Abstract
Flower development exists as a key period in the angiosperms life cycle and the proper development is considered with its reproductive success. Pistil abortion is one of the widely distributed aspects of berry plants and its basic mechanism in Japanese apricot is quite unclear and needs thorough investigation. The present study was carried out to get a deep insight into the pistil abortion mechanism in Japanese apricot using a transcriptomic approach. A large number of DEGs were identified from different development stages of normal and abortive pistils. Pair-wise comparison analysis was performed as LY1 vs DQD1, LY2 vs DQD2, and LY3 vs DQD3 and produced 3590, 2085, and 2286 transcripts, respectively. The Gene Ontology (GO) showed that different metabolic processes, plant hormones, developmental processes, and photosystem-related genes were involved in pistil abortion. The pathway analysis revealed significant enrichment of plant hormone's signal transduction and circadian rhythm pathways. Furthermore, transcription factors such as MYB, MADS-box, and NAC family showed lower expression in abortive pistils. The current study presents a new strategy for advanced research and understanding of the pistil abortion process in Japanese apricot and provides a possible reference for other deciduous fruit trees.
Keywords: Japanese apricot, Transcriptome, Pistil abortion, Plant hormone signal transduction, Gene expression
Introduction
Flower development is a key period for the reproductive success of angiosperm (Andrés and Coupland 2012; Huang, 2013). Pistil abortion is an extensive phenomenon that mostly occurs in fruit plants; regarding this, reports include Arabidopsis thaliana (Robinson-Beers et al. 1992), olive (Reale, 2009), Prunus armeniac (Lillecrapp et al. 1999), Xantoceras sorbifolia (Gao et al. 2002), and pomegranate (Chen, 2017). In Pongamia pinnata, only a few flowers and ovules begin to form fruits and seeds due to the reduced fertilization (Arathi et al. 1999). Several studies related to infertility and pistil abortion have shown that different environmental factors, such as light and temperature (Beppu and Kataoka 2011; Zinn et al. 2010), pathogenic effect (Kocsis and Jakab 2008), genetic factors (Causier et al. 2003; Peng et al. 2008), and phytohormones, such as auxin and gibberellin, just to mention a couple of examples (Kumar et al. 2011; Lim et al. 2010) are largely involved.
Japanese apricot (Prunus mume Sieb. et Zucc) is a prominent berry as well as an ornamental plant of the Rosaceae family, which originated from China about three centuries ago. Japanese apricot is regarded as famous deciduous fruit because of its economic importance, eye-catching red-skinned color, and used in value-added products (Ni et al. 2018; Sun et al. 2013; Wu et al. 2019). Two flower categories have been described in the Japanese apricot: perfect and imperfect. In general, the perfect flowers retain a proper pistil, while imperfect ones may exhibit the absence of the pistil, an aborted pistil, or pistils below the stamens, features that seriously affect fertility and subsequently leads to significant yield reduction (Gao et al. 2006; Hou et al. 2011). Earlier studies in 'Daqiandi' and 'Longyan' prominent cultivars of J. apricot, reported 76% and 5%, respectively, of imperfect flower occurrence (Gao et al. 2012). Moreover, morphological studies show that early December is the critical period for pistil development in 'Daqiandi', where instead of pistil differentiation and elongation, pistil abortion occurs (Shi et al. 2012). Several comparative proteomic analysis of perfect and imperfect flowers have been carried out and results indicate that besides flowering development associated genes, different metabolic pathways, involving starch, glucose, and photosynthesis are involved in pistil abortion (Wang 2008).
Transcriptome sequencing (RNA-seq) is a powerful tool aimed at genome-wide expression analysis, enabling elucidate the molecular mechanisms and defining putative gene functions (Jain 2011; Shi et al. 2012; Wang et al. 2009). Transcriptome sequencing related to flower bud development has been applied in litchi (Lu et al. 2014), P. pseudocerasus (Zhu et al. 2015), and pear (Bai et al. 2013). Recently, in Arabidopsis and other species, several key flowering regulators have been identified using extensive floral transcriptome analyses (Vining, 2015; Zhang, 2014). Various MADS-box family genes (Becker and Theißen 2003; Ó'Maoiléidigh et al. 2014), ABCDE model (Pelaz et al. 2000; Theissen and Melzer 2007), and some transcription factors (TFs) are involved in different floral parts and developmental stages. Several plant hormones, such as auxin (Cecchetti et al. 2008), gibberellin (Cheng, 2004), cytokinin (Han et al. 2014), brassinosteroids (Ye et al. 2010), and jasmonate (Yuan and Zhang 2015), have also been reportedly found to be involved in flower development, regulation, and fertility. "Gibberellins (GA)" stimulates cell growth by stimulating the destruction of growth-inhibiting DELLA proteins (Achard, 2009), while higher amounts of jasmonate cause sterility issues (Yuan and Zhang 2015). Therefore, in light of the above consequences, the present study was designed to elucidate pistil abortion mechanism based on high throughput transcriptome sequencing to identify key genes, analyze their expression profile, and enrich pathways involved in this phenomenon. Furthermore, we also analyzed various hormone contents at different developmental stages of normal and abortive pistils. Our results proposed a comprehensive regulation pattern of the molecular mechanism of pistil abortion and provide a novel insight aimed at future studies related to pistil abortion and sterility problems in fruit plants.
Materials and methods
Plant material
The Japanese apricot (P. mume) flower buds cv. ‘Daqiandi (Abortive pistils)’ and ‘Longyan (Normal pistils)’ were collected from the National Field Gene-Bank for Japanese apricot Nanjing, China from mid-November to mid-February with a one-week interval. Three key stages of pistil abortion (cv. Daqiandi pistils) i.e. stigma browning (DQD1), style browning (DQD2), and ovary browning (DQD3) were selected. At the same time, three parts of normal pistils i.e. stigma (LY1), style (LY2), and ovary (LY3) were also collected for stage-to-stage comparison. Each sample was collected in three biological replicates. Pistils were excised from the flower buds and used as experimental material, instantly frozen in liquid nitrogen, and kept at −80 °C for further analyses.
