Abstract
The SPIRIT 2013 statement aims to improve the completeness of clinical trial protocol reporting by providing evidence-based recommendations for the minimum set of items to be addressed. This guidance has been instrumental in promoting transparent evaluation of new interventions. More recently, there has been a growing recognition that interventions involving artificial intelligence (AI) need to undergo rigorous, prospective evaluation to demonstrate their impact on health outcomes. The SPIRIT-AI (Standard Protocol Items: Recommendations for Interventional Trials-Artificial Intelligence) extension is a new reporting guideline for clinical trial protocols evaluating interventions with an AI component. It was developed in parallel with its companion statement for trial reports: CONSORT-AI (Consolidated Standards of Reporting Trials-Artificial Intelligence). Both guidelines were developed through a staged consensus process involving literature review and expert consultation to generate 26 candidate items, which were consulted upon by an international multi-stakeholder group in a two-stage Delphi survey (103 stakeholders), agreed upon in a consensus meeting (31 stakeholders) and refined through a checklist pilot (34 participants). The SPIRIT-AI extension includes 15 new items that were considered sufficiently important for clinical trial protocols of AI interventions. These new items should be routinely reported in addition to the core SPIRIT 2013 items. SPIRIT-AI recommends that investigators provide clear descriptions of the AI intervention, including instructions and skills required for use, the setting in which the AI intervention will be integrated, considerations for the handling of input and output data, the human–AI interaction and analysis of error cases. SPIRIT-AI will help promote transparency and completeness for clinical trial protocols for AI interventions. Its use will assist editors and peer reviewers, as well as the general readership, to understand, interpret, and critically appraise the design and risk of bias for a planned clinical trial.
Introduction
A clinical trial protocol is an essential document produced by study investigators detailing a priori the rationale, proposed methods and plans for how a clinical trial will be conducted.1,2 This key document is used by external reviewers (funding agencies, regulatory bodies, research ethics committees, journal editors, peer reviewers, institutional review boards and, increasingly, the wider public) to understand and interpret the rationale, methodological rigour, and ethical considerations of the trial. Additionally, trial protocols provide a shared reference point to support the research team in conducting a high-quality study.
Despite their importance, the quality and completeness of published trial protocols are variable.1,2 The SPIRIT statement was published in 2013 to provide guidance for the minimum reporting content of a clinical trial protocol and has been widely endorsed as an international standard.3–5 The SPIRIT statement published in 2013 provides minimum guidance applicable for all clinical trial interventions but recognises that certain interventions may require extension or elaboration of these items.1,2 Artificial intelligence (AI) is an area of enormous interest, with strong drivers to accelerate new interventions through to publication, implementation, and market.6 While AI systems have been researched for some time, recent advances in deep learning and neural networks have gained considerable interest for their potential in health applications. Examples of such applications of these are wide ranging and include AI systems for screening and triage,7,8 diagnosis,9–12 prognostication,13,14 decision support,15 and treatment recommendation.16 However, in most recent cases, the majority of published evidence has consisted of in-silico, early-phase validation. It has been recognised that most recent AI studies are inadequately reported and existing reporting guidelines do not fully cover potential sources of bias specific to AI systems.17 The welcome emergence of randomised controlled trials seeking to evaluate the clinical efficacy of newer interventions based on, or including, an AI component (called “AI interventions” here)15,18–23 has similarly been met with concerns about design and reporting,17,24–26 This has highlighted the need to provide reporting guidance that is fit for purpose in this domain.
SPIRIT-AI (as part of the SPIRIT-AI and CONSORT-AI initiative) is an international initiative supported by SPIRIT and the EQUATOR (Enhancing the Quality and Transparency of Health Research) Network to extend or elaborate on the existing SPIRIT 2013 statement where necessary, to develop consensus-based AI-specific protocol guidance.27,28 It is complementary to the CONSORT-AI statement, which aims to promote high-quality reporting of AI trials. This Consensus Statement describes the methods used to identify and evaluate candidate items and gain consensus. In addition, it also provides the full SPIRIT-AI checklist, including new items and their accompanying explanations.
Methods
The SPIRIT-AI and CONSORT-AI extensions were simultaneously developed for clinical trial protocols and trial reports. An announcement for the SPIRIT-AI and CONSORT-AI initiative was published in October 2019,27 and the two guidelines were registered as reporting guidelines under development on the EQUATOR library of reporting guidelines in May, 2019. Both guidelines were developed in accordance with the EQUATOR Network’s methodological framework.29 The SPIRIT-AI and CONSORT-AI Steering Group, consisting of 15 international experts, was formed to oversee the conduct and methodology of the study. Definitions of key terms are provided in the glossary (panel).
Ethical approval
This study was approved by the ethical review committee at the University of Birmingham, UK (ERN_19–1100). Participant information was provided to Delphi participants electronically before survey completion and before the consensus meeting. Delphi participants provided electronic informed consent, and written consent was obtained from participants.
Literature review and candidate-item generation
An initial list of candidate items for the SPIRIT-AI and CONSORT-AI checklists was generated through review of the published literature and consultation with the Steering Group and known international experts. A search was performed on May 13, 2019, using the terms “artificial intelligence”, “machine learning”, and “deep learning” to identify existing clinical trials for AI interventions listed within the US National Library of Medicine’s clinical trial registry (ClinicalTrials.gov). There were 316 registered trials, of which 62 were completed and seven had published results.22,30–35 Two studies were reported with reference to the CONSORT statement,22,34 and one study provided an unpublished trial protocol.34 The Operations Team (XL, SCR, MJC, and AKD) identified AI-specific considerations from these studies and reframed them as candidate reporting items. The candidate items were also informed by findings from a previous systematic review that evaluated the diagnostic accuracy of deep-learning systems for medical imaging.17 After consultation with the Steering Group and additional international experts (n=19), 29 candidate items were generated, 26 of which were relevant for both SPIRIT-AI and CONSORT-AI and three of which were relevant only for CONSORT-AI. The Operations Team mapped these items to the corresponding SPIRIT and CONSORT items, revising the wording and providing explanatory text as required to contextualise the items. These items were included in subsequent Delphi surveys.
Delphi consensus process
In September, 2019, 169 key international experts were invited to participate in the online Delphi survey to vote upon the candidate items and suggest additional items. Experts were identified and contacted via the Steering Group and were allowed one round of “snowball” recruitment in which contacted experts could suggest additional experts. In addition, individuals who made contact following publication of the announcement were included.27 The Steering Group agreed that individuals with expertise in clinical trials and AI and machine learning (ML), as well as key users of the technology, should be well represented in the consultation. Stakeholders included health-care professionals, methodologists, statisticians, computer scientists, industry representatives, journal editors, policy makers, health “informaticists”, experts in law and ethics, regulators, patients, and funders. Participant characteristics are described in the appendix (p 1). Two online Delphi surveys were conducted. DelphiManager software (version 4.0), developed and maintained by the COMET (Core Outcome Measures in Effectiveness Trials) initiative, was used to undertake the e-Delphi surveys. Participants were given written information about the study and were asked to provide their level of expertise within the fields of (i) AI/ML and (ii) clinical trials. Each item was presented for consideration (26 for SPIRIT-AI and 29 for CONSORT-AI). Participants were asked to vote on each item using a 9-point scale, as follows: 1–3, not important; 4–6, important but not critical; and 7–9, important and critical. Respondents provided separate ratings for SPIRIT-AI and CONSORT-AI. There was an option to opt out of voting for each item, and each item included space for free text comments. At the end of the Delphi survey, participants had the opportunity to suggest new items. 103 responses were received for the first Delphi round, and 91 responses (88% of participants from round one) were received for the second round. The results of the Delphi surveys informed the subsequent international consensus meeting. 12 new items were proposed by the Delphi study participants and were added for discussion at the consensus meeting. Data collected during the Delphi survey were anonymised, and item-level results were presented at the consensus meeting for discussion and voting.
