Abstract
The nucleus pulposus (NP) of the intervertebral disc plays a critical role in distributing mechanical loads to the axial skeleton. Alterations in NP cells and, consequently, NP matrix are some of the earliest changes in the development of disc degeneration. Previous studies demonstrated a role for laminin-presenting biomaterials in promoting a healthy phenotype for human NP cells from degenerated tissue. Here we investigate the use of laminin-mimetic peptides presented individually or in combination on a poly(ethylene) glycol hydrogel as a platform to modulate the behaviors of degenerative human NP cells. Data confirm that NP cells attach to select laminin-mimetic peptides that results in cell signaling downstream of integrin and syndecan binding. Furthermore, the peptide-functionalized hydrogels demonstrate an ability to promote cell behaviors that mimic that of full-length laminins. These results identify a set of peptides that can be used to regulate NP cell behaviors toward a regenerative engineering strategy.
Keywords: Intervertebral disc, mechanobiology, integrin, syndecan, poly(ethylene) glycol
Introduction
Low back pain is a leading cause of years lived with disability (affecting ~80%–90% of people worldwide), and together with degenerative conditions of the intervertebral disc (IVD), represents a global socioeconomic and medical burden.1–3 The IVD is located between adjacent vertebrae in the spinal column and is made up of the nucleus pulposus (NP), anulus fibrosus, and cartilaginous endplates.4,5 These structures collectively distribute mechanical forces applied to the axial skeleton during activities of daily living and contribute flexibility to the spine.6–11 The degenerative cascade in the IVD is thought to initiate in the NP structure with changes including tissue dehydration and stiffening and a subsequent loss of disc height.1,4,12,13 Patients who present clinically with these changes may also experience impaired physical function, pain upon motion, and associated disability.1,14,15
When in a healthy, “juvenile” state, the NP is avascular and aneural and contains cells derived from the notochord embedded in a soft (~0.5–1 kPa), highly hydrated (~90% water by wet weight) matrix.16–20 This extracellular matrix contains proteoglycans (including aggrecan), collagens (largely type II), and laminins, amongst other proteins.11,17,21–23 Aging and degeneration, however, result in altered extracellular matrix biosynthesis for cells of the NP including increased collagen type I and reduced aggrecan production; the relative loss of proteoglycan results in decreased water content and a stiffening of the matrix (10–20 kPa).18,19,21,24–27 The NP cells also undergo changes with maturation and shift from an anabolic, notochord-like phenotype (characterized by large, circular, vacuolated cells in robust clusters) to an altered state in which the cells assume an elongated cell shape, lose their vacuoles, and have reduced biosynthetic capacities.11,17,18,25,26,28 As the NP has a limited intrinsic ability for self-repair, these alterations contribute to a feedforward degenerative cascade that results in progressive damage to the IVD across length scales, motivating an interest in regenerative engineering approaches for NP and IVD repair.7,15,24,25
Biomaterial and tissue engineering strategies for the IVD have been sought as a method to restore characteristics of the native tissues’ structure and function.29,30 A large body of work has demonstrated an ability to functionalize polymers with bio-adhesive ligands which are often either full-length extracellular matrix proteins, or else, small peptide sequences derived from a constituent of the extracellular matrix.31–37 These approaches offer the opportunity to engineer bioactive materials that promote cell attachment and modulate cell behaviors.29,38 While full-length proteins and short peptides have both been utilized for this purpose, the incorporation of peptides in biomaterial design has several advantages over full-length proteins - short peptides possess increased stability and can be more economic.39,40 Additionally, small peptide sequences offer the ability to provide controlled sites of biomaterial-cell interactions.39,40
Laminins are a compositionally minor component of the IVD extracellular matrix that have been shown to play important roles in facilitating cell attachment and mechanotransduction by interacting with cells through cell-membrane receptors including integrins and syndecans.41–43 NP cell interactions with soft (<1 kPa) biomaterials functionalized with full-length laminin proteins have been shown to promote cell attachment, cell clustering, increased biosynthesis of sulfated glycosaminoglycans, and increased expression of NP cell markers including integrin α3 and cytokeratin 8.44,45 Whereas, similarly soft biomaterials that were either unfunctionalized or conjugated with collagen type II did not elicit these same responses.44,45 Together these findings demonstrate that NP cells recognize cues from the laminin and modulate cellular behaviors in response. Full-length laminin proteins, however, contain hundreds of identified bioactive amino acid sequences across the α, β, and γ chains33,46 which can contribute to uncontrolled cellular interactions. Studies conducted over the past several decades have begun to elucidate the adhesive capabilities of laminin-mimetic peptides (LMPs), their role in promoting cell spreading or other cell behaviors, and the cell surface receptors that recognize each sequence.33,47–53 There is therefore an opportunity to use targeted LMPs to mimic the function of full-length laminins in biomaterial design for IVD regeneration.
In prior work, select LMPs (from the globular domain of the α laminin chain including AG73, IKVAV, AG10, and GD-6) were conjugated to polyacrylamide and screened for an ability to promote behaviors consistent with healthy NP cells – cell attachment, production of sulfated glycosaminoglycans (sGAGs), and expression of aggrecan, N-cadherin, GLUT1, and collagen I/II mRNA.40 In follow-on work, we recently developed an LMP-functionalized biomaterial using a poly(ethylene) glycol (PEG) backbone.54 PEG was chosen from amongst other hydrogel systems commonly used for NP repair29,55,56 as it is clinically relevant,57,58 offers independent control of substrate stiffness and peptide density,54 and allows for the bioactive component of the hydrogel to be controlled solely by the ligands functionalized to it.29,45,54 While gels made of 4% PEG (0.3–0.6 kPa) approximate the stiffness of the healthy human NP ECM16,19,24 and have demonstrated an ability to promote de-differentiation in degenerative NP cells,45,54,59 stiff gels may have advantages for clinical use including improved handling properties and better integration with the native ECM.60,61 Characterization of human NP cell attachment, morphology, cytoskeletal organization, and protein/gene expression profiles revealed an ability for stiff (15% PEG, 10.5 kPa) gels functionalized with 100 µM of peptide (50 µM AG73 in combination with 50 µM IKVAV) to recapitulate behaviors seen on soft PEG gels functionalized with full-length laminin (LM-111).54 This finding corroborates that stiffness alone is not the master variable controlling the NP cell phenotype, and that peptide density and sequence are important variables in designing a bioactive, clinically relevant, material for NP cell support.
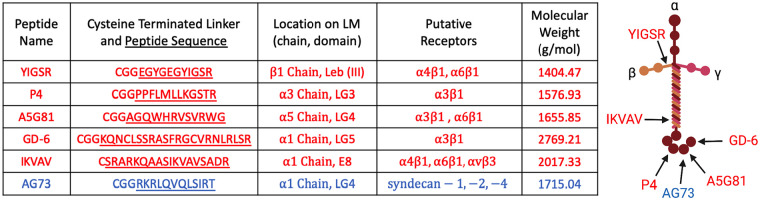
In the present study, a library of biomaterials was synthesized using the peptide-functionalized PEG hydrogel strategy to support development of biomaterials that present a range of LMPs in order to regulate NP cell behaviors. An initial list of candidate peptides from laminin isoforms present in the NP ECM21,23,37,62,63 was created and narrowed based on the reported ability of the peptide sequences to support cell attachment and in keeping with the cell surface receptors believed to mediate attachment for NP cells. The integrin-binding LMPs (YIGSR, P4, A5G81, GD-6, and IKVAV) used herein (Figure 1) were chosen based on prior literature which has validated their ability to promote attachment of NP cells or other cell types through integrins including α3 and α6,40,54,64–68 proteins known to be expressed in NP cells.41,42 The syndecan-binding peptide AG73 was also chosen for evaluation based on prior work suggesting that AG73 can facilitate NP cell attachment.40,54 The objective of this work was to evaluate the effects of presenting integrin-binding and syndecan-binding peptides individually or in combination upon the peptide-functionalized PEG hydrogels as measured by the ability of the biomaterial to promote NP cell attachment, modulate cell morphology, engage mechanosensitive signaling pathways, and regulate protein/gene expression.
Figure 1.
Laminin-mimetic peptides used to functionalize PEG hydrogels. (Left) Table of peptides, amino acid sequence, location on laminin protein, and putative cell surface receptors. (Right) Location of peptide sequences on the laminin protein, created with BioRender.com.
blue: syndecan-binding peptide; red: integrin-binding peptides.
Methods and materials
Primary human NP cell culture
NP tissue was obtained from to-be-discarded surgical waste tissues of anonymized patients (only sex, age and race were recorded; ages 16–75, male and female) receiving surgical treatment for degenerative conditions of the IVD. NP cells were enzymatically isolated from tissues as previously described.54,69,70 Briefly, NP tissue was digested at 37°C for 2–4 h (0.2% pronase (Roche; Basel, Switzerland), 0.4% collagenase type II (Worthington Biochemical; Lakewood, NJ), and 5% Fetal Bovine Serum (FBS); 23 ml per gram tissue). The resulting solution was passed through a 70 µm filter in order to isolate the NP cells which were then expanded in monolayer culture using Ham’s F12 media (Life Technologies; Carlsbad, CA) supplemented with 1% penicillin/streptomycin and 10% FBS under 5% CO2 and atmospheric O2 at 37°C. Expanded cells were used for experimentation at passages 0–4; all experiments made use of multiple samples (biological replicates) from at least three human subjects with assay-specific sample sizes as detailed below.
