Abstract
Purpose
To study the association between gut microbial abundance and sight-threatening diabetic retinopathy among patients with a history of type 2 diabetes mellitus.
Methods
An observational case-control study was performed using a sample population of diabetics referred to a tertiary eye institute. Sample subjects were identified as cases if they were diagnosed with sight-threatening diabetic retinopathy and controls if they were not but had at least a 10-year history of diabetes. Fecal swabs for all patients were collected for enumeration and identification of sequenced gut microbes. Statistical analyses were performed to associate the clinically relevant Bacteroidetes to Firmicutes relative abundance ratio (B/F ratio) with sight-threatening diabetic retinopathy and an optimal cutoff value for the ratio was identified using Youden's J statistics.
Results
A sample size of 58 diabetic patients was selected (37 cases, 21 controls). No statistically significant difference in the relative abundance among the predominant phyla between the groups were found. In our univariate analysis, the B/F ratio was elevated in cases compared to controls (cases, 1.45; controls, 0.94; P = 0.049). However, this statistically significant difference was not seen in our multivariate regression model. Optimal cutoff value of 1.05 for the B/F ratio was identified, and significant clustering of cases above this value was noted in beta diversity plotting.
Conclusions
No difference in gut microbial abundance for any particular phylum was noted between the control and diseased population. Increased gut microbial B/F ratio can be a potential biomarker for the development of sight-threatening diabetic retinopathy among type 2 diabetic patients.
Keywords: bacteroidetes; diabetes mellitus; firmicutes; gut dysbiosis, gut microbiome abundance; retinopathy
Diabetes mellitus (DM) is one of the world's fastest growing metabolic diseases.1 In 2017, 451 million individuals were assessed to have diabetes, more than 90% of whom had type 2 DM, with an extended increment to 693 million by 2045.2 Diabetic retinopathy (DR), a vision-threatening microvascular complications of DM, involves an interplay of angiogenic and inflammatory mediators in its pathogenesis.3 It progresses from mild nonproliferative diabetic retinopathy, characterized by increased vascular permeability, to moderate and severe nonproliferative diabetic retinopathy, characterized by vascular closure, and ultimately proliferative diabetic retinopathy (PDR).4 Sight-threatening diabetic retinopathy (STDR), which primarily comprises clinically significant macular edema (CSME), and PDR were observed in 14% and 23% of younger-onset patients, respectively. Although nonproliferative DR does not necessitate medical intervention besides close medical follow-up and strict glycemic management, these sight-threatening conditions that can present independently or together can result in severe visual loss if left untreated.5,6 Proper management can prevent more than 90% of cases of visual loss in patients suffering from PDR, diabetic macular edema (DME) or both.7 The established risk factors for the occurrence and progression of DR includes longer duration of DM, obesity, poor glycemic control, altered lipid profile, and hypertension.8 DM causes various metabolic and physiological variations from the norm in the retina; however, which of these abnormalities contributes to perceived anatomical changes that are seen in DR is less clearly understood.
Microbiome alludes to the multispecies network of microorganisms (bacteria, fungi, and viruses) that dwell within and over different parts of the human body varying in abundance and composition.9,10 It has been noted that aberrant microbial composition relating to the relative abundance of the two most abundant gut bacteria namely Bacteroidetes and Firmicutes is associated with gut dysbiosis through a process of disruption in the gut/vascular barrier. This disruption consequently leads to pathological translocation of gut bacteria and its metabolic byproducts inducing systemic inflammation. A recently performed animal model study did not note any significant difference between healthy individuals and patients with ocular manifestations in the form of DR with respect to Firmicutes/Bacteroidetes ratio.11 Further studies have also shown that type 2 diabetes, obesity, and high glycemic states are related with a modified gut microbiota resulting in some level of gut dysbiosis.11–16 The likely cause of this association as described by previous studies is likely linked to the gut microbiota's involvement in physiological functions such as energy metabolism, metabolic signaling, and regulation of integrity of the gut barrier.17–19 These variables also have a role in the presence and progression of DR, because DR accounts for more than 60% incidence in type 2 DM.20 In humans, there is only one report by Moubayed et al.14 studying the influence of gut dysbiosis based on abundance restricted to a small cohort from Saudi Arabia; they observed higher Bacteroidetes ratio in diabetic group, and no significant difference of bacterial strains in subjects with or without DR. No similar such clinical studies have been conducted to identify the association of alterations in the gut microbiome with respect to DR among a South Asian cohort.
