (Figure S3). Polymerase chain reaction (PCR) analyses was reliable to detect the transferred ZmSOC1 gene. Fourteen transgenic lines produced at least one transgenic seedling, and all the tested seedlings from the other seven lines were nontransgenic (Table S4). As anticipated, not all T0 transgenic lines were able to produce transgenic seeds due possibly to the chimeric nature of the transgenic plants produced from cotyledonary node explants. Unsurprisingly, only two lines showed a segregation rate of about 3:1 between transgenic and nontransgenic seedlings in the 14 transgenic lines (Table S4).
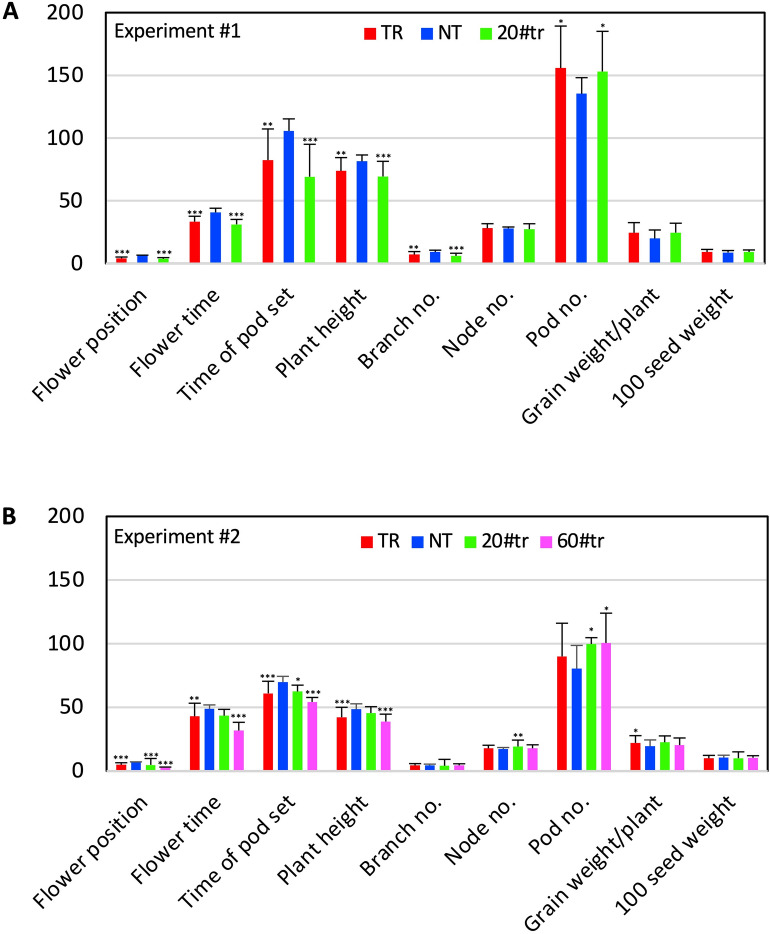
Plants from 10 transgenic lines were grown for phenotyping in each of the two experiments, including five transgenic evaluated in both experiments (Table S4). According to the local light length (https://www.timeanddate.com/sun/usa/lansing), the early and late-planting experiments were conducted by sowing the seeds on May 4th and June 4th, respectively, because soybean plants are photoperiod-sensitive for flowering. In fact, the variations in the environmental conditions (i.e., light and temperature) of the two experiments affected several traits of both transgenic and null segregants (hereafter: nontransgenic). For example, the early-planted nontransgenic plants in experiment #1, compared to the late-planted nontransgenic plants in experiment #2, were taller (81.5 cm vs. 48.5 cm), flowered earlier (41 days vs. 49 days), and set pods later (106 days vs. 70 days) (Fig. 4, Table S5). In both experiments, transgenic plants, compared to nontransgenic plants, showed an early reproductive phase with early flowering (i.e., 6 and 7 days earlier in the experiment #1 and #2, respectively), early pod set (i.e., 24 and 8 days earlier), and lower node positions (~ 2 nodes lower in both experiments) where the first flower appeared (Figs. 3B, 4). Additionally, the mature transgenic plants were 9.5% and 13.0% shorter than the nontransgenic ones in the experiment #1 and #2, respectively, but in each of the two experiments there was no difference in the total number of nodes between the transgenic and the nontransgenic plants, suggesting the reduced plant height was due to the reduced internode length (Fig. 4). This consistency suggested that expression of the ZmSOC1 was able to enhance reproductive production and reduce plant height. On the other hand, the early-planted plants in the experiment #1 were taller and had more seed pods that the late-planted plants in the experiment #2 due to the difference in photoperiod and maybe temperatures too. In both experiments, the transgenic plants had more seed pods and seed production, including a nonsignificant (P = 0.053) 23.2% of increase of the grain weight per plant in the experiment #1 and a significant (P = 0.040) 13.5% of increase in the experiment #2 (Figs. 3C,D, 4).
Figure 4.
Phenotypic changes in T1 transgenic (TR) soybean plants expressing the ZmSOC1 in the two experiments, where the seeds were sowed on May 4th in the experiment #1 and June 4th in the experiment #2, respectively. #20tr and #60tr are two independent TR lines. Y-axis showed mean value and STDEV of 16 nontransgenic (NT), 52 TR, and 16 #20tr plants in the experiment #1 (A) and 32 NT, 44 TR, 11 #20tr, and 7 #60tr lines in the experiment #2 (B). Flower position: The node number where the first flower appeared. Flowering time: Days when the first flowered appeared after the seeds were sowed. Time of pod set: Days when the first pod was set after the seeds were sowed. Plant height (cm): The height of the mature plant. Branch no., Node no., and Pod no.: The numbers for each mature plant were counted. Grain weight/plant (g): Weight of dry seed. 100 seed weight (g): 100 seed weight for each plant. For each of the nine traits, statistical analysis was conducted between NT and TR (or any of the individual TR line) separately. Significance codes: ***P < 0.001, **P < 0.01, *P < 0.05.
