Abstract
Due to their large size, charged surfaces, and environmental sensitivity, proteins do not naturally cross cell-membranes in intact form and, therefore, are difficult to deliver for both diagnostic and therapeutic purposes. Based upon the observation that clustered oligonucleotides can naturally engage scavenger receptors that facilitate cellular transfection, nucleic acid–metal organic framework nanoparticle (MOF NP) conjugates have been designed and synthesized from NU-1000 and PCN-222/MOF-545, respectively, and phosphate-terminated oligonucleotides. They have been characterized structurally and with respect to their ability to enter mammalian cells. The MOFs act as protein hosts, and their densely functionalized, oligonucleotide-rich surfaces make them colloidally stable and ensure facile cellular entry. With insulin as a model protein, high loading and a 10-fold enhancement of cellular uptake (as compared to that of the native protein) were achieved. Importantly, this approach can be generalized to facilitate the delivery of a variety of proteins as biological probes or potential therapeutics.
Proteins play key roles in living systems, and the ability to deliver active proteins to cells is attractive for both diagnostic and therapeutic purposes.1 Potential uses involve the evaluation of metabolic pathways,2 regulation of cellular processes,3 and treatment of disease involving protein deficiencies.4–6 During the past decade, a series of techniques have been developed to facilitate protein internalization by live cells, including the use of complementary transfection agents, nanocarriers,7–9 and protein surface modifications.10–13 Although each strategy has its own merit, none are perfect solutions; they can cause cytotoxicity, reduce protein activity, and suffer from low delivery payloads.14 For example, we have made the observation that one can take almost any protein and functionalize its surface with DNA to create entities that will naturally engage the cell-surface receptors involved in spherical nucleic acid (SNA) uptake.13,15–17 While this method is extremely useful in certain situations, it requires direct modification of the protein and large amounts of nucleic acid, on a per-protein basis, to effect transfection. Ideally, one would like to deliver intact, functional proteins without the need to chemically modify them, and to do so in a nucleic-acid efficient manner.
Metal organic frameworks (MOFs) have emerged as a class of promising materials for the immobilization and storage of functional proteins.18 Their mesoporous structures allow for exceptionally high protein loadings, and their framework architectures can significantly improve the thermal and chemical stabilities of the encapsulated proteins.19–24
However, although MOF NPs have been recognized as potentially important intracellular delivery vehicles for proteins,25–27 their poor colloidal stability and positively charged surfaces,28,29 inhibit their cellular uptake and have led to unfavorable bioavailabilities.30–33 Therefore, the development of general approaches for reducing MOF NP aggregation, minimizing positive charge (which can cause cytotoxicity), and facilitating cellular uptake is desirable.34,35
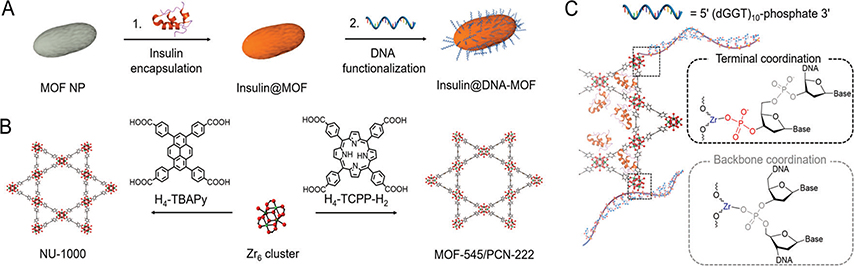
Herein, we report a new method for the intracellular delivery of proteins that relies on nucleic acid–MOF NP conjugates (Scheme 1A).34–37 In this protocol, two water stable zirconium mesoporous MOFs, NU-1000 (Zr6(μ3O)4(μ3OH)4(OH)4(H2O)4(TBAPy)2, H4-TBAPy = tetraethyl 4,4′,4″,4″′-(pyrene-1,3,6,8-tetrayl)tetrabenzoic acid) and P C N - 2 2 2 / M O F - 5 45 ( Z r 6 ( μ 3 - O ) 4 ( μ 3 OH)4(OH)4(H2O)4(TCPP-H2)2, H4-TCPP-H2 = tetrakis(4carboxyphenyl)porphyrin),38–40 were synthesized in nanoparticle form and used to encapsulate insulin, a model protein for the studies described herein (Scheme 1B).41,42 Next, via modification of literature procedures, these insulin@MOF NPs were surface functionalized with terminal phosphate-modified DNA to yield insulin@DNA-MOF NPs (Scheme 1C).36 The 3D oligonucleotide shell creates a steric and electrostatic barrier to stabilize MOF NPs in high dielectric media and renders them functional with respect to cellular entry.34 In principle, this strategy can be generalized to MOFs with different pore sizes and topologies, thereby creating an arsenal of nucleic acid–MOF-based delivery vehicles for transporting functional enzymes across cellular membranes with high payloads.