Library preparation and transcriptome sequencing
Total RNA was extracted from normal and abortive pistils at different developmental stages mentioned above followed the method previously described (Wu et al. 2019). Samples (1.0 g) were powdered in liquid nitrogen and added 5 mL of pre-cooled 80% chromatographic methanol. The liquid (3 mL) was transferred into another 5 mL tube and added 2 mL chromatographic methanol, rinsed well, and transferred into a new 5 mL centrifuge tube. After that, samples were purified from genomic DNA using RNase-free DNase I (TaKaRa, Japan), and RNA quality was checked by electrophoresis on 1% gel. Sequencing libraries were prepared using TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA) followed the company’s procedure. Briefly, Oligo-dT beads (Qiagen, Valencia, USA) were used to enrich mRNA. The mRNA fragments were transformed into short fragments using fragmentation buffer and then reverse transcribed into cDNA with random primers. Second strand cDNA was synthesized through DNA polymerase I, RNase H, and dNTPs. Then, the cDNA fragment was purified by Qiaquick PCR Purification Kits (Qiagen, Valencia, USA). The purified cDNA fragments were end-repaired, added to poly (A), and ligated to Illumina sequencing adapter. The selected fragments were amplified by Illumina HiSeq 4000 (BGI, Beijing, China) was used for pair-end sequencing. Eighteen libraries including three biological replicates of each sample were constructed for transcriptome sequencing (File S1).
Transcriptome sequencing, reads mapping, and quality control
After sequencing, millions of raw reads (FASTQ format) were obtained and filtered to eliminate low-quality reads and adopter sequences using the software SOAPnuke (v 1.4.0). The high-quality reads were mapped to P. mume genome using HISAT (v 2.1.0) (Kim et al. 2015) and the gene expression level was determined using RSEM (v 1.2.8) (Li and Dewey 2011). The complete flow chart for transcriptome analysis is shown in Fig S1.
Identification of differently expressed genes (DEGs) and functional enrichment analysis
DEGs analysis was carried out with an R package software DEGseq (Wang et al. 2009), and the gene expression level was calculated through the FPKM method. Due to large number of DEGs, the regulation of genes was determined by an absolute log2 fold change of ≥ + 1.5 (up-regulation) and ≤ −1.5 (down-regulation) and the p-value was less than 0.001. The threshold p-values were corrected to q-values by Benjamini et al., and the significance of differently expressed genes was determined through the FDR threshold ≤ 0.001 with a fold change of ≥ 2 (Benjamini and Yekutieli 2001).
Gene Ontology (GO) (Young et al. 2010) enrichment analysis was performed to classify the genes in the terms such as biological process (BP), cellular component (CC), and molecular function (MF) using the phyper function in R software. For pathway enrichment analysis, DEGs were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) with a p-value ≤ 0.05 accomplished standard of Bonferroni method (Bland and Altman 1995) to identify the significantly enriched pathway.
Plant transcription factor (TFs) prediction
To identify the potential transcription factors, all the identified DEGs were mapped to Plant Transcription Factor Database (PlantTFDB) using the HMMER (v 3.0) program and compared with Pfam 23.0 to find the protein domains encoding a transcription factor.
DEGs validation by RT-qPCR
Twelve genes were selected to verify the transcriptome sequencing results through Real-time quantitative PCR (RT-qPCR). Primers were designed using Beacon Designer software (Premier Biosoft, version 7.0). The RT-qPCR was performed according to the method followed by Guo and Iqbal (Guo et al. 2018; Iqbal et al. 2020a; Shi, 2020). Using RPII as a reference internal genes, each sample was taken thrice for biological repeat and the expression was calculated using the 2−△△CT method (Livak and Schmittgen 2001).
Measurement of endogenous hormones content
The hormone contents were determined using Liquid chromatography-Mass Spectrometry (LC–MS), USA as the method described by Balcke and Owen (Balcke, 2012; Owen and Abrams 2009). The 0.5–1.0 g of the samples identical ones to the transcriptome sequencing, powdered in liquid nitrogen, and added to 5 mL of pre-cooled 80% methanol. Thereafter, these were rinsed with 3 mL and 2 mL methanol respectively, transferred to 50 mL centrifuge tube placed in ice and put in dark for 12 h at 4 °C for leaching. Then, the samples were centrifuged at 10,000 × g for 10 min, the supernatant solution was transferred into a 50 mL centrifuge tube placed at 4 °C in darkness. After that, 5 mL pre-cooled 80% methanol was added to centrifuged residue, and put in dark for 12 h at 4 °C for leaching. Then, centrifugation was performed at 10,000 × g for 10 min, supernatant was collected and mixed with supernatant collected for the first time, and sample containing tubes were placed in ice box and shaken at 100 rpm/min in dark for 1 h. After that, samples were centrifuge at 10,000 × g for 10 min, and the supernatant solution was poured through the C18 SPE column, and the outflow phase solution was collected in new 50 mL centrifuge tubes. Then, the tubes were covered with preservatives, a precise hole was made in the middle using a toothpick, quickly freeze-dried in liquid nitrogen, and then transferred to a freezing dryer for more than 36 h. Thereafter, 1 mL pre-cooled methanol was added to fully dissolve the freeze-dried powder samples. Finally, the sample liquids were aspirated with a 2.5 mL syringe and passed through a 0.45 um organic ultrafiltration membrane, and different hormone contents were determined. Three technical repeats were performed for each biological repeat for hormone measurement.
Statistical analysis
The average of the values was calculated using Microsoft Excel (MS Office, 2016), and the graphs were prepared using the software Origin(Pro) 2019, and the data are shown as Mean ± SD.
Results
Transcriptome analysis overview
In the present study, a total of 379 million raw reads were obtained and filtered. About 373.4 million clean reads with a rate of 98.5% were achieved and mapped to the Prunus mume reference genome using HISAT (v 2.1.0) (Kim et al. 2015). As a result of the mapping, an average of 85.5% reads were mapped, from which 62.7% reads were uniquely mapped (Table 1; File S1).
Table 1.
