Abstract
Large size cell-laden hydrogel models hold great promise for tissue repair and organ transplantation, but their fabrication using 3D bioprinting is limited by the slow printing speed that can affect the part quality and the biological activity of the encapsulated cells. Here a fast hydrogel stereolithography printing (FLOAT) method is presented that allows the creation of a centimeter-sized, multiscale solid hydrogel model within minutes. Through precisely controlling the photopolymerization condition, low suction force-driven, high-velocity flow of the hydrogel prepolymer is established that supports the continuous replenishment of the prepolymer solution below the curing part and the nonstop part growth. The rapid printing of centimeter-sized hydrogel models using FLOAT is shown to significantly reduce the part deformation and cellular injury caused by the prolonged exposure to the environmental stresses in conventional 3D printing methods. Embedded vessel networks fabricated through multiscale printing allows media perfusion needed to maintain the high cellular viability and metabolic functions in the deep core of the large-sized models. The endothelialization of this vessel network allows the establishment of barrier functions. Together, these studies demonstrate a rapid 3D hydrogel printing method and represent a first step toward the fabrication of large-sized engineered tissue models.
Keywords: 3D bioprinting, continuous printing, endothelialization, hydrogels, stereolithography
1. Introduction
Large scale cell-laden hydrogel models hold great promise for tissue repair and organ transplantation, but their fabrication is faced with challenges in achieving clinically relevant size and hierarchical structures.[1] 3D bioprinting is an emerging technology for hydrogel fabrication and has been successfully used to create hydrogel models with biomimetic structures and functions;[2,3] however, its application in large, solid hydrogel fabrication has been limited by the slow printing speed that can affect the part quality and the biological activity of the encapsulated cells.[4,5]
Due to the point-by-point deposition process used in nozzle-based bioprinting techniques, extended printing time is required to fabricate a large-sized model with fine structures.[6,7] Prolonged exposure of the encapsulated cells to a variety of printing-induced environmental factors, such as the shear stress, the low oxygen level and the temperature shock, has been shown to cause serious cellular injury and cell death.[8,9] The effort to improve the printing resolution by using small diameter nozzles can cause further damage to the cells.[9,10] Additionally, due to the low mechanical strength of the hydrogel scaffold materials, it is very challenging for point-by-point deposition methods to create overhanging or hollow structures such as vascular channels inside solid parts. To address this limitation, Atala group utilized rigid polymeric scaffolds to support the printing of cell-laden hydrogel materials,[11] and Feinberg group extruded hydrogel material in a secondary supporting hydrogel to print biomimetic structures such as a heart chamber;[12] however, these approaches suffer from either the high rigidity of the supporting material or the complexity of the post-processing steps. Although extrusion printing of dissolvable templates composed of sacrificial materials such as fugitive inks and carbohydrate glass has enabled the creation of perfusable vascular channels in casted hydrogel constructs,[13–15] this approach has very limited capacity to create fine tissue structures other than vascular channels due to the simple casting method used.
Digital mask projection-stereolithography (MP-SLA) is a photopolymerization-based, layer-by-layer 3D printing technology that features multiscale fabrication capacity with high spatial resolution, allowing the bulk geometry and fine structure of a complex 3D model to be built through one single process.[16,17] The liquid resin provides natural self-support for the fabrication of hollow structures. This approach has been used to fabricate hydrogel models such as nerve conduits and muscle-powered biobots. Recently, multivascular networks have been created in hydrogels by controlling the spatial resolution of hydrogel photopolymerization using selected food dye photoabsorbers;[18–20] however, the layer-by-layer process used in these studies limited the printing speed, which can potentially cause dehydration-induced part deformation and reduced cell viability during the fabrication of large-sized hydrogel parts. Recently, the development of continuous liquid interface production (CLIP) technology drastically increased the fabrication speed of MP-SLA through continuously building the layers of a 3D part immediately above a “dead zone” formed by oxygen inhibition of photopolymerization.[21] In the dead zone, the flow of liquid water-insoluble-resin (WI-resin) enables continuous material replenishment at the polymerization interface. However, due to the low fluidity of the WI-resin material and the corresponding large suction force at the curing interface, the fabrication ability of the CLIP technology is limited to thin-walled parts.[21–23] The fabrication of a centimeter-sized solid hydrogel part has not yet been achieved using CLIP.
In this work, we established low suction force-driven, high-velocity flow of the hydrogel prepolymer for continuous MP-SLA printing through precisely controlling the photopolymerization condition. The high-velocity flow supports the continuous replenishment of the prepolymer solution below the curing part and the nonstop part growth. This method, the fast hydrogel stereolithography printing (FLOAT), allows the creation of a centimeter-sized, multiscale solid hydrogel model within several minutes. We showed that this process is unique to the hydrogel prepolymer solution and cannot be achieved using resin without externally supplemented oxygen. The rapid printing of centimeter-sized hydrogel models using FLOAT was shown to significantly reduce the part deformation and cellular injury caused by the prolonged exposure to the environmental stresses in layer-by-layer based printing methods. Media perfusion in the printed vessel network was shown to promote cell survival and metabolic function in the deep core of the large-sized hydrogel model over long term. The FLOAT is compatible with multiple photocurable hydrogel materials and the endothelialization of this vessel network allows the establishment of barrier functions. Together, these studies demonstrate a rapid hydrogel 3D printing method and represent a first step towards the fabrication of large-sized engineered tissue models.
2. Results and Discussion
2.1. Establish Low Suction Force-Driven, High-Velocity Flow of the Hydrogel Prepolymer to Enable FLOAT Method
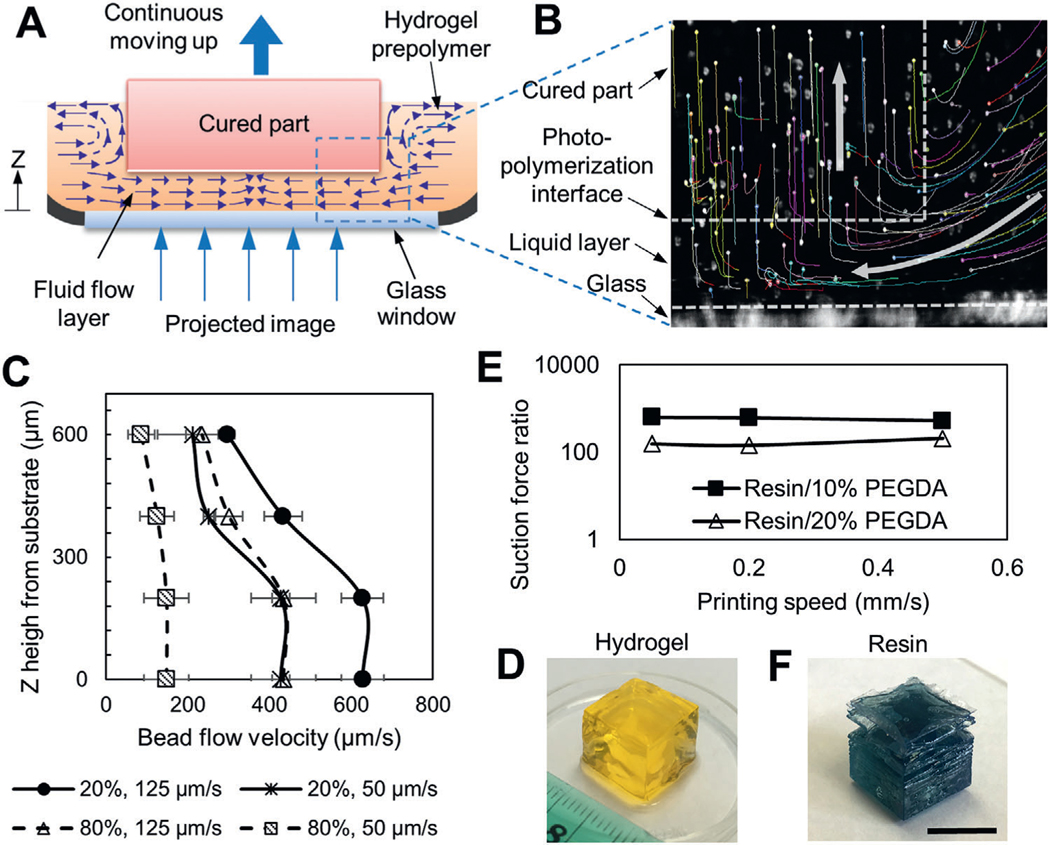
The hydrogel photopolymerization is affected by a number of parameters including exposure energy, printing speed, prepolymer concentration, photoinitiator absorption coefficient, and photoabsorber coefficient. We studied their effects and carefully adjusted the relative ratios of these parameters to formulate the optimal photopolymerization condition for desired printing performance. We first studied the effect of different photopolymerization conditions on the flow of the hydrogel prepolymer solution. Three different curing conditions were developed by varying the photoinitiator (lithium phenyl-2,4,6-trimethylbenzoylphosphinate, LAP) concentration while keeping the photoabsorber concentration and exposure energy constant. The motion of the fluorescence microbeads suspended in the prepolymer solution was tracked to understand the flow dynamics during printing (Figure 1A,B, Figure S1, Supporting Information). We showed the formation of a high-velocity directional flow of the prepolymer solution in an uncured liquid layer beneath the curing part with 0.6% LAP (Figure 1A,B, Movies S1 and S2, Supporting Information). This high-velocity flow supports the continuous replenishment of the prepolymer solution below the curing part and the nonstop part growth. Since there is no oxygen permeable window in the current setup, the formation of this uncured liquid layer is predominantly contributed by the natural high fluidity of the hydrogel prepolymer solution, which is fundamentally different from oxygen inhibition used in the previous methods.[21] Measured liquid flow velocity is in the order of several hundred micrometers per second and is affected by the upward motion speed of the part and the concentration (viscosity) of the prepolymer solution (Figure 1C, Figure S2B, Supporting Information). The highest flow velocity was observed in low concentration solution (20% PEGDA) under high printing speed (125 μm s−1), while the lowest flow velocity was observed in high concentration solution (80% PEGDA) under low printing speed (50 μm s−1). Regardless of the printing speed and liquid concentration, the flow velocity was found to decrease with increased z height from the glass substrate, likely because of the increased viscosity of the precursor fluid in the top region of the liquid layer where the photopolymerization occurs. The thickness of the uncured liquid layer was found to be around 600–800 μm (Figure 1B,C). With 0.1% LAP, only low-velocity and random flow was observed and the part did not form properly (Figure S2A, Supporting Information), suggesting an undercured condition. With 1% LAP, overly rapid curing caused the part bottom layer to stick to the glass window, thus blocking the fluid flow (Figure S2C, Supporting Information). In this overcured condition, later sudden detachment of the part from the glass window due to the upward motion caused the part delamination (Movie S3, Supporting Information). The comparison between the three photopolymerization conditions shows that 0.6% LAP is optimal for the formation of continuous prepolymer flow and nonstop part growth, and thus we chose this condition for the FLOAT printing.
