Abstract
Apolipoprotein E (apoE) (299 residues) is a highly helical protein that plays a critical role in cholesterol homeostasis. It comprises a four-helix bundle N-terminal (NT) and a C-terminal (CT) domain that can exist in lipid-free and lipid-associated states. In humans, there are two major apoE isoforms, apoE3 and apoE4, which differ in a single residue in the NT domain, with apoE4 strongly increasing risk of Alzheimer’s disease (AD) and cardiovascular diseases (CVD). It has been proposed that the CT domain initiates rapid lipid binding, followed by a slower NT domain helix bundle opening and lipid binding to yield discoidal reconstituted high density lipoprotein (rHDL). However, the contribution of the NT domain on the CT domain organization in HDL remains poorly understood. To understand this, we employed Cys-specific cross-linking and spatially-sensitive fluorophores in the NT and CT domains of apoE3 and apoE4, and in isolated CT domain. We noted that the helices in isolated CT domain are oriented parallel to those in the neighboring molecule in rHDL, whereas full length apoE3 and apoE4 adopt either an anti-parallel or hairpin-like organization. It appears that the bulky NT domain determines the spatial organization of its CT domain in rHDL, a finding that has significance for apoE4, which is more susceptible to proteolytic cleavage in AD brains, showing increased accumulation of neurotoxic NT and CT fragments. We envisage that the structural organization of HDL apoE would have profound functional consequences in its ability to regulate cholesterol homeostasis in AD and CVD.
Keywords: Apolipoprotein E3, Apolipoprotein E4, Lipoproteins, Excimer, Cross-linking, Nascent HDL
1. Introduction
Exchangeable apolipoproteins are critical components of lipoproteins that regulate cholesterol and triglyceride homeostasis in plasma and central nervous system (CNS). Their signature features are their small size (<40 kDa) and the abundance of amphipathic α-helices that allow them to exist in lipid-free (via intra- or inter-molecular helix-helix interactions) or lipid-associated (via helix-lipid interactions) states. Apolipoprotein E (apoE) is an archetypal apolipoprotein, comprised of 299 amino acids (~34 kDa) that are organized as a series of amphipathic α-helices. It is located on very low density lipoproteins (VLDL), chylomicron remnants, and sub-fractions of high density lipoprotein (HDL). The APOE gene is polymorphic in the human population, with three common alleles ε2, ε3, ε4 coding for isoforms apoE2, apoE3 and apoE4, respectively, which differ in amino acids at positions 112/158 (Cys/Cys, Cys/Arg and Arg/Arg, respectively). ApoE3 is considered anti-atherogenic [1,2], while apoE4 is considered a risk factor for cardiovascular disease (CVD) and Alzheimer’s disease (AD) [3–5].
The first two-thirds of apoE comprises the N-terminal (NT) domain (residues 1–191) that is linked to the C-terminal (CT) domain (201–299) via a flexible hinge (192–200) [6,7]. The CT domain has a higher lipid-binding affinity compared to the NT domain. The NT domain is folded as a 4-helix bundle wherein four long amphipathic α-helices (H1 (26–40), H2 (55–79), H3 (89–125) and H4 (131–164)) are organized in an up- and-down motif [8,9]. Helix H4 is rich in basic residues that play a vital role as a ligand for the low density lipoprotein receptor (LDLr) and related family of proteins [10], which participate in receptor-mediated endocytosis leading to lowering of plasma cholesterol levels. An a priori requirement for effective interaction with the LDLr is that the NT domain is in a lipid-associated state, wherein the helix bundle has undergone an opening and the non-polar faces of the helices are in contact with lipids [2]. The CT domain is composed of helices C1, C2 and C3 (encompassed within residues 210 and 276) that promote helix-helix interaction in lipid-free state, or helix-lipid interaction in lipid-associated state [11]. It anchors the entire protein to a lipoprotein particle, allowing the possibility for the NT domain to exist in either a lipid-free helix bundle state, or a lipid-bound open state [12,13]. The transition between the open and helix-bundle conformations of an anchored apoE appear to be regulated by the availability of hydrophobic lipid surface(s) on a lipoprotein particle. The anchoring of apoE to a lipid surface is an essential first step to facilitate helix bundle opening and receptor interaction of the NT domain, leading to eventual cellular clearance of the lipoprotein particle from plasma. It is important to understand factors that regulate this initial event in the generation of nascent HDL.
The NT domain of apoE is more stable when compared with other exchangeable apolipoproteins, with inter- and intra-helical salt bridges, and hydrophobic interactions such as aromatic stacking and leucine-zippers conferring resistance to unfolding and helix bundle opening [2,10,14]. It prefers helix-helix over helix-lipid interactions, a feature that is quintessential to its role as a ligand for the LDLr, allowing it to remain a helix bundle, thereby preventing premature clearance. The CT domain of apoE is less stable than the NT domain, with thermodynamic properties comparable to other apolipoproteins [7]. Structural predictions posit that C1 and C2 are folded as class A amphipathic α-helices (positively charged residues clustered at the polar-nonpolar interface and negatively charged residues at the center of the polar face), preferring helix-lipid over helix-helix interactions, while residues 268–289 form a class G* helix (random distribution of negative and positive residues around the perimeter of the polar face) [15]. Further, apoE CT truncation analysis, analytical ultracentrifugation and size-exclusion analyses revealed that residues 266–289 are involved in protein tetramerization [16–18] while NMR data of a monomeric mutant apoE3 (F257A/W264R/V269A/L279Q/V287E) revealed that helices C1, C2, and C3: (D271-W276) are wrapped around the NT domain helix bundle, an organization that may have been enforced by the multiple mutations at the dimer/tetramer interface [8,11,19,20]. Nevertheless, we propose that the apposition of the CT domain with the NT domain of apoE alters the lipid binding behavior of the CT domain.
In the current study, we examine the influence of the NT domain helix bundle on the spatial disposition of the CT domain in lipid associated state in reconstituted HDL (rHDL). We compare the behavior of the CT domain in isolation and in the context of the intact protein by subjecting various segments of the proteins to site-specific cross-linking and fluorescence spectroscopy to gauge spatial proximity. We show that the presence of the NT domain drives the organization and relative orientation of the CT segment of apoE in rHDL.
2. Materials and methods
2.1. Materials
Bis(maleimido)hexane (BMH) and 1,11-bismaleimido-triethyleneglycol (BM(PEG)3) were obtained from Thermo Scientific (Rockford, IL), N-(1-pyrene)maleimide (NPM), 5-((((2-iodoacetyl)amino)ethyl) amino)napthalene-1-sulfonic acid (IAEDANS) and tris-(2-carboxyethyl) phosphine hydrochloride (TCEP) from Invitrogen (Molecular Probes, Eugene, OR), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and cholesterol from Avanti Polar Lipids (Birmingham, AL), dithiothreitol (DTT) 99% purity from Acros Organics (Morris Plains, NJ), guanidine hydrochloride (GdnHCl) (99% purity) from Fisher Scientific (Fair Lawn, NJ). All solvents used were of analytical grade.
2.2. Constructs and proteins
A pET-20b(+) expression vector bearing ampicillin resistance and encoding the codon-optimized (Eurofins MWG Operon, Louisville, KY, USA) sequence of wild type (WT) apoE3(1–299), apoE4(1–299) or apoE (201–299), the latter also referred to as apoE CT domain, generated in previous studies was used. Single Cys constructs were generated using the QuikChange II Site Directed Mutagenesis kit (Agilent Technology, Stratagene, La Jolla, CA). For apoE3, which has a single Cys at position 112, the endogenous Cys was replaced by Ser (with no significant change in function or fold as described previously [21]) and will be referred to as apoE3. The selected sites for the various single Cys substitutions were determined from the X-ray structure of apoE3 NT (1–191) (PDB ID 1NFN) [9], and NMR structures of apoE3 NT (1–183) (PDB ID 2KC3) [8] and apoE3 rendered monomeric by multiple mutations at the interfacial site (F257A/W264R/V269A/L279Q/V287E) [11] (PDB ID 2L7B); these sites were distributed to sample different segments of the NT and CT domains. For apoE4 and isolated apoE CT domain (residues 201–299), sites closely corresponding to apoE3 were selected since their high resolution structures remain unknown. The probed sites for apoE3 and apoE4 (A29, A62, A102, A216 and A256) are referred by their secondary structural location (H1, H2, H3, C1 and C2, respectively) throughout the study, Table 1. Helix H4 (131–164) was not probed since it bears several critical residues for LDLr binding function. For the isolated CT domain, apoE(201–299), the selected sites (A209, S223, E255 and A277) probed the hinge loop (200–209), C1, C2 and Loop C (277–299), respectively. Since E255 in the CT domain was reported to form a salt bridge with R61 in the NT domain of apoE4 [22], the neighboring A256 was selected for sampling in both full length isoforms. All constructs bear a hexa-His tag that is linked to the N-terminal end of the protein via a TEV protease cleavage site. The proteins were recombinantly expressed in E. coli and purified using a Ni2+-affinity matrix (Hi-Trap chelating column, GE Healthcare, Piscataway, NJ).
Table 1.
The major secondary structural segments of apoE probed in the current study.
| Domain | Residues | Secondary structurea |
|---|---|---|
| NT | 26–40 | Helix H1 |
| NT | 55–79 | Helix H2 |
| NT | 89–125 | Helix H3 |
| Hinge | 200–209 | Hinge Loopb |
| CT | 210–223 | Helix C1 |
| CT | 236–266 | Helix C2 |
| CT | 277–299 | Loop Cb |
Secondary structure from NMR data of a monomeric form of apoE3 [11] (PDB ID 2L7B).
Hinge Loop and Loop C were probed for isolated CT domain only.
2.3. Labeling with AEDANS or NPM
ApoE3 and apoE4 single Cys variants were labeled with IAEDANS as described previously [23]. Briefly, single Cys apoE variants (5 mg in 10 mM ammonium bicarbonate, pH 7.4) were incubated with a two-fold molar excess of 50 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and a 50-fold molar excess of 8 M guanidine hydrochloride (GdnHCl) at 24 °C for 30 min to reduce disulfide bonds and unfold the protein, respectively. This was followed by incubation with a 2-fold molar excess of IAEDANS dissolved in 10 mM Tris-HCl, pH 8.0 containing 150 mM sodium chloride (Tris buffered saline, TBS) at 37 °C for 2 h. Labeled proteins were dialyzed extensively against 10 mM ammonium bicarbonate, pH 7.4 and stored at −20 °C. NPM labeling was carried out with ~5 mg of protein in 10 mM ammonium bicarbonate, pH 7.4 essentially as above and as described previously [24,25] using stock solution of 5 mM NPM in DMSO. The pyrene-labeled protein was applied to a 1 mL Ni-affinity Hi-Trap chelating columns (G.E. Healthcare, Piscataway, NJ) to remove excess unbound NPM, TCEP and GdnHCl. The eluted sample was dialyzed against 10 mM sodium phosphate, pH 7.4 containing 150 mM NaCl (phosphate buffered saline, PBS at 4 °C for 48 h). The stoichiometry of labeling was calculated using molar extinction coefficient (M−1 cm−1) of 44,460 for apoE3 and apoE4, and 16,500 for apoE(201–299) at 280 nm; 5700 for IAEDANS at 340 nm, and 40,000 for pyrene in methanol at 345 nm.