Figure 3.
Growth of nontransgenic (NT) and transgenic soybean plants from a representative, T1 transgenic (TR) line expressing the ZmSOC1. All the plants were grown under natural environmental conditions. (A) Flowering of the 35-day old plants in the experiment #1. (B) Pod formation in the 67-day old plants in the experiment #2. (C) Seed pod production in the 134-day old plants in the experiment #1. (D) Seed pod production in the harvested plants in the experiment #2.
Seed quality of nontransgenic and ten transgenic lines was evaluated by measuring eight quality parameters. When the ten transgenic lines were compared as one transgenic group to the nontransgenic seeds, no significant difference between the transgenic and the nontransgenic groups was found for all of the 10 seed quality parameters, suggesting that the expression of ZmSOC1 had little impact on grain quality (Table 1).
Table 1.
Effect of ZmSOC1 expression on seed quality. Protein, fiber and oil contents were measured using a Grain Analyzer. Fatty acids were determined by gas chromatography. The mean value (± STDEV) for each transgenic and nontransgenic (NT) line, were for the seeds from 2–4 transgenic plants except that all_tr is the average of all transgenic lines. Each transgenic line and all_tr were compared with the nt. NA not available. Signif. codes: ***P < 0.001, **P < 0.01, and *P < 0.05.
| Genotype | Protein (%) | Fiber (%) | Oil (%) | Palmitic (%) | Stearic (%) | Oleic (%) | Linoleic (%) | Linolenic (%) |
|---|---|---|---|---|---|---|---|---|
| #14tr | 42.03 ± 0.68 | 5.70 ± 0.17 | 16.13 ± 0.95 | 13.6 ± 0.95 | 3.86 ± 0.51 | 17.6 ± 0.69 | 55.83 ± 1.62 | 9.11 ± 1.33 |
| #16tr | 42.52 ± 2.52 | 5.62 ± 0.09 | 17.95 ± 0.32* | 11.96 ± 0.72 | 3.71 ± 0.27 | 18.21 ± 2.25 | 57.34 ± 1.99 | 8.77 ± 0.93 |
| #20tr | 41.96 ± 1.45 | 5.67 ± 0.19 | 17.44 ± 1.11 | 12.5 ± 0.59 | 3.75 ± 0.32 | 17.51 ± 2.71 | 57.11 ± 2.22 | 9.15 ± 1.28 |
| #21tr | 41.74 ± 0.27 | 5.61 ± 0.03 | 15.94 ± 1.59 | 13.81 ± 0.86 | 3.73 ± 0.35 | 17.82 ± 2.45 | 55.8 ± 2.46 | 8.85 ± 0.82 |
| #22tr | 41.27 ± 0.63 | 5.81 ± 0.07 | 16.21 ± 0.80 | 13.30 ± 2.27 | 3.96 ± 0.22 | 16.42 ± 4.43 | 59.07 ± 0.57 | 7.25 ± 1.38 |
| #25tr | 44.03 ± 0.33* | 5.59 ± 0.04 | 16.11 ± 0.30 | 15.41 ± 1.13 | 3.94 ± 0.27 | 17.63 ± 0.77 | 54.8 ± 0.35 | 8.21 ± 0.26 |
| #28tr | 42.28 ± 1.00 | 5.81 ± 0.03 | 18.13 ± 0.23** | NA | NA | NA | NA | NA |
| #35tr | 41.33 ± 1.51 | 5.95 ± 0.09 | 17.26 ± 0.68 | 15.59 ± 0.7 | 3.54 ± 0.35 | 16.58 ± 2.62 | 55.27 ± 1.84 | 9.00 ± 0.66 |
| #4tr | 42.37 ± 1.50 | 5.54 ± 0.17 | 15.29 ± 0.37 | 12.28 ± 0.68 | 3.78 ± 0.03 | 17.27 ± 2.1 | 56.97 ± 0.4 | 9.71 ± 1.06 |
| #60tr | 33.07 ± 2.32** | 6.28 ± 0.20* | 18.44 ± 0.37** | 15.91 ± 1.81 | 3.69 ± 0.22 | 16.27 ± 1.29 | 55.22 ± 0.98 | 8.91 ± 0.72 |
| all_tr | 41.41 ± 1.55 | 5.65 ± 0.27 | 17.30 ± 0.04 | 13.4 ± 1.56 | 3.72 ± 0.33 | 17.41 ± 2.33 | 56.53 ± 2.14 | 8.94 ± 1.04 |
| nt | 41.27 ± 3.07 | 5.75 ± 0.24 | 16.84 ± 1.24 | 13.47 ± 1.43 | 3.71 ± 0.32 | 17.10 ± 1.51 | 56.88 ± 1.34 | 8.85 ± 1.04 |
Phenotypic evaluation T2 transgenic plants
T2 plants were grown to identify homozygous transgenic lines and to bulk homozygous seeds for further tests. Meanwhile, flowering and pod formation time of the greenhouse-growing plants were evaluated. Early flowering and early pod formation were observed in the T2 transgenic plants. This provided further evidence that the expression of ZmSOC1 enhanced reproductive development.