Summary of transcriptome sequencing data in Japanese Apricot
| Type | LY1 | LY2 | LY3 | DQD1 | DQD2 | DQD3 |
|---|---|---|---|---|---|---|
| Total raw reads (M) | 61.0 | 62.5 | 62.7 | 64.2 | 63.9 | 64.8 |
| Total clean reads (M) | 60.6 | 61.2 | 61.7 | 63.5 | 63.5 | 62.8 |
| Total clean reads Q20 (%) | 97.2 | 97.6 | 97.5 | 97.2 | 97.1 | 97.6 |
| Total clean reads Q30 (%) | 92.6 | 93.4 | 93.4 | 89.4 | 89.1 | 90.1 |
| Total mapped reads (%) | 87.7 | 86.4 | 86.5 | 82.5 | 83.2 | 86.8 |
| Total unique mapped reads (%) | 64.2 | 63.4 | 62.0 | 60.0 | 60.6 | 65.9 |
Differentially expressed genes (DEGs) analysis
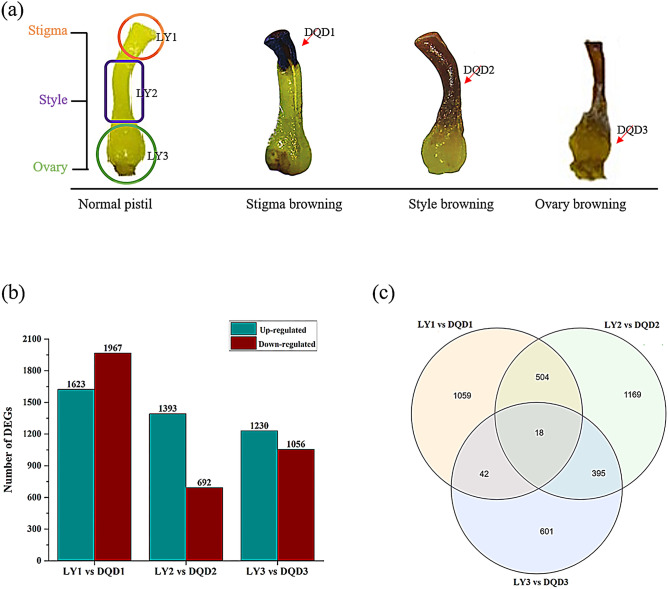
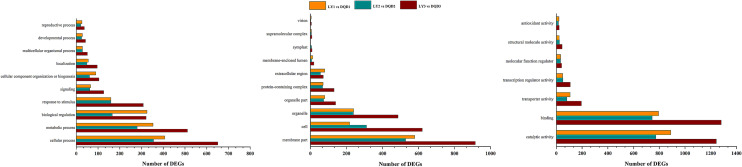
To understand the transcriptome sequence results, we selected abortive pistils from three developmental stages and compared with the same developmental stages of normal pistils (Fig. 1a) and their expression level were calculated using FPKM method. Based on differential expression, the comparison group “LY1 vs DQD1” produced 3590 genes (1623 up-regulated and 1967 down-regulated), while “LY2 vs DQD2” comparison group produced 2085 genes (1393 up-regulated and 692 down-regulated), whereas the comparison group "LY3 vs DQD3" produced 2286 genes (1230 up-regulated and 1056 down-regulated) (Fig. 1b). The Venn diagram showed both unique and common DEGs among these comparison groups (Fig. 1c).
Fig. 1.
Sample morphology and summary of the DEGs a Normal and abortive pistils from different developmental stages are shown. Stigma, style and ovary, three parts and their samples name used in this study are shown in figure b Regulation of DEGs are shown in gray (up-regulated) and dark red (down-regulated) among different comparison group c Venn diagram showed common and overlapped differentially expressed genes among different comparison group
Functional enrichment annotation of DEGs
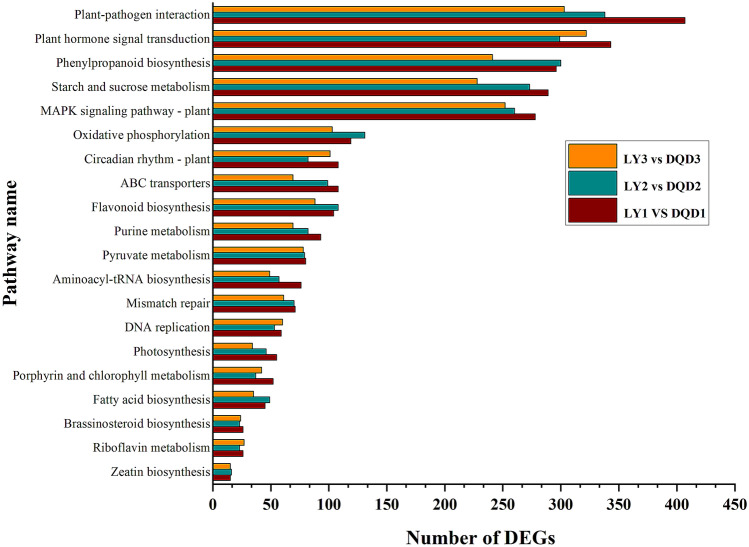
Gene Ontology analysis was carried out to further understand the major biological functions by categorizing them into three sub-classes. The comparison of LY1 vs DQD1, LY2 vs DQD2, and LY3 vs DQD3 represents 2661, 1514, and 1677 genes, respectively (Table 2). For the BP, biological regulation, cellular and metabolic processes were the most enriched terms; in the CC, membrane parts and cells were the more leading terms; while in MF, binding, catalytic and transporter activity were the most enriched terms (Fig. 2; File S2). Moreover, when KEGG pathway enrichment analysis was performed, a total of 134 significantly enriched pathways were found. In all comparison groups, plant hormone signal transduction (KO: 04,075; 343 DEGs), phenylpropanoid biosynthesis (KO: 00,940; 305 DEGs), plant-pathogen interaction (KO: 04,626; 407 DEGs), and circadian rhythm—plant (KO: 04,712; 108 DEGs) were found to be significantly enriched related to this study (Fig. 3; File S3).
Table 2.
Total number of DEGs in Gene Ontology (GO) among different comparison groups in Japanese apricot
| Comparison group | Total number of genes | Up-regulated | Down-regulated |
|---|---|---|---|
| LY1 vs DQD1 | 2661 | 1244 | 1417 |
| LY2 vs DQD2 | 1514 | 992 | 522 |
| LY3 vs DQD3 | 1677 | 905 | 772 |
Fig. 2.
GO functional enrichment analysis of DEGs among three comparison groups. The most enriched terms of ‘biological process, cellular component and molecular function’ are shown
Fig. 3.