Formation of hydrogels
Laminin-mimetic Peptide (LMP) Hydrogels 15% PEG hydrogels (% w/v; ~10.5 kPa) were formulated as previously described.54 Briefly, 8-arm star PEG terminated with maleimide (PEG-8MAL, MW 20 kDa) and PEG-dithiol (SH-PEG-SH, MW 600 Da) were obtained from Creative PEGWorks (Durham, NC). Cysteine-terminated lyophilized peptides (YIGSR, P4, A5G81, GD-6, IKVAV, and AG73; sequences listed in Figure 1, GenScript Biotech, Piscataway, NJ) and PEG-MAL were each dissolved in 1X PBS (pH 3.25; acidic conditions were used to control reaction speed54,71) and combined to achieve a final total peptide concentration of 100 µM and functionalization of the PEG backbone through a maleimide-thiol Michael-type addition reaction. Gels were functionalized with a single peptide (100 µM) or with two peptides (dual peptide; 50 µM of each to achieve a total of 100 µM). Separately, PEG-dithiol was also dissolved in 1X PBS pH 3.25. To form gels (at room temperature), the PEG-dithiol crosslinker was added to the PEG-MAL and peptide solution in order to initiate gelation through a second maleimide-thiol Michael-type addition reaction (Figure 2, Supplemental Figure 1). Gel solution was pipetted into chamber slides (Nunc Lab-Tek Chamber Slide Systems™, ThermoFisher Scientific, Waltham, MA) or well plates as needed for subsequent assays. Gels were neutralized in 1X PBS pH 7.4 and allowed to swell overnight at 4°C before use for experimentation.
Figure 2.
Schematic demonstrating gel formation and subsequent experimentation. PEG hydrogels can be functionalized with an integrin-binding peptide (red) and/or a syndecan-binding peptide (blue) using maleimide-thiol based Michael-type addition reactions. Image created with BioRender.com.
Hydrogels Functionalized with LM-111 4% PEG gels presenting full-length laminin (LM-111) were prepared as previously described (Supplemental Figure 1).45,54,59 PEGylated laminin (PEGLM) was formed by reacting Acrylate-PEG-hydroxysuccinimide (Ac-PEG-NHS, 10 kDa, Creative PEGWorks; Winston-Salem, NC) with laminin-111 (Trevigen; Gaithersburg, MD). Unreacted Ac-PEG-NHS was removed by dialyzing the PEGLM solution against PBS. The concentration of PEGLM was determined via 280 nm absorbance and then diluted in 1X PBS (pH 7.4) to 0.5 mg/ml and allowed to react with 8-arm PEG-Ac (4% w/v; 10 kDa; Creative PEGworks) and 10% (v/v) Irgacure 2959 (BASF, Ludwigshafen, Germany). Polymerization occurred upon exposure to UV light and gels were allowed to swell overnight in 1X PBS pH 7.4. Material characterization of both the PEG-peptide and PEGLM gels has been previously reported and the hydrogel stiffness has been shown to be a function of PEG incorporation rather than ligand density.45,54,59 15% PEG gels have been shown to produce gels with a bulk substrate stiffness of 10.5 kPa,54 while 4% PEG creates a hydrogel of 0.3–0.6 kPa.54,59
Characterization of NP attachment to hydrogels
Percent Cell Attachment 20,000 cells were seeded on hydrogels (LMP or PEGLM) or untreated chamber slides (negative control) and allowed to attach for 24 h. The cells were then fixed in 4% paraformaldehyde (PFA, 10 min), permeabilized with Triton-X (0.2% diluted in PBS+/+, 10 min), and blocked with 3.75% bovine serum albumin and 5% nonimmune goat serum for 30 min. Samples were then stained with phalloidin (1:200, Alexa Fluor-488, Invitrogen, Carlsbad, CA) to visualize the actin cytoskeleton; nuclei were counterstained with DAPI (2 µg/mL, Sigma Aldrich, St. Louis, MO). For each sample, a minimum of five regions of interest (ROI) were imaged across the gel using confocal microscopy (TCS-SPE with DM6 RGBV confocal microscope; Leica DFC7000T camera; using Leica LAS X core software; Leica Microsystems, Wetzlar, Germany).
Nuclei were counted in the ROIs and total cell attachment was calculated by extrapolating the cell number determined from the ROIs to the full area of the gel. Percent cell attachment was calculated as 100*(calculated cell attachment number/seeded cell number) that is, 100*(the extrapolated value/20,000 cells).
Cell Morphology From the imaged ROIs, cell morphology was categorized as single cells, small clusters (2–3 cells) or large cell clusters (4+ cells).44 Fiji software72 was used to quantify circularity and spread area for cells attaching as single cells or clusters.
Statistical Analysis of cell attachment and cell morphology Assays to measure cell attachment and morphology were performed for each of three separate human tissue samples (biological replicates) per hydrogel condition and a minimum of 70 cells were visualized per human subject. In order to test for differences in percent cell attachment and cell morphology amongst the single-peptide gels (YIGSR, P4, A5G81, GD-6, IKVAV, and AG73), one-way ANOVAs (factor (1) = peptide, levels (6) = peptide sequence) were performed with Tukey’s multiple comparisons tests with repeated measures. Additionally, in order to compare the effects of the LMP gels to biomaterials presenting the full-length laminin, repeated measures one-way ANOVAs with a Dunnett’s multiple comparison were used to test patient-matched data for differences in cell behaviors on the single-peptide gels compared to that observed on 4% PEGLM. Two-tailed t-tests were used to compare cell behavior metrics on gels with dual presentation of LMPs (ex. YIGSR + AG73) to gels functionalized with a single LMP (ex. YIGSR alone or AG73 alone).
Quantifying Effect of Integrin Blocking on Cell Attachment In order to assess the integrin subunits that mediate NP cell attachment to LMPs, integrin blocking was performed. Cells were serum-starved overnight before use for this assay. After being trypsinized and neutralized in trypsin soybean inhibitor, cells (10,000 cells per well) were incubated with integrin blocking or isotype control antibodies (1:50) for 30 min at 37°C before seeding on LMP gels in chamber slides (Table 1). After 2 h, cell attachment was quantified using the CellTiter-Glo (Promega, Madison, WI) assay according to manufacturer’s protocol and previous studies.40,54,58 Relative attachment to LMP gels was calculated as 100*(cell attachment in samples treated with integrin blocking antibodies/samples treated with IgG control antibody). A total of six technical replicates were quantified for each condition from three biological replicates. One-tailed paired t-tests were used to test for reductions in cell attachment between integrin blocked- and IgG-treated samples.
Table 1.
Primary antibodies utilized for immunocytochemistry and integrin blocking.
| Antibodies for Immunocytochemistry | ||
|---|---|---|
| Target | Species; Dilution | Manufacturer |
| YAP | Mouse; 1:100 | Santa Cruz Biotechnology |
| Paxillin | Rabbit; 1:100 | Abcam |
| Antibodies for live-cell integrin blocking (azide-free, 20 µg/mL) | ||
| Integrin | Anti-integrin antibody | Control IgG antibody |
| α3 | Clone P1B5 (MilliporeSigma) | Mouse IgG1 (ThermoFisher Scientific) |
Abcam, Cambridge, UK; MilliporeSigma, St. Louis, MO; Santa Cruz Biotechnology, Dallas, TX; Thermo Fisher Scientific, Waltham, MA.
Quantifying protein phosphorylation
Bioactivity of the gels was assessed by measuring phosphorylation of ERK 1/2 and GSK3β using AlphaLISA kits (PerkinElmer, Waltham, MA) per the manufacturer’s protocol with the following modifications. Serum-starved NP cells were seeded on LMP or PEGLM gels (10,000 cells per gel in 96 1/2 area well) and cultured at 37°C for 15 min (ERK 1/2) or 120 min (GSK3β). These timings were determined based on preliminary experiments (data not shown) and are consistent with previous protocols.58,73–75 After the appropriate culture time, the media was gently aspirated from the wells containing the gels and the lysis buffer was applied for 10 min to lyse adherent cells. The solution was then transferred to empty wells into which the acceptor and donor buffers were subsequently added; this was done in order to avoid interference from the gel. Six technical replicates were quantified for each gel and protein (ERK 1/2 and GSK3β) from three biological replicates. One-way ANOVAs were performed to test for differences in protein phosphorylation amongst single-peptide gels (Tukey’s multiple comparisons test) or compared to PEGLM (Dunnett’s multiple comparison test).
Characterization of gene expression of phenotypic markers
RT-qPCR was conducted to assess expression of genes characteristic of juvenile NP cells.28 A total of 300,000 cells were seeded on each gel condition. After 4 d of culture on 15% PEG LMP-functionalized or 4% PEGLM gels, RNA was extracted, reverse transcribed to cDNA, and qPCR was performed to quantify expression of ACAN, CDH2, GLUT1, COL2A1, and COL1A1 (Table 2). The ∆∆Ct method was used to compare expression of phenotype markers first to the housekeeping genes 18S and GAPDH and then to cells cultured on AG73-functionalized gels. This analysis allowed for the comparison of single integrin-binding LMP gels to the syndecan-binding gel and also allowed dual peptide gels to be compared to single peptide gels. To provide a reference by which to visualize the gene expression data from the LMP gels compared to the full-length protein, gene expression for cells cultured on 4% PEGLM was also quantified (using the same ∆∆Ct method – normalized first to housekeeping genes and then to the AG73-functionalized gels). Gene expression was quantified for each condition as obtained from 3 to 5 biological replicates. Radar plots were generated using LiveGap Charts (https://charts.livegap.com) in order to visualize patterns in gene expression between substrate conditions.
Table 2.
Primers for qRT-PCR.
| Gene | Common name | Product number (applied biosystems) |
|---|---|---|
| ACAN | Aggrecan | Hs00153936_m1 |
| CDH2 | N-Cadherin | Hs00983056_m1 |
| SLC2A1 (GLUT1) | Glucose transporter 1 | Hs00892681_m1 |
| COL2A1 | Collagen type II | Hs00156568_m1 |
| COL1A1 | Collagen type I | Hs00164004_m1 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase (Housekeeping Gene) | 4332649 |
| 18S | 18S Ribosomal RNA (Housekeeping Gene) | Hs99999901_s1 |
Characterization of protein expression and nascent protein production
Further characterization of protein expression on a subset of gels was utilized to confirm that the LMPs are able to support focal adhesion formation, biosynthesis, and intracellular signaling through an additional mechanotransducive pathway (YAP/TAZ). Expanded descriptions of immunolabeling processes are described below. Briefly, following 4 d of cell culture on A5G81- or IKVAV-functionalized gels (10,000 cells per gel in an 8-well chamber slide), cells were fixed using 4% PFA, permeabilized, and immunolabeled as described below with nuclei counterstained using DAPI. An average of 5 ROIs were imaged for each gel and each human subject such that a minimum of 35 cells were quantified for each protein respectively as obtained from three biological replicates. Two-tailed t-tests were performed to assess whether differences in these cell behaviors were observed between the two gel conditions.
