Abstract
Objectives
With the goal of facilitating the use of HIV-TRePS to optimize therapy in settings with limited healthcare resources, we aimed to develop computational models to predict treatment responses accurately in the absence of commonly used baseline data.
Methods
Twelve sets of random forest models were trained using very large, global datasets to predict either the probability of virological response (classifier models) or the absolute change in viral load in response to a new regimen (absolute models) following virological failure. Two ‘standard’ models were developed with all baseline variables present and 10 others developed without HIV genotype, time on therapy, CD4 count or any combination of the above.
Results
The standard classifier models achieved an AUC of 0.89 in cross-validation and independent testing. Models with missing variables achieved AUC values of 0.78–0.90. The standard absolute models made predictions that correlated significantly with observed changes in viral load with a mean absolute error of 0.65 log10 copies HIV RNA/mL in cross-validation and 0.69 log10 copies HIV RNA/mL in independent testing. Models with missing variables achieved values of 0.65–0.75 log10 copies HIV RNA/mL. All models identified alternative regimens that were predicted to be effective for the vast majority of cases where the new regimen prescribed in the clinic failed. All models were significantly better predictors of treatment response than genotyping with rules-based interpretation.
Conclusions
These latest models that predict treatment responses accurately, even when a number of baseline variables are not available, are a major advance with greatly enhanced potential benefit, particularly in resource-limited settings. The only obstacle to realizing this potential is the willingness of healthcare professions to use the system.
Introduction
While great progress had been made towards the UNAIDS targets for 2020 of 90% of infected people to be diagnosed, 90% of these to be on therapy and 90% of those treated having viral suppression (‘90-90-90’), this progress faltered and was blown off course by the COVID-19 pandemic.1 As of July 2020, it was estimated that 81% of the 38 million people living with HIV infection knew their status, 67% were on therapy and 59% had an undetectable viral load.
The target of 90% viral suppression is critical not only in order to prevent disease progression, morbidity and mortality but to curtail the spread of the virus.2 A continuing threat to this is the development of HIV drug resistance, often linked to poor adherence and interruptions to drug supplies in some settings.
A recent WHO report stated that 12 of 18 countries reporting survey data to WHO between 2014 and 2018 had levels of pre-treatment HIV drug resistance to efavirenz and/or nevirapine exceeding 10%.3 Only one-third of countries showed levels of viral suppression exceeding 90%. Across all the surveys, the prevalence of resistance among people receiving ART ranged from 3% to 29%. Among populations receiving NNRTI-based ART with viral non-suppression, the levels of NNRTI and NRTI resistance ranged from 50% to 97% and from 21% to 91%, respectively. Estimates of dual-class resistance (NNRTI and NRTI) ranged between 21% and 91% of individuals for whom NNRTI-based first-line ART failed.
When treatment fails, the combination of antiretroviral agents should be changed to resuppress the virus. In well-resourced countries the selection of a new combination is individualized by expert physicians using a comprehensive range of information, often including the results of a genotypic resistance test.4–6 However, resistance testing is relatively expensive and only moderately predictive of response to treatment.7
The challenge of individually optimized drug selection in low- and middle-income countries (LMICs) is much greater as resistance tests are typically unavailable or unaffordable, patient monitoring can be intermittent and drug options limited. In the absence of frequent viral load monitoring, therapy failure is often detected late and regimen switch decisions based on standard protocols rather than being individualized. The result can be suboptimal regimen selection, failure to achieve viral resuppression and further resistance, which may limit future therapeutic options and result in transmission to others.8
The HIV Resistance Response Database Initiative (RDI) has collected biological, clinical and treatment outcome data for more than 250 000 HIV-1 patients around the world. From these data, we have used machine learning to develop models to predict HIV-1 treatment outcomes and to identify optimal, individualized therapies.9–13 Models estimating the probability of a new regimen reducing plasma viral load to <50 copies HIV RNA/mL typically achieve accuracy of 80% or above in independent testing, without a genotype, or 85% if a genotype is available.14,15 Other models, developed for settings using different, higher thresholds to define virological response, predict the absolute change in viral load and correlate highly significantly with actual changes in independent testing.16
The models are used to power an online treatment decision support tool, the HIV Treatment Response Prediction System (HIV-TRePS). Clinicians in LMICs have commented that the utility of HIV-TRePS is limited because of its requirement for comprehensive baseline clinical and laboratory data. To make this system as useful as possible in the widest range of healthcare settings, including those with suboptimal patient monitoring and diagnostics, we therefore set out to develop models that can make accurate predictions despite missing some baseline information.