Differentially expressed (DE) transcripts (DETs) responding to ZmSOC1 expression
Transcriptome analysis of transgenic and nontransgenic plants was conducted for two major purposes, to verify the expression of the transgenes and to reveal the genes that responding to the expression of the ZmSOC1. In the leaves of six plants from the two transgenic lines, high expressions of the bar and the ZmSOC1 were found; in contrast, no sequence reads of the transgenes were detected in any of the three nontransgenic plants (Table 2). The results verified the expression of the two transgenes. Meanwhile, it showed that the expression of the transgenes in transgenic line #60tr was higher than that of transgenic line #20. The higher ZmSOC1 expression in the line #60tr may explain, at least partially, the phenotypic variations as well as the difference of the 1935 DETs identified between the two transgenic lines (Fig. 5A; Table 2; Figure S2; Table S6).
Table 2.
Differentially expressed genes (DEGs) overlapped in two transgenic lines (#20tr and #60tr), each compared to the nontransgenic line. #N/As represent a non-DEGs.
| Isoform ID | Log2(#60tr/nt) | Log2(#20tr/nt) | Log2(#60tr/#20tr) | Annotation_protein | e_value_annotation |
|---|---|---|---|---|---|
| DN18940_c0_g1_i5 | − 11.85 | − 11.78 | #N/A | MDHC_MEDSA | 0.0E+00 |
| DN17886_c0_g1_i15z | − 11.23 | − 11.16 | #N/A | ALFC5_ARATH | 3.7E−63 |
| DN22071_c0_g1_i12 | − 8.70 | − 8.63 | #N/A | TMK1_ARATH | 0.0E+00 |
| DN21378_c2_g2_i2 | − 8.41 | #N/A | − 7.65 | RB3GP_DROME | 7.75E−29 |
| DN14711_c0_g1_i5 | #N/A | 7.36 | #N/A | RB3GP_DANRE | 8.6E−24 |
| DN18476_c0_g3_i7 | − 8.33 | − 8.26 | #N/A | WRK70_SOLLC | 6.5E−37 |
| DN17384_c1_g4_i8 | − 8.14 | − 8.07 | #N/A | GID1B_ARATH | 0.0E+00 |
| DN20140_c1_g1 | − 8.01 | 1.75 | #N/A | MMSA_ARATH | 0.0E+00 |
| DN16057_c0_g1_i3 | − 7.54 | − 5.72 | #N/A | ADS3_ARATHz | 1.2E−172 |
| DN19626_c0_g2 | − 4.24 | − 9.37 | #N/A | RFP4B_DANRE | 9.4E+00 |
| DN13742_c0_g1 | − 2.41 | − 1.80 | #N/A | C93C1_SOYBN | 0.0E+00 |
| DN22021_c0_g1_i7z | − 1.96 | − 1.13 | #N/A | SUT31_ARATH | 0.0E+00 |
| DN16671_c0_g1_i7 | − 1.73 | − 11.11 | 9.52 | NRX3_ARATH | 2.2E−171 |
| DN14666_c0_g5_i2 | − 1.46 | − 0.99 | #N/A | MIP1B_ARATH | 4.2E−10 |
| DN19591_c0_g3_i2 | − 1.45 | − 1.06 | #N/A | PUB33_ARATH | 1.6E−12 |
| DN15732_c0_g1_i4z | − 1.24 | − 1.22 | #N/A | BOR4_ARATH | 0.0E+00 |
| DN16638_c0_g2_i7 | − 1.21 | − 1.19 | #N/A | MBCD_SOLTU | 0.0E+00 |
| DN19977_c0_g1 | − 1.20 | 11.22 | #N/A | FTSH2_ORYSJ | 1.6E−54 |
| DN20831_c0_g2_i9 | − 1.13 | − 1.38 | #N/A | WTR32_ARATH | 6.6E−171 |
| DN19207_c0_g4_i20 | − 1.10 | − 1.14 | #N/A | ATPA_NEOSM | 4.5E+00 |
| DN18355_c0_g1 | − 1.08 | − 0.78 | #N/A | KUA1_ARATH | 3.4E−35 |
| DN21645_c0_g1_i27 | − 1.06 | − 0.81 | #N/A | TPPA_ARATH | 0.0E+00 |
| DN18779_c1_g3_i3 | − 0.97 | − 0.97 | #N/A | RCA_VIGRR | 1.1E−102 |
