Abstract
Ossifying fibromas are very rare tumors that are sometimes seen as part of the hyperparathyroidism-jaw tumor syndrome (HPT-JT), which is caused by inactivating mutations of the HRPT2/CDC73 tumor suppressor gene. CDC73 mutations have been identified in a subset of sporadic cases but aberrant expression of the encoded protein, parafibromin, has not been demonstrated in ossifying fibroma. We sought to determine if loss of parafibromin regularly contributes to the development of sporadic, nonsyndromic ossifying fibroma. We examined a series of 9 ossifying fibromas, including ossifying, cemento-ossifying, and juvenile active variants, for parafibromin protein expression by immunohistochemistry and for CDC73 sequence abnormalities by Sanger sequencing and/or targeted AmpliSeq panel sequencing. Four ossifying fibromas showed a complete absence of nuclear parafibromin expression; loss of parafibromin expression was coupled with aberrant cytoplasmic parafibromin expression in 1 case. CDC73 mutations were detected in 2 cases with aberrant parafibromin expression. These results provide novel evidence, at the level of protein expression, that loss of the parathyroid CDC73/parafibromin tumor suppressor may play a role in the pathogenesis of a subset of ossifying fibromas.
Keywords: ossifying fibroma, jaw neoplasms, parafibromin, hyperparathyroidism-jaw tumor syndrome, CDC73
Ossifying fibromas belong to a group of histologically similar but etiologically diverse lesions of the craniofacial bones called benign fibro-osseous lesions. Definitive diagnosis, and therefore appropriate treatment, of the specific lesions within this group relies on the combination of microscopic findings with appropriate clinical and radiographic information. In the absence of any of these a noncommittal designation of “benign fibro-osseous lesion” is often, and appropriately, used [1]. Clinically, ossifying fibroma is a slow-growing, painless lesion most frequently found in the mandible and occasionally in the maxilla; proximity to the molar or premolar roots is common. Rarely, it can present in extragnathic bones of the craniofacial skeleton. Juvenile variants exhibit more rapid but noninvasive growth and frequently arise in extragnathic bones, especially surrounding the orbit. Also in contrast to the typical presentation of ossifying fibroma in the third or fourth decade of life (mean age 31 years), juvenile variants typically present in patients under the age of 15 [2]. Radiographically, ossifying fibromas are well-defined, expansile lesions, often beginning as a radiolucency, which may become progressively more radiopaque over time; surrounding bone exhibits normal trabecular architecture. Histologically, ossifying fibroma exhibits a fibrous stroma with varying amounts of mineralized material, which may resemble bony trabeculae or spherules of cementum (sometimes referred to as cemento-ossifying fibroma). The predominant matrix component has no apparent clinical significance [3]. Multinucleated giant cells are not generally observed [4]. Only a few studies have examined the molecular basis of ossifying fibroma and other benign fibro-osseous lesions [5-12].
Ossifying fibromas are rare tumors that, while generally sporadic and nonsyndromic, may be seen as part of the clinical spectrum of the familial hyperparathyroidism-jaw-tumor syndrome (HPT-JT). This autosomal dominant disorder is caused by germline loss-of-function mutations of the CDC73 (previously HRPT2) tumor suppressor gene and consists of a genetic predisposition to the development of ossifying fibromas and cemento-ossifying fibromas of the maxilla and/or mandible, benign or malignant parathyroid tumors, a variety of uterine tumors and renal hamartomas, or Wilm’s tumors [13-18]. CDC73 encodes parafibromin, an essential component of the PAF1 complex, an important transcriptional regulator with roles in maintenance of stem cell pluripotency [19]. Parafibromin inhibits cancer cell growth and causes G1 phase arrest in vitro [20], in part through regulation of cyclin D1 [21]. Studies in knockout animals also suggest a role for parafibromin in apoptosis [22]; effects on cell proliferation and apoptosis may be cell type–specific [23]. Parafibromin interacts directly with β-catenin [24] and likely plays a role in canonical Wnt signaling. Aberrant β-catenin expression has been described in ossifying fibroma [25]. Wnt signaling is a well-established regulator of cell proliferation, cell differentiation, stem cell “stemness,” and bone homeostasis; abnormalities in this pathway have been linked to a variety of human tumors and skeletal disease states (reviewed in [26-28]).