The two-day consensus meeting took place in January, 2020, and was hosted by the University of Birmingham, UK, to seek consensus on the content of SPIRIT-AI and CONSORT-AI. 31 international stakeholders from among the Delphi survey participants were invited to discuss the items and vote on their inclusion. Participants were selected to achieve adequate representation from all the stakeholder groups. 38 items were discussed in turn, comprising the 26 items generated in the initial literature review and item-generation phase (these 26 items were relevant to both SPIRIT-AI and CONSORT-AI; three extra items relevant only to CONSORT-AI were also discussed) and the 12 new items proposed by participants during the Delphi surveys. Each item was presented to the Consensus Group, alongside its score from the Delphi exercise (median and interquartile ranges) and any comments made by Delphi participants related to that item. Consensus meeting participants were invited to comment on the importance of each item and whether the item should be included in the AI extension. In addition, participants were invited to comment on the wording of the explanatory text accompanying each item and the position of each item relative to the SPIRIT 2013 and CONSORT 2010 checklists. After open discussion of each item and the option to adjust wording, an electronic vote took place, with the option to include or exclude the item. An 80% threshold for inclusion was pre-specified and deemed reasonable by the Steering Group to demonstrate majority consensus. Each stakeholder voted anonymously using Turning Point voting pads (Turning Technologies, version 8.7.2.14).
Checklist pilot
Following the consensus meeting, attendees were given the opportunity to make final comments on the wording and agree that the updated SPIRIT-AI and CONSORT-AI items reflected discussions from the meeting. The Operations Team assigned each item as an extension or elaboration item on the basis of a decision tree and produced a penultimate draft of the SPIRIT-AI and CONSORT-AI checklists (appendix p 6). A pilot of the penultimate checklists was conducted with 34 participants to ensure clarity of wording. Experts participating in the pilot included the following: (a) Delphi participants who did not attend the consensus meeting, and (b) external experts who had not taken part in the development process but who had reached out to the Steering Group after the Delphi study commenced. Final changes were made on wording only to improve clarity for readers, by the Operations Team (appendix p 7).
Recommendations
SPIRIT-AI checklist items and explanation
The SPIRIT-AI extension recommends that, in conjunction with existing SPIRIT 2013 items, 15 items (12 extensions and 3 elaborations) should be addressed for trial protocols of AI interventions. These items were considered sufficiently important for clinical trial protocols for AI interventions that they should be routinely reported in addition to the core SPIRIT 2013 checklist items. Figure 1 lists the SPIRIT-AI items.
Figure 1: SPIRIT-AI checklist.
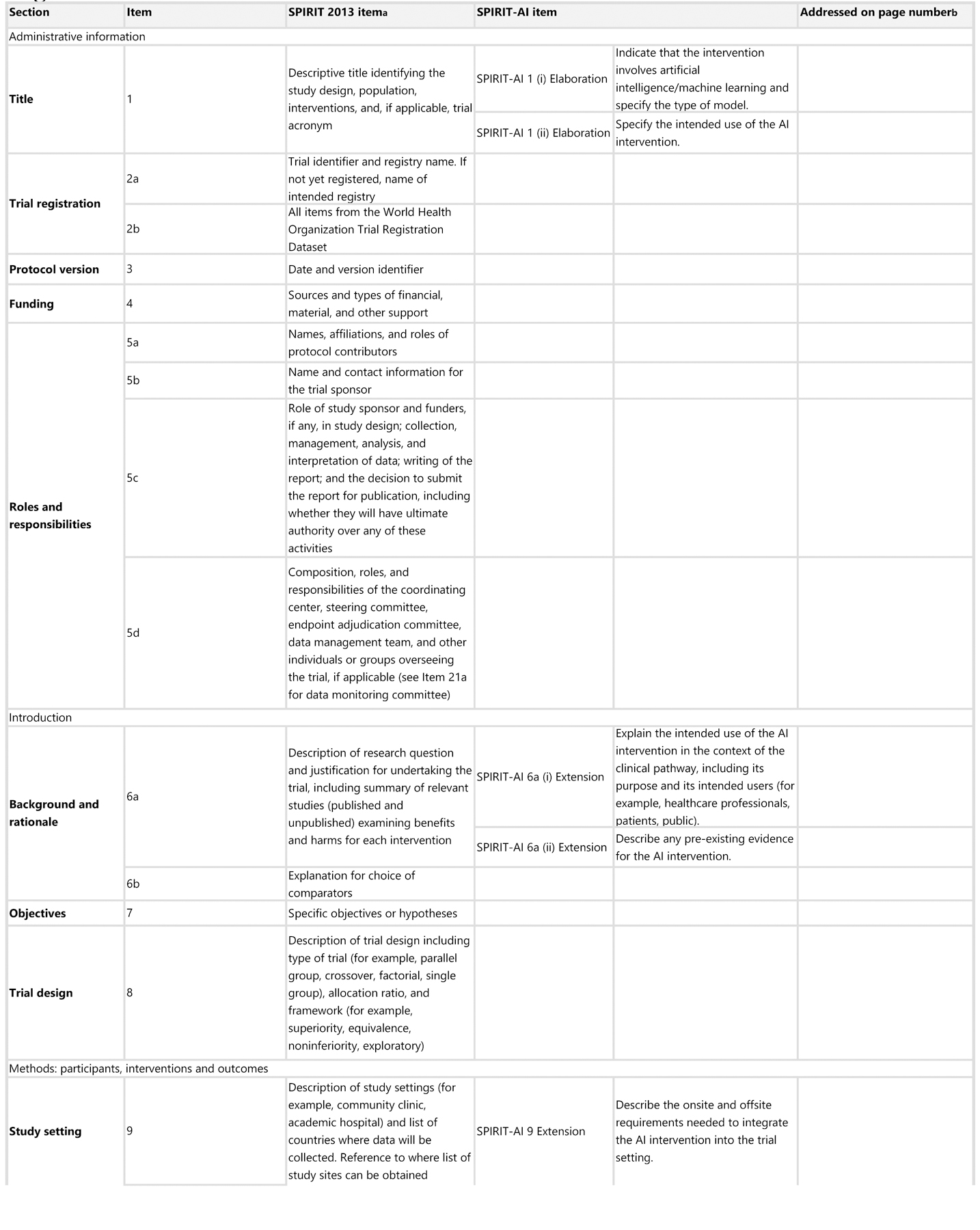
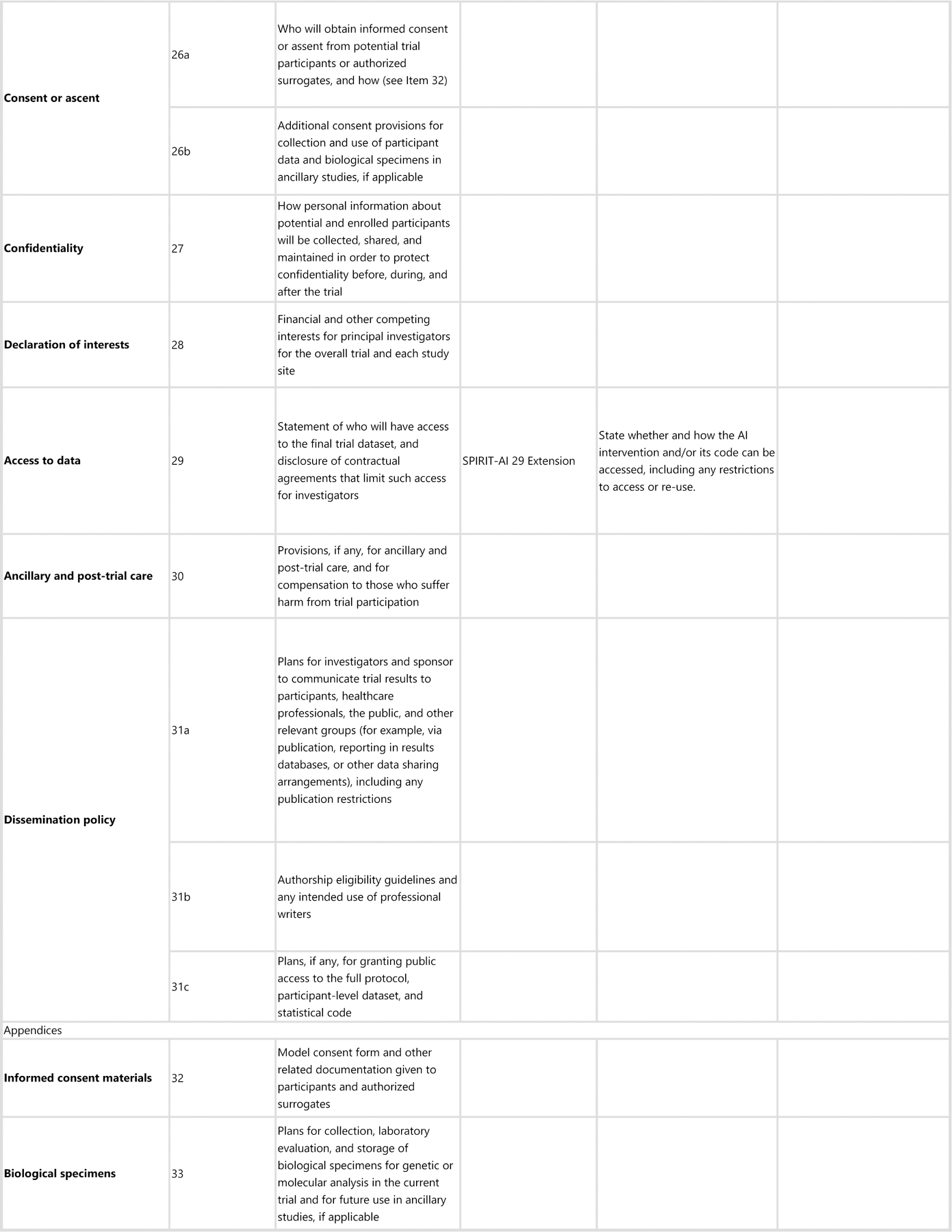
aIt is strongly recommended that this checklist be read in conjunction with the SPIRIT 2013 Explanation & Elaboration for important clarification on the items.