Scheme 1.
(A) Schematic Illustration of Insulin Encapsulation in the Mesoporous Channels of MOF NPs Followed by DNA Surface Functionalization; (B) Crystal Structures of Two Mesoporous Zr MOFs: NU-1000 and PCN-222/MOF-545 and Their Respective Organic Linkers; (C) DNA Functionalization of Insulin Encapsulated MOF NPs Using 3′ Terminal Phosphate Modified Nucleic Acids
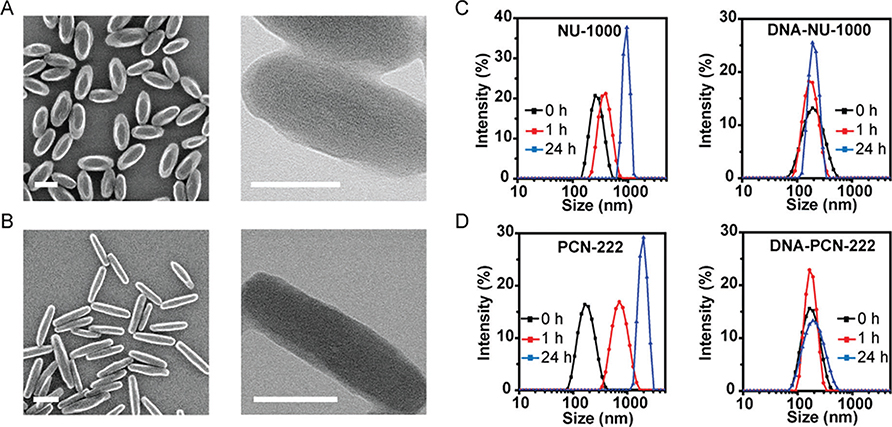
MOF NP syntheses43,44 and insulin encapsulations41 were realized via literature protocols. Specifically, NU-1000 MOF NPs [180(20) × 70(10) nm] were synthesized via a solvothermal reaction of zirconium chloride (ZrCl4) with H4TBAPy ligands, modulated by acetic acid in N,N-dimethylformamide (DMF) at 90 °C (Figure 1A). Similarly, PCN-222 NPs [210(30) × 50(10) nm] were synthesized via a solvothermal reaction between zirconyl chloride octahydrate (ZrOCl2·8H2O) and H4-TCPP-H2 ligands, modulated by dichloroacetic acid in DMF at 130 °C (Figure 1B). Next, the thermally activated crystals of NU-1000 were treated with a bis-tris-propane buffer (BTP, pH = 7) solution of insulin (0.4 mg/mL). The MOF NP insulin encapsulation efficiencies were determined by measuring the S [for insulin] and Zr (for MOFs) contents by inductively coupled plasma-optical emission spectroscopy. The maximum insulin loadings of 34 and 63 wt % were determined for NU-1000 and PCN-222 NPs, respectively, which are consistent with our previous report.41 The excess insulin in the supernatant was removed by sequential washing steps with DI water.
Figure 1.
Scanning electron microscopy (left) and transmission electron microscopy (right) images of as-synthesized NU-1000 NPs (A) and PCN-222 NPs (B). (C–D) Colloidal stability of NU-1000 and PCN-222 NPs in cell medium, as determined by DLS without (left) and with DNA surface modification (right). Scale bars = 100 nm.
