Abstract
Objective
Colon cancer has high morbidity and mortality rates, and proliferation, invasion and migration play an important role in colon cancer progression. Here, the effects of inhibin subunit beta A (INHBA) on cell proliferation, invasion and migration were investigated.
Methods
The UALCAN database was used to assess INHBA expression in colon cancer tissues and predict the survival of patients with high and low INHBA expression. The relevant proteins were detected by RT-qPCR and western blot. Cell transfection was performed to overexpress or inhibit INHBA and versican (VCAN). The high correlation between INHBA and VCAN found through LinkedOmics and StarBase databases was verified by immunoprecipitation assays. Cell proliferation was detected by cell counting kit-8 and colony formation assays. Wound healing and Transwell assays were used to assess migration and invasion.
Results
INHBA expression was upregulated in colon cancer tissues and cells. INHBA inhibition impaired the proliferation, migration and invasion of these cells. In addition, we confirmed the correlation between INHBA and VCAN in colon cancer cells. Finally, we found that INHBA interference inhibited the aggressive behavior of colon cancer cells by downregulating VCAN.
Conclusion
INHBA promotes the proliferation, migration and invasion of colon cancer cells through the upregulation of VCAN.
Keywords: Inhibin subunit beta A, colon cancer, proliferation, migration, invasion, versican
Introduction
Colon cancer is one of the most common malignant tumors, and it frequently occurs at the junction of the rectum and the sigmoid colon in the digestive system. 1 The clinical mortality and morbidity of colon cancer remain high, with 1.2 million new colon cancer cases each year and about 600,000 deaths due to colon cancer. 2 Therefore, an in-depth understanding of the mechanisms underlying the occurrence and development of colon cancer is of great significance to improve the prognosis of patients.
INHBA is a member of the transforming growth factor (TGF)-β superfamily and plays an important role in several cancers. A previous study reported that the abnormal methylation and differential expression of INHBA in gastric cancer were related to the prognosis of patients. 3 Furthermore, INHBA regulates the expression of IL13Ralpha2 to promote the metastasis of breast cancer cells, 4 and targeting the INHBA/TGF-β axis inhibits the formation and growth of prostate tumors. 5 These results indicate that INHBA is related to the occurrence, development and prognosis of various cancers. Previous studies have also identified INHBA as a prognostic predictor in patients with colorectal adenocarcinoma and colon cancer.6,7 Furthermore, miR-6785-5p targets INHBA to regulate the occurrence and progression of gastric cancer. 8 Therefore, we hypothesized that INHBA plays a certain role in regulating the biological activities of colon cancer cells.
Our analysis of the LinkedOmics database revealed that INHBA was positively correlated with most genes in colon cancer and highly correlated with versican (VCAN). VCAN is a large aggregated chondroitin sulfate proteoglycan involved in cell adhesion, proliferation, migration, angiogenesis and tissue morphogenesis and maintenance.9–12 VCAN is a potential prognostic biomarker for colon cancer recurrence and an important regulator of colon cancer cell proliferation and migration.13,14 However, the specific mechanisms by which VCNA mediates the proliferation, invasion and migration of colon cancer cells have not been reported. Therefore, we aimed to study the roles and mechanisms of INHBA in regulating the aggressive behavior of colon cancer cells to provide a theoretical basis for the treatment of colon cancer.
Materials and methods
Cell culture
The normal colonic mucosa cell line NCM460 and colon cancer cell lines CaCo2, SW1116, SW480, HCT-116 and LoVo were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin and streptomycin (all from Gibco; Thermo Fisher Scientific, Waltham, MA, USA) under standard culture conditions in a humidified atmosphere with 5% CO2 at 37°C. The study protocol did not need approval by an ethics committee or institutional review board because our article does not include animal or human experiments.
Database selection
We used the UALCAN database (http://ualcan.path.uab.edu/) to assess the expression of INHBA in normal intestinal tissues and colon cancer tissues and the survival of patients with colon cancer. The LinkedOmics (http://linkedomics.org/login.php) and StarBase (http://starbase.sysu.edu.cn/) databases were used to predict the genes associated with INHBA. Informed consent was not applicable because the patient data were from publicly available databases.
Western blot
Radioimmunoprecipitation assay buffer (Beyotime Technology, Jiangsu, China) was used for protein extraction. Protein concentration was measured using a BCA protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). Next, 40 µg of protein were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (12.5%) and transferred onto polyvinylidene difluoride membranes, which were subsequently blocked with 5% nonfat milk and incubated with relevant primary antibodies at 4°C overnight. Horseradish peroxidase-conjugated secondary antibodies were used to detect primary antibodies. The ECL detection system (Millipore, Billerica, MA, USA) was used to image protein bands, and a semiquantitative analysis was conducted using ImageJ 1.8.0 software (https://imagej.nih.gov/ij/). Primary antibodies included glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 1:1000, ab8226, Abcam, Cambridge, MA, USA), anti‐INHBA (1:1000, ab128958, Abcam), anti-proliferating cell nuclear antigen (PCNA, 1:1000, ab92552, Abcam), anti-Ki67 (1:1000, ab16667, Abcam), anti-matrix metalloproteinase 2 (MMP2, 1:1000, ab92536, Abcam), anti-MMP9 (1:1000, ab76003, Abcam) and anti-VCAN (1:1000, ab177480, Abcam).