bIndicates page numbers to be completed by authors during protocol development.
All 15 items included in the SPIRIT-AI extension passed the threshold of 80% for inclusion at the consensus meeting. SPIRIT-AI 6a (i), SPIRIT-AI 11a (v) and SPIRIT-AI 22 each resulted from the merging of two items after discussion. SPIRIT-AI 11a (iii) did not fulfil the criteria for inclusion on the basis of its initial wording (73% vote to include); however, after extensive discussion and rewording, the Consensus Group unanimously supported a re-vote, at which point it passed the inclusion threshold (97% to include).
Administrative information
SPIRIT-AI 1 (i) Elaboration: Indicate that the intervention involves artificial intelligence/machine learning and specify the type of model
Explanation. Indicating in the protocol title and/or abstract that the intervention involves a form of AI is encouraged, as it immediately identifies the intervention as an AI/ML intervention and also serves to facilitate indexing and searching of the trial protocol in bibliographic databases, registries, and other online resources. The title should be understandable by a wide audience; therefore, a broader umbrella term such as “artificial intelligence” or “machine learning” is encouraged. More-precise terms should be used in the abstract, rather than the title, unless they are broadly recognised as being a form of AI/ML. Specific terminology relating to the model type and architecture should be detailed in the abstract.
SPIRIT-AI 1 (ii) Elaboration: Specify the intended use of the AI intervention
Explanation. The intended use of the AI intervention should be made clear in the protocol’s title and/or abstract. This should describe the purpose of the AI intervention and the disease context.19,36 Some AI interventions may have multiple intended uses, or the intended use may evolve over time. Therefore, documenting this allows readers to understand the intended use of the algorithm at the time of the trial.
Introduction
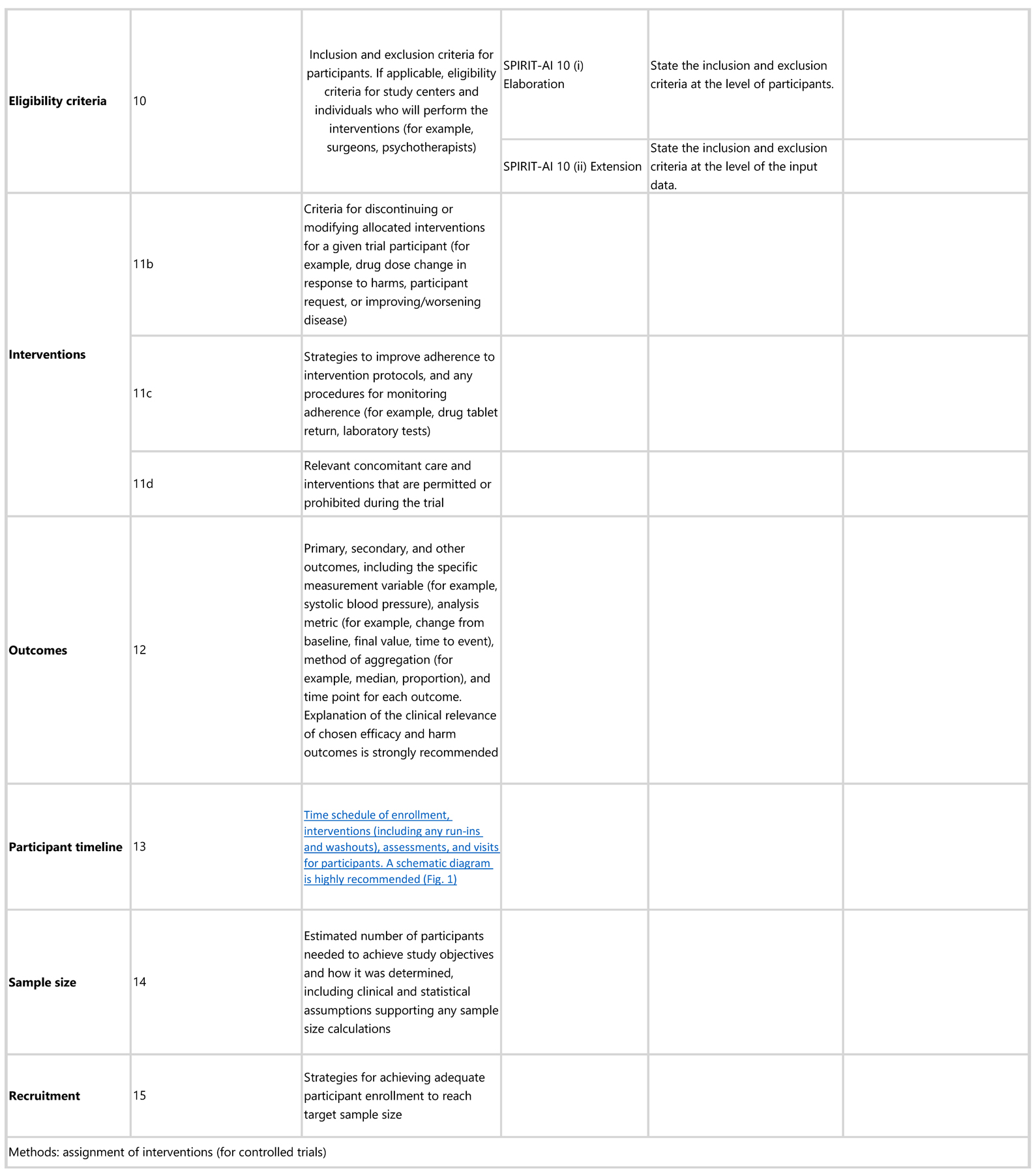
SPIRIT-AI 6a (i) Extension: Explain the intended use of the AI intervention in the context of the clinical pathway, including its purpose and its intended users (for example, health-care professionals, patients, public)
Explanation. In order to clarify how the AI intervention will fit into a clinical pathway, a detailed description of its role should be included in the protocol background. AI interventions may be designed to interact with different users, including health-care professionals, patients, and the public, and their roles can be wide-ranging (for example, the same AI intervention could theoretically be replacing, augmenting, or adjudicating components of clinical decision-making). Clarifying the intended use of the AI intervention and its intended user helps readers understand the purpose for which the AI intervention will be evaluated in the trial.
SPIRIT-AI 6a (ii) Extension: Describe any pre-existing evidence for the AI intervention
Explanation. Authors should describe in the protocol any pre-existing published evidence (with supporting references) or unpublished evidence relating to validation of the AI intervention or lack thereof. Consideration should be given to whether the evidence was for a use, setting, and target population similar to that of the planned trial. This may include previous development of the AI model, internal and external validations and any modifications made before the trial.
