Abstract
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has led to a severe pandemic and deeply affected the livelihood of people worldwide. In response to the pandemic, researchers have been rapidly studying different aspects of COVID‐19, such as virus detection, vaccinations, and epidemiological aspects of the disease. It has been reported that SARS‐CoV‐2 can induce uncontrolled inflammation and cause a lack of antiviral response, thereby aggravating the disease. Therefore, recovery of immune functions is key to COVID‐19 treatment. Many clinical trials are exploring suitable therapies, and some progress has been made. Early administration of interferons may prevent COVID‐19 exacerbation and/or promotes recovery from the diseases. Inhibitors of inflammation can prevent cytokine storms and multi‐organ damage. Convalescent plasma containing neutralizing antibodies has played an important role in therapeutic options at the beginning of the pandemic owing to the lack of other effective methods. To aid the development of treatment options for COVID‐19, this review focuses on immunotherapies, including treatment with interferons, inhibition of pro‐inflammatory mechanisms, and the use of convalescent plasma.
Keywords: convalescent plasma, coronavirus disease 2019, COVID‐19, cytokine storm, immunotherapy, interferon, severe acute respiratory syndrome coronavirus 2
Considerable progress has been made in the treatment of coronavirus disease 2019 (COVID‐19). Some vaccines have already been approved and are available on the market. Apart from vaccines, immunotherapy that induces a low or hyperimmune state in the body is also an effective method to combat viral infections. This review focuses on immunotherapy approaches to combat COVID‐19.
1. Introduction
During the last two decades, two large‐scale pandemics were related to coronaviruses, namely severe acute respiratory syndrome (SARS, 2002) and Middle East respiratory syndrome (MERS, 2012).[ 1 , 2 ] A consensus was reached that severe acute respiratory syndrome related to coronaviruses (SARSr‐CoV), most of which use bats as their natural hosts, may result in future disease outbreaks.[ 1 , 2 , 3 ] Similar to SARS‐CoV and MERS‐CoV, SARS‐CoV‐2 caused severe and fatal diseases after breaking out in 2019. Genome sequencing and alignment of the results revealed that SARS‐CoV‐2 nucleic acid sequence is 96% identical with the bat coronavirus RaTG13, 79% identical with SARS‐CoV, and 50% identical with MERS‐CoV.[ 1 , 4 ] It can be inferred that SARS‐CoV‐2 is a coronavirus of probable bat origin. Furthermore, compared to SARS‐CoV and MERS‐CoV, SARS‐CoV‐2 causes a more pathogenic disorder, coronavirus disease 2019 (COVID‐19).[ 5 ] Globally, as of 20 April 2021, there have been 141 754 944 confirmed cases of COVID‐19, including 30 25 835 deaths, according to the COVID‐19 Situation Dashboard of the World Health Organization (WHO).[ 6 ]
The common clinical manifestations of COVID‐19 are cough, shortness of breath, fever, and various other symptoms such as headache, hemoptysis, and diarrhea.[ 7 ] Lung infection is apparent, and ground‐glass‐like opacity areas can be found bilaterally in the lungs in computed tomography images of affected patients.[ 8 ] Moreover, low expression of type I/III interferons (IFNs) and high levels of pro‐inflammatory cytokines and chemokines (such as interleukin [IL]‐6) are observed, which may cause cytokine storms and uncontrolled inflammation. Excessive inflammation may worsen the course of the disease.[ 9 , 10 ] In severe cases, acute respiratory distress syndrome (ARDS), systemic inflammatory response syndrome, cardiac dysfunction, arrhythmias, viral sepsis, hypercoagulability, thrombotic complications, and multiple organ failure can be observed.[ 11 , 12 , 13 ]
According to the National Institutes of Health COVID‐19 treatment guidelines, standard treatment includes maintaining the hemodynamics of patients in‐balance and providing oxygenation and ventilation treatment, such as the employment of high‐flow nasal cannula and extracorporeal membrane oxygenation.[ 14 ] Except for these guidelines, no standard drug therapy is prescribed due to the uncertainty and safety of existing drugs. Vaccination is now the most effective way to prevent SARS‐CoV‐2 infection. Currently, researchers are mainly focusing on the development of vaccines against SARS‐CoV‐2 and its variants. Several vaccines have been and/or are in development, such as inactivated or live attenuated vaccines, subunit vaccines, vectored vaccines, and nucleic acid vaccines.[ 15 ] Inactivated vaccines, vector vaccines, and RNA‐based vaccines have already been approved and are available on the market.[ 16 ] Apart from vaccines, immunotherapy treatments that induce a low or hyperimmune state in the body are also an effective method to combat viral infections. Immunotherapy has been successfully used against SARS‐CoV and MERS‐CoV; thus, it is suggested that it can influence SARS‐CoV‐2 as well.[ 17 ] In this review, we mainly focus on IFN therapy, the inhibition of inflammatory responses, and convalescent plasma therapy against COVID‐19.
2. SARS‐CoV‐2 and SARS‐CoV‐2 Vaccines
2.1. Structure of SARS‐CoV‐2
SARS‐CoV‐2 belongs to the coronavirus family, the Betacoronavirus genus; it is a positive‐sense, single‐stranded enveloped RNA virus.[ 18 , 19 ] The coronavirus genome encodes several structural and nonstructural proteins (NSPs).[ 19 ] The NSPs promote viral transcription and replication by countering the host antiviral response and immune regulation.[ 20 , 21 ] Host infection, membrane fusion, virus assembly, morphogenesis, and viral particle release all rely on structural proteins.[ 19 ] Nucleocapsid (N), membrane (M), envelope (E), and spike (S) glycoproteins constitute structural proteins that are related to the viral envelope.[ 22 , 23 ] The N protein binds to the viral RNA and mediates viral replication, whereas the M, E, and S proteins mainly participate in envelope formation. The S protein protrudes from the viral envelope and plays a significant role in mediating the entry of the virus into the host cell.[ 23 , 24 ] The invasion of SARS‐CoV‐2 depends on the S protein binding to cellular receptors and its activation by host cell proteases, which are responsible for target cell adsorption and membrane fusion.[ 25 ]
2.2. SARS‐CoV‐2 Vaccines Phase 4 Clinical Trials
Similar to SARS‐CoV, SARS‐CoV‐2 uses the angiotensin‐converting enzyme II (ACE2) receptor to enter host cells.[ 1 ] Therefore, therapies effective against SARS‐CoV might also be effective against SARS‐CoV‐2. According to previous experience in developing SARS‐CoV vaccines, the S protein is a satisfactory vaccine antigen candidate owing to its high immunogenicity and its capability to induce neutralizing antibodies (NAbs).[ 26 ] As of March 2021, inactivated vaccines, vector vaccines, and RNA‐based SARS‐CoV‐2 vaccines containing S proteins have entered phase 4 clinical trials which means these vaccines are being administered worldwide at this time, beyond clinical trials and while trials continue (Figure 1 , Table 1 ).[ 16 ]
Figure 1.