Sporadically presenting tumors of the types commonly associated with HPT-JT, including parathyroid carcinomas and renal tumors, possess somatic (and occasionally germline) mutations of CDC73 [29-31]. Abnormalities of the encoded protein, parafibromin, have been clearly established in parathyroid carcinoma [32, 33]. Mutations of CDC73 have been identified in a subset of sporadically presenting ossifying fibromas; in one case, unexpected germline mutation was noted despite a negative family history and absence of any other features of HPT-JT. Aberrant protein expression, even in cases with CDC73 mutation, has not been previously demonstrated [5, 6]. We therefore sought to determine if loss of parafibromin immunoreactivity, in addition to CDC73 mutation, commonly contributes to the development of sporadic nonsyndromic ossifying fibroma.
Materials and Methods
Patients and Samples
Deidentified, archival, formalin-fixed, paraffin-embedded (FFPE) tumor samples were obtained from patients who underwent surgical excision of a jaw lesion. All samples were evaluated by an oral pathologist and diagnosed according to stringent clinicopathological criteria. Nine tumor samples, including 5 cemento-ossifying fibromas, 2 ossifying fibromas and 2 juvenile ossifying fibromas, were included in this study. No personal and/or family history suggestive of HPT-JT or a phenotypic variant was noted but complete clinical information was not available. Age at surgery was available for 5 cases and ranged from 8 to 50 years (mean 31). Thirty histologically similar, nonneoplastic benign fibro-osseous jaw lesions of other etiologies, consisting of 14 cemento-osseous dysplasias and 16 benign fibro-osseous lesions of unspecified origin, were also obtained as controls. All samples were obtained in accordance with Institutional Review Board-approved protocols and the Helsinki Declaration.
Immunohistochemistry
Parafibromin protein expression and localization was examined immunohistochemically using an anti-parafibromin antibody [34] (2H1, Santa Cruz Biotechnology, Dallas, TX, USA) and the ImmunoCruz mouse ABC Staining System (Santa Cruz Biotechnology), under conditions recommended by the manufacturer. Briefly, 5-micron paraffin sections were deparaffinized and hydrated. Antigen retrieval was performed by incubation at 95 °C in citrate buffer. Endogenous peroxidase activity blocked using BLOXALL (Vector Laboratories, Burlingame, CA, USA) and nonspecific binding reduced using Powerblock (Biogenex, Fremont, CA, USA), followed by serum blocking. Sections were then incubated with the 2H1 antibody (diluted 1:50 in phosphate buffered saline) for 30 minutes at room temperature, followed by incubation with a biotinylated secondary antibody, detection with Avidin Biotin Complex-Peroxidase and DAB substrate. Sections were counterstained with hematoxylin, dehydrated, and mounted. Sections were examined by 2 individuals and scored based on the percentage of tumor cells with positively stained nuclei; sections containing greater than 80% of nuclei staining positive across multiple fields were considered positive. In all cases, both individuals agreed on positive/negative scoring. Parathyroid adenoma sections were used as positive controls; no primary antibody was used for negative controls. Positive and negative controls were included on each run. For ossifying fibroma sections lacking nontumor cells to serve as an internal positive control, nuclear reactivity was confirmed by immunohistochemistry with an anti-TATA binding protein (TBP) antibody [35] (mAbcam 51841, Abcam, Cambridge, MA).
DNA Sequencing
To obtain genomic DNA, 20-μm thick paraffin sections were deparaffinized using xylene, followed by ethanol. DNA was extracted from deparaffinized sections using the QIAamp DNA FFPE Tissue Kit (Qiagen, Germantown, MD, USA). In samples which yielded sufficient DNA, the entire coding region and intron-exon boundaries of CDC73 were PCR amplified and sequenced as previously described [29]. When necessary, alleles were analyzed independently by subcloning PCR fragments using the TOPO TA Cloning kit for sequencing (Thermo Fisher Scientific, Asheville, NC, USA) and sequenced using T3 and T7 primers. All Sanger sequencing reactions were performed by Genewiz (South Plainfield, NJ, USA). Variations from the published sequence were confirmed with an independent PCR/sequencing reaction.
