Abstract
Many modern techniques employed to uncover the molecular fundamentals underlying biological processes require dissociated cells as their starting point/substrate. Investigations into ovarian endocrinology or folliculogenesis, therefore, necessitate robust protocols for dissociating the ovary into its constituent cell populations. While in the mouse, methods to obtain individual, mature follicles are well-established, the separation and isolation of single cells of all types from early mouse follicles, including somatic cells, has been more challenging. Herein we present two methods for the isolation of somatic cells in the ovary. These methods are suitable for a range of applications relating to the study of folliculogenesis and mouse ovarian development. First, an enzymatic dissociation utilising collagenase and a temporary, primary cell culture step using neonatal mouse ovaries which yields large quantities of granulosa cells from primordial, activating, and primary follicles. Second, a rapid papain dissociation resulting in a high viability single cell suspension of ovarian somatic cells in less than an hour, which can be applied from embryonic to adult ovarian samples. Collectively these protocols can be applied to a broad array of investigations with unique advantages and benefits pertaining to both.
Keywords: granulosa cells, granulosa cells, ovarian follicle, follicle development, dissociation
Introduction
The mammalian ovary undergoes dynamic changes during early embryonic and postnatal development to ensure fertility during the female reproductive lifespan. To facilitate these changes, the ovary contains multiple, heterogenous cell populations. The germ cell populations transition from primordial germ cells to oocytes subject to meiotic maturation. The somatic cell populations include pre-granulosa cells, granulosa cells, theca cell progenitors, theca cells and stromal cells, as well as luteal cell types after ovulation. In particular, granulosa cells have a range of different roles in the mature ovary, from the dormant, undifferentiated pregranulosa cell in the primordial follicle, to the steroidogenic, mitotic mural granulosa cell of the antral follicle (Matsuda et al., 2012). Detailed characterisation of these cell populations has been achieved in the adult mouse, once these cells have ceased differentiating (Liu et al., 2015). Less work has been performed in the developing ovary, where various important developmental processes and cell fate decisions occur. During early ovary development, granulosa cells represent the majority somatic cell type in the ovary. During gestation in the mouse, the granulosa cells in particular drive the process of sex determination, and then facilitate germ cell nest breakdown, primordial follicle formation and primordial follicle activation (Fig. 1) (Fu et al., 2018; Lv et al., 2019; Stevant and Nef, 2019). To support these numerous developmental pathways, there is heterogeneity within the granulosa cell population (Rastetter et al., 2014; Niu and Spradling, 2020; Wang et al., 2020). For example, two populations of primordial follicles are formed within the ovary, distinguished by their location in the ovary and the expression of either Forkhead box L2 (FOXL2) or Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 (LGR5) in the granulosa cells (Rastetter et al., 2014; Niu and Spradling, 2020). This heterogeneity is complex to study in the ovary, as the granulosa cells remain directly connected to the germ cells throughout development. New techniques and methodologies, like single-cell RNA-sequencing and CRISPR-CAS9 gene editing show huge potential in understanding the specific granulosa cell-regulated mechanisms throughout development.
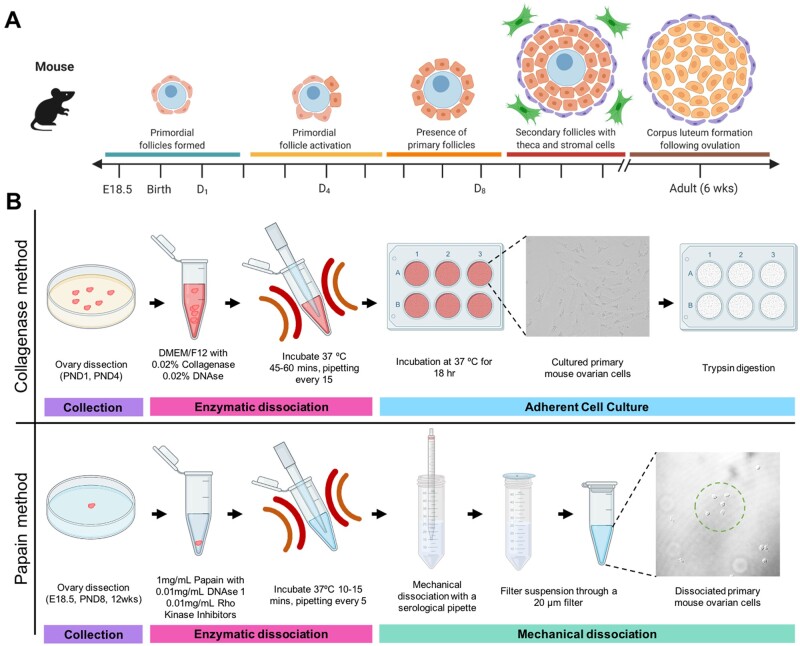
Figure 1.
Schematic diagram showing the timeline of ovarian follicle development in the mouse and the two dissociation protocols to retrieve somatic cells from these ovarian timepoints. (A) Relative timeline showing the key stages of folliculogenesis in embryonic mice (E = embryonic day) and post-natal mice (PND = days post birth). Germ cell cysts, also known as nests, breakdown to form primordial follicles (teal bar), containing an oocyte (blue) and pre-granulosa cells (orange), then the initial wave of primordial follicle activation occurs (yellow bar) which sets the ovarian reserve, leaving a proportion of primary follicles (orange bar) present in the ovary. Primary follicles then continue through folliculogenesis to form secondary follicles (red bar), which are surrounded by theca cells (purple) and stromal cells (green). Once the mice become sexually mature, around 6 weeks of age, corpus luteum structures form following ovulation, and the granulosa cells become luteinised and produce hormones (brown bar). Figure adapted from (Sarraj and Drummond, 2012), figure made using Biorender.com. (B) Schematic diagrams of the key steps in the collagenase and papain methods. Both methods involve collection steps (purple bar) and enzymatic dissociation steps (pink bar). The collagenase method uses differential cell adhesion in culture to select for granulosa cells (teal bar). In contract, the papain method using mechanical dissociation and filtration to enrich for somatic cells (blue bar). Figure made using Biorender.com.
There are several technical challenges to overcome relating to obtaining large quantities of viable granulosa cells for analysis, including the small size of the ovary during embryonic and postnatal development. Further technical challenges include the tight cellular association between granulosa cells and the oocyte and the lack of knowledge surrounding factors that distinguish the various pre-granulosa and granulosa cell populations. Common methods of tissue analysis like histological sections, or even whole tissue clearing studies (now achieved in ovaries; Feng et al., 2017) observe the cells at a static point in time and always in the context of the follicular structure. Additionally, the isolation of follicles from the ovary, and the use of laser capture microdissection (as in Oktay et al., 1997; Dolmans et al., 2006; Bonnet et al., 2011; Ernst et al., 2017, 2018; Steffensen et al., 2018) require significant and laborious manual effort into either classifying whole follicles by their diameter using pre-defined criteria (such as Pedersen and Peters, 1968), or by visually identifying each cell of interest to be isolated. All these methods require the tissue to be fixed first prior to isolation and are unable to yield a live cell population.
In other heterogenous cell populations, the isolation of a particular subset of cells is typically performed via fluorescence-activated cell sorting (FACS). However, there are currently no reliable cell surface markers or mice transgenic for fluorescent reporters such as green fluorescent protein (GFP) that are capable of distinguishing the primary follicle granulosa cells. Most recent protocols published on the mouse ovary detail the dissociation of ovarian tissue to retrieve oocytes or oogonial stem cells (OSCs) (Woods and Tilly, 2013; Navaroli et al., 2016; Zarate-Garcia et al., 2016; Clarkson et al., 2018). Thus, there is a demonstrated need for a method of separating granulosa cells from the follicle that requires less manual input achieving a sufficient yield for standard and advanced laboratory procedures, such as extraction of protein, DNA, and RNA, immunocytochemistry, discovery proteomics and mass spectrometry, and RNA sequencing. Such a technique should keep cells alive, ideally within minimal experimental steps and using no prior fixation or staining to maintain original cellular expression and other physiological characteristics.
A common solution involves the enzymatic dissociation of the ovarian tissue. Enzymatic dissociation represents an established procedure yet the primary outcomes of studies utilising this technique have largely centred on the isolation of whole follicles rather than individual cells (Dolmans et al., 2006), or isolating granulosa cells from mature ovaries (Morbeck et al., 1993). Enzymatic dissociation typically relies on the breakdown of the outer structure of the ovary (usually via collagenase, liberase or papain), to release the cells or follicular structures, followed by a temporary culture step to allow the granulosa cells to adhere to the surface while oocytes and debris remain suspended in the media. Collagenase is a more efficient enzyme for isolating individual cells, because it is known to break apart theca cells and disrupt follicular structure (Newton et al., 1999). Enzymatic dissociation methods, combined with mechanical dissociation, also allow for the preparation of a single cell suspension from ovarian tissue, necessary for the purpose of single-cell RNA-sequencing (scRNAseq). ScRNAseq requires sufficient tissue dissociation and high cell viability, to ensure the sequencing is of high quality and not contaminated by RNA from dying cells. Several recent studies have performed ovarian dissociation at various timepoints for sequencing studies, listed in Table I. All listed studies have different dissociation conditions, introducing variability between the data obtained from these studies. In this manuscript, we optimise the papain enzyme for obtaining suspensions of single cells from the mouse ovary. Papain is a cysteine protease and is frequently used in the dissociation of brain tissue (Carter et al., 2018; Ho et al., 2018; Yousef et al., 2018). It has been shown that extended dissociation in enzymatic solutions may reduce cell viability (Woods and Tilly, 2013), so short enzymatic dissociation coupled with mechanical dissociation is generally preferred to limit transcriptomic changes before scRNAseq (Nguyen et al., 2018). The papain dissociation protocol was adapted from three dissociation protocols commonly used for small tissue dissociations (Woods and Tilly, 2013; Williams et al., 2019; Weinberger et al., 2020).