Quantification of Focal Adhesions To assess the cell-gel interactions, cells on A5G81 or IKVAV gels were stained using anti-paxillin antibodies or respective isotype controls (Table 1) and an Alexa Fluor secondary antibody (1:200). The samples were imaged as previously described and quantification of focal adhesion area was performed in Fiji software according to previously reported protocols.54,72,76
Quantification of YAP Localization Cells on IKVAV or A5G81 gels were stained using a mouse anti-YAP primary antibody (1:100; Table 1) or respective isotype controls and an Alexa Fluor secondary antibody (1:200). Sample ROIs were then imaged as previously described. Nuclear and cytoplasmic localization was quantified in Fiji by measuring expression of YAP in regions with DAPI staining (nuclear YAP) and in the extra-nuclear space of the cell body (cytoplasmic YAP). YAP signal was reported as the ratio of nuclear to cytoplasmic protein expression.54,59
Characterization of Biosynthesis Additionally, the biosynthetic capacities of cells cultured on the A5G81 or IKVAV gels were quantified. Fluorescence non-canonical amino acid tagging (FUNCAT) imaging was used to quantify intracellular biosynthesis and extracellular protein deposition as previously described.77–79 Briefly, two media solutions were prepared for use in cell culture – DMEM (without HEPES, sodium pyruvate, L-methionine, L-cysteine, or L-glutamine) was supplemented with 10% FBS, ascorbic acid (Sigma Aldrich), sodium pyruvate (Thermo Fisher Scientific), glutamax (1:100, Thermo Fisher Scientific), 1% penicillin/streptomycin, L-cysteine (Sigma Aldrich), and either L-methionine (control media, Sigma Aldrich) or AHA (L-azidohomoalanine, an analog for L-methionine used for the labeling media, Click Chemistry Tools; Scottsdale, AZ). Cells (10,000 cells per gel in an 8-well chamber slide) were seeded on IKVAV- or A5G81-functionalized gels and cultured for 4 d in the media that containing either the amino acid methionine or the methionine analog (AHA). After this culture period, the media was removed and the samples were incubated for 40 min (37°C, 5% CO2) with DBCO-488 (5 mM DBCO-488 diluted 1:165 in PBS with 1% BSA; Click Chemistry Tools) to label the proteins which incorporated AHA. The samples were then washed, fixed with 4% PFA (10 min), permeabilized in 0.2% Triton-X (10 min), and stained with AlexaFluor phalloidin-633 and DAPI (2 µg/mL) to visualize cytoskeleton and nuclei, respectively. The samples were imaged as previously described for immunostaining (SPE DM6 Leica confocal microscopy) in order to obtain 2D confocal images. Nascent intracellular protein biosynthesis was assessed by quantifying the mean fluorescence intensity of the DBCO channel in the region within the cell body (as determined by co-localization with the phalloidin stain) and extracellular protein deposition was quantified by examining the fluorescence located outside of the cell body.
Dimensionality reduction
As previously described, cell behaviors were characterized on the LMP gels through multiple measures of NP cell phenotype. It is not well understood how distinct types of NP cell responses contribute to the overall NP cell phenotype. Therefore, analysis was performed to distill the multi-dimensional data presented in order to better understand the interplay between cell behaviors and the cues provided by the LMP gels. A data matrix was made representing the average value of the results of those cell behavior metrics which were assessed on all gel conditions (cell attachment, spread area of single cells, spread area of clustered cells, % of cells clustered, circularity of single cells, cell attachment following integrin α3 blocking, and gene expression (ACAN, CDH2, GLUT1, COL2A1, and COL1A1) from biological replicates obtained from three human patients.
Using this data matrix, correlation analysis was calculated in Prism GraphPad (v9 San Diego, CA). A correlation matrix (of correlation coefficient, r, values) was produced within the software as the result of pairwise linear correlations calculated between each of the metrics in order to determine cell behaviors which might be co-regulated in NP cells. Principal components analysis (PCA) was also performed in Prism GraphPad. In order to eliminate variability introduced by simultaneous study of variables with distinct ranges of values, the data were scaled for a mean of zero and a standard deviation of 1 for each variable. PCA was performed on this scaled data matrix and the eigenvalues were used to identify the variance captured by each principal component (PC). Furthermore, the PC scores were used to identify the clustering of LMP gel conditions, and the loadings provided insight into the relative contributions of measured cell behavior metrics to the variance captured in both PC 1 and 2.
Additionally, “leave one out” validation was performed within Prism GraphPad using the following method in order to identify the relative ability of a cell behavior to predict a given gene’s expression. First, a training set was constructed from the data collected on single peptide gels or PEGLM (cell behaviors = cell attachment, single cell area, large cluster area, % clustered, circularity, and attachment following integrin α3 blocking). Next, a single metric (ex. cell attachment) was removed from the training set and Principal Components Regression was performed (in Prism GraphPad) in order to predict the results of gene expression. R2 values were calculated as a metric of goodness of fit of the predicted gene expression values compared to the known gene expression; the removed data were then replaced in the training matrix and the process was repeated for all six-cell behavior metrics of the training data.
Results
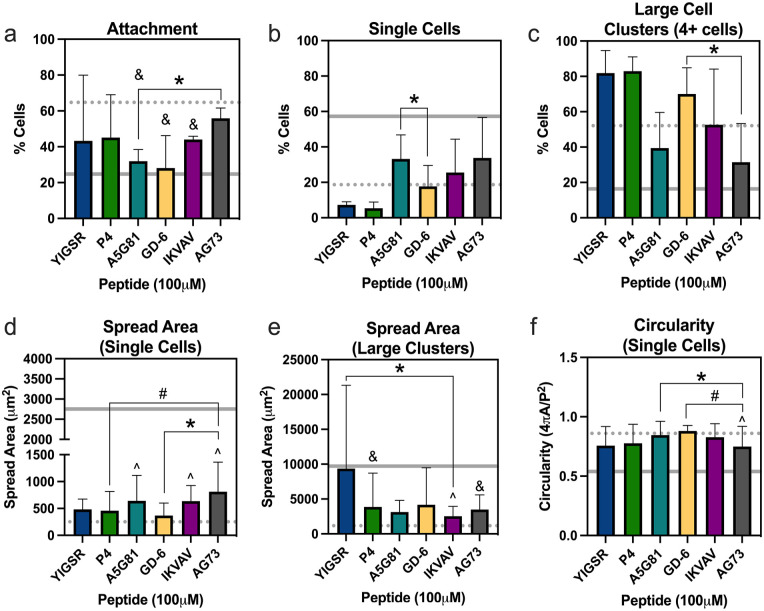
Laminin-mimetic peptide-functionalized gels promote NP cell attachment and rounded, clustered morphologies similar to PEGLM
NP cells attached to 15% LMP gels at levels greater than attachment to uncoated glass when gels were functionalized with one LMP; further cell attachment levels were not statistically different from 4% PEGLM (p > 0.05 for all comparisons; Figure 3(a)). Although cell attachment was similar between most gels functionalized with a single LMP, differences in cell attachment were observed between gels functionalized with A5G81 and those presenting AG73 (p = 0.042; Figure 3(a)).
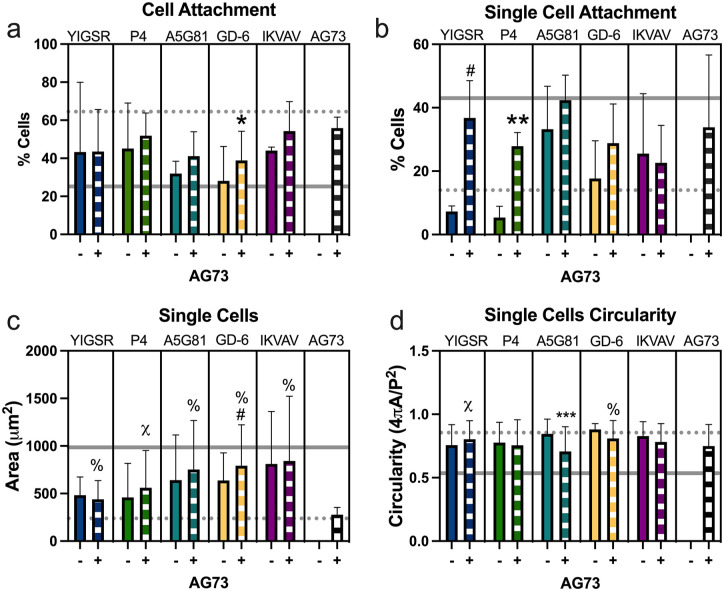
Figure 3.
Stiff gels (15% PEG) functionalized with laminin-mimetic peptides promote cell attachment and morphologies similar to those seen in cells in culture on 4% PEGLM: (a) total cell attachment to the LMP gels after 24 h. Percentage of cells that were adherent as (b) single cells or (c) large cell clusters (4+ cells). Spread area for (d) single cells or (e) large clusters. (f) Circularity of single cells.
For all plots: data represents samples as obtained from three human subjects.
bars: mean ± standard deviation; gray dotted line: average value on 4% PEGLM; gray solid line: average value on glass.
Comparing amongst single peptides: *p < 0.05, #p < 0.09; comparing to PEGLM: ^p < 0.05 and &p < 0.09.
Cell clustering is phenotypic of juvenile NP cells, so we next sought to explore the relative appearance of single cells and cell clusters (Supplemental Figure 2). The frequency of single cells and large clusters in adherent cells on PEGLM resembled those seen on the LMP gels (p > 0.21; Figure 3(b) and (c)). Furthermore, morphologies were generally similar amongst the LMP gel conditions; only A5G81 and GD-6 showed differences in the percentage of single cells (p = 0.028; Figure 3(b)) and fewer large clusters formed on AG73 compared to GD-6 (p = 0.045; Figure 3(c)).