Here we report 12 new sets of random forest (RF) models that accurately predict response to a change in ART without a baseline (at switch) genotype, CD4 count, time on therapy or, for the first time, none of the above. Classifier (C) models, which estimate the probability of a viral load of <50 copies/mL, and absolute (A) models, which predict the change in viral load, are reported.
The models were evaluated as potential additions to the RDI’s HIV-TRePS system. This paper represents the latest update alluded to in our previous published update.15
Methods
Clinical data
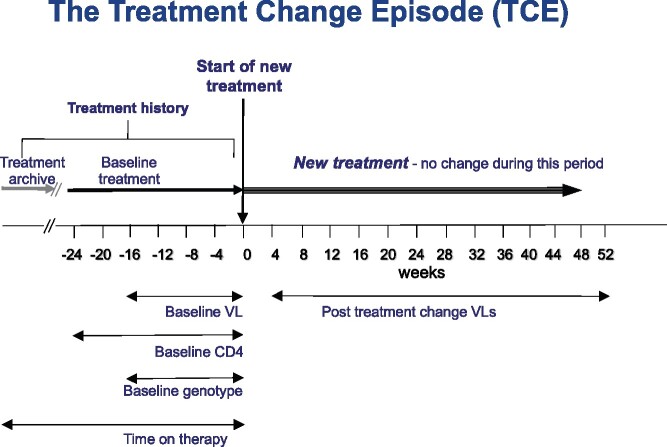
Treatment change episodes (TCEs), where ART was changed following virological failure (viral load >50 copies/mL) were collected from the RDI database.9 The change in therapy could have been for any reason, not necessarily the virological failure, although patients assessed by the clinics to have been non-adherent were excluded from the analysis. The gold-standard, complete set of data for a TCE was: on-treatment baseline plasma viral load (≤16 weeks prior to treatment change); CD4 cell count (≤24 weeks prior to treatment change); viral genotype (protease and reverse transcriptase sequence ≤16 weeks prior to treatment change); time on therapy (days since ART first introduced); the drugs in use prior to the change; the drugs used in the treatment history; the drugs in the new regimen; and follow-up plasma viral load obtained 4–52 weeks following introduction of the new regimen and time to that follow-up, as illustrated in Figure 1.
Figure 1.
The standard TCE. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
For each round of model development, TCEs were extracted from the RDI dataset that included all the variables required for that particular set of models. Consequently, the pools of qualifying data were different on each occasion and larger for those models requiring fewer baseline variables. Data were censored using rules, developed over the past 18 years and previously published, e.g. to exclude those rare TCEs from patients whose treatment involved the use of a single inhibitor (as opposed to combination therapy, typically approximately 1% of the available TCEs), were likely to have been non-adherent or whose data were likely to have been miscoded.9–15 The principle rules are summarized in Table 1. The remaining TCEs were partitioned at random, with 5% of patients extracted at random to provide an independent test set.
Table 1.