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells with TRIzol reagent (Invitrogen Inc., Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The cDNA was synthesized by reverse transcription with the M-MLV Reverse Transcriptase kit (Invitrogen, Inc.). RT-qPCR was performed with the SYBR Premix Ex Taq II (Takara, Dalian, China) and an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer’s protocol. The target gene expression level was normalized to GAPDH. Primer sequences were as follows: INHBA forward: 5ʹ-CCTCGGAGATCATCACGTTT-3ʹ and reverse: 5ʹ-CCCTTTAAGCCCACTTCCTC-3ʹ; PCNA forward: 5ʹ-CTGAAGCCGAAACCAGCTAGACT-3ʹ and reverse: 5ʹ-TCGTTGATGAGGTCCTTGAGTGC-3ʹ; Ki67 forward: 5ʹ-AATTCAGACTCCATGTGCCTGAG-3ʹ and reverse: 5ʹ-CTTGACACACACATTGTCCTCAGC-3ʹ; VCAN forward: 5ʹ-CAAACCCTGCCTCAACGGAGG-3ʹ and reverse: 5ʹ-CCTTCAGCAGCATCCCATGTGCGT-3ʹ; and GAPDH forward: 5ʹ-TGTTGCCATCAATGACCCC-3ʹ and reverse: 5ʹ-CTCCACGACGTACTCAGC-3ʹ. GAPDH was included as an internal control, and the 2−ΔΔCq method was used for gene expression analysis. 15
Cell transfection
Small interfering RNA (si)-INHBA-1, si-INHBA-2, overexpression VCAN (Ov-VCAN) and negative control (NC) plasmids at a concentration of 20 nM were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). siRNA and plasmids were transfected into cells using Lipofectamine RNAi max (Invitrogen Inc., 13778150) and Lipofectamine 3000 (Invitrogen Inc., L3000015), respectively, following the manufacturer’s instructions. Ov-VCAN or empty vector (Ov-NC) were transfected into cells with Lipofectamine® 2000 (Invitrogen Inc.) in accordance with the manufacturer’s protocol. After transfection for 48 hours at 37°C with 5% CO2, the transfection efficiency was measured via RT-qPCR analysis.
Cell Counting Kit-8 (CCK8) assay
Cell proliferation was examined using the CCK-8 Cell Counting Kit (Nanjing Vazyme Biotech Co., Ltd., Nanjing, China) in accordance with the manufacturer’s instructions. Briefly, HCT116 cells were grown in a 96-well plate for 24 hours, transfected and then cultured in normal medium. To measure cell proliferation at 0, 24, 48 or 72 hours after transfection, 10 µl of CCK-8 were added to each well, and cells were further incubated for 3 hours. Absorbance at 450 nm was measured using a microplate reader (VersaMax, San Francisco, CA, USA).
Colony formation assay
Cells were seeded into a six‐well plate at a density of 1 × 104 cells/well in RPMI 1640 containing 10% FBS for 12 hours. After treatment, cells were then re‐suspended in RPMI 1640 containing 10% FBS and cultured in 5% CO2 at 37°C for 15 days to allow colony formation. The plate was washed with cold phosphate-buffered saline (PBS). The colonies were fixed with 4% polyformaldehyde at room temperature and stained with 1% crystal violet for 30 minutes at room temperature. The colonies with more than 100 cells were counted using a microscope (Leica Microsystems, Wetzlar, Germany).
Wound healing assay
The cells were plated at a density of 1 × 105 cells/well in six-well culture plates. When the cells grew to 95% confluence, the cell monolayers were scraped with a sterile pipette tip, and then cells were washed with PBS several times to remove cell debris, followed by incubation with serum-free medium for 24 hours. Cells migrated to close the wound, representing in vitro healing. Wound healing was photographed using an inverted microscope and assessed by determining the rate of closure with the following formula: wound healing rate = [(the wound width at 0 hours – the wound width at 24 hours)/0-hour wound width] × 100%.
Transwell invasion assay
Cells (2 × 105 cells/well) were re-suspended in serum-free medium and seeded on the top side of filters with an 8-µm pore size (Millipore), and medium containing 10% FBS was added to the bottom side. Transwell invasion assays were performed following the manufacturer’s instructions. The images were taken using an inverted microscope.