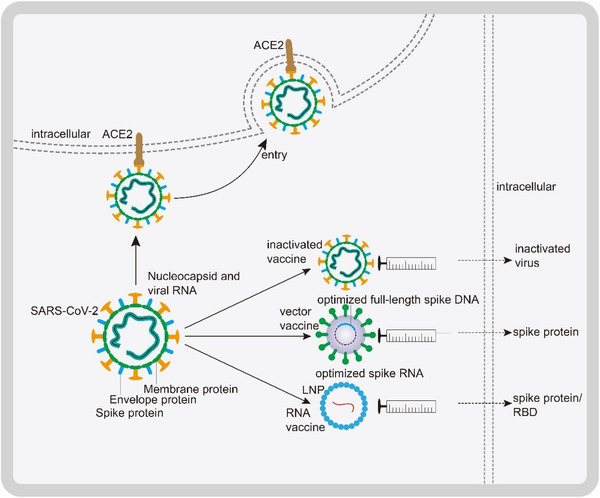
Mechanism of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) invasion and principles of representative vaccines. Nucleocapsid (N), spike (S), membrane (M), and envelope (E) glycoproteins constitute structural proteins of SARS‐CoV‐2 and are related to the envelope. The S protein binds angiotensin‐converting enzyme II (ACE2) to promote viral entry. Inactivated vaccines: SARS‐CoV‐2 is raised in Vero cells, inactivated through physical or chemical methods, and administered as a vaccine. Vectored vaccines: The full‐length S gene is cloned into a replication‐deficient adenovirus vector. After intramuscular injection of the vectored vaccine, the S protein is produced. RNA vaccines: Lipid nanoparticles (LNPs) that contain mRNA constitute RNA vaccines. Depending on the mRNA used, translation generates the S protein or the receptor‐binding domain (RBD) of the S protein in the recipient cell. Ultimately, the S protein or its RBD induce the production of NAbs.
Table 1.
Development of vaccine candidates at the clinical stage in phase 4 trials
| Vaccine type | Vaccine | Developer | No. of doses | Timing of doses [days] | Results of clinical trials | Ref. |
|---|---|---|---|---|---|---|
| Inactivated vaccines | Inactivated | Sinovac Research and Development Co., Ltd. | 2 | 0 and 14 | The phase 3 trial showed that two doses of 3 or 6 µg/0.5 mL of the vaccine were safe and well‐tolerated in adults aged 60 years and older. The seroconversion rates in those who received 3 or 6 µg doses were >95% after a two‐dose vaccination. | [ 27 ] |
| Inactivated | Sinopharm/China National Biotec Group Co./Beijing Institute of Biological Products | 2 | 0 and 21 | Humoral responses against SARS‐CoV‐2a) were induced in all vaccine recipients on day 42. | ||
| Non‐replicating vector vaccines | ChAdOx1 nCoV‐19b) | AstraZeneca/University of Oxford | ≈1–2 | 0 or 0 and 28 | The pooled analysis of four randomized trials showed that the vaccine efficacy after a single standard dose of the vaccine from day 22 to day 90 after vaccination was 76%. | [ 28 ] |
| RNA vaccines | mRNA‐1273 | Moderna/National Institute of Allergy and Infectious Diseases (NIAID) | 2 | 0 and 28 | The phase 3 trial showed that mRNA‐1273 had 94.1% efficacy for preventing COVID‐19 illness, including severe disease. | [ 29 ] |
| BNT162 | Pfizer/BioNTech/Fosun Pharma | 2 | 0 and 21 | The phase 3 trial showed that a two‐dose regimen of BNT162b2 elicited 95% protection against COVID‐19 in persons 16 years of age or older. | [ 30 ] |
a)SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; b)nCoV‐19 or COVID‐19, coronavirus disease 2019.
The use of inactivated viruses is the traditional method of vaccine development and has been proven to be safe and effective.[ 31 ] The advantages of inactivated vaccines are the ease of production and stable expression of antigenic epitopes; however, production requires biosafety level 3 growth of the pathogen.[ 15 , 32 ] Phase 3 trials of the inactivated vaccine developed by Sinovac showed that treatment was safe in healthy adults aged 60 years and older; seroconversion was observed in 96 of 98 (98%) participants in the 3 µg treatment group (Table 1 ).[ 27 ] Phase 1/2 trials of vaccines developed by Sinopharm showed that they were safe in adults aged 18–80 years old. Humoral responses against SARS‐CoV‐2 were induced in all vaccine recipients on day 42. Two dose treatments with 4 µg on days 0 and 21, or days 0 and 28, had better results compared with the single 8 µg dose or 4 µg dose on days 0 and 14.[ 33 ]
Vector vaccines contain a carrier virus construct, for example, an adenovirus, that carries genes of the target virus, such as the optimized full‐length S gene in the case of SARS‐CoV‐2. These vaccines express proteins in the host cells that can induce NAbs after vaccination. The advantages of vector vaccines are that they infect with antigen presenting cells directly and are genetically and physically stable, but prior immunity to the vector may be induced.[ 15 , 32 ] The pooled analysis of four randomized trials showed a 76% vaccine efficacy after a single standard dose of the Oxford SARS‐CoV‐2 vector vaccine from day 22 to day 90 post‐vaccination.[ 28 ]
RNA‐based vaccines are a relatively recent development whereby antigen information is delivered through LNPs that contain modified mRNA that produces the antigen by translation in the recipient cells.[ 34 ] An advantage of RNA vaccines is that they can be developed rapidly; however, to maintain stability, they should be kept in frozen storage which often requires high‐quality preservation technology.[ 15 , 32 ] The phase 3 trial showed a 94% efficacy for the Moderna mRNA‐1273 vaccine,[ 29 ] and the phase 3 trial of the Pfizer BNT162b2 vaccine showed that a two‐dose regimen was 95% effective in preventing COVID‐19.[ 30 ]
3. Role of IFNs in COVID‐19
3.1. IFN Signal Transduction Following RNA Virus Infection
Innate immunity is activated following the identification of pathogen‐associated molecular patterns by pattern recognition receptors; this is the first line of defense against a virus.[ 35 ] After binding to the surface of the host cell, the uncoated viral particle crosses through the cytomembrane and replicates as a double‐stranded RNA (dsRNA) intermediate. Subsequently, the viral particle is recognized by RNA pattern recognition receptors such as melanoma differentiation‐associated gene 5 (MDA‐5) in the cytoplasm and retinoic‐acid‐inducible gene I (RIG‐I).[ 36 , 37 ] After detecting the dsRNA, the antiviral signaling cascade is initiated through the mitochondrial antiviral signaling (MAVS) protein, which recruits tumor necrosis factor (TNF) receptor‐associated factors (TRAFs) and activates TANK binding kinase 1 (TBK1) and IκB kinase ε (IKKε). The TBK1/IKKε complex then activates, through phosphorylation, the IFN regulatory transcription factor (IRF)3 to induce the transcription of genes that encode IFNs.[ 38 ]
Type I and type III IFNs function via paracrine or autocrine mechanisms and interact with their respective receptors, type I IFN receptor (IFNAR) and type III IFN receptor (IFNLR), respectively. The receptor‐associated complex Janus‐activated kinase (JAK)1/tyrosine kinase (TYK)2 is activated, leading to the phosphorylation of signal transducer and activator of transcription (STAT)1 and STAT2.[ 37 ] JAK2 participates in type III IFN‐induced STAT phosphorylation.[ 39 ] Phosphorylated STAT1/STAT2 heterodimers combine with IRF9 to form an IFN‐stimulated gene factor 3 (ISGF3) complex. This complex is transferred into the nucleus and binds to IFN‐stimulated response elements (ISREs), which induce the expression of IFN‐stimulated genes (ISGs) to prompt an antiviral state.[ 40 ] Type I and III IFNs can induce similar ISG signatures and activate an antiviral state in infected and neighboring cells.[ 37 , 41 ]
3.2. SARS‐CoV‐2 Inhibits Innate Immune Responses
Recent research has demonstrated that IFN induced by SARS‐CoV‐2 may lead to inflammation when it is not produced in time.[ 42 ] In addition, SARS‐CoV‐2 antagonizes the antiviral response by inhibiting signal transduction and production of IFNs.[ 21 , 42 , 43 ]
Different proteins antagonize the production of IFNs through distinct mechanisms. The nsp6 can bind to TBK1 and inhibit its transduction. Similarly, nsp13 binds TBK1 and inhibits its phosphorylation. Open reading frame 6 (ORF6) binds karyopherin subunit α 2 (KPNA2), inhibiting IRF3 nuclear translocation,[ 21 ] and nsp1 can induce the degradation of mRNA to affect the production of IFNs.[ 21 , 42 ] In addition, nsp12, nsp14, ORF3, M, and N proteins have been shown to suppress IFN production, although the mechanism remains unclear.[ 42 , 44 ] Furthermore, signaling cascades downstream of IFN are inhibited. The phosphorylation of STAT1 is affected by nsp1, nsp6, nsp13, ORF3a, M, and ORF7b, whereas the phosphorylation of STAT2 is suppressed by nsp6, nsp13, ORF7a, and ORF7b. ORF6 associates with KPNA2 to prevent ISGF3 nuclear translocation.[ 21 ] ORF6 can also associate with Nup98‐Rae1 through its C‐terminal domain at the nuclear pore complex and directly impair ISGF3 nuclear translocation (Figure 2 ).[ 45 ]
Figure 2.