Samples were also processed for targeted sequencing using a 16-gene (including CDC73) custom AmpliSeq targeted sequencing panel that requires ~50 ng of input material, referred to as the ParThy panel, as previously described [36]. Briefly, Ion Torrent AmpliSeq library construction was performed according to the manufacturer’s (Thermo Fisher Scientific) protocol. Completed and equalized libraries were concentrated and loaded onto the 540 S5XL chip using the Ion Chef (Thermo Fisher Scientific); Torrent Chef and run conditions were set up for the ParThy cancer panel and the S5XL configuration was run according to the manufacturer’s protocol until completion. Variants from targeted panel sequencing were called using the command line TorrentSuite variantCaller module v5.2.2 using “Somatic—Low Stringency” settings and a custom forcecall VCF file, exported as VCFs, and loaded into a custom MySQL database. All variants from samples that passed quality control (>50% on-target reads, >80× mean depth, >50% uniformity, as defined in the TorrentSuite web interface) were manually reviewed in the Integrated Genomics Viewer [37].
Results
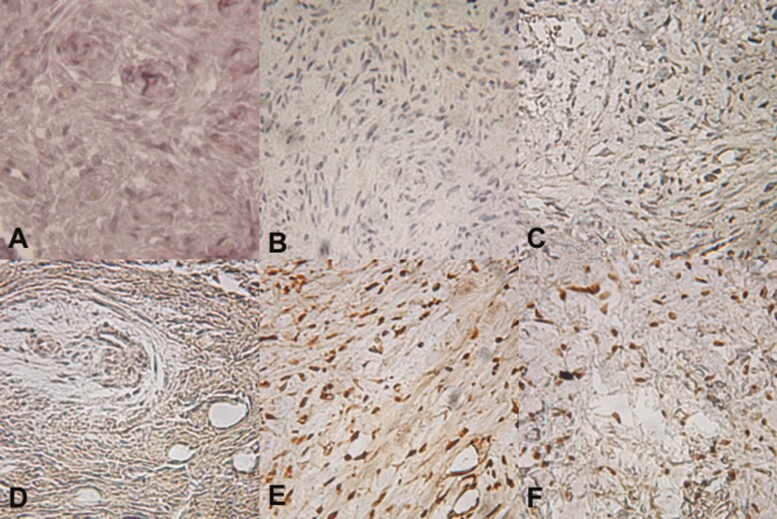
Immunohistochemical staining for parafibromin was performed on 9 ossifying fibroma samples. Four ossifying fibromas showed a complete absence of nuclear parafibromin expression (Table 1 and Fig. 1A-1D); loss of parafibromin expression was coupled with aberrant cytoplasmic parafibromin expression in 1 case (Fig. 1D). Nuclear reactivity to TATA binding protein was confirmed in all cases staining negative for parafibromin. Loss of staining or aberrant (nonnuclear) localization of parafibromin was not seen in any of 30 histologically similar benign fibro-osseous jaw lesions of other etiologies.
Table 1.
Tumor histologic subtype and parafibromin/CDC73 status
| Tumor number | Histologic subtype | Parafibromin staining | CDC73 mutation |
|---|---|---|---|
| 1 | ossifying fibroma | - | c.1A > G |
| 2 | Juvenile ossifying fibroma | + | |
| 3 | cemento-ossifying fibroma | - | c.44delA |
| 4 | cemento-ossifying fibroma | + | |
| 5 | cemento-ossifying fibroma | - nuclear + cytoplasmic |
|
| 6 | cemento-ossifying fibroma | + | |
| 7 | Juvenile ossifying fibroma | + | c.64G > A‡ |
| 8 | ossifying fibroma | + | |
| 9 | cemento-ossifying fibroma | - |
‡ low allelic fraction variant.
Figure 1.
Parafibromin Immunoreactivity in Sporadic Ossifying Fibroma. A-D, Four ossifying fibromas showed a complete absence of nuclear parafibromin expression; loss of parafibromin expression was coupled with aberrant cytoplasmic parafibromin expression in 1 case (D). E-F Normal, high nuclear parafibromin expression was noted in all other tumors examined in this study.