Top 20 enriched pathways and their number of genes are shown among three different comparison groups. The X-axis represents the enriched pathway name and Y-axis represents the number of DEGs
Identification of transcription factors (TF) encoding genes
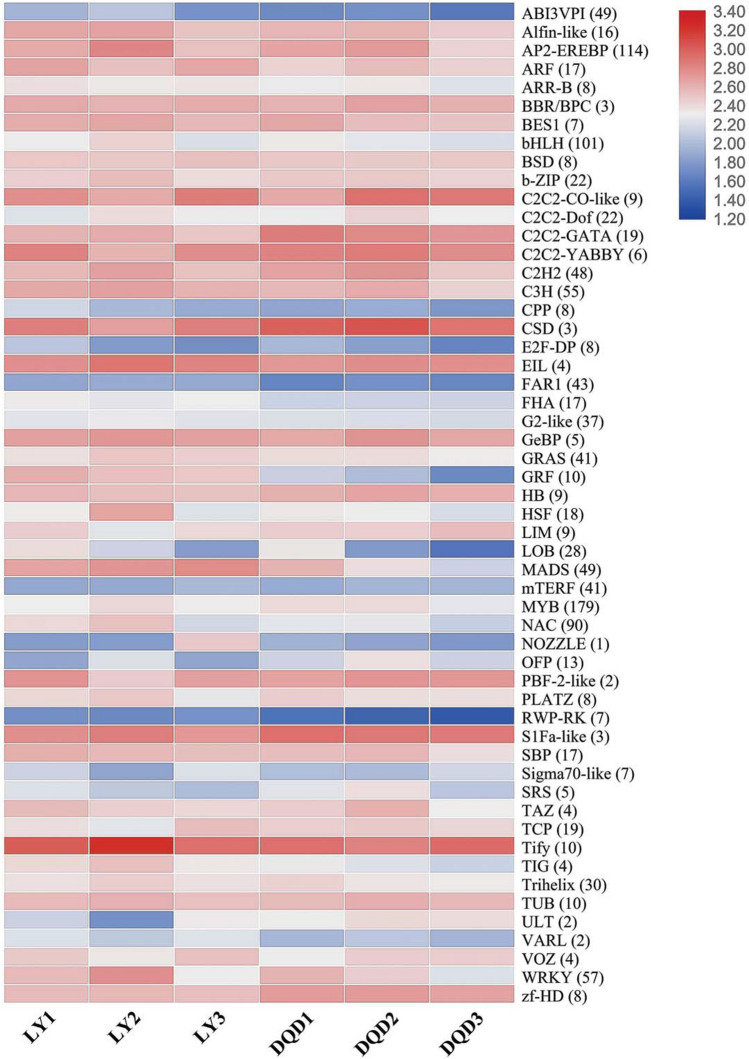
In this study, a total of 1435 TFs related genes were identified and classified into 59 different families, whose expression patterns are shown in Fig. 4 and File S4. From all these TF families, MYB (179) TF remained the most dominant TF family followed by AP2-EREBP (114), bHLH (101), NAC (90), MADS (49), WRKY (57), bZIP (22), TCP (19) and zf-HD (8). Moreover, numerous TFs related to hormonal activity such as ARF (17) and GRAS (41) TF were also found.
Fig. 4.
Identification and expression profiling of transcription factor genes during normal and abortive pistils from different developmental stages in Japanese apricot. Heatmap showing an average differential expression of each transcription factor family genes. The color scale represents the log2fc FPKM value
Identification of DEGs related to plant hormone
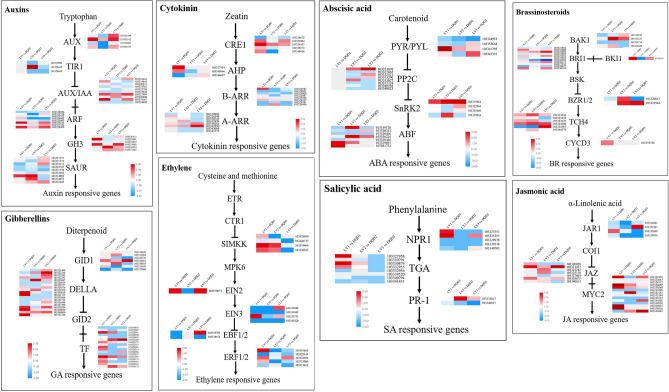
To determine the expression and regulation of various phytohormones related genes in normal and abortive pistils from different developmental stages, the genes related to plant hormones signaling transduction pathway were analyzed. In the present study, 60, 65, and 35 putative genes in auxin, GA and CK signaling pathway, while 40 genes in "BR", 23 genes in "ABA", 19 genes in "ETH", 38 genes in "JA" and 18 putative genes in SA signaling pathway were found and their expression are shown in Fig. 5.
Fig. 5.
Heatmap showing relative changes in the expression pattern related to plant hormone signaling pathway for eight hormones for different stages of pistil abortion in Japanese apricot. The color scale on each Heatmap showed their expression value
For auxin biosynthesis and metabolism, the DEGs were classified as AUX1 (4 genes), TIR1 (3 genes), AUX/IAA (14 genes), ARF (18 genes), GH3 (6 genes), and SAUR (15 genes), while for GA biosynthesis, DEGs were categorized as GID1 (15 genes), DELLA (23 genes) and TF (27 genes). For ABA biosynthesis, the DEGs were classified to PYR/PYL (4 genes), PP2C (8 genes), SnRK2 (4 genes), and ABF (7 genes), while putative genes involved in JA biosynthesis were characterized as JAR1 (4 genes), JAZ (7 genes) and MYC2 (27 genes). Most of the key regulatory genes showed a specific expression pattern, and the expression value of the genes related phytohormones are listed in File S5.
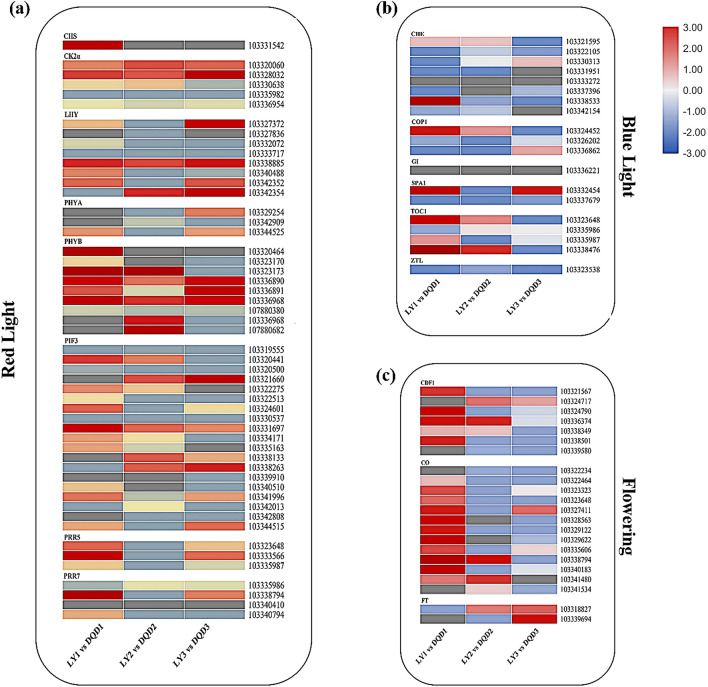
Identification of the genes related to the Circadian Rhythm pathway
In the present study, the circadian rhythm was also found to be the most enriched pathway and we analyzed the genes related to this pathway. Overall, 95 genes coding 17 key regulators were identified (Fig. 6) and the expression level of the genes related to each key regulator was analyzed and their FPKM values are shown in File S6. The genes encoding "CHS", "CK2a", "PHYB", "PIF3", and "PRR5" regulators were highly expressed in LY1 vs DQD1 comparison, while "LHY", "PHYA", and "PRR7″ were predominantly expressed in LY3 vs DQD3 comparison. Moreover, the genes encoding "CDF1″ and "CO" exhibited higher expression in LY1 vs DQD1 comparison, while "FT" showed a lower level of expression. Remarkably, "GI", "ZTL", and "CHE" also showed lower expression levels in all comparisons.