Co-immunoprecipitation (co-IP)
Cells were harvested, and an appropriate volume of lysis buffer was added to each sample. The cells were lysed on ice (4°C) for 30 minutes, after which the supernatants were harvested by centrifugation for 30 minutes at 300 × g. A small amount of each lysate was retained for western blot analysis, and the remainder was added to a tube containing 1 µg of the corresponding antibody. The samples were incubated overnight at 4°C. Next, 10 µL protein A agarose beads washed with lysate (Thermo Fisher Scientific) were added to the cell lysates and slowly shaken at 4°C for 2 to 4 hours to stimulate antibody/bead coupling. Following immunoprecipitation, the samples were centrifuged at 350 × g (4°C) for 30 minutes, and the supernatant was discarded. The samples were then washed three to four times with 1 mL buffer solution. Finally, 15 µL SDS (2X) was added, and the samples were boiled for 5 minutes prior to western blot analysis.
Statistical analysis
The data were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s test (GraphPad Prism software for Windows, v. 5.01, La Jolla, CA, USA). The prognostic value of INHBA was explored by Kaplan–Meier analysis. A value of P < 0.05 was considered statistically significant.
Results
INHBA expression in colon cancer cells
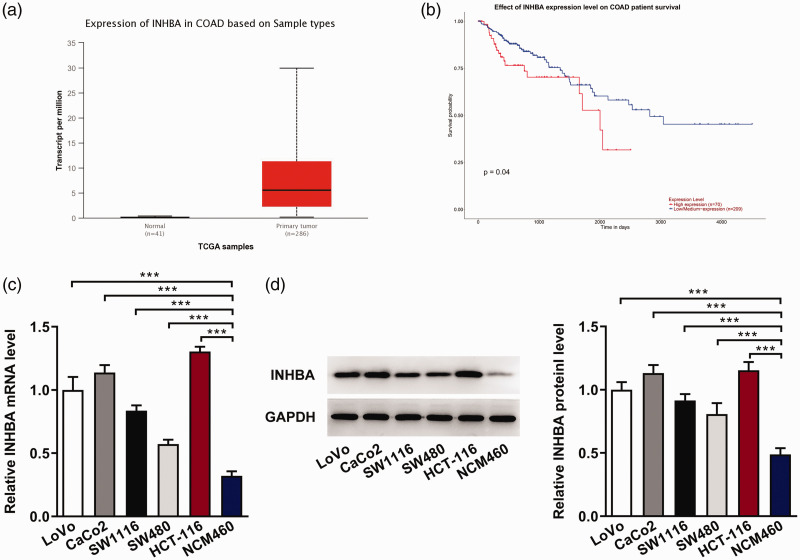
We analyzed the expression of INHBA in normal intestinal tissues (n = 41) and colon cancer tissues (n = 286) using the UALCAN database, and the results showed that the expression of INHBA was significantly increased in colon cancer tissues compared with normal intestinal tissues (Figure 1a). Patients with colon cancer who showed higher INHBA expression had a shorter overall survival time (Figure 1b). Subsequently, RT-qPCR (Figure 1c) and western blot (Figure 1d) were used to detect the expression of INHBA in normal colon mucosa cells and colon cancer cells. We found that the expression of INHBA was significantly increased in colon cancer cell lines, with HCT116 cells exhibiting the highest expression. Therefore, HCT116 cells were selected for subsequent studies.
Figure 1.
INHBA expression in colon cancer cells. The UALCAN database was used to assess INHBA expression levels in normal intestinal tissues (n = 41) and colon cancer tissues (n = 286) (a) and analyze the survival of patients with high (n = 70) and low/medium expression (n = 209) (b). (c) RT-qPCR detected the mRNA expression of INHBA in colon cancer cell lines. (d) Western blot detected the protein expression of INHBA. GAPDH was used as an internal control. ***P < 0.001.
COAD, colorectal adenocarcinoma; TCGA, The Cancer Genome Atlas; INHBA, inhibin subunit beta A; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; GAPDH, glycerol 3-phosphate dehydrogenase.