2.4. Preparation and characterization of rHDL containing apoE
Discoidal rHDL containing unlabeled or pyrene-labeled apoE single Cys variants and POPC or a mixture of POPC/cholesterol (9:1, w/w ratio) were prepared by the cholate dialysis method as described previously [26,27]. The initial lipid to protein w/w ratio was always maintained at 2.5:1, and the proteins pre-treated with DTT or TCEP where necessary prior to complex formation. Lipid-bound apoE was separated from unbound lipid-free protein and protein-free lipid by density gradient ultracentrifugation using KBr gradient as described earlier [28]. A qualitative assessment of presence of protein in the various fractions was carried out using the bicinchoninic acid (BCA) colorimetric assay (Thermo Scientific, Rockford, IL) and the presence of protein verified by SDS-PAGE analysis under reducing conditions. The concentration of phospholipids in the pooled fractions containing rHDL was determined using the phospholipid C assay kit (Wako Chemicals, VA USA) and the concentration of proteins by the DC protein assay (Fisher Scientific, Fair Lawn, NJ). The size of rHDL bearing apoE single Cys variants (20–50 μg) was estimated by native-PAGE using a 4–12% gradient Tris-glycine gel for 18 h at 120 V at 4 °C and visualized by staining with 0.5% Amido Black. High molecular weight standards (Amersham HMW Calibration Kit, G.E. Healthcare) were used to obtain an estimate of the mass and size of rHDL particles; the standards, their molecular masses and Stokes’ diameter are as follows: thyroglobulin (669 kDa, 17 nm); ferritin (440 kDa, 12.2 nm); catalase (250 kDa, 9.2 nm) and lactate dehydrogenase (140 kDa, 8.2 nm). The geometry of the rHDL particles was visualized by transmission electron microscopy (TEM) (JEOL 1200 EX electron microscope 90 keV) following negative staining by 1% sodium phosphotungstate [29].
2.5. Cross-linking studies
For sulfhydryl-specific cross-linking, discoidal rHDL containing apoE single Cys variants (10–15 μg) were pre-incubated with a 2–5× fold molar excess of DTT or TCEP over protein for 1–2 h at 37 °C, followed by the specified amount of bis(maleimido)hexane, BMH (Thermo Scientific, Rockford, IL). In a select case, lipid-free apoE3 or apoE4 bearing single Cys on H2 and C1 were mixed prior to reconstitution into discoidal particles to generate rHDL with one of each on a particle in the population. BMH was dissolved in DMSO and the volume added to the rHDL complex kept at <5% v/v; all samples were incubated at 4 °C for 16 h or at 24 °C for 4 h. The reaction was quenched by the addition of SDS-PAGE sample treatment buffer and the samples electrophoresed in a 4–20% acrylamide gradient Tris-glycine gel (Invitrogen, Carlsbad, CA) under reducing conditions and visualized with InstantBlue (Sigma-Aldrich, St. Louis, MO). In control reactions, rHDL was treated with DMSO alone. In select cases (A29C or A62C for NT and S223C or A256C for CT domain, and A62C/A216C mix for apoE3 and apoE4 discoidal particles), 1,11-bismaleimido-triethyleneglycol, BM(PEG)3 (Thermo Scientific, Rockford, IL) was used for cross-linking as described above. As a positive control to ensure cross-linking, an unrelated protein bearing Cys residues, lipid-free apoAI L126C/A158C was employed under similar conditions as those used for the rest of the experiments.
2.6. Fluorescence measurements
Steady state fluorescence analyses were performed on a Perkin-Elmer LS55B fluorometer (Perkin Elmer, Massachusetts) at 24 °C. Fluorescence emission spectra of AEDANS-labeled samples were recorded between 400 and 600 nm following excitation at 340 nm, with the excitation and emission slit widths set at 4.0 nm. Fluorescence polarization (FP) was measured using an excitation wavelength of 340 nm, an emission wavelength of 480 nm, an integration time of 15 s with polarizers in place, and a 5 nm slit width. Fluorescence emission spectra of pyrene-labeled samples were recorded between 350 and 550 nm following excitation at 345 nm, with the excitation and emission slit-widths set at 3–5 nm and a scan speed of 50 nm/min. An average of 10 scans were recorded. Excimer to monomer ratios were calculated by dividing the fluorescence intensity at 460 nm by that for the monomer emission peak at 375 nm. The microenvironment polarity of the fluorescent probe was assessed by the 375 nm to 395 nm monomer peak ratio. The fluorescence emission spectra were normalized with respect to the intensity at 375 nm. Throughout this work, e/m ratio is used as a convenient way to compare different spectra [25], rather than the m/e ratio that has been used in several reports [30–32]. All data for e/m and Py scale measurements are average of 3 experiments.
3. Results
In general, three sites in the NT domain (H1, H2, H3) and two sites in the CT domain (C1 and C2) of apoE3, apoE4 and isolated CT domain were probed, unless otherwise specified. In initial studies, the mobility of each probed site was assessed by FP measurements using AEDANS-labeled apoE3 and apoE4 to estimate the flexibility of each segment in lipid-free protein. FP is routinely used to track the changes in probe mobility, which is dependent upon the local environment, in this case tertiary contacts made by the protein. The magnitude of FP signal is proportional to its rotational relaxation time and depends on the hydrated volume of the protein-conjugated probe. FP values were obtained using AEDANS, a fluorescent probe that is routinely used for protein rotational analysis because of its long lifetime of 10–15 ns. The single Cys variants were labeled with AEDANS, yielding a labeling stoichiometry of ~1.0. The average FP values for the three sites probed in the NT domain in apoE3 and apoE4 are 0.37 and 0.39, respectively, while those in the CT domain for both isoforms was 0.35, Table 2. Previously, we reported FP values of 0.12 and 0.19 for isolated apoE3 NT domain bearing residues 1–191 in WT and single Cys constructs, respectively [21] and 0.35 for isolated CT domain (residues 201–299) [23]. The relatively lower values for isolated NT domain compared to similar sites probed in the full length proteins is expected since the former encompasses a helix bundle and is a compact, globular, monomeric protein. The higher values for the full length apoE are reflective of the tendency for the protein to oligomerize via the CT domain. Thus, in addition to the local mobility, oligomerization of apoE3 via the CT domain is expected to significantly decrease the tumbling of the protein, resulting in increased FP values for the probe located in the NT domain. Interestingly, the FP values for apoE(201–299) were similar to those of full length protein, confirming the tendency of the isolated CT domain to oligomerize.
Table 2.
AEDANS probe mobility in various segments in apoE3 and apoE4.
| Probe location | FP values* | |
|---|---|---|
| ApoE3 | ApoE4 | |
| H1 | 0.404 ± 0.006a,b,c | 0.386 ± 0.035 |
| H2 | 0.342 ± 0.012a | 0.377 ± 0.014 |
| H3 | 0.368 ± 0.012b | 0.411 ± 0.045 |
| C1 | 0.377 ± 0.068 | 0.361 ± 0.013 |
| C2 | 0.332 ± 0.030c | 0.354 ± 0.029 |
Values are means ± SD (n = 3).
P = 0.003 as calculated by Student’s t-test in Microsoft Excel.
P = 0.018 as calculated by Student’s t-test in Microsoft Excel.
P = 0.029 as calculated by Student’s t-test in Microsoft Excel.
Having confirmed that there were no major variances in the flexibility of the probed sites, the single Cys variants were then labeled with NPM under reducing conditions yielding labeling stoichiometry of ~0.7–1.0. NPM is a sulfhydryl-reactive fluorophore that displays enhanced emission upon covalent attachment to -SH group and is used for determining spatial proximity relationships in biomolecules. We exploit two major spectral features of pyrene for our studies: the presence of an excimer (e) band centered ~460 nm when one pyrene is spatially proximal to a neighboring pyrene molecule, followed as ratio of excimer to monomer band I (m) intensity (e/m ratio), and the prominence of monomer band III as an indicator of the polarity of the probe microenvironment, followed as Py scale value (ratio of fluorescence intensities at 375 and 385 nm). The variants were reconstituted with lipid to yield large discoidal rHDL that were on average ~ 16 nm diameter as described previously for WT protein [33] with lipid: protein molar ratio of 98.9 ± 22. Circular dichroism analysis revealed that all pyrene-labeled lipid-free and lipid-bound apoE variants adopted an α-helical structure characterized by a double trough at 208 and 222 nm similar to that of their WT counterparts [6,14,16,17], indicating that neither labeling with pyrene nor interaction with lipids significantly altered the overall fold of the protein (representative far-UV spectra shown in Supplementary materials, Fig. S1).
Fig. 1 shows fluorescence emission spectra of lipid-free and discoidal rHDL particles bearing pyrene-labeled single Cys apoE3 or apoE4 on helix H1, H2, H3, C1 or C2; Table 3 shows the e/m ratios for lipid-free and rHDL bound forms. There were several interesting observations: (i) for apoE3 significant excimer emission was noted for variants with pyrene on C1 and C2 (e/m ratio 0.9–1.5) in lipid-free state while those on H1, H2 and H3 had e/m of <0.5 (Fig. 1, left, black); (ii) in contrast for apoE4, significant excimer emission was noted for variants with pyrene not only on C1 and C2 (e/m 1.5–2.2) but also on H1 and H3 and to a notable extent on H2 (e/m 0.6–2.0) (Fig. 1, right, black); (iii) for both apoE3 and apoE4 a significant decrease in excimer emission was noted regardless of the location of pyrene in the rHDL bound form (Fig. 1, left and right, red). The decrease in excimer emission in rHDL state was particularly noteworthy for pyrenes located in the CT end (<15% of that noted in lipid-free state); the excimer emission decreased to 30–40% of that in lipid-free state for E3 variants, and 12–26% for apoE4 variants; (iv) Py scale values were <1 for all probed locations for lipid-free apoE4 compared to apoE3, indicative of a significantly non-polar micro-environment even in the absence of lipids, Table 4. Considering that all apolipoproteins are highly dynamic and flexible, there is possibility that the spectra of lipid-bound samples may have a minor contribution from that of lipid-free protein that has dissociated from rHDL.