The insulin@MOF NPs were functionalized with nucleic acids by coordinating the terminal phosphate-modified oligonucleotides to the surface Zr SBUs.36,45 The sequence used here, 5′ (dGGT)10-phosphate 3′, was chosen because it is known with SNAs that a G-rich shell, relative to poly dT shells, facilitates higher cellular uptake.46 In a typical NP functionalization experiment, excess oligonucleotides were added to a colloidal dispersion of MOF NPs and incubated for 4 h (Supporting Information). Particle DNA coverage was quantitively determined by measuring the P to Zr ratio by ICP-OES (8 ± 1 nmol/mg for NU-1000 NPs and 10 ± 1 nmol/mg for PCN-222 NPs). Powder X-ray diffraction (PXRD) and scanning electron microscopy (SEM) confirmed that the crystallinity and morphologies of the MOF NPs were maintained, post-DNA functionalization (Figures S2 and S3). Importantly, dynamic light scattering (DLS) verified that DNA surface functionalization significantly increases MOF NP colloidal stability in cellular media (90% DMEM buffer +10% fetal bovine serum) for at least 24 h; for comparison, unfunctionalized NU-1000 NPs aggregated in less than 1 h, hampering further in vitro use (Figure 1C,D, EM image of aggregated NPs: Figure S5).
In addition to colloidal stability, the intra- and extracellular stability of protein delivery vehicles in serum and serum free but biologically relevant matrices is important. Indeed, the ability to control degradation could be useful in the development of temporally controlled drug delivery applications. Under physiological conditions, intracellular fluid exhibits significantly higher inorganic phosphate concentration (5–10 mM) as compared to that of serum (∼1 mM).47,48
Therefore, the degradation profiles of insulin@DNA-NU-1000 NPs and insulin@DNA-PCN-222 NPs were evaluated by exposing them to solutions designed to emulate both extracellular and intracellular conditions (Supporting Information). To simulate serum, MOF NPs were incubated with 90% DMEM buffer +10% blood serum (pH = 7.0) at 37 °C with gentle shaking (400 rpm), where less than 5% of degradation occurred within 12 h for both vehicles, and less than 20% within 96 h, suggesting DNA-MOF NPs exhibit excellent stability and may be compatible with blood (Figure 2C, dashed). In contrast, when the same MOF NPs were incubated in an intracellular medium simulant (1 × phosphate buffered saline, pH = 7.0) at 37 °C with gentle shaking, the particles degrade at much faster rates (Figure 2C, solid) due to the high phosphate content, which competitively binds to Zr clusters. Interestingly, DNA-PCN-222 NPs exhibit a faster degradation rate (half-life = 1 h) when compared to that of DNA-NU-1000 NPs (half-life = 40 h). Such degradation kinetics could be useful for in vivo purposes by providing a means to control the temporal release of proteins from particles, once inside cells.