INHBA interference inhibits the proliferation of colon cancer cells
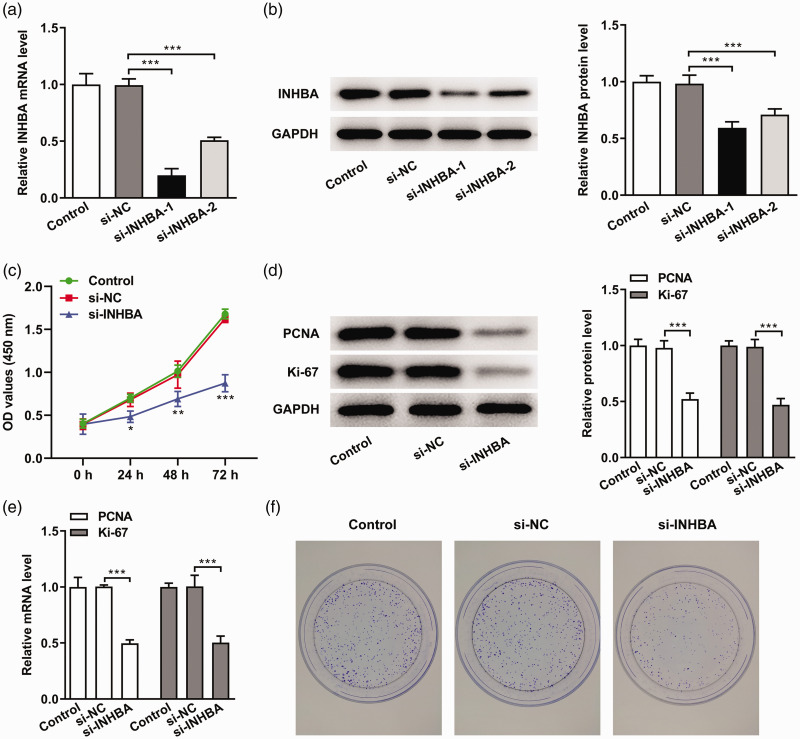
siRNA transfection was used to interfere with the expression of INHBA. RT-qPCR (Figure 2a) and western blot (Figure 2b) were used to detect the cellular expression of INHBA. Because si-INHBA-1 clearly interfered with INHBA expression, it was selected for subsequent experiments. We then measured cell proliferation in cells transfected with si-INHBA-1. The CCK-8 results showed that cell proliferation was significantly decreased in the si-INHBA group compared with the si-NC group (P < 0.05) (Figure 2c). Ki67 and PCNA are cell proliferation markers. 16 Next, we used RT-qPCR and western blot to detect the expression of Ki67 and PCNA. Compared with the si-NC group, the expression levels of PCNA and Ki67 in the si-INHBA group were significantly decreased (P < 0.001) (Figure 2d and e). In addition, we conducted colony formation assays to detect cell proliferation, and the results were consistent with the results of CCK-8 experiments (Figure 2f). These results indicated that INHBA knockdown significantly inhibited the proliferation of colon cancer cells.
Figure 2.
INHBA interference inhibited the proliferation of colon cancer cells. (a) RT-qPCR detected the mRNA expression of INHBA after cell transfection. (b) Western blot detected the protein expression of INHBA after cell transfection. GAPDH was used as an internal control. (c) CCK-8 assay detected cell viability. (d) Western blot detected the protein expression of PCNA and Ki67. (e) RT-qPCR detected the mRNA expression of PCNA and Ki67. (f) Colony formation assay detected cell proliferation. *P < 0.05, **P < 0.01, ***P < 0.001.
INHBA, inhibin subunit beta A; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; PCNA, proliferating cell nuclear antigen; CCK-8, cell counting kit-8; GAPDH, glycerol 3-phosphate dehydrogenase; NC, negative control.
INHBA interference inhibits the migration and invasion of colon cancer cells
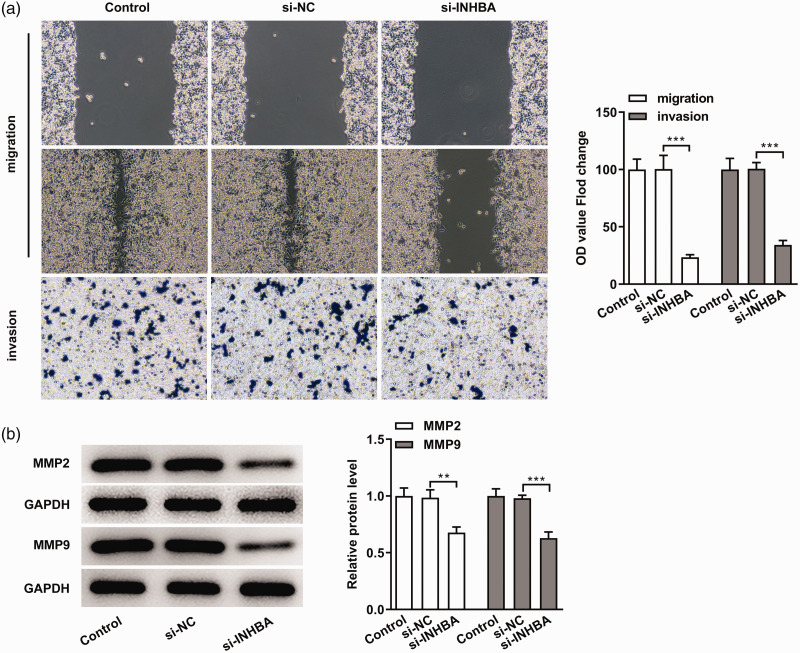
Next, we used wound healing and Transwell assays to detect cell migration and invasion. We found that the inhibition of INHBA expression significantly reduced the migratory and invasion abilities of colon cancer cells (P < 0.001) (Figure 3a). MMP2 and MMP9 are markers of tumor invasion and migration. 17 Western blot was used to detect the expression of MMP2 and MMP9, and we found that the MMP2 and MMP9 levels in the si-INHBA group were significantly decreased compared with the si-NC group (P < 0.01). These results indicated that INHBA interference inhibited the migration and invasion of colon cancer cells.