In contrast to degenerative NP cells which become elongated and demonstrate large cell spread areas, juvenile NP cells often assume a rounded cell morphology with relatively small spread areas. Therefore, the cell spread areas and circularity of cells was also quantified. While statistically significant differences were seen in cell spread area for single cells cultured on A5G81, IKVAV, and AG73 compared to PEGLM (p = 0.0018, p = 0.0022, and p < 0.00010 respectively; Figure 3(d)), all single cells on LMP gels showed areas that were reduced compared to cells on glass. Similar single cell areas were observed amongst most LMP gel conditions (Figure 3(d)), although single cell areas were larger on AG73 than they were on GD-6 (p = 0.045) and the difference between P4 and AG73 trended toward significance (p = 0.085). Likewise, the average size of large cell clusters on LMP gels were more similar to those on PEGLM than glass (Figure 3(e)). Large clusters on IKVAV had greater areas than those on PEGLM (p = 0.0092) and differences in large cluster size between PEGLM and P4 or AG73 trended toward significance (p = 0.069 and p = 0.053 respectively). YIGSR promoted the largest cluster size and showed altered cell cluster size compared to IKVAV (p = 0.032; Figure 3(f)). Only AG73 demonstrated circularity that differed compared to PEGLM (p = 0.0082) though circularity also differed between A5G81 and AG73 (p = 0.017) and trended toward significance for GD-6 and AG73 (p = 0.082). Understanding the morphological characteristics of cells on these substrates provides context for how the cells are sensing the underlying biomaterial.
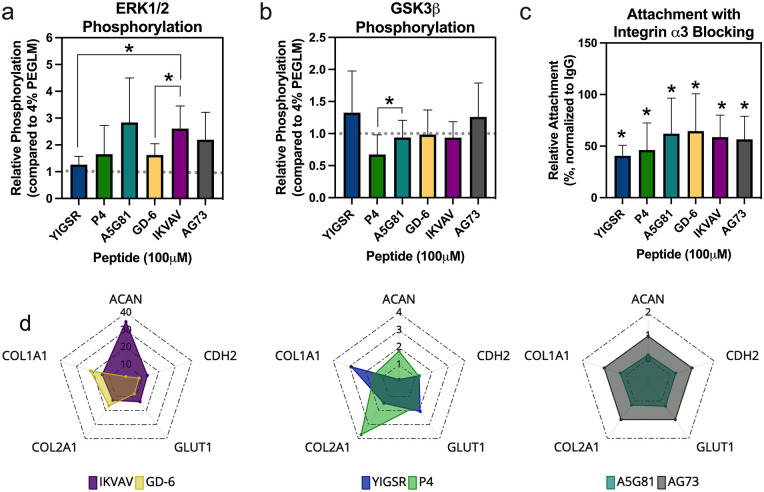
Laminin-mimetic peptides activate mechanosensitive pathways
Cell attachment, morphology, and spreading are known to be downstream of cell receptor interactions with extracellular ligands and subsequent activation of cell signaling pathways. Paxillin-positive focal adhesions were visualized on a subset of LMP gels and were observed to form in the adherent cells (data for A5G81 and IKVAV shown in Supplemental Figure 3(a)). Additionally, intracellular signaling cascades downstream of integrin binding and focal adhesion formation were shown to be activated by LMP gels. ERK 1/2 phosphorylation occurred on both 4% PEGLM and 15% LMP gels and was similar in both systems (Figure 4(a); p > 0.16 for all comparisons); further, activation of ERK1/2 was similar amongst most LMP gels, although IKVAV had increased ERK 1/2 phosphorylation compared to both YIGSR and GD-6 (p = 0.038 and p = 0.023 respectively). GSK3β phosphorylation showed similar trends; LMP gels promoted similar protein phosphorylation as PEGLM (Figure 4(b); p > 0.62 for all comparisons) and only differences between P4 and A5G81 reached levels of statistical significance (p = 0.022). Examination of another mechanosensitive pathway, YAP/TAZ, revealed that YAP was found to be present in both cytoplasmic and nuclear compartments when cells were cultured on a subset of LMP gels (IKVAV and A5G81; Supplemental Figure 3(b)). Additionally, quantification of nascent protein production in these same representative gels demonstrated that culture on LMP gels promoted both intracellular biosynthesis and extracellular protein deposition (Supplemental Figure 3(c)). Together, these findings provide evidence that the LMPs are bioactive and that cells cultured on single LMP-functionalized gels are able to signal through similar pathways as they do when they interact with full-length laminin. Furthermore, inhibition of integrin α3 function reduced cell attachment (p < 0.05) to all LMP gels (Figure 4(c)). Thus, the activation of these pathways is likely, at least in part, regulated by integrin-mediated mechanisms.
Figure 4.
Stiff LMP gels demonstrate similarities in bioactivity but differences in bio-inductive capabilities. Phosphorylation of major signaling proteins: (a) ERK 1/2 and (b) GSK3β. For (a) and (b) gray dotted line = average value on 4% PEGLM; comparing amongst single peptides: *p < 0.05. (c) Cell attachment following blocking of integrin α3; relative cell attachment = 100*(integrin-blocked/IgG control); *p < 0.05 compared to IgG-treated cells. (d) Radar plots depicting average expression for phenotypic markers relative (relative gene expression = 2−∆∆Ct comparing to housekeeping genes and syndecan-binding peptide AG73); scales adjusted as needed to show gene expression on LMP gels.
For all plots: data represents samples as obtained from at least three human subjects.
bars: mean ± standard deviation.
Laminin-mimetic peptides promote differential expression of markers of juvenile NP phenotype
Five markers of NP phenotype (ACAN, CDH2, GLUT1, COL1A1, and COL2A1) were quantified in cells cultured on the hydrogels to screen LMPs for their ability to shift adult degenerative NP cells toward a juvenile-like state. Patterns of gene expression differed between integrin-binding and syndecan-binding LMP gels. IKVAV and GD-6 promoted the greatest expression of all phenotypic markers (Figure 4(d) left). Expression of COL2A1 and COL1A1 were highest in GD-6, and IKVAV promoted the greatest expression of GLUT1, CDH2, and ACAN. YIGSR and P4 demonstrated intermediate gene expression profiles (Figure 4(d) middle). However, A5G81 promoted downregulations of the phenotypic markers (Figure 4(d) right). These findings indicate that amongst the LMPs tested, IKVAV and GD-6 may have the greatest effect in promoting the NP cell phenotype.
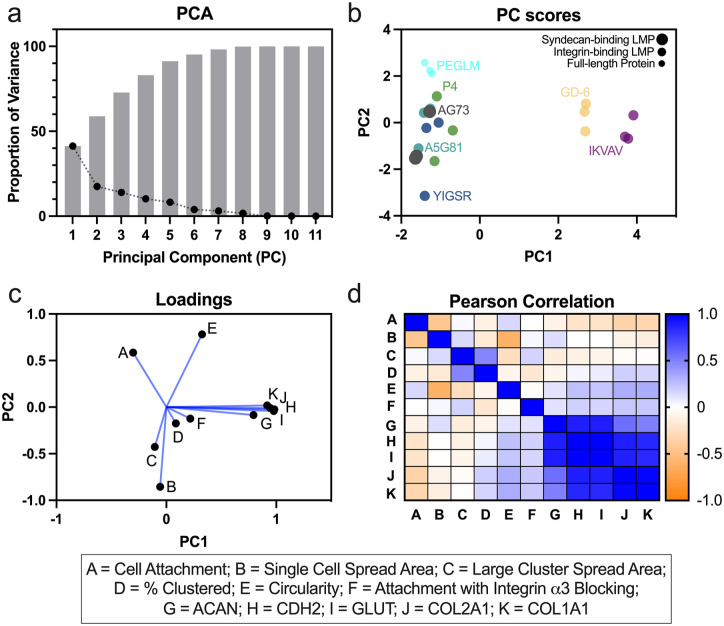
Principal components analysis (PCA) and Pearson correlations provide insights on co-regulated cell behaviors for NP cells cultured on gels functionalized with laminin-mimetic peptides
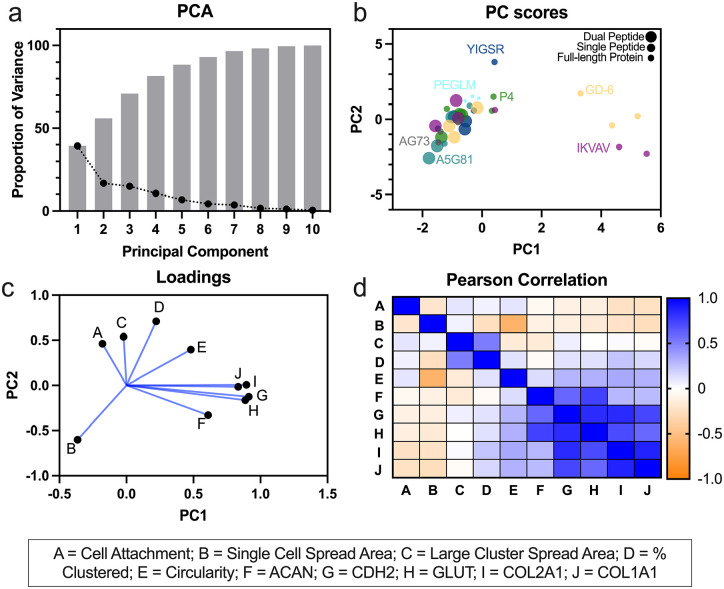
PCA was conducted to reduce dimensionality of the data and to identify clustering amongst LMP conditions and cell behaviors. 75% of total variance was captured by PCs 1–4; 41% and 17% of the variance were captured by PCs 1 and 2 respectively (Figure 5(a)). Examination of the resultant scores from PCA revealed separation of IKVAV and GD-6 from the other LMPs by PC1 (Figure 5(b)). Additionally, to a lesser degree, PC2 was able to separate PEGLM from the LMPs. The loadings from the PCA demonstrated that gene expression of phenotypic markers strongly correlated with PC1 and the gene expression metrics clustered together closely in both PC1 and PC2. Single cell spread area and cell circularity most strongly correlated with PC2, however, these two metrics were negatively correlated with each other (Figure 5(c)).
Figure 5.
Principal components analysis (PCA) and Pearson correlations were utilized to reduce dimensionality and observe co-regulated behaviors for single peptide gels: (a) percent of variance explained by each PC (black dots) and cumulative variance (gray bars), (b) PC Scores shows clustering by LMP; peptides were color coded, size of dot indicates syndecan-binding LMP (largest dot), integrin-binding LMP (medium dot), or full-length protein (smallest dot), (c) loadings show correlations between cell behavior metrics, and (d) Pearson correlation matrix (r values shown with the color map).
