Summary
The presence of personally identifiable information (PII) in natural language portions of electronic health records (EHRs) constrains their broad reuse. Despite continuous improvements in automated detection of PII, residual identifiers require manual validation and correction. Here, we describe an automated de-identification system that employs an ensemble architecture, incorporating attention-based deep-learning models and rule-based methods, supported by heuristics for detecting PII in EHR data. Detected identifiers are then transformed into plausible, though fictional, surrogates to further obfuscate any leaked identifier. Our approach outperforms existing tools, with a recall of 0.992 and precision of 0.979 on the i2b2 2014 dataset and a recall of 0.994 and precision of 0.967 on a dataset of 10,000 notes from the Mayo Clinic. The de-identification system presented here enables the generation of de-identified patient data at the scale required for modern machine-learning applications to help accelerate medical discoveries.
Keywords: mayo, nference, de-identification, ensemble, obfuscation, anonymization
Graphical abstract

Highlights
-
•
An ensemble approach to automated de-identification of unstructured clinical text
-
•
Our approach leverages advances in deep learning along with heuristics
-
•
Detected personally identifiable information is replaced with suitable surrogates
-
•
Patient data are de-identified at scale to accelerate medical discovery
The bigger picture
Clinical notes in electronic health records convey rich historical information regarding disease and treatment progression. However, this unstructured text often contains personally identifiable information such as names, phone numbers, or residential addresses of patients, thereby limiting its dissemination for research purposes. The removal of patient identifiers, through the process of de-identification, enables sharing of clinical data while preserving patient privacy. Here, we present a best-in-class approach to de-identification, which automatically detects identifiers and substitutes them with fabricated ones. Our approach enables de-identification of patient data at the scale required to harness the unstructured, context-rich information in electronic health records to aid in medical research and advancement.
Physician notes contain valuable information about patient health and treatment, but their broad reuse is constrained by the presence of personally identifiable information to protect patient confidentiality. Our approach automatically detects and hides these identifiers in plain sight by replacing them with suitable surrogates. The de-identification system presented outperforms other existing tools, allowing for the generation of de-identified patient data at the scale necessary to help accelerate medical discovery.
Introduction
The widespread adoption of electronic health records (EHRs) by health care systems has enabled digitization of patient health journeys. While the structured elements of EHRs (e.g., health insurance billing codes) have been relied upon to support the business of health care and front office applications for decades, the unstructured text (e.g., history and physical notes and pathology reports) contains far richer and nuanced information about patient care, supporting novel research.1, 2, 3, 4, 5 However, this text often contains personally identifiable information (PII) as defined in the Health Insurance Portability and Accountability Act of 1996 (HIPAA), such as personal name, phone number, or residential address.6 As a consequence, such data have limited reuse for secondary purposes.7 HIPAA permits data derived from EHRs to be widely shared and used when it is de-identified. Under the HIPAA Privacy Rule, de-identification can be accomplished in several ways. The most straightforward is the Safe Harbor implementation, which necessitates removal of an enumerated list of 18 categories of direct (e.g., Social Security number) and quasi-identifiers (e.g., date of service).
Implementing a scalable method for de-identification has several competing requirements. First, from a regulatory perspective, it must achieve extremely high recall, in that it needs to detect nearly all instances of PII. Second, from a clinical utility perspective, it must achieve extremely high precision, so that we maximize the correctness of the biomedical research performed. And, third, the approach needs to be cost effective, so that millions of records can be de-identified in a reasonable amount of time. The traditional approach of manual detection of PII is expensive, time consuming, and prone to human error,8,9 which makes automated de-identification a more promising alternative.10,11
Several recent advances in natural language processing (NLP) have created an opportunity to build accurate and scalable automated de-identification systems. First, transfer learning of autoregressive and autoencoder models12 for a supervised task such as named entity recognition (NER) requires very few labeled data, reducing human effort and error. Second, attention-based deep-learning models, such as transformers,13 allow for the non-sequential processing of text and enable the generation of rich contextualized word representations. Third, semantic segmentation algorithms generate a subword-based vocabulary,14,15 which can capture out-of-vocabulary words. Finally, the traditional transformer architecture has been improved upon through bidirectional encoder representations from transformers (BERT)16 and similar technologies that jointly train a masked language model (MLM) pre-training objective and a next sentence prediction task. BERT has set the stage for learning context-independent representations of terms in text and training context-sensitive models that transform those representations into context-aware representations based on the occurrence of a term in a sentence. We leverage these advances to support de-identification, which we formulate as an NER problem.
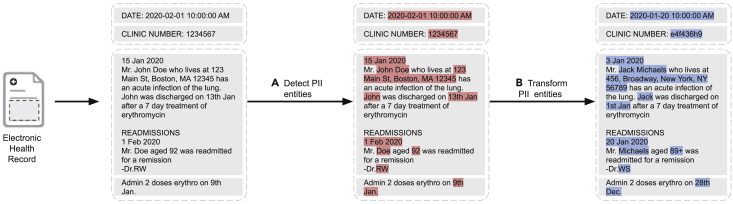
In this paper, we integrate a collection of approaches, blending the beneficial aspects of modern deep learning with rules and heuristics, to create a best-in-class approach to automated de-identification. The system transforms each detected PII instance into a suitable surrogate to mitigate the risk that any residual PII can be used to re-identify patients (Figure 1). The nference de-identification tool can be accessed at https://academia.nferx.com/deid/.
Figure 1.
Automated de-identification of electronic health records
Two steps in automated de-identification of EHRs: (A) detecting PII entities and (B) transforming them by replacement with suitable surrogates. PII, personally identifiable information.
Results
We first compare the performance of the nference de-identification system with other methods on the i2b2 2014 dataset.17 The resulting models are evaluated using precision, recall, and F1 scores (formulation provided in the supplemental methods) for NER on several groups of PII as defined in Table 1. We then compare the performance of these models on a substantially larger and diverse dataset from the Mayo Clinic and perform a deeper dive into the types of errors, distribution of errors per physician note, and distribution of errors per note type. It should be noted that this analysis focuses solely on the performance of detecting PII instances and does not address the risk of re-identification based on the semantics of any instances that the system fails to detect, an issue that is beyond the scope of this study.
Table 1.