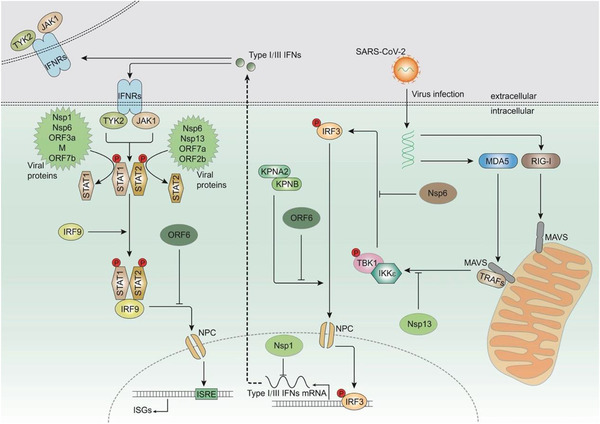
Production, transduction, and inhibition of type I/III interferons (IFNs). The intermediates (double‐stranded RNA, dsRNA) produced after infection with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) are recognized by melanoma differentiation‐associated gene 5 (MDA‐5) or retinoic‐acid‐inducible gene I (RIG‐I). Subsequently, MDA‐5 or RIG‐I is activated and interacts with mitochondrial antiviral signaling (MAVS) protein. MAVS recruits tumor necrosis factor (TNF) receptor‐associated factors (TRAFs) and activates TANK binding kinase 1 (TBK1) and IκB kinase ε (IKKε). They further activate IFN regulatory transcription factor 3 (IRF3) and trigger its nuclear entry to induce the expression of type I/III IFNs. Type I and type III IFNs function as autocrine or paracrine signaling molecules by binding to IFN receptors, which can activate tyrosine kinase 2 (TYK2) and Janus‐activated kinase 1 (JAK1). Activated TYK2 and JAK1 phosphorylate signal transducer and activator of transcription (STAT)1 and STAT2. Phosphorylated STAT1/STAT2 heterodimers combine with IRF9 to form an IFN‐stimulated gene factor 3 (ISGF3) complex. This complex translocates into the nucleus and interacts with IFN‐sensitive response elements (ISREs) to promote the transcription of IFN‐stimulated genes (ISGs). The SARS‐CoV‐2 nonstructural proteins (NSPs) can inhibit the production and function of IFNs. Nsp6 and nsp13 bind to TBK1 and inhibit its transduction and phosphorylation, respectively. Open reading frame 6 (ORF6) inhibits IRF3 nuclear translocation, and nsp1 induces the degradation of mRNA and inhibits translation, thereby affecting the production of IFNs. Phosphorylation of STAT1 is inhibited by ORF3a, ORF7b, nsp1, nsp6, nsp13, and M, whereas the phosphorylation of STAT2 is inhibited by ORF7a, ORF7b, nsp6, and nsp13. ORF6 prevents nuclear translocation of ISGF3.
3.3. Lessons from Other Coronaviruses
The function of IFNs in two related, highly pathogenic coronaviruses, SARS‐CoV and MERS‐CoV, is controversial in terms of their protective effects on the host.[ 37 ] However, the experience gained from studying SARS‐CoV and MERS‐CoV is valuable for exploring potential IFN treatment strategies against SARS‐CoV‐2. Recombinant therapy with IFN‐α and ribavirin has shown to improve the survival of patients with severe MERS‐CoV infection.[ 46 ] However, patients diagnosed with multiple comorbidities at the terminal stage of their disease did not benefit from this antiviral therapy.[ 47 ] In experiments with MERS‐CoV, mice treated with recombinant IFN‐β before the peak of viral replication (1 day after infection) showed better viral control and protection against fatal infection, whereas mice treated after the peak of viral replication (2–4 days after infection) showed increased infiltration and activation of inflammatory cells in the lungs and enhanced lethality, with no obvious change in virus replication.[ 48 ] From these results, it can be inferred that early IFN treatment can effectively limit viral replication.
3.4. IFN Actions against SARS‐CoV‐2
Research has demonstrated that pretreatment with IFN‐β inhibits the transcription and replication of SARS‐CoV‐2,[ 42 ] and that SARS‐CoV‐2 is sensitive to IFN.[ 49 ] Therefore, recombinant IFNs can potentially be used for immunotherapy. In a retrospective study, 77 patients with COVID‐19 received either nebulized IFN‐α2b, arbidol, or a combination of both. IFN‐α2b therapy significantly accelerated viral clearance and reduced the levels of C‐reactive protein and IL‐6, which are inflammatory markers.[ 50 ] In another retrospective multicenter cohort study, the early use of IFN significantly reduced mortality. Nevertheless, it was observed that patients with moderate COVID‐19 did not benefit from treatment with IFNs and that slower recovery was associated with delayed administration of IFN.[ 51 ] In addition, an investigator‐initiated open‐label study showed that recombinant IFN‐α nasal drops protected susceptible people and prevented COVID‐19 incidence without adverse effects.[ 52 ] In a double‐blind, placebo‐controlled trial, IFN‐λ accelerated viral decline. By day 7, 80% of the participants in the IFN‐λ group had an undetectable viral load, indicating the potential of IFN‐λ to avoid further deterioration and promote virus removal.[ 53 ] All of these results support the protective function of early treatment with IFNs in COVID‐19 patients.
Paradoxically, it has also been shown that the SARS‐CoV‐2 receptor ACE2 is an ISG. SARS‐CoV‐2 can promote infection through IFN‐stimulated upregulation of ACE2 in human airway epithelial cells.[ 54 ] Some of the inconsistent results in these reports can be explained by the limited number of patients in retrospective studies, the combination of drugs, and importantly, the timing of administration of these drugs. Additional results from ongoing clinical studies will provide more informative answers regarding IFN as a COVID‐19 therapy.