Participants, interventions, and outcomes
SPIRIT-AI 9 Extension: Describe the onsite and offsite requirements needed to integrate the AI intervention into the trial setting
Explanation. There are limitations to the generalisability of AI algorithms, one of which is when they are used outside of their development environment.37,38 AI systems are dependent on their operational environment, and the protocol should provide details of the hardware and software requirements to allow technical integration of the AI intervention at each study site. For example, it should be stated if the AI intervention requires vendor-specific devices, if there is a need for specialised computing hardware at each site, or if the sites must support cloud integration, particularly if this is vendor specific. If any changes to the algorithm are required at each study site as part of the implementation procedure (such as fine-tuning the algorithm on local data), then this process should also be clearly described.
SPIRIT-AI 10 (i) Elaboration: State the inclusion and exclusion criteria at the level of participants
Explanation. The inclusion and exclusion criteria should be defined at the participant level as per usual practice in protocols of non-AI interventional trials. This is distinct from the inclusion and exclusion criteria made at the input-data level, which are addressed in item 10 (ii).
SPIRIT-AI 10 (ii) Extension: State the inclusion and exclusion criteria at the level of the input data
Explanation. “Input data” refers to the data required by the AI intervention to serve its purpose (for example, for a breast cancer diagnostic system, the input data could be the unprocessed or vendor-specific post-processing mammography scan upon which a diagnosis is being made; for an early-warning system, the input data could be physiological measurements or laboratory results from the electronic health record). The trial protocol should pre-specify if there are minimum requirements for the input data (such as image resolution, quality metrics, or data format) that would determine pre-randomisation eligibility. It should specify when, how, and by whom this will be assessed. For example, if a participant met the eligibility criteria for lying flat for a CT scan as per item 10 (i), but the scan quality was compromised (for any given reason) to such a level that it is no longer fit for use by the AI system, this should be considered as an exclusion criterion at the input-data level. Note that where input data are acquired after randomisation (addressed by SPIRIT-AI 20c), any exclusion is considered to be from the analysis, not from enrolment (figure 2).
Figure 2: CONSORT 2010 flow diagram, adapted for AI clinical trials.
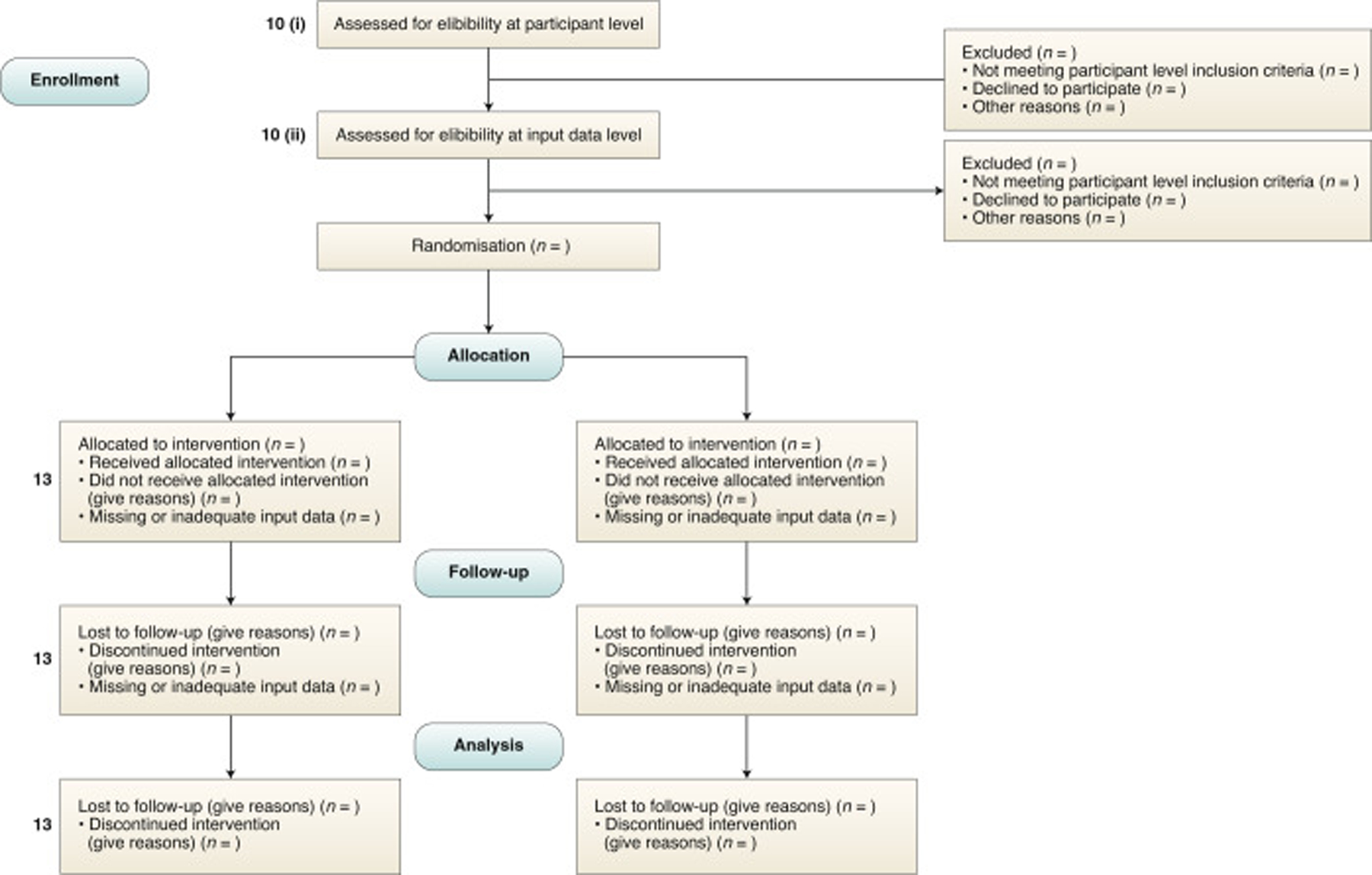
AI=artificial intelligence. SPIRIT-AI 10 (i): State the inclusion and exclusion criteria at the level of participants. SPIRIT-AI 10 (ii): State the inclusion and exclusion criteria at the level of the input data. SPIRIT 13 (core CONSORT item): Time schedule of enrollment, interventions (including any run-ins and washouts), assessments, and visits for participants. A schematic diagram is highly recommended.