The list of entities covered by each group of direct and quasi-identifiers
| Group name | Included entities |
|---|---|
| A (entities to be detected for a HIPAA Safe Harbor implementation) | (1) age over 89, (2) phone/fax numbers, (3) email addresses, (4) websites and URLs, (5) IP addresses, (6) dates, (7) Social Security number, (8) medical record numbers, (9) vehicle/device numbers, (10) account/certificate/license numbers, (11) health plan number, (12) street address, (13) city, (14) ZIP code, (15) employer name, and (16) personal names of patients and family members |
| B | Group A and (17) provider (doctor/nurse) names, (18) user IDs (of care providers) |
| C | Group B and (19) health care organization/facility names, (20) country, (21) state |
It should be noted that groups B and C encompass entities beyond HIPAA Safe Harbor.
Performance on the 2014 i2b2 de-identification dataset
The i2b2 2014 De-identification and Heart Disease Risk Factors challenge17 is a publicly available dataset of clinical documents with annotated PII elements. This dataset consists of a training set of 792 clinical notes and a test set of 515 clinical notes.
We compared the performance of our approach on the 2014 i2b2 test set with six other established de-identification tools: the method proposed by Dernoncourt et al. that blends conditional random fields (CRFs) and artificial neural networks (ANNs),18 Scrubber,19 Physionet,8 Philter,20 MIST,21 and NeuroNER.22
The results are provided in Table 2. First, we cite the CRF and ANN approach (CRF + ANN)18 scores against the group A entities (HIPAA only) as reported in their paper. We also directly report the results for Scrubber, Physionet, and Philter from prior publications20 without performing an empirical analysis because the dataset (2014 i2b2) and the set of PII entities are the same as that used in our investigation. We trained MIST using sentences from the i2b2 training corpus (see supplemental methods and Table S3). We downloaded and used a pre-trained model for NeuroNER (see supplemental methods). We present the performance of these methods on group B (see Table 1) entities, which we use as the basis of our comparison.
Table 2.
Performance of de-identification methods on the 2014 i2b2 test corpus
| Method name | Group | Precision | Recall | F1 | Basis of results |
|---|---|---|---|---|---|
| CRF + ANN (Dernoncourt et al.) | A | 0.979 | 0.978 | 0.978 | Dernoncourt et al.18 |
| Physionet | B | 0.894 | 0.698 | 0.784 | Norgeot et al.20 |
| Scrubber | B | 0.762 | 0.878 | 0.815 | Norgeot et al.20 |
| Philter | B | 0.785 | 0.999∗ | 0.879 | Norgeot et al.20 |
| MIST (trained on i2b2) | B | 0.907 | 0.879 | 0.893 | N/A |
| NeuroNER | B | 0.979 | 0.950 | 0.964 | N/A |
| nference (fine-tuned on Mayo) | B | 0.961 | 0.988 | 0.974 | N/A |
| nference (fine-tuned on Mayo + i2b2) | B | 0.979∗ | 0.992 | 0.985∗ | N/A |
The results for Scrubber, Physionet, Philter, and the CRF + ANN method are based on previous publications. The MIST method required training and, thus, was trained on the 2014 i2b2 training dataset. We used a pre-trained model for NeuroNER. The two versions of the nference approach were fine-tuned on (1) only the Mayo dataset and (2) both the Mayo and the i2b2 datasets.
Best performance for the metric.
We present two versions of the nference system. The first version was fine-tuned only on Mayo data and did not utilize any characteristics of the i2b2 training data. When evaluated with group B, this model achieved a precision, recall, and F1 score of 0.961, 0.988, and 0.974, respectively. The second version of our system involved fine-tuning our model with sentences from the i2b2 training set. We could not incorporate inclusion lists and sentence templates associated with the i2b2 data since the dataset is small (see “methods” for details). The precision, recall, and F1 score increased to 0.979, 0.992, and 0.985, respectively. Precision and recall per identifier type are provided in Table S4.
Performance on the Mayo test dataset
The Mayo Clinic dataset consisted of 10,000 randomly sampled notes from a corpus of 104 million notes corresponding to 477,000 patients' EHR records.
The evaluation performed on the Mayo test dataset was based on identifiers defined by group C since this group best represented the distribution of PII in the dataset. The performance of the de-identification methods (in terms of precision, recall, and F1) are presented in Table 3. The nference method performed best, with precision, recall, and F1 scores of 0.967, 0.994, and 0.979, respectively. Compared with the performance on the i2b2 dataset, we see improved recall (increase of 0.01) and a reduced precision value (decrease of 0.021). NeuroNER achieves precision, recall, and F1 scores of 0.928, 0.933, and 0.931, respectively. The F1 scores of Scrubber, Physionet, and Philter were lower than those achieved on the i2b2 dataset. Among these three methods, Philter demonstrates a relatively high recall of 0.918. Closely following Philter, the MIST model achieves a recall of 0.889 with overall performance similar to that on the i2b2 dataset.
Table 3.
Performance of de-identification methods on the Mayo test dataset
| Method | Precision | Recall | F1 |
|---|---|---|---|
| Scrubber | 0.756 | 0.677 | 0.715 |
| Philter | 0.709 | 0.918 | 0.800 |
| Physionet | 0.837 | 0.772 | 0.803 |
| MIST (trained on Mayo) | 0.818 | 0.889 | 0.852 |
| NeuroNER (trained on Mayo) | 0.928 | 0.933 | 0.931 |
| nference (fine-tuned on Mayo) | 0.967∗ | 0.994∗ | 0.979∗ |
These methods were evaluated against group C entities.
Best performance for the metric.
Error analysis on the Mayo dataset
We further investigated cases in the Mayo dataset where the nference de-identification model failed to successfully detect the PII element completely (i.e., false negatives). This occurred at a rate of 0.6% (see Table 4). Across the 10,000 notes considered in the test set, there were 848 error instances that contained these false negative errors. Accounting for duplicate occurrences of the same sentence, there were 797 unique error instances. We grouped these instances based on the type of identifier. The prevalence of the error category is shown in the second column, while the third column in the table represents the contribution of each category to the error in recall (sums to 0.6%).
Table 4.