Figure 3.
INHBA interference inhibited the migration and invasion of colon cancer cells. (a) Wound healing and Transwell assays detected cell migration and invasion. (b) Western blot detected the protein expression of MMP2 and MMP9. GAPDH was used as an internal control. **P < 0.01 and ***P < 0.001.
INHBA, inhibin subunit beta A; MMP, matrix metalloproteinase; GAPDH, glycerol 3-phosphate dehydrogenase; NC, negative control.
INHBA and VCAN expression are correlated in colon cancer cells
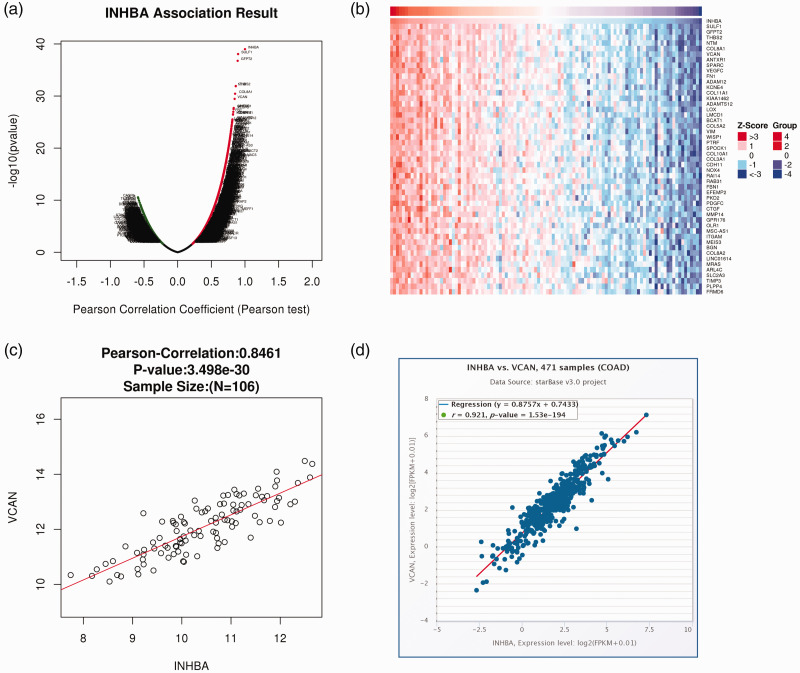
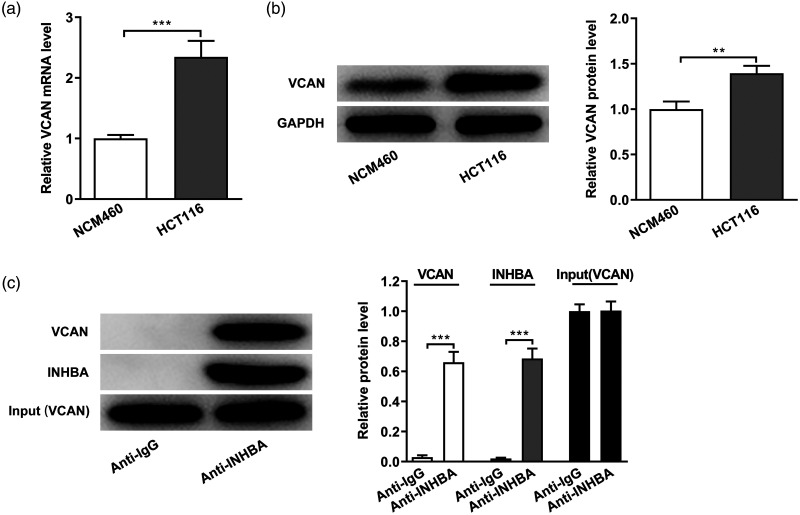
The LinkedOmics database predicted a correlation between INHBA and VCAN. The volcano plot in Figure 4a shows the genes with positive and negative correlations with INHBA. The top 50 genes with significant positive and negative correlations with INHBA were shown in the heat map (Figure 4b). The heat map demonstrates a diverse influence of INHBA on the transcriptome in colon cancer. VCAN showed the strongest positive correlation with INHBA (Figure 4c). Subsequently, their correlation was verified using the StarBase database (Figure 4d). Then, the cellular expression of VCAN was detected by RT-qPCR (Figure 5a) and western blot (Figure 5b), which revealed significantly increased expression in HCT-116 cells compared with normal colonic mucosa cells (P < 0.01). Subsequently, the interaction between VCAN and INHBA was verified by co-IP experiments (Figure 5c). Together, these experiments show that INHBA and VCAN expression are correlated in colon cancer cells.
Figure 4.