In our present study, we studied the gut bacterial microbiomes of people with previously diagnosed type 2 DM from a primarily South Indian population to ascertain whether dysbiosis in the gut microbiome is associated with STDR. In addition, Bacteroidetes to Firmicutes ratio (B/F ratio) has been associated in previous literature as a potential diagnostic biomarker for diabetes,11,14,21,22 and, hence, we wanted to assess its utility as a screening biomarker in STDR as well.
Materials and Methods
Between April 2019 and October 2019, patients with pre-existing type 2 DM presenting to a tertiary referral eye care hospital to undergo ophthalmic evaluation were recruited for the study. All patients who met the above inclusion criteria underwent comprehensive ophthalmic evaluation including best-corrected visual acuity, slit-lamp biomicroscopy, and fundus examination and an exclusion criteria that included presence of any other vascular retinopathy other than DR, use of systemic antibiotic during the past six months, and any other systemic or ocular inflammatory or degenerative or neoplastic disorder that could independently affect the retina was applied. In total, 58 patients were recruited who were identified into two groups, the control group who were patients with at least a 10-year history of previously diagnosed type 2 DM on the basis of existing clinical criteria and no diagnosis of DR on eye examination and the cases group who were type 2 diabetics diagnosed with sight-threatening DR such CSME or PDR.
The classification of sight-threatening DR was based on Early Treatment Diabetic Retinopathy Study Research Group and International clinical diabetic retinopathy and diabetic macular edema disease severity scales.23,24
-
•
CSME was defined as thickening of the retina at or within 500 µm of the center of the macula; hard exudate at or within 500 µm of the center of the macula associated with thickening of adjacent retina; or a zone of retinal thickening 1-disc area or larger, any part of which is within 1-disc diameter of the center of the macula
-
•
PDR is characterized by neovascularization of the disc, neovascularization of the retina, neovascularization of the iris, neovascularization of the angle, vitreous hemorrhage, or tractional retinal detachment. With regard to macular edema, it should be noted whether macular edema is present or absent.
Qualitative data regarding baseline demographic information including age, duration of DM, pre-existing comorbidities, antiglycemic medications, body mass index, HbA1c, fasting, and postprandial blood glucose levels, blood group type and dietary patterns were collected using patient questionnaire.
The study protocol was approved by the Institutional Review Board at Vision Research Foundation, Sankara Nethralaya, Chennai. Written informed consent was obtained from all the studied subjects for sample collection and subsequent analyses, and it adhered to the tenets of the Declaration of Helsinki.
Stool Collection and Next-Generation Sequencing
A clean catch of fecal sample was collected from each patient using fecal swabs (Norgen Biotek Corp, Thorold, Canada). Norgen's Microbiome DNA Isolation Kit was used to isolate the bacterial DNA from the stool sample. Purification was based on spin column chromatography. Initially, Lysis Additive A was added to the swab collection tube and incubated at 65˚C to efficiently and rapidly homogenize the sample. The sample was then spun in a centrifuge, and the supernatant is transferred to a DNAse-free microcentrifuge tube. Binding Buffer I was added, and the lysate was incubated for 10 minutes on ice. The lysate was then spun for two minutes to pellet any cell debris, the supernatant was collected, an equal volume of 70% ethanol was added to the lysate and the solution was loaded onto a spin-column. Norgen's spin column binds nucleic acids in a manner that depends on ionic concentrations, thus only the DNA will bind to the column while the proteins and other contaminants are removed in the flow through or retained on top of the resin. The bound DNA was then washed using the provided Binding Buffer B and Wash Solution A, and the purified DNA was eluted using the Elution Buffer B.
Library Preparation and Next Generation Sequencing
The standard Illumina protocol was used to generate the microbiome library. In all sequencing runs V4 region of 16 rRNA gene were amplified. The primer pair used were 515F (5’-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) targeting V4 region of the 16S rRNA gene.
The 16s rRNA Bacterial Sequencing was conducted by Npedia technologies, in conjunction with National Centre for Cell Science, Pune on the stool samples and for this purpose Illumina next generation sequencing (NGS) Miseq platform was used with 250 × 2 paired end chemistry as their approved procedure. The raw output from Illumina MiSeq were demultiplexed to give FASTQ output that was used for downstream analysis.