Figure 1.
FLOAT printing is enabled by high-velocity fluid flow and low fluid suction force. A) Schematic of the printing interface in FLOAT. Continuous replenishment of prepolymer solution below the curing part supports nonstop part growth. B) Tracked trajectories of fluorescence microbeads during printing. White arrows show the direction of microbead motion. Microbeads were carried by the flow into a liquid layer, trapped in the hydrogel upon photocuring and then carried upwards by the cured part. C) The flow velocity profile of 20% and 80% PEGDA prepolymer solution in uncured liquid layer under 50 and 125 μm s−1 printing speed. D) A FLOAT-printed 20% PEGDA 4 kDa hydrogel cube. E) Experimentally measured suction force ratios (WI-resin/PEGDA) at various printing speeds. F) A resin cube printed using continuous MP-SLA. Note the smooth surface and sharp edges of the hydrogel cube comparing to the rough surface and delaminated layers of the resin cube. Scale bar is 1 cm.
To understand the mechanism of the prepolymer flow, we experimentally measured the suction force at the printing interface. The suction force generated during continuous printing of a hydrogel cube under optimal curing condition was measured (Figure S3A, Supporting Information). The suction forces were steady and remained at a low level between 10 mN and 38 mN for 10% PEGDA part and between 41 mN and 96 mN for 20% PEGDA part under varied printing speeds (Figure S3B,C, Supporting Information). The steady printing process led to good part quality, as demonstrated by the printed 1 cm hydrogel cube that is featured by the smooth surface and sharp edges (Figure 1D, Figure S3D,E, Supporting Information). To show that this process is unique to the hydrogel material, we printed a resin cube using the same experimental setup. The suction force was unsteady and fluctuated significantly with the peak suction forces in the range of 6 to 20 N (Figure S4, Supporting Information), which were several hundred times higher than those measured in hydrogel printing (Figure 1E). Highly disturbed printing process significantly impaired the quality of the resin part, causing layer delamination and part breakage (Figure 1F). Together, these suggest that the low fluid suction force in FLOAT printing helped to maintain a steady printing process, thus ensuring good part quality.
In addition to print with PEGDA, we also tested the compatibility of FLOAT printing with several other hydrogel materials including gelatin methacrylate (GelMA) and the blends of GelMA and PEGDA at different concentrations. We examined part quality during printing and evaluated the mechanical property of printed parts using compression test. The elastic modulus of pure PEGDA hydrogel models increases with increased precursor concentration (Figure S5, Supporting Information). The addition of PEGDA helps to improve the printability of GelMA prepolymer and increases the stiffness of the printed part. Due to the less amount of total energy used in FLOAT printing, the part stiffness is slightly lower than the stiffness of layer-by-layer printed parts but is slightly higher than the stiffness of casted parts (Figure S5, Supporting Information). However, regardless of the prepolymer composition and printing method, all printed parts have a stiffness less than 8 kPa, which agreed well with the elastic modulus of the soft tissues.[24,25]
The ability of the FLOAT method to print biodegradable materials was tested by printing a composite hydrogel scaffold composed of 8% GelMA blended with 5% eight-arm PEG norbornene (PEGNB), which is hydrolytically degradable. The degradation rate of 8% GelMA + 5% PEGNB was compared to that of 8% GelMA + 5% PEGDA. We observed 51% dry weight reduction in GelMA/PEGNB scaffold and 28% dry weight reduction in GelMA/PEGDA scaffold after 20 d incubation in PBS (Figure S6, Supporting Information). The faster degradation rate in GelMA/PEGNB scaffold is due to the hydrolytic degradation of the ester linkages between degradable norbornene DTT groups. It is expected that the inclusion of cells will further accelerate the degradation of these scaffold due to the proteolytic degradation of the GelMA.
2.2. Multiscale Printing by the FLOAT
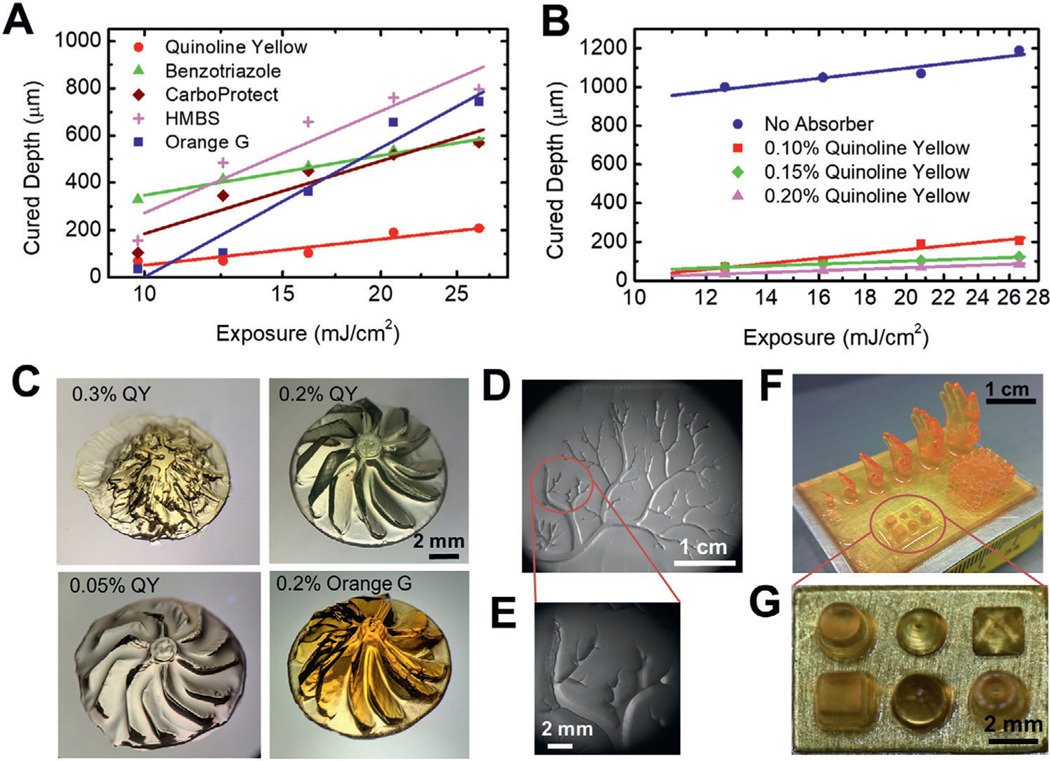
Since the photoabsorber has been shown to be critical to the printing resolution in the previous studies,[26,27] we explored the photoabsorbers that can be compatible with the FLOAT printing process. We compared commonly used photoabsorbers and studied their effects on the curing depth, which serves as a measurement of the vertical resolution. We showed that quinoline yellow (QY) produced the smallest curing layer thickness of 40–150 μm, thus offering the best control of the vertical resolution, due to its maximum absorption at 412 nm which is very close to the 405 nm wavelength of the curing light source (Figure 2A,B). Since the maximum absorptions of widely used 4-hydroxy benzophenone (HMBS) and benzotriazole are between 365 and 385 nm, they were not effective in controlling the curing layer thickness. Although tinuvin carboprotect has a maximum absorption close to 400 nm, its poor solubility in aqueous solution reduced its efficiency in FLOAT process (Figure 2A). Increasing photoabsorber concentration was shown to reduce curing layer thickness (Figure 2B). The use of QY and other photoabsorbers such as orange G in an optimal range was not found to significantly alter the high-velocity prepolymer flow and the continuous printing process of the FLOAT.