Figure 2.
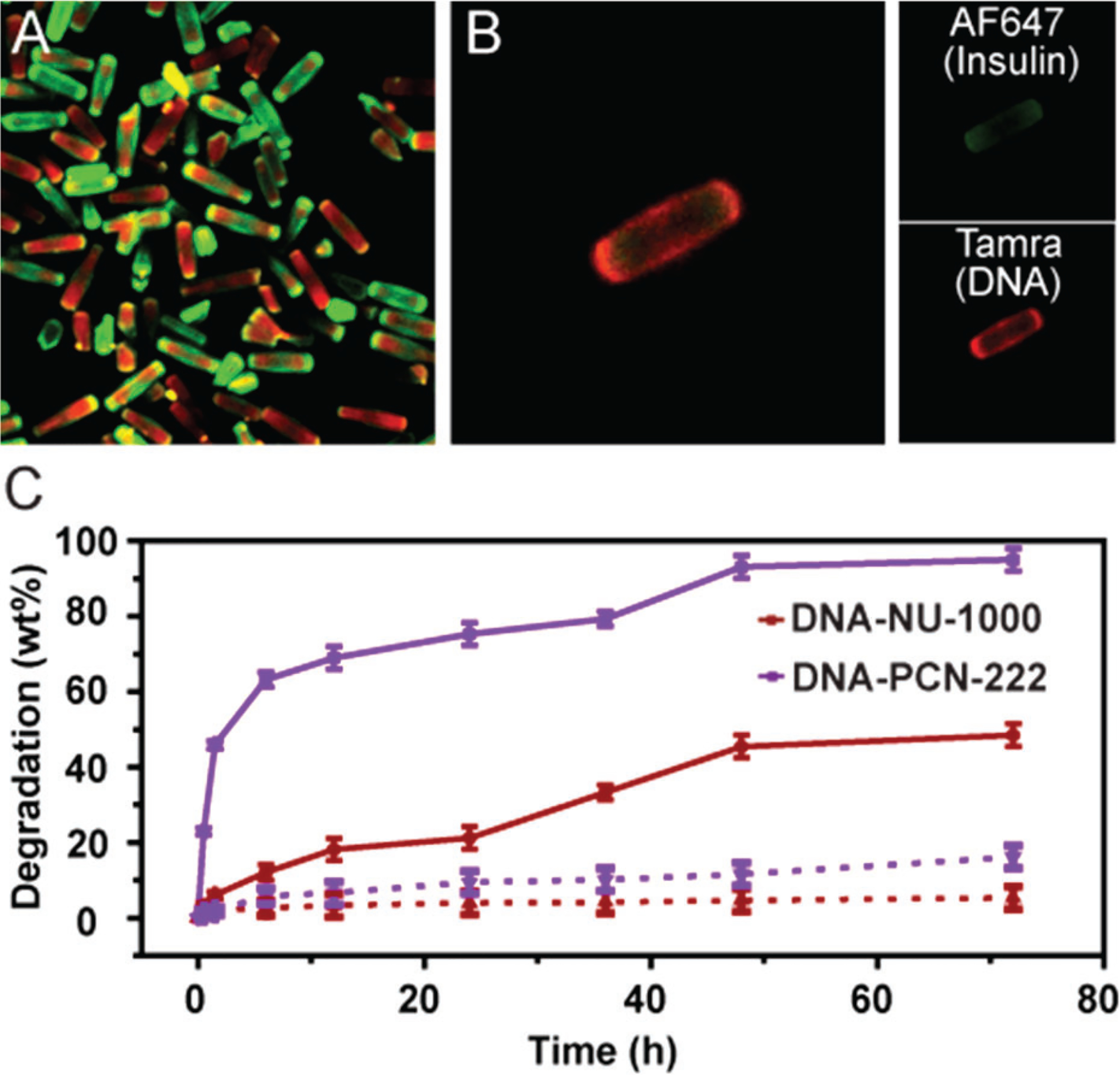
(A) Representative confocal fluorescence micrographs of 10 μm insulin@DNA-NU-1000 particles verified the colocalization of insulin (AF647 channel) and DNA (TAMRA channel). (B) Z-stack image of a single 10 μm insulin@DNA-NU-1000 crystal. (C) Degradation profiles of DNA-NU-1000 NPs and DNA-PCN-222 NPs incubated in extracellular medium (dashed lines) and in simulated intracellular medium (solid lines) at 37 °C with 400 rpm shaking.
To directly visualize nucleic acid-modified, insulin encapsulated MOF NPs, we employed confocal laser scanning microscopy to image them. Due to the resolution limits of confocal microscopy, larger particles (2.8 μm × 10 μm for NU1000), AlexaFluor 647 dye (AF647)-labeled insulin, and TAMRA-labeled DNA were used. With such particles, the colocalization of AF647 and TAMRA signals can be clearly observed, verifying the encapsulation of insulin and DNA surface functionalization of the MOF (Figure 2A). To obtain detailed information regarding relative distribution of insulin and DNA, Z-stack images of a single MOF particle were taken, where TAMRA signal (DNA) was observed to preferentially occupy the periphery while AF647 (insulin) was present throughout the particle (Figure 2B and Figure S7). Brighter AF647 signals were observed at both ends of the particle as compared to the center section of the MOF, consistent with the previous observation that proteins diffuse into NU-1000 through its 1D channels.42 Due to the large diameter of the MOF pores (3.2 nm for NU-1000 and 3.7 nm for PCN-222),40 single stranded DNA was also expected to penetrate through the MOF pores and functionalize the internal surface, leading to fluorescence signal inside the particles. As verified by N2 adsorption isotherms, reduced N2 uptake capacity was observed postinsulin encapsulation for both MOFs, and further loss of porosity was observed post-DNA functionalization (Figures S9 and S10). Furthermore, an enzyme-linked immunosorbent assay (ELISA) was employed to determine whether insulin would leach from the MOF NP pores and/or lose catalytic activity during the DNA functionalization process. In both cases, no appreciable leaching and/or insulin activity loss was observed for insulin@DNA-NU-1000 and insulin@DNA-PCN-222 constructs (Figure S8).