Data Analysis
Taxonomic Classification
The Illumina demultiplexed paired-end sequenced dataset was processed by the R package DADA2(1.14.1)25 to correct for amplicon errors, to identify chimeras, and to merge paired-ends reads. The paired ends were filtered and trimmed at 220 for forward run and 160 for reverse run, on the basis of the quality profile of the reads.
The end product of DADA2 yielded a total of 10,129 unique ASVs (amplicon sequence variants) and tallied as the number of times each exact ASV was observed for each sample. A phyloseq R object26 comprised of all 10129 ASVs was generated, with a lookup table of taxonomy assignments to each ASVs obtained by using the SILVA v132 reference database, along with a phylogenetic tree and subject sample metadata.
Statistical Analysis
A microbiome R package27 and phyloseq R package were used to analyze the abundance, relative abundance, alpha diversity of samples. R 3.6.3 was used for all the analysis. All the R visualization was done using ggplot2 (v 3.3.2). Further statistical analyses were performed using a standard software package (Stata, version 16.1, StataCorp). Descriptive statistics on patients’ characteristics and summary statistics of gut microbiome relative abundance comparing controls and cases were performed. Two-sample t-testing was performed to compare the B/F abundance ratio between cases and controls. Multivariate linear regression analysis with adjusted cofactors decided a priori was performed to determine the significance of B/F ratio in differentiating cases from controls. Youden's J statistics method was used to identify an optimal B/F ratio cutoff point to differentiate between the two study groups. Bray-Curtis dissimilarity using principal co-ordinate analysis to differentiate clustering between cases and controls against this cutoff point was performed.
Results
Baseline Characteristics of Cases and Controls
In our study cohort of 58 patients, based on eye examination 21 patients were found to be in the control group, and 37 were diagnosed with STDR. Of these 37 cases, 21 cases were found to be diagnosed as PDR with or without DME with a median disease duration of 0.50 (interquartile range, 0.50–1.00) years, and 16 cases were noted to be diagnosed as CSME alone with a median disease duration of 0.75 (interquartile range, 0.50–2.63) years.
Table 1 illustrates the baseline characteristics of cases and controls. All of the variables—age, gender, vegetarian diet, body mass index, duration of DM, duration of hypertension, glycemic controls, Log MAR best-corrected visual acuity of the worse eye, use of metformin, and associated systemic diseases such and coronary artery disease and dyslipidemia—were similar in both of the groups; systemic hypertension, however, was present in higher proportion of subjects in cases than controls (70.27% vs. 42.86%; P = 0.04).
Table 1.
Baseline Characteristics of Cases and Controls
| Variables | Cases N = 37 | Controls N = 21 | P |
|---|---|---|---|
| Mean age ± SD (y) | 57.45 ± 8.08 | 57.50 ± 7.60 | 0.982 |
| Men, N (%) | 25 (67.56) | 13 (61.90) | 0.665 |
| Vegetarian, N (%) | 16 (43.24) | 7 (33.33) | 0.462 |
| Body mass index, mean ± SD | 26.44 ± 5.23 | 26.53 ± 5.52 | 0.951 |
| Median duration of diabetes (y) (IQR) | 12 (8–20) | 12 (10–20) | 0.671 |
| Median duration of hypertension (years) (IQR) | 2 (0–8) | 0 (0–10) | 0.280 |
| HbA1c, mean ± SD | 7.48 ± 1.44 | 7.49 ± 1.48 | 0.980 |
| Use of metformin, N (%) | 4 (10.81) | 2 (9.52) | 0.877 |
| Median Log MAR BCVA of the worse eye (IQR) | 0.20 (0.50–2.50) | 0.00 (0.00–0.10) | <0.0001 |
| Associated systemic diseases (based on history & medications) | |||
| Hypertension, N (%) | 26 (70.27) | 9 (42.86) | 0.042 |
| Coronary artery disease, N (%) | 9 (24.32) | 6 (28.57) | 0.725 |
| Dyslipidemia, N (%) | 3 (8.11) | 3 (14.29) | 0.462 |
BCVA, best-corrected visual acuity; Cases, subjects with sight-threatening diabetic retinopathy; controls, subjects with diabetes mellitus, but no diabetic retinopathy; HbA1c, glycated hemoglobin; IQR, interquartile range; SD, standard deviation.