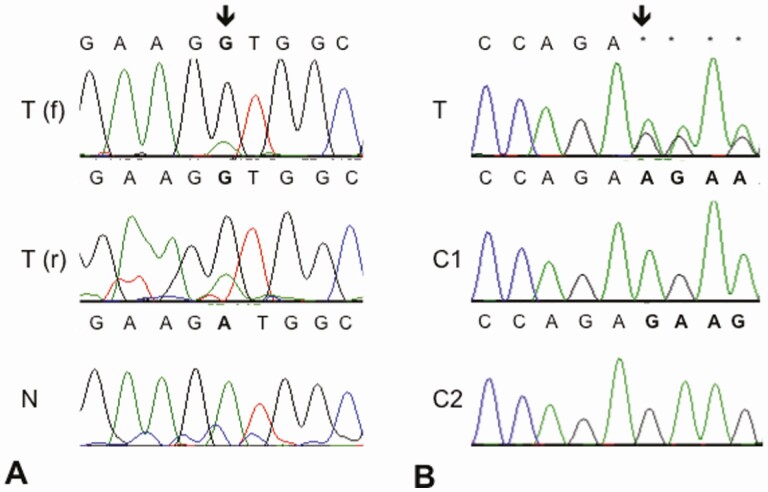
CDC73 sequence analysis was performed on available genomic DNA. Two samples yielded sufficient DNA to perform full Sanger sequencing of the entire CDC73 coding region and intron/exon boundaries. Eight samples, including 1 that had been subjected to Sanger sequencing, were processed for targeted AmpliSeq panel sequencing but only 2 passed quality control. In total, 3 of 9 tumor samples were fully sequenced for CDC73. Information on demineralization status was not available for individual samples but demineralization may have affected DNA yield and/or quality [38]. One tumor contained an adenine to guanine transition mutation at the first position of the CDC73 coding sequence (c.1A > G). This mutation would be expected to result in loss of the start codon and no functional parafibromin protein production, as the next potential start codon is 104 basepairs downstream and out of frame. While nontumor DNA was not available from this case for thorough loss of heterozygosity analysis, the mutation appears to be homozygous (Fig. 2A). One tumor contained a heterozygous, single base pair deletion, c.44delA (Fig. 2B), predicted to produce in a p.Lys15fs*5 mutation in the resultant protein. For more accurate visualization of the sequence variant, the 2 alleles were analyzed independently by subcloning of the PCR fragment. One additional CDC73 sequence variant, c.64G > A, resulting in a predicted novel p.Gly22Arg change, was identified on the targeted sequencing panel in 1 ossifying fibroma with normal parafibromin expression. This variant was present in a very small allelic fraction (0.07), likely representing a subclonal change, and was not independently confirmed by Sanger sequencing. None of the observed variants were present among the more than 230 000 general population alleles in the Genome Aggregation Database (https://gnomad.broadinstitute.org/gene/ENSG00000134371?dataset = gnomad_r2_1, accessed April 21, 2021). Germline DNA was not available from any of the patients to determine germline/somatic status of the identified mutations.
Figure 2.
CDC73 Sequence Analysis. A, In 1 tumor (T), Sanger sequence results show an adenine to guanine transition mutation at position 1 of the CDC73 coding sequence in both the forward (f) and reverse (r) directions, as compared with a nontumor control (N) sample. B, Another tumor (T) contained a heterozygous, single base pair deletion, c.44delA, predicted to produce in a p.Lys15fs*5 mutation in the resultant protein. The 2 alleles were also analyzed independently by subcloning of PCR fragments prior to sequencing; the normal reference sequence was present in Clone 1 (C1) while the deletion mutation was present in Clone 2 (C2).
Discussion
In this study, we examined a series of 9 sporadically presenting tumors, including cemento-ossifying fibroma, ossifying fibroma, and juvenile active ossifying fibroma variants, for parafibromin protein expression via immunohistochemistry, and CDC73 mutation status by direct sequencing. Four ossifying fibromas showed a complete absence of nuclear parafibromin expression; loss of parafibromin expression was coupled with aberrant cytoplasmic parafibromin expression in 1 case. In 2 cases with loss of parafibromin expression, inactivating mutations of CDC73 could be detected. These results provide novel evidence at the level of protein expression for the suggestion that loss of CDC73/parafibromin may play an important role in the pathogenesis of a subset of ossifying fibroma cases.