These trends were further confirmed through the correlation matrix (Figure 5(d)) which demonstrated strong positive correlations (r > 0.5) amongst the phenotypic markers, moderate negative correlations between single cell area and circularity (r = −0.61) as well as cell attachment and single cell area (r = −0.45), and weak correlations between cell attachment and expression of collagens (r = −0.35). These findings corroborate that LMP gels IKVAV and GD-6 were most different from other substrates based on the gene expression profiles they promote in NP cells. In order to determine the ability of subsets of the data to predict NP phenotypic markers, “leave one out” validation was performed in combination with principal components regression. These data demonstrated that removing the integrin blocking data reduced the ability to predict ACAN expression (Supplemental Figure 4). Further, the ability to predict CDH2, GLUT1, COL2A1, and COL1A1 expression was reduced upon removal of the single cell spread area and single cell circularity data (Supplemental Figure 4). These findings suggest that ACAN expression may be most strongly related to integrin-mediated mechanotransduction, while the other markers tested may be more strongly regulated by cell shape and cellular contractility.
Dual presentation of integrin-binding and syndecan-binding peptides promote similar morphologies as gels functionalized with a single peptides
Having characterized gels functionalized with a single integrin-binding or syndecan-binding peptide, next gels were formulated to present both an integrin-binding peptide and the syndecan-binding peptide AG73 (dual peptide gels). Dual peptide gels demonstrated patterns of cell attachment that largely replicated those seen for the gels functionalized with a single integrin-binding or syndecan-binding peptide (Figure 6(a)) and showed patterns of attachment that were between those seen on glass and PEGLM. Differences in cell attachment between single and dual peptides was only observed for the integrin-binding peptide GD-6; co-presentation of GD-6 and AG73 demonstrated increased cell attachment compared to GD-6 alone (p = 0.028), but not to AG73 alone (p = 0.13).
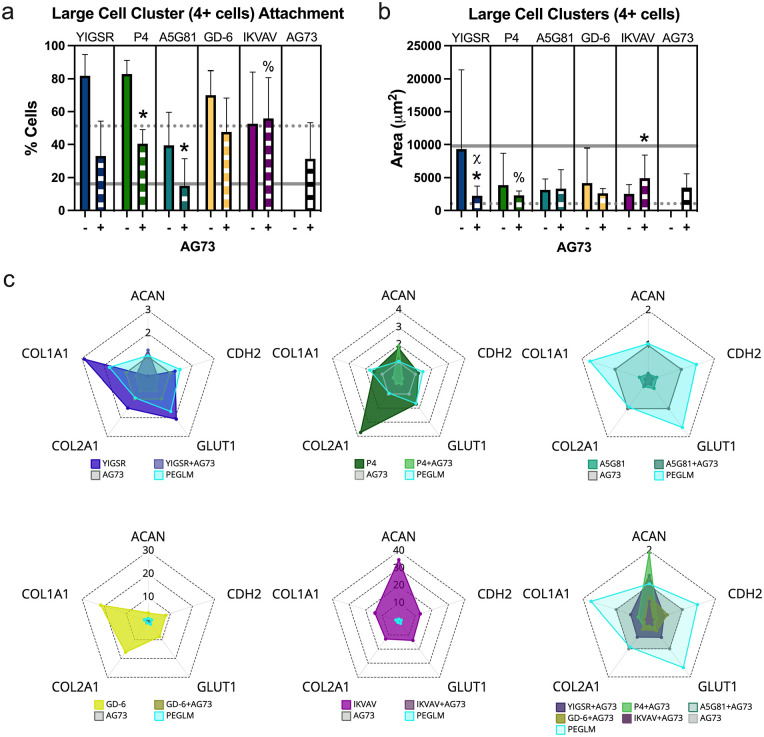
Figure 6.
Characterization of PEG gels with integrin-binding and syndecan-binding peptides and single cell behaviors: (a) total cell attachment for cells seeded on single or dual peptide gels, (b) cell attachment to single or dual peptide gels as single cells, (c) cell spread area for single cells, (d) single cell circularity.
For all plots: data obtained from at least three human subjects.
bars: mean ± standard deviation; dotted line: average value on 4% PEGLM; solid line: average value on glass.
t-Tests comparing integrin binding peptide to dual peptide gels: *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.09; comparing dual peptide gels to syndecan-binding peptide: %p < 0.05, χp < 0.09.
The morphology of attached cells to dual peptide gels was quantified and compared to gels functionalized with a single LMP. Dual presentation of YIGSR and AG73 or P4 and AG73 increased single cell attachment compared to YIGSR (p = 0.063) or P4 (p = 0.0025) alone (Figure 6(b), Supplemental Figure 2). In contrast, all dual peptide conditions promoted attachment of single cells at levels similar to AG73 alone (p > 0.26; Figure 6(b)). For all dual peptide gels, single cell attachment was greater than PEGLM and below glass (Figure 6(b)). Attachment as large clusters (4+ cells) to dual peptide gels was reduced for P4 and AG73 (p = 0.0170) and for A5G81 and AG73 (p = 0.044), compared to the respective single integrin-binding peptide (Figure 7(a)). However, only IKVAV + AG73 showed differential large cell cluster formation compared to AG73 alone (p = 0.0065; Figure 7(a)). Large cell cluster formation was most similar to PEGLM for dual peptide gels that contained IKVAV and AG73 or GD-6 and AG73.
Figure 7.
Characterization of PEG gels with integrin-binding and syndecan-binding peptides on the behavior of cells in large clusters and gene expression profiles: (a) attachment to gels as large clusters, (b) quantification of spread area for large cell clusters, (c) radar plots of relative gene expression for NP phenotypic markers for cells cultured on single peptide gels, dual peptide gels, or PEGLM (relative gene expression = 2−∆∆Ct comparing to housekeeping genes and syndecan-binding peptide AG73); scales adjusted to capture the respective data on a single axis.
For all plots: data from at least three human subjects.
bars: mean ± standard deviation; dotted line: average value on 4% PEGLM; solid line: average value on glass.
t-Tests comparing integrin binding peptide to dual peptide gels: *p < 0.05; comparing dual peptide gels to syndecan-binding peptide: %p < 0.05, χp < 0.09.
We next quantified the cell spread areas for the single cells or large cell clusters. Spread area for single cells was increased for all dual peptide conditions compared to AG73 alone (p < 0.062 for all comparisons; Figure 6(c)), however, compared to gels functionalized with a single integrin-binding peptide, only GD-6 + AG73 showed different single cell spread area (p = 0.059). Large cell cluster area was decreased on dual peptide gels compared to both single peptide gels for YIGSR (p = 0.058 compared to AG73 alone, p = 0.015 compared to YIGSR alone, Figure 7(b)). Cells cultured on gels functionalized with P4 and AG73 had similar cell areas compared to gels which presented P4 only (p = 0.17), but reduced cell cluster area compared to the AG73 gels (p = 0.029). Additionally, gels with IKVAV + AG73 had increased cell areas compared to IKVAV (p = 0.010) but not AG73 (p = 0.20). For both single cells and clusters adherent to dual peptide gels, spread cell areas were observed to be between the size of cells/clusters on soft PEGLM and glass.
All single cells on LMP gels assumed a geometry that was generally round (circularity > 0.7), though several statistically significant differences were observed as a function of LMP presentation (Figure 6(d)). Functionalization of gels with both integrin and syndecan binding peptides promoted significantly reduced cell circularity for A5G81 (p = 0.0001) compared to the gel condition with A5G81 alone. Additionally, gels presenting YIGSR + AG73 and GD-6 + AG73 promoted increased cell circularities compared to gels with only the syndecan binding peptide AG73 (p = 0.08 and p = 0.037).
Gene expression of phenotypic markers was generally highest on gels functionalized with a single integrin-binding peptide (Figure 7(c)). For example, expression of COL1A1, COL2A1, and GLUT1 was highest on YIGSR gels compared to AG73, YIGSR + AG73, or PEGLM gels. Expression of ACAN was similar between YIGSR + AG73, PEGLM, and AG73 gels, and higher on these gels than on gels functionalized with YIGSR alone. In contrast, CDH2 expression was similar between AG73, YIGSR, and PEGLM gels and lowest on YIGSR + AG73.
Dimensionality reduction of dual and single peptide data together demonstrated that PC1 and PC2 accounted for 39% and 16% of total variance respectively (Figure 8(a)). As with the single peptide data (Figure 5), it was observed that IKVAV and GD-6 (without AG73) both separated from the rest of the data along PC1 (Figure 8(b)). Neither PC1 nor PC2 was able to separate the other single and dual LMP conditions. Gene expression for CDH2, GLUT1, COL2A1, and COL1A1 showed strong correlations with PC1 (Figure 8(c)); whereas percentage of clustered cells was the most strongly correlated to PC2 (Figure 8(c)). The correlation matrix confirmed that while most genes were associated, only weak correlations were seen between ACAN and COL2A1 and COL1A1 (Figure 8(d)). The data from both single and dual peptide gels also demonstrated negative correlations between cell circularity and cell area, corroborating that a greater degree of circularity (rounder cells) is associated with decreased cell spread areas.
Figure 8.
Principal components analysis (PCA) and Pearson correlations were utilized to reduce dimensionality and observe co-regulated behaviors for single and dual peptide gels: (a) percent of variance explained by each PC (black dots) and cumulative variance (gray bars), (b) PC Scores shows clustering by LMP; peptides were color coded, size of dot indicates full-length protein (smallest dot), single LMP peptide (medium sized dots), dual LMP peptide (largest dot), (c) Loadings show correlations between cell behavior metrics, and (d) Pearson correlation matrix (r values shown with the color map).