Relative Abundance of Gut Microbiome in Cases and Controls
The distribution of various gut microbiome after genomic sequencing were identified into 17 different phyla in this study. When analyzing the microbial abundance for the study population as a whole, most sequences were affiliated predominantly with four phyla. Bacteroidetes occupied the largest portion (40.27% [35.3%–45.26%]) followed by Firmicutes (41.27% [37.38%–45.21%]), Proteobacteria (8.72% [5.92%–11.47%]), and Actinobacteria (8.00% [5.97%–10.03%]).
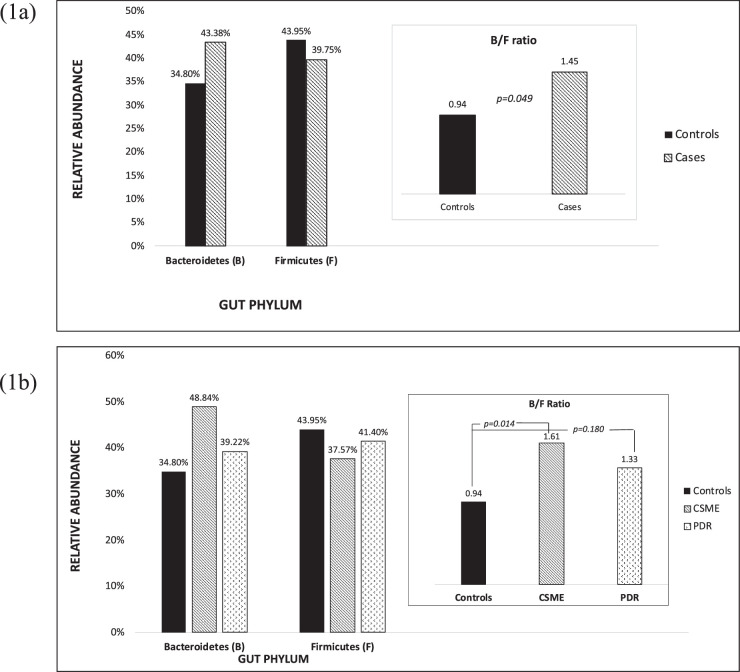
Based on t-test statistics comparing control and cases (Table 2), a similar distribution within the individual groups was noted with the most common phyla being Bacteroidetes (cases, 43.38%; controls, 34.80%) and Firmicutes (cases, 39.75%; controls, 43.95%), and less common ones were Proteobacteria (cases, 8.53%; controls, 9.05%) and actinobacteria (cases, 6.63%; controls, 0.61%). However, no statistically significant difference in the distribution of these phyla between cases and controls were identified.
Table 2.
Relative Abundance of Gut Microbiome in Cases and Controls
| Phyla | Cases (N = 37) | Control (N = 21) | P |
|---|---|---|---|
| Most common phyla | |||
| Overall | 83.13% | 78.75% | 0.923 |
| Bacteroidetes | 43.38% | 34.80% | 0.526 |
| Firmicutes | 39.75% | 43.95% | 0.757 |
| Less common phyla | |||
| Overall | 15.16% | 9.66% | 0.885 |
| Proteobacteria | 8.53% | 9.05% | 0.946 |
| Actinobacteria | 6.63% | 0.61% | 0.290 |
| Rare phyla | |||
| Overall | 2.622% | 1.714% | 0.876 |
| Verrucomicrobia | 0.377% | 0.585% | 0.910 |
| Tenericutes | 0.451% | 0.290% | 0.926 |
| Epsilonbacteraeota | 0.562% | 0.001% | 0.734 |
| Cyanobacteria | 0.315% | 0.202% | 0.938 |
| Lentisphaerae | 0.224% | 0.172% | 0.967 |
| Kiritimatiellaeota | 0.008% | 0.257% | 0.773 |
| Elusimicrobia | 0.224% | 0.000% | 0.830 |
| Chloroflexi | 0.162% | 0.000% | 0.855 |
| Euryarchaeota | 0.169% | 0.008% | 0.861 |
| Spirochaetes | 0.005% | 0.165% | 0.817 |
| Synergistetes | 0.107% | 0.021% | 0.910 |
| Fusobacteria | 0.018% | 0.013% | 0.989 |
| Abditibacteriota | 0.000% | 0.000% | 0.996 |
Comparing B/F Ratio in Cases and Controls
When we compared the ratio of the most common identified gut phyla, Bacteroidetes (B) and Firmicutes (F), in cases and controls, the B/F ratio was significantly higher in cases than controls (cases, 1.45; controls, 0.94; P = 0.049) (Fig. 1a) was noted in our t-test analysis; however, no significant difference between the groups was seen in our linear regression model. Additional subgroup analysis exploring the diseased cohort individually as patients diagnosed with CSME alone or PDR (with or without DME) noted that both disease groups in univariate analysis had a higher B/F ratio compared to the control group. However, this difference was found to be statistically significant only in the CSME-alone group (CSME, 1.61; controls, 0.94; P = 0.014) and not statistically significant in the PDR group (PDR, 1.33; controls, 0.94; P = 0.180) (Fig. 1b). Multivariate linear regression models subgrouping the diseased cohort and analyzing this correlation once again did not reveal any significant difference against the control group (Table 3).