Prevalence and examples of types of false negatives encountered by the nference de-identification system when applied on the Mayo test set
| Category | Number of error instances (n = 797) | Contribution to recall error (E = 0.6%) | Example (the PII presented in these examples is fictitious) |
|---|---|---|---|
| Clinic location | 208 | 0.1461% | He had a DWI in January and was required to do treatment through Samson rehab in St. Louis, Missouri |
| Dates | 183 | 0.1285% | CPL dated 4/27/04 |
| Doctor/nurse name/initial | 169 | 0.1187% | Sent: 2020-10-20 10:00 a.m.. Subject: RE: Consumer/Pat |
| Pharmacy name | 54 | 0.0379% | S: fax received from Trioki Rx with request for new RX for Viread (tenofovir) |
| Phone number | 50 | 0.0351% | Phone number patient/caller is calling from or the number of the provider: 724.161.1754 |
| Organization/company | 35 | 0.0246% | Last we talked about her involvement in a group called GO GIRLS! |
| Health care organization | 22 | 0.0154% | Jane is brought in by a Minerva female attendant and said Jane has been like this for "weeks and weeks." |
| Numeric identifier | 9 | 0.0063% | Manufactured by Merck lot number 78-32-DK, expiration date 2020/10/20 |
| Location (address or partial address) | 8 | 0.0056% | 500 State Highway 72 |
| Patient name | 4 | 0.0028% | PLOF: X was independent with self cares living |
The entities highlighted in italics indicate the word or phrase that the system failed to detect.
The most prevalent error was in the recognition of entities pertaining to clinic locations (208 of 797). Many of these were due to partially identified phrases (e.g., “Room 7A” was missed in “out of Southwest Building Room 7A”). The second most prevalent error type was in dates, with 183 false negatives. The third most prevalent error category was in doctor/nurse names and initials, with 169 false negatives. Abbreviations and shorthand used by providers (typically while signing off on a clinical note) contributed to the errors in this category.
Ambiguous instances of PII also resulted in false negatives. These were cases that a human reader would have difficulty/uncertainty in deeming as PII. An example of this is the word “tp” in the phrase “Comment: 03-12-2005 08:04:12—verified tp.” We found that 26% of errors were those in which the nurse abstractors themselves did not agree on the characterization of PII (Cohen's κ for errors was lower than for non-errors, at 0.7453), pointing to the inherent ambiguity.
Distribution of errors per note
We further investigated the rate at which errors in detecting PII (false negatives) occurred on a per-note level. As shown in Table 5, the error instances were distributed across 637 notes. Furthermore, we see that a majority of false negatives are spread evenly across the notes (525 of 637 notes, or 82.4%, contain a single error). For each subsequent error rate, we computed the coverage of PII entities. Here, coverage represents the fraction of PII present in the subset of notes up to the corresponding error rate.
Table 5.
Distribution of number of errors per note
| Errors per note | Number of notes | Total errors | Cumulative errors | PII coverage | Average number of error types |
|---|---|---|---|---|---|
| 0 | 9,363 | 0 | 0 | 0.9940 | 0 |
| 1 | 525 | 525 | 525 | 0.9978 | 1.00 |
| 2 | 80 | 160 | 685 | 0.9989 | 1.56 |
| 3 | 10 | 30 | 715 | 0.9991 | 1.75 |
| 4 | 6 | 24 | 739 | 0.9992 | 2.30 |
| 5 | 6 | 30 | 769 | 0.9994 | 2.75 |
| 6 | 2 | 12 | 781 | 0.9995 | 2.5 |
| 7 | 2 | 14 | 795 | 0.9996 | 2.25 |
| 8 | 2 | 16 | 811 | 0.9997 | 2.25 |
| 9 | 3 | 27 | 838 | 0.9999 | 2.33 |
| 10 | 1 | 10 | 848 | 1.0000 | 2.00 |
PII coverage represents the fraction of PII present in the subset of notes up to the corresponding error rate. Average number of error types denotes the number of distinct error types (such as date errors or name errors) per note.
Even for notes with a large number of errors (more than 6), the number of distinct error types is between 2 and 3. This illustrates that most of the errors are of the same type and an artifact of repetition of text within a note. For example, in the note with 10 errors, eight of the instances were related to location, while the remaining two were related to date. Examples of the errors pertaining to location here are “Location of INR sample: Other: Smallville Other: Smallville Other: Smallville”, “Recommend Recheck: Other: 04/01/2017 Smallville Other: 04/01/2017 Smallville”, “Recommend Recheck: Other: 04/01/2017 Smallville Other: 04/01/2017 Smallville Other: 04/01/2017 Other: 04/01/2017 Smallville”. Here, the location errors all pertain to the same location “Smallville,” which illustrates how the effective amount of identifiable content is substantially smaller than suggested by the raw count. The date presented (“04/01/2017”) was successfully detected. Both the date and the location have been replaced with synthetic values for the purpose of this example.
Distribution of note types
In the Mayo test set, a physician note is associated with a note type (e.g., progress note, emergency visit, telephone encounter). Given that the structure and semantics of these note types vary greatly from one another, we analyze the enrichment of errors across them. From the 637 notes with errors, we found 134 distinct note types with at least 1 error. The top 14 note types with highest error content are listed in Table 6. Notes of the type “Anti-coag service visit summary” contain the highest rate of errors (22 of 26 sampled notes), followed by “Electrocardiogram” (19 of 30 sampled notes).
Table 6.
Distribution of number of errors per note type
| Note type | No. of error instances | No. of PII instances | No. of notes with at least one error | Total No. of notes | Fraction of notes with at least one error |
|---|---|---|---|---|---|
| Phone message/call | 60 | 7,466 | 54 | 605 | 0.09 |
| Ambulatory patient summary | 59 | 14,502 | 49 | 334 | 0.15 |
| Physician office/clinic message | 42 | 8,352 | 36 | 661 | 0.05 |
| Report | 50 | 3,173 | 36 | 131 | 0.27 |
| Medication renewal/refill | 36 | 4,626 | 31 | 358 | 0.09 |
| Progress note, family practice | 27 | 4,975 | 24 | 237 | 0.10 |
| Ambulatory discharge medication list | 27 | 8,109 | 23 | 226 | 0.10 |
| Anti-coag service visit summary | 24 | 1,189 | 22 | 26 | 0.85 |
| Electrocardiogram | 19 | 411 | 19 | 30 | 0.63 |
| Anticoagulation patient intake—text | 49 | 5,777 | 18 | 50 | 0.36 |
| Letter | 15 | 3,519 | 14 | 157 | 0.09 |
| Ambulatory depart summary | 12 | 3,938 | 12 | 163 | 0.07 |
| Progress notes | 14 | 3,943 | 11 | 199 | 0.06 |
| Telephone encounter | 12 | 2,034 | 11 | 273 | 0.04 |
The proportion of sampled notes for a given type that contain at least one error is presented in the last column. This indicates in which note type an error is more likely to occur.