The biological effects of IFNs need to be thoroughly examined to safely evaluate their clinical efficacy and implement sound treatment strategies.[ 37 ] Recombinant and pegylated IFN‐α and IFN‐β have been authorized for use in the treatment of multiple sclerosis and viral hepatitis, bringing hope for the development of IFN treatment against COVID‐19. Compared to type I IFNs, IFN‐λ has a higher or similar virological response rate and fewer adverse events.[ 55 ] In a study on influenza A virus, IFN‐α treatment restricted viral replication but increased pulmonary secretion of pro‐inflammatory cytokines, as well as epithelial cell death. By contrast, IFN‐λ treatment lowered the viral load and did not drive immunopathological responses.[ 56 ] Thus, IFN‐λ may be a more attractive therapeutic strategy for COVID‐19. Regrettably, recombinant IFN‐λs are currently only used in clinical trials and have not yet been approved for any indication.[ 57 ] To date, type I and type III IFNs are being evaluated in several clinical trials for their potential as therapies for COVID‐19.
4. The Role of Anti‐Inflammation
4.1. Inflammatory Changes Caused by SARS‐CoV‐2
Infection and replication of SARS‐CoV‐2 can lead to pyroptosis and vascular leakage, which can cause subsequent inflammatory responses.[ 9 ] At the same time, SARS‐CoV‐2 can infect alveolar macrophages and monocytes, cells that produce cytokines and chemokines.[ 58 , 59 ] Under the recruitment of chemokines, inflammatory cells are attracted to the site of inflammation from the blood vessels, thereby driving persistent alveolar inflammation.[ 60 ] Uncontrolled inflammation induces multi‐organ damage (such as myocardial damage), leading to multi‐organ failure, especially in the renal, hepatic, and cardiac systems.[ 9 ] In a cohort study, higher levels of IL‐6, IL‐1β, CCL8, CCL2, CXCL16, CXCL9, CXCL8, and CXCL2 were observed in patients with SARS‐CoV‐2 infection, compared to patients with other respiratory issues.[ 10 ] Another clinical study demonstrated that intensive care unit (ICU) patients had higher levels of chemokines and cytokines (TNF‐α, IL‐2, IL‐10, GSCF, and MCP1) than those of non‐ICU patients.[ 7 ] The production of a large number of pro‐inflammatory chemokines and cytokines may result in cytokine storms, which promote ARDS and sepsis symptoms (the causes of death in 69.5% and 28.0% of fatal COVID‐19 cases, respectively).[ 59 , 61 ] Therefore, it is important to consider how to counter uncontrolled inflammation induced by SARS‐CoV‐2.
4.2. Inhibition of Inflammation
Inhibition of inflammation may be a treatment option in COVID‐19. Several reports and ongoing clinical trials have explored the therapeutic potential of targeting pro‐inflammatory factors to avoid cytokine storms in severe cases (Table 2 , Figure 3 ).
Table 2.
Promising immunotherapy approaches
| Type | Target | Description | Results of clinical trials | Planned clinical trials |
|---|---|---|---|---|
| Interferons | ||||
| IFN‐α2ba) | IFNAR | Enhances ISGb) expression and induces an antiviral state | A retrospective multicenter cohort study showed that early administration (≤5 days after admission) of IFN‐α2b was associated with reduced in‐hospital mortality | |
| IFN‐α | IFNAR | Enhances ISG expression and induces an antiviral state | The 28‐day incidence of COVID‐19c) was zero in both high‐ and low‐risk groups. IFN‐α nasal drops may prevent COVID‐19 and enhance the protection based on standard physical isolation | NCT04320238 |
| IFN‐λ | IFNLR | Enhances ISG expression and induces an antiviral state | By day 7, 80% of the participants in the peginterferon lambda group had an undetectable viral load | NCT04354259 |
| Inhibition of inflammation | ||||
| Colchicine | IL‐1βd), neutrophils | Inhibits IL‐1β production, as well as neutrophil chemotaxis and activity | Colchicine significantly improved clinical manifestations | NCT04326790 |
| Tocilizumab | IL‐6R | Blocks IL‐6 transduction | Shorter time to clinical improvement and lower mortality were observed | NCT02735707 |
| Sarilumab | IL‐6R | Blocks IL‐6 transduction | Prognosis was improved | NCT02735707 |
| Siltuximab | IL‐6 | Blocks IL‐6 transduction | The 30‐day mortality rate of persons treated with siltuximab was significantly lower | NCT04322188 |
| Anakinra | IL‐1R | Blocks IL‐1 transduction | Improved clinical features and higher survival rates were observed | NCT04318366 |
| Canakinumab | IL‐1β | Blocks IL‐1 transduction | Improvement of oxygen support category was observed | NCT04348448 |
| Baricitinib | AAK1e), JAKf) | Inhibits cell entry of SARS‐CoV‐2g) and the JAK/STATh) pathway | Recovery time was reduced and improvement in clinical status was accelerated among patients with COVID‐19 | NCT04401579 |
| Convalescent plasma | ||||
| Convalescent plasma | Spike protein | Nabsi) bind to viral proteins, thereby inhibiting viral entry, limiting viral amplification | Clinical symptoms and paraclinical criteria rapidly improved | |
a)IFN, interferon; b)ISG, IFN‐stimulated gene; c)COVID‐19, coronavirus disease 2019; d)IL, interleukin; e)AAK1, adaptor‐associated protein kinase 1; f)JAK, Janus‐activated kinase; g)SARS‐CoV, severe acute respiratory syndrome coronavirus; h)STAT, signal transducer and activator of transcription; i)NAbs, neutralizing antibodies.
Figure 3.
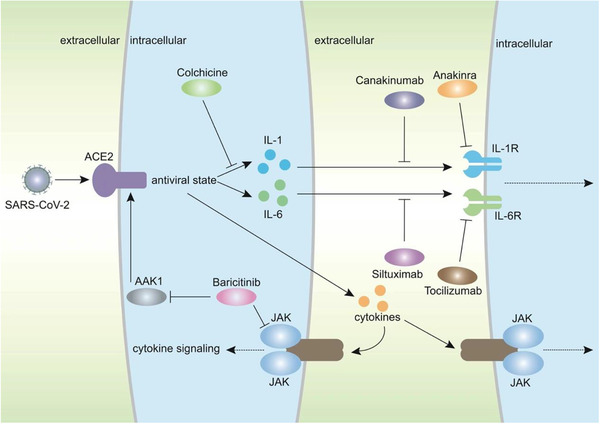
Potential methods to inhibit inflammation in COVID‐19 patients. Infection with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) activates cytokine signaling and induces inflammation and cytokine storms. Colchicine, tocilizumab, siltuximab, anakinra, canakinumab, and baricitinib can inhibit the transduction of cytokine signaling in different ways. Colchicine inhibits the production of interleukin‐1β (IL‐1β). Tocilizumab binds to the interleukin 6 receptor (IL‐6R) to prevent the function of IL‐6. Siltuximab acts by targeting IL‐6. Anakinra targets IL‐1R to inhibit inflammation. Canakinumab is an anti‐IL‐1β antibody that can block transduction. Baricitinib inhibits the JAK/STAT pathway, which mediates cytokine signaling. Furthermore, baricitinib interacts with adaptor‐associated protein kinase 1 (AAK1) to inhibit the entry of SARS‐CoV‐2.