Figure 1.
(a) Comparison of B/F ratio among cases and controls. (b) Comparison of B/F ratio among controls and cases subgroup as CSME or PDR.
Table 3.
Multivariate Linear Regression Model Comparing the Association of B/F Ratio to Sight-Threatening Diabetic Retinopathy
| 95% CI | |||||
|---|---|---|---|---|---|
| Variables | Category | Coefficient | Lower | Upper | P Value |
| Sight-threatening Diabetic Retinopathy | — | 0.379 | −0.216 | 0.976 | 0.206 |
| Glycated hemoglobin - HbA1c | >7.0% | −0.162 | −0.742 | 0.417 | 0.575 |
| Body mass index | < 25.0 (Reference) | ||||
| 25.0–29.9 | −0.437 | −1.112 | 0.237 | 0.198 | |
| > 30.0 | −0.093 | −0.763 | 0.577 | 0.780 | |
| Duration of diabetes (y) | — | −0.017 | −0.055 | 0.020 | 0.365 |
| Age (y) | — | −0.006 | −0.044 | 0.032 | 0.747 |
| Gender (male) | — | −0.348 | −0.935 | 0.239 | 0.238 |
| Hypertension | — | 0.361 | −0.280 | 1.002 | 0.263 |
| Cardiovascular disease | — | 0.335 | −0.296 | 0.967 | 0.291 |
| Dyslipidemia | — | −0.629 | −1.546 | 0.288 | 0.174 |
| Diet | Nonvegetarian | −0.216 | −0.767 | 0.335 | 0.434 |
CI, confidence interval.
B/F Ratio Optimum Cutoff Point
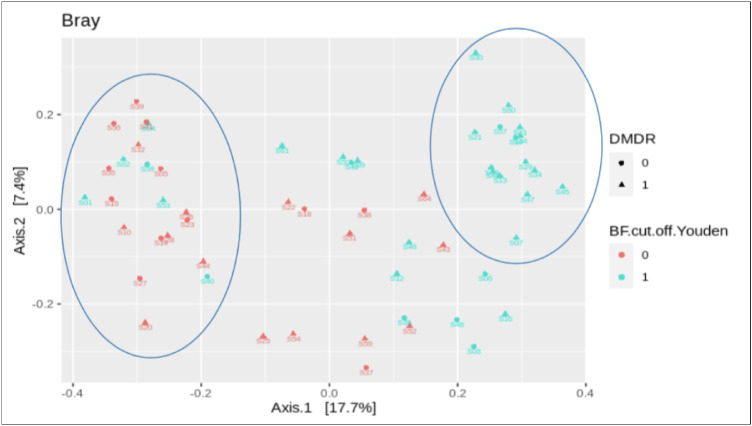
Youden's J statistics method, provided an optimal cutoff point of 1.05 (sensitivity, 62%; specificity, 57%), with controls being identified below this value and cases above. Difference in clustering within our sample population on the basis of taxonomic abundance profile using principle co-ordinate analysis, Bray-Curtis dissimilarity found a high percentage of cases clustered above this cutoff point (Fig. 2).
Figure 2.