Discussion
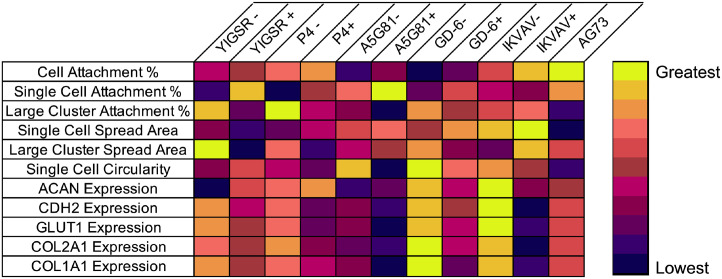
Data from the present study demonstrate the ability of an array of LMPs conjugated to a 15% PEG biomaterial system to regulate NP cell behaviors in a manner similar to 4% PEGLM. The use of peptides offers advantages over full-length proteins in both economic value and ligand specificity. Furthermore, this polymer system is able to rapidly crosslink in situ while retaining the ability to encapsulate and deliver cells to an IVD defect.80 Herein we have screened these peptides for their ability to regulate NP cell attachment, morphology, and phenotype, confirmed the bioactivity of the LMPs, and have identified cell receptors that facilitate mechanotransduction between the cells and the biomaterials (Table 3). Additionally, we have shown that integrin-mediated mechanisms may be most able to de-differentiate degenerative human NP cells toward a juvenile-like state.
Table 3.
Summary of cell behaviors induced by gels functionalized with an integrin binding peptide without (-) or with (+) co-presentation of the syndecan binding peptide AG73. The colormap indicates ranking from the lowest value (purple) to greatest value (yellow)
|
Previous studies have identified the ability for LMPs to modulate cell behaviors in peptide sequence- and cell source-dependent manners. Data from the present study similarly demonstrated that NP cell attachment and morphology metrics such as cell spread area and circularity showed variability amongst the peptide-conjugated hydrogels. Notably, stiff gels functionalized with 100 µM of a single LMP promoted behaviors that generally resembled those of cells on soft gels functionalized with full-length LM-111 and corroborate prior findings of studies which have conjugated LMPs to additional polymeric backbones including polyacrylamide40 and alginate (unpublished data). This is likely in part associated with the observation that interactions between NP cells and the LMP-functionalized gel activated mechanosensitive pathways ERK 1/2 and GSK3β at levels comparable to that seen in soft PEGLM. Engagement of these pathways is known to contribute to regulation of gene expression,81,82 focal adhesion formation,83 and cytoskeletal remodeling84–86 at early time points, all of which ultimately result in cell viability,83–85,87 cell cycle progression,85,88 and motility.84 Additionally, these cellular processes modulate cell differentiation or phenotype.84,89 Prior research has demonstrated that gene expression of NP cell markers can be regulated through microenvironmental conditions such as substrate stiffness, ligand presentation and density, and cell shape as well as relative contributions of cell-cell and cell-matrix interactions (Figure 9).59,70,90–92 Statistical modeling in the present study corroborates these findings and further suggests that engagement of integrin α3 may specifically promote ACAN expression, while CDH2, GLUT1, COL2A1, and COL1A1 transcription may be promoted by processes further downstream of integrin activation including cell shape and cytoskeletal regulation. Despite all the single-LMP gels promoting protein phosphorylation at levels comparable to the full-length laminin, differences were observed between peptides and the full protein for expression of the phenotypic markers. Gene expression of phenotypic markers was greatest on the gels functionalized with IKVAV (maximum expression for ACAN, CDH2, and GLUT1) and GD-6 (maximum expression for COL2A1 and COL1A1) and the lowest expression of phenotypic markers was seen on the A5G81 gels. This finding indicates that specific LMPs may be particularly well-suited to drive biosynthetic activity in NP cells.
Figure 9.
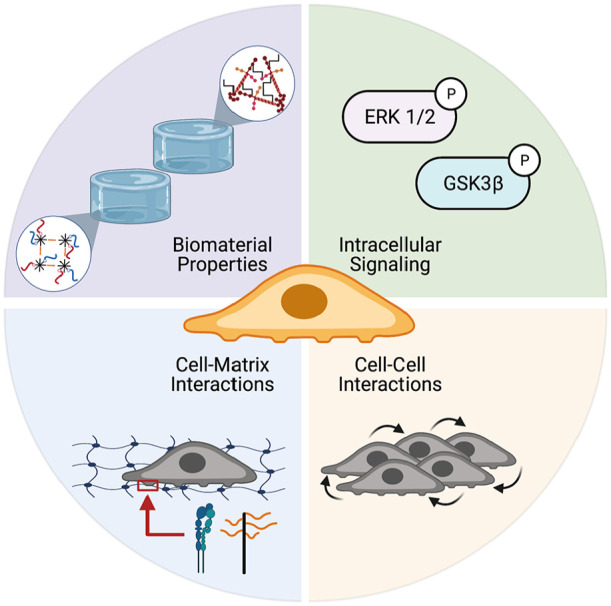
NP cell morphology, biosynthesis, and phenotype can be modulated through biomaterials which present full-length laminins or LMPs that promote intracellular signaling and
development of cell-cell and cell-matrix interactions. Figure created with BioRender.com.
Having characterized biomaterials functionalized with a single LMP, gels were next made that presented an integrin-binding LMP in combination with the syndecan-binding LMP, AG73. Prior reports have indicated a synergistic interaction between integrins and syndecans93–96 in driving cell attachment and intracellular signaling. Thus, a hypothesis for the present study was that dual presentation of an integrin-binding peptide with AG73 would promote increased NP cell attachment and expression of phenotypic markers as compared to single peptide presentation alone. The metrics for cell attachment, cell/cluster morphology, and gene expression generally demonstrated that stiff gels functionalized with 100 µM of total LMP were able to promote similar behaviors as the soft PEGLM. However, when comparing between the single and dual peptide gels, the presence of AG73 tended to be antagonistic to the effects observed for the integrin-mediated modulation of NP cell behavior on dual peptide-functionalized hydrogels. For example, gene expression of NP markers was generally greatest on the single peptide gel, and lowest on the dually-functionalized hydrogel. This effect may be in part due to syndecan phosphorylation resulting in integrin endocytosis and trafficking away from the membrane and thereby altered integrin expression and a decreased ability to interact with the integrin-binding LMPs in the gel, as has been reported in prior studies.93,95,97–99 Future experimentation would be needed to characterize the kinetics of expression of adhesive proteins, focal adhesion turnover, and intracellular signaling in order to elucidate the relative roles of integrins and syndecans in mediating degenerative NP cells interactions with the extracellular environment.86,93,95,99–101
While the dual presentation of integrin-binding and syndecan-binding peptides did not elicit a synergistic response in the present study, combinations of other peptides may have the beneficial effects not specifically observed here.102 For example, combining IKVAV and GD-6 which each promoted the greatest expression of different phenotypic markers may result in an additive or synergistic effect and warrants future study. While we examined a subset of integrin-binding LMPs, the full-length laminin protein contains many other integrin-binding peptides which have not yet been screened for use in NP cell culture. Further characterization of the LMP conditions used herein (or the introduction of other LMPs) should expand on the results of the present study to include the quantification of a broad range of phenotypic markers at the protein level and the results of focal adhesion formation, nascent protein production, and YAP localization that were quantified for A5G81- or IKVAV-functionalized gels should be expanded to include all gel conditions. Another limitation of the present study was the use of 2D culture; additional experimentation will be needed to explore the integrin and syndecan mediated mechanisms in 3D and under physiologic mechanical loading conditions. Lastly, here only degenerative cells were used, however, characterizing juvenile NP cell interactions with LMP gels may demonstrate differences in mechanotransduction between healthy and pathological samples.
Overall the data presented demonstrate characterization of a library of bioactive materials that present either a single LMP or a combination of an integrin- and syndecan-binding peptide on a PEG-hydrogel platform. The results from assessments of cell attachment, morphology, and phenotypic measures validate the ability of this engineered biomaterial constructed from 15% PEG to recapitulate behaviors elicited by soft gels prepared from 4% PEG and functionalized with full-length proteins. Use of a 15% PEG hydrogel platform, with a measured compressive stiffness of 10.5 kPa and rapid gelation time (~10 min), has demonstrated success with regulating the NP cell phenotype and global transcriptome54,103 and can be injected into an IVD defect.80 These findings are clinically relevant as stiffer, injectable and in situ crosslinking biomaterials have advantages as a vehicle to deliver cells in a regenerative engineering strategy to the degenerative disc space.
Supplementary Material
Acknowledgments
The authors would also like to thank Dr. Bailey Fearing and Dr. Chelsey Dunham for their assistance with the PEGLM and FUNCAT techniques respectively.
Footnotes
Authorship: J.E.S designed and performed research, analyzed data, and wrote the manuscript; M.N.B, M.Y.L, Z.Z. and L.J. performed research and analyzed data; J.M.B, M.C.G, and M.P.K. contributed to data collection; L.A.S. contributed to study conception and design and data interpretation. All authors provided approval for the submitted manuscript.
Availability of data and materials: Data and materials are available upon written request to the corresponding author.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Science Foundation (DGE-1745038), National Institutes of Health (R01AR069588, R01AR077678), and the Spencer T. and Ann W. Olin Fellowship for Women in Graduate Study. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of these funding sources.
Informed consent, ethical approval, and human rights: Tissue samples were obtained from to-be-discarded surgical waste tissue (exempt from pathology review, with approval of the Washington University in St. Louis Institutional Review Board). Patients were de-identified and only sex, age and race were obtained for experimental purposes. This was classified as not human subjects research and informed consent of patients was not performed.
Citation diversity statement: Efforts in diverse disciplines have identified that citation bias exists and that the research of woman and other minority scholars are often under-cited relative to the number of published manuscripts in a given research area.104–106 The authors of this paper recognize this bias, its harmful impacts, and have made efforts to reference prior literature that reflect diversity of thought, gender, race, ethnicity, and other factors.
ORCID iD: Julie E Speer  https://orcid.org/0000-0002-2948-4550
https://orcid.org/0000-0002-2948-4550
Supplemental material: Supplemental material for this article is available online.