Table I.
Comparison of published protocols used to dissociate mouse ovarian tissue.
| Year | Timepoint | Mouse | Mincing of tissue? | Enzymes used | Cell types isolated | Filtration used? | Enzyme dissociation time | Reference |
|---|---|---|---|---|---|---|---|---|
| 2012 | Adult (8wk) | Wild-type C57BL6 | Mince using a scalpel |
|
All ovarian cells | 70 µm | 15min | White et al. (2012) |
| 2013 | Adult (6–8 weeks) | Wild-type—no specific strain | Mince using a scalpel |
|
Oogonial stem cells | 100 µm | 30 min | Woods and Tilly, (2013) |
| 2019 | E10.5—E13.5, E16.5, PND6 | Tg (Nr5a1-GFP) CD1 mice | No |
|
Somatic cells | 40 µm | 10 min | Stévant et al. (2019) |
| 2019 | PND2 and PND6 | Wild-type F1 mice | No |
|
All ovarian cells | No | 30 min | Lo et al. (2019) |
| 2020 | E11.5, E12.5, E14.5, E16.5, E18.5, PND1 and PND5 | Wild-type—no specific strain | No | 0.25% Trypsin | All ovarian cells | 70 µm then 30 µm |
|
Niu and Spradling, (2020) |
| 2020 | E16.5, P0, P6 | Wild-type C57BL6 | Cut into small pieces |
|
All ovarian cells | 40 µm | 6–8 min | Wang et al. (2020) |
| 2021 | E12.5, E13.5, E16.5, E18.5 | Tg (Nr5a1-GFP) CD1 mice | No |
|
All ovarian cells | 70 µm | 15 min | Mayère et al. (2021) |
Here we present two methods for the dissociation of the mouse ovary, which are appropriate for the main molecular biology techniques used to characterise and investigate the somatic cell populations present across ovary development. The collagenase dissociation method yields a largely pure granulosa cell population from the early postnatal mouse ovary and is suitable for primary cell culture and techniques requiring a large amount of sample. Due to the culture step, this protocol is primarily recommended for downstream experiments including in vitro genetic manipulation studies, like CRISPR and transfection, as well as in vitro treatment experiments for the testing of inhibitors and toxicological compounds in granulosa cells (Fig. 1). In contrast, the papain dissociation protocol captures all the somatic cell types from the embryonic through to adult ovary and is thus a useful protocol for sequencing studies to characterise gene expression in multiple cell linages across a diverse developmental range. The papain protocol allows for consistency in studies where multiple ovarian timepoints are to be studied, and experiments can be performed with one ovary per replicate, limiting the number of mice required for experimental techniques. The papain dissociation protocol was optimised for high-throughput experiments, including single-cell and bulk RNA-sequencing, so it is also a suitable protocol for the validation of novel genes from these experiments, including quantitative PCR and immunocytochemistry (Fig. 1). In this study detailing the two dissociation methods, we characterise the cells retrieved from both methods and show that key granulosa genes are expressed, and cell identity is maintained using both methods. These methods allow for further investigations into granulosa and somatic cell function, and the identification of potential markers to identify the various sub-classes of granulosa cells throughout development.
Materials and methods: Part 1
Reagents for collagenase dissociation method
Antiseptic solution (i.e. iodine solution; Perrigo Australia, Belrose, NSW, Australia, RIO00802F, used in this protocol)
Ascorbic acid (Sigma, St. Louis, MO, USA, A0278) made up to 5 mg/ml in MilliQ water
Bovine serum albumen (BSA) (Sigma, A7906)
Collagenase type II (Sigma, C6885)
Dulbecco's Modified Essential Medium/Ham's F-12 Medium (DMEM/F12) (Sigma, D6421)
DNase I (Roche, Basel, Switzerland, 11284932001)
Fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA, 10099141)
Hank’s Balanced Salt Solution (HBSS) (Life Technologies, 14025-092)
Hepes solution, 1 M (Thermofisher, Waltham, MA, USA, SH30237.01)
Insulin-Transferrin-Selenium (ITS -G) (Life Technologies, 41400-045)
Leibovitz medium (L-15) (Life Technologies, 21083-027)
L-Glutamine, 200 mM (Thermofisher, 25030-81)
Penicillin-streptomycin, 10,000 U/ml (Thermofisher, 15140-122)
Trypan blue (Invitrogen, Carlsbad, CA, USA, T10282)
Trypsin/Ethylenediaminetetraacetic acid (EDTA) (Sigma, T4049)
Equipment for collagenase dissociation method
The following list includes necessary equipment and consumables are presented. The product details of the consumables that were utilised in the collection of this data are presented for transparency, but similar products should be sufficient to replicate these procedures.
Benchtop centrifuge compatible with 15 ml tubes and capable of 800g
Biosafety cabinet
Cell counter
Cell culture plate, 6-well (Greiner Bio-one, Kremsmünster, Austria, Cellstar)
Centrifuge tubes, 15 mL (Greiner Bio-one, 188261)
Filter, 0.22 µm size (Sarstedt, Newton, NC, USA, 83.1826.001)
Haemocytometer
Heated microscope stage
Incubator with 5% CO2 in air
Microcentrifuge
Microcentrifuge tube, 1.5 mL (Sarstedt, 72.690.001)
Round bottom polypropylene tube with cap for aeration (BD, Franklin Lakes, NJ, USA, 352059)
Serological pipettes, filtered, 10 mL and 25 mL volumes (from Sarstedt)
Small petri dish, IVF petri dish (Thermofisher, 150255) used in this protocol
Stereo microscope
Water bath
Optional: Cell culture plates, 24 or 48-well
Optional: Glass pipette tips, 20 μl, 200 μl, 1000 μl volumes
Optional: Glass serological pipettes, 1 ml, 10 ml volumes
Optional: Paraformaldehyde diluted to 4% (w/v) in phospho-buffered saline solution
Solutions for collagenase dissociation method
Dissociation working solution: prepare 1 ml of DMEM/F12 in a 1.5 ml microfuge tube and heat to 37°C. Dissolve 0.02% (2 mg) Collagenase II, and 0.02% (2 mg) DNase I. To be made fresh each time.
Hank’s solution: under sterile conditions, prepare a 50 ml container, adding 125 mg of BSA, 0.5 ml 1 M Hepes (pH 8.0), and bring to 50 ml with HBSS. Filter sterilise and store at 4°C until use.
Ovary culture media: under sterile conditions, in a 50 ml container, add 25 mg BSA, 1 ml penicillin–streptomycin, 1.25 ml FBS, 125 µl ITS-G, and 78 µl l-glutamine. Make the solution up to 25 ml with DMEM/F12 media. To be stored at 4°C and heated to 37°C before use.
Protocol for collagenase dissociation method
Isolation of granulosa cells (Day 1)
1. For collecting ovaries of sizes up to 20 mg (approximately 1 adult ovary, or 4–8 neonatal ovaries which are the subject of this protocol), prior to specimen collection, prepare a small petri dish containing L-15 media, supplemented with 1% FBS and allow to heat to 37°C.
Note: where >20 mg of the specimen is to be dissociated, it is optimal to set up multiple dissociations in parallel rather than to increase the reaction sizes in a single dish or tube.
2. Add harvested ovary (or ovaries) to the dish (see Appendix 1 for ovary dissection details). Transfer the dish to a heated stage (at 37°C) under a stereo microscope, remove the ovarian bursa and oviduct keeping only ovary tissue for dissociation.
3. Prepare 1 ml of the dissociation working solution (pre-warmed to 37°C in a water bath) in a round-bottom tube (with a loose-fitting cap for gas exchange), transferring the tissue from L-15 into the dissociation solution.
4. Incubate at 37°C in 5% CO2 for 45–60 min depending on the size and degradation level of the tissue, until the tissue has released a majority of its contents. Occasionally some clumps of tissue remain, as fibrous tissue does not completely disintegrate. Pipette the solution using a P200 pipette tip that has been pre-coated with Hank’s solution every 15 min to mechanically facilitate the dissociation of the tissue.
Note: once the ovary has started to break down, all pipette tips for subsequent steps should be pre-coated with Hank’s solution, where the BSA helps to prevent cells adhering to pipette walls. Glass pipette tips are an optional addition and when pre-coated in Hank’s solution, further reduce cell loss.
5. Transfer the solution to a microcentrifuge tube, using pre-coated pipette tips, and centrifuge for 1500g for 3 min.
6. Remove the supernatant and add Trypsin-EDTA. Incubate at 37°C for 20 min (for tissue sizes <10 mg, incubate for 15 min).