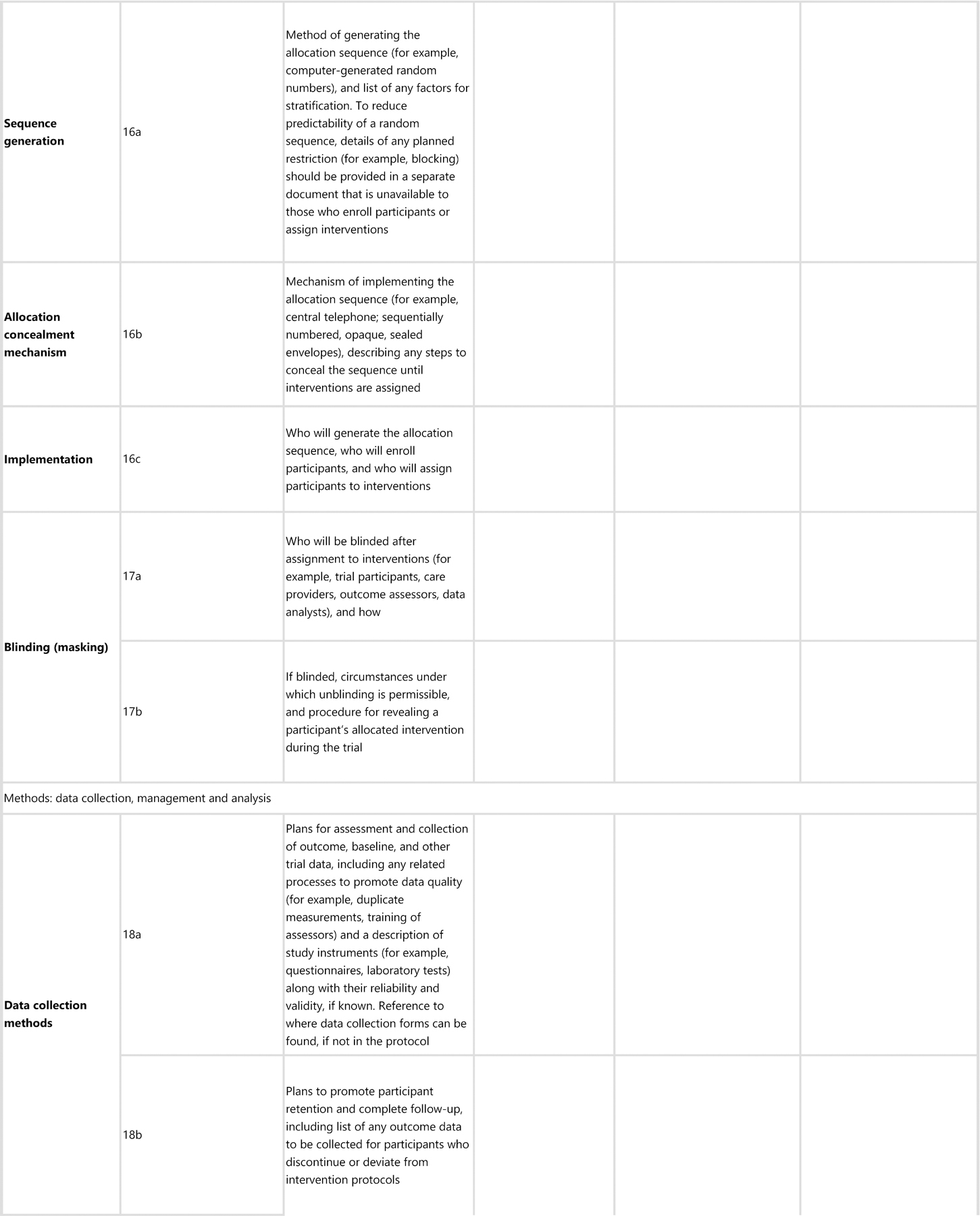
SPIRIT-AI 11a (i) Extension: State which version of the AI algorithm will be used
Explanation. Similar to other forms of software as a medical device, AI systems are likely to undergo multiple iterations and updates in their lifespan. The protocol should state which version of the AI system will be used in the clinical trial and whether this is the same version that was used in previous studies that have been used to justify the study rationale. If applicable, the protocol should describe what has changed between the relevant versions and the rationale for the changes. Where available, the protocol should include a regulatory marking reference, such as a unique device identifier, that requires a new identifier for updated versions of the device.39
SPIRIT-AI 11a (ii) Extension: Specify the procedure for acquiring and selecting the input data for the AI intervention
Explanation. The measured performance of any AI system may be critically dependent on the nature and quality of the input data.40 The procedure for how input data will be handled, including data acquisition, selection and pre-processing before analysis by the AI system, should be provided. Completeness and transparency of this process are integral to feasibility assessment and to future replication of the intervention beyond the clinical trial. It will also help to identify whether input-data-handling procedures will be standardised across trial sites.
SPIRIT-AI 11a (iii) Extension: Specify the procedure for assessing and handling poor-quality or unavailable input data
Explanation. As with SPIRIT-AI 10 (ii), “input data” refers to the data required by the AI intervention to serve its purpose. As noted in SPIRIT-AI 10 (ii), the performance of AI systems may be compromised as a result of poor-quality or missing input data41 (for example, excessive-movement artifact on an electrocardiogram). The study protocol should specify if and how poor-quality or unavailable input data will be identified and handled. The protocol should also specify a minimum standard required for the input data and the procedure for when the minimum standard is not met (including the impact on, or any changes to, the participant care pathway).
Poor-quality or unavailable data can also affect non-AI interventions. For example, suboptimal quality of a scan could affect a radiologist’s ability to interpret it and make a diagnosis. It is therefore important that this information is reported equally for the control intervention, where relevant. If this minimum quality standard is different from the inclusion criteria for input data used to assess eligibility pre-randomisation, this should be stated.
SPIRIT-AI 11a (iv) Extension: Specify whether there is human–AI interaction in the handling of the input data, and what level of expertise is required for users
Explanation. A description of the human–AI interface and the requirements for successful interaction when input data are handled should be provided. Examples include clinician-led selection of regions of interest from a histology slide that is then interpreted by an AI diagnostic system,42 or an endoscopist’s selection of a colonoscopy video clips as input data for an algorithm designed to detect polyps.21 A description of any planned user training and instructions for how users will handle the input data provides transparency and replicability of trial procedures. Poor clarity on the human–AI interface may lead to a lack of a standard approach and may carry ethical implications, particularly in the event of harm.43,44 For example, it may become unclear whether an error case occurred due to human deviation from the instructed procedure, or if it was an error made by the AI system.
SPIRIT-AI 11a (v) Extension: Specify the output of the AI intervention
Explanation. The output of the AI intervention should be clearly defined in the protocol. For example, an AI system may output a diagnostic classification or probability, a recommended action, an alarm alerting to an event, an instigated action in a closed-loop system (such as titration of drug infusions), or another output. The nature of the AI intervention’s output has direct implications on its usability and how it may lead to downstream actions and outcomes.
SPIRIT-AI 11a (vi) Extension: Explain the procedure for how the AI intervention’s outputs will contribute to decision-making or other elements of clinical practice
Explanation. Since health outcomes may also critically depend on how humans interact with the AI intervention, the trial protocol should explain how the outputs of the AI system are used to contribute to decision-making or other elements of clinical practice. This should include adequate description of downstream interventions that can impact outcomes. As with SPIRIT-AI 11a (iv), any elements of human–AI interaction on the outputs should be described in detail, including the level of expertise required to understand the outputs and any training and/or instructions provided for this purpose. For example, a skin-cancer-detection system that produces a percentage likelihood as output should be accompanied by an explanation of how this output should be interpreted and acted upon by the user, specifying both the intended pathways (eg, skin-lesion excision if the diagnosis is positive) and the thresholds for entry to these pathways (eg, skin-lesion excision if the diagnosis is positive and the probability is >80%). The information produced by comparator interventions should be similarly described, alongside an explanation of how such information was used to arrive at clinical decisions for patient management, where relevant.
