Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized primarily by progressive loss of motor neurons. Although ALS occurs worldwide and the frequency and spectrum of identifiable genetic causes of disease varies across populations, very few studies have included African subjects. In addition to a hexanucleotide repeat expansion (RE) in C9orf72, the most common genetic cause of ALS in Europeans, REs in ATXN2, NIPA1 and ATXN1 have shown variable associations with ALS in Europeans. Intermediate range expansions in some of these genes (e.g. ATXN2) have been reported as potential risk factors, or phenotypic modifiers, of ALS. Pathogenic expansions in NOP56 cause spinocerebellar ataxia-36, which can present with prominent motor neuron degeneration. Here we compare REs in these genes in a cohort of Africans with ALS and population controls using whole genome sequencing data. Targeting genotyping of short tandem repeats at known loci within ATXN2, NIPA1, ATXN1 and NOP56 was performed using ExpansionHunter software in 105 Southern African (SA) patients with ALS. African population controls were from an in-house SA population control database (n = 25), the SA Human Genome Program (n = 24), the Simons Genome Diversity Project (n = 39) and the Illumina Polaris Diversity Cohort (IPDC) dataset (n = 50). We found intermediate RE alleles in ATXN2 (27–33 repeats) and ATXN1 (33–35 repeats), and NIPA1 long alleles (≥8 repeats) were rare in Africans, and not associated with ALS (p > 0.17). NOP56 showed no expanded alleles in either ALS or controls. We also compared the differences in allele distributions between the African and n = 50 European controls (from the IPDC). There was a statistical significant difference in the distribution of the REs in the ATXN1 between African and European controls (Chi-test p < 0.001), and NIPA1 showed proportionately more longer alleles (RE > 8) in Europeans vs. Africans (Fisher’s p = 0.016). The distribution of RE alleles in ATXN2 and NOP56 were similar amongst African and European controls. In conclusion, repeat expansions in ATXN2, NIPA1 and ATXN1, which showed associations with ALS in Europeans, were not replicated in Southern Africans with ALS.
Keywords: Repeat expansion, Mutation, Motor neuron disease, Africa
Highlights
-
•
No associations with ATXN2, ATXN1, NIPA1 expansion mutations, and ALS in Africans.
-
•
Repeat expansion allele distribution in ATXN1 and NIPA1 were different in Africans compared with European populations.
-
•
ALS cases with diverse genetic ancestry should included in ALS gene discovery studies.
Abbreviations
- RE
Repeat expansion
- SA
South Africa
- ALS
Amyotrophic lateral sclerosis
- SCA
Spinocerebellar ataxia
- HSP
Hereditary spastic paraplegia
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease, characterized primarily by progressive loss of motor neurons in the brain and spinal cord, resulting in death within 2–5 years of symptom onset. ALS occurs worldwide although the incidence and genetic causes may differ among different populations, which may have a biological basis (Vucic et al., 2020, Henning et al., 2020). A family history of ALS is present in about 10% of cases with an identifiable genetic cause of disease present in ~15% of patients, including a small proportion without a family history (Taylor et al., 2016). More than 16 different genes harbor mutations considered to be causative of ALS, and most of these are missense mutations. However, the most common mutation in Europeans with ALS is a large hexanucleotide repeat expansion (RE) mutation in the non-coding region of C9orf72 (Renton et al., 2011, DeJesus-Hernandez et al., 2011). While genetic studies which include Africans are few, we have previously reported a similar frequency of C9orf72 expansion mutations in South African ALS patients who self-categorized as having mixed-African ancestry (probable European admixture), compared with European-ancestry cases; notably, however, no C9orf72 repeat expansions were identified in the small number of black South Africans included in this cohort (Nel et al., 2019a). Similarly, Asian ALS populations also have a very low frequency of the C9ORF72 expansion mutation (Liu et al., 2018).
Non-coding large repeat expansions are known to be the primary pathogenic mutation in several inherited neurological diseases such as the autosomal dominant spinocerebellar ataxias (SCA) and hereditary spastic paraplegia (HSP), but intermediate expansions of these non-coding repeats may also be risk factors for diseases such as ALS, or may exert their effect by modifying the phenotype. In Europeans, intermediate REs in ATXN2 (Wang et al., 2014), NIPA1 (Blauw et al., 2012) and ATXN1 (Tazelaar et al., 2020) have shown variable associations with ALS. REs in the NOP56 gene cause SCA type 36, which can present with prominent motor neuron degeneration, have also been investigated for association with ALS. Replicating genetic associations in different populations strengthens the pathogenetic hypothesis, and if not replicated, may open new questions regarding pathogenetic mechanisms.