7. Inactivate Trypsin by the addition of FCS and mix the solution by gently pipetting. Then centrifuge solution at 1500g for 3 min.
8. Under sterile conditions in a biosafety cabinet to avoid contamination, remove the supernatant and resuspend the pellet in 100 μl of ovary culture media pre-heated to 37°C.
9. Remove an aliquot of cells and dilute 1:1 with Trypan Blue for cell count and viability measurements using haemocytometer or other cell counting method.
Note: cell count on Day 1 is a homogenised mix of the assortment of cell types in the ovary and cell count numbers may not necessarily reflect the number of cells obtained on Day 2.
When the purpose of the dissociation is to extract protein or RNA, proceed with Step 10 and Day 2, or see protocol: ‘preparation for immunocytochemistry’.
10. Where cell count is ≤3 million in total, prepare a 6-well plate for primary culture by adding 2 ml of ovary culture media per well, with 10 μl ascorbic acid. Add up to 50 000 cells per cm2 (or around 500 000 cells per well of a 6-well plate) then place in 37°C incubator for 18 h. Scale up reagents as necessary where cell count exceeds 3 million by adding cells to a larger flask or by including a second 6-well plate.
Day 2 of collagenase dissociation method
1. In a biosafety cabinet and using a serological pipette, remove the media from the plate (granulosa cells adhere to the base of the plate) and dispose of in antiseptic solution.
Note: avoid disturbing the base or side of the plate as much as possible so as to not disrupt the granulosa cells.
2. Rinse adhered cells with 2 ml HBSS per well by gently dropping the liquid and swirling the plate for a few rotations to thoroughly coat the surface of the wells. Withdraw the wash liquid and discard in antiseptic solution.
Note: for all steps where liquid is being exchanged on the plate of cells, do not let the cells dry out. Proceed through these steps quickly, and cover with the lid to reduce drying when unattended, even momentarily.
3. Pre-treat the cells with a short trypsin digestion by adding 1 ml of trypsin-EDTA to each well (again, gently dropping the liquid to avoid disruption) and swirling the plate for 10–20 s. Then, withdraw and discard the trypsin.
4. Add 1 ml of trypsin to each well of the plate, and incubate at 37°C for 5 min, or until cell projections are removed from the surface of the plate (Fig. 2).
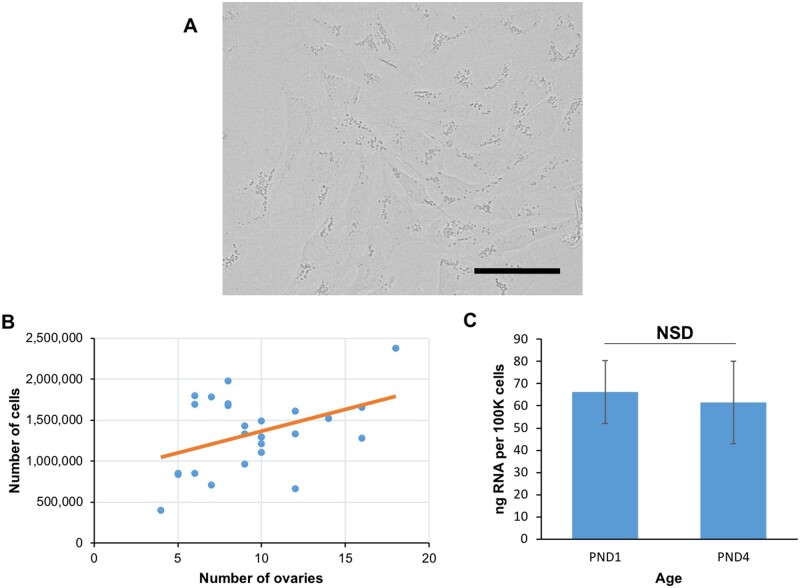
Figure 2.
Granulosa cell characteristics after collagenase dissociation. (A) Bright field images of granulosa cells adhering to plate surface following enzymatic dissociation and overnight culture. Images taken at 40× magnification, scale bar equivalent to 50 μm, (B) granulosa cell yield by number of ovaries dissociated in a single replicate with linear fit, and (C) RNA yield (ng per 100 000 cells) by age of ovaries dissociated. Data presented is from 25 dissociation replicates, 11 utilising PND1 ovaries, and 14 replicates of PND4 ovaries. NSD, no significant difference; PND, days post birth.
5. Inactivate by adding FBS to the trypsin at a ratio of 1:5 and mix mechanically by pipetting.
6. Collect the cells by repeatedly pipetting the trypsin/FBS solution over the surface of the plate to lift any remaining adherent cells and transfer the liquid into a 15 ml centrifuge tube.
Note: in a 6-well plate, a high proportion of cells adhere along the edge of the wells and the fine tip of a p20 pipette tip can lift these cells with a concentrated ejection of liquid.
7. Do a final wash of the plate with 1 ml of HBSS per well, swirling the liquid and pipetting over the surface of the plate, then add the liquid to the 15 ml cell suspension.
Note: using a microscope on bright field setting, inspect the plate between washes for any remaining cells. An additional wash of 1 ml HBSS can be performed if necessary.
8. Spin the tube containing cells in a benchtop centrifuge for 5 min at 800g.
Note: the final steps may be performed outside of the biosafety cabinet. For final steps, it is crucial that pipette tips coming into contact with cells are pre-coated with Hank’s solution to avoid cell loss in the plastic tips. Additionally, the pellet may be difficult to visualise by eye, so ensure the supernatant is carefully removed by pipetting from the opposing side of the tube to the pellet.
9. Discard the supernatant and resuspend the pellet in 1 ml of HBSS to wash the cells (transferring to a 1.5 ml tube for efficiency). Spin the cells in a microfuge for 5 min at 1200g.
10. Remove the supernatant and repeat the washes by resuspending in HBSS and centrifugation for a total of three washes.
11. Discard the supernatant of the final wash and resuspend the cells in a small volume of HBSS for a final cell count as described in Day 1, Step 9.
12. Centrifuge cells at 1200g for 5 min and proceed with preparation for either protein or RNA extraction. Alternatively, cell pellets may be stored as bulk samples at −80°C for later use.
Preparation for immunocytochemistry
1. Working in a biosafety cabinet, prepare a plate or series of plates (24- or 48-well depending on the number of replicates or cell numbers required) for primary culture by adding an appropriate volume of ovary culture media per well (500 μl for 24-well, and 200 μl for 48-well), with a volume of (5 mg/ml) ascorbic acid totalling 0.5% of the solution. Add up to 50 000 live cells per cm2, then place in 37°C incubator for 18 h.
2. Continuing in a biosafety cabinet after incubation, spike the growth media with 4% paraformaldehyde (at a ratio of 1 part 4% paraformaldehyde to 4 parts media) and incubate at room temperature for 2 min.
3. Discard liquid into antiseptic solution and replace with an equivalent volume of 4% paraformaldehyde for 10 min at room temperature to fix cells.
Note: for all steps where liquid is being exchanged on the plate of cells, do not let the cells dry out. Proceed through these steps quickly, and cover with the lid to reduce drying when unattended. Additionally, avoid disturbing the base or side of the plate as much as possible so as to not disrupt the granulosa cells.
4. Discard the leftover paraformaldehyde and rinse cells with HBSS to remove excess fixative by gently dropping the liquid and swirling the plate for a few rotations to thoroughly coat the surface of each well. Withdraw the wash liquid and discard in antiseptic solution. Repeat for a second wash.
5. Cover each well with HBSS, seal the plate with the lid. Can be stored for up to 3 months at 5°C.
Materials and methods: Part 2
Reagents for papain dissociation method
Bovine serum albumin (BSA) (Sigma, A7906)
DNase I (Merck, Kenilworth, NJ, USA, 10104159001)
10 x Hank’s Balanced Salt Solution (HBSS) (Thermofisher, 14185052)
Papain (Roche, 10108014001)
1 x Phosphate Buffered Saline (PBS) (Sigma, P4417)
Rho associated kinase inhibitor Y-27632 (Abmole Bioscience, Houston, TX, USA, M1817)
1 M TRIS pH 8.0 (Thermofisher, AM9855G)
Trypan Blue (Invitrogen, T10282)
Equipment papain dissociation method
Low speed centrifuge with tissue culture rotors (compatible with 15 ml and 50 ml Falcon tubes)
Biosafety tissue culture hood
Microcentrifuge
Stereo microscope
37°C water bath
IVF Petri Dish, 35 mm, with Lid (Thermofisher, 150255)
3-Well glass plate dish
Syringe and filter, 0.22 μm size
15 ml Falcon tubes
50 ml Falcon tubes
20 μm Pre-separation Filters (Miltenyi Biotec, Bergisch Gladbach, Germany, 130-101-812)
10 mL Pyrex® glass disposable serological pipettes (Merck, CLS707710N)
1.5 mL DNA LoBind tubes (Eppendorf, Framingham, MA, USA, EP0030108051)
20 μL Axygen Low binding filter tips (Thermofisher, 14-222-799)
200 μL Axygen Low binding filter tips (Thermofisher, 14-222-777)
1000 μL Axygen Low binding filter tips (Thermofisher, 14-222-765)
12-well Thermo Scientific™ PTFE Diagnostic Slides (Thermofisher, 10028210)
Optional: Countess II Cell Counter (Invitrogen, AMQAX1000)Optional: Countess™ Cell Counting Chamber Slides (Invitrogen, C10312)Optional: Haemocytometer
Solutions for papain dissociation method
Hank’s solution: In a 50 ml Falcon tube, add 5 ml of 10 × HBSS, 125 mg of BSA and 0.5 ml of 1 M TRIS pH 8.0. Bring the solution to 50 ml with distilled water, and filter sterilise using a 0.22 μm filter. To be made fresh each time.