Fig. 1.
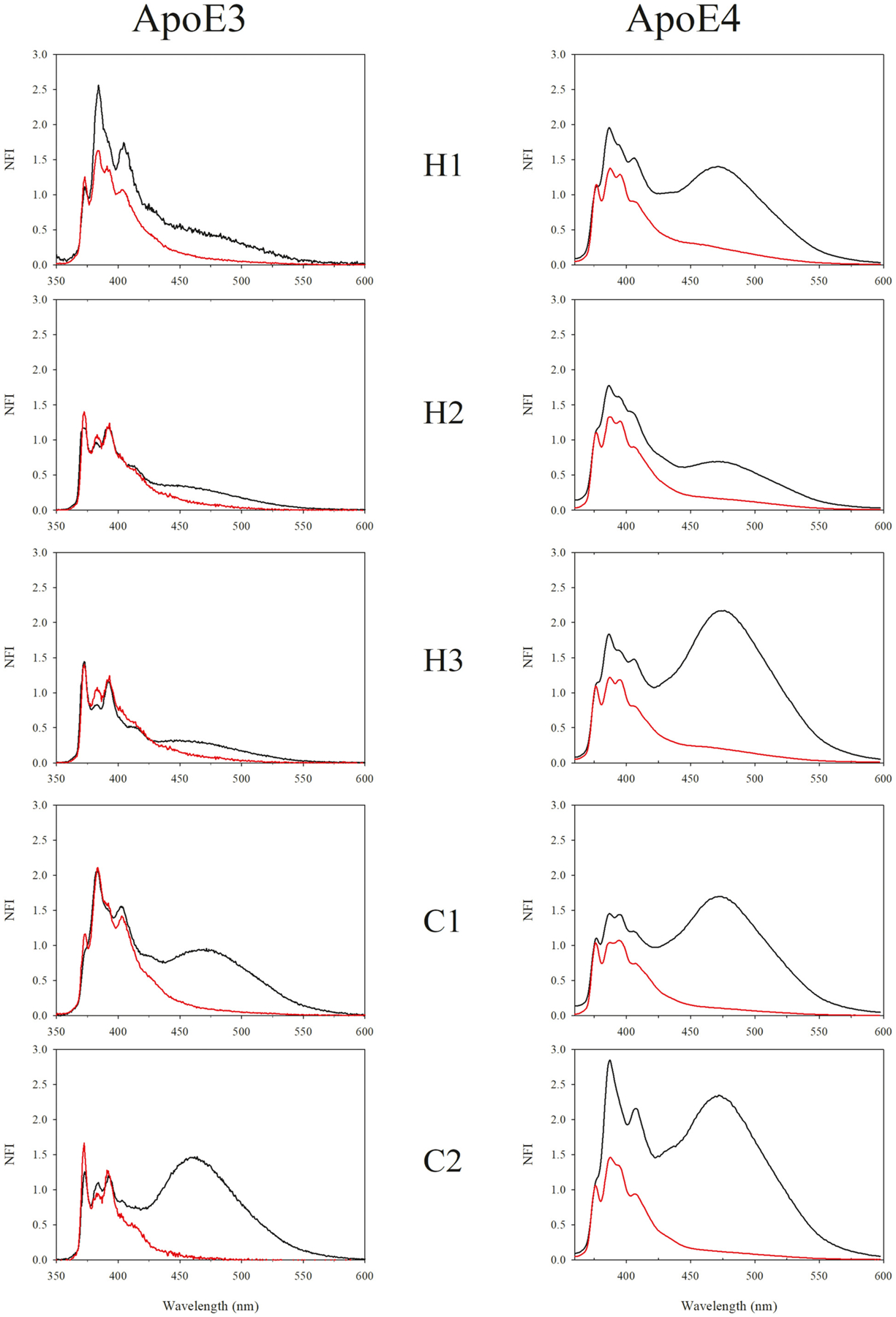
Fluorescence emission spectra of pyrene labeled apoE3 and apoE4 single Cys variants in lipid free and discoidal rHDL bound states. The probe’s location in the different segments is depicted. Emission spectra of all samples (10 μg/mL protein) were recorded at an excitation wavelength at 345 nm. Each spectrum was normalized with respect to the fluorescence emission intensity of the first monomer peak at 375 nm. Black, lipid-free state; red, rHDL bound state. NFI: normalized fluorescence intensity.
Table 3.
Excimer/monomer ratios of pyrene labeled apoE3, apoE4 and isolated CT domain in lipid-free and rHDL associated states.
| Site probed | apoE3 | apoE4 | CT(201–299) | |||
|---|---|---|---|---|---|---|
| Lipid-free | rHDL | Lipid-free | rHDL | Lipid-free | rHDL | |
| Ratio of fluorescence intensities at 460 and 375 nm (e/m)a | ||||||
| H1 | 0.46 | 0.14 | 1.33 | 0.29 | – | – |
| H2 | 0.32 | 0.13 | 0.67 | 0.18 | – | – |
| H3 | 0.29 | 0.11 | 1.96 | 0.23 | – | – |
| Hinge loop | – | – | – | – | 0.55 | 0.28 |
| C1 | 0.91 | 0.14 | 1.56 | 0.13 | 3.33 | 0.28 |
| C2 | 1.45 | 0.06 | 2.22 | 0.14 | 2.63 | 0.13 |
| Loop C | – | – | – | – | 1.82 | 0.34 |
Values shown are derived from spectra that are average of 3 independent experiments.
Table 4.
Py scale (band I/III ratio) of pyrene labeled apoE3, apoE4 and isolated CT domain in lipid-free and rHDL bound states.
| Site probed | apoE3 | apoE4 | CT(201–299) | |||
|---|---|---|---|---|---|---|
| Lipid-free | rHDL | Lipid-free | rHDL | Lipid-free | rHDL | |
| Py scale (ratio of fluorescence intensities at 375 and 385 nm)a | ||||||
| H1 | 0.43 | 0.69 | 0.53 | 0.80 | – | – |
| H2 | 1.10 | 1.02 | 0.58 | 0.81 | – | – |
| H3 | 1.30 | 1.03 | 0.56 | 0.88 | – | – |
| Hinge loop | – | – | – | – | 1.14 | 0.58 |
| C1 | 0.56 | 0.53 | 0.72 | 1.00 | 0.56 | 0.58 |
| C2 | 0.99 | 1.12 | 0.38 | 0.76 | 0.62 | 0.90 |
| Loop C | – | – | – | – | 0.55 | 0.51 |
Values shown are derived from spectra that are average of 3 independent experiments.
Previous studies have shown that the isolated CT domain of apoE likely forms an intermolecular 4-helix bundle, with helices C1 and C2 from one molecule juxtaposed to corresponding segments from neighboring molecules, an inference derived using the pyrene excimer feature [25]. To assess the organization of this isolated CT domain on rHDL, fluorescence emission spectra of rHDL associated pyrene-labeled apoE (201–299) with probes located on helix C1 or C2 were compared with those of corresponding lipid-free protein reported earlier [25], Fig. 2. Significant excimer emission was noted in lipid-free state for pyrenes located on C1, C2 which decreased in lipid-associated state in rHDL. Further, Loop C was probed as an additional precaution, confirming findings obtained with C1 and C2. In contrast, the excimer emission of pyrene located in the Hinge Loop was significantly lower than the other sites for lipid-free protein but elicited a similar trend of decrease upon lipid association. The Py scale values were <1 for probes located on C1, C2 and Loop C in lipid-free and rHDL-associated states.
Fig. 2.
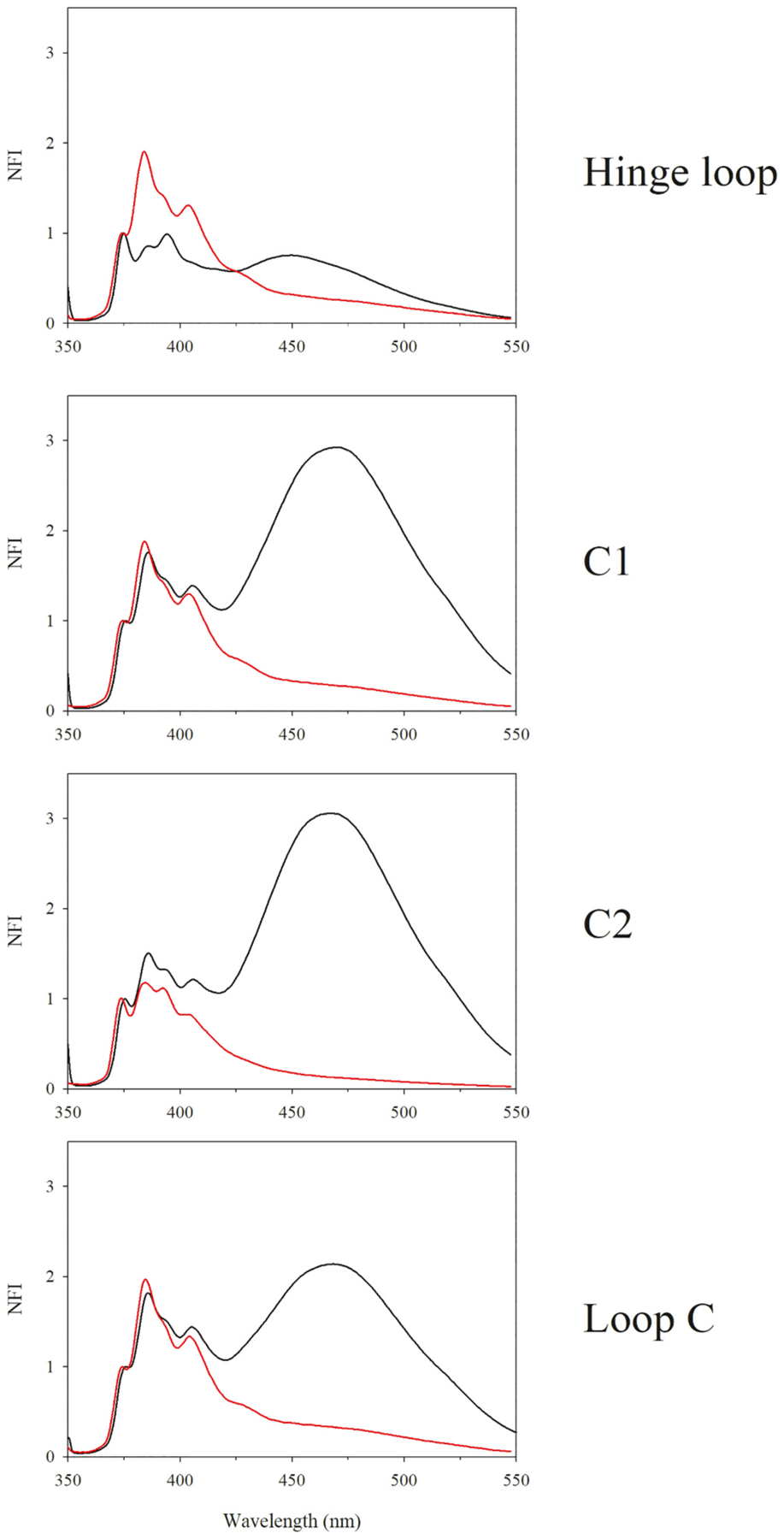
Fluorescence emission spectra of pyrene labeled apoE(201–299) single Cys variants in lipid free and discoidal rHDL bound states. The probe’s location in the different segments is depicted. Emission spectra of all samples (10 μg/mL protein) were recorded at an excitation wavelength at 345 nm. Each spectrum was normalized with respect to the fluorescence emission intensity of the first monomer peak at 375 nm. Black, lipid-free state; red, rHDL bound state. Fluorescence emission spectra of lipid-free pyrene-labeled apoE(201–299) is reproduced with permission from [25]. Copyright {2010} American Chemical Society.