Methods
Usage of Mayo clinic dataset
The Mayo EHR dataset is based on data from 477,000 patients that originated from multiple EHR data systems (including Epic and Cerner) spanning over 20 years. The dataset includes 104 million physician notes that capture the health care journey of patients in addition to structured tables containing lab test measurements, diagnosis information, orders, and medicine administration records. This research was conducted with approval from the Mayo Clinic Institutional Review Board.
We randomly sampled 10,000 notes, which were reduced to a set of unique sentences. This yielded a test set of 172,102 sentences. These were subsequently annotated by six Mayo Clinic nurse abstractors to create a ground truth label for every word and/or phrase. Each sentence was annotated by at least two different nurse abstractors. The interannotator agreement on labeling a token as PII had a Cohen's κ of 0.9694 (see supplemental methods and Table S5 for details).
An additional set of 10,000 notes was selected to fine-tune the models. We manually annotated 61,800 unique sentences from these notes to create a tagged fine-tuning set. See supplemental methods for more details.
Detection of PII entities
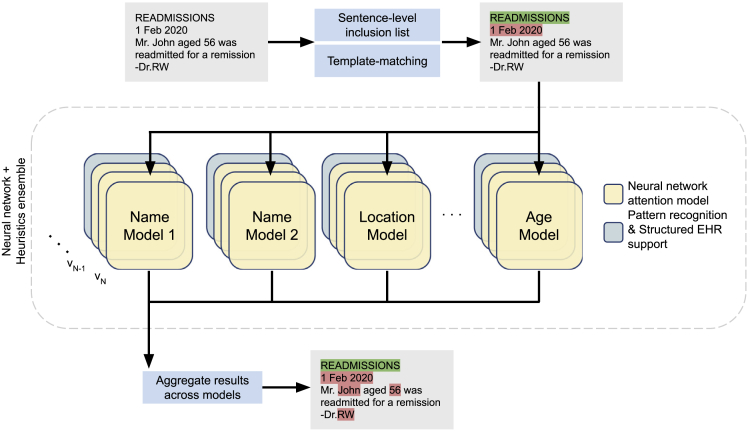
The ensemble architecture described in this section leverages state-of-the-art attention-based deep-learning models in conjunction with rules harvested from the data (each of which is described below) to handle semi-structured text (Figure 2). There are several salient features of this approach that are worth noting.
Figure 2.
Ensemble architecture for de-identification of unstructured text
Sentence-based inclusion lists and template matching prune out sentences that either (1) lack PII or (2) contain PII in specific well-defined patterns. An ensemble of attention-based neural networks identifies complementary features across different PII types. For each entity type, multiple model versions (v1, v2, …, vN) are used in tandem. In addition, pattern recognition modules and structured EHR content from matched patients support the anonymization process. The results from each component of the ensemble are aggregated to yield the original note labeled with PII tags.
Hybrid deep-learning models
The newer breed of attention-based deep-learning models, in conjunction with transfer learning, allow for faster tuning of these models with significantly smaller sets of labeled data for detecting PII identifiers. We use pre-trained language models based on the BERT16 architecture that are then fine-tuned for detecting (1) personal names, (2) organizations, (3) locations, and (4) ages. We employed the BERT-base-cased model (https://huggingface.co/bert-base-cased) through the HuggingFace/Transformers (https://github.com/huggingface/transformers) library. This is a case-sensitive English language pre-trained model based off of the BERT architecture trained using an MLM objective. The fine-tuning process involves training the pre-trained language model on an NER task using a training set of annotated sentences. We used a total of 61,800 tagged example sentences to fine-tune the models. We fine-tuned each transformer model with a maximum sequence length of 256 (after tokenization) over four epochs. We used a training batch size of 32 and a learning rate of 5 × 10−5 with a warm-up proportion of 0.4. We then evaluated the model on a validation dataset and computed the accuracy. We performed the fine-tuning and model validation processes in an iterative manner (see supplemental methods, Figure S1, and Table S1 for complete implementation details). Identifiers such as names, locations, organizations, and ages are well suited to a statistical entity recognition method because they can use the context of the surrounding text to disambiguate the entity type of a word. By contrast, pattern-matching rules are significantly hampered in this respect. It would be hard, for instance, to detect “Glasgow” as a medical term in “He had no helmet and his Glasgow score was 6” and as a location in “Mr. Smith had visited his family in Glasgow” using look-up dictionaries.
However, we use patterns to deterministically tag reasonably well-defined PII identifiers, which are almost entirely context independent and unambiguous. This category includes dates and times, phone and pager numbers, clinical IDs and numeric identifiers, email, URLs, IP addresses, and vehicle numbers. In addition, harvested sentence templates (described further below) are relied upon to deterministically tag PII instances matched by the template patterns. Our methods apply to content in both structured (e.g., lab comments) and free-form text (e.g., progress notes).
In addition, it should be noted that we designed our method to detect and transform information about those who provide care, such as physicians, nurses, and pharmacies. Although this is not required by HIPAA Safe Harbor, it allows health care organizations to protect the identities of their employees as well.
Ensemble of models framework and iterative fine-tuning
Given the regulatory necessity of extremely high recall for de-identification, we aggregate the results of multiple models trained for the same PII type. Our ensemble involved employing at least one individual model for names, organizations, locations, and ages (see Table S2). An additional text-normalized model was also trained and utilized for names. In this respect, if a term is detected as PII in any of the models for that type, then it is tagged. A divide-and-conquer approach has been implemented that harnesses the power of multiple models to identify PII or extract meaningful entities (Figure 2). In contrast to a “one size fits all” model, this approach enables each individual model to be fine-tuned to learn different (and complementary) features of the unstructured EHR data, as has been shown to be used in prior de-identification systems23. For instance, one model focuses on identifying peoples' names, while another is geared toward addresses and locations.