Dissociation solution: Prepare 900 µl of Hank’s solution in a 1.5 ml tube. Add 100 µl of papain, 1 µl of DNAse I and 1 µl of Rho associated kinase inhibitor to the 1.5 ml containing Hank’s solution. Prewarm dissociation solution at 37°C until tissue is ready for dissociation.
Ethics
Australia: C57BL/6J mice were supplied by the University of Newcastle Animal Services Unit under ethics approval number A-2018-803. All experiments involving the use of animals were conducted in accordance with the Institutes’ Animal Care and Ethics Committee guidelines in accordance with Australian NHMRC Guidelines on Ethics in Animal Experimentation.
UK: All experiments carried out on mice were approved under the UK Animal (Scientific procedures) Act (Project licence 80/2405).
Protocol for papain dissociation method
This protocol was developed for the isolation of somatic cells from one neonatal mouse ovary across a range of time points (embryonic day (E)18.5, post-natal day (PND) 8 and 12 weeks (WKS)).
1. Pre-warm a water bath to 37°C.
2. Prepare a small petri dish containing 1 × PBS.
3. Pre-warm 500 μl of papain dissociation in a 1.5 ml LoBind microcentrifuge tube.
4. Carefully dissect the ovaries from the female mouse, by dissecting with the uterus and ovarian bursa.
5. Add harvested ovary to the PBS dish. Transfer the dish to the stereo microscope, gently remove the ovarian bursa and oviduct, keeping the ovary tissue for dissociation.
If collecting embryonic or early post-natal ovaries, proceed straight to Step 11. If collecting somatic cells from an adult ovary, proceed to Step 6.
6. Place 500 μl of papain/HBSS/DNase 1/Rho inhibitors dissociation solution into a glass plate.
7. Once ovary is cleaned of fat in PBS, transfer ovary to the well containing 500 μl papain.
8. Mince the ovary using mincing scissors or a scalpel, and transfer liquid containing cells to 1.5 ml Eppendorf tube, max 1 ovary per tube.
9. Wash the glass well with 500 μl of papain dissociation solution and add to 1.5 ml Eppendorf tube (should total 1 ml of dissociation solution).
10. Once the ovary is minced, transfer the ovary tissue into the LoBind microcentrifuge tube containing the papain dissociation solution.
Note: once the ovary is dissociated, all pipette tips for subsequent steps should be pre-coated with Hank’s solution to prevent cells adhering to pipette walls. Pre-coating involves pipetting the Hank’s solution up and down with the pipette tip several times.
11. Incubate the tissue in the papain dissociation solution for 10 min in the 37°C water bath.
12. Homogenise the tissue in the papain dissociation solution after 5 min, with a P200 tip that has been pre-coated with Hank’s solution. Gently pipette up and down 10 times against the wall of the tube to disrupt the tissue.
13. Stop the reaction by adding the dissociated ovarian cells to 4 ml of Hank’s solution in a 50 ml falcon tube. Rinse the 1.5 ml microcentrifuge tube with another 1 ml of Hank’s solution and re-unite with the reminder of the cells in the falcon tube.
14. Pre-coat a 10 ml glass serological pipette with Hank’s solution.
15. Use the 10 ml glass serological pipette to pipette the cells up and down 10–15 times against the bottom of the 50 ml tube containing the homogenised material.
16. In a low-speed centrifuge, spin down the dissociated cells at 500g for 5 min at room temperature.
17. Remove the supernatant using a glass pipette and dispose of the waste. Resuspend the cells in 4 ml of Hank’s solution (glass pipettes should also be pre-coated with Hank’s solution).
18. Pass the dissociated cells through a 20 mm filter, placed into a clean 15 ml tube. Rinse the 50 ml falcon tube with 1 ml of Hank’s solution and add this to the filter.
19. Once the liquid has passed through the filter, remove the filter and discard, and place the lid on the 15 ml tube.
20. Centrifuge the cell suspension at 500g for 5 min.
21. Remove the supernatant using a glass pipette and resuspend the cells in 1 ml of Hank’s solution. Transfer the 1 ml of cell suspension to a 1.5 ml LoBind Eppendorf tube and take a 10 μl aliquot for cell counting.
22. Centrifuge the 1 ml cell suspension at 500g for 5 min.
23. Perform a cell and viability count with the 10 μl aliquot of cells, by diluting the aliquot 1:1 with the live/dead stain Trypan Blue and pipetting the solution into the Cell Chamber slides for analysis by the Countess II. A haemocytometer can also be used for a count and viability measurement.
24. Once the centrifugation is complete, remove the supernatant using a P1000 pipette. Cell pellets can be stored at −80°C, prepared immediately for protein or RNA extraction or fixed for immunocytochemistry (protocol below).
Preparation for immunocytochemistry
1. Following the cell dissociation, resuspend the cells in 4% PFA in PBS and fix cells for 20 min at room temperature with gentle nutation.
2. Spin cells down at 800 rpm for 5 min to pellet.
3. Remove the supernatant using a P1000 pipette and resuspend the cells in PBS.
4. Spin cells down at 800 rpm for 5 min to pellet.
5. Wash the cells 2 × with PBS, spinning the cells down at 800 rpm for 5 min to pellet each time.
6. For the final wash, resuspend cells in PBS so that the concentration is 1 × 106 cells/ml.
7. Spot the cells onto 12-well glass slides by pipetting 10 μl of cell suspension into each well, and either dry at room temperature or on a heat block (50°C) until the slides are dry.
8. Store the slides at −20°C for up to 2 months, until needed for immunocytochemistry.
Anticipated results
Anticipated results for collagenase dissociation method
Following the temporary culture step, the structure of granulosa cells resembles cultured fibroblasts (Fig. 2A) (Sadowska et al., 2015; Parvari et al., 2016), with adherent cytoplasmic projections and visible speckled lysosomes (numerous in early follicles, see Ndiaye et al., 2015). For an average litter of C57BL/6J (between 6 and 8 ovaries), a sufficient yield of granulosa cells can be accomplished, protocol yields (mean ± SD) 1.07 ± 0.7 × 106 total granulosa cells from a PND1 dissociation, with 1.98 ± 1.8 × 106 from PND4 (Table II). The use of additional ovaries increases the potential cell yield in a weak yet positive linear correlation (r2 = 0.24, P = 0.0131), because the enzyme approaches saturation point (Fig. 2B). We typically achieve a yield of at least 70 ng of RNA per 100 000 cells (Table II), with no significant differences (N = 28, P = 0.842) between RNA yield in PND1 and PND4 ovaries (Fig. 2C).
Table II.
Collagenase dissociation collection characteristics by ovarian timepoint.
| Age of dissociated ovaries (n = 5) | Granulosa cell yield per dissociation, mean ± SD | RNA yield (ng RNA per 100,000 cells), mean ± SD | RIN score, mean ± SD |
|---|---|---|---|
| PND1 | 1.07 ± 0.7 × 106 | 91 ± 94 | 9.6 ± 0.4 |
| PND4 | 1.98 ± 1.8 × 106 | 70 ± 57 | 10 ± 0 |
PND, post-natal day; RIN, RNA integrity number.
Thus, for a dissociation using a typical litter size for C57BL6/J mice (giving between 4 and 8 ovaries), ∼1.3 × 106 cells and approximately 845 ng of total RNA can be obtained. The quality of RNA from dissociated granulosa cells is exceptionally pure as determined via quality control instrument Agilent 2100 bioanalyzer, achieving RNA integrity number (RIN) scores above 9 (Table II), making these samples ideal for next-generation RNA sequencing which typically requires RIN scores > 6 (Kukurba and Montgomery, 2015). Differences in RNA yield and quality may be due to many factors including, but not limited to loss of viability of cells obtained after culture, starting cell numbers and losses at each stage, variation in preparation of reagents and buffers. See Table III for a troubleshooting guide.
Table III.