Although the e/m ratio in the rHDL bound states did not reach 0.02, which is the ratio observed for free pyrene in DMSO [25], the significant decrease during the transition from lipid-free to lipid-bound state suggests that pyrenes on two neighboring molecules of intact apoE or isolated CT domain are either farther than 10 Å from each other on discoidal rHDL or that the pyrene rings are not favorably stacked to yield excimer emission.
To distinguish between these possibilities and to obtain independent information about the relative alignment and orientation of apoE3 and apoE4 in discoidal particles, cross-linking analysis was employed using BMH, a sulfhydryl-specific bifunctional reagent with a hydrocarbon spacer-arm of 13 Å between two reactive maleimide groups. The rationale was that if Cys residues on neighboring apoE molecules surrounding the periphery of the phospholipid bilayer in rHDL are within 13 Å of each other, we can expect that the maleimide groups of BMH will react with the two Cys to form stable irreversible covalent thioether cross-links that cannot be cleaved by reducing agents, yielding dimeric bands in reducing SDS-PAGE. To test this, discoidal particles bearing single Cys on helix H1, H2, H3, C1 or C2 in apoE3 or apoE4 were treated with increasing molar concentrations of BMH and electrophoresed by SDS-PAGE under reducing conditions, Fig. 3. A dominant band around 36 kDa corresponding to monomeric apoE was noted in all cases regardless of the location of Cys or the isoform. A very faint band around 72 kDa corresponding to a dimeric form was noted in apoE3 variants, even in the absence of cross-linker, and in lipid-free protein (Fig. 3, left, all lane 2), suggesting presence of an SDS-resistant dimeric species, noted previously in other studies [34]. The absence of a robust dimeric band suggested that the Cys from two neighboring molecules on discoidal rHDL are farther than 13 Å from each other. To investigate the possibility that the Cys on neighboring apoE molecules are located farther than 13 Å, BM(PEG)3, a PEG-based bis-maleimide cross-linker with a spacer arm of 17.8 Å, was employed. Spot checks of BM(PEG)3 cross-linking on the NT and CT domains of apoE3 and apoE4, Fig. 4 revealed no significant cross-linking.
Fig. 3.
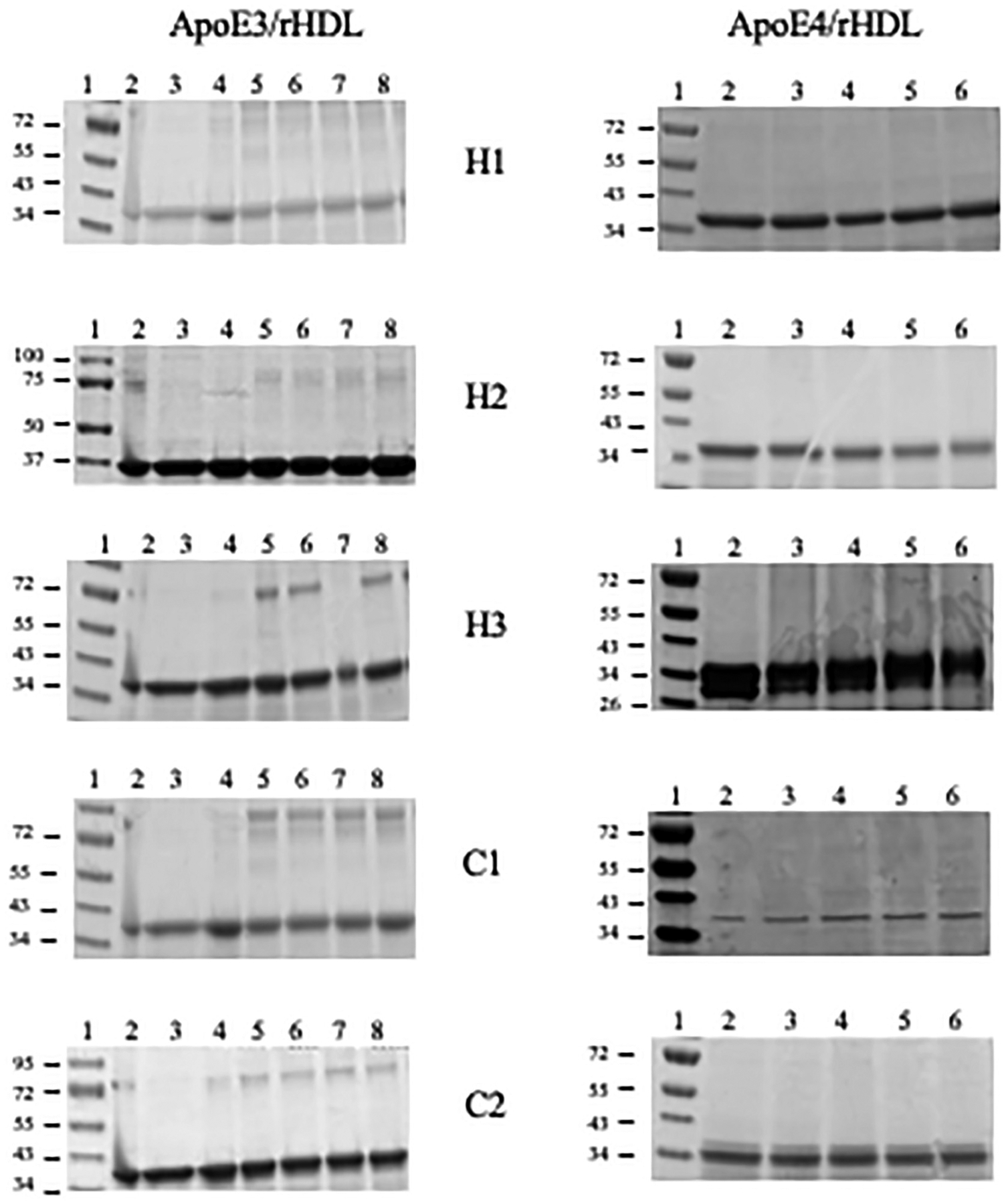
BMH cross-linking analysis of rHDL with apoE3 and apoE4 Cys variants. Discoidal rHDL bearing 10–15 μg apoE3 (left) or apoE4 (right) single Cys variants were treated with BMH in DMSO and electrophoresed on a 4–20% acrylamide gradient SDS-PAGE gel as described under Methods. The lane assignments for apoE3 variants (left) are as follows: Lane 1, molecular mass standards in kDa; lane 2, lipid-free protein; lane 3, rHDL; lane 4, rHDL + DMSO; lanes 5, 6, 7, and 8, rHDL+10-, 25-, 50- and 100- fold molar excess of BMH over apoE3, respectively. The lane assignments for apoE4 variants (right) are: Lane 1, molecular mass standards in kDa; lane 2, rHDL + DMSO; lane 3, 4, 5 and 6, rHDL+10-, 25-, 50- and 100-fold molar excess of BMH over apoE4, respectively.
Fig. 4.
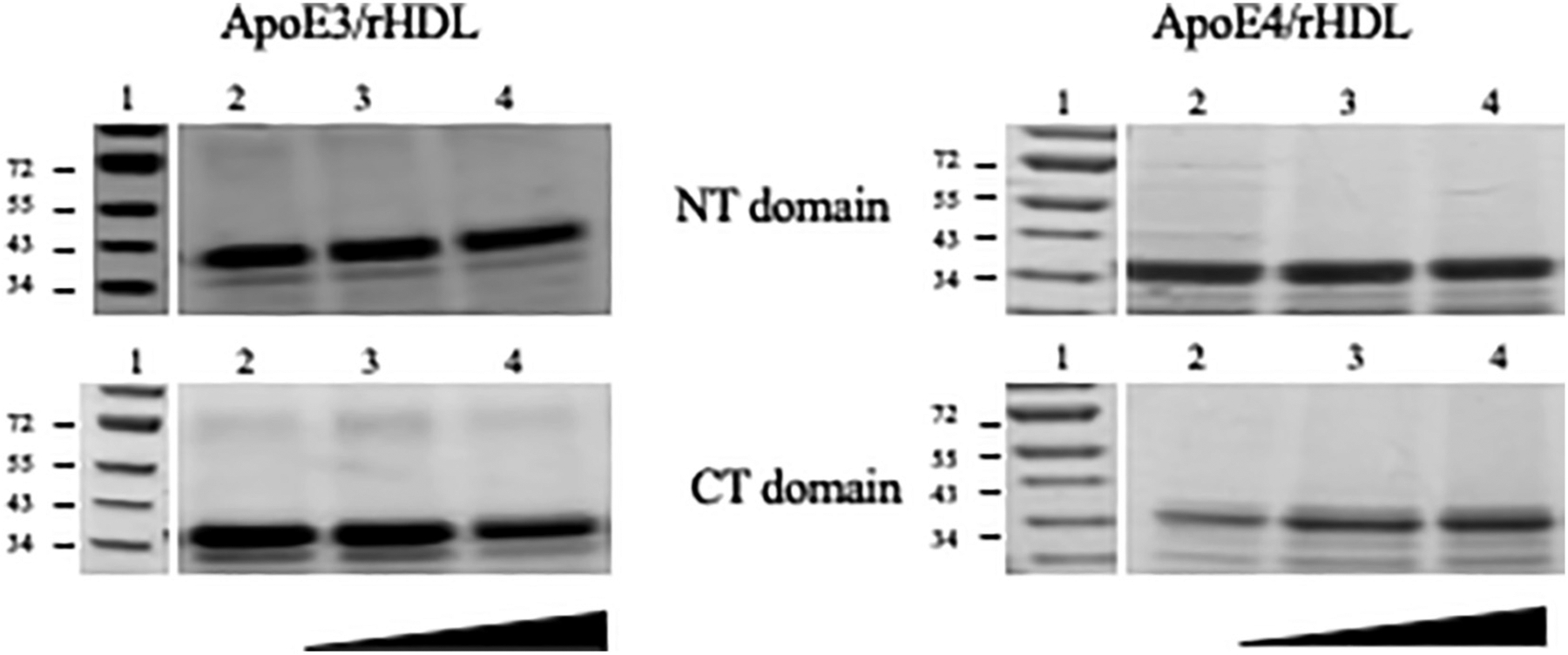
BM(PEG)3 cross-linking analysis of rHDL with apoE3 and apoE4 Cys variants. Discoidal rHDL bearing 10–15 μg apoE3 (left) or apoE4 (right) were treated with increasing concentration of BM(PEG)3 (in the direction shown) in DMSO and electrophoresed on a 4–20% acrylamide gradient SDS-PAGE gel as described under Methods. Two single Cys variants were selected to represent the NT (top) and CT (bottom) domains for each isoform. The lane assignments are as follows: Lane 1, molecular mass standards in kDa; lane 2, rHDL + DMSO; lanes 3 and 4, rHDL+10×–25× and 50×–100× molar excess of BM(PEG)3 over apoE, respectively.