The involvement of CDC73/parafibromin in ossifying fibroma has previously been assessed in only a few cases [5-7], reflecting the rarity of this disease. In one study, 2 of 4 ossifying fibromas examined were positive for inactivating CDC73 mutations that would be predicted to result in loss of parafibromin expression. However, strong immunostaining was noted in all tumors, even in a case with biallelic frameshift mutations [5]. Similar results were observed in another study, which identified somatic, inactivating CDC73 mutations in 2 of 40 ossifying fibromas but demonstrated strong, nuclear and cytoplasmic parafibromin expression in all cases [6]. The reasons for this discrepancy are unclear but might be due, at least in part, to the relationship between the location of the particular mutations observed and the recognition site of the antibody. All 3 studies used anti-parafibromin antibodies raised against a peptide corresponding to amino acids 87–100 of the parafibromin protein (encoded entirely within exon 3), although in the case of Pimenta et al, the antibody was obtained from a different source. While it has generally been accepted that CDC73 frameshift mutations result in loss of parafibromin immunoreactivity, positive immunoreactivity in mutation-positive cases has been previously described [39]. Two of the 3 mutations observed by Pimenta et al occurred late in the coding sequence (exons 13 and 14). It is possible that these mutations resulted in production of parafibromin protein that was stable enough to be recognized by the antibody, while likely impairing normal parafibromin function, as these mutations would affect the highly conserved C-terminal Cdc73 core domain. In contrast, the predicted inactivating mutations observed in our study occurred much earlier in the coding sequence (both in exon 1) and would be expected to completely abolish the epitope recognized by the antibody. Alternately, the differences in our results could be due to protocol differences [40], subtle differences in case selection and/or population differences between Brazil, China, and the United States.
While loss of parafibromin immunoreactivity was not uniformly seen in the ossifying fibromas examined in this study, parafibromin staining is routinely positive in histologically similar benign fibro-osseous jaw lesions of other etiologies, both in the present study and another study [7]. Parafibromin staining therefore could be useful as a diagnostic tool in equivocal cases, in a similar manner to its use in parathyroid tumors with atypical features suggestive of, but not unequivocal for, carcinoma [32, 33, 41].
In one case in our study, without a detectable CDC73 mutation, we observed loss of nuclear staining but aberrant cytoplasmic staining for parafibromin. While the precise mechanisms underlying tumorigenesis subsequent to CDC73 loss remain unclear, missense mutations affecting nuclear localization have been previously described in HPT-JT families [42] (reviewed in [43]), suggesting a central role for loss of parafibromin nuclear localization in tumorigenesis.
Interestingly, targeted deep sequencing on the ParThy panel revealed a novel, low allelic fraction CDC73 sequence variant. This variant of uncertain significance, c.64G > A (p.Gly22Arg), is not located within any known functional domains of parafibromin and is predicted as “possibly damaging” by PolyPhen-2 and to “affect protein function” by SIFT. As the source of tumor DNA for these studies was from FFPE tissue, and we were able to histologically examine the tissue sections immediately adjacent to those used for DNA extraction, we can be reasonably confident that this low allelic fraction variant represents a subclonal tumor change, as opposed to a clonal change masked by a large number of nontumor cells within the sample. The presence of this variant as a subclonal change within one ossifying fibroma suggests that in addition to its role as a primary driver of ossifying fibroma development, CDC73 mutation could also play a role in clonal evolution or progression of ossifying fibroma. Several low allelic fraction variants were identified in other ParThy panel genes, which merit further investigation.
Further studies are needed to determine the extent of involvement of CDC73/parafibromin abnormalities in ossifying fibroma and to identify additional molecular contributors to the pathogenesis of these tumors. In particular, analysis of a large series of tumors accompanied by germline DNA, detailed clinical histories, and follow-up, which were not available for our series, allowing for genotype-phenotype correlations, would significantly enhance the understanding of the pathogenesis of ossifying fibroma.
Acknowledgments
We wish to thank Drs. Ellen Eisenberg and Easwar Natarajan for their pathology expertise and aid in sample procurement, and Justin Bellizzi for expert technical assistance.
Financial Support: This work was supported by grant DHHS/NIDCR 1F32-DE021307 (to J.C.G.) from the National Institutes of Health and by the Murray-Heilig Fund in Molecular Medicine.
Glossary
Abbreviations
- FFPE
formalin-fixed, paraffin-embedded
- HPT-JT
hyperparathyroidism-jaw tumor syndrome
Additional Information
Disclosures: The authors have no conflicts of interest, including financial or personal relationships, to declare.