Table of troubleshooting tips for collagenase dissociation protocol.
| Issue | Possible cause(s) | Advice for solution |
|---|---|---|
| Ovary tissue not breaking down during enzymatic dissociation step | Enzymatic failure | Ensure reagents stored and used correctly, that solution is completely combined and that the total quantity of tissue is ≤20 mg |
| Tissue rigidity | Older ovaries are more fibrous and may require more forceful mechanical agitation when pipetting puncture/tearing with a needle while in the dissociation solution. Additionally, an extra 15-min incubation in dissociation solution for a total 75 min may suffice | |
| Cells adhering to plate after trypsinisation and wash | Insufficient digestion | Check cells before and after 5-minute incubation with trypsin to ensure they have lifted from the plate. Incubate for a further minute if required. Additionally, tapping plate firmly against a solid surface and pipetting directly onto the plate with force may dislodge any rounded cells remaining on the bottom of the plate |
| Loss of cell viability after overnight culture | Insufficient nutrients | Ensure ovary culture media is kept at 4°C when not in use, and always heated to 37°C before use. Doubling the proportion of FCS in media to 10% may also increase viability |
| Overcrowding on plate | Dilute cell suspension before plating and seed multiple plates, or switch seeding cells in a flask (T25 or T75) | |
| Small/no pellet after collection | Cell loss | Ensure pipette tips and serological pipettes are all coated with Hank’s solution before coming into contact with cells. When not in use, store Hank’s solution at 4°C, equilibrate to room temp before use, and do not use following storage exceeding 2 weeks |
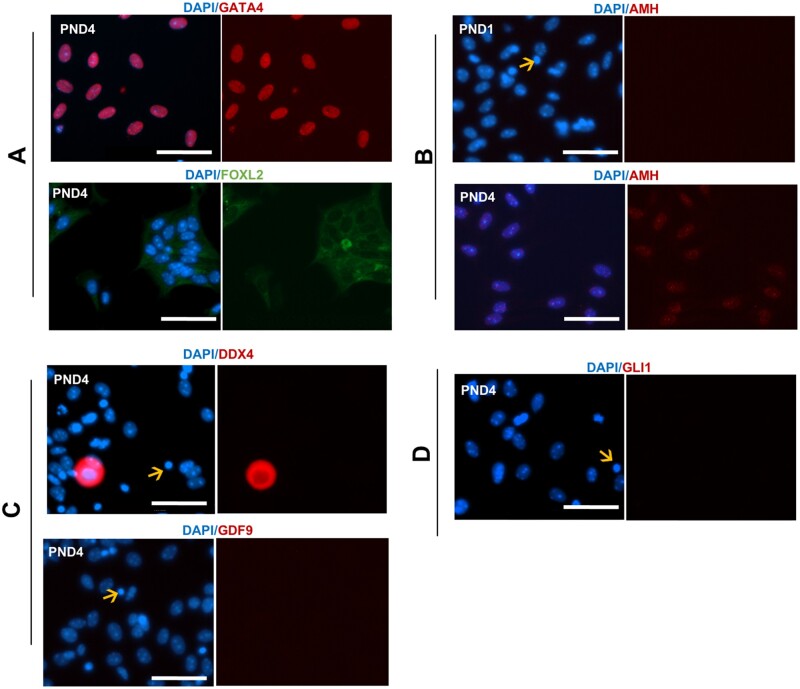
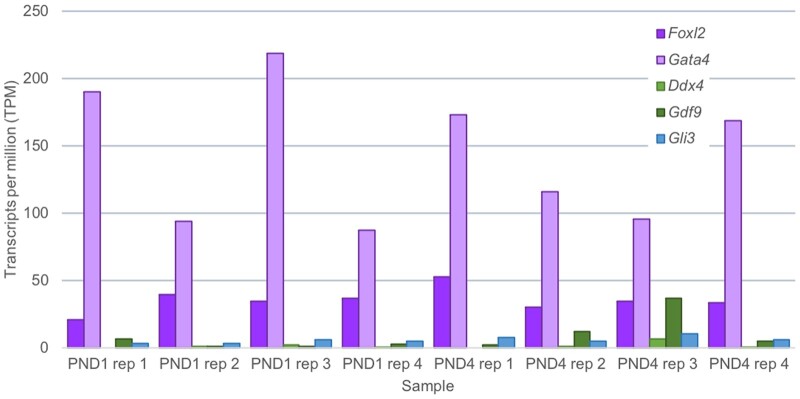
The purity of cells obtained from performing this dissociation can be determined by immunofluorescent detection of a range of markers indicative of ovarian cell types (see Appendix 1 for experimental conditions). Through staining for the granulosa cell markers GATA4 and FOXL2 (Pisarska et al., 2011; Efimenko et al., 2013), the majority of cells post-culture were validated as granulosa cells (Fig. 3A). It was possible to distinguish primordial and primary granulosa cell populations via localisation of AMH, which is expressed in growing granulosa cells from the primary to small antral stage (Visser et al., 2006); thus, it is expressed in cells dissociated from PND4 ovaries, yet not expressed in cells originating from PND1 ovaries (Fig. 3B). In PND4 dissociated cells, markers for oocytes (GDF9 and DDX4; Aaltonen et al., 1999; Raz, 2000) indicated very minimal contamination (Fig. 3C) at a proportion of 0.4% compared to granulosa-positive cells (99.6%) from counts of 300 cells repeated across n = 4 biological replicates (P < 0.001). This was mirrored in counts from PND1 dissociated cells, where 0.25% of GDF9 and DDX4 positive cells were identified compared to 99.75% of GATA4 positive granulosa cells from counts of 300 cells repeated across n = 4 biological replicates (P < 0.001). Confirming the isolation of a pure granulosa cell population, there was no expression of theca cell marker, GLI1 (Fig. 3D), which is to be expected as theca cells are not observed in the neonatal ovary, but appear once follicles have two or more layers of granulosa cells (Young and McNeilly, 2010), which is not until around PND6 in the mouse. It was observed that following overnight culture, there is some loss in viability of the cells as identified by apoptotic cells with condensed nuclei (denoted by arrows in Fig. 3). These results were confirmed at the gene level by RNA sequencing (see Appendix 1 for method). While a culture step has the capacity to alter the physiology and gene expression of the cell, RNA transcripts of dissociated cells after culture reveal classic granulosa cell expression (Foxl2 and Gata4) compared to very minimal or no detection of other ovarian cell markers, specifically oocytes (Ddx4, Gdf9) and theca cells (Gli3) (Fig. 4). In summary, the described technique is an efficient way to collect mass quantities of live granulosa cells from primordial, activating, and primary follicles for downstream functional analyses without the need for manual cell sorting.
Figure 3.
Fluorescence images for collagenase dissociation method. (A) Immunofluorescent localisation of ovarian cell markers counterstained with blue nuclear marker (4′‐6‐diamidino‐2‐phenylindole (DAPI)) and the granulosa cell markers (GATA4 and FOXL2), (B) differentiating granulosa cell marker (AMH) in post-natal day (PND) 1 and PND4 cells dissociated via collagenase, (C) oocyte markers (DDX4 and GDF9), and (D) theca cell marker (GLI1). Arrows indicates apoptotic cells. All experiments performed in PND1 and PND4 n = 3 for each group, representative images taken at PND4 stage except where indicated. Images taken at 40× magnification, scale bar equivalent to 50 μm.
Figure 4.
Gene expression of dissociated cells using the collagenase dissociation method. Expression of selected ovarian cell-specific markers determined via RNA sequencing of collagenase-dissociated cells originating from both postnatal day (PND) 1 and postnatal day 4 ovaries (n = 4 biological replicates per timepoint). Granulosa cell markers (Foxl2 and Gata4) in purple, oocyte markers (Ddx4 and Gdf9) in green, and theca cell marker (Gli3) in blue. TPM stands for transcripts per million mapped reads and refers to the gene expression value generated via RNA sequencing.
Anticipated results for papain dissociation method
This protocol has been optimised for the dissociation of one ovary per replicate to minimise the need for large amounts of tissue. From one single representative ovary at E18.5, 6.49 ± 2.0 × 104 total cells can be retrieved (Table IV). The average viability of these cells (determined by Trypan Blue staining) is 75% ± 0.1, suitable for any subsequent analyses which require live cells (Table IV). The number of cells obtained increases with increasing ovarian age; 1.25 ± 0.7 × 105 total cells can be retrieved from one single ovary at PND8, with a high cell viability 85% ± 0.0 (Table IV). In the adult ovary, represented by 12 weeks in this manuscript, 3.00 ± 1.0 × 105 total cells are present following dissociation, with a high cell viability of 67% ± 0.1 (Table IV). We typically achieve a yield of 81 ng of RNA per 100 000 cells at E18.5, 78 ng of RNA per 100 000 cells at PND8 and 83 ng of RNA per 100 000 cells at 12 weeks, although this number is variable (Table IV). Variability can be due to a number of factors, the main ones listed in the troubleshooting guide in Table V. RNA integrity was confirmed as high quality by using the Agilent TapeStation instrument, with RIN above 9 for all three timepoints (Table V).
Table IV.
Papain dissociation collection characteristics by ovarian timepoint (N = 4).
| Age of ovary | Cell yield from one ovary, mean ± SD | Cell viability from one ovary, mean ± SD | RNA yield (ng RNA per 100,000 cells), mean ± SD | RIN score, mean ± SD |
|---|---|---|---|---|
| E18.5 | 6.49 ± 2.0 × 104 | 75% ± 0.1 | 81 ± 49 | 9.5 ± 0.9 |
| PND8 | 1.25 ± 0.7 × 105 | 85% ± 0.0 | 78 ± 25 | 9.7 ± 0.3 |
| 12WKS | 3.00 ± 1.0 × 105 | 67% ± 0.1 | 83 ± 43 | 9.5 ± 0.4 |
PND, post-natal day; RIN, RNA Integrity number; WKS, weeks.
Table V.