Figure 2.
Multiscale printing by FLOAT. A) Measured curing depths with different types of photoabsorbers. The smaller the curing depth, the greater the vertical resolution of the printed part. B) Increasing photoabsorber concentration reduces curing depth. C) Printing resolution demonstrated by a turbine rotor model printed under different types and concentrations of photoabsorbers. 0.3% and 0.05% QY correspond to undercured and overcured conditions respectively. 0.2% QY represents optimal curing, which results in better part quality than 0.2% Orange G. D) FLOAT-printed vascular tree structure in a PEGDA slab with an area of 3.5 cm ×2.5 cm. E) Enlarged view shows micrometer-scale fine structure of the vascular tree branches. F) FLOAT-printed human hand models of different sizes, a truss model and primitives on a 3.5 cm × 2.5 cm PEGDA slab. G) Enlarged view of the primitives. All models in this figure were printed using 20% PEGDA 4 kDa.
The printing resolution and part quality of the FLOAT method were validated through printing a series of models with different geometries. We printed a turbine rotor model with an overall diameter of 1cm and individual blade thickness of 650 μm (Figure 2C, Figure S7A–C, Supporting Information). Since the turbine rotor blades are overhang structures with a relatively large curvature, the geometric fidelity of the blade is used as a verification of the printing quality. Undercured condition using 0.3% QY and overcured conditions using 0.05% QY were shown to lead to poor part quality as compared to the good part printed using 0.2% QY. Orange G at 0.2% allowed the printing of the overall geometry of the turbine model, but the blades are less sharp and smooth as compared to those printed with 0.2% QY (Figure 2C). HMBS is not effective in controlling the printing resolution, resulting in a heavily overcured part (Figure S7C, Supporting Information). Figure 2D shows a 3 cm × 2.5 cm hydrogel slab containing a printed vascular tree structure with numerous branches of different widths. The widths of the largest and smallest branches are 1760 and 50 μm respectively (Figure 2E). A variety of 3D models were printed on a 3.5 cm × 2.5 cm rectangular hydrogel substrate that include an array of different-sized hands, a truss and an array of miniature primitives (Figure 2F, Figure S7D, Supporting Information). The smallest hand is 4.5 mm high with little finger diameter of 150 μm and largest hand is 17 mm high. The truss has 1 cm edge length and contains 5 struts equally distributed on each side. The primitives (cube, cone, pyramid, cylinder and dome) are about 2 mm high (Figure 2G, Figure S7E, Supporting Information).
2.3. The Effects of Rapid Printing on the Part Quality and Cellular Functions in Large-Sized Models
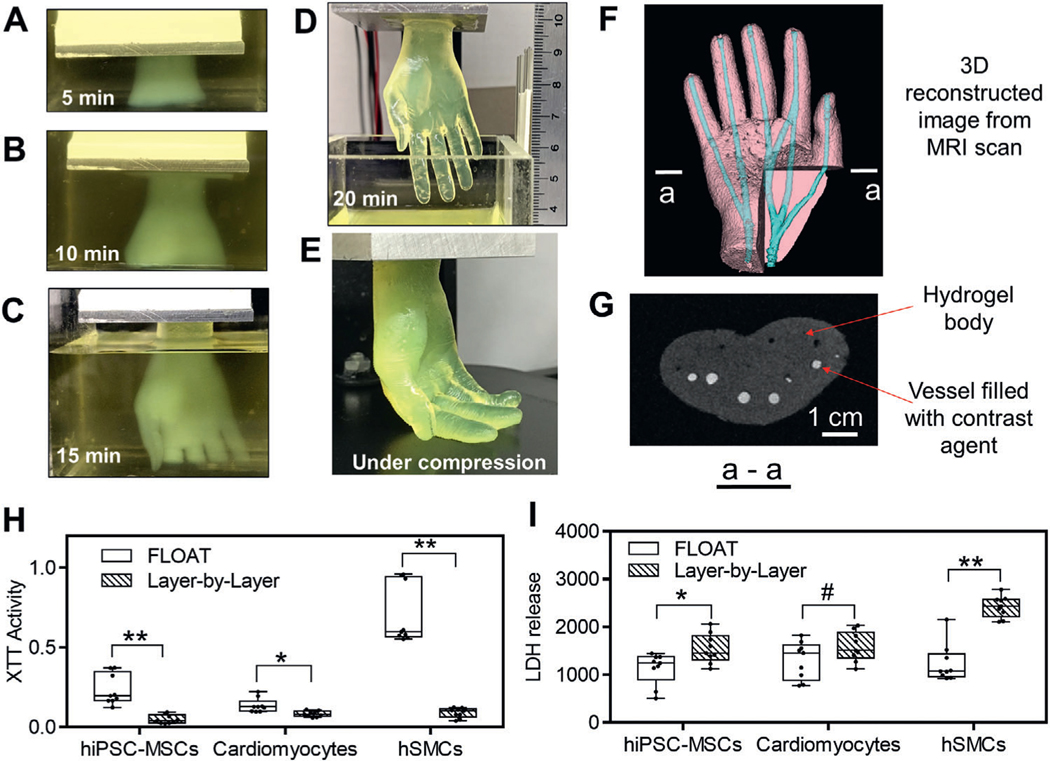
Here, we tested the effect of the rapid printing process of the FLOAT on the part quality and cellular health in large-sized hydrogel models. We first tested the ability of the FLOAT to rapidly fabricate centimeter-sized hydrogel models. A several centimeter-sized (2.6 × 1.7 × 5.6 cm) hydrogel human hand model was FLOAT-printed very rapidly in 20 min (Figure 3A–D, Movie S4, Supporting Information). This hand model is very compliant, as demonstrated by the bending of the fingers under compression and the free recovery upon release (Figure 3E, Figure S8, Movie S5, Supporting Information). In contrast, it took 2 h and 6.5 h to print the same model using the traditional layer-by-layer SLA process at 150 and 50 μm layer thickness, respectively. During this process, hydrogel dehydration occurred, causing distortion and detachment of the model (Figure 3F, Figure S9, Supporting Information). The shape fidelity of the computer model is maintained quite well in the printed hand model (Figure S10A,B, Supporting Information). The material composition and geometrical features such as the vascular channel structure of this hand model is compatible with magnetic resonance imaging (MRI), as demonstrated by the 3D reconstructed MRI scan image showing clearly the vascular channels (Figure 3G, Figure S10C,D, Supporting Information).
Figure 3.
The effects of rapid printing on part quality and cellular function in large-sized models. A–E) Demonstration of FLOAT printing process of a centimeter-sized human hand model. Sequential images of the hand model that was continuously formed in 10% PEGDA 400 Da preploymer pool at 5 min, 10 min, and 15 min and completed at 20 min. It would take 6.5 h to print the same model using traditional layer-by-layer printing method. D) The length of the completed hand model is 5.6 cm. E) Fingers were easily bent under compression, showing the compliance of the hydrogel hand model. F) Hand model printed in 2 h using the traditional layer-by-layer SLA process. Severe layer detachment and finger distortion occurred due to dehydration. G) 3D reconstructed image of FLOAT-printed hand model from MRI scanning. The channels were visualized with the aid of a contrast agent. Metabolic activity and cytotoxicity measured by H) XTT assay and I) LDH assay shortly after the printing of centimeter-sized, cell-laden samples. Cells in FLOAT-printed samples experienced much less printing-induced injury and have much higher metabolic activity than those in layer-by-layer printed samples. The above cell-laden samples were printed using 7% GelMA plus 2% PEGDA 8 kDa. n = 9. All box plots with whiskers represent the data distribution based on five number summary (maximum, third quartile, median, first quartile, minimum). **, p < 0.001; *, p < 0.05; #, p = 0.1 determined by two-tailed t-test.
We then tested the health, metabolism, and viability of a variety of cell types in FLOAT-printed centimeter-sized hydrogel models (6 min print time), and compared the results with same-sized, cell-laden hydrogel model printed using layer-by-layer SLA (2 h print time). Cell types that are often used in tissue engineering including human induced pluripotent stem cell-derived mesenchymal stem cells (hiPSC-MSCs), neonatal mouse cardiomyocytes and primary human skeletal muscle cells (hSMCs) were used in the tests. We showed that the averaged metabolic activities in FLOAT printed model are two to ten times higher than those in conventional layer-by-layer SLA printed model for all three cell types, as measured by the XTT assay (Figure 3H). The averaged cytotoxicity in FLOAT printed model is two to six times lower than that in conventional layer-by-layer printed model, as measured by lactate dehydrogenase (LDH) assay (Figure 3I). Together, these data suggest that cells in FLOAT-printed parts experienced much less printing-induced injury and have much higher metabolic activity than those in layer-by-layer SLA printed parts.