References
- 1. Hoy D, March L, Brooks P, et al. The global burden of low back pain: estimates from the Global Burden of Disease 2010 study. Ann Rheum Dis 2014; 73: 968–974. [DOI] [PubMed] [Google Scholar]
- 2. Ravindra VM, Senglaub SS, Rattani A, et al. Degenerative lumbar spine disease: estimating global incidence and worldwide volume. Glob Spine J 2018; 8: 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg 2006; 88-A: 21–24. [DOI] [PubMed] [Google Scholar]
- 4. Chan WCW, Sze KL, Samartzis D, et al. Structure and biology of the intervertebral disk in health and disease. Orthop Clin North Am 2011; 42: 447–464. [DOI] [PubMed] [Google Scholar]
- 5. Urban JPG, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther 2003; 5: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fearing BV, Hernandez PA, Setton LA, et al. Mechanotransduction and cell biomechanics of the intervertebral disc. JOR Spine 2018; 1: e1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology 2008; 48: 5–10. [DOI] [PubMed] [Google Scholar]
- 8. Setton LA, Chen J. Mechanobiology of the intervertebral disc and relevance to disc degeneration. J Bone Joint Surg 2006; 88: 52–57. [DOI] [PubMed] [Google Scholar]
- 9. Neidlinger-Wilke C, Galbusera F, Pratsinis H, et al. Mechanical loading of the intervertebral disc: from the macroscopic to the cellular level. Eur Spine J 2014; 23: S333–S343. [DOI] [PubMed] [Google Scholar]
- 10. Nachemson AL. Disc pressure measurements. Spine (Phila Pa 1976) 1981; 6: 93–97. [DOI] [PubMed] [Google Scholar]
- 11. Roughley PJ. Biology of intervertebral disc aging and degeneration. Spine (Phila Pa 1976) 2004; 29: 2691–2699. [DOI] [PubMed] [Google Scholar]
- 12. Kandel R, Roberts S, Urban JPG. Tissue engineering and the intervertebral disc: the challenges. Eur Spine J 2008; 17: S480–S491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weber KT, Jacobsen TD, Maidhof R, et al. Developments in intervertebral disc disease research: pathophysiology, mechanobiology, and therapeutics. Curr Rev Musculoskelet Med 2015; 8: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim C-H, Jee W-H, Son BC, et al. Discogenic lumbar pain: association with MR imaging and CT discography. Eur J Radiol 2005; 54: 431–437. [DOI] [PubMed] [Google Scholar]
- 15. Pfirrmann CWA, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001; 26: 1873–1878. [DOI] [PubMed] [Google Scholar]
- 16. Iatridis JC, Weidenbaum M, Setton LA, et al. Is the nucleus pulposus a solid or a fluid? Mechanical behaviors of the nucleus pulposus of the human intervertebral disc. Spine (Phila Pa 1976) 1996; 21: 1174–1184. [DOI] [PubMed] [Google Scholar]
- 17. Bowles RD, Masuda K, Bonassar LA, et al. Tissue engineering for regeneration and replacement of the intervertebral disc. In: Lanza R, Langer R, Vacanti J. (eds) Principles of tissue engineering. London: Academic Press, pp.1223–1251. [Google Scholar]
- 18. Gilchrist CL, Cao L, Setton LA. Intervertebral disc cell mechanics and mechanobiology. In: Winkelstein BA. (ed.) Orthopaedic biomechanics. Boca Raton: CRC Press, 2013, pp.75–100. [Google Scholar]
- 19. Cloyd JM, Malhotra NR, Weng L, et al. Material properties in unconfined compression of human nucleus pulposus, injectable hyaluronic acid-based hydrogels and tissue engineering scaffolds. Eur Spine J 2007; 16: 1892–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walter C, Davis JT, Mathur J, et al. Physical defects in basement membrane-mimicking collagen-IV matrices trigger cellular EMT and invasion. Integr Biol 2018; 10: 342–355. [DOI] [PubMed] [Google Scholar]
- 21. Chen J, Jing L, Gilchrist CL, et al. Expression of laminin isoforms, receptors, and binding proteins unique to nucleus pulposus cells of immature intervertebral disc. Connect Tissue Res 2009; 50: 294–306. [PMC free article] [PubMed] [Google Scholar]
- 22. Hayes A, Benjamin M, Ralphs JR. Extracellular matrix in development of the intervertebral disc. Matrix Biol 2001; 20: 107–121. [DOI] [PubMed] [Google Scholar]
- 23. Tang X, Richardson WJ, Fitch RD, et al. A new non-enzymatic method for isolating human intervertebral disc cells preserves the phenotype of nucleus pulposus cells. Cytotechnology 2014; 66: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iatridis JC, Setton LA, Weidenbaum M, et al. Alterations in the mechanical behavior of the human lumbar nucleus pulposus with degeneration and aging. J Orthop Res 1997; 15: 318–322. [DOI] [PubMed] [Google Scholar]
- 25. Urban JPG, Roberts S, Ralphs JR. The nucleus of the intervertebral disc from development to degeneration. Am Zool 2000; 40: 53–61. [Google Scholar]
- 26. Hwang PY, Setton LA, Chen J, et al. The role of extracellular matrix elasticity and composition in regulating the nucleus pulposus cell phenotype in the intervertebral disc: a narrative review. J Biomech Eng 2014; 136: 021010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walter BA, Mageswaran P, Mo X, et al. MR elastography–derived stiffness: a biomarker for intervertebral disc degeneration. Radiology 2017; 285: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Risbud MV, Schoepflin ZR, Mwale F, et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res 2015; 33: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitz TC, Salzer E, Crispim JF, et al. Characterization of biomaterials intended for use in the nucleus pulposus of degenerated intervertebral discs. Acta Biomater 2020; 114: 1–15. [DOI] [PubMed] [Google Scholar]
- 30. O’Halloran DM, Pandit AS. Tissue-engineering approach to regenerating the intervertebral disc. Tissue Eng 2007; 13: 1927–1954. [DOI] [PubMed] [Google Scholar]
- 31. Xing H, Lee H, Luo L, et al. Extracellular matrix-derived biomaterials in engineering cell function. Biotechnol Adv 2020; 42: 107421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rahmany MB, Van Dyke M. Biomimetic approaches to modulate cellular adhesion in biomaterials: a review. Acta Biomater 2013; 9: 5431–5437. [DOI] [PubMed] [Google Scholar]
- 33. Katagiri F, Takagi M, Nakamura M, et al. Screening of integrin-binding peptides in a laminin peptide library derived from the mouse laminin b chain short arm regions. Arch Biochem Biophys 2014; 550–551: 33–41. [DOI] [PubMed] [Google Scholar]
- 34. Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol 2005; 23: 47–55. [DOI] [PubMed] [Google Scholar]
- 35. Hamley IW. Small bioactive peptides for biomaterials design and therapeutics. Chem Rev 2017; 117: 14015–14041. [DOI] [PubMed] [Google Scholar]
- 36. Hubbell JA. Bioactive biomaterials. Curr Opin Biotechnol 1999; 10: 123–129. [DOI] [PubMed] [Google Scholar]
- 37. Sun Y, Wang T, Toh W, et al. The role of laminins in cartilaginous tissues: from development to regeneration. Eur Cells Mater 2017; 34: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu WF, Chen CS. Engineering biomaterials to control cell function. Mater Today 2005; 8: 28–35. [Google Scholar]
- 39. Collier JH, Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials 2011; 32: 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bridgen DT, Fearing BV, Jing L, et al. Regulation of human nucleus pulposus cells by peptide-coupled substrates. Acta Biomater 2017; 55: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gilchrist CL, Chen J, Richardson WJ, et al. Functional integrin subunits regulating cell–matrix interactions in the intervertebral disc. J Orthop Res 2007; 25: 829–840. [DOI] [PubMed] [Google Scholar]
- 42. Bridgen DT, Gilchrist CL, Richardson WJ, et al. Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc. J Orthop Res 2013; 31: 1661–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki N, Ichikawa N, Kasai S, et al. Syndecan binding sites in the laminin α1 chain G domain. Biochemistry 2003; 42: 12625–12633. [DOI] [PubMed] [Google Scholar]
- 44. Gilchrist CL, Darling EM, Chen J, et al. Extracellular matrix ligand and stiffness modulate immature nucleus pulposus cell-cell interactions. PLoS One 2011; 6: e27170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francisco AT, Hwang PY, Jeong CG, et al. Photo-crosslinkable laminin-functionalized polyethylene glycol hydrogel for intervertebral disc regeneration. Acta Biomater 2014; 10: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kikkawa Y, Hozumi K, Katagiri F, et al. Laminin-111-derived peptides and cancer. Cell Adhes Migr 2013; 7: 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hozumi K, Ishikawa M, Hayashi T, et al. Identification of cell adhesive sequences in the N-terminal region of the laminin α2 chain. J Biol Chem 2012; 287: 25111–25122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hozumi K, Akizuki T, Yamada Y, et al. Cell adhesive peptide screening of the mouse laminin a 1 chain G domain. Arch Biochem Biophys 2010; 503: 213–222. [DOI] [PubMed] [Google Scholar]
- 49. Kikkawa Y, Sugawara Y, Harashima N, et al. Identification of laminin α5 short arm peptides active for endothelial cell attachment and tube formation. J Pept Sci 2017; 23: 666–673. [DOI] [PubMed] [Google Scholar]
- 50. Siqueira AS, Pinto MP, Cruz MC, et al. Laminin-111 peptide C16 regulates invadopodia activity of malignant cells through β1 integrin, Src and ERK 1/2. Oncotarget 2016; 7: 47904–47917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuratomi Y, Nomizu M, Tanaka K, et al. Laminin gamma 1 chain peptide, C-16 (KAFDITYVRLKF), promotes migration, MMP-9 secretion, and pulmonary metastasis of B16-F10 mouse melanoma cells. Br J Cancer 2002; 86: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nomizu M, Kim WH, Yamamura K, et al. Identification of cell binding sites in the laminin α1 chain carboxyl-terminal globular domain by systematic screening of synthetic peptides. J Biol Chem 1995; 270: 20583–20590. [DOI] [PubMed] [Google Scholar]
- 53. Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J 1990; 4: 148–160. [DOI] [PubMed] [Google Scholar]
- 54. Barcellona MN, Speer JE, Fearing B V, et al. Control of adhesive ligand density for modulation of nucleus pulposus cell phenotype. Biomaterials 2020; 250: 120057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henry N, Clouet J, Le Bideau J, et al. Innovative strategies for intervertebral disc regenerative medicine: From cell therapies to multiscale delivery systems. Biotechnol Adv 2018; 36: 281–294. [DOI] [PubMed] [Google Scholar]