Table of troubleshooting tips for papain dissociation protocol.
| Issue | Possible cause(s) | Advice for solution |
|---|---|---|
| Poor tissue dissociation | Enzymatic failure |
|
| Tissue rigidity |
|
|
| Loss of cell viability during the dissociation | Excessive tissue dissociation |
|
| Cell clumping/a failure to get a single cell suspension | Insufficient tissue dissociation |
|
| Small/no pellet after dissociation | Cell loss |
|
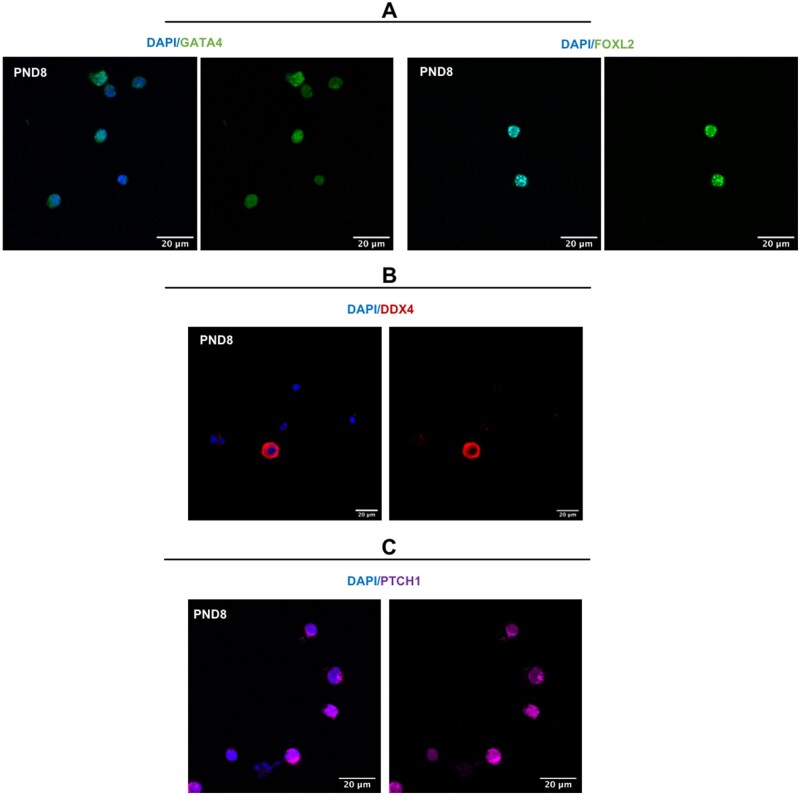
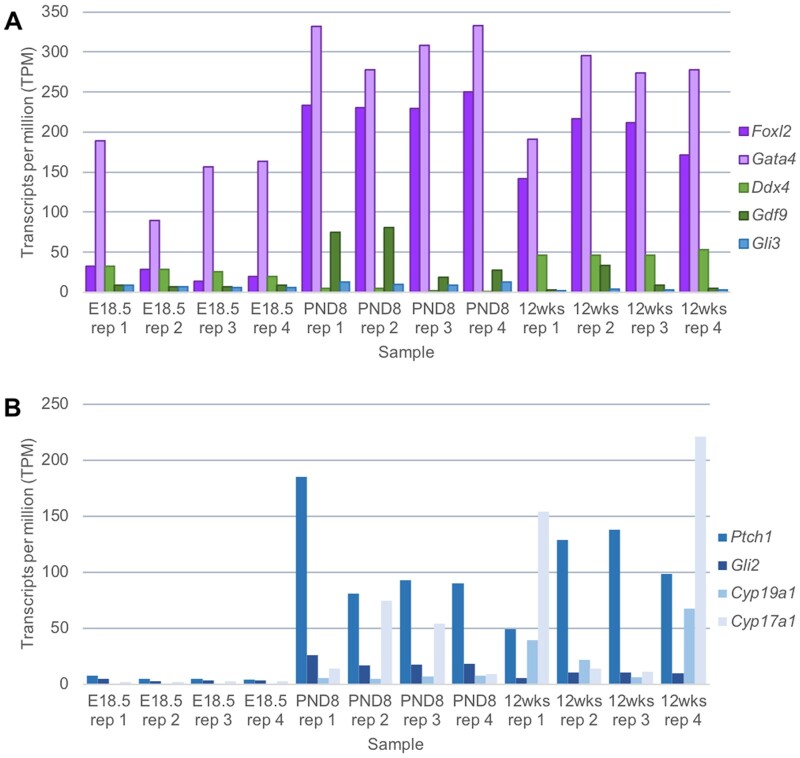
To determine the percentages of cell types isolated from the PND8 ovary using the papain dissociation protocol, we performed immunofluorescence for several protein markers including the granulosa cell markers GATA4 (Efimenko et al., 2013) and FOXL2 (Schmidt et al., 2004), granulosa cell and theca cell marker PTCH1 (Liu et al., 2015) and oocyte marker DDX4 (Zarate-Garcia et al., 2016) and counted positively stained cells to determine the relative proportions of the different cell types (see Appendix 2 for experimental conditions). These were calculated from counts of 200 cells repeated across 3 biological replicates. 84% of the cells stained positively for both GATA4 and FOXL2, indicating the majority of dissociated cells are granulosa cells (Fig. 5A). The use of a 20 µm filter at the end of the papain dissociation protocol removed most germ cells from the cell suspension, with approximately 1% of cells staining positive for DDX4 (Fig. 5B). 9% of the cells were PTCH1 positive and FOXL2 negative, which is indicative of theca cells (Fig. 5C). The proportion of DDX4 positive cells and PTCH1 positive cells in relation to GATA4/FOXL2 positive granulosa cells is time point dependent. At E18.5, 72% of cells stained positive for GATA4/FOXL2, with 1.7% of cells stained with the oocyte marker DDX4 from counts of 200 cells repeated across n = 3 biological replicates. Dissociated cells from this timepoint showed no PTCH1 expression, consistent with a lack of theca cells present in the embryonic mouse ovary (Abedel-Majed et al., 2019). From cells dissociated from the 12 weeks ovary timepoint, 64% of cells stained positive for GATA4/FOXL2, with 1.6% of cells stained with the oocyte marker DDX4. PTCH1 positive cells were more abundant, with 20% of cells expressing both PTCH1 and FOXL2, indicative of theca cells. These results were confirmed at the gene level, by RNA sequencing (see Appendix 2 for methods). High levels of Foxl2 and Gata4 transcripts were detected at all three timepoints (E18.5, PND8 and 12 weeks), with lower levels of the oocyte transcripts Ddx4 and Gdf9 and theca cell marker Gli3 (Fig. 6A). We do observe a higher expression of the oocyte transcripts Ddx4 and Gdf9 compared with the collagenase method, potentially due to the presence of oocytes smaller than 20 μm passing through the filter into the dissociated cell population, particularly at the E18.5 timepoint. However, upregulation of other theca markers Ptch1 and Cyp17a1 were observed at 12wks, when substantial amounts of theca cells are present in the ovary (Fig. 6B) (Young and McNeilly, 2010; Abedel-Majed et al., 2019; Liu et al., 2015; Richards et al., 2018).
Figure 5.
Fluorescence images for papain dissociation method. Immunofluorescent localisation of ovarian cell markers counterstained with blue nuclear marker (4′‐6‐diamidino‐2‐phenylindole (DAPI)): (A) granulosa cell markers (GATA4 and FOXL2), (B) oocyte marker (DDX4), and (C) theca cell marker (PTCH1). All experiments performed on dissociated ovarian cells from PND8 ovary (n = 3), and representative images shown. Images taken at 60× magnification, scale bar 20 μm.
Figure 6.
Gene expression of dissociated cells using the papain dissociation method. (A) Expression of selected ovarian cell-specific markers determined via RNA sequencing of papain-dissociated cells originating from embryonic day 18.5, postnatal Day 8 and postnatal Day 12 week-old ovaries (n = 4 samples per group). Granulosa cell markers (Foxl2 and Gata4) in purple, Oocyte markers (Ddx4 and Gdf9) in green, and theca cell marker (Gli3) in blue. (B) Expression of selected theca cell-specific markers (Ptch1, Gli2, Cyp19a1, and Cyp17a1) determined via RNA sequencing of papain-dissociated cells originating from both embryonic Day 18.5, postnatal Day 8 and postnatal Day 12 week-old ovaries (n = 4 samples per group). TPM stands for transcripts per million mapped reads and refers to the gene expression value generated via RNA sequencing.
Following dissociation, these cells can be used for a variety of experimental techniques, including fixation steps to perform immunofluorescence protocols. This protocol allows for the dissociation of mouse ovarian tissue at numerous timepoints, to capture all somatic cell lineages for detailed analysis and characterisation.
Conclusion
These two protocols enable the isolation of multiple cell types from various stages of mouse ovarian development. The collagenase dissociation method is an efficient method for collecting a pure granulosa cell population from the neonatal ovary, whereas the papain dissociation protocol captures all the somatic cell types from the ovary at different developmental stages. There are limitations with both described protocols; namely the extended culture time for the collagenase protocol and the presence of small contaminating cells following filtration using the papain protocol. These limitations should be taken into consideration and used to select an appropriate protocol for the subsequent downstream technologies. To date, it is difficult to isolate pure populations of somatic cell types from the developing mouse ovary, due to a lack of markers to label the various heterogenous cell populations. We present these two methods to facilitate further research into identifying novel markers of mouse ovarian cells and the use of suitable antibodies and fluorescently tags for cell sorting applications.