2. Method
The ALS cohort included 105 African ancestry subjects, satisfying either El Escorial criteria for clinically definite ALS in almost all cases, or clinically probable ALS in one (Brooks et al., 2000). Participants provided an EDTA sample from which DNA was extracted for whole genome sequencing (WGS) as previously described (Nel et al., 2019a, Nel et al., 2019b). Thirty-four of these were participants in the CReATe Consortium’s Phenotype-Genotype-Biomarker study (clinicaltrials.gov: NCT02327845). Participants categorized themselves according to the following SA racial census categories (http://www.statssa.gov.za): black African (n = 27), Cape mixed-African ancestry (M/A) (n = 78). Cape mixed-African ancestry (M/A) refers to the individuals with admixture (European and South East Asian) but predominantly represents genetic ancestry from Khoisan and black Africans (De Wit et al., 2010). The study was approved by the University of Cape Town Research Ethics Committee.
The WGS datasets of non-ALS African ancestry population controls (n = 138) were collected from various studies and included individuals from South-, East-, West- and North Africa: n = 24 healthy individuals from the South African (SA) Human Genome Program (Choudhury et al., 2017), n = 25 SA individuals with myasthenia gravis, an immune-mediated neuromuscular disease (Nel et al., 2019b), n = 39 healthy individuals from the Simons Genome Diversity Cohort (SGDP) (Mallick et al., 2016) and n = 50 healthy individuals from the Illumina Polaris Diversity Cohort (IPDC) represented in the 1000 Genomes project (Genomes Project et al., 2015). To compare the African control population architecture of these RE loci with European controls, we included 50 individuals from Northern and Southern Europe from the IPDC dataset. All samples (ALS and non-ALS controls) were analyzed together using the same pipeline (https://github.com/grbot/varcall) which included alignment to the NCBI GRCh38 reference genome (with al contigs) and joint variant calling according to the Genome Analysis Toolkit (GATK) best practice guidelines (https://gatk.broadinstitute.org/hc/en-us). Since the data was obtained from different studies, sequencing libraries were prepared using different kits (which resulted in different read lengths (100 or 150 bp)) and samples were sequenced using different instruments (all Illumina). All samples were sequenced to a read depth ≥ 30x (i.e. high coverage).
Repeat alleles sizes in ATXN2, NIPA1, ATXN1 and NOP56 were determined using the Illumina ExpansionHunter v4.0.2 tool (Dolzhenko et al., 2017). Relevant reads were extracted from the WGS BAM file and aligned to an appropriate sequence graph representing each locus. The ExpansionHunter algorithm uses a combination of spanning, flanking and in-repeat reads to determine the genotype (including confidence interval). Ambiguous genotypes were either resolved or excluded from the analysis following visual inspection of the pileup of read alignments at the RE locus using the Illumina GraphAlignmentViewer (https://github.com/Illumina/GraphAlignmentViewer/). With the exception of ATXN1, the distribution of RE allele lengths between groups could not be compared statistically as the conditions to perform an overall Chi-test were not met. Instead, we used the Fisher’s exact test to assess the proportion of intermediate or short RE lengths between groups. A Bonferroni correction was performed on the Chi-test assessing the exploratory comparison of control populations. Statistical analysis was performed in GraphPad Prism v8.4.3.
3. Results
The Southern African ALS cohort comprised 105 individuals (53% male, 19% bulbar onset) with an average age of onset of 51 years (Table 1). At clinical presentation, the majority of patients (81%) had clinical evidence of both upper and lower motor neuron involvement, which is defined as the ALS-variant motor neuron disease, whereas the remainder presented initially with other clinical phenotypes (i.e. pure upper motor neuron syndrome or pure lower motor neuron syndrome), before progressing to the ALS phenotype. Two participants had frontotemporal dementia and ALS at presentation. Three participants had a family history of ALS.
Table 1.