| DN16210_c0_g4_i5 | − 0.95 | − 0.75 | #N/A | DJC76_ARATH | 1.3E−29 |
| DN16424_c0_g1_i2 | − 0.86 | − 2.41 | #N/A | CO9_TAKRU | 1.4E+00 |
| DN16521_c0_g2_i3z | 1.29 | 2.10 | #N/A | PUP18_ARATH | 9.9E−01 |
| DN19407_c0_g5_i1 | 1.65 | 5.28 | 1.55 | 14312_ARATH | 5.4E−162 |
| DN18907_c0_g1_i10 | 1.73 | 2.05 | #N/A | SSG1_IPOBA | 0.0E+00 |
| DN17259_c0_g1 | 4.23 | 3.79 | 3.35 | 5MMP_ARATH | 2.3E−38 |
| DN15402_c0_g1_i2 | 4.71 | #N/A | #N/A | CB23_SOYBN | 0.0E+00 |
| DN15174_c0_g1_i14 | #N/A | − 1.17 | #N/A | CB23_PEA | 1.3E−175 |
| DN20172_c0_g1_i11 | 4.87 | 4.76 | #N/A | APA2_ARATH | 0.0E+00 |
| DN21013_c0_g1_i3 | 4.94 | 5.18 | #N/A | CHLD_PEA | 0.0E+00 |
| DN18221_c0_g1 | 5.11 | − 0.85 | 4.41 | BBD1_ARATH | 1.1E−141 |
| DN15371_c1_g2_i9 | 5.12 | 4.47 | #N/A | BH068_ARATH | 1.5E−67 |
| DN15819_c1_g1_i18 | 5.12 | 4.92 | #N/A | RPP2B_ARATH | 2.1E−04 |
| DN21013_c0_g1_i1 | 5.90 | 5.59 | #N/A | CHLD_PEA | 0.0E+00 |
| DN20112_c0_g1_i5 | 6.35 | #N/A | #N/A | GIL1_ARATH | 1.3E−40 |
| DN19249_c0_g1_i14 | #N/A | − 0.96 | 1.30 | GIL1_ARATH | 1.9E−67 |
| DN14475_c1_g1_i3 | 6.72 | 6.16 | #N/A | SEOA_ARATH | 2.4E−102 |
| DN16965_c0_g1 | 7.49 | 8.14 | #N/A | PAP7_ARATH | 2.0E−37 |
| DN21148_c0_g3_i6 | 7.97 | 7.60 | #N/A | VOZ1_ARATH | 1.0E−56 |
| DN19870_c0_g1_i5 | 8.09 | 7.90 | #N/A | HSF24_SOLPE | 6.2E−73 |
| DN20857_c0_g1_i7 | 8.35 | 8.80 | #N/A | RF298_ARATH | 8.6E−02 |
| DN19124_c0_g1_i3 | 8.45 | 8.00 | #N/A | BABA2_DANRE | 3.8E−18 |
| DN22183_c0_g1_i13 | 8.60 | #N/A | #N/A | FUCO2_ARATH | 1.6E−96 |
| DN22183_c1_g1_i16 | #N/A | − 8.27 | #N/A | FUCO2_ARATH | 1.3E−163 |
| DN19377_c0_g1_i6 | 8.66 | 8.87 | #N/A | TSTC_DICDI | 4.8E−06 |
| DN17242_c0_g1_i10 | 8.88 | 8.38 | #N/A | RS31_ARATH | 1.9E−90 |
| DN21974_c0_g1_i11 | 9.03 | 8.77 | #N/A | DNLI4_ARATH | 0.0E+00 |
| DN15112_c1_g2_i5z | 9.15 | 9.93 | #N/A | APRR3_ARATH | 4.7E−07 |
| DN19594_c1_g4_i5 | 9.86 | 7.80 | #N/A | SYP22_ARATH | 3.5E−145 |
| DN21349_c0_g1_i10 | 10.05 | 9.81 | #N/A | TI100_ARATH | 2.3E−134 |
| DN18077_c0_g1_i8 | 10.07 | 9.24 | #N/A | PAB_ARATH | 1.8E−160 |
| DN14378_c1_g1 | 10.08 | 7.41 | #N/A | Y9096_DICDI | 3.9E−06 |
| DN14171_c0_g5_i1z | 10.10 | 9.29 | #N/A | PER31_ARATH | 4.1E−153 |
| DN21622_c0_g1_i26 | 10.17 | 10.89 | #N/A | ACR3_ARATH | 2.3E−166 |
| DN14254_c0_g2 | 10.62 | 9.10 | #N/A | FULL_VITVI | 2.6E−89 |
| DN7082_c0_g1_i1y | 10.77 | 7.81 | 2.97 | MAD50_ORYSJ, ZmSOC1 | 3.0E−119 |
| DN20211_c1_g1_i9 | 10.91 | 11.01 | #N/A | ESYT1_HUMAN | 7.2E−13 |
| DN16691_c0_g6_i5y | 11.27 | 8.08 | 3.14 | bar_gene | |
| DN21779_c0_g1_i16 | 11.28 | 10.91 | #N/A | Y005_SYNY3 | 4.2E−39 |
| DN19935_c0_g1_i1 | 11.51 | #N/A | 11.58 | FERON_ARATH | 0.0E+00 |
| DN21094_c1_g10_i2 | #N/A | 6.78 | #N/A | FERON_ARATH | 2.9E−129 |
zThe transcript verified by RT-qPCR. yTransgenes.
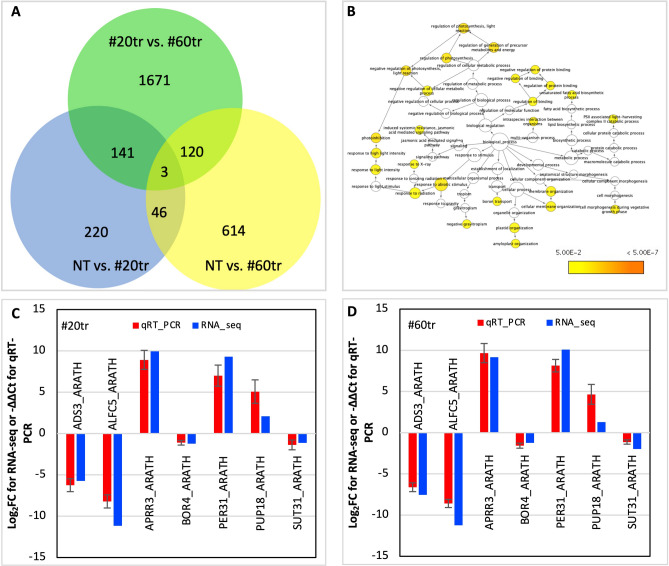
Figure 5.