Both protocols have been shown to allow the preparation of high-quality samples with excellent viability for RNA sequencing, with the advantage of producing a single cell suspension necessary for scRNAseq. In summary, these techniques provide an opportunity to explore mouse ovarian cell function at numerous developmental timepoints.
Data availability
The data underlying this article are available within it and in Appendices.
Acknowledgements
The authors gratefully acknowledge Karine Rizzoti for her reagents and insight on the papain dissociation protocol.
Authors’ roles
All authors were involved in experimental design and conception, and the interpretation of data. E.A.F. and E.R.F. performed the experiments and subsequent data analysis. E.A.F., E.R.F. and J.M.S. wrote the manuscript. All authors revised and approved the final manuscript.
Funding
The authors acknowledge the financial assistance to J.M.S. by the Australian National Health and Medical Research Council (G1600095) and the Hunter Medical Research Institute (G1501433 and G1801335). This work was also supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001107), the UK Medical Research Council (FC001107), and the Wellcome Trust (FC001107). The authors acknowledge the generous financial assistance to R.L.B. by the UK Biotechnology and Biological Sciences Research Council (BB/N018680/1). E.R.F. is the recipient of an Australian Government Research Training Program Scholarship.
Conflict of interest
The authors declare no conflict of interest.
Appendix 1. Collagenase dissociation protocol
Animal details
C57BL/6J mice were supplied by the University of Newcastle Animal Services Unit under ethics approval number A-2018-803. All experiments involving the use of animals were conducted in accordance with the Institutes’ Animal Care and Ethics Committee guidelines in accordance with Australian NHMRC Guidelines on Ethics in Animal Experimentation.
Mice were housed under a controlled lighting regime (16 L: 8 D) at 21–22°C and supplied with food and water ad libitum. Neonatal mice (PND1 and PND4) were euthanized by asphyxiation with carbon dioxide, followed by decapitation. Ovaries were collected immediately after euthanasia, and bursa and uterus remnants removed under a dissecting microscope.
Immunocytochemistry
Granulosa cells fixed in 24-well plates (as described in materials and methods) were blocked for non-specific binding by an incubation of 3%BSA, 10% donkey serum in PBS for 1 h at room temperature. Granulosa cells were probed with antibodies specific for GATA4 (ab84593; Abcam, Cambridge, UK), FOXL2 (ab5096; Abcam), AMH (130233; Abcam), DDX4 (ab13840; Abcam), GDF9 (ab93892; Abcam), and GLI1 (ab92611; Abcam) at a dilution of 1:300. Primary antibodies were visualised using either a donkey anti-goat Alexa 555-conjugated secondary antibody (a21432; Invitrogen) or a goat anti-rabbit Alexa 555-conjugated secondary antibody (a21428; Invitrogen) at a dilution of 1:200; 4′‐6‐diamidino‐2‐phenylindole (DAPI) was used as a nuclear counterstain. Slides were imaged using the EVOS FL (AMF4300; Thermofisher).
RNA extraction and quantity
Extractions for granulosa cell RNA were performed using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany; cat no. 74034) as per the manufacturer’s instructions. Post-extraction, the synthesis of cDNA was completed using the SuperScript IV VILO system (Invitrogen, Carlsbad, CA, USA; cat no. 11766050). The RNA purity, quality, and concentration was determined using the Agilent Bioanalyser 2100. The samples were prepared for this machine via the Agilent RNA 6000 Nano Assay (5067-1511; Agilent, Palo Alto, CA, USA).
RNA sequencing
Eight, qRNA-seq libraries were sequenced on the NextSeq 2 × 75 bp high output via Auckland Genomics at the University of Auckland. These RNA samples consisted of two groups of mouse granulosa cells (isolated from postnatal Day 1 and postnatal Day 4), with four replicates each. From the output of the RNA sequencing, the bioinformatics team at Auckland Genomics Facility performed the following analyses. The overall quality of the data was visualized using the program FastqQC (Andrews, 2014). TrimGalore (Lindgreen, 2012), was used to check and remove adapters from the sequences. Sequences with a Phred score less than 30 were trimmed. Reads with a length shorter than 25 base pairs were discarded. Hisat2 (Kim et al., 2015; Pertea et al., 2016) was used to map the cleaned reads to the mouse transcriptome (grcm38). Output sam files from hisat2 were then sorted by genomic position and converted to bam format using samtools sort (Li et al., 2009). To generate expression estimates for each sample, the mapped reads were then assembled into transcripts using StringTie. Differential expression analysis was performed using the R software package Ballgown (Fu et al., 2018). Low abundance transcripts with a variance less than one across the samples were removed. To view the similarity between the samples, the Pearson correlation and distance between the samples were calculated. Differential gene expression between the two treatments were calculated using FPKM values. The data were then sorted by their adjusted p-value (false discovery rate) and absolute log2 fold change values.
Appendix 2. Papain dissociation protocol
Animal details
C57BL/6J mice were used in all experiments. All experiments carried out on mice were approved under the UK Animal (Scientific Procedures) Act (Project licence 70/8560). Mice were housed in individually ventilated cages under a controlled lighting regime and supplied with food and water ad libitum. Mouse husbandry, breeding, ear biopsies and vaginal plug checks were performed by the Biological Research Facility team at the Francis Crick Institute. The day of vaginal plug was considered as E0.5 from the time of conception and the day of birth was termed P0. For E18.5 collections, the mum was euthanised via cervical dislocation, and the E18.5 embryos were dissected from the uterus and euthanised by decapitation. PND8 females were euthanised by decapitation. Twelve-week-old female mice were euthanised by cervical dislocation. At all three timepoints, ovaries were collected immediately following euthanasia and the ovarian bursa was removed under a dissecting microscope.
Immunocytochemistry
Granulosa cells fixed on glass slides (as described in Materials and methods) were permeabilised in PBS with 0.01% Triton X-100 for 10 min. The cells were blocked with 10% donkey serum in PBS for 1 h at room temperature. Following blocking, granulosa cells were probed with antibodies specific for GATA4 (ab84593; Abcam), FOXL2 (ab5096; Abcam), DDX4 (ab27591; Abcam) and PTCH1 (ab39266; Abcam). Primary antibodies were visualised using either a donkey anti-goat Alexa 555-conjugated secondary antibody (a21432; Invitrogen), a donkey anti-rabbit Alexa 555-conjugated secondary antibody (a32794; Invitrogen) or a donkey anti-mouse Alexa 488-conjugated secondary antibody (a32766; Invitrogen) at a 1:100 dilution and 4′‐6‐diamidino‐2‐phenylindole (DAPI) was used as a nuclear counterstain. Cells were imaged using the Olympus FV1000 Confocal microscope (Olympus America, Center Valley, PA, USA) with a 60× oil immersion lens.
RNA extraction and quantity
RNA extractions were performed using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany; cat no. 74034) as per the manufacturer’s instructions. cDNA synthesis was performed using the Superscipt III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA; cat no. 18080-044). The RNA purity, quality and concentration were determined using the Agilent TapeStation.
RNA sequencing
Twelve libraries were sequenced on an Illumina HiSeq 4000 machine (Illumina, San Diego, CA, USA). The ‘Trim Galore!’ utility version 0.4.2 was used to remove sequencing adaptors and to quality trim individual reads with the q-parameter set to 20 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The sequencing reads were then aligned to the mouse genome and transcriptome (Ensembl GRCm38release-89) using RSEM version 1.3.0 (Li and Dewey, 2011) in conjunction with the STAR aligner version 2.5.2 (Dobin et al., 2013). Sequencing quality of individual samples was assessed using FASTQC version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and RNA-SeQC version 1.1.8 (DeLuca et al., 2012). Differential gene expression was determined using the R-bioconductor package DESeq2 version 1.14.1 (Development RC Team, 2008; Love et al., 2014). Gene set enrichment analysis (GSEA) was conducted as described (Subramanian et al., 2005).