Furthermore, there are additional models corresponding to cased and uncased variants of the raw data (referred to as “Name Model 1” and “Name Model 2” in Figure 2). Each model here corresponds to an attention-based deep neural network. One advantage of carving out the entity space to be handled individually by separate models is that each model needs to learn only the distribution of entities of a specific type as opposed to all entities. However, this introduces a challenge in resolving terms in a sentence that have conflicting and/or ambiguous entity types. These conflicts are resolved in the aggregation phase of our ensemble, where a simple voting threshold of one claim is employed (i.e., an entity is considered PII even if one model in the system tags it as such). Since the majority of the components in the ensemble are designed to detect complementary features, we are able to improve recall without much loss of precision.
Integrating databases as part of the core model
We use publicly available databases of names, locations, and addresses to supplement the model fine-tuning process. First names with supporting gender information were downloaded from the US Census database. Cities across the United States as well as lists of hospitals were obtained from Wikipedia. These public databases were used to augment training of our models. In addition, patient-specific information from structured EHRs, including patient names and residential addresses, are used to augment the model training and match against PII in the text.
Sentence-based inclusion list
Clinical note corpora contain a large number of repeated sentences. These stem from various processes, including automated reminders (e.g., “Please let your doctor know if you have problems taking your medications”), repeated phrases in the writing style of physicians (e.g., “Rubella: yes”; “Pain symptoms: no”), or shared elements in the clinical notes, such as section headers (e.g., “History of Present Illness”). From the corpus of physician notes from the Mayo Clinic, a set of 1,600 sentences, which did not contain PII, was incorporated into an “inclusion list.” This inclusion list was further expanded with a set of 25,000 sentences containing medically relevant entities, such as disease or drug names (see supplemental methods for details on how the inclusion list was constructed). This has the added benefit of improving the precision of the de-identification system because it reduces the risk of misclassifying these important entities as PII by the neural network models. In addition, sentences marked as being devoid of PII during the validation phase in the iterative fine-tuning process were also added to the inclusion list (see supplemental methods).
Autogenerating templates using statistical NER models
In addition to exact sentences with high prevalence there are also a large number of PII-containing sentences that can be mapped to a template (e.g., “Electronically signed by: SMITH, JOHN C on 01/02/1980 at 12:12 PM CST” maps to a template of the form “Electronically signed by: <LAST NAME>, <FIRST NAME> <INITIAL> on <DATE> at <TIME>”). While machine-learning NER models can be trained and/or fine-tuned to learn these patterns, there are instances where entity recognition fails. So, although a name of the form “SMITH, JOHN C” might be detected, “DEWEY” in “DEWEY, JONES K” may not be detected. By contrast, regular expression rules faithfully match every PII for these cases.
The problem, however, is that the process of identifying such templates and generating the corresponding regular expressions is an arduous task because it involves manual inspection of a sufficiently large sample of sentences in the corpus. Here, we use the NER ensemble models designed for the detection of PII to aid in the harvesting of these pattern templates. Sentences from our fine-tuning set of 10,000 notes are passed through the ensemble, and detected PII is transformed to its corresponding IOB2 (inside-outside-beginning) mask (e.g., “Electronically signed by: B-PER I-PER I-PER on B-DATE at B-TIME PM CST”), generating a potential NER template. In addition, a “syntax template” for these sentences is also generated, such that any term that was detected as an entity is mapped to its syntactic representation—one of “W” for alphabets only, “N” for numbers only, and “A” for alphanumeric (e.g., “Electronically signed by: W, W W on N/N/N at N:N PM CST”). Finally, for each unique syntax template, if there exists only one NER template among all instances of the syntax template, a regular expression rule is generated (e.g., “Electronically signed by: [A-Za-z]+, [A-Za-z]+ [A-Za-z]+ on \d+/\d+/\d + at \d+:\d + PM CST”) by mapping each syntax token to its corresponding regular expression pattern: “W” to “[A-Za-z]+”, “N” to “\d+”, and “A” to “\w”.
Transformation of tagged PII entities
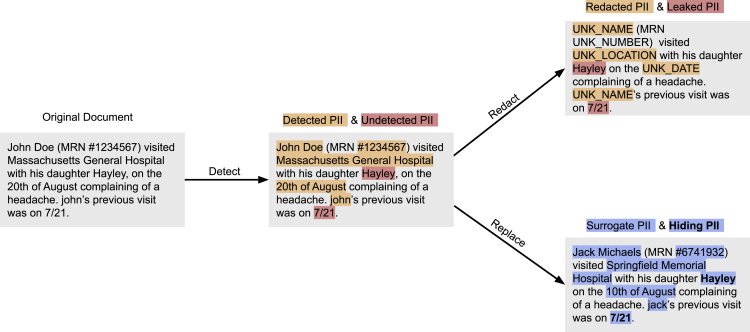
The de-identification process is designed to recognize words and phrases that represent PII and other sensitive elements with high recall. However, if the input text is transformed to the de-identified version by redacting detected PII, undetected PII (e.g., “Hayley” and the date “7/21” in Figure 3) is obviously leaked to any person who reads the document. As such, the obfuscation process aims to conceal these residual PII by replacing detected PII with suitable surrogates so it is difficult to distinguish between the residual PII and the surrogates.21,24,25 This method has been implemented in several de-identification approaches.26,27 As highlighted in Figure 3, it is difficult for a human to determine which of “Jack Michaels” or “Hayley” is a leaked instance of PII in the output of the replacement strategy using this mechanism of hiding in plain sight (HIPS).28 Evidence with human readers has shown that when the recall of a NLP tool is high (i.e., when most real identifiers are detected), the rate of distinguishing real from filler identifiers is no better than what one would encounter by random chance. It has further been shown, however, that under highly controlled conditions, it is possible for a machine-learning system to replicate the behavior of the natural language de-identification tool to remove fillers and leave real identifiers in place.28,29
Figure 3.
Obfuscation of tagged PII entities
An illustration of the hiding in plain sight mechanism to highlight the utility of the detect → replace strategy. After obfuscation, distinguishing real PII from surrogates is no better than what one would expect by random chance. PII, personally identifiable information.
In addition to employing the HIPS method, we apply entity-specific rules and heuristics to improve the fidelity of the surrogate. We further improve interpretability of the output by ensuring that every unique PII token in all EHR records for a patient has the same transformation.