In contrast, when rHDL containing isolated apoE CT domain (residues 201–299) with Cys on helix C1 or C2 was cross-linked with increasing concentration of BMH, robust dimeric bands with apparent molecular mass of ~27 kDa were observed, Fig. 5. Cys located on two other segments on apoE CT domain (Hinge Loop and Loop C) yielded similar results with strong dimeric bands. For A209C/rHDL, a second dimeric band was noted, likely a folding conformer.
Fig. 5.
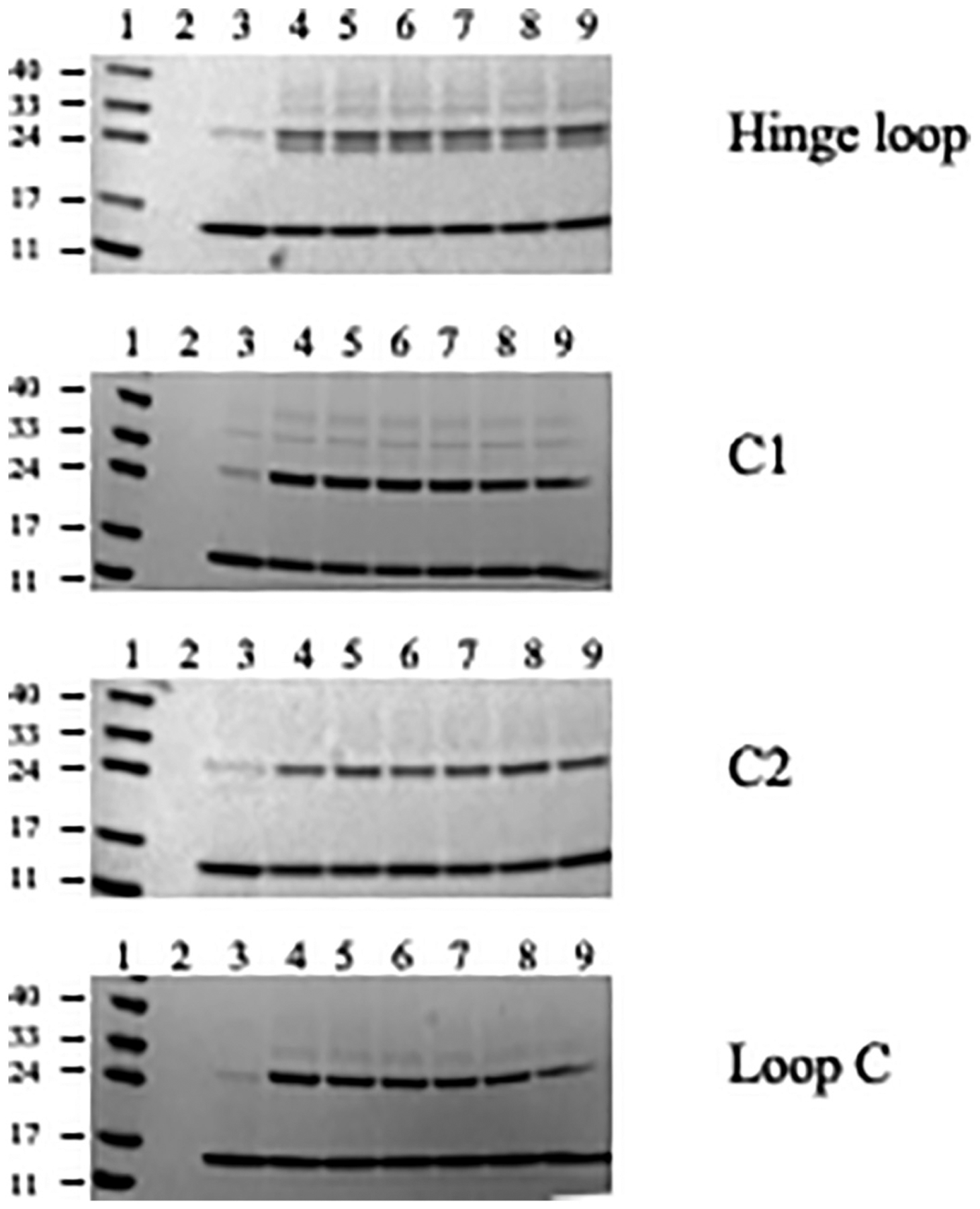
Cross-linking analysis of rHDL with apoE(201–299) single Cys variants. Discoidal rHDL bearing 10–15 μg apoE(201–299) single Cys variants were treated with BMH in DMSO and electrophoresed on a 4–20% acrylamide gradient gel as described under Methods. The lane assignments are as follows: Lane 1, molecular mass standards in kDa; lane 2, DMSO; rHDL + DMSO; lanes 4, 5, 6, 7, 8 and 9, rHDL +2×, 5×, 10×, 25×, 50×, 100× molar excess BMH over apoE(201–299), respectively.
4. Discussion
Site-directed probing using Cys-specific cross-linkers and spatially-sensitive fluorophores are classical approaches to study protein-protein interaction and are particularly versatile alternatives to investigate proteins in the context of large molecular complexes that defy high resolution structural analysis [35]. In the current study, these were employed to understand the influence of the bulky 4-helix bundle NT domain of apoE bearing the LDLr binding sites on the relative orientation of its CT domain in reconstructed HDL that resembles nascent HDL generated in vivo. The photophysical characteristics of pyrene fluorescence emission lend itself well to study spatial proximity between specified sites as demonstrated by conformational studies of several proteins [36,37] including apolipoproteins [25,30–33]. The notable features of the fluorescence emission spectrum of pyrene include an ensemble of five major vibronic bands designated bands I-V with peaks at ~375, 379, 385, 395 and 410 nm. Termed monomeric bands, they arise due to pi-pi* transitions; the peak at 375 nm is attributed to the first vibronic band with a 0–0 transition and the peak at 385 nm to the 3rd vibronic band with a 0–2 transition [38–41]. Since the electronic and vibronic states are coupled, the intensity of band III is sensitive to the polarity of the probe microenvironment, showing increased intensity relative to band I in non-polar environments, and decreased intensity in polar environments [42,43]. The ratio between the intensities at 375 and 385 nm (termed Py scale) has been a useful metric of probe location.
In addition to the monomeric family of peaks, the presence of a broad featureless band at longer wavelengths centered ~460 nm is indicative of the occurrence of excited state dimer (excimer) formation. Excimer formation occurs when an excited state and a ground state pyrene are ~10–15 Å from each other and are favorably aligned in a precise configuration to allow stacking interaction, a feature made possible given the long lifetime of pyrene emission (~50 ns) [25,36,42,43]. In the current study, we exploited this unique spectral feature to monitor spatial proximity between two neighboring apoE molecules on rHDL. It is important to note that two pyrene rings may be proximal but not in favorable configuration for excimer formation; in such a situation a non-fluorescent complex may occur. Lack of or decrease in excimer band must therefore be interpreted with caution and should be accompanied by an independent assessment as noted in the current study. The decrease in excimer emission noted in lipid-associated state of apoE3, apoE4 and isolated CT domain suggests that pyrenes on two neighboring molecules of intact apoE or isolated CT domain are either farther than 10–15 Å from each other on discoidal rHDL or that the pyrene rings are not favorably stacked to yield excimer emission. Site-specific cross-linking data using short and long spacer arms indicated that two Cys on identical locations on neighboring molecules on rHDL are farther than cross-linking distance for intact apoE3 and apoE4, but that they are spatially proximal in isolated CT domain. From the robust dimeric bands noted for all apoE CT variants regardless of the location of Cys residues, we inferred that apoE CT domain adopts a parallel in-sync orientation with respect to each other on rHDL. Although the e/m ratio did not reach 0.02 noted for pyrene in DMSO, to reconcile with the decrease in excimer emission in case of pyrene-labeled isolated apoE CT domain, we propose that neighboring pyrene rings are aligned non-optimally yielding non-fluorescent complexes. The intensity of the band III peak at 385 nm is higher than that of band I at 375 nm in all probed sites, and particularly noteworthy at positions 209, 223 and 277 in CT domain, indicating that the pyrene rings are in a hydrophobic environment such as that in the interior of the rHDL with the non-polar faces of the helices facing the fatty acyl chain of phospholipid. It is possible that the plane of the pyrene rings are parallel to the axis of the fatty acyl chains, which would prevent stacking, as schematically represented in Fig. 6.
Fig. 6.