As previously stated, a key characteristic of SNA-NP conjugates is their ability to effectively enter cells. Therefore, we tested whether insulin@DNA-MOF NPs exhibited enhanced cellular uptake. Specifically, NU-1000 and PCN-222 NPs were encapsulated with AF647-labeled insulin and functionalized with TAMRA-labeled DNA and incubated with human ovarian adenocarcinoma cells, SKOV-3, for 0.5, 2, 6, and 24 h (Supporting Information). As a control group, a mixture of free TAMRA-labeled DNA and AF-647-labeled insulin was incubated with cells at the same concentration. Confocal laser scanning microscopy confirms the enrichment of insulin in cellular vesicles, as evidenced by strong colocalization of AF647 and TAMRA signals in cellular vesicles (Figures 3A–C). The Z-stack images confirm that the insulin@DNA-MOF NPs are internalized by the cells, as opposed to attached to their membranes. Consistent with this conclusion, flow cytometry showed a 10-fold increase in fluorescence in cells treated with insulin@DNA-MOF NPs as compared to those treated with the free insulin + DNA control group (Figure 3D). The insulin@DNA-MOF NPs exhibits similar levels of enhancement in cellular uptake, as compared to that of conventional SNA-NP conjugates.49 Finally, MTT assays show that the particles result in no apparent cytotoxicity or antiproliferative effects (Figures 3E).
Figure 3.

(A–C) Flow cytometry plots and confocal fluorescence micrographs of SK-OV cells after treatment with free insulin + DNA (A), insulin@DNA-NU-1000 (B), and insulin@DNA-PCN-222 (C). (D) Cellular uptake of insulin delivered in different constructs as determined by flow cytometry. Fluorescence at 647 nm was measured in SK-OV cells after treatment with insulin at various incubation time (0.5 and 2 h). (E) MTT assay verifies no appreciable cytotoxicity induced by insulin@DNA-PCN-222 and insulin@DNA-NU-1000 NPs. Scale bar = 10 μm.
In conclusion, we have developed a facile strategy for using nucleic-acid modified MOF NPs to deliver proteins across cell membranes at high payloads and negligible cytotoxicity. This work is important since it highlights how clustered surface oligonucleotides on these modular materials can be used to make them colloidally stable in physiological environments and useful for intracellular biological applications. Future design iterations will allow for encapsulating various proteins by tuning the MOF pore sizes,42,50,51 and potentially codelivery of protein and nucleic acid targets that are important for many purposes, including in vivo imaging,2 gene regulation,35 therapeutics,5 and the study of fundamental cellular processes.4
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported by the following awards: Air Force Office of Scientific Research FA9550-14-10274, National Science Foundation’s MRSEC program (DMR1121262) and made use of its Shared Facilities at the Materials Research Center of Northwestern University. O.K.F. gratefully acknowledge the Defense Threat Reduction Agency for financial support (HDTRA1-14-1-0014). Shunzhi.W. acknowledges support from the PPG Fellowship. Shuya.W. acknowledges support from the Ryan Fellowship at Northwestern University and the Chemistry of Life Processes (CLP) Predoctoral Training Program at Northwestern University.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b12705.