Demographic and clinical characteristics of African ALS cohort (n = 105).
| ALS African genetics n = 105 | |
|---|---|
| Age at onset, years, mean (± SD) | 51 (±14) |
| Male, n (%) | 56 (53%) |
| Familial ALS, n (%) | 3 (3%) |
| Region of symptom onset, n (%) | |
| Cognitive | 3 (3%) |
| Bulbar | 20 (19%) |
| Cervical | 37 (35%) |
| Diaphragm | 2 (2%) |
| Lumbosacral | 43 (41%) |
| Phenotype at presentation | |
| Mixed upper motor and lower motor neuron | 85 (81%) |
| Pure upper motor neuron | 11 (10%) |
| Pure lower motor neuron | 7 (7%) |
| ALS-FTD | 2 (2%) |
Familial ALS refers to families in which there are at least 2 biologically related individuals with ALS. FTD -frontotemporal dementia.
3.1. ATXN2
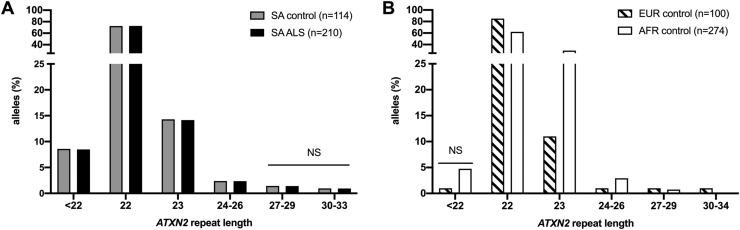
Pathogenic expansions of ≥ 35 CAG repeats in the 5′ untranslated region of the ATXN2 gene, result in SCA2. Assessing intermediate expansions of 27–33 CAG repeats among South Africans, only 2 (0.9%) ALS cases had an RE of ≥30, compared to no African controls (0/114) with intermediate expansions in the range 30–33 (Fig. 1A). We found no statistical difference in the allele distribution among ALS cases and controls (range 2–30 alleles; p = 0.17). The repeat allele distribution among the SA population controls (n = 114 alleles; range between 2 and 25, Fig. 1A) were similar to an independent SA sample which assessed Huntington disease phenocopies (n = 178 alleles; range between 6 and 25 (Baine et al., 2018)). Although the most frequent allele of 22 repeats was similar between African and European controls (62% vs 85%, respectively), we observed more very short repeats (<22) in Africans compared to the Europeans (5% vs 1%, respectively), whereas intermediate alleles (RE 27–33) were very rare in Africans (Africans <1%; European controls 2%). However, these results were not statistically significant (Fig. 1B).
Fig. 1.
ATXN2 repeat allele frequencies in A. a Southern African (SA) cohort with ALS vs matched SA ancestry controls, and B. controls from across Africa (AFR) compared to Europe (EUR). % refers to the proportion of alleles in the groups. NS refers to Fisher exact test which was not significant.
3.2. NIPA1
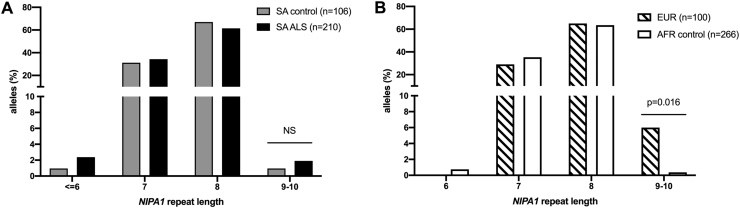
Exon 1 of the Non-imprinted in Prader-Willi/Angelman syndrome region protein 1. (NIPA1) encodes a GCG repeat which most frequently results in an allele length of 7 or 8 in healthy controls. In South Africans, the repeat allele distribution between ALS cases vs controls were similar (p = 0.67) (Fig. 2A). Four cases and a control each carried a long allele (≥9 GCG repeats). Although the NIPA1 allele size range is narrow among controls, the distribution of longer alleles with 9–10 GCG repeats, was extremely rare in Africans (0.4%) whereas 6% of European controls had alleles of 9–10 GCG repeats (p = 0.016) (Fig. 2B).
Fig. 2.
NIPA1 repeat allele frequencies in A. a Southern African (SA) cohort with ALS vs matched Southern African ancestry controls, and B. controls from across Africa (AFR) compared to Europe (EUR). % refers to the proportion of alleles in the groups. Fisher exact tests were performed; NS refers to not significant.