Monitoring
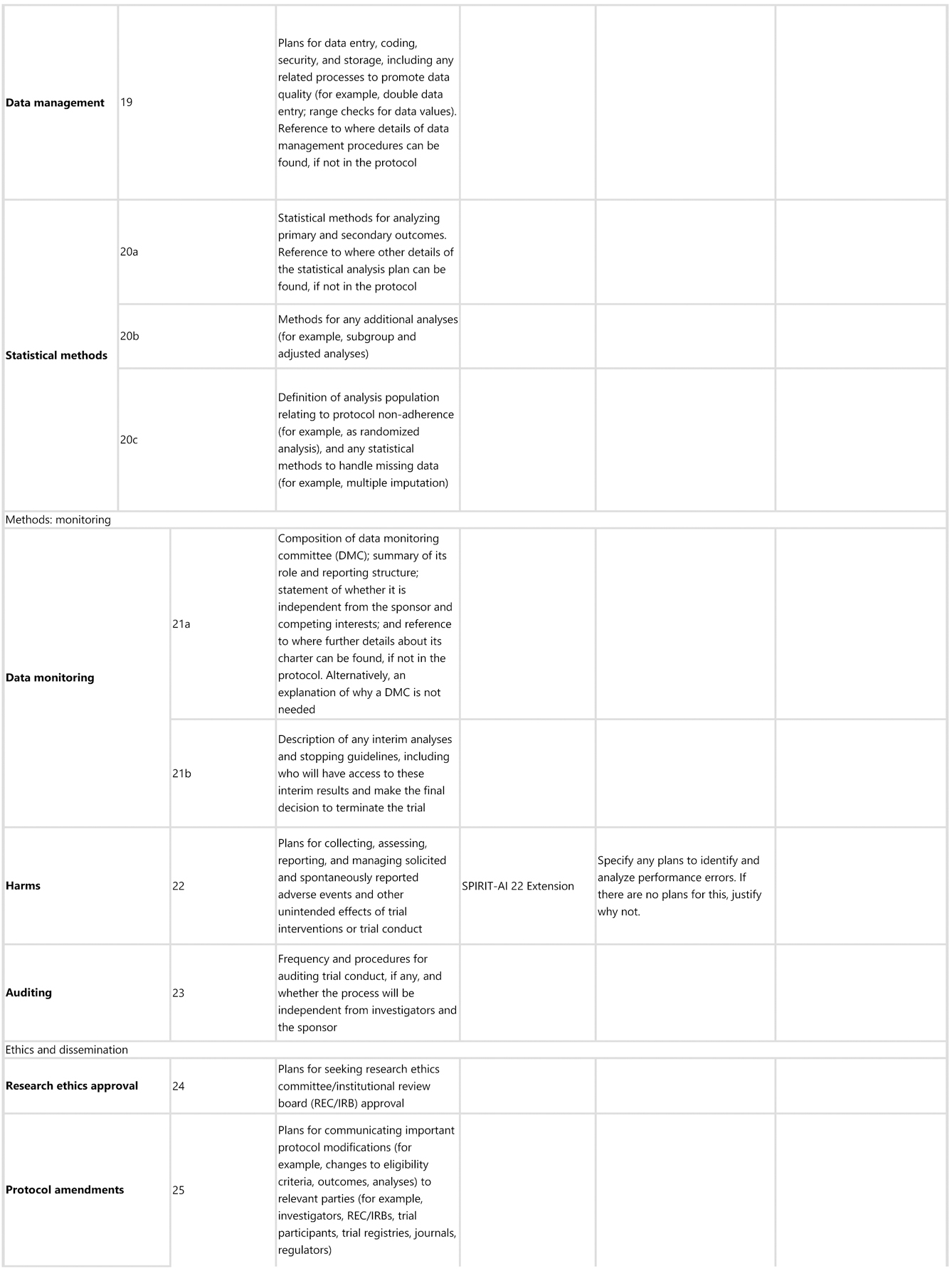
SPIRIT-AI 22 Extension: Specify any plans to identify and analyse performance errors. If there are no plans for this, explain why not
Explanation. Reporting performance errors and failure case analysis is especially important for AI interventions. AI systems can make errors that may be hard to foresee but that, if allowed to be deployed at scale, could have catastrophic consequences.45 Therefore, identifying cases of error and defining risk-mitigation strategies is important for informing when the intervention can be safely implemented, and for which populations. The protocol should specify whether there are any plans to analyse performance errors. If there are no plans for this, a justification should be included in the protocol.
Ethics and dissemination
SPIRIT-AI 29 Extension: State whether and how the AI intervention and/or its code can be accessed, including any restrictions to access or re-use
Explanation. The protocol should make clear whether and how the AI intervention and/or its code can be accessed or re-used. This should include details about the license and any restrictions to access.
Discussion
The SPIRIT-AI extension provides international consensus-based guidance on AI-specific information that should be reported in clinical trial protocols, alongside SPIRIT 2013 and other relevant SPIRIT extensions.4,46 It comprises of 15 items: three elaborations to the existing SPIRIT 2013 guidance in the context of AI trials, and 12 new extensions. The guidance does not aim to be prescriptive about the methodological approach to AI trials; instead, it aims to promote transparency in reporting the design and methods of a clinical trial to facilitate understanding, interpretation, and peer review.
A number of extension items relate to the intervention (items 11 [i]–11 [vi]), its setting (item 9) and intended role (item 6a [i]). Specific recommendations were made pertinent to AI systems related to algorithm version, input and output data, integration into trial settings, expertise of the users, and protocol for acting upon the AI system’s recommendations. It was agreed that these details are critical for independent evaluation of the study protocol. Journal editors reported that despite the importance of these items, they are currently often missing from trial protocols and reports at the time of submission for publication, which provides further weight to their inclusion as specifically listed extension items.
A recurrent focus of the Delphi comments and Consensus Group discussion was the safety of AI systems. This is in recognition that these systems, unlike other health interventions, can unpredictably yield errors that are not easily detectable or explainable by human judgement. For example, changes to medical imaging that are invisible, or appear random, to the human eye may change the likelihood of the resultant diagnostic output entirely.47,48 The concern is that given the theoretical ease with which AI systems could be deployed at scale, any unintended harmful consequences could be catastrophic. Two extension items were added to address this. SPIRIT-AI item 6a (ii) requires specification of the prior level of evidence for validation of the AI intervention. SPIRIT-AI item 22 requires specification of any plans to analyse performance errors, to emphasise the importance of anticipating systematic errors made by the algorithm and their consequences.
One topic that was raised in the Delphi survey responses and consensus meeting that is not included in the final guidelines is “continuously evolving” AI systems (also known as “continuously adapting” or “continuously learning” AI systems). These are AI systems with the ability to continuously train on new data, which may cause changes in performance over time. The group noted that, while of interest, this field is relatively early in its development without tangible examples in health-care applications, and that it would not be appropriate for it to be addressed by SPIRIT-AI at this stage.49 This topic will be monitored and revisited in future iterations of SPIRIT-AI. It is worth noting that incremental software changes, whether continuous or iterative, intentional or unintentional, could have serious consequences on safety performance after deployment. It is therefore of vital importance that such changes are documented and identified by software version and that a robust post-deployment surveillance plan is in place.
This study is set in the current context of AI in health; therefore, several limitations should be noted. First, at the time of SPIRIT-AI development, there were only seven published trials and no published trial protocols in the field of AI for health care. Thus, the discussion and decisions made during the development of SPIRIT-AI are not always supported by existing real-world examples. This arises from our stated aim of addressing the issues of poor protocol development in this field as early as possible, recognising the strong drivers in the field and the specific challenges of study design and reporting for AI. As the science and study of AI evolves, we welcome collaboration with investigators to co-evolve these reporting standards to ensure their continued relevance. Second, the literature search for AI randomised controlled trials used terminology such as “artificial intelligence”, “machine learning”, and “deep learning”, but not terms such as “clinical decision support systems” and “expert systems”, which were more commonly used in the 1990s for technologies underpinned by AI systems and share risks similar to those of recent examples.50 It is likely that such systems, if published today, would be indexed under ”artificial intelligence” or ”machine learning”; however, clinical-decision support systems were not actively discussed during this consensus process. Third, the initial candidate-items list was generated by a relatively small group of experts consisting of Steering Group members and additional international experts. However, additional items from the wider Delphi group were taken forward for consideration by the Consensus Group, and no new items were suggested during the consensus meeting or post-meeting evaluation.
As with the SPIRIT statement, the SPIRIT-AI extension is intended as a minimum reporting guidance, and there are additional AI-specific considerations for trial protocols that may warrant consideration (appendix pp 2–5). This extension is aimed particularly at investigators planning or conducting clinical trials; however, it may also serve as useful guidance for developers of AI interventions in earlier validation stages of an AI system. Investigators seeking to report studies developing and validating the diagnostic and predictive properties of AI models should refer to TRIPOD-ML (Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis–Machine Learning)24 and STARD-AI (Standards For Reporting Diagnostic Accuracy Studies–Artificial Intelligence),51, both of which are currently under development. Other potentially relevant guidelines, which are agnostic to study design, are registered with the EQUATOR Network.52 The SPIRIT-AI extension is expected to encourage careful early planning of AI interventions for clinical trials and this, in conjunction with CONSORT-AI, should help to improve the quality of trials for AI interventions.
There is widespread recognition that AI is a rapidly evolving field, and there will be the need to update SPIRIT-AI as the technology, and newer applications for it, develop. Currently, most applications of AI/ML involve disease detection, diagnosis, and triage, and this is likely to have influenced the nature and prioritisation of items within SPIRIT-AI. As wider applications that utilise “AI as therapy” emerge, it will be important to re-evaluate SPIRIT-AI in the light of such studies. Additionally, advances in computational techniques and the ability to integrate them into clinical workflows will bring new opportunities for innovation that benefits patients. However, they may be accompanied by new challenges of study design and reporting to ensure transparency, minimise potential biases and ensure that the findings of such a study are trustworthy and the extent to which they may be generalisable. The SPIRIT-AI and CONSORT-AI Steering Group will continue to monitor the need for updates.