References
- Aaltonen J, Laitinen MP, Vuojolainen K, Jaatinen R, Horelli-Kuitunen N, Seppä L, Louhio H, Tuuri T, Sjöberg J, Bützow R.. et al. Human growth differentiation factor 9 (GDF-9) and its novel homolog GDF-9B are expressed in oocytes during early folliculogenesis. J Clin Endocrinol Metabol 1999;84:2744–2750. [DOI] [PubMed] [Google Scholar]
- Abedel-Majed MA, Romereim SM, Davis JS, Cupp AS.. Perturbations in lineage specification of granulosa and theca cells may alter corpus luteum formation and function. Front Endocrinol 2019;10:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. Version 0.11. 2. Babraham Institute, Cambridge, UK http://www.bioinformatics.babraham.ac.uk/projects/fastqc2014. [Google Scholar]
- Bonnet A, Bevilacqua C, Benne F, Bodin L, Cotinot C, Liaubet L, Sancristobal M, Sarry J, Terenina E, Martin P.. et al. Transcriptome profiling of sheep granulosa cells and oocytes during early follicular development obtained by Laser Capture Microdissection. BMC Genomics 2011;12:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RA, Bihannic L, Rosencrance C, Hadley JL, Tong Y, Phoenix TN, Natarajan S, Easton J, Northcott PA, Gawad C.. A single-cell transcriptional atlas of the developing murine cerebellum. Curr Biol 2018;28:2910–2920.e2912. [DOI] [PubMed] [Google Scholar]
- Clarkson YL, McLaughlin M, Waterfall M, Dunlop CE, Skehel PA, Anderson RA, Telfer EE.. Initial characterisation of adult human ovarian cell populations isolated by DDX4 expression and aldehyde dehydrogenase activity. Sci Rep 2018;8:6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, Getz G.. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 2012;28:1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Development RC Team. R: A Language and Environment for Statistical Computing;2008. Vienna, Austria: R Foundation for Statistical Computing.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmans M-M, Michaux N, Camboni A, Martinez-Madrid B, Van Langendonckt A, Annarita Nottola S, Donnez J.. Evaluation of Liberase, a purified enzyme blend, for the isolation of human primordial and primary ovarian follicles. Hum Reprod 2006;21:413–420. [DOI] [PubMed] [Google Scholar]
- Efimenko E, Padua MB, Manuylov NL, Fox SC, Morse DA, Tevosian SG.. The transcription factor GATA4 is required for follicular development and normal ovarian function. Dev Biol 2013;381:144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst EH, Franks S, Hardy K, Villesen P, Lykke-Hartmann K.. Granulosa cells from human primordial and primary follicles show differential gene expression profiles. Hum Reprod 2018;33:666–679. [DOI] [PubMed] [Google Scholar]
- Ernst EH, Grondahl ML, Grund S, Hardy K, Heuck A, Sunde L, Franks S, Andersen CY, Villesen P, Lykke-Hartmann K.. Dormancy and activation of human oocytes from primordial and primary follicles: molecular clues to oocyte regulation. Human Reprod (Oxford, England) 2017;32:1684–1700. [DOI] [PubMed] [Google Scholar]
- Feng Y, Cui P, Lu X, Hsueh B, Moller Billig F, Zarnescu Yanez L, Tomer R, Boerboom D, Carmeliet P, Deisseroth K.. et al. CLARITY reveals dynamics of ovarian follicular architecture and vasculature in three-dimensions. Sci Rep 2017;7:44810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A, Oberholtzer SM, Bagheri-Fam S, Rastetter RH, Holdreith C, Caceres VL, John SV, Shaw SA, Krentz KJ, Zhang X.. et al. Dynamic expression patterns of Irx3 and Irx5 during germline nest breakdown and primordial follicle formation promote follicle survival in mouse ovaries. PLoS Genet 2018;14:e1007488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H, Both MD, Siniard A, Sharma S, Notwell JH, Wallace M, Leone DP, Nguyen A, Zhao E, Lee H.. et al. A guide to single-cell transcriptomics in adult rodent brain: the medium spiny neuron transcriptome revisited. Front Cell Neurosci 2018;12:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukurba KR, Montgomery SB.. RNA sequencing and analysis. Cold Spring Harb Protoc 2015;2015:951–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 2012;5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Peng J, Matzuk MM, Yao HH.. Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat Commun 2015;6:6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo BKM, Sheikh S, Williams SA.. In vitro and in vivo mouse follicle development in ovaries and reaggregated ovaries. Reproduction 2019;157:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, He C, Huang C, Wang H, Hua G, Wang Z, Zhou J, Chen X, Ma B, Timm BK.. et al. Timely expression and activation of YAP1 in granulosa cells is essential for ovarian follicle development. Faseb J 2019;33:10049–10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Inoue N, Manabe N, Ohkura S.. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev 2012;58:44–50. [DOI] [PubMed] [Google Scholar]
- Mayère C, Neirijnck Y, Sararols P, Rands CM, Stévant I, Kühne F, Chassot AA, Chaboissier M-C, Dermitzakis ET, Nef S.. Single-cell transcriptomics reveal temporal dynamics of critical regulators of germ cell fate during mouse sex determination. The FASEB Journal 2021;35:e21452. [DOI] [PubMed] [Google Scholar]
- Morbeck D, Flowers W, Britt J.. Response of porcine granulosa cells isolated from primary and secondary follicles to FSH, 8-bromo-cAMP and epidermal growth factor in vitro. Reproduction 1993;99:577–584. [DOI] [PubMed] [Google Scholar]
- Navaroli DM, Tilly JL, Woods DC.. Isolation of mammalian oogonial stem cells by antibody-based fluorescence-activated cell sorting. Methods Mol Biol (Clifton, NJ) 2016;1457:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye K, Carrière PD, Sirois J, Silversides DW, Lussier JG.. Differential expression of lysosome-associated protein transmembrane-4 beta (LAPTM4B) in granulosa cells of ovarian follicles and in other bovine tissues. J Ovarian Res 2015;8:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton H, Picton H, Gosden R.. In vitro growth of oocyte–granulosa cell complexes isolated from cryopreserved ovine tissue. Reproduction 1999;115:141–150. [DOI] [PubMed] [Google Scholar]
- Nguyen QH, Pervolarakis N, Nee K, Kessenbrock K.. Experimental considerations for single-cell RNA sequencing approaches. Front Cell Dev Biol 2018;6:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Spradling AC.. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci USA 2020;117:20015–20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktay K, Nugent D, Newton H, Salha O, Chatterjee P, Gosden RG.. Isolation and characterization of primordial follicles from fresh and cryopreserved human ovarian tissue. Fertil Steril 1997;67:481–486. [DOI] [PubMed] [Google Scholar]
- Parvari S, Yazdekhasti H, Rajabi Z, Gerayeli Malek V, Rastegar T, Abbasi M.. Differentiation of mouse ovarian stem cells toward oocyte-like structure by coculture with granulosa cells. Cell Reprogram 2016;18:419–428. [DOI] [PubMed] [Google Scholar]
- Pedersen T, Peters H.. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil 1968;17:555–557. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650–1667.27560171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarska MD, Barlow G, Kuo F-T.. Minireview: roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology 2011;152:1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastetter RH, Bernard P, Palmer JS, Chassot AA, Chen H, Western PS, Ramsay RG, Chaboissier MC, Wilhelm D.. Marker genes identify three somatic cell types in the fetal mouse ovary. Dev Biol 2014;394:242–252. [DOI] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol 2000;1:reviews1017.1. reviews1017. 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JS, Ren YA, Candelaria N, Adams JE, Rajkovic A.. Ovarian follicular theca cell recruitment, differentiation, and impact on fertility: 2017 update. Endocr Rev 2018;39:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowska A, Nynca A, Korzeniewska M, Piasecka-Srader J, Jablonska M, Orlowska K, Swigonska S, Ciereszko RE.. Characterization of porcine granulosa cell line AVG-16. Folia Biol (Praha) 2015;61:184–194. [PubMed] [Google Scholar]
- Sarraj MA, Drummond AE.. Mammalian foetal ovarian development: consequences for health and disease. Reproduction 2012;143:151–163. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M.. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 2004;131:933–942. [DOI] [PubMed] [Google Scholar]
- Steffensen LL, Ernst EH, Amoushahi M, Ernst E, Lykke-Hartmann K.. Transcripts encoding the androgen receptor and IGF-related molecules are differently expressed in human granulosa cells from primordial and primary follicles. Front Cell Dev Biol 2018;6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevant I, Nef S.. Genetic control of gonadal sex determination and development. Trends Genet 2019;35:346–358. [DOI] [PubMed] [Google Scholar]
- Stévant I, Kühne F, Greenfield A, Chaboissier M-C, Dermitzakis ET, Nef S.. Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep 2019;26:3272–3283.e3. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES.. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JA, de Jong FH, Laven JS, Themmen AP.. Anti-Müllerian hormone: a new marker for ovarian function. Reproduction 2006;131:1–9. [DOI] [PubMed] [Google Scholar]
- Wang J-J, Ge W, Zhai Q-Y, Liu J-C, Sun X-W, Liu W-X, Li L, Lei C-Z, Dyce PW, De Felici M. et al. Single-cell transcriptome landscape of ovarian cells during primordial follicle assembly in mice. PLoS Biol 2020;18:e3001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M, Simões FC, Patient R, Sauka-Spengler T, Riley PR.. Functional heterogeneity within the developing zebrafish epicardium. Dev Cell 2020;52:574–590.e576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White YAR, Woods DC, Takai Y, Ishihara O, Seki H, Tilly JL.. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med 2012;18:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Candido-Ferreira I, Repapi E, Gavriouchkina D, Senanayake U, Ling ITC, Telenius J, Taylor S, Hughes J, Sauka-Spengler T.. Reconstruction of the global neural crest gene regulatory network in vivo. Dev Cell 2019;51:255–276.e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DC, Tilly JL.. Isolation, characterization and propagation of mitotically active germ cells from adult mouse and human ovaries. Nat Protoc 2013;8:966–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, McNeilly AS.. Theca: the forgotten cell of the ovarian follicle. Reproduction 2010;140:489–504. [DOI] [PubMed] [Google Scholar]
- Yousef H, Czupalla CJ, Lee D, Butcher EC, Wyss-Coray T.. Papain-based single cell isolation of primary murine brain endothelial cells using flow cytometry. Bio Protoc 2018;8:e3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate-Garcia L, Lane SI, Merriman JA, Jones KT.. FACS-sorted putative oogonial stem cells from the ovary are neither DDX4-positive nor germ cells. Sci Rep 2016;6:27991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available within it and in Appendices.