Main criteria for data censoring
| Objective | Exclusion definition |
|---|---|
| Remove non-adherent patients | All TCEs with a measure of adherence associated with the TCE or the patient indicating non-adherence (based on site-specific cut-offs). |
| TCEs with baseline genotypes available that were predicted by the current RDI models and the Stanford HIVdb genotype interpretation system to respond to their new regimen but failed in the clinic. | |
| Removal of TCEs involving single drugs | Any TCEs with a single drug at baseline or as the new regimen. |
| Removal of TCEs with drugs for which we had inadequate data | This varies from model to model according to the data available, but any TCEs including drugs that appear in fewer than 250 cases in a training set. |
| Removal or conversion of indeterminate or unrealistic viral load values | Viral load values of 0 or 50 are assumed to be <50 copies/mL but the precise value is unknown and they are coded as 50 copies/mL. |
| Viral load data of the form ‘<X’ where X is >50 or 1.7 log10 copies/mL or ‘>X’ because the true value is unknown. | |
| Any TCEs with a viral load that is exactly 2.60 log10 or 400 copies/mL on the assumption that the values were from an assay with this as the lower limit of detection and that the true value is unknown. | |
| Removal of unrealistic viral load values or CD4 counts |
Plasma follow-up viral load values >9 log10 copies HIV RNA/mL. CD4 counts >2000 copies/mm3. |
The development of computational models
Each set of RF models was developed using methodology described in detail elsewhere.10,12 The following comprises the full set of the latest 115 input variables used for modelling (new variables underlined):
1: Baseline CD4 count value (cells/mm3);
2: Time on therapy (days since first ART was introduced);
3–64: 62 mutations, detected in the baseline genotype: HIV reverse transcriptase mutations (n = 33): M41L, E44D, A62V, K65R, D67N, 69 insert, T69D/N, K70R, L74V, V75I, F77L, V90I, A98G, L100I, L101I/E/P, K103N, V106A/M, V106I, V108I, Y115F, F116Y, V118I, 138A/G/K, Q151M, V179D/F/T, Y181C/I/V, M184V/I, Y188C/L/H, G190S/A, L210W, T215F/Y, K219Q/E, P236L; protease mutations (n = 29): L10F/I/R/V, V11I, K20M/R, L24I, D30N, V32I, L33F, M36I, M36L/V, M46I/L, I47V, G48V, I50V, I50L, F53L, I54 (any change), Q58E, L63P, A71 (any change), G73 (any change), T74P, L76V, V77I, V82A/F/S, V82T, I84V/A/C, N88D/S, L89V, L90M). The mutations were selected on the basis of the IAS–USA mutation list as well as previous modelling studies;16
65–89: Drugs in the new regimen (25 binary variables; present = 1, not present = 0):
zidovudine, didanosine, stavudine, abacavir, lamivudine, emtricitabine, tenofovir disoproxil fumarate, efavirenz, nevirapine, etravirine, indinavir, nelfinavir, saquinavir, amprenavir, fosamprenavir, lopinavir, atazanavir, darunavir, enfuvirtide, raltegravir, tipranavir, maraviroc, elvitegravir, rilpivirine and dolutegravir;
90–114: Treatment history variables (as above);
115: Time to follow-up viral load (days).
The output of each classifier model was the estimated probability of the follow-up plasma viral load being less than 50 copies HIV RNA/mL. The output for each absolute model was the estimated change in follow-up viral load from baseline.
Validation and independent testing
Each of the committees of five RF models was developed using a 5× cross-validation scheme. For each partition, the model’s output for the validation cases was compared with the actual response observed in the clinic and the best-performing model selected for the final committee. For each of the five final classifier models, the optimum operating point (OOP) was identified (the cut-off for the probability of response being classed as response versus failure that gave the best performance overall).
The performance of the models as predictors of response was then evaluated using the independent test cases. The average of the classifier models’ estimates of probability of response and the responses observed in the clinics for these cases were used to plot receiver operating characteristic (ROC) curves and assess the AUC. In addition, the average OOP was used to obtain the overall accuracy, sensitivity and specificity of the models. The absolute models’ estimates of the change in viral load from baseline and the responses observed in the clinics were correlated using Pearson’s product moment method, a scatterplot produced and the correlation of determination (r2) and mean absolute difference between predicted and observed changes in viral load compared.
Comparison of the accuracy of the classifier models versus rules-based interpretation of the genotype
Genotypic sensitivity scores (GSSs) were obtained for test cases with baseline genotypes available using three interpretation systems in common use: ANRS (v30), REGA (v10.0) and Stanford HIVdb (v8.9-1). The GSS was calculated by adding the score for each drug in the regimen (full susceptibility = 1, partial susceptibility = 0.5 and no response = 0). These scores were then used as predictors of response and the performance compared with that of the models.