- 56. Choi Y, Park MH, Lee K. Tissue engineering strategies for intervertebral disc treatment using functional polymers. Polymers (Basel) 2019; 11: 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bakaic E, Smeets NMB, Hoare T. Injectable hydrogels based on poly(ethylene glycol) and derivatives as functional biomaterials. RSC Adv 2015; 5: 35469–35486. [Google Scholar]
- 58. Francisco AT, Mancino RJ, Bowles RD, et al. Injectable laminin-functionalized hydrogel for nucleus pulposus regeneration. Biomaterials 2013; 34: 7381–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fearing BV, Jing L, Barcellona MN, et al. Mechanosensitive transcriptional coactivators MRTF-A and YAP/TAZ regulate nucleus pulposus cell phenotype through cell shape. FASEB J 2019; 33: 14022–14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen MH, Wang LL, Chung JJ, et al. Methods to assess shear-thinning hydrogels for application as injectable biomaterials. ACS Biomater Sci Eng 2017; 3: 3146–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bao Q-B, Yuan HA. Prosthetic disc replacement: the future? Clin Orthop Relat Res 2002; 394: 139–145. [DOI] [PubMed] [Google Scholar]
- 62. Gilchrist C, Francisco A, Plopper G, et al. Nucleus pulposus cell-matrix interactions with laminins. Eur Cells Mater 2011; 21: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Riester SM, Lin Y, Wang W, et al. RNA sequencing identifies gene regulatory networks controlling extracellular matrix synthesis in intervertebral disk tissues. J Orthop Res 2018; 36: 1356–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boateng SY, Lateef SS, Mosley W, et al. RGD and YIGSR synthetic peptides facilitate cellular adhesion identical to that of laminin and fibronectin but alter the physiology of neonatal cardiac myocytes. Am J Physiol Physiol 2004; 288: C30–C38. [DOI] [PubMed] [Google Scholar]
- 65. Nomizu M, Kuratomi Y, Ponce ML, et al. Cell adhesive sequences in mouse laminin β1 chain. Arch Biochem Biophys 2000; 378: 311–320. [DOI] [PubMed] [Google Scholar]
- 66. Katagiri F, Ishikawa M, Yamada Y, et al. Screening of integrin-binding peptides from the laminin α4 and α5 chain G domain peptide library. Arch Biochem Biophys 2012; 521: 32–42. [DOI] [PubMed] [Google Scholar]
- 67. Gehlsen K, Sriramarao P, Furcht L, et al. A synthetic peptide derived from the carboxy terminus of the laminin A chain represents a binding site for the alpha 3 beta 1 integrin. J Cell Biol 1992; 117: 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim J-M, Park WH, Min B-M. The PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5 α3 chain is crucial for integrin α3β1 binding and cell adhesion. Exp Cell Res 2005; 304: 317–327. [DOI] [PubMed] [Google Scholar]
- 69. Baer AE, Wang JY, Kraus VB, et al. Collagen gene expression and mechanical properties of intervertebral disc cell-alginate cultures. J Orthop Res 2001; 19: 2–10. [DOI] [PubMed] [Google Scholar]
- 70. Fearing BV, Speer JE, Jing L, et al. Verteporfin treatment controls morphology, phenotype, and global gene expression for cells of the human nucleus pulposus. JOR Spine 2020; 3: e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Darling NJ, Hung Y-S, Sharma S, et al. Controlling the kinetics of thiol-maleimide Michael-type addition gelation kinetics for the generation of homogenous poly(ethylene glycol) hydrogels. Biomaterials 2016; 101: 199–206. [DOI] [PubMed] [Google Scholar]
- 72. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ferletta M, Kikkawa Y, Yu H, et al. Opposing roles of integrin α6Aβ1 and dystroglycan in laminin-mediated extracellular signal-regulated kinase activation. Mol Biol Cell 2003; 14: 2088–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grzesiak JJ, Smith KC, Burton DW, et al. GSK3 and PKB/Akt are associated with integrin-mediated regulation of PTHrP, IL-6 and IL-8 expression in FG pancreatic cancer cells. Int J Cancer 2005; 114: 522–530. [DOI] [PubMed] [Google Scholar]
- 75. Blaustein M, Pérez-Munizaga D, Sánchez MA, et al. Modulation of the Akt pathway reveals a novel link with PERK/eIF2α, which is relevant during hypoxia. PLoS One 2013; 8: e69668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Horzum U, Ozdil B, Pesen-Okvur D. Step-by-step quantitative analysis of focal adhesions. MethodsX 2014; 1: 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dunham C, Havlioglu N, Chamberlain A, et al. Adipose stem cells exhibit mechanical memory and reduce fibrotic contracture in a rat elbow injury model. FASEB J 2020; 34: 12976–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McLeod CM, Mauck RL. High fidelity visualization of cell-to-cell variation and temporal dynamics in nascent extracellular matrix formation. Sci Rep 2016; 6: 38852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. tom Dieck S, Kochen L, Hanus C, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods 2015; 12: 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Barcellona MN. Development of a laminin-mimetic peptide-functionalized hydrogel system for the phenotypic modulation of nucleus pulposus cells of the intervertebral disc. St. Louis, Missouri: Washington University in St. Louis. Dissertation. [Google Scholar]
- 81. Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci 2010; 35: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu F, Yang X, Geng M, et al. Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta Pharm Sin B 2018; 8: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Saleem S, Li J, Yee S-P, et al. β1 integrin/FAK/ERK signalling pathway is essential for human fetal islet cell differentiation and survival. J Pathol 2009; 219: 182–192. [DOI] [PubMed] [Google Scholar]
- 84. Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev 2009; 23: 397–418. [DOI] [PubMed] [Google Scholar]
- 85. Larsen M, Artym VV, Green JA, et al. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol 2006; 18: 463–471. [DOI] [PubMed] [Google Scholar]
- 86. Yamada K, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol 1995; 7: 681–689. [DOI] [PubMed] [Google Scholar]
- 87. Risbud MV, Fertala J, Vresilovic EJ, et al. Nucleus pulposus cells upregulate PI3K/Akt and MEK/ERK signaling pathways under hypoxic conditions and resist apoptosis induced by serum withdrawal. Spine (Phila Pa 1976) 2005; 30: 882–889. [DOI] [PubMed] [Google Scholar]
- 88. Moreno-Layseca P, Streuli CH. Signalling pathways linking integrins with cell cycle progression. Matrix Biol 2014; 34: 144–153. [DOI] [PubMed] [Google Scholar]
- 89. Du J, Zu Y, Li J, et al. Extracellular matrix stiffness dictates Wnt expression through integrin pathway. Sci Rep 2016; 6: 20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu Z, Li C, Meng X, et al. Hypoxia-inducible factor-lα mediates aggrecan and collagen Π expression via NOTCH1 signaling in nucleus pulposus cells during intervertebral disc degeneration. Biochem Biophys Res Commun 2017; 488: 554–561. [DOI] [PubMed] [Google Scholar]
- 91. Zhang Y-H, Zhao C-Q, Jiang L-S, et al. Substrate stiffness regulates apoptosis and the mRNA expression of extracellular matrix regulatory genes in the rat annular cells. Matrix Biol 2011; 30: 135–144. [DOI] [PubMed] [Google Scholar]
- 92. Peck SH, McKee KK, Tobias JW, et al. Whole transcriptome analysis of notochord-derived cells during embryonic formation of the nucleus pulposus. Sci Rep 2017; 7: 10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Roper JA, Williamson RC, Bass MD. Syndecan and integrin interactomes: large complexes in small spaces. Curr Opin Struct Biol 2012; 22: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hozumi K, Kobayashi K, Katagiri F, et al. Syndecan-and integrin-binding peptides synergistically accelerate cell adhesion. FEBS Lett 2010; 584: 3381–3385. [DOI] [PubMed] [Google Scholar]
- 95. Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans. Nat Rev Mol Cell Biol 2007; 8: 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Afratis NA, Nikitovic D, Multhaupt HAB, et al. Syndecans–key regulators of cell signaling and biological functions. FEBS J 2017; 284: 27–41. [DOI] [PubMed] [Google Scholar]
- 97. De Franceschi N, Hamidi H, Alanko J, et al. Integrin traffic-the update. J Cell Sci 2015; 128: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Elfenbein A, Simons M. Syndecan-4 signaling at a glance. J Cell Sci 2013; 126: 3799–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Brooks R, Williamson RC, Bass MD. Syndecan-4 independently regulates multiple small GTPases to promote fibroblast migration during wound healing. Small GTPases 2012; 3: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Valdivia A, Cárdenas A, Brenet M, et al. Syndecan-4/PAR-3 signaling regulates focal adhesion dynamics in mesenchymal cells. Cell Commun Signal 2020; 18: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Costa P, Scales TME, Ivaska J, et al. Integrin-specific control of focal adhesion kinase and RhoA regulates membrane protrusion and invasion. PLoS One 2013; 8: e74659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ali S, Saik JE, Gould DJ, et al. Immobilization of cell-adhesive laminin peptides in degradable PEGDA hydrogels influences endothelial cell tubulogenesis. Biores Open Access 2013; 2: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Barcellona MN, Speer JE, Jing L, et al. Engineered peptide-functionalized hydrogels modulate the RNA transcriptome of human nucleus pulposus cells in vitro. bioRxiv. Epub ahead of print 6 March 2021. DOI: 10.1101/2021.03.05.434094. [DOI] [Google Scholar]
- 104. Dworkin JD, Linn KA, Teich EG, et al. The extent and drivers of gender imbalance in neuroscience reference lists. Nat Neurosci 2020; 23: 918–926. [DOI] [PubMed] [Google Scholar]
- 105. Zurn P, Bassett DS, Rust NC. The citation diversity statement: a practice of transparency, a way of life. Trends Cogn Sci 2020; 24: 669–672. [DOI] [PubMed] [Google Scholar]
- 106. Rowson B, Duma SM, King MR, et al. Citation diversity statement in BMES journals. Ann Biomed Eng 2021; 49: 947–949. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.