Consider the input text “John Smith, a pleasant 67 year old presented with his son Jack. John complains of breathing difficulty,” which was transformed to “Jane Kate, a pleasant 67 year old presented with his son Matt. Ryan complains of breathing difficulty.” In this example, “Jane Kate” as a surrogate is an obvious giveaway that it is a fake name and therefore lends itself to be distinguished from any true PII that may have leaked. Furthermore, it appears that a third completely different person is complaining of breathing difficulty. So an ideal transformation would have maintained the format of first name followed by last name, and the gender for “John Smith” and every instance of “John” or “Smith” in the input would be transformed to the same output; something like “Jacob Hamilton, a pleasant 67 year old presented with his son David. Jacob complains of breathing difficulty.”
As discussed, we manage the replacement of surrogates per entity type (see Table S2). Names are transformed in a manner that is consistent with format, gender, and ethnicity of the original (i.e., “Ms. Lopez visited New York General Hospital for her routine checkup” becomes “Ms. Hernandez visited Mass General Hospital for her routine checkup”). Dates are handled in a way to preserve their formatting (i.e., “March 5th, 2014” becomes “February 27th, 2014” and “03-05-2014” becomes “02-27-2014”). The shift in the date is a patient-specific random number. This ensures that dates are shifted consistently for a given patient. Locations and organizations are replaced with suitable surrogates chosen from a predefined dictionary. PII entities that contain numeric digits (such as phone number or patient ID) involve replacing these numbers randomly while maintaining overall length and format.
While the transformation output of an input token is the same for all instances of its occurrence for a given patient, it would be different across patients. That is, while all instances of “John” in one patient might be transformed to “Jacob,” for another patient it could be “Aaron.”
Discussion
Numerous approaches to de-identification have been developed. Automated de-identification systems can broadly be segmented into four categories: (1) rule-based systems, (2) traditional machine-learning systems, (3) deep-learning systems, and (4) hybrid and ensemble systems.
Rule-based systems8,19,20,30,31 use pattern matching rules, regular expressions, and dictionary and public database look-ups to identify PII elements. These are simple to implement and usually deterministic; however, these systems have several drawbacks. First, pattern-matching rules for identifiers are typically not robust for handling variance in input due to typographical errors (spelling, punctuation, casing, etc.). A rule that matches “Dr. John” may not be able to match “Dr john.” Second, creating template patterns to match sentence fragments like “Provider Name: Dr. John” that tag any term after “Provider Name: Dr.” as a name, for example, requires manual effort to understand the data to create these templates. Doing this for large datasets with notes for millions of patients is time consuming and intractable. Third, dictionary-based systems may not be complete, resulting in increased false negatives (i.e., true PII that is not detected). Fourth, blindly using dictionary/database look-ups induces false positives because they tag phrases that are not identifiers in the context in which they are used that need to be disambiguated.32 For example, in “The doctor determined his Braden score as normal”, the term “Braden” might be flagged as PII, when it is only a clinical term.
Traditional machine-learning systems21,33, 34, 35 use traditional machine-learning algorithms, such as support vector machines and CRFs, to perform NER classification as PII for each word in a sentence. The classification task involves creating labeled data and defining features based on properties like part-of-speech tags, typography (e.g., capitalization, casing, spacing, font weights, or font types), punctuation, and frequency of words and/or their neighbors. These methods, in addition to requiring significant effort in encoding the feature vectors, may not generalize across datasets.
Deep-learning systems18 have become the state of the art for a wide variety of application domains, including vision (e.g., image classification) and speech (e.g., voice recognition and generation). In language-related tasks (e.g., machine translation), these approaches have surpassed human-level performance.36 Deep learning has proven beneficial in numerous NLP tasks, including predicting the next word (language modeling), tagging tasks such as part of speech tags, entities in a sentence (entity recognition), and dependency parsing. This has enabled applications that traditionally required custom rules and hand-crafted features to be solved without any feature engineering. Modern deep-learning approaches for de-identification have been shown to outperform their predecessors,18 but they require very large quantities of domain-specific labeled training data to perform well. Specifically, the challenges include, but are not limited to, the presence of long and highly descriptive sentences, usage of clinical shorthand (that varies across physicians and medical specialties), and a variety of semi-structured machine-generated content. Moreover, publicly available datasets for de-identification (including the popular i2b2 2014 dataset)17 lack diversity, often focusing on only a few types of notes or areas of disease. Training and benchmarking with such datasets are likely to bias the resulting models and fail to capture the nuanced and complex nature of physician notes. Recently, attention-based neural network (transformer) models have also been implemented for de-identification but have shown limited generalizability in the absence of support from encoded rules.37
Hybrid38 and ensemble systems39,40 use combinations of rule-based and machine-learning-based components in tandem to improve PII detection efficacy. With these approaches, the choice of components, finding the right split of tasks between them, and the optimal strategy for combining results from them become crucial. Some approaches41 invoke engineering post-processing layers that fix the errors that are introduced by other (earlier) components. In cases where there is, by design, overlap in the type of PII being predicted (e.g., multiple components detecting people’s names), considerable effort is spent measuring and choosing a method, like a stacked meta classifier or voting scheme, to pick a winning component.39 The nference de-identification system presented here addresses the limitations of prior methods11 and achieves high levels of recall and precision.
There are several opportunities to further improve the performance of de-identification systems. First, existing knowledge graphs and language models trained on biomedical corpora can be leveraged. For example, if a patient's note contains the sentences “Patient diagnosed with lung cancer” and “ECOG performance status was determined to be 2,” ECOG would not be treated as PII, since it has a strong biological association with lung cancer based on the knowledge graph. In the de-identification process, this could be used to recover biological terms incorrectly tagged as PII (false positives). Second, the quality of sentences that are provided to the model can be improved. Unstructured clinical text does not always contain well-formatted text, commonly due to missing punctuation and incorrect casing. A case-sensitive pre-trained model along with an MLM objective can be used to train a system capable of correctly introducing punctuation in the right location. Another challenge with the quality of clinical documents is the prevalence of short fragments and bullet points, giving rise to sentences with poor context. The context of a single sentence can be expanded using preceding and succeeding sentences or employing document-level transformer models such as Transformer-XL.42 Third, unsupervised methods can be incorporated to accelerate the annotation process of the NER task. Grouping the word representations generated by a transformer model yields informative clusters (e.g., a cluster of names) that can be annotated according to the nature of words present in the cluster. The NER task can then be formulated as a masked language task, where the overlap of the list of potential candidates for a missing word with the clusters can inform the entity type of the missing word.