Experimental materials, methods, MOF NP syntheses, PXRD patterns, DNA sequences, nitrogen sorption isotherms, and supplemental figures (PDF)
REFERENCES
- (1).D’Astolfo DS; Pagliero RJ; Pras A; Karthaus WR; Clevers H; Prasad V; Lebbink RJ; Rehmann H; Geijsen N Efficient Intracellular Delivery of Native Proteins. Cell 2015, 161, 674–690. [DOI] [PubMed] [Google Scholar]
- (2).Hoffman RM Green fluorescent protein imaging of tumour growth, metastasis, and angiogenesis in mouse models. Lancet Oncol. 2002, 3, 546–556. [DOI] [PubMed] [Google Scholar]
- (3).Torchilin V Intracellular delivery of protein and peptide therapeutics. Drug Discovery Today: Technol. 2008, 5, e95–e103. [DOI] [PubMed] [Google Scholar]
- (4).Desnick RJ; Schuchman EH Enzyme replacement and enhancement therapies: Lessons from lysosomal disorders. Nat. Rev. Genet 2002, 3, 954–966. [DOI] [PubMed] [Google Scholar]
- (5).Leader B; Baca QJ; Golan DE Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discovery 2008, 7, 21–39. [DOI] [PubMed] [Google Scholar]
- (6).Petros RA; DeSimone JM Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discovery 2010, 9, 615–627. [DOI] [PubMed] [Google Scholar]
- (7).Ghosh P; Yang XC; Arvizo R; Zhu ZJ; Agasti SS; Mo ZH; Rotello VM Intracellular Delivery of a Membrane-Impermeable Enzyme in Active Form Using Functionalized Gold Nanoparticles. J. Am. Chem. Soc 2010, 132, 2642–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gu Z; Biswas A; Zhao M; Tang Y Tailoring nanocarriers for intracellular protein delivery. Chem. Soc. Rev 2011, 40, 3638–3655. [DOI] [PubMed] [Google Scholar]
- (9).Xu XM; Costa A; Burgess DJ Protein Encapsulation in Unilamellar Liposomes: High Encapsulation Efficiency and A Novel Technique to Assess Lipid-Protein Interaction. Pharm. Res 2012, 29, 1919–1931. [DOI] [PubMed] [Google Scholar]
- (10).Schwarze SR; Ho A; Vocero-Akbani A; Dowdy SF In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science 1999, 285, 1569–1572. [DOI] [PubMed] [Google Scholar]
- (11).Lawrence MS; Phillips KJ; Liu DR Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc 2007, 129, 10110–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cronican JJ; Thompson DB; Beier KT; McNaughton BR; Cepko CL; Liu DR Potent Delivery of Functional Proteins into Mammalian Cells in Vitro and in Vivo Using a Supercharged Protein. ACS Chem. Biol 2010, 5, 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Brodin JD; Sprangers AJ; McMillan JR; Mirkin CA DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc 2015, 137, 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fu AL; Tang R; Hardie J; Farkas ME; Rotello VM Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjugate Chem. 2014, 25, 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- (16).Patel PC; Giljohann DA; Daniel WL; Zheng D; Prigodich AE; Mirkin CA Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjugate Chem. 2010, 21, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Choi CHJ; Hao LL; Narayan SP; Auyeung E; Mirkin CA Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lian XZ; Fang Y; Joseph E; Wang Q; Li JL; Banerjee S; Lollar C; Wang X; Zhou HC Enzyme-MOF (metal-organic framework) composites. Chem. Soc. Rev 2017, 46, 3386–3401. [DOI] [PubMed] [Google Scholar]
- (19).Lykourinou V; Chen Y; Wang XS; Meng L; Hoang T; Ming LJ; Musselman RL; Ma SQ Immobilization of MP-11 into a Mesoporous Metal-Organic Framework, MP-11@mesoMOF: A New Platform for Enzymatic Catalysis. J. Am. Chem. Soc 2011, 133, 10382–10385. [DOI] [PubMed] [Google Scholar]
- (20).Shih YH; Lo SH; Yang NS; Singco B; Cheng YJ; Wu CY; Chang IH; Huang HY; Lin CH Trypsin-Immobilized Metal-Organic Framework as a Biocatalyst In Proteomics Analysis. ChemPlusChem 2012, 77, 982–986. [Google Scholar]