Corticosteroid treatment slows down inflammation, but it is a double‐edged sword.[ 62 ] The lessons from the previous SARS outbreak do not favor this approach;[ 62 ] treatment of COVID‐19 with corticosteroids may induce long‐term complications, prolonged virus shedding, and secondary infections.[ 63 ] In a multicenter, single‐blind, randomized controlled trial, the use of corticosteroids also prolonged the shedding of SARS‐CoV‐2 in mild cases.[ 64 ] In addition, treatment with high‐dose corticosteroids potentially increases the mortality of patients with severe COVID‐19.[ 65 ] Overall, no study supports that patients with COVID‐19 benefit from corticosteroids; on the contrary, corticosteroids are more likely to be harmful.[ 66 ]
Colchicine is another drug that controls inflammation and is traditionally used for the treatment of rheumatic diseases. It can inhibit the production of IL‐1β, as well as neutrophil chemotaxis and activity. In a randomized, multicenter clinical trial, patients with COVID‐19 treated with colchicine showed improved clinical manifestations.[ 67 ]
Tocilizumab is a recombinant humanized anti‐human IL‐6 receptor monoclonal antibody that binds to both membrane‐bound interleukin‐6 receptor (mIL‐6R) and soluble interleukin‐6 receptor (sIL‐6R), thereby blocking IL‐6 transduction. Notably, tocilizumab has been approved for rheumatoid arthritis and cytokine storms caused by chimeric antigen receptor T‐cell therapy. In an ongoing international, multifactorial, adaptive platform trial, critically ill patients were treated with tocilizumab. A shorter time to clinical improvement and lower mortality rates were observed.[ 68 ] Sarilumab, another antibody against IL‐6R and similar to tocilizumab, also improved the prognosis of patients with COVID‐19. Tocilizumab and sarilumab had similar 90 day survival outcomes, time to ICU, hospital discharge, and improvement based on the WHO ordinal scale at day 14. In addition, adverse reactions were rarely observed.[ 68 ] Siltuximab, a monoclonal antibody targeting IL‐6, was used in an observational cohort study. The 30‐day mortality rate was significantly lower in siltuximab‐treated patients with COVID‐19 than in matched control cohorts.[ 69 ]
Anakinra is an IL‐1R antagonist that binds to IL‐1R and inhibits its signal transduction. Anakinra has also been approved for the treatment of rheumatoid arthritis. In a retrospective cohort study, 29 patients received high‐dose intravenous anakinra, and improved clinical features and higher survival rates were observed.[ 70 ] In another study, improvements at the clinical and laboratory levels were observed in eight of nine moderate‐to‐severe COVID‐19 cases.[ 71 ] Canakinumab, an anti‐IL‐1β antibody, has also been explored; patients treated with canakinumab showed improvement in oxygen support in a prospective observational study.[ 72 ]
JAK is an intracellular tyrosine kinase that mediates cytokine signaling, for example, via the JAK/STAT pathway. Baricitinib, a JAK inhibitor, has been used to treat rheumatic arthritis.[ 73 ] Baricitinib also blocks the cell entry of SARS‐CoV‐2 in the early stage of infection by targeting adaptor‐associated protein kinase 1 (AAK1), which is involved in mediating endocytosis and intracellular transport through ACE2.[ 74 , 75 ] In a pilot study on safety and clinical impact, the percentage of discharges at week 2 was 58% in baricitinib‐treated patients compared with 8% in controls. The evidence that most patients were discharged and none of the patients required ICU support showed that treatment with baricitinib improved the clinical outcome.[ 76 ]
Furthermore, antagonists against TNF‐α (ChiCTR2000030089), IFN‐γ (NCT04324021), and IL‐17 have the potential to be used in the therapy against COVID‐19. Clinical trials targeting some of these pro‐inflammatory cytokines are under way.[ 11 ]
In addition to the abovementioned antagonists, continuous renal replacement therapy (a type of hemofiltration or hemodiafiltration method) and therapeutic plasma exchange also have the potential to inhibit inflammation by removing pro‐inflammatory molecules.[ 74 ] Similarly, the artificial liver blood purification system is a promising treatment option that can quickly block cytokine storms and remove pro‐inflammatory mediators. This method is also conducive to the balance of electrolytes, acid‐base, and body fluids, thereby improving the treatment effect in critically ill patients.[ 12 ] With the progress in technology and methods, there will be more ways to defend against the pandemic by controlling inflammation.
5. Convalescent Plasma Therapy
5.1. History of Treatment with Convalescent Plasma
Convalescent plasma is a passive immunization strategy used for the prevention and treatment of infectious diseases since the beginning of the 20th century.[ 77 ] It refers to plasma collected from individuals who have recovered from a disease and produced NAbs. Transfusion of convalescent plasma involves the passive administration of substantial amounts of antibodies and offers instant immunity to infected individuals. In many cases, convalescent plasma has been used successfully to prevent and/or treat infectious diseases after exposure, such as SARS, MERS, and H1N1.[ 78 ] In therapy against SARS, treatment with convalescent plasma resulted in shorter hospital stays and lower mortality in affected patients. No immediate adverse effects were observed after the infusion of this plasma.[ 79 ] Patients treated with convalescent plasma before 14 days of illness had a better prognosis.[ 80 ]
5.2. Possible Mechanisms Involved in Convalescent Plasma Therapy
A possible mechanism for the observed effects of convalescent plasma therapy is that NAbs bind to spike proteins, thereby inhibiting the entry and amplification of the virus. This has been observed in both SARS and MERS.[ 26 ] Apart from NAbs, other plasmatic components obtained from donors may also be involved, such as anti‐inflammatory cytokines, clotting factors, natural antibodies, defensins, and other yet undefined proteins. Thus, transfusion of convalescent plasma may offer additional benefits for infected patients, such as an improved immunological regulation via amelioration of a severe inflammatory response.[ 81 ]
5.3. Clinical Trials Using Convalescent Plasma
Many clinical reports have demonstrated the efficacy of convalescent plasma. In a trial of five critically ill patients with COVID‐19 and ARDS, their clinical status improved after treatment with convalescent plasma. ARDS was resolved in four of the patients after 12 days, and three of the patients were weaned from mechanical ventilation within 2 weeks. Three patients were discharged from the hospital ≈50 days after this treatment.[ 82 ] Another trial involving 10 adults with severe COVID‐19 showed that convalescent plasma significantly increased and maintained NAb levels, terminating viremia within 7 days.[ 83 ] Within 3 days, rapid improvements in clinical symptoms and paraclinical criteria were detected. In radiological examinations, varying degrees of absorption of lung lesions were observed over 7 days. Similar to IFNs, early treatment with convalescent plasma within 14 days had a better outcome.[ 83 ] These results suggest that convalescent plasma is a promising option for severe COVID‐19 patients. Notably, the timely treatment appears to be a vital factor. The virus load often reaches its peak in the first week; thus, treatment in the early stage would presumably be better.[ 80 ]
5.4. Safety of Convalescent Plasma
Additional points need to be addressed relative to convalescent plasma treatment; the safety of this treatment in patients with COVID‐19 needs to be confirmed.[ 84 ] Mild adverse effects, such as fever, skin erythema, and nausea, may be observed after convalescent plasma infusion.[ 83 ] Antibody‐mediated pro‐inflammatory or antibody‐dependent enhancement of infection effects may also occur in the treatment with convalescent plasma.[ 85 ] Despite these reports, the administration of convalescent plasma is generally considered safe for COVID‐19 treatment, and it is not associated with any major adverse events. The first study with 5000 COVID‐19 patients treated with convalescent plasma showed that the incidence of all serious adverse events was <1% within the first 4 h after transfusion (mortality rate ≈0.3%).[ 85 ]
6. Conclusions and Future Perspectives
Considerable progress has been made in our knowledge of COVID‐19. The pathogenesis of COVID‐19 mainly involves two processes: Initially, the disease is driven by the replication of SARS‐COV‐2 and later in the process, the body's hyperimmune state induces tissue damage.[ 10 ] Immunotherapies using IFNs, inhibition of inflammation, and convalescent plasma function in different ways and have a synergistic effect.