Supplementary Material
Acknowledgments
We thank the participants who were involved in the Delphi study and Pilot study (Supplementary Note), Eliot Marston for providing strategic support (University of Birmingham, Birmingham, UK), and Charlotte Radovanovic (University Hospitals Birmingham NHS Foundation Trust, UK) and Anita Walker (University of Birmingham, UK) for administrative support. The views expressed in this publication are those of the authors, Delphi participants and stakeholder participants and may not represent the views of the broader stakeholder group or host institution. This work was funded by a Wellcome Trust Institutional Strategic Support Fund: Digital Health Pilot Grant, Research England (part of UK Research and Innovation), Health Data Research UK, and the Alan Turing Institute. The study was sponsored by the University of Birmingham, UK. The study funders and sponsors had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; preparation, review or approval of the manuscript; or decision to submit the manuscript for publication. MJC is a National Institute for Health Research (NIHR) Senior Investigator and receives funding from the NIHR Birmingham Biomedical Research Centre; the NIHR Surgical Reconstruction and Microbiology Research Centre and NIHR ARC West Midlands at the University of Birmingham and University Hospitals Birmingham NHS Foundation Trust; Health Data Research UK; Innovate UK (part of UK Research and Innovation); the Health Foundation; Macmillan Cancer Support; and UCB Pharma. ADa and JJD are also NIHR Senior Investigators. The views expressed in this article are those of the author(s) and not necessarily those of the NIHR, or the Department of Health and Social Care. DM is supported by a University of Ottawa Research Chair. MKEZ is supported by the US Food and Drug Administration (FDA), and DP is supported in part by the Office of the Director at the National Library of Medicine (NLM), US National Institutes of Health (NIH). AB is supported by an NIH award 7K01HL141771-02. PAK received grants from UKRI Future Leaders Fellowship and from Moorfields Eye Charity Career Development Award. SJV received funding from the Engineering and Physical Sciences Research Council, UK Research and Innovation (UKRI), Accenture, Warwick Impact Fund, Health Data Research UK, and European Regional Development Fund. SR is an employee of the UKRI. This article may not be consistent with NIH and/or FDA’s views or policies. It reflects only the views and opinions of the authors.
Panel: Glossary
- Artificial Intelligence
The science of developing computer systems which can perform tasks normally requiring human intelligence
- AI intervention
A health intervention that relies upon an AI/ML component to serve its purpose
- CONSORT
Consolidated Standards of Reporting Trials
- CONSORT-AI extension item
An additional checklist item to address AI-specific content that is not adequately covered by CONSORT 2010
- Class-activation map
Class-activation maps are particularly relevant to image classification AI interventions. Class-activation maps are visualisations of the pixels that had the greatest influence on predicted class, by displaying the gradient of the predicted outcome from the model with respect to the input. They are also referred to as “saliency maps” or “heat maps”
- Health outcome
Measured variables in the trial that are used to assess the effects of an intervention
- Human–AI interaction
The process of how users (humans) interact with the AI intervention, for the AI intervention to function as intended
- Clinical outcome
Measured variables in the trial that are used to assess the effects of an intervention
- Delphi study
A research method that derives the collective opinions of a group through a staged consultation of surveys, questionnaires, or interviews, with an aim to reach consensus at the end
- Development environment
The clinical and operational settings from which the data used for training the model are generated. This includes all aspects of the physical setting (such as geographical location, physical environment), operational setting (such as integration with an electronic record system, installation on a physical device) and clinical setting (such as primary, secondary and/or tertiary care, patient disease spectrum)
- Fine-tuning
Modifications or additional training performed on the AI intervention model, done with the intention of improving its performance
- Input data
The data that need to be presented to the AI intervention to allow it to serve its purpose
- Machine learning
A field of computer science concerned with the development of models/algorithms that can solve specific tasks by learning patterns from data, rather than by following explicit rules. It is seen as an approach within the field of AI
- Operational environment
The environment in which the AI intervention will be deployed, including the infrastructure required to enable the AI intervention to function
- Output data
The predicted outcome given by the AI intervention based on modelling of the input data. The output data can be presented in different forms, including a classification (including diagnosis, disease severity or stage, or recommendation such as referability), a probability, a class-activation map, etc. The output data typically provide additional clinical information and/or trigger a clinical decision
- Performance error
Instances in which the AI intervention fails to perform as expected. This term can describe different types of failures, and it is up to the investigator to specify what should be considered a performance error, preferably based on prior evidence. This can range from small decreases in accuracy (compared to expected accuracy) to erroneous predictions or the inability to produce an output, in certain cases
- SPIRIT
Standard Protocol Items: Recommendations for Interventional Trials
- SPIRIT-AI
An additional checklist item to address AI-specific content that is not adequately covered by SPIRIT 2013
- SPIRIT-AI elaboration item
Additional considerations to an existing SPIRIT 2013 item when applied to AI interventions
- AI
artificial intelligence
- ML
machine learning
Footnotes
Declaration of interests
MJC has received personal fees from Astellas, Takeda, Merck, Daiichi Sankyo, Glaukos, GlaxoSmithKline, and the Patient-Centered Outcomes Research Institute (PCORI), outside the submitted work. ADa is an advisor for Google DeepMind, outside the submitted work. LF reports personal fees from Allergan, Bayer, and Novartis, outside the submitted work. JF reports personal fees from British Medical Journal, during the conduct of the study. HH reports that he is Managing Director at Hardian Health, consultancy for health technology firms. PAK reports personal fees from DeepMind Technologies, Roche, Novartis, Apellis, Bayer, Allergan, Topcon, and Heidelberg Engineering, outside the submitted work. AYL reports personal fees from Genentech, US Food and Drug Administration, and Verana Health, grants from Microsoft, NVIDIA, Carl Zeiss Meditec, and Santen, outside the submitted work. CSL reports grants from National Institute of Health/National Institute on Aging, outside the submitted work. CJK is an employee of Google and owns Alphabet stock. AE is an employee of Salesforce CRM. RiS is an employee of Pinpoint Science. JMa was an employee of AstraZeneca PLC at the time of this study. RuS is Editor-in-Chief of The Lancet Digital Health and reports personal fees from The Lancet Group, during the conduct of the study. JMo is Chief Editor of the journal Nature Medicine; he has recused himself from any aspect of decision-making on this manuscript and played no part in the assignment of this manuscript to in-house editors or peer reviewers, and was also separated and blinded from the editorial process from submission inception to decision. SJV reports funding from IQVIA. All other authors declare no competing interests.
Data sharing
Data requests should be made to the corresponding author and release will be subject to consideration by the SPIRIT-AI and CONSORT-AI Steering Group.
Contributor Information
Samantha Cruz Rivera, Centre for Patient Reported Outcome Research, Institute of Applied Health Research, University of Birmingham, Birmingham, UK; Birmingham Health Partners Centre for Regulatory Science and Innovation, University of Birmingham, Birmingham, UK.
Xiaoxuan Liu, Academic Unit of Ophthalmology, Institute of Inflammation and Ageing, College of Medical and Dental Sciences, University of Birmingham, Birmingham, UK; Birmingham Health Partners Centre for Regulatory Science and Innovation, University of Birmingham, Birmingham, UK; Department of Ophthalmology, University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK; Moorfields Eye Hospital NHS Foundation Trust, London, UK; Health Data Research UK, London, UK.
An-Wen Chan, Department of Medicine, Women’s College Research Institute, Women’s College Hospital, University of Toronto, Toronto, ON, Canada.
Alastair K Denniston, Centre for Patient Reported Outcome Research, Institute of Applied Health Research, University of Birmingham, Birmingham, UK; Academic Unit of Ophthalmology, Institute of Inflammation and Ageing, College of Medical and Dental Sciences, University of Birmingham, Birmingham, UK; Birmingham Health Partners Centre for Regulatory Science and Innovation, University of Birmingham, Birmingham, UK; Department of Ophthalmology, University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK; National Institute of Health Research Biomedical Research Centre for Ophthalmology, Moorfields Hospital London NHS Foundation Trust and University College London, Institute of Ophthalmology, London, UK; Health Data Research UK, London, UK.
Melanie J Calvert, Centre for Patient Reported Outcome Research, Institute of Applied Health Research, University of Birmingham, Birmingham, UK; Birmingham Health Partners Centre for Regulatory Science and Innovation, University of Birmingham, Birmingham, UK; Health Data Research UK, London, UK; National Institute of Health Research Surgical Reconstruction and Microbiology Centre, and National Institute of Health Research Birmingham Biomedical Research Centre, University of Birmingham and University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK; National Institute of Health Research Applied Research Collaborative West Midlands, Birmingham, UK.