To further demonstrate the cell viability and function in FLOAT-printed complex structures, we printed gyroid models (2 cm edge length) with encapsulated human mesenchymal stem cells (hMSCs). The hMSCs were shown to remain viable, spread well and undergo osteogenic differentiation during a 9 d culture period, as demonstrated by the ALP positive staining (Figure S11, Supporting Information). The abilities of the FLOAT to print with uniform cell distribution and high cell density were also validated, as demonstrated in the Figures S12 and 13 (Supporting Information). The high cell density part was printed using 50 million cells per mL.
2.4. Embedded Vessel Network Is Critical to Long-Term Cell Functions in Large-Sized Models
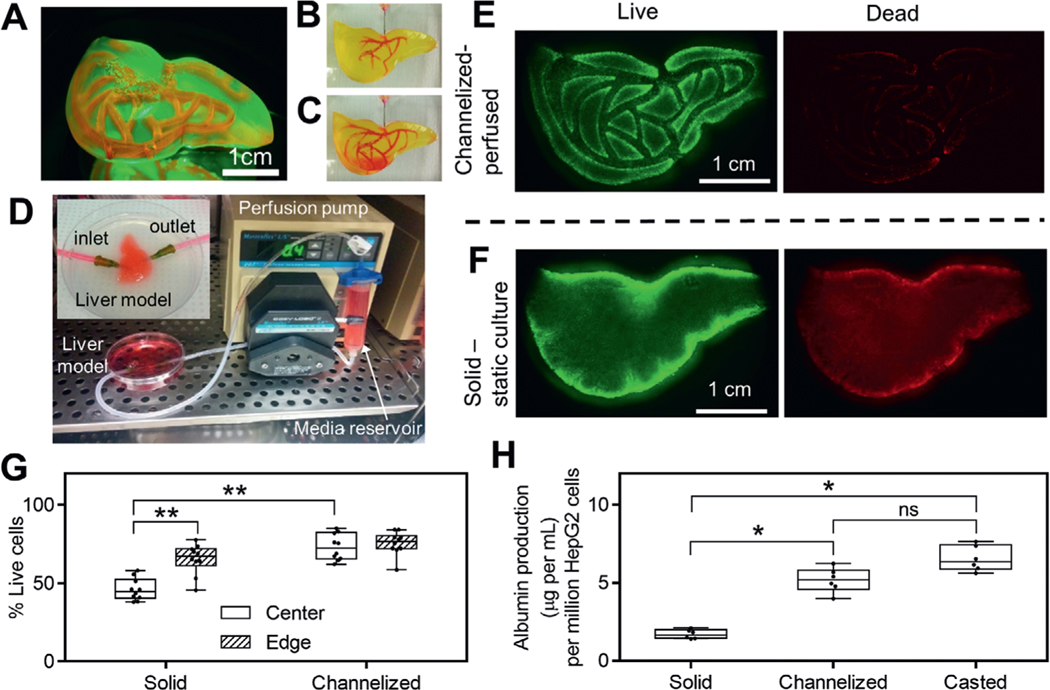
We tested the long-term cell survival and functions in FLOAT-printed, centimeter-sized hydrogel models. A liver-shaped model (3.5 cm × 2.5 cm × 1.5 cm) containing perfusable channels was FLOAT-printed in 5 min using cell-laden PEGDA prepolymer conjugated with RGD peptide (Movie S6, Supporting Information). The size of the liver model is larger than that of the rat and is comparable to the median liver size of a 26 weeks fetus.[28] Visual inspection of the liver model shows smooth surface and monolithic, translucent hydrogel body without clump, bubble or delamination (Figure 4A). Perfusable channel network was fabricated in the liver model to improve the mass transport in the bulk hydrogel body. Through judiciously limiting the light exposure in designed channel areas, we fabricated the bulk body of the liver model and perfusable channels together through a single FLOAT printing process. To examine the inter-connectivity and printing fidelity of the channel network, a small molecule dye (rhodamine B) was injected into the channels and allowed to flow through the network (Figure 4B,C, Movie S7, Supporting Information). The cross-sectional view of the liver model shows open lumen of the channels with a height of 3 mm and width of 800 μm, which is consistent with the design value and show no sign of overcuring (Figure S14A,B, Supporting Information). Immediately after injection, the channels contained the dye solution well and there was no dye leakage into the hydrogel interstitial space (Figure S14C, Supporting Information). Later, time-dependent dye diffusion into the interstitial space was observed. Full diffusion of the dye throughout the entire liver model was achieved over a 30 min period (Figure S14D, Supporting Information), as demonstrated by the gradual equilibration of the fluorescent dye intensity between the vascular channels and adjacent interstitial spaces during this period (Figure S14E, Supporting Information).
Figure 4.
Media perfusion in printed channels helps to maintain long-term cell functions. A) A liver model with smooth surface and monolithic, translucent hydrogel body was printed using 15% PEGDA 4 kDa. Vascular channel network was filled with rhodamine B and visualized under fluorescence. At the beginning B) and at the end C) of rhodamine B injection into prefabricated, vascular-like channels. D) Experimental setup of the perfusion system in an incubator. Inset shows enlarged view of the perfusion chamber for liver model. Inlet and outlet of the channel network are connected to perfusion tubing through 18 gauge needles. Fluorescent images of live and dead cells in channelized, perfused liver model (E) and solid, F) statically cultured liver model after 3 d of culture. G) Measured cell viability in channelized and solid liver models at both the center and edge regions, n = 10. H) Albumin production for channelized liver model, solid liver model and casted model after 6 d of culture, n = 6. Cell-laden liver models in E–G) were printed using 15% PEGDA 4 kDa conjugated with 10 × 10−3m RGD and liver model in H was printed using 8% GelMA plus 5% PEGDA 8 kDa. All box plots with whiskers represent the data distribution based on five number summary (maximum, third quartile, median, first quartile, minimum). **, p < 0.001; *, p < 0.05; determined by nonparametric unpaired t-test.
To support the survival of cells encapsulated in the bulk hydrogel body of the liver model, the inlet and outlet of the channel network were connected to a recirculating flow system (Figure 4D). Media perfusion in the channel network was maintained for up to 6 d and cell viability and albumin secretion by encapsulated HepG2 cells were analyzed. As a comparison, a non-channelized liver model of identical size and scaffold composition was fabricated and maintained in static culture. At the end of culture period, both liver models were sliced horizontally and vertically to allow the examination of cell viability throughout the bulk hydrogel body. Gross observation showed uniform cell distribution in the liver model without obvious clumps or cavities, confirming the good cell deposition ability of the FLOAT process (Figure 4E,F). Results of live/dead assay showed that perfused model has relatively high cell viability of approximately 80% in both the core and the edge regions (Figure 4G); however, the cell viability in the core region of the nonchannelized model is only 50%. The cell viability in the edge region of the nonchannelized model is reasonably high at ≈70%, possibly due to the direct contact with the culture media (Figure 4G). Albumin secretion of the perfused liver model was 5.2 μg per million cells which is nearly three times higher than that of the nonperfused liver model (Figure 4H). The casted hydrogel thin layer model was observed to secrete a slightly higher amount of albumin (6.5 μg per million cells) than the perfused liver model, possibly due to its small thickness (1 mm) that does not limit media diffusion. Together, these results suggested that the viability and physiological functions of cells encapsulated in FLOAT-printed, large-sized hydrogel model were maintained at reasonably high level under efficient media perfusion.