In silico analysis to evaluate the potential of the models to help avoid treatment failure
In order to evaluate further the potential clinical utility of the models, we assessed their ability to identify alternative, practical regimens that were predicted to be effective (probability of virological response above the OOP for classifier models or the follow-up viral load below the threshold for response for the absolute models). Lists of regimens in regular clinical use were identified from the RDI database. The baseline data for test TCEs were entered into the models and predictions obtained for the regimens on the drug lists that had no more drugs than the regimen used in the clinic.
HIV-TRePS is potentially of most utility in LMICs where genotyping, monitoring and some drugs may be scarce or unavailable. It was important therefore to assess the ability of the models to identify effective alternative regimens using combinations of drugs that are commonly available in LMICs. In silico analyses were therefore performed, modelling alternative regimens comprising only those relatively inexpensive drugs that have been widely used in LMICs over the past 10 years, namely zidovudine, abacavir, lamivudine, emtricitabine, tenofovir disoproxil fumarate, efavirenz, nevirapine, lopinavir, etravirine, atazanavir and darunavir.
Results
Characteristics of the datasets
Each set of models used a different dataset according to the specification of the models. As an example, the baseline, treatment and response characteristics of the datasets used in the most recent modelling without a genotype are summarized in Tables 2 and 3. As a result of random partitioning the training and test sets in each case were well matched.
Table 2.
Demographic characteristics of the TCEs with no genotype (NG)
| Characteristic | Training set | Test set |
|---|---|---|
| TCEs, n | 62 940 | 3260 |
| Patients, n | 20 513 | 1080 |
| Gender, n (%) | ||
| Male | 40 019 (64) | 2102 (64) |
| Female | 13 121 (21) | 632 (19) |
| Unknown | 9800 (15) | 526 (16) |
| Median age (years) | 42 | 43 |
| Geographical sources of TCEs | ||
| Argentina | 199 | 12 |
| Australia | 233 | 16 |
| Brazil | 57 | 9 |
| Canada | 3460 | 193 |
| Germany | 6546 | 364 |
| India | 645 | 42 |
| Italy | 2535 | 94 |
| Japan | 117 | 1 |
| Mexico | 509 | 19 |
| Netherlands | 9539 | 553 |
| Romania | 772 | 64 |
| Serbia | 35 | 0 |
| South Africa | 5209 | 274 |
| Spain | 13 283 | 647 |
| Other sub-Saharan Africa | 70 | 4 |
| UK | 9318 | 440 |
| USA | 4263 | 261 |
| Unknown | 6150 | 267 |
| Total | 62 940 | 3260 |
Table 3.
Clinical and laboratory data
| Parameter | Training set | Test set |
|---|---|---|
| Baseline data | ||
| Median (IQR) baseline VL (log10 copies/mL) | 3.93 (2.73–4.76) | 3.96 (2.8–4.79) |
| Median (IQR) days since first treatment | 1669 (709–3010) | 1700 (741–3036) |
| Median (IQR) number of previous regimens | 4 (1–8) | 4 (1–8) |
| Treatment history | ||
| Median (IQR) number of previous drugs | 5 (3–8) | 5 (3–8) |
| NRTI experience, n (%) | 62 780 (99.7) | 3257 (99.9) |
| NNRTI experience, n (%) | 41 957 (66.7) | 2228 (68.3) |
| PI experience, n (%) | 43 227 (68.7) | 2203 (67.6) |
| Integrase inhibitor experience, n (%) | 2850 (5) | 142 (4) |
| CCR5 inhibitor experience, n (%) | 731 (1) | 30 (1) |
| New regimens | ||
| 2 NRTI + 1 PI, n (%) | 22 598 (35.9) | 1089 (33.4) |
| 2 NRTI + 1 NNRTI, n (%) | 11 658 (18.5) | 584 (17.9) |
| 3 NRTIs + 1 PI, n (%) | 4063 (6.5) | 253 (7.8) |
| 3 NRTIs, n (%) | 2511 (4.0) | 113 (3.5) |
| 3 NRTIs + 1 NNRTI, n (%) | 1609 (2.6) | 71 (2.2) |
| 2 NRTIs, n (%) | 2154 (3.4) | 109 (3.3) |
| 2 NRTIs + 1 NNRTI + 1 PI, n (%) | 1833 (2.9) | 111 (3.4) |
| 1 PI + 1 integrase inhibitor, n (%) | 0 (0) | 0 (0) |
| 4 NRTIs, n (%) | 899 (1.4) | 50 (1.5) |
| 1 NRTI + 1 NNRTI + 1 PI, n (%) | 1355 (2.2) | 61 (1.9) |
| 1 NRTI + 1 PI, n (%) | 1037 (1.6) | 74 (2.3) |
| Other, n (%) | 13 226 (21.0) | 745 (22.9) |
VL, viral load.