IFN therapy focuses on the early course of infection, and plays an important role in antiviral host responses, inducing ISGs leading to an antiviral state.[ 40 ] However, infection with SARS‐CoV‐2 and translation of its proteins can lead to low IFN levels.[ 10 ] Therefore, treatment with IFNs may be a promising therapeutic strategy. Clinical trial results have shown that type I and type III IFNs can contribute to the recovery from COVID‐19, but the timing of administration is an important factor.[ 50 , 51 , 52 , 53 ] Early treatment with IFNs may play a protective role, whereas delayed treatment may be correlated with worse progress.[ 51 ]
Therapy targeting inhibition of inflammation focuses on the later course of infection. One of the sequelae SARS‐CoV‐2 infection is the subsequent inflammatory response, which can result in cytokine storms. Therefore, it is important to assess how inflammation induced by SARS‐CoV‐2 can be inhibited. The mechanisms of this uncontrolled inflammation have been extensively explored. Drugs such as tocilizumab and anakinra, originally designed to treat other inflammatory diseases, can be used to explore the treatment of uncontrolled inflammation in COVID‐19. However, the experiences gained from treatments with corticosteroids call attention to unwanted effects, such as secondary infections.[ 63 ] Therefore, treatment with immunomodulatory drugs requires more data from clinical trials.
Convalescent plasma played an important role at the beginning of the pandemic when there were no effective vaccines or treatments because of the novelty of SARS‐COV‐2. Convalescent plasma containing NAbs is an immediately available strategy to prevent and treat COVID‐19, although there may be associated risks of mild adverse events.[ 78 ]
Many clinical immunotherapy trials are ongoing, and new findings are shared via the WHO Solidarity Trial (Table 2). Considering other potential and pathogenic SARSr‐CoVs and the lessons of SARS and MERS, it is of great significance and potential to continue to explore immunotherapies against SARS‐CoV‐2.
In summary, immunotherapy is an effective and promising therapy for COVID‐19; however, there are still many unresolved issues that require further investigation. Most immunotherapies against COVID‐19 have been shown to induce mild adverse effects in clinical trials. Thus, the safety of immunotherapies requires further investigation. Moreover, the timing of administration is important. Early treatment with IFNs improves the outcome, whereas delayed treatment aggravates the disease. Therefore, the method of administration needs to be further explored.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
H.G., L.Z., and Z.M. contributed equally to this work. H.G. conceived and drafted the manuscript. L.Z., Z.M., Z.T., and F.Z. discussed the concepts of the manuscript. Z.M. drew the figures. F.Z. approved the version to be submitted.
Acknowledgements
The authors would like to apologize to those researchers whose related works were not able to be cited in this review. This work was supported by the National Natural Science Foundation of China (31871405 to FZ), National Science Foundation of Jiangsu (BK20180043 and 19KJA550003 to FZ).
Biographies
Haodong Guo obtained his Bachelor's degree in Biotechnology from Soochow University in 2020. He is currently pursuing his Master's degree under the supervision of Prof. Fangfang Zhou at the Institutes of Biology and Medical Science, Soochow University, studying on the mechanisms of antiviral innate immunity.

Lili Zhou obtained her Bachelor's degree in Biotechnology and Master's degree in Immunity from Soochow University and began to purchase her Doctorate degree in Immunity under the supervision of Prof. Fangfang Zhou at the Institutes of Biology and Medical Science, Soochow University in 2020. Her current research focuses on antiviral innate immunity and anti‐tumor immunity.

Zhenyu Ma received his Bachelor's degree in Biotechnology from Jiangsu Ocean University and began to study Immunity for his Master's degree under the supervision of Prof. Fangfang Zhou at the Institutes of Biology and Medical Science, Soochow University in 2020. His current research focuses on mechanisms of antiviral innate immunity.

Fangfang Zhou obtained a Ph.D. degree from Tsinghua University in 2008. She then worked as a postdoctoral fellow with Prof. Peter ten Dijke in Leiden University Medical Center. In 2014, she joined the Institutes of Biology and Medical Sciences, Soochow University. Her research interest is the molecular mechanism of innate immunity in viral infection and cancer development.

References
- 1. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fan Y., Zhao K., Shi Z. ‐. L., Zhou P., Viruses 2019, 11, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui J., Li F., Shi Z. L., Nat. Rev. Microbiol. 2019, 17, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., et al., Lancet 2020, 395, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naqvi A. A. T., Fatima K., Mohammad T., Fatima U., Singh I. K., Singh A., Atif S. M., Hariprasad G., Hasan G. M., Hassan M. I., Biochim. Biophys. Acta, Mol. Basis Dis. 2020, 1866, 165878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO COVID‐19 Dashboard. Geneva: World Health Organization, https://covid19.who.int/ (accessed: April 2021).
- 7. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Lancet 2020, 395, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C., Lancet Infect. Dis. 2020, 20, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tay M. Z., Poh C. M., Rénia L., MacAry P. A., Ng L. F. P., Nat. Rev. Immunol. 2020, 20, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanco‐Melo D., Nilsson‐Payant B. E., Liu W. C., Uhl S., Hoagland D., Møller R., Jordan T. X., Oishi K., Panis M., Sachs D., Wang T. T., Schwartz R. E., Lim J. K., Albrecht R. A., tenOever B. R., Cell 2020, 181, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Copaescu A., Smibert O., Gibson A., Phillips E. J., Trubiano J. A., J. Allergy Clin. Immunol. 2020, 146, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo J., Xia H., Wang S., Yu L., Zhang H., Chen J., Shi D., Chen Y., Zhang Y., Xu K., Xu X., Sheng J., Qiu Y., Li L., Front. Immunol. 2020, 11, 586073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haimei M. A., Clin. Appl. Thromb./Hemostasis 2020, 26, 1076029620944497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.COVID‐19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID‐19) Treatment Guidelines, https://www.covid19treatmentguidelines.nih.gov/ (accessed: April 2021).
- 15. Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F., Signal Transduct Target Ther. 2020, 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draft Landscape of COVID‐19 Candidate Vaccines, https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed: April 2021).