Transcriptome analysis in leaves among T1 nontransgenic (NT) and two soybean transgenic lines of #20tr and #60tr. (A) Venn diagram illustrating overlap of the three transcriptomic comparisons of the annotated, differentially expressed transcripts (DETs). (B) Gene networks of differentially expressed genes shared in leaf tissues of transgenic lines of #20tr and #60tr. The ontology file of GO_full in BiNGO was used to identify overrepresented GO terms (p < 0.05). Bubble size and color indicate the frequency of the GO term and the P-value, respectively. (C,D) Comparison of the RT-qPCR analysis result and the RNA-seq data of the selected DETs. − ∆∆Ct is an average of three biological and three technical replicates for each DET. GmActin 1 (SAC1_ARATH) was used to normalize the RT-qPCR results.
410 and 782 DETs were identified in the two comparisons between each of the two transgenic lines (i.e., #20tr and #60tr) and the nontransgenic line, respectively (Fig. 5A). Of the annotated soybean genes, 58 DE genes (DEGs), including 31 upregulated, 25 downregulated DEGs and two other DEGs were shared in the two comparisons; the two DEGs (i.e., FUCO2_ARATH and FTSH2_ORYSJ) showed opposite changes in the two transgenic lines (Table 2). Using regular RT-PCR, expression of both the bar and the ZmSOC1 was detected in the transgenic plants but was absent in the nontransgenic ones. In addition, the results of RT-qPCR analysis of seven selected DEGs were consistent with those from RNA-sequencing data (Table 2, Fig. 5).
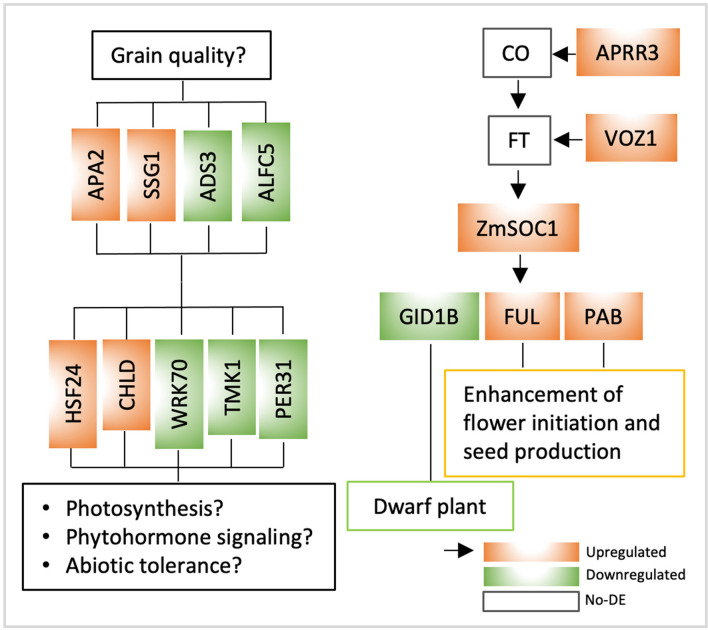
Of the 31 upregulated genes detected in both the #20tr and #60tr lines, the FUL-like MADS-box gene (FULL_VITVI) is a downstream gene of the SOC122; as expected, the expression of the transgene ZmSOC1 promoted flowering in the transgenic soybean plants by enhanced the expression of a soybean’s FUL-like gene. Transcription factor VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1_ARATH) is a positive regulator of plant flowering48,49, and it also binds to the cis-acting region of the PYROPHOSPHATE-ENERGIZED VACUOLAR MEMBRANE PROTON PUMP 1 gene (AVP1) which regulates auxin-mediated organ development and enhance NaCl and drought tolerance50,51. The upregulated VOZ1 resulted in an increased expression of the AVP1 in the #60tr but not in the #20tr line (Table S4). PEROXIDASE 31 (PER31_ARATH) has multifunction related to lignin, auxin catabolism, abiotic stresses. Protein IN CHLOROPLAST ATPASE BIOGENESIS (PAB_ARATH) is required for plant flowering and seed production52. TWO-COMPONENT RESPONSE REGULATOR-like APRR3 (APRR3_ARATH) regulates photoperiodic flowering response53. Chloroplastic, magnesium-chelatase subunit ChlD (CHLD_PEA) is involved in chlorophyll biosynthesis. Protein TIC 100 (TI100_ARATH) plays an important role in chloroplast biogenesis and embryo development in ending seed dormancy54. ASPARTIC PROTEINASE A2 (APA2_ARATH) is involved in lipid metabolic process. GRANULE-BOUND STARCH SYNTHASE 1(SSG1_IPOBA) catalyzes the synthesis of amylose. HEAT SHOCK FACTOR PROTEIN HSF24 (HSF24_ARATH) is a DNA binding protein. The 21 downregulated DETs included six genes expressed only in the nontransgenic line (i.e., MDHC_MEDSA, ALFC5_ARATH, TMK1_ARATH, WRK70_SOLLC, RL10_EUPES, GID1B_ARATH), 15 expressed in both the transgenic and nontransgenic lines (Table 2). CYTOPLASMIC MALATE DEHYDROGENASE (MDHC_MEDSA) is involved in carbohydrate metabolic process. FRUCTOSE-BISPHOSPHATE ALDOLASE 5 (ALFC5_ARATH) hydrolyzes the fructose 1–6-bisphosphate to fructose 6-phosphate. Receptor protein kinase TMK1 (TMK1_ARATH) functions in auxin signal transduction55. WRKY DNA-binding transcription factor 70 (WRK70_SOLLC) modulates various phytohormones signals and affects senescence, biotic and abiotic stress responses56. Gibberellin receptor GID1B (GID1B_ARATH) is a soluble gibberellin (GA) receptor redundant with GID1A and GID1C that control root growth, seed germination, and flower development through GA signaling; loss-of-function of these receptors can lead to plant dwarfing57–59. CHLOROPLASTIC PALMITOYL-MONOGALACTOSYLDIACYLGLYCEROL DELTA-7 DESATURASE (ADS3_ARATH) catalyzes desaturation of fatty acid60,61 (Table 2). 60S RIBOSOMAL PROTEIN L10 (RL10_EUPES) is a component of the small ribosomal subunit. The 58 upregulated and downregulated DEGs could be responsible, at least in part, for the phenotypic changes of the ZmSOC1-expressing plants. The other two shared DEGs along with those non-shared DETs likely contributed to the differences between the two transgenic lines (Table 2, Table S6).