Results of the classifier modelling
The performance characteristics of the models during cross-validation and independent testing are summarized in Table 4. During cross-validation the AUC values ranged from 0.79 for the models developed without information on baseline genotype, CD4 count or time on therapy, up to 0.90 for models developed with information on baseline genotype and CD4 count but without time on therapy, the most accurate models for the prediction of virological response to date. Overall accuracy (the percentage of cases correctly predicted as responders or failures) ranged from 72% to 82%. Sensitivity (the proportion of responses correctly predicted) ranged from 71% up to 80%, while specificity (the proportion of the predictions of response that were correct) ranged from 73% to 84%.
Table 4.
Summary of the results of classifier (C) models
| Missing baseline data | Model name | Training data (n) | CV/test | AUC | Overall accuracy (%) | Sens (%) |
Spec (%) |
OOP |
|---|---|---|---|---|---|---|---|---|
| None | CG | 13 936 | CV | 0.89 | 81 | 77 | 83 | 0.42 |
| test | 0.89 | 81 | 75 | 85 | 0.42 | |||
| Genotype | CNG | 56 717 | CV | 0.84 | 77 | 71 | 80 | 0.43 |
| test | 0.84 | 76 | 71 | 79 | 0.43 | |||
| Time on therapy | CG(−ToT) | 19 080 | CV | 0.90 | 82 | 80 | 84 | 0.42 |
| test | 0.90 | 82 | 80 | 83 | 0.42 | |||
| Genotype/time on therapy | CNG(−ToT) | 56 717 | CV | 0.83 | 76 | 71 | 80 | 0.44 |
| test | 0.82 | 76 | 71 | 79 | 0.44 | |||
| Genotype/CD4 count | CNG(−CD4) | 62 940 | CV | 0.83 | 76 | 72 | 79 | 0.44 |
| test | 0.82 | 75 | 73 | 77 | 0.44 | |||
| Genotype/time on therapy/ CD4 count | CNG(−ToT −CD4) | 50 270 | CV | 0.79 | 72 | 71 | 73 | 0.38 |
| test | 0.78 | 72 | 74 | 70 | 0.38 |
CV, cross-validation; test, independent testing; sens, sensitivity; spec, specificity.
Independent testing
Independent testing resulted in AUC values ranging from 0.78 for the models developed without information on baseline genotype, CD4 count or time on therapy, up to 0.90 for models developed with information on baseline genotype and CD4 count but without time on therapy. Overall accuracy ranged from 72% to 82%. Sensitivity ranged from 71% up to 80%, while specificity ranged from 70% to 85%.
Results of the absolute modelling
The results of the models developed to predict the absolute change in viral load from baseline are presented in Table 5.
Table 5.
Summary of the results of absolute (A) models
| Missing baseline data | Model name | Training data (n) | CV/test | r | r 2 | P value | MAE |
|---|---|---|---|---|---|---|---|
| None | AG | 13 936 | CV | 0.75 | 0.56 | 0.0001 | 0.65 |
| test | 0.74 | 0.55 | 0.0001 | 0.69 | |||
| Genotype | ANG | 56 717 | CV | 0.70 | 0.49 | 0.0001 | 0.71 |
| test | 0.68 | 0.46 | 0.0001 | 0.73 | |||
| Time on therapy | AG(−ToT) | 19 080 | CV | 0.74 | 0.55 | 0.0001 | 0.65 |
| test | 0.72 | 0.52 | 0.0001 | 0.70 | |||
| Genotype/time on therapy | ANG(−ToT) | 50 270 | CV | 0.69 | 0.48 | 0.0001 | 0.72 |
| test | 0.67 | 0.45 | 0.0001 | 0.74 | |||
| Genotype/CD4 count | ANG(−CD4) | 62 940 | CV | 0.70 | 0.49 | 0.0001 | 0.71 |
| test | 0.69 | 0.48 | 0.0001 | 0.75 | |||
| Genotype/time on therapy/CD4 count | ANG(−ToT −CD4) | 56 717 | CV | 0.68 | 0.46 | 0.0001 | 0.74 |
| test | 0.66 | 0.44 | 0.0001 | 0.75 |
CV, cross-validation; test, independent testing; MAE, mean absolute error - the difference between predicted and actual change in plasma viral load (log10 copies HIV RNA/mL).