Fig. 6.
Expression patterns of the genes related to the circadian rhythm pathway are shown. These genes are involved in a red-light signaling b blue-light signaling c key genes related to flowering
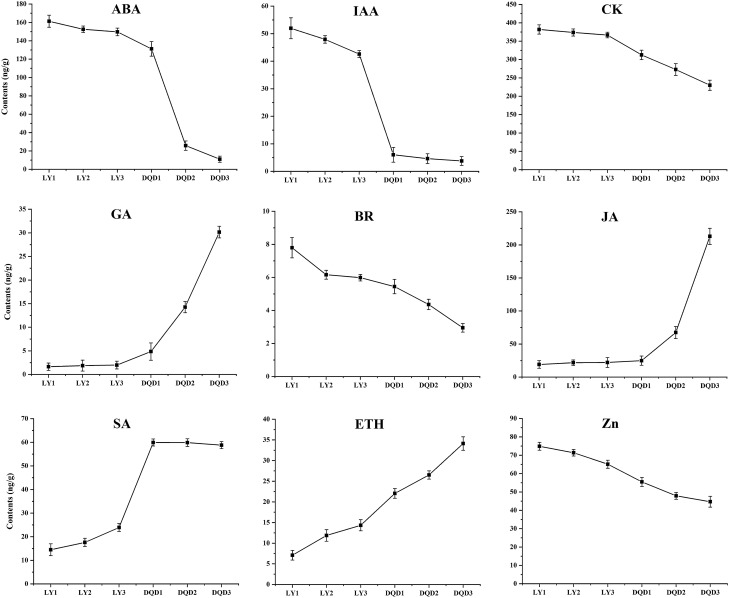
Phytohormones contents in normal and abortive pistils from different developmental stages in Japanese Apricot
To determine the phytohormone contents, tissue samples identical to the ones used for transcriptome analysis were collected. The contents of "ABA", "IAA", "CK", "BR" and "Zeatin (Zn)" were higher in normal pistil development stages and showed a decreasing trend onward to DQD3 stage in abortive pistils. In contrast, the contents of "GA", "JA", "SA", and "ETH" were lower in normal pistil development stages while showed an increasing trend in abortive pistil stages from DQD1 to DQD3 (Fig. 7). The contents of "ABA", "IAA", "CK", "BR", and "Zeatin" were recorded as high in normal pistils i.e. 161.32 ng/g, 51.95 ng/g, 381.90 ng/g, 7.79 ng/g, and 74.88 ng/g, respectively, while the contents of "GA", "JA", "SA", and "ETH" were recorded as high at DQD3 stage of the abortive pistils i.e. 30.15 ng/g, 212.97 ng/g, 58.81, and 34.12 ng/g, respectively.
Fig. 7.
Phytohormones contents in normal and abortive pistils from different developmental stages in Japanese apricot. Standard deviation is shown in error bars
Confirmation of transcriptome results through RT-qPCR
To validate the differential expression of the genes identified from the transcriptome data, we randomly selected 12 genes to perform RT-qPCR (primers and descriptions of the genes are listed in File S7). The result showed that the genes exhibited similar expression profiles to those observed from the transcriptomic data (Fig S2).
Discussion
Transcriptome sequencing
Flower development is a unique process for fruit-bearing plants and is very important for their reproductive success. Gene expression and its regulation have an important role in plant development and necessary for understanding the molecular mechanism of any developmental process (Krizek and Fletcher 2005). Several studies revealed that Illumina sequencing remains a dynamic tool for DEGs analysis in different flower development processes (Wong et al. 2013; Zhang et al. 2014). Therefore, we performed transcriptome sequencing to compare the expression profiles of normal and abortive pistils from different developmental stages to find out the putative genes and pathways associated with the pistil abortion problem in Japanese apricot. In general, the functional annotation analysis revealed that all the DEGs were related to a series of functions and different biological processes, such as flower development and regulation, secondary metabolites processes as well as cellular ones, indicating that pistil abortion might be controlled by several mechanisms involving many genes. In brief, the transcriptome data provide an important resource for understanding the molecular mechanism and different biological processes related to the pistil abortion problem in Japanese apricot, providing a reference for further studies related to this issue.
Identification of transcription factors genes related to pistil abortion in Japanese apricot
TFs are the proteins that bind to specific DNA sequences leading to the transcriptional regulation of the target gene (Caarls et al. 2015; Kim et al. 2016). In the present study, various TF families containing large numbers of TFs genes were identified and showed variant expression patterns between normal and abortive pistil development stages. Among these TF families, MADS-box, C2C2-CO like, EIL, PBF2-like, MYB, bHLH, zf-HD, AP2-EREBP, C2H2, TCP, C2C2-YABBY, GRF, and CSD were found to be important during pistil abortion. These identified families have a strong role in reproductive processes in several plant species (Sharma, 2012; Singh et al. 2013). From the bHLH family genes, involved in regulating numerous flower developmental processes in Arabidopsis (Zhang et al. 2006), 101 were identified in this study exhibiting consistent expression patterns in all stages. The bZIP transcription factors are the key regulators for flower development (Uno et al. 2000) and through which we also identified 22 of them in our study.
From YABBY family genes e.g. AtYABBY1, AtYABBY2, and AtYABBY3, which are involved in the growth and development of different floral organs (Bowman 2000; Lugassi et al. 2010), one was identified as an up-regulated gene in all stages, indicating that YABBY TFs are closely associated with pistil abortion. MADS-box gene family plays an important role in ovule and pistil development and a decrease in the expression cause pistil and ovule abortion in Arabidopsis (Alvarez-Buylla et al. 2010; Cucinotta et al. 2014). Here, we identified the MADS-box family genes in normal and abortive pistils as the higher expression of the genes was in normal pistils while lower expression in abortive pistils, decreasing from DQD1 to DQD3. Growth-regulating factors (GRFs) e.g. AtGRF5 and AtGRF8 are the plant-specific genes involved in different developmental processes of flower formation (Lee et al. 2009; Omidbakhshfard et al. 2015). In our data, we identified GRF TFs genes showing higher expression in normal pistils, and decreasing expression from DQD1 to DQD3 stage of abortive pistils. Recently, Liang et al. (Liang et al. 2014) reported that a decrease in GRF expression level causes pistil abnormalities, which indicates that GRF might have an important role in pistil abortion. Based on these findings, our results are consistent with the previous results in Arabidopsis (Liang et al. 2014), providing more evidence regarding the involvement of these TFs in pistil abortion of Japanese apricot.