3.3. ATXN 1
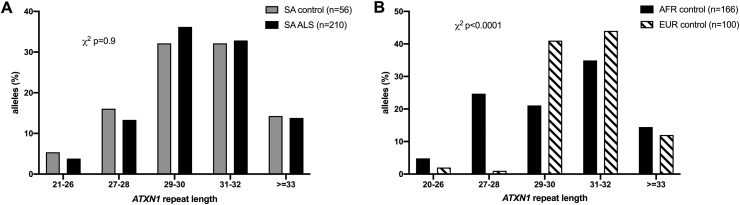
The pathogenic range of the CAG expansion in Ataxin-1 (ATXN1), which results in SCA1, is ≥39 repeats, whereas the intermediate range REs ≥ 33 was reported to associate modestly with European ALS patients (Lattante et al., 2018, Conforti et al., 2012, Tazelaar et al., 2020). In this SA cohort, the CAG repeats in ALS cases ranged from 21 to 37, compared to 24–35 in controls; the allele distribution was not different between the two groups (p = 0.99), including a similar proportion of intermediate ATXN1 alleles (REs 33–35) amongst SA ALS and SA controls (both ~14%) (Fig. 3A). The overall distribution of RE alleles was significantly different between African and European controls (corrected p < 0.001); this difference appears to result from a higher frequency of short alleles < 29 amongst Africans (30% vs 3% in Europeans) while the frequency of intermediate RE ≥ 33 alleles was similar in both groups.
Fig. 3.
ATXN 1 repeat allele frequencies in A. a Southern African (SA) cohort with ALS vs SA ancestry controls, and B. controls from across Africa (AFR) compared to Europe (EUR). % refers to the proportion of alleles in the groups. X2 refers to uncorrected Chi-test.
3.4. NOP56
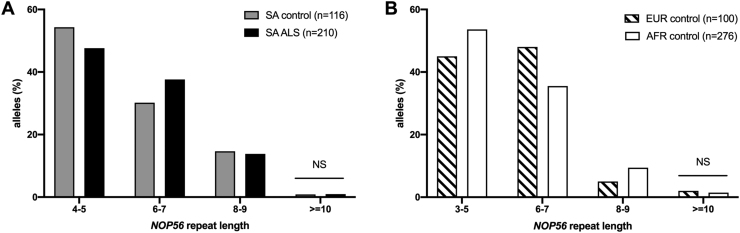
An abnormal hexanucleotide repeat expansion (>650 repeats) located in intron 1 of nucleolar protein 56 (NOP56), results in SCA36 (García-Murias et al., 2012, Kobayashi et al., 2011). The interest in ALS arose from observing individuals with SCA36 developing prominent features of motor neuron degeneration in up to two-thirds of cases (Garcia-Murias et al. 2012). In the SA sample the NOP56 repeat allele size ranged between 4 and 11 repeats, with a similar distribution in ALS and controls (p = 0.99) (Fig. 4A). Similarly, there was no difference found between the distribution of NOP56 RE alleles (≥10) comparing African with European controls (p = 0.66) (Fig. 4B).
Fig. 4.
NOP56 repeat allele frequencies in A. a Southern African (SA) cohort with ALS vs SA ancestry controls, and B. controls from across Africa (AFR) compared to Europe (EUR). % refers to the proportion of alleles in the groups. Fisher exact tests were performed; NS refers to not significant.
4. Discussion
Expanded motifs in ATXN2, NIPA1, ATXN1 and NOP56 genes in South Africans with African genetic ancestry, did not show associations with ALS. However, the associations of intermediate range repeat expansions in ATXN2, NIPA1 and ATXN1 were modest in Europeans with ALS, and this sample may have been too small to be informative of low frequency association signals. We also evaluated the genetic architecture of these loci in a modest sample of population controls and found a non-significant tendency for Africans to have more shorter repeat expansion alleles compared to their European counterparts.