Bioinformatics analysis of the relationship between INHBA and VCAN. (a and b) The LinkedOmics database predicted a correlation between INHBA and VCAN. (c and d) The correlation between INHBA and VCAN was verified using the StarBase database.
COAD, colorectal adenocarcinoma; INHBA, inhibin subunit beta A; VCAN, versican.
Figure 5.
INHBA and VCAN expression are correlated in colon cancer cells. (a) RT-qPCR detected the mRNA expression of VCAN. (b) Western blot detected the protein expression of VCAN. GAPDH was used as an internal control. (c) Co-IP assays detected the interaction between INHBA and VCAN. **P < 0.01 and ***P < 0.001.
INHBA, inhibin subunit beta A; VCAN, versican; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; Co-IP, co-immunoprecipitation; GAPDH, glycerol 3-phosphate dehydrogenase.
INHBA interference inhibits the proliferation of colon cancer cells through VCAN
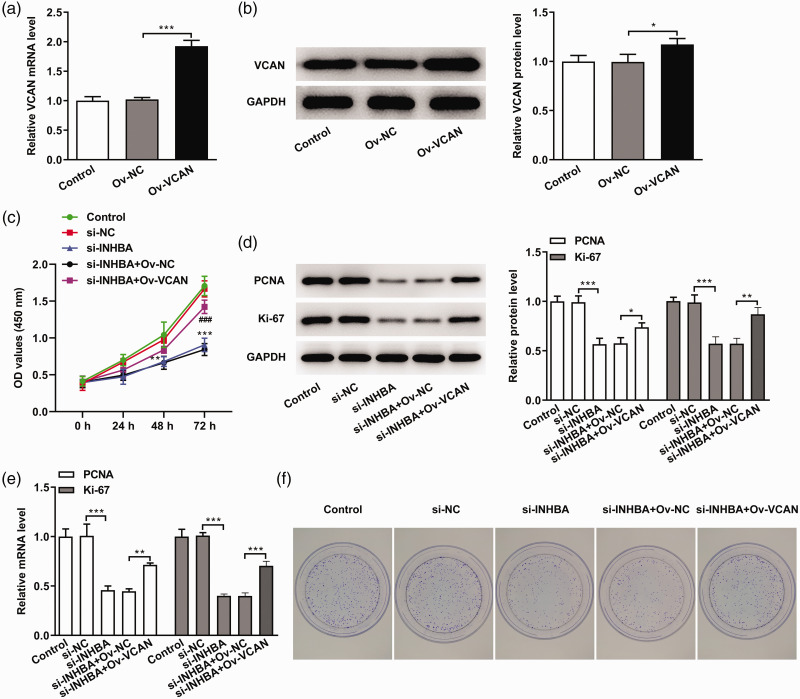
VCAN-overexpressing cells were constructed using cell transfection, and the transfection efficiency was determined by RT-qPCR and western blot (Figure 6a and b). The cells were divided into the following groups: control (untransfected), si-NC, si-INHBA, si-INHBA+OV-NC and si-INHBA+OV-VCAN. The CCK-8 results showed that the cell survival rate in the si-INHBA+OV-VCAN group was significantly increased compared with the si-INHBA+OV-NC group (P < 0.001) (Figure 6c). The expression levels of proliferation-related genes and proteins were detected by RT-qPCR and western blot. The results showed that the expression levels of PCNA and Ki67 in the si-INHBA+OV-VCAN group were significantly increased compared with the si-INHBA+OV-NC group (P < 0.01) (Figure 6d and e). The results of the clonal formation experiments were consistent with those of the CCK-8 experiment (Figure 6f).
Figure 6.
INHBA interference inhibited the proliferation of colon cancer cells through VCAN. (a) RT-qPCR assays detected the mRNA expression of VCAN. (b) Western blot detected the protein expression of VCAN. GAPDH was used as an internal control. (c) CCK-8 assays detected cell viability. (d) Western blot detected the protein expression of PCNA and Ki67. (e) RT-qPCR detected the mRNA expression of PCNA and Ki67. (f) Colony formation assay detected cell proliferation. *P < 0.05, **P < 0.01, ***P < 0.001.
INHBA, inhibin subunit beta A; VCAN, versican; Ov, overexpression; NC, negative control; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; CCK-8, cell counting kit-8; PCNA, proliferating cell nuclear antigen; GAPDH, glycerol 3-phosphate dehydrogenase.