Concluding remarks
Overall, this work implemented an ensemble approach to de-identification of unstructured EHR data incorporating transformer models supported by heuristics for automatically identifying PII across diverse clinical note types. Upon detection, suitable surrogates replaced PII in the processed text, thereby concealing residual identifiers (HIPS). The system demonstrates high precision and recall on both publicly available datasets and a large and diverse dataset from the Mayo Clinic.
Experimental procedures
Resource availability
Lead contact
Sankar Ardhanari is the lead contact for this study and can be reached at sankar@nference.net.
Materials availability
There are no physical materials associated with this study.
Data and code availability
The 2014 i2b2 data are publicly available subject to signed safe usage and for research only. The Mayo EHR clinical notes are not publicly available at this time. The source code is not currently available, but the nference de-identification tool can be accessed at https://academia.nferx.com/deid/.
Acknowledgments
We would like to thank the Mayo Clinic and the Mayo Clinic IRB, under whose auspices the development of the de-identification methods and testing against real-world datasets were made possible. We thank the nurse abstractors—Wendy Gay, Kathy Richmond, Denise Herman, Sandra Severson, Dawn Pereda, and Jane Emerson—for annotating the ground truth for the 172,102 sentences in the Mayo dataset that was used for testing the performance of the system; the Mayo Data Team of Ahmed Hadad, Connie Nehls, and Salena Tong for preparing and helping us understand the Mayo EHR data; and Andy Danielsen for supporting the collaboration. Finally, we thank Murali Aravamudan, Rakesh Barve, and A.J. Venkatakrishnan for their thoughtful review and feedback on the manuscript.
Author contributions
Conceptualization, K.M., A.R., and S.A.; methodology, K.M., A.R., V.A., and S.A.; validation, K.M., B.M., V.A., S.B., J.A., J.R., and S.A.; formal analysis, K.M., B.M., and S.A.; data curation, K.M., V.A., and S.B.; writing – original draft, K.M. and A.R.; writing – review & editing, B.M., J.A., J.R., W.F., J.H., V.S., and S.A.; supervision, V.S. and S.A.; resources, V.S. and S.A.
Declaration of interests
J.A., J.H., and W.F. do not have any competing interests in this project. B.M. is a contracted consultant of the Mayo Clinic. The authors on this article from nference have equity in nference and have a financial interest in nference. A patent application has been submitted by K.M., A.R., and S.A. Mayo Clinic and nference may stand to gain financially from the successful outcome of the research.
Published: May 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100255.
Contributor Information
Venky Soundararajan, Email: venky@nference.net.
Sankar Ardhanari, Email: sankar@nference.net.
Supplemental information
References
- 1.Wagner T., Shweta F., Murugadoss K., Awasthi S., Venkatakrishnan A.J., Bade S., Puranik A., Kang M., Pickering B.W., O’Horo J.C. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. Elife. 2020;9:e58227. doi: 10.7554/eLife.58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iqbal E., Mallah R., Rhodes D., Wu H., Romero A., Chang N., Dzahini O., Pandey C., Broadbent M., Stewart R. ADEPt, a semantically-enriched pipeline for extracting adverse drug events from free-text electronic health records. PLoS One. 2017;12:e0187121. doi: 10.1371/journal.pone.0187121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K., LePendu P., Chen W.S., Iyer S.V., Readhead B., Dudley J.T., Shah N.H. Automated detection of off-label drug use. PLoS One. 2014;9:e89324. doi: 10.1371/journal.pone.0089324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzal N., Sohn S., Scott C.G., Liu H., Kullo I.J., Arruda-Olson A.M. Surveillance of Peripheral Arterial Disease cases using natural language processing of clinical notes. AMIA Jt. Summits Transl Sci. Proc. 2017;2017:28–36. [PMC free article] [PubMed] [Google Scholar]
- 5.Finlayson S.G., LePendu P., Shah N.H. Building the graph of medicine from millions of clinical narratives. Sci. Data. 2014;1:140032. doi: 10.1038/sdata.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Office for Civil Rights H.H.S. Standards for privacy of individually identifiable health information. Final rule. Fed. Regist. 2002;67:53181–53273. [PubMed] [Google Scholar]
- 7.Berg, H., Henriksson, A., and Dalianis, H. (2020). The Impact of De-identification on Downstream Named Entity Recognition in Clinical Text. Proceedings of the 11th International Workshop on Health Text Mining and Information Analysis.
- 8.Neamatullah I., Douglass M.M., Lehman L.-W.H., Reisner A., Villarroel M., Long W.J., Szolovits P., Moody G.B., Mark R.G., Clifford G.D. Automated de-identification of free-text medical records. BMC Med. Inform. Decis. Mak. 2008;8:32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglass M., Clifford G.D., Reisner A., Moody G.B., Mark R.G. Computer-assisted de-identification of free text in the MIMIC II database. Computers in Cardiology. 2004:341–344. doi: 10.1109/CIC.2004.1442942. [DOI] [Google Scholar]
- 10.Leevy J.L., Khoshgoftaar T.M., Villanustre F. Survey on RNN and CRF models for de-identification of medical free text. J. Big Data. 2020;7:73. [Google Scholar]
- 11.Yogarajan V., Pfahringer B., Mayo M. A review of automatic end-to-end de-identification: is high accuracy the only metric? Appl. Artif. Intelligence. 2020;34:251–269. [Google Scholar]
- 12.Yang Z., Dai Z., Yang Y., Carbonell J., Salakhutdinov R.R., Le Q. Advances in Neural Information Processing Systems. arXiv; 2019. XLNet: generalized autoregressive pretraining for language understanding. 1906.08237. [Google Scholar]
- 13.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Advances in Neural Information Processing Systems. arXiv; 2017. Attention is all you need. 1706.03762. [Google Scholar]
- 14.Sennrich, R., Haddow, B., and Birch, A. (2016). Neural Machine Translation of Rare Words with Subword Units. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers).
- 15.Kudo, T., and Richardson, J. (2018). SentencePiece: A simple and language independent subword tokenizer and detokenizer for Neural Text Processing. Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing: System Demonstrations.