4.1. ATXN2
Unlike Africans with ALS, in whom we found no association with intermediate repeat expansions (CAG 27–33), in Europeans this occurred twice as often in ALS patients compared to controls (Elden et al., 2010, Wang et al., 2014). However, the ATXN2 expansion architecture is complex because, despite African controls having relatively more shorter ATXN2 repeats compared to Europeans [this study, (Baine et al., 2018)], SCA2, which is due to a larger ATXN2 RE (>36 repeats), is more frequent among South Africans with African genetic ancestry compared to those with European ancestry (Smith et al., 2012). Similarly, Cuba also has a high frequency of SCA2, but without evidence of increased frequencies of intermediate alleles as pre-mutations (Laffita-Mesa et al., 2012) or an increased frequency of ALS (Ryan et al., 2019). Our results, and those of others, suggest that the association of ATXN2 intermediate RE amongst non-European populations is either weak (Chinese individuals with ALS) (Chen et al., 2011) or absent (Africans with ALS). Therefore, while there are consistent association signals of ATXN2 intermediate RE in Europeans with ALS (Wang et al., 2014), emerging indications of a role in ALS pathogenesis (Yang et al., 2019, Elden et al., 2010, Lagier-Tourenne and Cleveland, 2010) and the increasing focus on its therapeutic potential in ALS (van den Heuvel et al., 2014), our results suggest that ATXN2 may be less important in ALS populations without European genetic ancestry.
4.2. NIPA1
In this study we found no difference between long alleles (≥9 GCG repeats) among South African ALS and controls; this contrasts with previous reports in northern Europeans in which the long alleles were >1.5x more likely in cases with ALS (Blauw et al., 2012, Tazelaar et al., 2019). However, our results are similar to a recent multicohort analysis from ProjectMinE, which included substantial numbers of cases with ALS (and controls) from the United Kingdom and the United States, and which found similar frequencies of NIPA1 long alleles between ALS cases and controls (4.7 vs 4.4%) (Tazelaar et al., 2019, van Blitterswijk et al., 2014).
4.3. ATXN1
A modest association between ATXN1 intermediate REs 33–38 and ALS in Europeans has been reported (Tazelaar et al., 2020), but this was not replicated in this African sample. Although the allele frequencies amongst African and European controls differed overall, there were no significant differences in the frequencies of the intermediate alleles (14% vs 13%, respectively).
4.4. NOP56
We found no association between NOP56 allele frequencies and ALS cases in this SA cohort, which is similar to the results in Chinese and Japanese cohorts (Lee et al., 2016, Kobayashi et al., 2011). The normal range of allele sizes amongst African controls ranged between 3 and 10 repeats whereas Europeans and Chinese controls carried between 4 and 14 (this study; García-Murias et al., 2012, Lee et al., 2016).
Within the last decade, a large repeat expansion in C9orf72 was found to be the most common genetic mutation in both sporadic (7%) and familial (38%) ALS cases with European genetic ancestry (Rademakers and van Blitterswijk, 2013). A founder C9orf72 expansion mutation was suggested to have arisen in Northern Europe (Rademakers and van Blitterswijk, 2013). Populations with 17th century European admixture, which may include South Africans with mixed-African ancestry and Chinese individuals from Taiwan, and who have ALS, appear to have higher frequencies of the C9orf72 expansion mutation than ALS populations without European admixture, such as Asians and black Africans (Liu et al., 2018, Nel et al., 2019a). The modest association signals of co-inheritance of ATXN2 intermediate alleles with large C9orf72 expansions in Europeans with ALS, have suggested that ATXN2 could be acting as a disease modulator (van Blitterswijk et al., 2014). This postulate is supported by experimental evidence that expanded repeats within ATXN2 (Elden et al., 2010, Yang et al., 2019) and ATXN1 (Tazelaar et al., 2020) may impact TDP-43 biology. The aforementioned pathogenic pathways may be important in Europeans with ALS, but not in Africans, and support the different oligogenic multi-step hypothesis of ALS in East Asians (and possibly Africans) vs. Europeans (Vucic et al., 2020).
Herein we determined genotypes using computational tools, and these results have not been validated by repeat-primed PCR (RP-PCR). However, benchmarking of ExpansionHunter results against RP-PCR results for the C9orf72 hexanucleotide repeat locus showed that the algorithm is highly specific for detecting pathogenic expansions (Dolzhenko et al., 2017). In our previous report we found 100% concordance between pathogenic C9orf72 expansions detected by ExpansionHunter from sequences generated by PCR-free methods, and RP-PCR (Nel et al., 2019a). The relatively small size of this African ALS cohort compared with the larger European and Chinese cohorts, has to be considered a limitation. However, South Africans have substantial ancient KhoiSan genetic contributions adding to their rich genetic diversity and short linkage disequilibrium blocks (Choudhury et al., 2017); these differences in genetic architecture enhances the informative value of including Africans in the search for pathogenic ALS pathways.