2.5. Endothelialize the Vessel Network in FLOAT-Printed Hydrogel Model
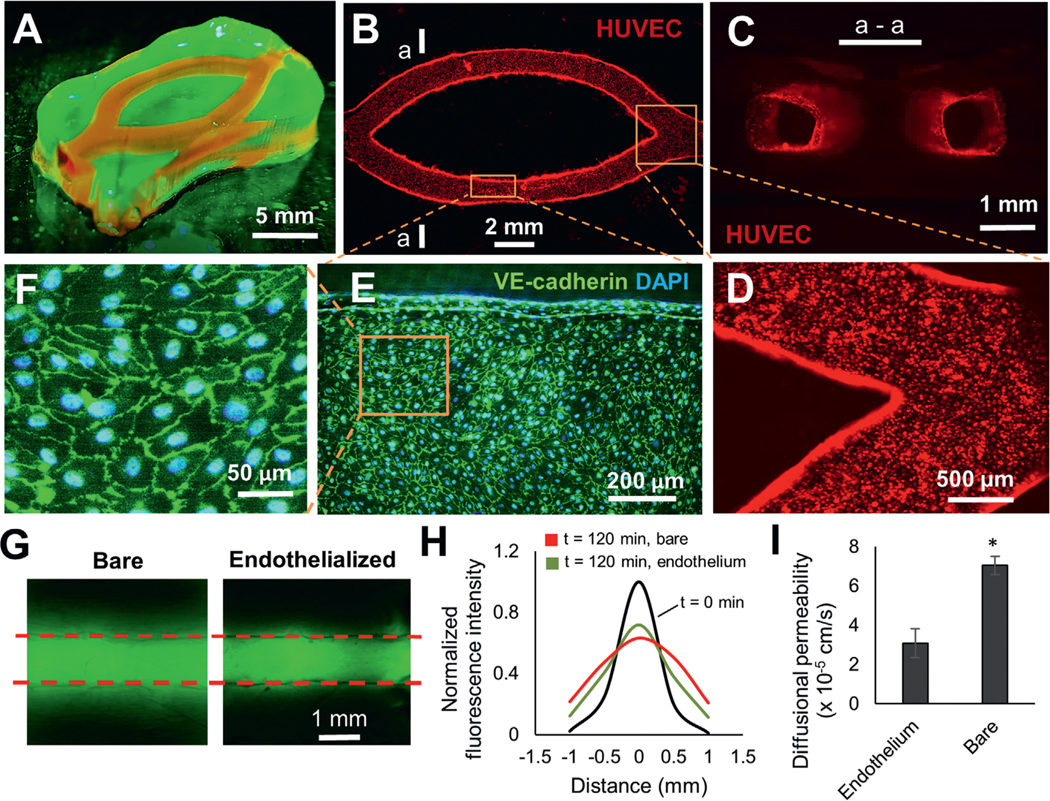
Since vascularization is critical for the long-term survival of engineered tissues in vivo,[29] we explored the potential to endothelialize the prefabricated channels in FLOAT-printed thick hydrogel models. In an initial trial, HUVECs were found to adhere poorly on the channel wall in PEGDA models even with the precoating of fibronectin. To improve endothelial cell adhesion and spreading, we blended GelMA with PEGDA prepolymer and found that increased GelMA component along with reduced PEGDA component helped to improve endothelial adhesion. Using 7% GelMA blended with 3% PEGDA 8000 Da, we printed a tissue patch model (2 cm × 1 cm × 4 mm) consisting of two channels for endothelialization demonstration (Figure 5A). After seeding, a large amount of endothelial cells adhered to the channel walls and formed patches of aggregates that smoothened out in the first 2 d (Figure S15, Supporting Information). A uniform and confluent layer of endothelial cells formed by day 3 and remained stable up to day 9 (Figure 5B; Figure S15, Supporting Information). The cross-sectional view of the channels showed that endothelial cells fully lined the lumen of the microchannels (Figure 5C). The endothelial layer is of good quality at both the straight segment and junction areas of the channels with well-established endothelial cell junctions (Figure 5D–F).
Figure 5.
Endothelialize the printed vessel network in FLOAT-printed model. A) A centimeter-size, channelized hydrogel model is illuminated under fluorescence. Channels are shown by injected rhodamine dye. B) A fluorescence image of HUVEC-lined channels 9 d after endothelial seeding. C) The cross-sectional view of endothelialized channels at section a—a shown in (B). D) Zoom-in view of endothelialized corner region of the channels. E) Immunofluorescent co-staining of the VE-cadherin and DAPI shows the formation of a confluent endothelial monolayer in the channel. F) A zoom-in view of the cell layer shows well-formed endothelial cell junctions. G) Fluorescence images show the diffusion of FITC–dextran (3—5 kDa) in bare and endothelialized channels 120 min after injection. H) The plot of normalized fluorescence intensity change due to dextran dye diffusion in the vicinity of a bare and an endothelialized channel over a 120 min period. I) Calculated diffusional permeability of bare and endothelialized channels. Models in this figure were printed using 7% GelMA + 3% PEGDA 8 kDa. Data are reported as mean ± SD, n = 3, * p < 0.05.
Endothelial barrier function was assessed by measuring the rate of solute permeation from the lumen to the surrounding scaffold. The FITC-dextran (3–5 kDa) in a bare channel was found to diffuse more rapidly and extensively than that in an endothelialized channel over a 120 min time period (Figure 5G,H). The diffusional permeability coefficient in bare channels was measured to be 7 × 10−5 cm s−1, and this number decreased to 3 × 10−5 cm s−1 in the presence of an endothelium (Figure 5I), which confirms the barrier function of the endothelium.
3. Discussion
Hydrogel fabrication has been one of the main focused areas in bioprinting, but much of the efforts have been made on improving the printing resolution.[29,30] Although the spatial resolution limited by nozzle size and material morphology control has been improved recently for extrusion-printing, which allows the creation of tissue models with biomimetic features such as vascular channels,[11,14] such point-by-point material deposition method is still limited by its inability to print at multiple length scales and its slow printing speed that exposes cells to prolonged environmental stresses. The addition of colored dyes as photoabsorbers has allowed the fabrication of multivascular network using the layer-by-layer SLA method; however, this method still suffers from the slow printing speed, which requires antisettling agents such as Xanthan gum and glycerol to prevent cell settlement during the fabrication of large-sized cell-laden tissue models.[20] To address these challenges, FLOAT method combines high printing speed with multiscale printing capacity to allow the fabrication of a centimeter-sized hydrogel model in several minutes, as compared to several hours needed in extrusion-based printing and layer-by-layer SLA printing for a similar-sized part.[11,31] The development of the FLOAT method is achieved through studying the interactive effect of the process parameters and precisely controlling the prepolymer formula to enable high-velocity flow. Our studies demonstrated the different photopolymerization conditions that permit and inhibit the high-velocity flow, which allowed the formulation of the optimal prepolymer composition. The low suction force occurred in the optimal curing condition was shown to be critical to drive the prepolymer flow and to maintain good printing quality, thus unveiling the fundamental mechanism of the process.
In the current study, we showed that the continuous SLA process is unique to the hydrogel prepolymer solutions and cannot be achieved in WI-resins. It is possible that continuous hydrogel printing can also be achieved with the O2 permeable window and external oxygen supply as described in the CLIP setup,[21] though the experimental setup will be more complicated. The current studies on the prepolymer flow and suction forces under controlled photopolymerization conditions should provide guidelines for such studies. Suction force is one of the limiting factors for the continuous printing of large-size models. We showed that the suction force in FLOAT hydrogel printing is several hundred times less than that in continuous resin printing. This can potentially be explained by the fluid mechanics theory of Stefan’s adhesion where separation force is inversely proportional to the cube of the separation distance and linearly proportional to the fluid viscosity.[23,32] Although we did not measure the dead zone thickness in our continuous resin printing, it should be smaller than that in the CLIP method (30μm). Since the liquid flow layer thickness is more than 650 μm in the FLOAT method, the more than five times difference in the separation distance can lead to several hundred times difference in the separation force between the two methods. The difference in the viscosity further contributes to the difference in the separation force. The viscosity of the WI-resin is in the range of 100–10000 cp, but that of the hydrogel prepolymer solution is in between 6.1 and 38.5 cp for 10%–20% PEG 3500 and 8000 and in between 5 and 22 cp for 10% GELMA.[33,34] These polymer solution concentrations are consistent with those used in the current study. The low suction force in FLOAT method permits the fabrication of large, solid parts with good quality, thus improving over continuous resin printing where only small-size, thin-walled parts can be fabricated due to the large suction force of the WI resin.[21–23]
In the current study, we performed material optimization to seek hydrogel formula with both good printability and good biocompatibility. One of the major factors affecting the printability is the mechanical strength of the material. Sufficient mechanical strength is needed to avoid layer delamination or part breakage during continuous printing. Owing to the very low suction force in FLOAT, we show that a low, soft tissue-like mechanical strength (1–8 kPa) of the printed part is sufficient to support the continuous printing. In the current work, we added PEGDA to the GelMA mixture (total polymer less than 13% v/v) to enhance its mechanical strength and showed that this mixture is sufficient to support the metabolic function of the encapsulated liver cells. Future strategies to enhance the mechanical strength of polymeric materials may include improving photo-crosslinking chemistry[35] or developing nanomaterial doped polymer composites.[36] However, it should be noted that high mechanical strength out of the stiffness range of the soft tissues (1–100 kPa) should be avoided for tissue engineering applications, since it will not support the growth and function of encapsulated cells. In studies that do not involve cell encapsulation, such as the nerve conduits fabricated by Zhu et al.,[37] high stiffness hydrogel parts (300–3000 kPa) have been produced by using high concentration precursor solutions (total polymer 32.5% v/v). Relatively high concentration precursor solutions (total polymer 20% v/v) was used to build cell-laden constructs using layer-by-layer SLA, but they only supported short term cell viability and function within 24 h.[20]
4. Experimental Section
Printing System Setup:
The printing system presented in the current study is based on MP-SLA setup with significant modification to the hardware and software systems to enable a continuous printing process. The system utilized a bottom-up configuration where a digital light processor (DLP) device dynamically projects a digital mask image onto photosensitive material at the bottom of a transparent liquid tank. A high-resolution (1080P) dynamic micromirror device (DMD, Texas Instruments) was used as a dynamic digital mask generator, a blue LED (405 nm) or UV LED (385 nm) was used as the light source, and a high-speed FPGA chip and on-board RAM was used to store large volume of images and display the images with ultrafast speed to enable reliable and smooth continuous printing. A holistic framework was developed with customized and integrated hardware, software and firmware to enable the ultrafast and multiscale printing by leveraging the knowledge on hydrogel material property, process dynamics and light–matter interaction. In particular, dynamic synchronization of the image generation and the motion of build-platform were achieved using custom software, which allowed for high-speed continuous printing and eliminated layer-by-layer process. Grayscale mask image optimization and light calibration were conducted to achieve the highest accuracy and precision, which enables multiscale printing. A custom-made tank of dimensions 6 cm × 4 cm × 2cm was fabricated using a transparent glass as the base. The glass was coated with polydimethylsiloxane (PDMS) to render the surface hydrophobic, thereby preventing attachment of the cured part to the bottom of the tank.