Effect of plant-hormones and related genes on pistil abortion of Japanese apricot
Phytohormones are important regulators for plant growth and development. The flower development process is strongly controlled by the regulation of hormones (Chandler 2011). Auxins are considered to be important hormone involved in various plant development processes, such as floral organ identification and floral primordia initiation (Alabadí et al. 2009). Auxin is closely associated with its biosynthesis, polar transportation, or auxin signaling disruption, which leads toward the failure of flower formation (Aloni et al. 2006). In litchi, during floral bud initiation, the expression level of auxin signaling transduction related genes was changed (Zhang et al. 2014). In ARF family, various genes are involved in the flower development process. In Arabidopsis, ARF6 and ARF8 are considered to be essential for controlling vegetative and floral organ growth and development (Liu, 2014). In the present study, ARF1, ARF4, ARF9, and ARF18 homologous to ARF6 and ARF8 from Arabidopsis and tomato are were highly expressed, proposing their immersion in pistil abortion of Japanese apricot. Moreover, other phytohormones such as ABA, GA, and CK are also involved in promoting flower development. The effect of GA on flowering was mediated by different DELLA proteins, RGL1, 2, and 3, REPRESSOR OF ga1-3 (RGA), and GIBBERELLIC ACID INSENSITIVE (GAI) (Porri et al. 2012). In woody plants, abscisic acid (ABA) stimulates floral initiation and differentiation process (Shan, 2012), while CK stimulates flowering through transcriptional activation of FT paralog TSF in Arabidopsis (D’Aloia, 2011). The homologous of these phytohormones-related key genes (ABA, FT, CO, etc., and other hormone classes genes) were also identified in Japanese apricot. The significant changes in the expression level of these hormone-related genes confirmed the potential role of these hormones in pistil abortion. ETH is a significant hormone in plant vegetative and reproductive organ abscission. Pistil abortion is considered to be an evolutionary adaptation mechanism to conserve resources in andromonoecious species, maintaining a balance between pistil number and available resources. Therefore, the rate of pistil abortion of large-fruited are higher (Rosati et al. 2011).
ABA is an essential hormone that plays a regulatory role in plant development, involving seed development, flower and phase transition, and plant response to diverse environmental stress (Li, 2015). ABA can also inhibit flower organ formation and has been reported in olive and apple (Meng et al. 2012; Ulger et al. 2004). This study also confirmed the significant changes in ABA-related genes in normal and abortive pistil stages. The expression of ABA-responsive genes related to SnRK2 and ABF was higher in normal pistils and decreased in abortive pistils from DQD1 to DQD3. In addition, we also found that ABA content in normal pistils was higher than abortive pistils. Therefore, the decrease in ABA content may be one of the reasons for pistil abortion.
Ethylene promotes pistil development, and the EBF transcription factor is a member of the AP2 gene family, and its coding protein has a negative impact on AGAMOUS gene regulation (Perata 2013). Studies in tobacco showed ethylene as an upstream regulator for ovule development (Kaur-Sawhney et al. 1988). During the process of pistil abortion, changes in the expression of ethylene response signal molecules were recorded between normal and abortive pistils. Meanwhile, the expression of ethylene response signal factor ERF 1/2 showed lower expression at DQD3 stage of abortive pistils, this indicated that ethylene may be an important regulator of pistil abortion in Japanese apricot. In the whole differentiate stage of pistil, pistil abortion represents versatile style. In this process, the changes of hormones play an important role in pistil abortion, especially IAA, ABA, and CTK. Related genes involved in hormone synthesis expression regulate the content of hormones and adapt to the occurrence of pistil abortion under adversity.
Gene identification related to floral transition and development
Genetic network controlling flower development depends on different important pathways such as photoperiod, vernalization, and GA-induction (Blázquez et al. 2003; Iqbal et al. 2020b; Srikanth and Schmid 2011). The photoperiodic pathway includes three main parts: circadian clock, photoreceptors, and clock to flowering output pathway (Simpson 2003). Phytochromes and cryptochromes are initially activated by the light signal (Fowler, 1999; Simpson and Dean 2002), which can coordinate the internal biological processes and external rhythm changes (Digel et al. 2015; Imaizumi 2010). The circadian clock consists of three linking loops that measure the changes in day length and regulate CYCLING DOF FACTOR (CDF), FKF1, and GI (Imaizumi 2010). FKF1 and GI simplify the CO expression, a TF that promotes flowering through inducing the direct downstream expression of the genes such as FT and SOC1 (Kardailsky, 1999; Liu, 2008). In our study, both CDF1 and FT expression in Japanese apricot is quite lower suggesting that these might have their putative role in pistil abortion. The overall expression of the genes in this pathway showed a specific expression pattern, which suggested that these genes may play an important role in pistil abortion and the reproductive growth of other flower organs.
Conclusion
The current study was done to explore the pistil abortion mechanism in Japanese apricot using transcriptome sequencing. An abundance of the DEGs was recognized and analyzed among different comparison groups. Most of the DEGs were involved in various biological process such as flower development and their regulation. Phytohormones signaling and circadian rhythm were the most significantly enriched pathways. Furthermore, different TF families such as NAC, YABBY, and GRF showed their specific expression in pistil abortion. The presents results will further be used for the detection of several QTLs for the proportion of perfect flowers in Japanese apricot, is of great value for future genetic study and marker-assisted breeding, which can provide useful information for the identification of candidate genes responsible for pistil abortion, as well as for marker-assisted breeding. This study provides a foundation for further understanding of the molecular mechanism of pistil abortion and also provides a possible future direction for other fruit trees.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31772282 and 31971703), the National Key Research and Development Program of China (2018YFD1000107), China Postdoctoral Science Foundation (2018 M640497), and Jiangsu Postdoctoral Science Research Foundation (2018K216C) for funding this research in materials collection, data analysis, and experiment.
Authors’ contribution statement
ZG and SI conceived and designed the study. SI performed the experiment, analyzed data and wrote the whole manuscript. NX, BY. FH and ZP assist in sample collection and data analysis. ST and SA and DC revised the manuscript. All authors read and approved the final version of the manuscript.
Data availability
The sequencing data were deposited to NCBI-GEO under the accession numbers GSE141096.
Declaration
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shahid Iqbal, Email: shahidiqbalpak@hotmail.com.
Zhenpeng Pan, Email: 2017104013@njau.edu.cn.
Faisal Hayat, Email: maken_faisal@yahoo.com.
Yang Bai, Email: 2019204009@njau.edu.cn.
Daouda Coulibaly, Email: 2019204061@njau.edu.cn.
Sajid Ali, Email: sajidali@bzu.edu.pk.
Xiaopeng Ni, Email: 2017204013@njau.edu.cn.