5. Conclusion
This report sheds light on population-specific genetic differences in individuals with ALS. Intermediate length repeat expansions in ATXN2, NIPA1 and ATXN1, which showed associations with ALS in Europeans, were not replicated in a small sample of Southern Africans with ALS. Our results underscore the importance of including individuals with different genetic ancestries to comprehensively dissect the pathogenetic mechanisms underlying ALS and to uncover the missing heritability contributing to large proportions of cases with ALS.
Ethics comment
We have read and abided by all the ethical standards for manuscripts submitted to IBRO reports.
CRediT authorship contribution statement
Melissa Nel: Data analysis, Methodology, Conceptualization. Thandeka Mavundla: Data analysis. Kayleigh Gultig: Data analysis. Jeannine M. Heckmann: Methodology, Conceptualization, Project administration, Funding acquisition. Gang Wu: Project administration, Funding acquisition. Joanne Wuu: Project administration, Funding acquisition. Anne Cooley: Project administration, Funding acquisition. Jason Myers: Project adminsitration. Evadnie Rampersaud: Project adminstration. Michael Benatar: Project administration, Funding acquisition. Gerrit Botha: Project administration, Funding acquisition. Nicola Mulder: Project administration, Funding acquisition.
Conflicts of interest
None.
Acknowledgements
We thank Egor Dolzhenko, Illumina Incorporated, San Diego, California USA, for his advice. We thank the CReATe consortium and Paul Taylor’s laboratory who sequenced 34 genomes at St Jude’s funded by Amoytrophic Lateral Sclerosis Association (ALSA), and St Jude American Lebanese Syrian Associated Charities (ALSAC), and 25 genomes sequenced by the National Human Genome Research Institute (U24HG006941). The Clinical Research in ALS and related disorders for Therapeutic Develoment (CReATe), Consortium (U54NS092091) is part of Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advancing Translational Sciences (NCATS). This consortium is funded through collaboration between NCATS, and the National Institute of Neurological Disorders and Stroke (NINDS). We also thank the Southern African Human Genome Programme (SAHGP) participants. The SAHGP WGS dataset was generated by the national SAHGP initiative funded by the Department of Science and Technology of South Africa. MN is funded by a UCT Neuroscience Institute postdoctoral fellowship and a CReATe scholars award, TM is funded by the South African National Research Foundation grant.
References
- Baine F.K., Peerbhai N., Krause A. A study of Huntington disease-like syndromes in black South African patients reveals a single SCA2 mutation and a unique distribution of normal alleles across five repeat loci. J. Neurol. Sci. 2018;390:200–204. doi: 10.1016/j.jns.2018.04.031. [DOI] [PubMed] [Google Scholar]
- Blauw H.M., van Rheenen W., Koppers M., Van Damme P., Waibel S., Lemmens R., van Vught P.W.J., Meyer T., Schulte C., Gasser T., Cuppen E., Pasterkamp R.J., Robberecht W., Ludolph A.C., Veldink J.H., van den Berg L.H. NIPA1 polyalanine repeat expansions are associated with amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21:2497–2502. doi: 10.1093/hmg/dds064. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M., Mullen B., Heckman M.G., Baker M.C., DeJesus-Hernandez M., Brown P.H., Murray M.E., Hsiung G.Y. Ataxin-2 as potential disease modifier in C9ORF72 expansion carriers. Neurobiol. Aging. 2014;35:2421.e13–2421.e17. doi: 10.1016/j.neurobiolaging.2014.04.016. e13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B.R. Brookes, R.G. Miller, M. Swash, T.L. Munsat, World Federation of Neurology Research Group on Motor Neuron Diseases., 2000. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. ALS and other motor neuron disorders 1, 293-299. [DOI] [PubMed]