Gene ontology (GO) analysis of the 58 shared DEGs, which were consistently upregulated or downregulated in both transgenic lines, resulted in a total of 41 overrepresented GO terms (P < 0.05) in three networks, including 26 in “biological process”, 18 in “molecular function”, and six in “cellular component” (Fig. 5B, Figure S4). These overrepresented GO terms in the three networks revealed the multi-faceted effect of ZmSOC1 expression in the two soybean transgenic lines.
Discussion
In this study, a maize MADS-box gene ZmSOC1, which has a 54.8% identity to the soybean’s GmSOC1 at the protein level, was constitutively expressed in soybean cultivar Jack. We found that the constitutively expressed ZmSOC1 promoted flowering and reduced plant height, suggesting that the monocot-derived ZmSOC1 functions similarly to the SOC1 in Arabidopsis as a flowering pathway integrator21. Furthermore, we found that the transgenic plants had a 23.2% and a 13.5% of higher grain weight per plant than the nontransgenic plants in the two experiments, respectively. The DEGs responding to the expression of the ZmSOC1 suggested broader effects of the ZmSOC1 on plant height, flowering time, and the other unmeasured traits (e.g., photosynthesis efficiency and abiotic tolerance), which could contribute to yield increase. This is the first report that a constitutively expressed SOC1 gene was evaluated in soybean. We used the ZmSOC1 instead of the GmSOC1 in this study because it was easier to detect integration and expression of the transgene ZmSOC1 than that of a transferred endogenous GmSOC1 in transgenic soybean plants.
ZmSOC1 expression hastens soybean flowering
This is the first demonstration that a major flowering pathway integrator SOC1 from maize functioned in soybean. Unsurprisingly, the expression of the ZmSOC1 promoted soybean plant flowering because the ZmSOC1 is similar to the GmSOC1, which is a SOC1 homology that functions as a positive regulator for plant initiation and flowering21,29,30,33,38. Interestingly, more phenotypic diversities were observed in the T0 plants than the T1 plants because of the differences in the genetic background of different transgenic lines as well as the culture condition for the plants. Remarkably, the type of the altered flowers appeared only in T0 transgenic plants although these abnormal flowers produced no seeds. In this particular case, the abnormal flowers could be caused by the transgene expression because none of the T0 nontransgenic plants had similar abnormal flowers. In general, T0 transformants of soybean obtained through the transformation of half-seed explants were often chimeric, which made the T0 transformants not convincing transgenic targets for phenotyping.
Overexpression of MADS-box gene GmAGL1 was more effective in hastening soybean plant (cv. Jack) flowering under long-day than short-day conditions32. In this study, regardless of the photoperiod variations in the two experiments, the transgenic plants flowered earlier than the nontransgenic plants. The plants grown under short-day conditions in experiment #1 flowered earlier and set pods much later than those under the long-day conditions in experiment #2. The low temperatures during the early development stage in the experiment #1 could have delayed pod set, which could in turn contribute to taller and more branches for each plant observed in the experiment #1 than those in the experiment #2.
ZmSOC1 expression reduces soybean size
ZmSOC1 overexpression reduced the height of maize transgenic plants38. Similarly, the ectopic expression of the ZmSOC1 resulted in the reduction of the transgenic soybean plants. This provides further evidence to demonstrate that manipulation of the expression of a SOC1 homolog is an effective approach to change plant architecture.
Effects of the ZmSOC1 expression on soybean yield and grain quality
Both yield and seed quality are determined by the interactions of genetic and environmental factors. Overexpression of the GmAGL1 in two transgenic lines promoted plant maturity but had no trade-off of yield and grain quality32. In this study, T1 transgenic plants from ten transgenic lines were used to expand the genetic diversity of the transgenic plants, meanwhile, the plants were evaluated under two environmental. It was exciting and convincing that the transgenic plants had a higher yield than the nontransgenic plants grown in the pots although field trials are still needed to evaluate homozygous transgenic plants.
Genes and gene networks responding to the expression of the ZmSOC1
We used RNA sequencing data of two transgenic lines to identify the differentially expressed genes associated with the expression of the ZmSOC1 expression. Although the data only represented the transcriptome at the moment of sampling, the information of the DEGs was very useful to reveal the molecular mechanism that underpinned the phenotypic changes in the transgenic plants (Table 2, Fig. 6). For example, in addition to the upregulated expression of the APRR3 and VOZ1, and PAB, the upregulated FUL gene (FULL_VITVI) was a direct molecular evidence to support the enhanced floral initiation and early flowering in the transgenic plants. The increased expressions of APRR3 and VOZ1 were not associated with the increased CO or FT due possible to the strong expression of both genes in the leaf tissues harvested for sequencing at noon when the sunshine was strong. The downregulated GID1B was the best indicator for the reduced plant size of the transgenic plants because of the known function of the GID1B as a GA receptor in regulating plant height, and flower development57–59. The associations among the expressed ZmSOC1, the upregulated expression of FUL gene, the downregulated GID1B, the promoted flowering, and reduced plant height suggested that the GA signaling pathway (e.g., GID1B) had involved in ZmSOC1 expression-induced flowering and plant dwarfing. This provides a new insight to elucidate the pathway of an overexpressed SOC1 in reducing plant height.