- 16.Devlin J., Chang M., Lee K., Toutanova K. Association for Computational Linguistics); 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding; pp. 4171–4186. [Google Scholar]
- 17.Stubbs A., Uzuner Ö. Annotating longitudinal clinical narratives for de-identification: the 2014 i2b2/UTHealth corpus. J. Biomed. Inform. 2015;58:S20–S29. doi: 10.1016/j.jbi.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dernoncourt F., Lee J.Y., Uzuner O., Szolovits P. De-identification of patient notes with recurrent neural networks. J. Am. Med. Inform. Assoc. 2017;24:596–606. doi: 10.1093/jamia/ocw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry A.J., Fitch B., Savova G., Kohane I.S., Reis B.Y. Improved de-identification of physician notes through integrative modeling of both public and private medical text. BMC Med. Inform. Decis. Mak. 2013;13:112. doi: 10.1186/1472-6947-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norgeot B., Muenzen K., Peterson T.A., Fan X., Glicksberg B.S., Schenk G., Rutenberg E., Oskotsky B., Sirota M., Yazdany J. Protected Health Information filter (Philter): accurately and securely de-identifying free-text clinical notes. NPJ Digit Med. 2020;3:57. doi: 10.1038/s41746-020-0258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aberdeen J., Bayer S., Yeniterzi R., Wellner B., Clark C., Hanauer D., Malin B., Hirschman L. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int. J. Med. Inform. 2010;79:849–859. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Dernoncourt F., Lee J.Y., Szolovits P. NeuroNER: an easy-to-use program for named-entity recognition based on neural networks. arXiv. 2017 1705.05487. [Google Scholar]
- 23.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc. AMIA Annu. Fall Symp. 1996:333–337. [PMC free article] [PubMed] [Google Scholar]
- 24.Yeniterzi R., Aberdeen J., Bayer S., Wellner B., Hirschman L., Malin B. Effects of personal identifier resynthesis on clinical text de-identification. J. Am. Med. Inform. Assoc. 2010;17:159–168. doi: 10.1136/jamia.2009.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meystre S., Shen S., Hofmann D., Gundlapalli A. Can physicians recognize their own patients in de-identified notes? Stud. Health Technol. Inform. 2014;205:778–782. [PubMed] [Google Scholar]
- 26.Heider P.M., Obeid J.S., Meystre S.M. A comparative analysis of speed and accuracy for three off-the-shelf de-identification tools. AMIA Jt. Summits Transl Sci. Proc. 2020;2020:241–250. [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrández O., South B.R., Shen S., Friedlin F.J., Samore M.H., Meystre S.M. BoB, a best-of-breed automated text de-identification system for VHA clinical documents. J. Am. Med. Inform. Assoc. 2013;20:77–83. doi: 10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrell D., Malin B., Aberdeen J., Bayer S., Clark C., Wellner B., Hirschman L. Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text. J. Am. Med. Inform. Assoc. 2013;20:342–348. doi: 10.1136/amiajnl-2012-001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrell D., Cronkite D.J., Li M., Nyemba S., Malin B., Aberdeen J., Hirschman L. The machine giveth and the machine taketh away: a parrot attack on clinical text deidentified with hiding in plain sight. J. Am. Med. Inform. Assoc. 2019;26:1536–1544. doi: 10.1093/jamia/ocz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison F.P., Li L., Lai A.M., Hripcsak G. Repurposing the clinical record: can an existing natural language processing system de-identify clinical notes? J. Am. Med. Inform. Assoc. 2009;16:37–39. doi: 10.1197/jamia.M2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzuner O., Luo Y., Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J. Am. Med. Inform. Assoc. 2007;14:550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruch P., Baud R.H., Rassinoux A.M., Bouillon P., Robert G. Medical document anonymization with a semantic lexicon. Proc. AMIA Symp. 2000:729–733. [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrández O., South B.R., Shen S., Friedlin F.J., Samore M.H., Meystre S.M. Evaluating current automatic de-identification methods with Veteran’s health administration clinical documents. BMC Med. Res. Methodol. 2012;12:109. doi: 10.1186/1471-2288-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meystre S.M., Friedlin F.J., South B.R., Shen S., Samore M.H. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med. Res. Methodol. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M., Scaiano M., El Emam K., Malin B. Efficient Active learning for electronic medical record de-identification. AMIA Jt. Summits Transl Sci. Proc. 2019;2019:462–471. [PMC free article] [PubMed] [Google Scholar]
- 36.Popel M., Tomkova M., Tomek J., Kaiser Ł., Uszkoreit J., Bojar O., Žabokrtský Z. Transforming machine translation: a deep learning system reaches news translation quality comparable to human professionals. Nat. Commun. 2020;11:4381. doi: 10.1038/s41467-020-18073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, A.E.W., Bulgarelli, L., and Pollard, T.J. (2020). Deidentification of free-text medical records using pre-trained bidirectional transformers. In Proceedings of the ACM Conference on Health, Inference, and Learning, (New York, NY, USA: Association for Computing Machinery), pp. 214–221. [DOI] [PMC free article] [PubMed]
- 38.Liu Z., Chen Y., Tang B., Wang X., Chen Q., Li H., Wang J., Deng Q., Zhu S. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. J. Biomed. Inform. 2015;58(Suppl):S47–S52. doi: 10.1016/j.jbi.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y., Heider P., Meystre S.M. Ensemble-based methods to improve de-identification of electronic health record narratives. AMIA Annu. Symp. Proc. 2018;2018:663–672. [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y., Meystre S.M. Ensemble method-based extraction of medication and related information from clinical texts. J. Am. Med. Inform. Assoc. 2020;27:31–38. doi: 10.1093/jamia/ocz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H.-J., Wu Y., Zhang Y., Xu J., Xu H., Roberts K. A hybrid approach to automatic de-identification of psychiatric notes. J. Biomed. Inform. 2017;75S:S19–S27. doi: 10.1016/j.jbi.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai, Z., Yang, Z., Yang, Y., Carbonell, J., Le, Q., and Salakhutdinov, R. (2019). Transformer-XL: Attentive Language Models beyond a Fixed-Length Context. Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 2014 i2b2 data are publicly available subject to signed safe usage and for research only. The Mayo EHR clinical notes are not publicly available at this time. The source code is not currently available, but the nference de-identification tool can be accessed at https://academia.nferx.com/deid/.