- Chen Y., Huang R., Yang Y., Chen K., Song W., Pan P., Li J., Shang H.F. Ataxin-2 intermediate-length polyglutamine: a possible risk factor for Chinese patients with amyotrophic lateral sclerosis. Neurobiol. Aging. 2011;32(1925):e1–e5. doi: 10.1016/j.neurobiolaging.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Choudhury A., Ramsay M., Hazelhurst S., Aron S., Bardien S., Botha G., Chimusa E.R., Christoffels A. Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat. Commun. 2017;8:2062. doi: 10.1038/s41467-017-00663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti F.L., Spataro R., Sproviero W., Mazzei R., Cavalcanti F., Condino F., Simone I.L., Logroscino G., Patitucci A., Magariello A., Muglia M., Rodolico C., Valentino P., Bono F., Colletti T., Monsurro M.R., Gambardella A., La Bella V. Ataxin-1 and ataxin-2 intermediate-length PolyQ expansions in amyotrophic lateral sclerosis. Neurology. 2012;79:2315–2320. doi: 10.1212/WNL.0b013e318278b618. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G.Y.R., Karydas A., Seeley W.W., Josephs K.A., Coppola G., Geschwind D.H., Wszolek Z.K., Feldman H., Knopman D.S., Petersen R.C., Miller B.L., Dickson D.W., Boylan K.B., Graff-Radford N.R., Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E., van Vugt J.J.F.A., Shaw R.J., Bekritsky M.A., van Blitterswijk M., Narzisi G., Ajay S.S., Rajan V., Lajoie B.R., Johnson N.H., Kingsbury Z., Humphray S.J., Schellevis R.D., Brands W.J., Baker M., Rademakers R., Kooyman M., Tazelaar G.H.P., van Es M.A., McLaughlin R., Sproviero W., Shatunov A., Jones A., Al Khleifat A., Pittman A., Morgan S., Hardiman O., Al-Chalabi A., Shaw C., Smith B., Neo E.J., Morrison K., Shaw P.J., Reeves C., Winterkorn L., Wexler N.S., Housman D.E., Ng C.W., Li A.L., Taft R.J., van den Berg L.H., Bentley D.R., Veldink J.H., Eberle M.A. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., Padmanabhan A., Clay-Falcone D., McCluskey L., Elman L., Juhr D., Gruber P.J., Rüb U., Auburger G., Trojanowski J.Q., Lee V.M.Y., Van Deerlin V.M., Bonini N.M., Gitler A.D. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Murias M., Quintáns B., Arias M., Seixas A.I., Cacheiro P., Tarrío R., Pardo J., Millán M.J., Arias-Rivas S., Blanco-Arias P., Dapena D., Moreira R., Rodríguez-Trelles F., Sequeiros J., Carracedo Á., Silveira I., Sobrido M.J. ‘Costa da Morte’ ataxia is spinocerebellar ataxia 36: clinical and genetic characterization. Brain. 2012;135:1423–1435. doi: 10.1093/brain/aws069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project, Consortium, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning F., Heckmann J.M., Naidu K., Vlok L., Cross H.M., Marin B. Incidence of motor neuron disease/amyotrophic lateral sclerosis in South Africa: a 4‐year prospective study. Eur. J. Neurol. 2020;28:81–89. doi: 10.1111/ene.14499. [DOI] [PubMed] [Google Scholar]
- van den Heuvel D.M., Harschnitz O., van den Berg L.H., Pasterkamp R.J. Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis? Trends Mol. Med. 2014;20:25–35. doi: 10.1016/j.molmed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Abe K., Matsuura T., Ikeda Y., Hitomi T., Akechi Y., Habu T., Liu W., Okuda H., Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffita-Mesa J.M., Velázquez-Pérez L.C., Santos Falcón N., Cruz-Mariño T., González Zaldívar Y., Vázquez Mojena Y., Almaguer-Gotay D., Almaguer Mederos L.E., Rodríguez Labrada R. Unexpanded and intermediate CAG polymorphisms at the SCA2 locus (ATXN2) in the Cuban population: evidence about the origin of expanded SCA2 alleles. Eur. J. Hum. Genet. 2012;20:41–49. doi: 10.1038/ejhg.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Cleveland D.W. Neurodegeneration: an expansion in ALS genetics. Nature. 2010;466:1052–1053. doi: 10.1038/4661052a. [DOI] [PubMed] [Google Scholar]