Figure 6.
Potential effects of the expression of the ZmSOC1 on soybean plant growth and development revealed by the shared differential expressed genes (DEGs) identified in leaves of two transgenic lines (i.e., #20tr and #60tr) (Table 2).
In addition, five DEGs (i.e., HSF24, CHLD, WRK70, TMK1, and PER31) involved in plant photosynthesis, phytohormone signaling, or abiotic tolerance (Table 2, Fig. 6) had impact on yield potential in the transgenic plants, and more studies are still needed to investigate photosynthetic efficiency, phytohormone types and contents, and abiotic tolerance.
Conclusion
Soybean is a photoperiod-sensitive crop. Both plant height and days to flowering and maturity are of importance for soybean adaptability and yield. We demonstrated that expression of the ZmSOC1 in soybean was very effective in promoting flowering and reducing plant height. Remarkably, the expression of the ZmSOC1 could increase grain production. Transcriptome analysis of two transgenic lines revealed the genes underpinning the potential phenotypic changes driven by the expression of the ZmSOC1 in the transgenic plants. Overall, the results provided new information to understand SOC1-mediated flowering in soybean. Most importantly, both the phenotypic and transcriptome data of the transgenic plants suggest that modulating expression of a MADS-box gene (e.g., ZmSOC1) is a powerful approach to hasten plant development and thus provides a great potential to enhance plant adaptability and yield.
Supplementary Information
Acknowledgements
We thank Dr. Vance Baird for providing us the soybean cultivar Jack, Dr. Kan Wang at Iowa State University for providing us the pTF101.1 plasmid, and Mr. Pete Callow and John Ryner for phenotypic data collection, Dr. Zhaoming Qi and Ms. Cuihua Gu for seed quality analysis.
Author contributions
G.S. conceived and supervised the study; X.H., G.S., and D.W. conducted the experiments; G.S. and D.W. analyzed data; G.S. wrote the manuscript.
Funding
This research was partially supported by MTRAC Program at Michigan State University, East Lansing, MI.
Data availability
The datasets analyzed during the current study are available from the corresponding author on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92215-x.
References
- 1.Pastor AV, et al. The global nexus of food-trade-water sustaining environmental flows by 2050. Nat. Sustain. 2019;2:499–507. doi: 10.1038/s41893-019-0287-1. [DOI] [Google Scholar]
- 2.Bhattarai MD, Secchi S, Schoof J. Projecting corn and soybeans yields under climate change in a Corn Belt watershed. Agric. Syst. 2017;152:90–99. doi: 10.1016/j.agsy.2016.12.013. [DOI] [Google Scholar]
- 3.Ray DK, et al. Climate change has likely already affected global food production. PLoS One. 2019;14:e0217148. doi: 10.1371/journal.pone.0217148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pareek A, Dhankher OP, Foyer CH. Mitigating the impact of climate change on plant productivity and ecosystem sustainability. J. Exp. Bot. 2020;71:451–456. doi: 10.1093/jxb/erz518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koester RP, Skoneczka JA, Cary TR, Diers BW, Ainsworth EA. Historical gains in soybean (Glycinemax Merr.) seed yield are driven by linear increases in light interception, energy conversion, and partitioning efficiencies. J. Exp. Bot. 2014;65:3311–3321. doi: 10.1093/jxb/eru187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koester RP, Nohl BM, Diers BW, Ainsworth EA. Has photosynthetic capacity increased with 80 years of soybean breeding? An examination of historical soybean cultivars. Plant Cell Environ. 2016;39:1058–1067. doi: 10.1111/pce.12675. [DOI] [PubMed] [Google Scholar]
- 7.Lopez MA, Xavier A, Rainey KM. Phenotypic variation and genetic architecture for photosynthesis and water use efficiency in soybean (Glycinemax L. Merr) Front. Plant Sci. 2019;10:680. doi: 10.3389/fpls.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP. Accelerating yield potential in soybean: Potential targets for biotechnological improvement. Plant Cell Environ. 2012;35:38–52. doi: 10.1111/j.1365-3040.2011.02378.x. [DOI] [PubMed] [Google Scholar]
- 9.Du Y, et al. Shoot/root interactions affect soybean photosynthetic traits and yield formation: A case study of grafting with record-yield cultivars. Front. Plant Sci. 2019;10:445. doi: 10.3389/fpls.2019.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Zonneveld M, et al. Mapping patterns of abiotic and biotic stress resilience uncovers conservation gaps and breeding potential of Vigna wild relatives. Sci. Rep. (U.K.) 2020;10:2111. doi: 10.1038/s41598-020-58646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran LS, Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics. 2010;10:447–462. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- 12.Homrich MS, Wiebke-Strohm B, Weber RL, Bodanese-Zanettini MH. Soybean genetic transformation: A valuable tool for the functional study of genes and the production of agronomically improved plants. Genet. Mol. Biol. 2012;35:998–1010. doi: 10.1590/s1415-47572012000600015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dezar CA, Gago GM, Gonzalez DH, Chan RL. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsisthaliana plants. Transgenic Res. 2005;14:429–440. doi: 10.1007/s11248-005-5076-0. [DOI] [PubMed] [Google Scholar]