- 17. Sariol A., Perlman S., Immunity 2020, 53, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., N. Engl. J. Med. 2020, 382, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mittal A., Manjunath K., Ranjan R. K., Kaushik S., Kumar S., Verma V., PLoS Pathog. 2020, 16, e1008762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snijder E. J., Decroly E., Ziebuhr J., Adv. Virus Res. 2016, 96, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia H., Cao Z., Xie X., Zhang X., Chen J. Y., Wang H., Menachery V. D., Rajsbaum R., Shi P. Y., Cell Rep. 2020, 33, 108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Lee J. Y., Yang J. S., Kim J. W., Kim V. N., Chang H., Cell 2020, 181, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y., Liu Q., Guo D., J. Med. Virol. 2020, 92, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Li F., Annu. Rev. Virol. 2016, 3, 237; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C. L., Abiona O., Graham B. S., McLellan J. S., Science 2020, 367, 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann M., Kleine‐Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., Cell 2020, 181, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Du L., He Y., Zhou Y., Liu S., Zheng B. J., Jiang S., Nat. Rev. Microbiol. 2009, 7, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Z., Hu Y., Xu M., Chen Z., Yang W., Jiang Z., Li M., Jin H., Cui G., Chen P., Wang L., Zhao G., Ding Y., Zhao Y., Yin W., Lancet Infect. Dis. 2021, 21, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voysey M., Costa Clemens S. A., Madhi S. A., Weckx L. Y., Folegatti P. M., Aley P. K., Angus B., Baillie V. L., Barnabas S. L., Bhorat Q. E., Bibi S., Briner C., Cicconi P., Clutterbuck E. A., Collins A. M., Cutland C. L., Darton T. C., Dheda K., Dold C., Duncan C. J. A., Emary K. R. W., Ewer K. J., Flaxman A., Fairlie L., Faust S. N., Feng S., Ferreira D. M., Finn A., Galiza E., Goodman A. L., et al., Lancet 2021, 397, 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., et al., N. Engl. J. Med. 2021, 384, 403.33378609 [Google Scholar]
- 30. Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., R. W. Frenck, Jr. , Hammitt L. L., Ö T., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C., N. Engl. J. Med. 2020, 383, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C., Science 2020, 369, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krammer F., Nature 2020, 586, 516. [DOI] [PubMed] [Google Scholar]
- 33. Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G. F., Tan W., Wu G., Xu M., Lou Z., Huang W., Xu W., Huang B., Wang H., Wang W., Zhang W., Li N., Xie Z., Ding L., You W., Zhao Y., Yang X., Liu Y., Wang Q., Huang L., Yang Y., Xu G., Luo B., Wang W., Liu P., Guo W., Yang X., Lancet Infect. Dis. 2021, 21, 39.33069281 [Google Scholar]
- 34. Walsh E. E., R. W. Frenck, Jr. , Falsey A. R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M. J., Bailey R., Swanson K. A., Li P., Koury K., Kalina W., Cooper D., Fontes‐Garfias C., Shi P. Y., Tureci O., Tompkins K. R., Lyke K. E., Raabe V., Dormitzer P. R., Jansen K. U., Şahin U., Gruber W. C., N. Engl. J. Med. 2020, 383, 2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.a) Park A., Iwasaki A., Cell Host Microbe 2020, 27, 870; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Totura A. L., Baric R. S., Curr. Opin. Virol. 2012, 2, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yin X., Riva L., Pu Y., Martin‐Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P. D., Yang C. ‐. C., Herbert K. M., Yoh S., Hultquist J. F., García‐Sastre A., Chanda S. K., Cell Rep. 2021, 34, 108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park A., Iwasaki A., Cell Host Microbe 2020, 27, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y. T., Grishin N. V., Chen Z. J., Science 2015, 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- 39. Odendall C., Kagan J. C., Curr. Opin. Virol. 2015, 12, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ivashkiv L. B., Donlin L. T., Nat. Rev. Immunol. 2014, 14, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Z., Hamming O. J., Ank N., Paludan S. R., Nielsen A. L., Hartmann R., J. Virol. 2007, 81, 7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J., Nat. Commun. 2020, 11, 3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuen C. K., Lam J. Y., Wong W. M., Mak L. F., Wang X., Chu H., Cai J. P., Jin D. Y., To K. K., Chan J. F., Yuen K. Y., Kok K. H., Emerging Microbes Infect. 2020, 9, 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Zheng Y., Zhuang M. W., Han L., Zhang J., Nan M. L., Zhan P., Kang D., Liu X., Gao C., Wang P. H., Signal Transduct Target Ther. 2020, 5, 299; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen K., Xiao F., Hu D., Ge W., Tian M., Wang W., Pan P., Wu K., Wu J., Viruses 2020, 13, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miorin L., Kehrer T., Sanchez‐Aparicio M. T., Zhang K., Cohen P., Patel R. S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K. M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin‐Sancho L., Krogan N. J., Chanda S. K., Schotsaert M., Wozniak R. W., Ren Y., Rosenberg B. R., Fontoura B. M. A., García‐Sastre A., Proc. Natl. Acad. Sci. USA 2020, 117, 28344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Omrani A. S., Saad M. M., Baig K., Bahloul A., Abdul‐Matin M., Alaidaroos A. Y., Almakhlafi G. A., Albarrak M. M., Memish Z. A., Albarrak A. M., Lancet Infect. Dis. 2014, 14, 1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Al‐Tawfiq J. A., Momattin H., Dib J., Memish Z. A., Int. J. Infect. Dis. 2014, 20, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Channappanavar R., Fehr A. R., Zheng J., Wohlford‐Lenane C., Abrahante J. E., Mack M., Sompallae R., P. B. McCray, Jr. , Meyerholz D. K., Perlman S., J. Clin. Invest. 2019, 129, 3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lokugamage K. G., Hage A., de Vries M., Valero‐Jimenez A. M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V. D., J. Virol. 2020, 94, e01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Q., Chen V., Shannon C. P., Wei X. S., Xiang X., Wang X., Wang Z. H., Tebbutt S. J., Kollmann T. R., Fish E. N., Front. Immunol. 2020, 11, 1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., Qian X., Wang S., Guo Y., Yu H., Cui M., Tong G., Xu Y., Zheng Z., Lu Y., Hong P., Cell Host Microbe 2020, 28, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meng Z., Wang T., Chen L., Chen X., Li L., Qin X., Li H., Luo J., medRxiv:2020.04.11.20061473 2020. [Google Scholar]
- 53. Feld J. J., Kandel C., Biondi M. J., Kozak R. A., Zahoor M. A., Lemieux C., Borgia S. M., Boggild A. K., Powis J., McCready J., Tan D. H. S., Chan T., Coburn B., Kumar D., Humar A., Chan A., O'Neil B., Noureldin S., Booth J., Hong R., Smookler D., Aleyadeh W., Patel A., Barber B., Casey J., Hiebert R., Mistry H., Choong I., Hislop C., Santer D. M., Lorne Tyrrell D., Glenn J. S., Gehring A. J., Janssen H. L. A., Hansen B. E., Lancet Respir. Med. 2021. [Google Scholar]
- 54. Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., Cao Y., Yousif A. S., Bals J., Hauser B. M., Feldman J., Muus C., M. H. Wadsworth, 2nd , Kazer S. W., Hughes T. K., Doran B., Gatter G. J., Vukovic M., Taliaferro F., Mead B. E., Guo Z., Wang J. P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J. M. S., Taylor C. J., Lin B., et al., Cell 2020, 181, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muir A. J., Arora S., Everson G., Flisiak R., George J., Ghalib R., Gordon S. C., Gray T., Greenbloom S., Hassanein T., Hillson J., Horga M. A., Jacobson I. M., Jeffers L., Kowdley K. V., Lawitz E., Lueth S., Rodriguez‐Torres M., Rustgi V., Shemanski L., Shiffman M. L., Srinivasan S., Vargas H. E., Vierling J. M., Xu D., Lopez‐Talavera J. C., Zeuzem S., J. Hepatol. 2014, 61, 1238. [DOI] [PubMed] [Google Scholar]