Hydrogel Material Preparation:
Polyethylene glycol diacrylate (PEGDA) MW 4000 Da and 8000 Da were synthesized following published method.[38] Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), a visible light photoinitiator, was synthesized following published method by Anseth group.[39] To enhance cell adhesion onto PEGDA scaffold, cell adhesive arginine-glycine-aspartic acid-serine (RGDS) peptide (Bachem) was conjugated to monoacrylated polyethylene glycol succinimidyl valerate (a-PEG-SVA, LaysanBio) following the method described by Turturro et al.[40] Gelatin from porcine skin was purchased from Sigma. Gelatin methacrylate was synthesized using previously published protocol.[41] The final product was dialyzed and freeze dried before use. A detailed list of hydrogel material composition for all printed parts is listed in Table S1 (Supporting Information).
Cell Culture and Maintenance:
Human liver cell line HepG2/C3A (ATCC) was maintained in minimum essential medium (MEM) supplemented with 10% FBS and 1% penicillin/streptomycin. RFP-tagged human umbilical vein endothelial cells (RFP-HUVECs, Angio-proteomie) were cultured in endothelial cell growth medium (EGM-2, Promocell). Human dermal fibroblasts were maintained using F12K media supplemented with 2% FBS. The human primary skeletal muscle cells were cultured in proliferation medium composed of high glucose Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 20% fetal bovine serum in a humidified 37 °C incubator at 5% CO2. hiPSC-MSCs were thawed in DMEM, high glucose, GlutaMAX Supplement, pyruvate media (Thermofisher, 10569010) with 10% FBS containing 10 × 10−6 m ROCK inhibitor Y-27632 (MedChemExpress, HY-10583). After 24 h, the cells were cultured in fresh media without the ROCK inhibitor. To obtain GFP-tagged HF-MSCs, lentivirus encoding GFP was produced using plasmid for GFP expression (CMV-GFP) purchased from Addgene (Cambridge MA). Transduction was carried out using CMV-GFP lentiviruses in growth media with 8 μg mL−1 polybrene (Sigma-Aldrich) for 3–4 h. A detailed list of printed hydrogel parts with cells is listed in Table S2 (Supporting Information).
Cardiomyocyte Isolation:
All animal experiments were performed according to the protocol approved by the University at Buffalo Institutional Animal Care and Use Committee. Primary mouse cardiomyocytes were harvested following manufacturer recommended procedure. Briefly, day 1–4 neonatal mice were euthanized by cervical dislocation and their hearts dissected. Cardiac tissue was minced into 1–3 mm3 pieces and digested using the Pierce Primary Cardiomyocyte Isolation Kit (ThermoScientific). For each experimental replicate, primary cardiomyocytes were pooled from seven neonatal mouse hearts (yielding approximately 5–10 × 106 cells). The entire isolation protocol was accomplished within 2 h of animal euthanasia to ensure high cell yield and reproducible viability. Isolated neonatal mouse cardiomyocytes were cultured in DMEM for primary cell isolation (Thermofisher, 88287) supplemented with 10% heat inactivated FBS. A detailed list of printed hydrogel parts with cells is listed in Table S2 (Supporting Information).
Measurement of Liquid Flow Velocity and Uncured Liquid Layer Thickness:
The flow properties of the hydrogel prepolymer solution in the uncured liquid layer during FLOAT printing was studied by tracking the movement of the fluorescent microbeads in the solution. Green fluorescent microbeads (d ≈ 25 μm, Fluoresbrite, Polysciences) were mixed with 20% or 80% PEGDA prepolymer solution (MW 4000 Da) containing 0.1%, 0.6% and 1% photoinitiator LAP and 0.03% photoabsorber Orange G. Both 20% PEGDA and 80% PEGDA solutions were used, and two printing speeds of 50 and 125 μm s−1 were tested. The video of the microbead movement during continuous printing of a 4 mm wide part was recorded through a side-mounted inspection microscope at five frames per second. The bead flow trajectories were tracked using ImageJ, and the flow velocity was determined as the travel distance divided by the travel time. The uncured liquid layer thickness was defined as the distance between the bottom surface of the cured part and the glass tank base and was measured in the video. The bead flow velocity at different heights in uncured liquid layer was measured and compared.
Measurement of Suction Force at the Curing Interface:
The suction forces was measured at the curing interface during FLOAT printing of 10% and 20% PEGDA 4000 Da hydrogel pre-polymer solutions containing 0.6% LAP photoinitiator using an UXcell load cell sensor with a maximum capacity of 100 g (model # a11112800ux0213). The suction force during MP-SLA printing of acrylate-based resin part (Makerjuice Standard, MakerJuice Labs) was measured using an UXcell load cell sensor with a maximum capacity of 2 kg (Model # a14071900ux0072). The load cell weighing sensor was mounted as a cantilever beam structure, with one end fixed to the Z stage and the other end fixed to the build platform. The suction force is recorded at three different platform moving velocities: 0.05, 0.2, and 0.5 mm s−1. A solid cube model with 1 cm edge length was used in the printing of both the PEGDA hydrogel and resin materials.
Curing Depth Study:
Curing depth is the thickness of the layer that is cured by light irradiation at critical exposure, and it is significantly affected by the exposure energy and the light absorbing property of the photosensitive material. Both the exposure energy and the type and concentration of the photoabsorbers were varied to study their effects on the curing depth during FLOAT printing of PEGDA hydrogel. The base prepolymer solution contains 20% PEGDA 4000 Da, 0.6% LAP photoinitiator and 0.01% TEMPO. Five different photoabsorbers including 0.1% Orange G (Sigma), 0.1%, 0.15% and 0.2% Quinoline yellow (Sigma), 0.1% Tinuvin Carboprotect (a gift from BASF), 0.1% 4-hydroxy benzophenone (Sigma) and 0.1% benzotriazole (Sigma) were mixed with the base solution separately.
Exposure energy was calculated as the production of energy density and the exposure time and was varied between 8 to 28 mJ cm−2. A rectangular image (4.7 × 9.4 mm) was projected for various exposure times, and the thickness of the cured layer was measured using a Verasonics ultrasound system coupled with an L22–8 linear transducer array at the frequency of 15 MHz. Furthermore, the effect of curing depth control on the printing resolution by printing a turbine rotor model under various photoabsorber conditions was assessed. The base of the turbine model is 10 mm and the thickness of the individual blades is 0.65 mm. The base prepolymer solution is the same as the one used in curing depth study. Quinoline yellow was tested at 0.05%, 0.2%, and 0.3% and orange G and HMBS at 0.2%.
Optimization of Hydrogel Materials for FLOAT Printing:
To understand the compatibility of FLOAT printing with various hydrogel materials, pure PEGDA 4000 Da prepolymer was prepared with varying concentrations at 10%, 15%, 20%, and 30% and pure GelMA prepolymer with two concentrations at 10% and 15%. Blended prepolymer containing GelMA and PEGDA 8000 Da at following concentrations was also prepared: 6% + 2%, 7% + 3%, 8% + 5%, 10% + 2%, 10% + 5%, and 11% + 4%. Results show that blended prepolymers with less than 2% PEGDA were too soft to print, as demonstrated by the peeling of parts during printing.
Compression Testing to Measure Part Stiffness:
Unconfined compression tests were performed on 3D printed hydrogel samples with varying composition and monomer concentrations. Briefly, PEGDA 4000 Da at 10%, 15%, 20%, 30% and 5% PEGDA 8000 Da plus 8% GelMA composite hydrogel prepolymers were prepared with 0.6% LAP photoinitiator. 6 mm cubes were printed using either FLOAT method or layer-by-layer method and allowed to swell for 24 h before compression testing. Compression test was performed on a Mark-10 motorized test stand equipped with a Mark-10 Series 5 force gauge (force range 0.0005–1 N) at a loading speed of 5 mm min−1. The loading force and sample deformation were recorded in real time and were later used to calculate stress and strain. Young’s modulus was determined from the slope of the stress–strain curve. Young’s moduli of samples fabricated using FLOAT method and layer-by-layer method were compared to that of nonprinted casted hydrogels.