- 14.Ribichich KF, et al. Successful field performance in warm and dry environments of soybean expressing the sunflower transcription factor HB4. J. Exp. Bot. 2020;71:3142–3156. doi: 10.1093/jxb/eraa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preuss SB, et al. Expression of the Arabidopsisthaliana BBX32 gene in soybean increases grain yield. PLoS One. 2012;7:e30717. doi: 10.1371/journal.pone.0030717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. U.S.A. 2017;114:172–177. doi: 10.1073/pnas.1616459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycinemax) germplasm. BMC Genomics. 2015;16:217. doi: 10.1186/s12864-015-1441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtan HE, et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011;156:2109–2123. doi: 10.1104/pp.111.177139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19:460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari SB, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010;61:2247–2254. doi: 10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- 22.Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell. 2010 doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 24.Kong F, et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010;154:1220–1231. doi: 10.1104/pp.110.160796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai YP, et al. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 2020;18:298–309. doi: 10.1111/pbi.13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- 27.Huang F, et al. A soybean MADS-box protein modulates floral organ numbers, petal identity and sterility. BMC Plant Biol. 2014;14:89. doi: 10.1186/1471-2229-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan CM, et al. Genome-wide expression analysis of soybean MADS genes showing potential function in the seed development. PLoS One. 2013;8:e62288. doi: 10.1371/journal.pone.0062288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong X, et al. Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol. Biol. Rep. 2012;39:6967–6974. doi: 10.1007/s11033-012-1524-0. [DOI] [PubMed] [Google Scholar]
- 30.Na X, et al. Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean. Plant Cell Rep. 2013;32:1219–1229. doi: 10.1007/s00299-013-1419-0. [DOI] [PubMed] [Google Scholar]
- 31.Ehlers K, et al. The MADS box genes ABS, SHP1, and SHP2 are essential for the coordination of cell divisions in ovule and seed coat development and for endosperm formation in Arabidopsisthaliana. PLoS One. 2016;11:e0165075. doi: 10.1371/journal.pone.0165075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng XR, et al. Soybean MADS-box gene GmAGL1 promotes flowering via the photoperiod pathway. BMC Genomics. 2018;19:51. doi: 10.1186/s12864-017-4402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi YJ, et al. GmAGL1, a MADS-box gene from soybean, is involved in floral organ identity and fruit dehiscence. Front. Plant Sci. 2017;8:175. doi: 10.3389/fpls.2017.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song G-Q, Han X. K-domain technology: Constitutive expression of a blueberry keratin-like domain mimics expression of multiple MADS-box genes in enhancing maize grain yield. Front. Plant Sci. 2021 doi: 10.3389/fpls.2021.664983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song GQ, Walworth A, Zhao DY, Hildebrandt B, Leasia M. Constitutive expression of the K-domain of a Vacciniumcorymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 2013;32:1819–1826. doi: 10.1007/S00299-013-1495-1. [DOI] [PubMed] [Google Scholar]
- 36.Song G-Q, Chen Q. Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 2018;276:10. doi: 10.1016/j.plantsci.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, et al. Overexpression of zmm28 increases maize grain yield in the field. Proc. Natl. Acad. Sci. U.S.A. 2019 doi: 10.1073/pnas.1902593116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alter P, et al. Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiol. 2016;172:389–404. doi: 10.1104/pp.16.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 2006;25:206–213. doi: 10.1007/s00299-005-0048-7. [DOI] [PubMed] [Google Scholar]
- 40.Bubeck DM, Fehr WR, Hammond EG. Inheritance of palmitic and stearic acid mutants of soybean. Crop Sci. 1989;29:5. doi: 10.2135/cropsci1989.0011183X002900030021x. [DOI] [Google Scholar]
- 41.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:5. [Google Scholar]
- 42.Jian B, et al. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008;9:59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamboni A, Pierantoni L, De Franceschi P. Total RNA extraction from strawberry tree (Arbutusunedo) and several other woody plants. Iforest. 2008;1:122–125. doi: 10.3832/Ifor0465-0010122. [DOI] [Google Scholar]
- 44.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/Nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, et al. Assessment of conditions affecting Agrobacterium-mediated soybean transformation and routine recovery of transgenic soybean. Dev. Plant. Gen. 2000;5:88–94. [Google Scholar]
- 48.Yasui Y, et al. The phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 redundantly regulate flowering in arabidopsis. Plant Cell. 2012;24:3248–3263. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Choudhary P, Gupta M, Nath U. VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2 interact with CONSTANS and promote photoperiodic flowering transition. Plant Physiol. 2018;176:2917–2930. doi: 10.1104/pp.17.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li JS, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 51.Gaxiola RA, et al. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao J, et al. PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chloroplast ATP synthase coupling factor 1. Proc. Natl. Acad. Sci. U.S.A. 2015;112:4152–4157. doi: 10.1073/pnas.1413392111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 54.Liang QJ, et al. EMB1211 is required for normal embryo development and influences chloroplast biogenesis in Arabidopsis. Physiol. Plant. 2010;140:380–394. doi: 10.1111/j.1399-3054.2010.01407.x. [DOI] [PubMed] [Google Scholar]
- 55.Xu T, et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atamian HS, Eulgem T, Kaloshian I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta. 2012;235:299–309. doi: 10.1007/s00425-011-1509-6. [DOI] [PubMed] [Google Scholar]
- 57.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakajima M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 59.Iuchi S, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 2007;50:958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- 60.Heilmann I, Pidkowich MS, Girke T, Shanklin J. Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10266–10271. doi: 10.1073/pnas.0402200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 2004;136:4237–4245. doi: 10.1104/pp.104.052951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on request.