References
- 1.Chan A-W, Tetzlaff JM, Altman DG, et al. SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med 2013; 158: 200–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan A-W, Tetzlaff JM, Gøtzsche PC, et al. SPIRIT 2013 explanation and elaboration: guidance for protocols of clinical trials. BMJ 2013; 346: e7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarkis-Onofre R, Cenci MS, Demarco FF, et al. Use of guidelines to improve the quality and transparency of reporting oral health research. J Dent 2015; 43: 397–404. [DOI] [PubMed] [Google Scholar]
- 4.Calvert M, Kyte D, Mercieca-Bebber R, et al. Guidelines for inclusion of patient-reported outcomes in clinical trial protocols: the SPIRIT-PRO Extension. JAMA 2018; 319: 483–94. [DOI] [PubMed] [Google Scholar]
- 5.Dai L, Cheng CW, Tian R, et al. Standard protocol items for clinical trials with traditional Chinese medicine 2018: recommendations, explanation and elaboration (SPIRIT-TCM Extension 2018). Chin J Integr Med 2019; 25: 71–79. [DOI] [PubMed] [Google Scholar]
- 6.He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med 2019; 25: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020; 577: 89–94. [DOI] [PubMed] [Google Scholar]
- 8.Abràmoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci 2016; 57: 5200–06. [DOI] [PubMed] [Google Scholar]
- 9.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 2018; 24: 1342–50. [DOI] [PubMed] [Google Scholar]
- 10.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018; 15: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleuren LM, Klausch TLT, Zwager CL, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med 2020; 46: 383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yim J, Chopra R, Spitz T, et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med 2020; 26: 892–99. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Goo JM, Lee KH, Kim YT, Park CM. Preoperative CT-based deep learning model for predicting disease-free survival in patients with lung adenocarcinomas. Radiology 2020; 296: 216–24. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Berzin TM, Glissen Brown JR, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut 2019; 68: 1813–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyler NS, Mosquera-Lopez CM, Wilson LM, et al. An artificial intelligence decision support system for the management of type 1 diabetes. Nat Metab 2020; 2: 612–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health 2019; 1: e271–97. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Zhang J, Zhou W, et al. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut 2019; 68: 2161–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wijnberge M, Geerts BF, Hol L, et al. Effect of a machine learning-derived early warning system for intraoperative hypotension vs standard care on depth and duration of intraoperative hypotension during elective noncardiac surgery: The HYPE randomized clinical trial. JAMA 2020; 323: 1052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong D, Wu L, Zhang J, et al. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol 2020; 5: 352–61. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Liu X, Berzin TM, et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol 2020; 5: 343–51. [DOI] [PubMed] [Google Scholar]
- 22.Lin H, Li R, Liu Z, et al. Diagnostic efficacy and therapeutic decision-making capacity of an artificial intelligence platform for childhood cataracts in eye clinics: a multicentre randomized controlled trial. EClinicalMedicine 2019; 9: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su J-R, Li Z, Shao XJ, et al. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc 2020; 91: 415–24.e4. [DOI] [PubMed] [Google Scholar]
- 24.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet 2019; 393: 1577–79. [DOI] [PubMed] [Google Scholar]
- 25.Gregory J, Welliver S, Chong J. Top 10 reviewer critiques of radiology artificial intelligence (AI) articles: qualitative thematic analysis of reviewer critiques of machine learning/deep learning manuscripts submitted to JMRI. J Magn Reson Imaging 2020; 52: 248–54. [DOI] [PubMed] [Google Scholar]
- 26.Nagendran M, Chen Y, Lovejoy CA, et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 2020; 368: m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CONSORT-AI and SPIRIT-AI Steering Group. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat Med 2019; 25: 1467–68. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Faes L, Calvert MJ, Denniston AK. Extension of the CONSORT and SPIRIT statements. Lancet 2019; 394: 1225. [DOI] [PubMed] [Google Scholar]
- 29.Moher D, Schulz KF, Simera I, Altman DG. Guidance for developers of health research reporting guidelines. PLoS Med 2010; 7: e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caballero-Ruiz E, García-Sáez G, Rigla M, Villaplana M, Pons B, Hernando ME. A web-based clinical decision support system for gestational diabetes: Automatic diet prescription and detection of insulin needs. Int J Med Inform 2017; 102: 35–49. [DOI] [PubMed] [Google Scholar]
- 31.Kim TWB, Gay N, Khemka A, Garino J. Internet-based exercise therapy using algorithms for conservative treatment of anterior knee pain: a pragmatic randomized controlled trial. JMIR Rehabil Assist Technol 2016; 3: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labovitz DL, Shafner L, Reyes Gil M, Virmani D, Hanina A. Using artificial intelligence to reduce the risk of nonadherence in patients on anticoagulation therapy. Stroke 2017; 48: 1416–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolae A, Morton G, Chung H, et al. Evaluation of a machine-learning algorithm for treatment planning in prostate low-dose-rate brachytherapy. Int J Radiat Oncol Biol Phys 2017; 97: 822–29. [DOI] [PubMed] [Google Scholar]
- 34.Voss C, Schwartz J, Daniels J, et al. Effect of wearable digital intervention for improving socialization in children with autism spectrum disorder: a randomized clinical trial. JAMA Pediatr 2019; 173: 446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendes-Soares H, Raveh-Sadka T, Azulay S, et al. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Netw Open 2019; 2: e188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi KJ, Jang JK, Lee SS, et al. Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the liver. Radiology 2018; 289: 688–97. [DOI] [PubMed] [Google Scholar]
- 37.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med 2019; 17: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pooch EHP, Ballester PL, Barros RC. Can we trust deep learning models diagnosis? The impact of domain shift in chest radiograph classification. arXiv 2019; 1909.01940. http://arxiv.org/abs/1909.01940.
- 39.International Medical Device Regulators Forum. Unique Device Identification System (UDI System) Application Guide. 2019. http://www.imdrf.org/documents/documents.asp (accessed March 24, 2020).
- 40.Sabottke CF, Spieler BM. The effect of image resolution on deep learning in radiography. Radiol Artif Intell 2020; 2: e190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heaven D Why deep-learning AIs are so easy to fool. Nature 2019; 574: 163–66. [DOI] [PubMed] [Google Scholar]
- 42.Kiani A, Uyumazturk B, Rajpurkar P, et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med 2020; 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25: 1337–40. [DOI] [PubMed] [Google Scholar]
- 44.Habli I, Lawton T, Porter Z. Artificial intelligence in health care: accountability and safety. Bull World Health Organ 2020; 98: 251–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakden-Rayner L, Dunnmon J, Carneiro G, Ré C. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. arXiv 2019; 1909.12475. http://arxiv.org/abs/1909.12475. [DOI] [PMC free article] [PubMed]
- 46.SPIRIT. Publications & Downloads. https://www.spirit-statement.org/publications-downloads/ (accessed March 24, 2020).
- 47.Zech JR, et al. Confounding variables can degrade generalization performance of radiological deep learning models. arXiv 2018; 1807.00431. http://arxiv.org/abs/1807.00431.
- 48.Finlayson SG, Bowers JD, Ito J, Zittrain JL, Beam AL, Kohane IS. Adversarial attacks on medical machine learning. Science 2019; 363: 1287–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CS, Lee AY. Clinical applications of continual learning machine learning. Lancet Digital Health 2020; 2: e279–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutton RT, Pincock D, Baumgart DC, Sadowski DC, Fedorak RN, Kroeker KI. An overview of clinical decision support systems: benefits, risks, and strategies for success. NPJ Digit Med 2020; 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sounderajah V, Ashrafian H, Aggarwal R, et al. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: The STARD-AI Steering Group. Nat Med 2020; 26: 807–08. [DOI] [PubMed] [Google Scholar]
- 52.Talmon J, Ammenwerth E, Brender J, de Keizer N, Nykänen P, Rigby M. STARE-HI--Statement on reporting of evaluation studies in Health Informatics. Int J Med Inform 2009; 78: 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