Measure the Effect of the Printing Speed on Cellular Metabolic Activity and Cytotoxicity:
The effect of the printing speed on cellular metabolic activity and cytotoxicity was assessed on cell-laden hydrogel models printed using either FLOAT method or layer-by-layer SLA method. Cell-laden solid hydrogel blocks (3 mm × 3 mm × 2 cm) were printed using both methods. A colorimetric XTT assay (2,3-bis-(2-methoxy4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide disodium salt, biotium, 10060; 5-methylphenazinium methyl sulfate (coupling agent), VWR, 200 000–760) was performed to determine the metabolic activity of cells, and an absorbance LDH assay (Lactate dehydrogenase assay, CytoTox-ONE Homogeneous Membrane Integrity Assay kit from Promega, G7890) was used to determine the level of cytotoxicity caused by membrane compromisation, three different cell types that are commonly used in tissue engineering including hiPSC-MSCs, neonatal mouse cardiomyocytes and primary human skeletal muscle cells (hSMCs) were used in the printing and analyses. A prepolymer solution containing 7% GelMA plus 2% PEGDA 8000 Da, 0.6% LAP and 0.015% QY was mixed with the cell suspension. A solid hydrogel part with dimensions of 3 × 3 × 20 mm was 3D printed using either layer-by-layer SLA method (2 h printing time) or FLOAT method (6 min printing time). The part printed using FLOAT method was maintained in an incubator for nearly 2 h to wait for the completion of the part printed using layer-by-layer method. The two parts were then analyzed at the same time. The parts were sliced and digested with 5 mg mL−1 collagenase for 30 min to collect the cells for metabolic activity measurement using XTT assay. The supernatant was used to determine LDH activity.
Centimeter-Size Liver Model Fabrication:
The 3D computer model of the liver containing channel network was created in Rhinoceros 3D software and sliced. The channel network was designed based on the distribution of the main hepatic blood vessels but with modifications to facilitate media perfusion and nutrient delivery. The cross-section of a single channel is rectangular with a width of 0.8–1 mm and a height of 3 mm. To print the liver model of 3.5 cm × 2.5 cm × 1.5 cm, 15 mL of 15% PEGDA 4000 Da prepolymer containing 0.6% LAP photoinitiator and 0.15%–0.2% Quinoline yellow was prepared, with the supplement of 10 × 10−3 m a-PEG-RGDS for cell adhesion. 8 × 106 cells mL−1 of HepG2 cells were suspended in the above solution. Cell-laden liver models with or without vasculature channels were printed in a laminar flow biosafety cabinet and transferred to a P-100 dish containing culture media for long-term culture in a CO2 incubator. Both channelized and solid liver models were submerged in 10 mL of culture media in the P-100 dish, but the channelized model was further supplied with media perfusion. A detailed list of printed hydrogel parts with cells is listed in Table S2 (Supporting Information).
Dye Injection and Diffusion in the Liver Model:
To examine the interconnectivity of the channel network and the mass transport of small molecule solutes in the liver model, rhodamine B solution (1 mg mL−1) was injected through the inlet of the channel network using an 18-gauge needle. Dye diffusion over a period of 30 min was imaged under fluorescence at 5 min interval using a MVX10 MacroZoom microscope (Olympus) equipped with 1X Apo objective. Obvious diffusion of the rhodamine into the bulk hydrogel scaffold was observed 5 min after injection. Time-dependent dye diffusion in the vicinity of a channel was quantified by measuring the fluorescence intensity.
Recirculating Flow System Setup:
During media perfusion of the channelized liver model, inlet and outlet of the vascular channels were connected to 1/16 in. sterile peristaltic tubings (PharMed BPT) through 18-gauge needles. Tubings were then hooked up to a Masterflex peristaltic pump (Cole Parmer). A 30 mL syringe (Nordson EFD Inc.) was used in the fluidic circuit as a media reservoir. The total media volume in the fluidic circuit was about 20–25 mL. The entire fluid circuit operated inside a CO2 incubator at 37° under 0.4 mL min−1 flow rate to avoid disturbance to the construct. The channelized liver model was submerged in culture media in a P-100 dish during perfusion.
Encapsulated Cell Viability and Albumin Production in Cultured Liver Model:
The relatively long-term cell viability in the liver models was assessed after 72 h of culture using live/dead viability kit (L-3224, ThermoFisher). To ensure diffusion of staining dyes into the center of the centimeter-size liver model and also to better visualize the cells in the center region, models were sliced horizontally at the mid-height before staining. Live and dead cells were visualized on a fluorescence microscope, and the images from six representative areas at the edge and center of the liver models were used for quantification. The results were compared between channelized and solid liver models.
Albumin secretion by HepG2 cells encapsulated in liver models with or without vasculature channels was quantified using ELISA (Human Albumin ELISA kit, Ab 108788, Abcam). Liver models (polymer composition: 8% GelMA plus 5% PEGDA 8000 Da and 0.6% LAP) were FLOAT printed with a mixed cell population containing HepG2 cell to normal human dermal fibroblast at a ratio of 2:1 and a final density of 8 million cells per mL. The channelized model was maintained under media perfusion, and the solid model was maintained under static culture. Casted hydrogel thin layer of 5 mm × 5 mm × 1 mm was used as a nonprinted control. After 6 d in culture, the liver models were sliced through the horizontal and vertical axis, and sliced hydrogel parts were incubated for 3 h to allow the release of the albumin from hydrogel core into the culture media. Cell culture supernatants were collected afterwards and used in ELISA assay. The absorbance of the samples was measured on a Sapphire microplate reader (TECAN) at 450 nm. Albumin standards were used to calculate the amount of albumin secreted by the cells. The concentration of albumin was then normalized based on HepG2 cell number.
Endothelialization of Vascular Channel Network:
To demonstrate endothelialization of vascular-like channels, a tissue patch model (2 cm × 1 cm × 4 mm) was FLOAT-printed containing two microchannels, using 7% GelMA + 3% PEGDA 8000 Da mixed prepolymer solution composed of 0.6% LAP and 0.015% QY. Prior to HUVEC injection, 0.01 mg mL−1 fibronectin solution was injected into the channels and incubated for 45 min. A 50 μL suspension of RFP-HUVECs containing 10 × 106 cells per mL was injected into the channels, and the cells were allowed to adhere for 5 h and then placed on a shaker vibrating at 60 rpm for long-term culture in the incubator. The adhesion, growth, and morphology of HU-VECs in the channels were evaluated over a 9 d culture period. A detailed list of printed hydrogel parts with cells is listed in Table S2 (Supporting Information).
Diffusional Permeability Testing:
FITC-conjugated dextran (MW 3–5 kDa) was used to study the diffusional permeability in the absence and presence of endothelial layer in the printed channels. EGM-2 media containing 100 μg mL−1 FITC dextran was injected into the channels with or without endothelial lining and the dye diffusion over a period of 2 h was imaged under fluorescence at 30 min interval using a MVX10 MacroZoom microscope (Olympus) equipped with 1× Apo objective. The changes of the intensity profile over time were quantified using ImageJ.
Diffusional permeability, Pd (cm s−1) of FITC-dextran for a channel with diameter, d is quantified by using the equation[14,42]:
| (1) |
where ΔI is the increase in the fluorescence intensity in the lumen right after injection of the dextran dye, dI/dt is the slope of the fluorescent intensity increase in the surrounding region with respect to time. Diffusional permeability of channels with and without endothelial lining were calculated and compared.
Immunofluorescence Staining and Microscopy:
Immunofluorescence staining was used to assess the uniformity of the endothelial layer in the vascular channels. After 9 d in culture, the printed models were fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 for 30 min at RT. The constructs were washed three times with PBS. Prior to incubation with primary antibody for VE-cadherin (ab33168, 1:500, Abcam), the constructs were placed in a blocking solution containing 3% BSA solution for 3 h at RT. Primary antibodies to VE-cadherin was incubated overnight concentration in 1% BSA. The constructs were then washed with PBS three times to remove unbound primary antibodies followed by secondary blocking step with 10% Goat serum for 3 h at RT. Alexa Fluor 488-conjugated anti-rabbit IgG (Abcam, ab150077, 1:400) was used as the secondary antibody. Samples were counterstained with DAPI to visualize nuclei. Confocal microscopy was performed on an Andor Technology DSD2 confocal unit coupled to an Olympus IX-81 motorized inverted microscope equipped with Plan-Apochromat × 10 air objective. Images were taken with an optical slice of 2μm and 2D projected images were obtained using Z-stack tool on ImageJ.
Magnetic Resonance Imaging (MRI):
MRI was performed on the printed hydrogel hand model using a 4.7 Tesla preclinical scanner incorporating the ParaVision 3.02 imaging platform and a 72 mm I.D. quadrature radiofrequency coil (Bruker BioSpin, Billerica MA). Prior to imaging, printed vascular channels were filled with mineral oil to aid in visualization of the channel morphology during MRI. Following scout scans, a three-dimensional, spoiled gradient echo scan was acquired encompassing the entire model. Acquisition parameters include: echo/repetition time = 3/15 ms, flip angle = 40°, field of view = 64 × 50 × 40 mm, averages = 2, acquisition matrix = 256 × 200 × 160.
Following acquisition, regions of interest were created for both the hand and channels using Analyze 10.0 (AnalyzeDirect, Overland Park, KS). A transparent, volumetric rendering was performed within Analyze using opacity values of 0.6 and 1.0 for the hand and channels, respectively.