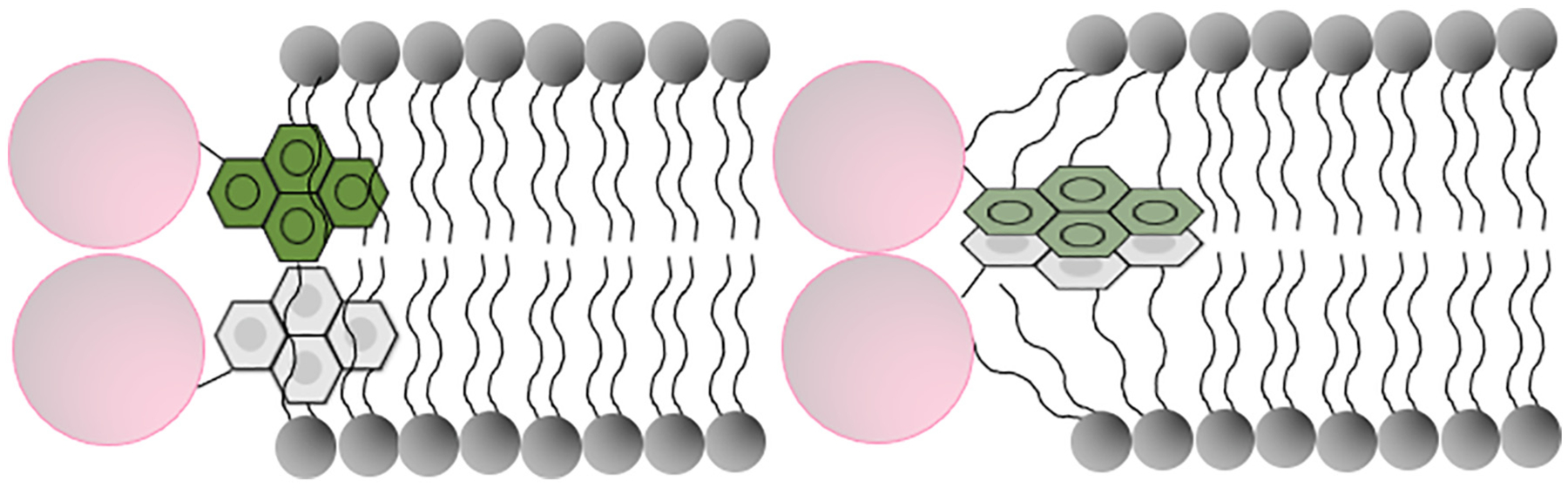
Cross-sectional view of discoidal lipoprotein showing pyrene-bearing apoE helices juxtaposed to the lipid bilayer. The axes of the helices (pink) are oriented perpendicular to the axes of the fatty acyl chains of phospholipids in the bilayer (grey). Details of the attachment of the pyrene rings to the -SH of Cys via maleimide ring are not shown for simplicity. Left: Proximal but non-optimal arrangement of pyrene rings (no excimer formation), requires rotation of -S-CH-bond (not shown) to accommodate interdigitation of pyrene rings (green and grey) between the fatty acyl chains of the phospholipids. Right: Proximal and optimal arrangement of pyrene rings (allows excimer formation) that would require significant distortion of the fatty acyl chains of the phospholipid. Not to scale.
Taken together, the decreased excimer emission coupled to lack of cross-linking suggests that intact apoE molecules are oriented in an anti-parallel, out-of-sync parallel or hairpin-loop orientation. Previous studies suggest an anti-parallel orientation of neighboring molecules in apoE3 in discoidal complexes and a hairpin loop for apoE4 [33,44–46]. Further, the hairpin model of apoE4 was proposed to adopt two possible conformations in solution, where the NT domain four-helix helix bundle was either partially open or remained intact [47]. In contrast, we posit an in-sync parallel orientation for the isolated CT domain on rHDL based on the strong cross-linking pattern, attributing the decreased excimer emission to formation of non-fluorescent complexes, Fig. 7. This model takes into consideration the number of apoE molecules that was estimated to be 4–6 per discoidal rHDL based on the following experimental observations and theoretical restrictions, and the assumption that lipid-bound apoE exists in an extended helical conformation: molecular mass and Stokes’ diameter of rHDL ~500 kDa and ~16 nm, respectively from native PAGE (data not shown), discoidal particle geometry from electron microscopy imaging (data not shown), lipid: protein molar ratio of 100:1 from compositional analysis, α-helical conformation as noted previously [33,46,47], estimation of POPC bilayer thickness to be ~50 Å [48] and previous FTIR data that indicate that the helical axes of lipid associated apoE are perpendicular to the axes of the fatty acyl chains of phospholipids in discoidal rHDL [47,49]. Further attempts were made to distinguish between anti-parallel and hairpin conformation by mixing lipid-free apoE3 (or apoE4) bearing single Cys on H2 and C1 in a 1:1 molar ratio prior to reconstitution into discoidal particles, followed by cross-linking as described. The rationale was that it would be possible to cross link if the Cys on H2 and C1 on two different molecules are proximal on the discoidal particle. No significant cross-linking was noted in apoE3 or apoE4 with either BMH or BM(PEG)3, Fig. S2, indicating that the two Cys are >13–18 Å from each other. Taken together, these data suggest that the two molecules are aligned anti-parallel out-of-sync with each other and/or in a hairpin conformation for intact apoE.
Fig. 7.
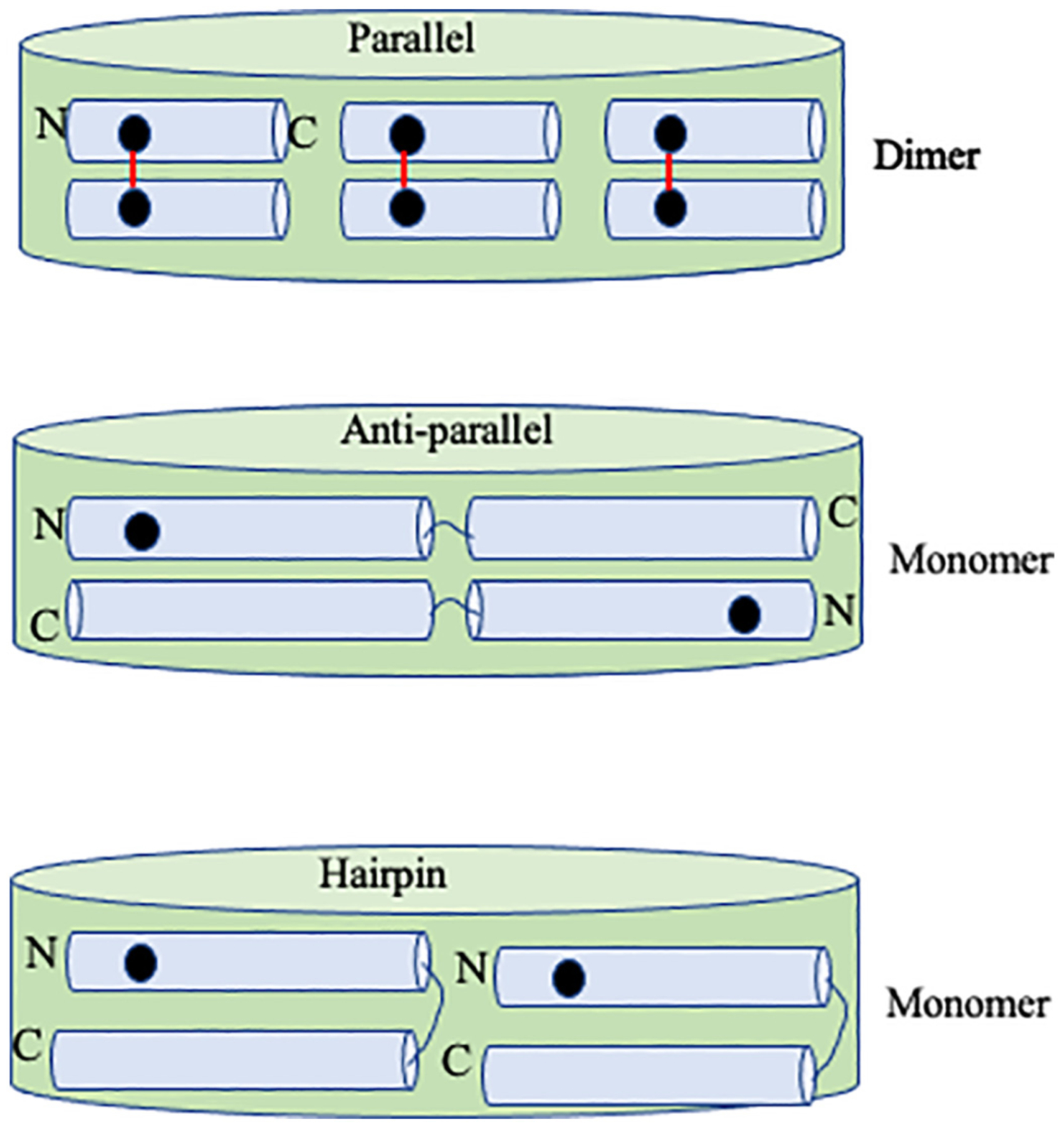
Schematic illustration of single Cys location in parallel, anti-parallel and hairpin orientation of apoE in rHDL. Top, Six isolated apoE CT domains (apoE (201–299)) shown in rHDL, with black dot representing a single Cys, and red line showing BMH cross-linking that would result in a dimeric band in SDS-PAGE. Middle, Two full length apoE (1–299) shown in anti-parallel, or bottom, hairpin conformation, with single Cys farther than cross-linking distance that would yield a monomeric band in both cases. The number of apoE molecules per discoidal particle has no influence on the outcome. Not to scale.
The observation that the isolated CT domain adopts an alternate orientation compared to the intact apoE3 or apoE4 suggests that the presence of the NT domain 4-helix bundle may impose a steric hindrance especially during the initial phase of lipid binding, which is proposed to be initiated by the C- and N-terminal segments [12,13]. It is noteworthy that the 4-helix bundle of apoE3 and apoE4 are 2–3 fold more stable ( ~ 10–12 kcal/mol) [6,14,50], offering resistance to chemical-induced unfolding (concentration of GdnHCl at mid-point of transition [GdnHCl]1/2 ~2.5–3.0 M) compared to the CT domain ( ~ 4.0 kcal/mol and [GdnHCl]1/2 ~ 1.0 M), and other helix bundles in apolipoproteins, such as apoAI, apoCII ( ~ 1.0–4.0 kcal/mol and [GdnHCl]1/2 ~ 1.0 M) [7], insect apolipophorins ( ~ 1.0–3.0 kcal/mol and [GdnHCl]1/2 ~ 0.5 M) [51]. The NT domains also have correspondingly lower lipid binding affinity compared to the CT domain [52–55]. In general, the ease of unfolding is correlated with ease of binding to lipids, since less structured segments tend to seek stability by lipid binding interaction. Further, hydrogen-deuterium exchange coupled to mass spectrometry in conjunction with fluorescence polarization data suggest that lipid binding, as predicted by ease of exchange and ease of unfolding, involve multiple steps [21,23]. It was proposed that upon encountering lipids, the oligomeric assembly of apoE dissociates and likely anchors to the surface via the CT domain, with the short and relatively polar helix C3 (residues 271–276, DMQRQW) likely seeking stability prior to C1 and C2. It was also suggested that the lipid binding of the NT domain is a complex process [13,55,56] involving disruption of the inter-domain interaction, an initial anchoring of the helix bundle via the flexible hinge segments (close to the CT domain) and loop 3 (the 80’s loop between H2 and H3) [56], which in turn triggers opening and binding of helix bundle.