- 56. Davidson S., McCabe T. M., Crotta S., Gad H. H., Hessel E. M., Beinke S., Hartmann R., Wack A., EMBO Mol. Med. 2016, 8, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lazear H. M., Schoggins J. W., Diamond M. S., Immunity 2019, 50, 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grant R. A., Morales‐Nebreda L., Markov N. S., Swaminathan S., Querrey M., Guzman E. R., Abbott D. A., Donnelly H. K., Donayre A., Goldberg I. A., Klug Z. M., Borkowski N., Lu Z., Kihshen H., Politanska Y., Sichizya L., Kang M., Shilatifard A., Qi C., Lomasney J. W., Argento A. C., Kruser J. M., Malsin E. S., Pickens C. O., Smith S. B., Walter J. M., Pawlowski A. E., Schneider D., Nannapaneni P., Abdala‐Valencia H., et al., Nature 2021, 590, 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jafarzadeh A., Chauhan P., Saha B., Jafarzadeh S., Nemati M., Life Sci. 2020, 257, 118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song P., Li W., Xie J., Hou Y., You C., Clin. Chim. Acta 2020, 509, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang B., Zhou X., Qiu Y., Song Y., Feng F., Feng J., Song Q., Jia Q., Wang J., PLoS One 2020, 15, e0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stockman L. J., Bellamy R., Garner P., PLoS Med. 2006, 3, e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Paassen J., Vos J. S., Hoekstra E. M., Neumann K. M. I., Boot P. C., Arbous S. M., Crit. Care 2020, 24, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang X., Feng Y. M., Ni J. X., Zhang J. Y., Liu L. M., Hu K., Wu X. Z., Zhang J. X., Chen J. W., Zhang J. C., Su J., Li Y. L., Zhao Y., Xie J., Ding Z., He X. L., Wang W., Jin R. H., Shi H. Z., Sun B., Respiration 2021, 100, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., Shi J., Zhou M., Wu B., Yang Z., Zhang C., Yue J., Zhang Z., Renz H., Liu X., Xie J., Xie M., Zhao J., J. Allergy Clin. Immunol. 2020, 146, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Russell C. D., Millar J. E., Baillie J. K., Lancet 2020, 395, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deftereos S. G., Giannopoulos G., Vrachatis D. A., Siasos G. D., Giotaki S. G., Gargalianos P., Metallidis S., Sianos G., Baltagiannis S., Panagopoulos P., Dolianitis K., Randou E., Syrigos K., Kotanidou A., Koulouris N. G., Milionis H., Sipsas N., Gogos C., Tsoukalas G., Olympios C. D., Tsagalou E., Migdalis I., Gerakari S., Angelidis C., Alexopoulos D., Davlouros P., Hahalis G., Kanonidis I., Katritsis D., Kolettis T., et al., JAMA Network Open 2020, 3, e2013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gordon A. C., Mouncey P. R., Al‐Beidh F., Rowan K. M., Nichol A. D., Arabi Y. M., Annane D., Beane A., van Bentum‐Puijk W., Berry L. R., Bhimani Z., Bonten M. J. M., Bradbury C. A., Brunkhorst F. M., Buzgau A., Cheng A. C., Detry M. A., Duffy E. J., Estcourt L. J., Fitzgerald M., Goossens H., Haniffa R., Higgins A. M., Hills T. E., Horvat C. M., Lamontagne F., Lawler P. R., Leavis H. L., Linstrum K. M., Litton E., et al., N. Engl. J. Med. 2021, 384, 1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gritti G., Raimondi F., Ripamonti D., Riva I., Landi F., Alborghetti L., Frigeni M., Damiani M., Micò C., Fagiuoli S., Cosentini R., Lorini F. L., Gandini L., Novelli L., Morgan J. P., Owens B. M. J., Kanhai K. J. K., Reljanovic G. T., Rizzi M., Di Marco F., Rambaldi A., medRxiv: 2020.04.01.20048561 2020. [Google Scholar]
- 70. Cavalli G., De Luca G., Campochiaro C., Della‐Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere‐Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L., Lancet Rheumatol. 2020, 2, e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aouba A., Baldolli A., Geffray L., Verdon R., Bergot E., Martin‐Silva N., Justet A., Ann. Rheum. Dis. 2020, 79, 1381. [DOI] [PubMed] [Google Scholar]
- 72. Landi L., Ravaglia C., Russo E., Cataleta P., Fusari M., Boschi A., Giannarelli D., Facondini F., Valentini I., Panzini I., Lazzari‐Agli L., Bassi P., Marchionni E., Romagnoli R., De Giovanni R., Assirelli M., Baldazzi F., Pieraccini F., Rametta G., Rossi L., Santini L., Valenti I., Cappuzzo F., Sci. Rep. 2020, 10, 21775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Keystone E. C., Taylor P. C., Drescher E., Schlichting D. E., Beattie S. D., Berclaz P. Y., Lee C. H., Fidelus‐Gort R. K., Luchi M. E., Rooney T. P., Macias W. L., Genovese M. C., Ann. Rheum. Dis. 2015, 74, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim J. S., Lee J. Y., Yang J. W., Lee K. H., Effenberger M., Szpirt W., Kronbichler A., Shin J. I., Theranostics 2021, 11, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.a) Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P. J., Monteil V., Lauschke V. M., Mirazimi A., Youhanna S., Tan Y. J., Baldanti F., Sarasini A., Terres J. A. R., Nickoloff B. J., Higgs R. E., Rocha G., Byers N. L., Schlichting D. E., Nirula A., Cardoso A., Corbellino M., EMBO Mol. Med. 2020, 12, 12697; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J., Lancet 2020, 395, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D., J. Infect. 2020, 81, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Marano G., Vaglio S., Pupella S., Facco G., Catalano L., Liumbruno G. M., Grazzini G., Blood Transfus. 2016, 14, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bloch E. M., Shoham S., Casadevall A., Sachais B. S., Shaz B., Winters J. L., van Buskirk C., Grossman B. J., Joyner M., Henderson J. P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P. G., Thomas D., Sullivan D. J., Paneth N., Gehrie E., Spitalnik S., Hod E. A., Pollack L., Nicholson W. T., Pirofski L. A., Bailey J. A., Tobian A. A., J. Clin. Invest. 2020, 130, 2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Soo Y. O., Cheng Y., Wong R., Hui D. S., Lee C. K., Tsang K. K., Ng M. H., Chan P., Cheng G., Sung J. J., Clin Microbiol. Infect. 2004, 10, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheng Y., Wong R., Soo Y. O., Wong W. S., Lee C. K., Ng M. H., Chan P., Wong K. C., Leung C. B., Cheng G., Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rojas M., Rodríguez Y., Monsalve D. M., Acosta‐Ampudia Y., Camacho B., Gallo J. E., Rojas‐Villarraga A., Ramírez‐Santana C., Díaz‐Coronado J. C., Manrique R., Mantilla R. D., Shoenfeld Y., Anaya J. M., Autoimmun Rev. 2020, 19, 102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L., J. Am. Med. Assoc. 2020, 323, 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., et al., Proc. Natl. Acad. Sci. USA 2020, 117, 9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chai K. L., Valk S. J., Piechotta V., Kimber C., Monsef I., Doree C., Wood E. M., Lamikanra A. A., Roberts D. J., McQuilten Z., et al., Cochrane 2020, 7, CD013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Joyner M. J., Wright R. S., Fairweather D., Senefeld J. W., Bruno K. A., Klassen S. A., Carter R. E., Klompas A. M., Wiggins C. C., Shepherd J. R., Rea R. F., Whelan E. R., Clayburn A. J., Spiegel M. R., Johnson P. W., Lesser E. R., Baker S. E., Larson K. F., Ripoll J. G., Andersen K. J., Hodge D. O., Kunze K. L., Buras M. R., Vogt M. N., Herasevich V., Dennis J. J., Regimbal R. J., Bauer P. R., Blair J. E., Van Buskirk C. M., et al., J. Clin. Invest. 2020, 130, 4791. [DOI] [PMC free article] [PubMed] [Google Scholar]