INHBA interference inhibits the migration and invasion of colon cancer cells through VCAN
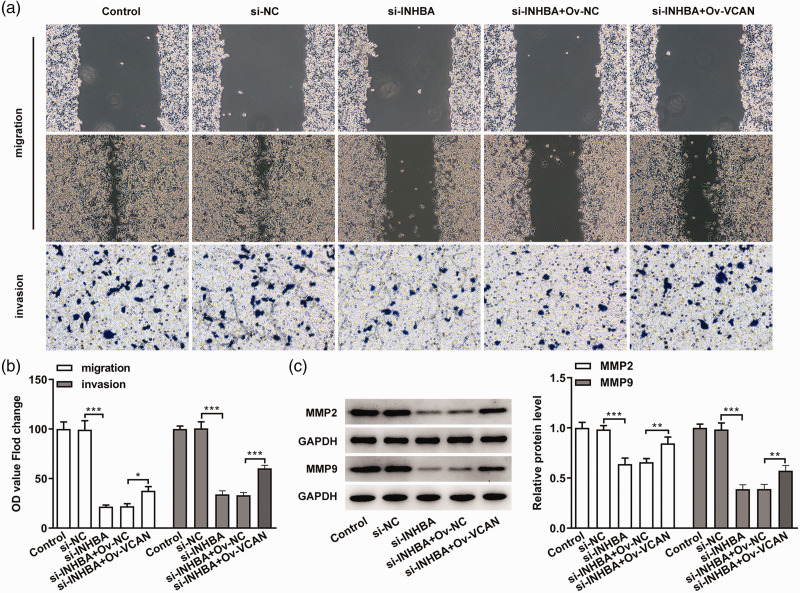
Next, we examined migration and invasion in VCAN-overexpressing cells. The results showed that the cell migration and invasion abilities of the si-INHBA+OV-VCAN group were significantly increased compared with the si-INHBA+OV-NC group (P < 0.01) (Figure 7a and b), which was accompanied by increased protein expression of MMP2 and MMP9 (Figure 7c). These results indicated that the inhibitory effects of INHBA interference on the migration and invasion of colon cancer cells are mediated through VCAN.
Figure 7.
INHBA interference inhibited the migration and invasion of colon cancer cells through VCAN. (a) Wound healing and Transwell experiments detected cell migration and invasion. (b) Statistical analysis of cell migration and invasion. (c) Western blot detected the protein expression of MMP2 and MMP9. GAPDH was used as an internal control. **P < 0.01 and ***P < 0.001.
INHBA, inhibin subunit beta A; VCAN, versican; MMP, matrix metalloproteinase; Ov, overexpression; NC, negative control; GAPDH, glycerol 3-phosphate dehydrogenase.
Discussion
According to epidemiological data, the incidence of colon cancer is increasing, and the onset can occur at any age, with the highest incidence observed in the age group of 40 to 50 years old. 18 The incidence and mortality of colon cancer are high, and it poses a significant risk to people’s health. 19
Through a literature review, we found that INHBA is abnormally expressed in colon cancer, and its expression can be used as a prognostic predictor, suggesting that it plays a key role in the occurrence and development of colon cancer. 6 In this paper, we determined that INHBA was abnormally elevated in patients with colon cancer and negatively correlated with the overall survival rate of these patients. At the cellular level, we also confirmed that INHBA was aberrantly expressed in different colon cancer cell lines, including LoVo, CaCo2, SW1116, SW480 and HTC-116. Our experimental results were consistent with the results of bioinformatics analysis. However, the specific role and mechanisms of INHBA in colorectal cancer have not yet been reported.
The proliferation, invasion and migration of tumor cells play an important role in the occurrence and development of cancer. 20 Chen et al. 21 showed that silencing the expression of INHBA inhibited the migration and invasion of gastric cancer cells. Therefore, we speculated that INHBA has a certain regulatory effect on the proliferation, invasion and migration of colon cancer cells. We found that the proliferation, invasion and migration of colon cancer cells were significantly decreased after interfering with INHBA expression, suggesting that the increased expression of INHBA in colon cancer cells enhances their tumor-promoting abilities. In addition, our results provide evidence for the potential use of INHBA as a new biomarker for molecular diagnosis and target for drug therapy in colon cancer.
We found a high correlation between INHBA and VCAN expression in colon cancer through LinkedOmics and StarBase database analysis. We also confirmed the interaction between VCAN and INHBA using co-IP assays. These results suggest that INHBA transcriptionally regulates the expression of VCAN. VCAN plays an important role in the occurrence and development of cancer. Zhang et al. 10 found that the inhibition of VCAN impaired the migratory activity of breast cancer cells. Additionally, VCAN expression is abnormally elevated in gastric cancer, and VCAN knockdown inhibits the proliferation, invasion and migration of gastric cancer cells. 22 In our study, we found that the expression of VCAN was significantly increased in colon cancer cells. A previous study reported that VCAN is a potential prognostic biomarker for colon cancer recurrence and plays an important regulatory role in the proliferation and migration of colon cancer cells. 13 Therefore, we speculated that INHBA might influence the aggressive behavior of colon cancer cells through the regulation of VCAN. To address this question, we overexpressed VCAN in cells with inhibited INHBA expression, and we found that VCAN overexpression reversed the inhibitory effects of INHBA on the proliferation, invasion and migration of colon cancer cells. Therefore, we concluded that interfering with the expression of INHBA impairs the proliferation, invasion and migration of colon cancer cells by inhibiting the expression of VCAN.