- Lattante S., Pomponi M.G., Conte A., Marangi G., Bisogni G., Patanella A.K., Meleo E., Lunetta C., Riva N., Mosca L., Carrera P., Bee M., Zollino M., Sabatelli M. ATXN1 intermediate-length polyglutamine expansions are associated with amyotrophic lateral sclerosis. Neurobiol. Aging. 2018;64:157 e1–57 e5. doi: 10.1016/j.neurobiolaging.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Tsai P.C., Guo Y.C., Hsiao C.T., Liu G.T., Liao Y.C., Soong B.W. Spinocerebellar ataxia type 36 in the Han Chinese. Neurol. Genet. 2016;2 doi: 10.1212/NXG.0000000000000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., He J., Gao F.B., Gitler A.D., Fan D. The epidemiology and genetics of amyotrophic lateral sclerosis in China. Brain Res. 2018;1693:121–126. doi: 10.1016/j.brainres.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538(7624):201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel M., Agenbag G.M., Henning F., Cross H.M., Esterhuizen A., Heckmann J.M. C9orf72 repeat expansions in South Africans with amyotrophic lateral sclerosis. J. Neurol. Sci. 2019;401:51–54. doi: 10.1016/j.jns.2019.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel M., Mulder N., Europa T.A., Heckmann J.M. Using whole genome sequencing in an African subphenotype of myasthenia gravis to generate a pathogenetic hypothesis. Front. Genet. 2019;10:136. doi: 10.3389/fgene.2019.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R., van Blitterswijk M. Motor neuron disease in 2012: novel causal genes and disease modifiers. Nat. Rev. Neurol. 2013;9:63–64. doi: 10.1038/nrneurol.2012.276. [DOI] [PubMed] [Google Scholar]
- Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H. A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M., Zaldívar Vaillant T., McLaughlin R.L., Doherty M.A., Rooney J., Heverin M., Gutierrez J., Lara-Fernández G.E., Pita Rodríguez M., Hackembruch J., Perna A., Vazquez M.C., Musio M., Ketzoian C.N., Logroscino G., Hardiman O. Comparison of the clinical and genetic features of amyotrophic lateral sclerosis across Cuban, Uruguayan and Irish clinic-based populations. J. Neurol. Neurosurg. Psychiatry. 2019;90:659–665. doi: 10.1136/jnnp-2018-319838. [DOI] [PubMed] [Google Scholar]
- Smith D.C., Bryer A., Watson L.M., Greenberg L.J. Inherited polyglutamine spinocerebellar ataxias in South Africa. S. Afr. Med. J. 2012;102:683–686. doi: 10.7196/samj.5521. [DOI] [PubMed] [Google Scholar]
- Taylor J.P., Brown R.H., Jr., Cleveland D.W. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazelaar G.H.P., Boeynaems S., De Decker M., Van Vugt J.J.F.A., Kool L., Goedee H.S., McLaughlin R.L., Sproviero W. ATXN1 repeat expansions confer risk for amyotrophic lateral sclerosis and contribute to TDP-43 mislocalization. Brain Communications. 2020;2 doi: 10.1093/braincomms/fcaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazelaar G.H.P., Dekker A.M., van Vugt J.J.F.A., van der Spek R.A., Westeneng H.J., Kool L.J.B.G., Kenna K., van Rheenen P.W. Association of NIPA1 repeat expansions with amyotrophic lateral sclerosis in a large international cohort. Neurobiol. Aging. 2019;74:234.e9–234.e15. doi: 10.1016/j.neurobiolaging.2018.09.012. 234 e9-34 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic S., Higashihara M., Sobue G., Atsuta N., Doi Y., Kuwabara S., Kim S.H., Kim I., Oh K.W., Park J., Kim E.M., Talman P., Menon P., Kiernan M.C. ALS is a multistep process in South Korean, Japanese, and Australian patients. Neurology. 2020;94:e1657–e1663. doi: 10.1212/WNL.0000000000009015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.D., Gomes J., Cashman N.R., Little J., Krewski D. Intermediate CAG repeat expansion in the ATXN2 gene is a unique genetic risk factor for ALS–A systematic review and meta-analysis of observational studies. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Delport W., Chimusa E.R., Meintjes A., Moller M., van Helden P., Seoighe C., Hoal E.G. Genome-wide analysis of the Southern African Coloured population in the Western Cape. Hum. Genet. 2010;128:145–153. doi: 10.1007/s00439-010-0836-1. [DOI] [PubMed] [Google Scholar]
- Yang Y., Halliday G.M., Kiernan M.C., Tan R.H. TDP-43 levels in the brain tissue of ALS cases with and without C9ORF72 or ATXN2 gene expansions. Neurology. 2019;93:e1748–e1755. doi: 10.1212/WNL.0000000000008439. [DOI] [PubMed] [Google Scholar]