- (21).Liang K; Ricco R; Doherty CM; Styles MJ; Bell S; Kirby N; Mudie S; Haylock D; Hill AJ; Doonan CJ; Falcaro P Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun 2015, 6, 7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li P; Moon SY; Guelta MA; Harvey SP; Hupp JT; Farha OK Encapsulation of a Nerve Agent Detoxifying Enzyme by a Mesoporous Zirconium Metal-Organic Framework Engenders Thermal and Long-Term Stability. J. Am. Chem. Soc 2016, 138, 8052–8055. [DOI] [PubMed] [Google Scholar]
- (23).Doonan C; Riccò R; Liang K; Bradshaw D; Falcaro P Metal–Organic Frameworks at the Biointerface: Synthetic Strategies and Applications. Acc. Chem. Res 2017, 50, 1423–1432. [DOI] [PubMed] [Google Scholar]
- (24).Gkaniatsou E; Sicard C; Ricoux R; Mahy JP; Steunou N; Serre C Metal-organic frameworks: a novel host platform for enzymatic catalysis and detection. Mater. Horiz 2017, 4, 55–63. [Google Scholar]
- (25).Liu WL; Lo SH; Singco B; Yang CC; Huang HY; Lin CH Novel trypsin-FITC@MOF bioreactor efficiently catalyzes protein digestion. J. Mater. Chem. B 2013, 1, 928–932. [DOI] [PubMed] [Google Scholar]
- (26).Cao Y; Wu ZF; Wang T; Xiao Y; Huo QS; Liu YL Immobilization of Bacillus subtilis lipase on a Cu-BTC based hierarchically porous metal-rganic framework material: a biocatalyst for esterification. Dalton Trans. 2016, 45, 6998–7003. [DOI] [PubMed] [Google Scholar]
- (27).Lian XZ; Erazo-Oliveras A; Pellois JP; Zhou HC High efficiency and long-term intracellular activity of an enzymatic nanofactory based on metal-organic frameworks. Nat. Commun 2017, 8, DOI: 10.1038/s41467-017-02103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sindoro M; Yanai N; Jee AY; Granick S Colloidal-Sized Metal-Organic Frameworks: Synthesis and Applications. Acc. Chem. Res 2014, 47, 459–469. [DOI] [PubMed] [Google Scholar]
- (29).Morris W; Wang SZ; Cho D; Auyeung E; Li P; Farha OK; Mirkin CA Role of Modulators in Controlling the Colloidal Stability and Polydispersity of the UiO-66 Metal–Organic Framework. ACS Appl. Mater. Interfaces 2017, 9, 33413. [DOI] [PubMed] [Google Scholar]
- (30).Frohlich E The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed 2012, 7, 5577–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Baati T; Njim L; Neffati F; Kerkeni A; Bouttemi M; Gref R; Najjar MF; Zakhama A; Couvreur P; Serre C; Horcajada P In depth analysis of the in vivo toxicity of nanoparticles of porous iron(III) metal-organic frameworks. Chem. Sci 2013, 4, 1597–1607. [Google Scholar]
- (32).He CB; Liu DM; Lin WB Nanomedicine Applications of Hybrid Nanomaterials Built from Metal-Ligand Coordination Bonds: Nanoscale Metal-Organic Frameworks and Nanoscale Coordination Polymers. Chem. Rev 2015, 115, 11079–11108. [DOI] [PubMed] [Google Scholar]
- (33).Ruyra A; Yazdi A; Espin J; Carne-Sanchez A; Roher N; Lorenzo J; Imaz I; Maspoch D Synthesis, Culture Medium Stability, and In Vitro and In Vivo Zebrafish Embryo Toxicity of Metal-Organic Framework Nanoparticles. Chem. - Eur. J 2015, 21, 2508–2518. [DOI] [PubMed] [Google Scholar]
- (34).Morris W; Briley WE; Auyeung E; Cabezas MD; Mirkin CA Nucleic Acid-Metal Organic Framework (MOF) Nanoparticle Conjugates. J. Am. Chem. Soc 2014, 136, 7261–7264. [DOI] [PubMed] [Google Scholar]
- (35).He CB; Lu KD; Liu DM; Lin WB Nanoscale Metal–Organic Frameworks for the Co-Delivery of Cisplatin and Pooled siRNAs to Enhance Therapeutic Efficacy in Drug-Resistant Ovarian Cancer Cells. J. Am. Chem. Soc 2014, 136, 5181–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang SZ; McGuirk CM; Ross MB; Wang SY; Chen PC; Xing H; Liu Y; Mirkin CA General and Direct Method for Preparing Oligonucleotide-Functionalized Metal–Organic Framework Nanoparticles. J. Am. Chem. Soc 2017, 139, 9827–9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang ZJ; Fu Y; Kang ZZ; Liu XG; Chen N; Wang Q; Tu YQ; Wang LH; Song SP; Ling DS; Song HY; Kong XQ; Fan CH Organelle-Specific Triggered Release of Immunostimulatory Oligonucleotides from Intrinsically Coordinated DNA-Metal-Organic Frameworks with Soluble Exoskeleton. J. Am. Chem. Soc 2017, 139, 15784–15791. [DOI] [PubMed] [Google Scholar]