In future studies, we will verify our results in vivo using animal models. Based on the conclusion of this article, we aim to develop targeted drugs for the treatment of colon cancer. Overall, our study confirmed that INHBA interference decreases the proliferation, invasion and migration of colon cancer cells, and this effect may be mediated through the inhibition of VCAN expression. Our study provides a theoretical basis for molecular diagnosis and targeted therapy in colon cancer.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
ORCID iD: Yuan Liu https://orcid.org/0000-0003-1817-6170
References
- 1.Labianca R, Beretta GD, Kildani B, et al. Colon cancer. Crit Rev Oncol Hematol 2010; 74:106–133. doi: 10.1016/j.critrevonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Bo H, Fan L, Li J, et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer 2018; 9: 4677–4683. doi: 10.7150/jca.26461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C, Liang Y, Ma MH, et al. KRT15, INHBA, MATN3, and AGT are aberrantly methylated and differentially expressed in gastric cancer and associated with prognosis. Pathol Res Pract 2019; 215: 893–899. doi: 10.1016/j.prp.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Kalli M, Mpekris F, Wong CK, et al. Activin A Signaling Regulates IL13Ralpha2 Expression to Promote Breast Cancer Metastasis. Front Oncol 2019; 9: 32. doi: 10.3389/fonc.2019.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z, Wang K, Tan S. microRNA-211-mediated targeting of the INHBA-TGF-beta axis suppresses prostate tumor formation and growth. Cancer Gene Ther 2020. doi: 10.1038/s41417-020-00237-w. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Yu W, Liang C, et al. INHBA is a prognostic predictor for patients with colon adenocarcinoma. BMC Cancer 2020; 20: 305. doi: 10.1186/s12885-020-06743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Wu J, Zhang J, et al. Integrated bioinformatics analysis of key genes involved in progress of colon cancer. Mol Genet Genomic Med 2019; 7: e00588. doi: 10.1002/mgg3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Xie Y, Chen W, et al. microRNA-6785-5p-loaded human umbilical cord mesenchymal stem cells-derived exosomes suppress angiogenesis and metastasis in gastric cancer via INHBA. Life Sci 2021: 119222. doi: 10.1016/j.lfs.2021.119222. [DOI] [PubMed] [Google Scholar]
- 9.Ricciardelli C, Sakko AJ, Ween MP, et al. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009; 28: 233–245. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Zou X, Qian W, et al. Enhanced PAPSS2/VCAN sulfation axis is essential for Snail-mediated breast cancer cell migration and metastasis. Cell Death Differ 2019; 26: 565–579. doi: 10.1038/s41418-018-0147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salem M, O'Brien JA, Bernaudo S, et al. miR-590-3p Promotes Ovarian Cancer Growth and Metastasis via a Novel FOXA2-Versican Pathway. Cancer Res 2018; 78: 4175–4190. doi: 10.1158/0008-5472.CAN-17-3014. [DOI] [PubMed] [Google Scholar]
- 12.Jiang K, Liu H, Xie D, et al. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol Lett 2019; 17: 3191–3202. doi: 10.3892/ol.2019.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long X, Deng Z, Li G, et al. Identification of critical genes to predict recurrence and death in colon cancer: integrating gene expression and bioinformatics analysis. Cancer Cell Int 2018; 18: 139. doi: 10.1186/s12935-018-0640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chida S, Okayama H, Noda M, et al. Stromal VCAN expression as a potential prognostic biomarker for disease recurrence in stage II-III colon cancer. Carcinogenesis 2016; 37: 878–887. doi: 10.1093/carcin/bgw069. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Jurikova M, Danihel L, Polak S, et al. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem 2016; 118: 544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Kalhori V, Tornquist K. MMP2 and MMP9 participate in S1P-induced invasion of follicular ML-1 thyroid cancer cells. Mol Cell Endocrinol 2015; 404: 113–122. doi: 10.1016/j.mce.2015.01.037. [DOI] [PubMed] [Google Scholar]
- 18.Colon Cancer. Am Fam Physician 2018; 97: Online. [PubMed]
- 19.Cappell MS. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol Clin North Am 2008; 37: 1–24, v. doi: 10.1016/j.gtc.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal 2017; 35: 250–255. doi: 10.1016/j.cellsig.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Chen ZL, Qin L, Peng XB, et al. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF-beta signaling pathway. J Cell Physiol 2019; 234: 18065–18074. doi: 10.1002/jcp.28439. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y, Sun H, Wu L, et al. VUp-Regulation of VCAN Promotes the Proliferation, Invasion and Migration and Serves as a Biomarker in Gastric Cancer. Onco Targets Ther 2020; 13: 8665–8675. doi: 10.2147/OTT.S262613. [DOI] [PMC free article] [PubMed] [Google Scholar]