The Bray-Curtis principal coordinates analysis (PCoA) beta diversity plotting of cases versus controls against B/F ratio (1 = B/F ratio >1, 0 = B/F ratio < 1). The above PCoA plot groups microbial communities characterized by similar sets of 16S rRNA gene sequences. Cases have been labeled as triangles and the control group as circles along with blue coloration depicting patients with a B/F ratio >1 and red with a B/F ratio < 1. Within the PCoA plot, cases with a corresponding B/F ratio > 1 are grouped together primarily as depicted by the right circle. In similar fashion, majority of the control group with a corresponding B/F ratio < 1 tend to be grouped together based on gene sequencing as described by the left circle. This grouping based on gene sequencing is in line with our study findings of a positive correlation between elevated B/F ratio and patients with a diagnosis of STDR.
Discussion
Hippocrates, the father of modern medicine, suggested nearly more than 2000 years ago, “All diseases begin in the gut.” If not all diseases, at least many chronic metabolic diseases such as diabetes mellitus do.13,28,29 The Human Microbiome Project (National Institute of Health) and Metagenomics of the Human Intestinal Tract (MetaHIT, the European Project) were initiated to characterize microbial communities from different parts of the body.30–32 In our cross-sectional study, we observed that the most common phyla in the gut microbiome were Bacteroidetes (Gram-negative) and Firmicutes (Gram-positive), accounting for nearly 83% in cases and 79% in controls. The percentage value of Bacteroidetes abundance was noted to be higher, and the percentage value of Firmicutes abundance was noted to be lower in cases versus controls, respectively. Hence, we noted that the ratio of Bacteroidetes to Firmicutes was also found to be significantly higher in cases than controls. Although this difference did not provide statistical significance when accounting for multiple covariables, we speculate that this difference that we initially found could be due to the fact that gut dysbiosis, which is a multifactorial pathology, could be involved in chronic inflammation and retinal ischemia, both of which play an important role in the pathophysiology of STDR.33–38 It was interesting to note that when subgroup analysis of different disease diagnosis within STDR (CSME vs. PDR) were performed, the same increased B/F ratio was found in both disease conditions against diabetics without retinopathy. However, statistical significance of this correlation was only noted among the CSME only group. Because these two STDR disease conditions have different pathologic mechanisms,39,40 this variation in our findings might shed light into narrowing down which metabolic pathways might be contributing to the increased B/F ratio among those with STDR.
Normally, the intestinal lining prevents the migration of microbes and their metabolites from the gut lumen to the bloodstream. However, a change in the intestinal milieu, intestinal dysbiosis, may deregulate the barrier effect of gut lining and cause leaky gut syndrome. Predominant gram-negative bacterial phyla such as Bacteroidetes releases bacterial endotoxin, lipopolysaccharides and triggers an innate or natural immunity and thereby proinflammatory pathways resulting in vascular dysfunction. A study done on a mouse with diabetes (db/db mice) showed its fecal bacterial composition (predominantly in Bacteroidetes and Firmicutes), presenting with impaired intestinal barrier function and replicating some of the features of diabetic retinopathy—acellular capillaries, activation of retina microglia, and infiltration of peripheral immune cells into the retina.41
Taking into consideration the previously researched significance of B/F ratio in systemic diseases such as diabetes, we wanted to explore its association with STDR in a clinical setting. In addition, on the basis of the observed potential association with STDR among diabetics from our study, we wanted to further explore the possibility of its role as a potential biomarker in the diagnosis of STDR.11,14 We were able to formulate an optimal B/F ratio cutoff point of 1.05, above which we can predict with high accuracy the probability of STDR being present among type 2 diabetic patients. This was evidently noted in our beta diversity analysis where the plotted clustering above the 1.0 cutoff predominantly (86.67%) comprised patients diagnosed with STDR (15 patients in total, 13 cases, 2 controls), whereas the clustering below the 1.0 cutoff did not provide any significant discrimination between cases and controls.
Although our study was able to identify a positive correlation between gut dysbiosis and developing STDR, further prospective studies comprising varied geographic populations and possibly a larger sample size would be required to strengthen this clinically relevant association. There are several questions yet to be answered with respect to gut microbiome, such as the influence of racial differences, diet, socioeconomic status, antibiotic use, role of antiglycemics, as well as prebiotic/probiotic supplementation. As our understanding of gut dysbiosis and its influences on health and disease expands, we will have a new direction to explore newer therapeutics to manage diabetes and its microvascular complications such as diabetic retinopathy.