Ting Shi, Email: shiting@njau.edu.cn.
Zhihong Gao, Email: gaozhihong@njau.edu.cn.
References
- Achard P, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19(14):1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Blázquez MA, Carbonell J, Ferrándiz C, Pérez-Amador MA. Instructive roles for hormones in plant development. Int J Develop Biology. 2009;53(8):1597. doi: 10.1387/ijdb.072423da. [DOI] [PubMed] [Google Scholar]
- Aloni R, Aloni E, Langhans M, Ullrich CI. Role of auxin in regulating Arabidopsis flower development. Planta. 2006;223(2):315–328. doi: 10.1007/s00425-005-0088-9. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER et al. (2010) Flower development The Arabidopsis Book/American Society of Plant Biologists 8 [DOI] [PMC free article] [PubMed]
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Arathi H, Ganeshaiah K, Shaanker RU, Hegde S. Seed abortion in Pongamia pinnata (Fabaceae) Am J Bot. 1999;86(5):659–662. doi: 10.2307/2656574. [DOI] [PubMed] [Google Scholar]
- Bai S, Saito T, Sakamoto D, Ito A, Fujii H, Moriguchi T. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013;54(7):1132–1151. doi: 10.1093/pcp/pct067. [DOI] [PubMed] [Google Scholar]
- Balcke GU, et al. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Method. 2012;8(1):47. doi: 10.1186/1746-4811-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theißen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogen Evol. 2003;29(3):464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annal Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- Beppu K, Kataoka I. Studies on pistil doubling and fruit set of sweet cherry in warm climate. J Japan Soc Hortic Sci. 2011;80(1):1–13. doi: 10.2503/jjshs1.80.1. [DOI] [Google Scholar]
- Bland JM, Altman DG. Multiple significance tests. Bonferroni Method Bmj. 1995;310(6973):170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33(2):168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Bowman JL. The YABBY gene family and abaxial cell fate. Current Opin Plant Biol. 2000;3(1):17–22. doi: 10.1016/S1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- Caarls L, Pieterse CM, Van Wees S. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015;6:170. doi: 10.3389/fpls.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Cook H, Davies B. An antirrhinum ternary complex factor specifically interacts with C-function and SEPALLATA-like MADS-box factors. Plant Mol Biol. 2003;52(5):1051–1062. doi: 10.1023/A:1025426016267. [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell. 2008;20(7):1760–1774. doi: 10.1105/tpc.107.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. The hormonal regulation of flower development. J Plant Growth Regul. 2011;30(2):242–254. doi: 10.1007/s00344-010-9180-x. [DOI] [Google Scholar]
- Chen L, et al. Transcriptomic analysis reveals candidate genes for female sterility in pomegranate flowers. Front Plant Sci. 2017;8:1430. doi: 10.3389/fpls.2017.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, et al. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131(5):1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- Cucinotta M, Colombo L, Roig-Villanova I. Ovule development, a new model for lateral organ formation. Front Plant Sci. 2014;5:117. doi: 10.3389/fpls.2014.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aloia M, et al. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 2011;65(6):972–979. doi: 10.1111/j.1365-313X.2011.04482.x. [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M. Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell. 2015;27(9):2318–2334. doi: 10.1105/tpc.15.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18(17):4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SM, Ma K, Du XH, Li FL. Advances in Research on Xanthoceras Sorbifolia. Chin Bull Bot. 2002;19(3):296–301. [Google Scholar]
- Gao Z, Wang S, Zhang Z. Comparative study on flower and fruit characteristics of 29 varieties in Japanese apricot (Prunus mume Sieb et Zucc) Jiangsu Agri Sci. 2006;6:231–233. [Google Scholar]
- Gao Z, Shi T, Luo X, Zhang Z, Zhuang W, Wang L. High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot. BMC Genom. 2012;13(1):371. doi: 10.1186/1471-2164-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Iqbal S, Ma R, Song J, Yu M, Gao Z. High-density genetic map construction and quantitative trait loci analysis of the stony hard phenotype in peach based on restriction-site associated DNA sequencing. BMC Genom. 2018;19(1):612. doi: 10.1186/s12864-018-4952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Nat Acad Sci. 2014;111(18):6840–6845. doi: 10.1073/pnas.1318532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J-H, Gao Z-H, Zhang Z, Chen S-M, Ando T, Zhang J-Y, Wang X-W. Isolation and characterization of an AGAMOUS homologue PmAG from the Japanese apricot (Prunus mume Sieb et Zucc) Plant Mol Biol Rep. 2011;29(2):473–480. doi: 10.1007/s11105-010-0248-3. [DOI] [Google Scholar]
- Huang Y-J, et al. Use of transcriptome sequencing to understand the pistillate flowering in hickory (Carya cathayensis Sarg) BMC Genom. 2013;14(1):691. doi: 10.1186/1471-2164-14-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13(1):83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S, et al. Genome-wide analysis of PmTCP4 transcription factor binding sites by ChIP-Seq during pistil abortion in Japanese apricot. Plant Genome. 2020;13(3):e20052. doi: 10.1002/tpg2.20052. [DOI] [PubMed] [Google Scholar]
- Iqbal S, et al. Identification and expression profiling of sugar transporter genes during sugar accumulation at different stages of fruit development in apricot. Gene. 2020 doi: 10.1016/j.gene.2020.144584. [DOI] [PubMed] [Google Scholar]
- Jain M. Next-generation sequencing technologies for gene expression profiling in plants. Brief Funct Genom. 2011;11(1):63–70. doi: 10.1093/bfgp/elr038. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, et al. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kaur-Sawhney R, Tiburcio A, Galston A. Spermidine and flower-bud differentiation in thin-layer explants of tobacco. Planta. 1988;173(2):282–284. doi: 10.1007/BF00403022. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016;33:48–56. doi: 10.1016/j.pbi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Kocsis M, Jakab G. Analysis of BABA (β-aminobutyric acid)-induced female sterility in Arabidopsis flowers. Acta Biol Szegediensis. 2008;52(1):247–249. [Google Scholar]
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6(9):688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genom. 2011;285(3):245–260. doi: 10.1007/s00438-011-0602-7. [DOI] [PubMed] [Google Scholar]
- Lee BH, Ko J-H, Lee S, Lee Y, Pak J-H, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009;151(2):655–668. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. Transcriptomic insights into antagonistic effects of gibberellin and abscisic acid on petal growth in Gerbera hybrida. Front Plant Sci. 2015;6:168. doi: 10.3389/fpls.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014;164(1):249–258. doi: 10.1104/pp.113.225144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillecrapp A, Wallwork M, Sedgley M. Female and male sterility cause low fruit set in a clone of theTrevatt'variety of apricot (Prunus armeniaca) Sci Hortic. 1999;82(3–4):255–263. doi: 10.1016/S0304-4238(99)00061-8. [DOI] [Google Scholar]
- Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot. 2010;61:1419–1430. doi: 10.1093/jxb/erq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, et al. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135(8):1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Liu N, et al. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J Exp Bot. 2014;65(9):2507–2520. doi: 10.1093/jxb/eru141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu X, Kim H, Zhong S, Chen H, Hu Z, Zhou B. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn and identification of differentially expressed genes in response to reactive oxygen species. BMC Genom. 2014;15(1):805. doi: 10.1186/1471-2164-15-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugassi N, Nakayama N, Bochnik R, Zik M. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 2010;10(1):131. doi: 10.1186/1471-2229-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Ma S, Shao J, Sun J, Ma B, Wang H. Effects of spraying 6-BA on axillary bud growth and the dynamic changes of endogenous hormones in'Tianhong 2'Fuji nursery apple trees. Acta Horticulturae Sinica. 2012;39(5):837–844. [Google Scholar]
- Ni X, Xue S, Iqbal S, Wang W, Ni Z, Khalil-ur-Rehman M, Gao Z (2018) Candidate genes associated with red colour formation revealed by comparative genomic variant analysis of red-and green-skinned fruits of Japanese apricot (Prunus mume). PeerJ 6:e4625 [DOI] [PMC free article] [PubMed]
- Ó'Maoiléidigh DS, Graciet E, Wellmer F. Gene networks controlling A rabidopsis thaliana flower development. New Phytol. 2014;201(1):16–30. doi: 10.1111/nph.12444. [DOI] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant. 2015;8(7):998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Origin(Pro), Version Number (2019) OriginLab Corporation, Northampton, MA, USA
- Owen SJ, Abrams SR (2009) Measurement of plant hormones by liquid chromatography–mass spectrometry. In: Plant Hormones. Springer, pp 39–51 [DOI] [PubMed]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405(6783):200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Peng S, Luo T, Zhou J, Niu B, Lei N, Tang L, Chen F. Cloning and quantification of expression levels of two MADS-box genes from Momordica charantia. Biol Plant. 2008;52(20):222–230. doi: 10.1007/s10535-008-0049-9. [DOI] [Google Scholar]
- Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199(3):639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012;139(2):2198–2209. doi: 10.1242/dev.077164. [DOI] [PubMed] [Google Scholar]
- Reale L, et al. Morphological and cytological development and starch accumulation in hermaphrodite and staminate flowers of olive (Olea europaea L) Sex Plant Reprod. 2009;22(3):109–119. doi: 10.1007/s00497-009-0096-1. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4(10):1237–1249. doi: 10.2307/3869410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati A, Caporali S, Paoletti A, Famiani F. Pistil abortion is related to ovary mass in olive (Olea europaea L) Sci Hortic. 2011;127(4):515–519. doi: 10.1016/j.scienta.2010.12.002. [DOI] [Google Scholar]
- Shan H, et al. Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotechnol. 2012;51(2):160–173. doi: 10.1007/s12033-011-9451-1. [DOI] [PubMed] [Google Scholar]
- Sharma R, et al. Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct Integr Genomics. 2012;12(2):229–248. doi: 10.1007/s10142-012-0274-3. [DOI] [PubMed] [Google Scholar]
- Shi T, Zhuang W, Zhang Z, Sun H, Wang L, Gao Z. Comparative proteomic analysis of pistil abortion in Japanese apricot (Prunus mume Sieb et Zucc) J Plant Physiol. 2012;169(13):1301–1310. doi: 10.1016/j.jplph.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Shi T, et al. Analyzing Differentially Expressed Genes and Pathways Associated with Pistil Abortion in Japanese Apricot via RNA-Seq. Genes. 2020;11(9):1079. doi: 10.3390/genes11091079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. Evolution of flowering in response to day length: flipping the CONSTANS switch. BioEssays. 2003;25(9):829–832. doi: 10.1002/bies.10330. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. Arabidopsis, the Rosetta stone of flowering time? Science. 2002;296(5566):285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Singh VK, Garg R, Jain M. A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol J. 2013;11(6):691–701. doi: 10.1111/pbi.12059. [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. Regulation of flowering time: all roads lead to rome. Cell Mol Life Sci. 2011;68(12):2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, et al. Genome-wide characterization and linkage mapping of simple sequence repeats in mei (Prunus mume Sieb et Zucc) PLoS ONE. 2013;8(3):e59562. doi: 10.1371/journal.pone.0059562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Annal Bot. 2007;100(3):603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulger S, Sonmez S, Karkacier M, Ertoy N, Akdesir O, Aksu M. Determination of endogenous hormones, sugars and mineral nutrition levels during the induction, initiation and differentiation stage and their effects on flower formation in olive. Plant Growth Regul. 2004;42(1):89–95. doi: 10.1023/B:GROW.0000014897.22172.7d. [DOI] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci. 2000;97(21):11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining KJ, et al. The floral transcriptome of E ucalyptus grandis. New Phytol. 2015;206(4):1406–1422. doi: 10.1111/nph.13077. [DOI] [PubMed] [Google Scholar]
- Wang S (2008) Preliminary studies on differences of related characteristics between perfect flower and imperfect flower and protemics in Japanese apricot Nanjing: Nanjing Agricultural University
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process. PLoS ONE. 2013;8(6):e65319. doi: 10.1371/journal.pone.0065319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shi T, Iqbal S, Zhang Y, Liu L, Gao Z. Genome-wide discovery and characterization of flower development related long non-coding RNAs in Prunus mume. BMC Plant Biol. 2019;19(1):64. doi: 10.1186/s12870-019-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci. 2010;107(13):6100–6105. doi: 10.1073/pnas.0912333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Zhang D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr Opin Plant Biol. 2015;27:44–51. doi: 10.1016/j.pbi.2015.05.024. [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133(16):3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- Zhang H-N, et al. Transcriptomic analysis of floral initiation in litchi (Litchi chinensis Sonn) based on de novo RNA sequencing. Plant Cell Rep. 2014;33(10):1723–1735. doi: 10.1007/s00299-014-1650-3. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li Y, Xin D, Chen W, Shao X, Wang Y, Guo W. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus) Gene. 2015;555(2):362–376. doi: 10.1016/j.gene.2014.11.032. [DOI] [PubMed] [Google Scholar]
- Zinn KE, Tunc-Ozdemir M, Harper JF. Temperature stress and plant sexual reproduction: uncovering the weakest links. J Exp Bot. 2010;61(7):1959–1968. doi: 10.1093/jxb/erq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data were deposited to NCBI-GEO under the accession numbers GSE141096.