Data Availability
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Eversole R, Su L, ElMofty S. Benign fibro-osseous lesions of the craniofacial complex. A review. Head Neck Pathol. 2008;2(3):177-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khanna J, Ramaswami R. Juvenile ossifying fibroma in the mandible. Ann Maxillofac Surg. 2018;8(1):147-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wright JM, Vered M. Update from the 4th edition of the world health organization classification of head and neck tumours: odontogenic and maxillofacial bone tumors. Head Neck Pathol. 2017;11(1):68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacDonald-Jankowski DS. Ossifying fibroma: a systematic review. Dentomaxillofac Radiol. 2009;38(8):495-513. [DOI] [PubMed] [Google Scholar]
- 5. Pimenta FJ, Gontijo Silveira LF, Tavares GC, et al. HRPT2 gene alterations in ossifying fibroma of the jaws. Oral Oncol. 2006;42(7):735-739. [DOI] [PubMed] [Google Scholar]
- 6. Chen Y, Hu DY, Wang TT, et al. CDC73 gene mutations in sporadic ossifying fibroma of the jaws. Diagn Pathol. 2016;11(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Mesquita Netto AC, Gomez RS, Diniz MG, et al. Assessing the contribution of HRPT2 to the pathogenesis of jaw fibrous dysplasia, ossifying fibroma, and osteosarcoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;115(3):359-367. [DOI] [PubMed] [Google Scholar]
- 8. Pereira TDSF, Diniz MG, França JA, et al. The Wnt/β-catenin pathway is deregulated in cemento-ossifying fibromas. Oral Surg Oral Med Oral Pathol Oral Radiol. 2018;125(2):172-178. [DOI] [PubMed] [Google Scholar]
- 9. Gollin SM, Storto PD, Malone PS, et al. Cytogenetic abnormalities in an ossifying fibroma from a patient with bilateral retinoblastoma. Genes Chromosomes Cancer. 1992;4(2):146-152. [DOI] [PubMed] [Google Scholar]
- 10. Dal Cin P, Sciot R, Fossion E, Van Damme B, Van den Berghe H. Chromosome abnormalities in cementifying fibroma. Cancer Genet Cytogenet. 1993;71(2):170-172. [DOI] [PubMed] [Google Scholar]
- 11. Sawyer JR, Tryka AF, Bell JM, Boop FA. Nonrandom chromosome breakpoints at Xq26 and 2q33 characterize cemento-ossifying fibromas of the orbit. Cancer. 1995;76(10):1853-1859. [DOI] [PubMed] [Google Scholar]
- 12. Wang TT, Zhang R, Wang L, Chen Y, Dong Q, Li TJ. Two cases of multiple ossifying fibromas in the jaws. Diagn Pathol. 2014;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carpten JD, Robbins CM, Villablanca A, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32(4):676-680. [DOI] [PubMed] [Google Scholar]
- 14. Williamson C, Cavaco BM, Jauch A, et al. Mapping the gene causing hereditary primary hyperparathyroidism in a Portuguese kindred to chromosome 1q22-q31. J Bone Miner Res. 1999;14(2):230-239. [DOI] [PubMed] [Google Scholar]
- 15. Haven CJ, Wong FK, van Dam EW, et al. A genotypic and histopathological study of a large Dutch kindred with hyperparathyroidism-jaw tumor syndrome. J Clin Endocrinol Metab. 2000;85(4):1449-1454. [DOI] [PubMed] [Google Scholar]
- 16. van der Tuin K, Tops CMJ, Adank MA, et al. CDC73-related disorders: clinical manifestations and case detection in primary hyperparathyroidism. J Clin Endocrinol Metab. 2017;102(12):4534-4540. [DOI] [PubMed] [Google Scholar]
- 17. Chiofalo MG, Sparaneo A, Chetta M, et al. A novel CDC73 gene mutation in an Italian family with hyperparathyroidism-jaw tumour (HPT-JT) syndrome. Cell Oncol (Dordr). 2014;37(4):281-288. [DOI] [PubMed] [Google Scholar]
- 18. Aldred MJ, Talacko AA, Savarirayan R, et al. Dental findings in a family with hyperparathyroidism-jaw tumor syndrome and a novel HRPT2 gene mutation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;101(2):212-218. [DOI] [PubMed] [Google Scholar]
- 19. Ding L, Paszkowski-Rogacz M, Nitzsche A, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4(5):403-415. [DOI] [PubMed] [Google Scholar]
- 20. Zhang C, Kong D, Tan MH, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006;350(1):17-24. [DOI] [PubMed] [Google Scholar]