Measure the Degradation of Printed Scaffold:
Eight-arm PEG Norbornene (tripentaerythritol) (PEGNB) was purchased from Jenkem Technology (Plano, TX). Dithiothreitol was purchased from Thermofisher Scientific. Eight-arm PEG norbornene (tripentaerithrytol) is a hydrolytically degradable PEG molecule that forms crosslinks under light exposure.[43,44] Herein FLOAT printing was used to compare the degradation rates of polymer blends of 8% GelMA and 5% PEGDA or 5% PEGNB. Prepolymer solutions containing 8% GelMA plus 5% PEGDA or PEGNB along with 0.6% LAP were prepared and 5 mm × 5 mm × 2 mm parts were FLOAT printed and incubated in PBS for up to 20 d. The dry weights of the parts were measured on Day 0, 10, and 20 to observe the polymer weight change with respect to time. A detailed list of printed hydrogel parts with cells is listed in Table S1 (Supporting Information).
Osteogenic Differentiation:
For demonstration of osteogenic differentiation in 3D printed models, human mesenchymal stem cells (hMSCs) were encapsulated in a gyroid structure printed using FLOAT using a prepolymer solution containing 7% GelMA plus 3% PEGDA and 0.6% LAP as photoinitiator. One day after printing, the media was changed to osteogenic differentiation media (growth media supplemented with 0.1 × 10−6 m dexamethasone, 50 × 10−6 m ascorbic acid and 10 × 10−3 m 2-glycerol phosphate). After 8 d in differentiation media, the models were tested for viability using live/dead viability kit (L-3224, ThermoFisher) and osteogenic differentiation using Fast Blue RR Salt/Naphtol (Sigma). A detailed list of printed hydrogel parts with cells is listed in Table S2 (Supporting Information).
Statistics:
Data are presented as the mean with error bars showing the standard deviation (SD). Nonparametric unpaired t-test or one-way ANOVA was used to determine statistical significance for two-group comparisons or multiple-group comparisons, respectively. Values of p < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
Research reported in this study was supported by National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number R01EB019411 (R.Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also acknowledge the funding support from the School of Engineering and Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The technology described in the current work is included in a pending US patent application.
Contributor Information
Nanditha Anandakrishnan, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Hang Ye, Department of Industrial and Systems Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Zipeng Guo, Department of Industrial and Systems Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Zhaowei Chen, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Kyle I. Mentkowski, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA Department of Medicine Division of Cardiology Jacobs School of Medicine and Biomedical Sciences University at Buffalo Buffalo, NY 14203, USA.
Jennifer K. Lang, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA Department of Medicine Division of Cardiology Jacobs School of Medicine and Biomedical Sciences University at Buffalo Buffalo, NY 14203, USA; VA WNY Healthcare System Buffalo, NY 14215, USA.
Nika Rajabian, Department of Chemical and Biological Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Stelios T. Andreadis, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA Department of Chemical and Biological Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Zhen Ma, Department of Biomedical and Chemical Engineering Syracuse Biomaterials Institute Syracuse University Syracuse, NY 13244, USA
Joseph A. Spernyak, Department of Cell Stress Biology Roswell Park Comprehensive Cancer Center Buffalo, NY 14263, USA
Jonathan F. Lovell, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA
Depeng Wang, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA.
Jun Xia, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA
Chi Zhou, Department of Industrial and Systems Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA
Ruogang Zhao, Department of Biomedical Engineering University at Buffalo The State University of New York Buffalo, NY 14260, USA
References
- [1].Atala A, Kasper FK, Mikos AG, Sci. Transl. Med 2012, 4, 160rv12. [DOI] [PubMed] [Google Scholar]
- [2].Murphy S, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [3].Sears NA, Seshadri DR, Dhavalikar PS, Cosgriff-Hernandez E, Tissue Eng., Part B 2016, 22, 298. [DOI] [PubMed] [Google Scholar]
- [4].Varkey M, Visscher DO, van Zuijlen PP, Atala A, Yoo JJ, Burns Trauma 2019, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ding S, Feng L, Wu J, Zhu F, Tan Z, Yao R, ACS Biomater. Sci. Eng 2018, 4, 3108. [DOI] [PubMed] [Google Scholar]
- [6].Ozbolat IT, Hospodiuk M, Biomaterials 2016, 76, 321. [DOI] [PubMed] [Google Scholar]
- [7].Cui H, Nowicki M, Fisher JP, Zhang LG, Adv. Healthcare Mater 2017, 6, 1601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens MM, Fischer H, Adv. Healthcare Mater 5, 326 2016. [DOI] [PubMed] [Google Scholar]
- [9].Aguado BA, Mulyasasmita W, Su J, Lampe KJ, Heilshorn SC, Tissue Eng., Part A 2011, 18, 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nair K, Gandhi M, Khalil S, Yan KC, Marcolongo M, Barbee K, Sun W, Biotechnol. J 2009, 4, 1168. [DOI] [PubMed] [Google Scholar]
- [11].Kang HW, Lee SJ, Ko I, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [12].Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Finberg AW, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [13].Miller J, Stevens K, Yang M, Baker B, Nguyen D, Cohen D, Chen C, Nat. Mater 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kolesky D, Homan K, Skylar-Scott M, Lewis J, Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bertassoni L, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino A, Khademhosseini A, Lab Chip 2014, 14, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang AP, Qu X, Soman P, Hribar KC, Lee JW, Chen S, He S, Adv. Mater 2012, 24, 4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhou C, Chen Y, Yang Z, Khoshnevis B, Rapid Prototyping J 2013, 19, 153. [Google Scholar]
- [18].Ma X, Qu X, Zhu W, Li Y, Yuan S, Zhang H, Chen S, Proc. Natl. Acad. Sci. USA 2016, 113, 2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cvetkovic C, Raman R, Chan V, Williams BJ, Tolish M, Bajaj P, Bashir R, Proc. Natl. Acad. Sci. USA 2014, 111, 10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Grigoryan B, Paulsen SJ, Corbett D, Sazer D, Fortin C, Zaita A, Miller J, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tumbleston JR, Shirvanyants D, Ermoshkin N, Janusziewicz R, Johnson AR, Kelly D, DeSimone JM, Science 2015, 347, 1349. [DOI] [PubMed] [Google Scholar]
- [22].Wang Q, Sun Y, Guo B, Li P, Li Y, AIP Adv. 2019, 9, 015225. [Google Scholar]
- [23].Pan Y, He H, Xu J, Feinerman A, Rapid Prototyping J 2017, 23, 353. [Google Scholar]
- [24].Asmani M, Velumani S, Li Y, Wawrzyniak N, Hsia I, Chen Z, Zhao R, Nat. Commun 2018, 9, 2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Discher DE, Janmey P, Wang YL, Science 2005, 310, 1139. [DOI] [PubMed] [Google Scholar]
- [26].Zhang R, Larsen NB, Lab Chip 2017, 17, 4273. [DOI] [PubMed] [Google Scholar]
- [27].Lin H, Zhang D, Alexander PG, Yang G, Tan J, Cheng AW, Tuan RS, Biomaterials 2013, 34, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tongprasert F, Srisupundit K, Luewan S, Tongsong T, Clin J. Ultrasound 2011, 39, 74. [DOI] [PubMed] [Google Scholar]
- [29].Griffith C, Miller C, Sainson R, Calvert J, Jeon N, Hughes C, George S, Tissue Eng. 2005, 11, 257. [DOI] [PubMed] [Google Scholar]
- [30].Shandalov Y, Egozi D, Koffler J, Dado-Rosenfeld D, Ben-Shimol D, Freiman A, Levenberg S, Proc. Natl. Acad. Sci. USA 2014, 111, 6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hinton T, Jallerat Q, Palchesko R, Park J, Grodzicki M, Shue HJ, Feinberg AW, Sci. Adv 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liravi F, Das S, Zhou C, Comput.-Aided Des 2015, 99, 134. [Google Scholar]
- [33].Gonzalez-Tello P, Camacho F, Blazquez F, J. Chem. Eng. Data 1994, 39, 611. [Google Scholar]
- [34].Wang Z, Tian Z, Menard F, Kim K, Biofabrication 2017, 9, 044101. [DOI] [PubMed] [Google Scholar]
- [35].Pawar AA, Saada G, Cooperstein I, Larush L, Jackman JA, Tabaei SR, Magdassi S, Sci. Adv 2016, 2, e1501381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].O’Brien CM, Holmes B, Faucett S, Zhang LG, Tissue Eng., Part B 2015, 21, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhu W, Tringale KR, Woller SA, You S, Johnson S, Shen H, Xu W, Mater. Today 2018, 21, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mann B, Gobin A, Tsai A, Schmedlen R, West J, Biomaterials 2001, 22, 3045. [DOI] [PubMed] [Google Scholar]
- [39].Fairbanks B, Schwartz M, Bowman C, Anseth K, Biomaterials 2009, 30, 6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Turturro M, Sokic S, Larson J, Papavasiliou G, Biomed. Mater 2013, 8, 025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen Y, Lin R, Qi H, Yang Y, Bae H, Melero-Martin J, Khademhosseini A, Adv. Funct. Mater 2012, 22, 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wong AD, Russell LM, Katt ME, Searson PC, ACS Biomater. Sci. Eng 2018, 5, 633. [DOI] [PubMed] [Google Scholar]
- [43].Aimetti AA, Machen AJ, Anseth KS, Biomaterials 2009, 30, 6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS, Adv. Mater 2009, 21, 5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.