- (38).Mondloch JE; Bury W; Fairen-Jimenez D; Kwon S; DeMarco EJ; Weston MH; Sarjeant AA; Nguyen ST; Stair PC; Snurr RQ; Farha OK; Hupp JT Vapor-Phase Metalation by Atomic Layer Deposition in a Metal-Organic Framework. J. Am. Chem. Soc 2013, 135, 10294–10297. [DOI] [PubMed] [Google Scholar]
- (39).Morris W; Volosskiy B; Demir S; Gandara F; McGrier PL; Furukawa H; Cascio D; Stoddart JF; Yaghi OM Synthesis, Structure, and Metalation of Two New Highly Porous Zirconium Metal-Organic Frameworks. Inorg. Chem 2012, 51, 6443–6445. [DOI] [PubMed] [Google Scholar]
- (40).Feng DW; Gu ZY; Li JR; Jiang HL; Wei ZW; Zhou HC Zirconium-Metalloporphyrin PCN-222: Mesoporous Metal-Organic Frameworks with Ultrahigh Stability as Biomimetic Catalysts. Angew. Chem., Int. Ed 2012, 51, 10307–10310. [DOI] [PubMed] [Google Scholar]
- (41).Chen Y; Li P; Modica JA; Drout RJ; Farha OK Acid-Resistant Mesoporous Metal-Organic Framework toward Oral Insulin Delivery: Protein Encapsulation, Protection, and Release. J. Am. Chem. Soc 2018, 140, 5678–5681. [DOI] [PubMed] [Google Scholar]
- (42).Li P; Modica JA; Howarth AJ; Vargas EL; Moghadam PZ; Snurr RQ; Mrksich M; Hupp JT; Farha OK Toward Design Rules for Enzyme Immobilization in Hierarchical Mesoporous Metal-Organic Frameworks. Chem. 2016, 1, 154–169. [Google Scholar]
- (43).Li P; Klet RC; Moon SY; Wang TC; Deria P; Peters AW; Klahr BM; Park HJ; Al-Juaid SS; Hupp JT; Farha OK Synthesis of nanocrystals of Zr-based metal-organic frameworks with csq-net: significant enhancement in the degradation of a nerve agent simulant. Chem. Commun 2015, 51, 10925–10928. [DOI] [PubMed] [Google Scholar]
- (44).Kelty ML; Morris W; Gallagher AT; Anderson JS; Brown KA; Mirkin CA; Harris TD High-throughput synthesis and characterization of nanocrystalline porphyrinic zirconium metal-organic frameworks. Chem. Commun 2016, 52, 7854–7857. [DOI] [PubMed] [Google Scholar]
- (45).Deria P; Bury W; Hupp JT; Farha OK Versatile functionalization of the NU-1000 platform by solvent-assisted ligand incorporation. Chem. Commun 2014, 50, 1965–1968. [DOI] [PubMed] [Google Scholar]
- (46).Narayan SP; Choi CHJ; Hao LL; Calabrese CM; Auyeung E; Zhang C; Goor OJGM; Mirkin CA The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates. Small 2015, 11, 4173–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Libanati CM; Tandler CJ The distribution of the water-soluble inorganic orthophosphate ions within the cell: accumulation in the nucleus. Electron probe microanalysis. J. Cell Biol 1969, 42, 754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bansal VK Serum Inorganic Phosphorus. In Clinical Methods: The History, Physical, and Laboratory Examinations; Walker HK, Hall WD, Hurst JW, Eds.; Butterworth Publishers: Boston, 1990. [PubMed] [Google Scholar]
- (49).Cutler JI; Auyeung E; Mirkin CA Spherical nucleic acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [DOI] [PubMed] [Google Scholar]
- (50).Feng DW; Liu TF; Su J; Bosch M; Wei ZW; Wan W; Yuan DQ; Chen YP; Wang X; Wang KC; Lian XZ; Gu ZY; Park J; Zou XD; Zhou HC Stable metal-organic frameworks containing single-molecule traps for enzyme encapsulation. Nat. Commun 2015, 6, DOI: 10.1038/ncomms6979. [DOI] [PubMed] [Google Scholar]
- (51).Li P; Chen Q; Wang TC; Vermeulen NA; Mehdi BL; Dohnalkova A; Browning ND; Shen D; Anderson R; GomezGualdron DA; Cetin FM; Jagiello J; Asiri AM; Stoddart JF; Farha OK Hierarchically Engineered Mesoporous Metal-Organic Frameworks toward Cell-free Immobilized Enzyme Systems. Chem. 2018, 4, 1022–1034. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.