Lastly, previous apoE3/apoAI domain swapping studies, wherein the NT domain of apoAI was appended to the CT domain of apoE, support our current findings of the influence of apoE NT domain on the lipid binding behavior of the CT domain [57]. ApoAI shares the same ancestral gene as apoE3 and shows similar structural characteristics with Pro-punctuated tandem repeats of 22-mers that form amphipathic α-helices [15,58]. Although high resolution structural information of intact apoAI is lacking, biophysical studies in combination with modeling based on partial X-ray crystallographic analyses suggests that the NT domain bears a 4-helix bundle [59,60] similar to apoE3. However, because the NT domain of apoAI is not as stable as that of apoE3, swapping it for apoE3 NT domain (yielding apoAI-NT/apoE-CT) significantly enhanced the vesicle solubilization rate of the chimera, which otherwise shows a very poor ability to bind DMPC vesicles to yield rHDL [23,21,61]. Taken together with the current study, it appears that the stable NT domain helix bundle of apoE3 not only mitigates the lipid binding rate of its CT tail, but that it also influences the relative orientation of one monomer with respect to the other on rHDL. Similar structural flexibility has been noted in apoAI, which adapts its orientation, registry and number per particle depending on the lipid composition and particle size, varying between in-sync, out-of-sync anti-parallel and a hairpin loop on a rHDL particle [62,63]. Overall, our current observations indicate that in absence of the NT domain, the CT domain of apoE adopts an alternate orientation in rHDL; further studies are needed to determine if both configurations of the CT domain (in isolation and in the context of the intact protein) are able to sustain similar ability to promote cholesterol efflux and undergo particle maturation to form spherical lipoproteins.
The current study yielded additional confirmatory information about the conformational organization of lipid-free apoE4. In lipid-free state both apoE3 and apoE4 exist predominantly as tetramers with some dimeric and higher state oligomeric forms [16,64,65]. The tetramerization occurs at the CT domain between neighboring molecules via helix-helix interactions [16,18,66]. Although both lipid-free apoE3 and apoE4 show significant excimer peaks for pyrenes located on all the selected sites (as a point of comparison, the e/m ratio of free pyrene in DMSO is ~0.02 [25]), the e/m ratios for those on apoE4 are significantly higher and the Py scale lower in 3 of the 5 probed sites compared to apoE3. This suggests that apoE4 adopts an altered conformation involving extensive protein-protein interactions, as also noted by others [52], who proposed that a salt bridge interaction between R61 on H2 and E255 on C2 juxtaposes the NT and CT domains in apoE4 [22,67]. The NT-CT domain interaction in apoE4 would position the NT domains from neighboring molecules in spatial proximity in the context of a tetramer. Nevertheless, NMR studies of apoE3 also propose an NT-CT domain interaction in lipid-free state via a salt bridge between K95 (on H3) and E255 (on C2) [11]. In both isoforms, the NT-CT domain interaction likely juxtaposes the NT domain from neighboring molecules in the context of a tetrameric protein as inferred by the presence of excimer peaks for both. The observation is further supported by cross-linking data (Supplementary materials, Fig. S3) where BMH-mediated dimer formation was noted in both apoE3 and apoE4 for Cys located in H1, H2 and H3. Taken together, it appears that although both apoE3 and apoE4 elicit NT-CT domain interaction and display juxtaposed CT-CT and NT-NT domains, the overall conformational organization of apoE4 is notably different from apoE3, an observation that requires further structural and conformational analysis.
Using the excimer approach with pyrenes located at 161, 169, 176 or 181, previous studies revealed that self-association via the CT domain brings the hinge loop between neighboring molecules in closer proximity in apoE3 [25]. The spatial disposition between the NT and CT domains likely renders the linker segment more susceptible to proteolytic cleavage, a feature that is particularly relevant in AD where NT and CT fragments have been identified in postmortem brain tissues and in mouse models [68–71]. ApoE4 appears to be more susceptible to proteolytic cleavage leading to increased accumulation of fragments seen in post-mortem brains.
In the CNS, apoE is located on HDL-like particles, which play a critical role in ABCA1- and ABCG1-mediated cholesterol efflux from astrocytes to form nascent discoidal and mature spherical particles for the delivery of cholesterol to the neurons via the LDLr family of proteins. While the CT domain plays a major role in the initial step of cholesterol efflux, the NT domain bearing the LDLr binding sites is responsible for cellular uptake of the cholesterol-laden lipoprotein via the lipoprotein receptor family of proteins [72,73]. Since the two domains can function in isolation, identification of isolated CT domain fragments of apoE implies that it may promote robust cholesterol efflux [74] from the astrocytes; however, this process is likely ineffective without the successful uptake of HDL by the neurons, thereby affecting synaptic plasticity and function [73]. This suggests that both domains must be present in their functional orientation for efficient delivery of cholesterol. Our study revealed that the orientation of the CT domain of apoE on rHDL particles depends on the presence of the NT domain, potentially influencing cholesterol efflux and neuronal uptake. Additional studies are needed to further our understanding of the influence of one domain over the other in apoE and the extent to which this affects cholesterol homeostasis in the CNS.
Supplementary Material
Acknowledgements
This work was funded by NIH-GM105561 (VN) and McAbee-Over-street Scholarship (SK and AP) and NIH-GM071638 (AD). We thank Wendy Beck for generating apoE3 and apoE4 single Cys constructs. We thank Kyle Meyer for generously sharing apoAI L126C/A158C as spot checks for cross-linking.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
This work was funded in part by the NIH (NIH-GM105561) to VN AD was a trainee in the NIH training grant (GM071638)
AP is currently working in Novartis Institutes for BioMedical Research but not at the time the work was carried out.
Abbreviations:
- IAEDANS
5-((((2-iodoacetyl)amino)ethyl)amino)napthalene-1-sulfonic acid
- apoE3
apolipoprotein E3
- BMH
bis(maleimide)hexane
- BM(PEG)3
1,11-bismaleimido-triethyleneglycol
- CT
C-terminal
- DTT
dithiothreitol
- GdnHCl
guanidine hydrochloride
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- LDLR
low density lipoprotein receptor
- NT
N-terminal
- NPM
N-(1-pyrene)maleimide
- PBS
phosphate-buffered saline
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- TBS
Tris-buffered saline
- WT
wild type
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamem.2021.183618.
References
- [1].Mahley RW, Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders, J Mol Med Berl Ger 94 (2016) 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Saito H, Lund-Katz S, Phillips MC, Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins, Prog. Lipid Res 43 (2004) 350–380. [DOI] [PubMed] [Google Scholar]
- [3].de Chaves EP, Narayanaswami V, Apolipoprotein E and cholesterol in aging and disease in the brain, Future Lipidol 3 (2008) 505–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Getz GS, Reardon CA, Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall, J. Lipid Res 50 (2009) S156–S161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Huang Y, Mahley RW, Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases, Neurobiol Dis 72PA (2014) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT, Human apolipoprotein E3 in aqueous solution, II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem 263 (1988) 6249–6258. [PubMed] [Google Scholar]
- [7].Wetterau JR, Aggerbeck LP, Rall SC, Weisgraber KH, Human apolipoprotein E3 in aqueous solution, I. Evidence for two structural domains. J Biol Chem 263 (1988) 6240–6248. [PubMed] [Google Scholar]
- [8].Sivashanmugam A, Wang J, A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity, J. Biol. Chem 284 (2009) 14657–14666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA, Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E, Science 252 (1991) 1817–1822. [DOI] [PubMed] [Google Scholar]
- [10].Weisgraber KH, Apolipoprotein E: structure-function relationships, Adv. Protein Chem 45 (1994) 249–302. [DOI] [PubMed] [Google Scholar]
- [11].Chen J, Li Q, Wang J, Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Narayanaswami V, Ryan RO, Molecular basis of exchangeable apolipoprotein function, Biochim. Biophys. Acta 1483 (2000) 15–36. [DOI] [PubMed] [Google Scholar]
- [13].Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Molecular mechanism of apolipoprotein E binding to lipoprotein particles, Biochemistry 48 (2009) 3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH, Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain, Biochemistry 39 (2000) 11657–11666. [DOI] [PubMed] [Google Scholar]
- [15].Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM, The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function, J. Lipid Res 33 (1992) 141–166. [PubMed] [Google Scholar]
- [16].Sakamoto T, Tanaka M, Vedhachalam C, Nickel M, Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H, Contributions of the carboxyl-terminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms, Biochemistry 47 (2008) 2968–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H, Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4, Biochemistry 45 (2006) 4240–4247. [DOI] [PubMed] [Google Scholar]
- [18].Westerlund JA, Weisgraber KH, Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization, J. Biol. Chem 268 (1993) 15745–15750. [PubMed] [Google Scholar]
- [19].Fan D, Li Q, Korando L, Jerome WG, Wang J, A monomeric human apolipoprotein E carboxyl-terminal domain, Biochemistry 43 (2004) 5055–5064. [DOI] [PubMed] [Google Scholar]
- [20].Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J, A monomeric, biologically active, full-length human apolipoprotein E, Biochemistry 46 (2007) 10722–10732. [DOI] [PubMed] [Google Scholar]
- [21].Yang L, Hernandez RV, Tran TN, Nirudodhi S, Beck WHJ, Maier CS, Narayanaswami V, Ordered opening of LDL receptor binding domain of human apolipoprotein E3 revealed by hydrogen/deuterium exchange mass spectrometry and fluorescence spectroscopy, Biochim Biophys Acta BBA - Proteins Proteomics 1866 (2018) 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dong LM, Weisgraber KH, Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins, J. Biol. Chem 271 (1996) 19053–19057. [DOI] [PubMed] [Google Scholar]