Conclusion
There is no significant difference in the relative abundance of individual phylum between patients with STDR and no retinopathy within patients with type 2 diabetes. Gut dysbiosis with respect to difference in B/F relative abundance ratio displays significant involvement when differentiating diabetic patients with or without STDR, and this B/F ratio can play a potential role as a biomarker in the diagnosis of STDR.
Acknowledgments
The authors thank Vijay Vaidyanathan, Npedia Tech; Biohub Data Science Pvt. Ltd, Chennai, Tamil Nadu, India.
Supported by Novartis health care Pvt. Ltd, India.
Disclosure: R. Khan, None; A. Sharma, None; R. Ravikumar, None; A. Parekh, None; R. Srinivasan, None; R.J. George, None; R. Raman, None
References
- 1. Sami W, Ansari T, Butt NS, Ab Hamid MR. Effect of diet on type 2 diabetes mellitus: a review. Int J Health Sci. 2017; 11: 65–71. [PMC free article] [PubMed] [Google Scholar]
- 2. Cho N, Shaw JE, Karuranga S, et al.. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018; 138: 271–281. [DOI] [PubMed] [Google Scholar]
- 3. Chawla A, Chawla R, Jaggi S.. Microvascular and macrovascular complications in diabetes mellitus: distinct or continuum?. Indian J Endocrinol Metab. 2016; 20(4): 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fong DS, Aiello L, Gardner TW, et al.. Retinopathy in diabetes. Diabetes Care. 2004; 27(Suppl 1): s84–s87. [DOI] [PubMed] [Google Scholar]
- 5. Porta M, Bandello F.. Diabetic retinopathy. Diabetologia. 2002; 45: 1617–1634. [DOI] [PubMed] [Google Scholar]
- 6. Wu L, Fernandez-Loaiza P, Sauma J, Hernandez-Bogantes E, Masis M.. Classification of diabetic retinopathy and diabetic macular edema. World J Diabetes. 2013; 4: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferris FL. How effective are treatments for diabetic retinopathy?. JAMA. 1993; 269: 1290–1291. [PubMed] [Google Scholar]
- 8. Wat N, Wong RL, Wong IY.. Associations between diabetic retinopathy and systemic risk factors. Hong Kong Med J. 2016; 22: 589–599. [DOI] [PubMed] [Google Scholar]
- 9. Shivaji S. We are not alone: a case for the human microbiome in extra intestinal diseases. Gut Pathog. 2017; 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakravarthy SK, Jayasudha R, Prashanthi GS, et al.. Dysbiosis in the gut bacterial microbiome of patients with uveitis, an inflammatory disease of the eye. Indian J Microbiol. 2018; 58: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Floyd JL, Grant MB.. The gut–eye axis: lessons learned from murine models. Ophthalmol Ther. 2020; 9: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin J, Li Y, Cai Z, et al.. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490(7418): 55–60. [DOI] [PubMed] [Google Scholar]
- 13. Karlsson FH, Tremaroli V, Nookaew I, et al.. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013; 498(7452): 99–103. [DOI] [PubMed] [Google Scholar]
- 14. Moubayed NM, Bhat RS, Al Farraj D, Al Dihani N, El Ansary A, Fahmy RM. Screening and identification of gut anaerobes (Bacteroidetes) from human diabetic stool samples with and without retinopathy in comparison to control subjects. Microb Pathog. 2019; 129: 88–92. [DOI] [PubMed] [Google Scholar]
- 15. Schwiertz A, Taras D, Schäfer K, et al.. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010; 18(1): 190–5. [DOI] [PubMed] [Google Scholar]
- 16. Larsen N, Vogensen FK, Van Den Berg FW, et al.. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010; 5(2): e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clavel T, Desmarchelier C, Haller D, et al.. Intestinal microbiota in metabolic diseases: from bacterial community structure and functions to species of pathophysiological relevance. Gut microbes. 2014; 5: 544–551. [DOI] [PubMed] [Google Scholar]
- 18. Rial SA, Karelis AD, Bergeron KF, Mounier C.. Gut microbiota and metabolic health: the potential beneficial effects of a medium chain triglyceride diet in obese individuals. Nutrients. 2016; 8(5): 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levy M, Thaiss CA, Zeevi D, et al.. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015; 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fong DS, Aiello L, Gardner TW, et al.. Retinopathy in diabetes. Diabetes Care. 2004; 27(Suppl 1): s84–s87. [DOI] [PubMed] [Google Scholar]