- 21. Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24(7):1272-1276. [DOI] [PubMed] [Google Scholar]
- 22. Wang P, Bowl MR, Bender S, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol Cell Biol. 2008;28(9):2930-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwata T, Mizusawa N, Taketani Y, Itakura M, Yoshimoto K. Parafibromin tumor suppressor enhances cell growth in the cells expressing SV40 large T antigen. Oncogene. 2007;26(42):6176-6183. [DOI] [PubMed] [Google Scholar]
- 24. Takahashi A, Tsutsumi R, Kikuchi I, et al. SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a tumor suppressor to an oncogenic driver. Mol Cell. 2011;43(1):45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvai AE, Jordan RC. Erratum to: fibro-osseous lesions of the craniofacial bones: beta-catenin immunohistochemical analysis and CTNNB1 and APC mutation analysis. Head Neck Pathol. 2014;8(3):369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4(5):a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wend P, Holland JD, Ziebold U, Birchmeier W. Wnt signaling in stem and cancer stem cells. Semin Cell Dev Biol. 2010;21(8):855-863. [DOI] [PubMed] [Google Scholar]
- 28. Regard JB, Zhong Z, Williams BO, Yang Y. Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb Perspect Biol. 2012;4(12):a007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shattuck TM, Välimäki S, Obara T, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349(18):1722-1729. [DOI] [PubMed] [Google Scholar]
- 30. Zhao J, Yart A, Frigerio S, et al. Sporadic human renal tumors display frequent allelic imbalances and novel mutations of the HRPT2 gene. Oncogene. 2007;26(23):3440-3449. [DOI] [PubMed] [Google Scholar]
- 31. Siu WK, Law CY, Lam CW, et al. Novel nonsense CDC73 mutations in Chinese patients with parathyroid tumors. Fam Cancer. 2011;10(4):695-699. [DOI] [PubMed] [Google Scholar]
- 32. Tan MH, Morrison C, Wang P, et al. Loss of parafibromin immunoreactivity is a distinguishing feature of parathyroid carcinoma. Clin Cancer Res. 2004;10(19):6629-6637. [DOI] [PubMed] [Google Scholar]
- 33. Cetani F, Ambrogini E, Viacava P, et al. Should parafibromin staining replace HRTP2 gene analysis as an additional tool for histologic diagnosis of parathyroid carcinoma? Eur J Endocrinol. 2007;156(5):547-554. [DOI] [PubMed] [Google Scholar]
- 34. RRID:AB_628102. [Google Scholar]
- 35. RRID:AB_945758. [Google Scholar]
- 36. Pandya C, Uzilov AV, Bellizzi J, et al. Genomic profiling reveals mutational landscape in parathyroid carcinomas. JCI Insight. 2017;2(6):e92061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77(21):e31-e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caputo M, Irisarri M, Alechine E, Corach D. A DNA extraction method of small quantities of bone for high-quality genotyping. Forensic Sci Int Genet. 2013;7(5):488-493. [DOI] [PubMed] [Google Scholar]
- 39. Juhlin CC, Villablanca A, Sandelin K, et al. Parafibromin immunoreactivity: its use as an additional diagnostic marker for parathyroid tumor classification. Endocr Relat Cancer. 2007;14(2):501-512. [DOI] [PubMed] [Google Scholar]
- 40. Delellis RA. Challenging lesions in the differential diagnosis of endocrine tumors: parathyroid carcinoma. Endocr Pathol. 2008;19(4):221-225. [DOI] [PubMed] [Google Scholar]
- 41. Juhlin CC, Erickson LA. Genomics and epigenomics in parathyroid neoplasia: from bench to surgical pathology practice. Endocr Pathol. 2021;32(1):17-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pazienza V, la Torre A, Baorda F, et al. Identification and functional characterization of three NoLS (nucleolar localisation signals) mutations of the CDC73 gene. Plos One. 2013;8(12):e82292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cardoso L, Stevenson M, Thakker RV. Molecular genetics of syndromic and non-syndromic forms of parathyroid carcinoma. Hum Mutat. 2017;38(12):1621-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.