- [23].Fabilane CS, Nguyen PN, Hernandez RV, Nirudodhi S, Duong M, Maier CS, Narayanaswami V, Mechanism of lipid binding of human apolipoprotein E3 by hydrogen/deuterium exchange/mass spectrometry and fluorescence polarization, Protein Pept Lett 23 (2016) 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bains GK, Kim SH, Sorin EJ, Narayanaswami V, The extent of pyrene excimer fluorescence emission is a reflector of distance and flexibility: analysis of the segment linking the LDL receptor-binding and tetramerization domains of apolipoprotein E3, Biochemistry 51 (2012) 6207–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Patel AB, Khumsupan P, Narayanaswami V, Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E, Biochemistry 49 (2010) 1766–1775. [DOI] [PubMed] [Google Scholar]
- [26].Jonas A, [32] Reconstitution of high-density lipoproteins, in: Methods in Enzymology, Elsevier, 1986, pp. 553–582. [DOI] [PubMed] [Google Scholar]
- [27].Kim SH, Kothari S, Patel AB, Bielicki JK, Narayanaswami V, A pyrene based fluorescence approach to study conformation of apolipoprotein E3 in macrophage-generated nascent high density lipoprotein, Biochem. Biophys. Res. Commun 450 (2014) 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Khumsupan P, Ramirez R, Khumsupan D, Narayanaswami V, Apolipoprotein E LDL receptor-binding domain-containing high-density lipoprotein: a nanovehicle to transport curcumin, an antioxidant and anti-amyloid bioflavonoid, Biochim. Biophys. Acta 1808 (2011) 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gursky O, Ranjana DL Gantz, Complex of human apolipoprotein C−1 with phospholipid: thermodynamic or kinetic stability? Biochemistry 41 (2002) 7373–7384. [DOI] [PubMed] [Google Scholar]
- [30].Sahoo D, Narayanaswami V, Kay CM, Ryan RO, Fluorescence studies of exchangeable apolipoprotein-lipid interactions: superficial association of apolipophorin III with lipoprotein surfaces, J. Biol. Chem 273 (1998) 1403–1408. [DOI] [PubMed] [Google Scholar]
- [31].Sahoo D, Narayanaswami V, Kay CM, Ryan RO, Pyrene excimer fluorescence: a spatially sensitive probe to monitor lipid-induced helical rearrangement of apolipophorin III, Biochemistry 39 (2000) 6594–6601. [DOI] [PubMed] [Google Scholar]
- [32].Sahoo D, Weers PMM, Ryan RO, Narayanaswami V, Lipid-triggered conformational switch of apolipophorin III helix bundle to an extended helix organization, J. Mol. Biol 321 (2002) 201–214. [DOI] [PubMed] [Google Scholar]
- [33].Drury J, Narayanaswami V, Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence, J. Biol. Chem 280 (2005) 14605–14610. [DOI] [PubMed] [Google Scholar]
- [34].Cruz S, Narayanaswami V, Cellular uptake and clearance of oxidatively-modified apolipoprotein E3 by cerebral cortex endothelial cells, Int. J. Mol. Sci 20 (2019) 4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bains G, Patel AB, Narayanaswami V, Pyrene: a probe to study protein conformation and conformational changes, Mol Basel Switz 16 (2011) 7909–7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lavis LD, Raines RT, Bright ideas for chemical biology, ACS Chem. Biol 3 (2008) 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lehrer SS, Pyrene excimer fluorescence as a probe of protein conformational change, in: Biswas BB, Roy S (Eds.), Proteins: Structure, Function, and Engineering, Springer US, Boston, MA, 1995, pp. 115–132. [DOI] [PubMed] [Google Scholar]
- [38].Hara K, Ware WR, Influence of solvent perturbation on the radiative transition probability from the 1B1u state of pyrene, Chem. Phys 51 (1980) 61–68. [Google Scholar]
- [39].Karpovich DS, Blanchard GJ, Relating the polarity-dependent fluorescence response of pyrene to vibronic coupling, Achieving a fundamental understanding of the py polarity scale. J Phys Chem 99 (1995) 3951–3958. [Google Scholar]
- [40].Peter Geigle K, Wolf J, Hohlneicher G, Franck-Condon/Herzberg-Teller interferences in the 1Lb transitions of pyrene and chrysene, J Photochem Photobiol Chem 105 (1997) 183–187. [Google Scholar]
- [41].Kalyanasundaram K, Thomas JK, Environmental effects on vibronic band intensities in pyrene monomer fluorescence and their application in studies of micellar systems, J. Am. Chem. Soc 99 (1977) 2039–2044. [Google Scholar]
- [42].Nakajima A, Intensity enhancement induced by solute—solvent interaction between pyrene and polar solvents, Spectrochim Acta Part Mol Spectrosc 38 (1982) 693–695. [Google Scholar]
- [43].Somerharju P, Pyrene-labeled lipids as tools in membrane biophysics and cell biology, Chem. Phys. Lipids 116 (2002) 57–74. [DOI] [PubMed] [Google Scholar]
- [44].Fisher CA, Narayanaswami V, Ryan RO, The lipid-associated conformation of the low density lipoprotein receptor binding domain of human apolipoprotein E, J. Biol. Chem 275 (2000) 33601–33606. [DOI] [PubMed] [Google Scholar]
- [45].Henry N, Krammer E-M, Stengel F, Adams Q, Van Liefferinge F, Hubin E, Chaves R, Efremov R, Aebersold R, Vandenbussche G, Pŕevost M, Raussens V, Deroo S, Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach, PLoS Comput. Biol 14 (2018), e1006165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Narayanaswami V, Szeto SS, Ryan RO, Lipid association-induced N- and C-terminal domain reorganization in human apolipoprotein E3, J. Biol. Chem 276 (2001) 37853–37860. [DOI] [PubMed] [Google Scholar]
- [47].Raussens V, Fisher CA, Goormaghtigh E, Ryan RO, Ruysschaert JM, The low density lipoprotein receptor active conformation of apolipoprotein E, Helix organization in n-terminal domain-phospholipid disc particles. J Biol Chem 273 (1998) 25825–25830. [DOI] [PubMed] [Google Scholar]
- [48].Shahane G, Ding W, Palaiokostas M, Orsi M, Physical properties of model biological lipid bilayers: insights from all-atom molecular dynamics simulations, J. Mol. Model 25 (2019) 76. [DOI] [PubMed] [Google Scholar]
- [49].Raussens V, Narayanaswami V, Goormaghtigh E, Ryan RO, Ruysschaert J-M, Alignment of the apolipophorin-III α-helices in complex with dimyristoylphosphatidylcholine: a unique spatial orientation, J. Biol. Chem 270 (1995) 12542–12547. [DOI] [PubMed] [Google Scholar]
- [50].Weers PMM, Narayanaswami V, Ryan RO, Modulation of the lipid binding properties of the N-terminal domain of human apolipoprotein E3: ApoE-NT lipid binding interactions, Eur. J. Biochem 268 (2001) 3728–3735. [DOI] [PubMed] [Google Scholar]
- [51].Weers P, Ryan R, Apolipophorin III: role model apolipoprotein, Insect Biochem. Mol. Biol 36 (2006) 231–240. [DOI] [PubMed] [Google Scholar]
- [52].Mizuguchi C, Hata M, Dhanasekaran P, Nickel M, Phillips MC, Lund-Katz S, Saito H, Fluorescence analysis of the lipid binding-induced conformational change of apolipoprotein E4, Biochemistry 51 (2012) 5580–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mizuguchi C, Hata M, Dhanasekaran P, Nickel M, Okuhira K, Phillips MC, Lund-Katz S, Saito H, Fluorescence study of domain structure and lipid interaction of human apolipoproteins E3 and E4, Biochim Biophys Acta BBA - Mol Cell Biol Lipids 1841 (2014) 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Lund-Katz S, Phillips MC, Lipid binding-induced conformational change in human apolipoprotein E, Evidence for two lipid-bound states on spherical particles. J Biol Chem 276 (2001) 40949–40954. [DOI] [PubMed] [Google Scholar]
- [55].Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC, Lund-Katz S, Effects of polymorphism on the lipid interaction of human apolipoprotein E, J. Biol. Chem 278 (2003) 40723–40729. [DOI] [PubMed] [Google Scholar]
- [56].Segelke BW, Forstner M, Knapp M, Trakhanov SD, Parkin S, Newhouse YM, Bellamy HD, Weisgraber KH, Rupp B, Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding, Protein Sci Publ Protein Soc 9 (2000) 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lek MT, Cruz S, Ibe NU, Beck WHJ, Bielicki JK, Weers PMM, Narayanaswami V, Swapping the N- and C-terminal domains of human apolipoprotein E3 and AI reveals insights into their structure/activity relationship, PLoS One 12 (2017), e0178346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM, The amphipathic α helix: a multifunctional structural motif in plasma apolipoproteins, in: Advances in Protein Chemistry, Elsevier, 1994, pp. 303–369. [DOI] [PubMed] [Google Scholar]
- [59].Melchior JT, Walker RG, Cooke AL, Morris J, Castleberry M, Thompson TB, Jones MK, Song HD, Rye K-A, Oda MN, Sorci-Thomas MG, Thomas MJ, Heinecke JW, Mei X, Atkinson D, Segrest JP, Lund-Katz S, Phillips MC, Davidson WS, A consensus model of human apolipoprotein A-I in its monomeric and lipid-free state, Nat. Struct. Mol. Biol 24 (2017) 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Thomas MJ, Bhat S, Sorci-Thomas MG, Three-dimensional models of HDL apoAI: implications for its assembly and function, J. Lipid Res 49 (2008) 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vedhachalam C, Duong PT, Nickel M, Nguyen D, Dhanasekaran P, Saito H, Rothblat GH, Lund-Katz S, Phillips MC, Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles, J. Biol. Chem 282 (2007) 25123–25130. [DOI] [PubMed] [Google Scholar]
- [62].Silva RAGD, Hilliard GM, Li L, Segrest JP, Davidson WS, A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins, Biochemistry 44 (2005) 8600–8607. [DOI] [PubMed] [Google Scholar]
- [63].Silva RAGD, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye K-A, Davidson WS, Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Garai K, Frieden C, The association–dissociation behavior of the ApoE proteins: kinetic and equilibrium studies, Biochemistry 49 (2010) 9533–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hubin E, Verghese PB, van Nuland N, Broersen K, Apolipoprotein E associated with reconstituted high-density lipoprotein-like particles is protected from aggregation, FEBS Lett. 1873–3468 (2019) 13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Choy N, Raussens V, Narayanaswami V, Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain, J. Mol. Biol 334 (2003) 527–539. [DOI] [PubMed] [Google Scholar]
- [67].Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA, Human apolipoprotein E Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms, J. Biol. Chem 269 (1994) 22358–22365. [PubMed] [Google Scholar]
- [68].Brecht WJ, Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice, J. Neurosci 24 (2004) 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW, Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons, Proc. Natl. Acad. Sci 98 (2001) 8838–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Marques MA, Tolar M, Harmony JAK & Crutcher KA, A thrombin cleavage fragment of apolipoprotein E exhibits isoform-specific neurotoxicity: NeuroReport 7 (1996) 2529–2532. [DOI] [PubMed] [Google Scholar]
- [71].Tolar M, Keller JN, Chan S, Mattson MP, Marques MA, Crutcher KA, Truncated apolipoprotein E (ApoE) causes increased intracellular calcium and may mediate ApoE neurotoxicity, J. Neurosci 19 (1999) 7100–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Holtzman DM, Herz J, Bu G, Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease, Cold Spring Harb Perspect Med 2 (2012) a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lane-Donovan C, Philips GT, Herz J, More than cholesterol transporters: lipoprotein receptors in CNS function and neurodegeneration, Neuron 83 (2014) 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, Bielicki JK, The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins, Biochemistry 46 (2007) 2583–2593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