- 21. Beli E, Yan Y, Moldovan L, et al.. Restructuring of the gut microbiome by intermittent fasting prevents retinopathy and prolongs survival in db/db mice. Diabetes. 2018; 67(9): 1867–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belizário JE, Faintuch J. Microbiome and gut dysbiosis. In Metabolic Interaction in Infection. Cham: Springer; 2018: 459–476. [DOI] [PubMed] [Google Scholar]
- 23. Early Treatment Diabetic Retinopathy Study Research Group. Grading diabetic retinopathy from stereoscopic color fundus photographs—an extension of the modified Airlie House classification: ETDRS report number 10. Ophthalmology. 1991; 98: 786–806. [PubMed] [Google Scholar]
- 24. Wilkinson CP, Ferris FL 3rd, Klein RE, et al.. Global Diabetic Retinopathy Project Group. Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology. 2003; 110: 1677–1682. [DOI] [PubMed] [Google Scholar]
- 25. Callahan BJ, McMurdie PJ, Rosen MJ, et al.. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016; 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013; 8(4): e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lahti L, Shetty S, Blake T, Salojarvi J. Tools for microbiome analysis in R. Version 2.1.26. Available at: http://microbiome.github.com/microbiome. Accessed September 25, 2020.
- 28. Liang D, Leung RK, Guan W, Au WW.. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018; 10(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vajro P, Paolella G, Fasano A.. Microbiota and gut-liver axis: a mini-review on their influences on obesity and obesity related liver disease. J Pediatr Gastroenterol Nutr. 2013; 56(5): 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huttenhower C, Gevers D, Knight R, et al.. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486(7402): 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI.. The human microbiome project. Nature. 2007; 449(7164): 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin J, Li R, Raes J, et al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464(7285): 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romero-Aroca P, Baget-Bernaldiz M, Pareja-Rios A, Lopez-Galvez M, Navarro-Gil R, Verges R.. Diabetic macular edema pathophysiology: vasogenic versus inflammatory. J Diabetes Res. 2016; 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noma H, Mimura T, Yasuda K, Shimura M.. Role of inflammation in diabetic macular edema. Ophthalmologica. 2014; 232: 127–135. [DOI] [PubMed] [Google Scholar]
- 35. Das A, McGuire PG, Rangasamy S.. Diabetic macular edema: pathophysiology and novel therapeutic targets. Ophthalmology. 2015; 122: 1375–1394. [DOI] [PubMed] [Google Scholar]
- 36. Bresnick GH, De Venecia G, Myers FL, Harris JA, Davis MD.. Retinal ischemia in diabetic retinopathy. Arch Ophthalmol. 1975; 93: 1300–1310. [DOI] [PubMed] [Google Scholar]
- 37. Takagi H, Watanabe D, Suzuma K, et al.. Novel role of erythropoietin in proliferative diabetic retinopathy. Diabetes Res Clin Pract. 2007; 77(3): S62–S64. [DOI] [PubMed] [Google Scholar]
- 38. Aouiss A, Idrissi DA, Kabine M, Zaid Y.. Update of inflammatory proliferative retinopathy: ischemia, hypoxia and angiogenesis. Curr Res Transl Med. 2019; 67(2): 62–71. [DOI] [PubMed] [Google Scholar]
- 39. Scholl S, Kirchhof J, Augustin AJ.. Pathophysiology of macular edema. Ophthalmologica. 2010; 224(Suppl. 1): 8–15. [DOI] [PubMed] [Google Scholar]
- 40. Gündüz K, Bakri SJ.. Management of proliferative diabetic retinopathy. Compr Ophthalmol Update. 2007; 8: 245–256. [PubMed] [Google Scholar]
- 41. Fernandes R, Viana SD, Nunes S, Reis F.. Diabetic gut microbiota dysbiosis as an inflammaging and immunosenescence condition that fosters progression of retinopathy and nephropathy. Biochim Biophys Acta Mol Basis Dis. 2019; 1865: 1876–1897. [DOI] [PubMed] [Google Scholar]