During cross-validation the models achieved highly statistically significant correlations between actual and predicted change in viral loads from baseline, with r values ranging from 0.68 for the models without genotype, CD4 count or time on therapy, to 0.75 for those models with all baseline variables present. This equates to r2 values ranging from 0.46 to 0.56. The mean absolute error ranged from 0.74 log10 HIV RNA/mL for the models without genotype, CD4 count or time on therapy to 0.65 log10 HIV RNA/mL for those with all variables.
In independent testing, the models again achieved highly significant correlations between actual and predicted change in viral loads from baseline with r values ranging from 0.66 for the models without genotype, CD4 count or time on therapy, to 0.74 for those models with all baseline variables present (r2 = 0.44 to 0.55). The mean absolute error ranged from 0.75 to 0.69 log10 HIV RNA/mL for models with all variables.
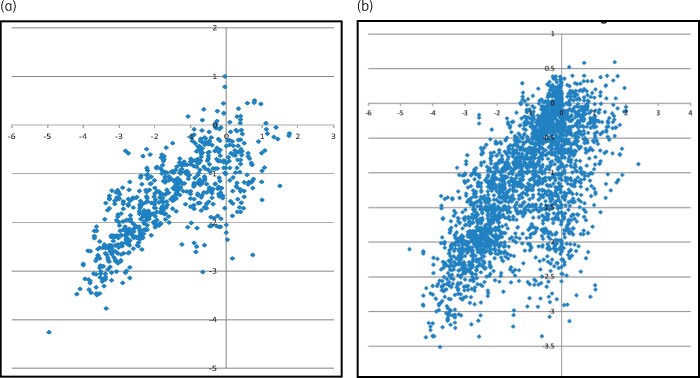
Scatterplots of the performance of the models with all variables and those missing baseline genotypes, CD4 counts and time on therapy are presented in Figure 2.
Figure 2.
Predicted versus observed change in log10-transformed viral load for absolute (A) models with (a) all variables (AG) and (b) without genotype, CD4 count or time on therapy [ANG(−ToT −CD4)]. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Comparing the predictive accuracy of the classifier models versus genotyping
In every case the performance of the models, including those models that do not use a genotype in their predictions, was highly significantly superior to that of genotyping with rules-based interpretation. For example, the classifier models that do not require a genotype achieved an AUC of 0.84 in independent testing. Of the test cases, 652 had baseline genotypes and the models achieved an AUC of 0.84 for this subset. The genotype systems achieved AUC values of 0.53–0.54 (Table 6). All three genotype interpretation systems were significantly poorer at predicting responses than the models (P < 0.00001).
Table 6.
Comparison of predictive accuracy for genotyping versus the models
| Model | AUROC | Comparison | P value |
|---|---|---|---|
| ANG models | 0.84 | ||
| ANRS | 0.54 | ANG models versus ANRS | <0.00001 |
| HIVDB | 0.54 | ANG models versus HIVDB | <0.00001 |
| REGA | 0.53 | ANG models versus REGA | <0.00001 |
In silico analysis
For the classifier models, the percentage of all test cases for which the models were able to find effective alternatives from the standard list of regimens in common use ranged from 91% to 98% (Table 7). They identified alternative regimens with a higher probability of response than the regimen used in the clinic (but not necessarily above the OOP) for 98%–100%. For the subset of test cases that failed their new regimen in the clinic, the models identified effective alternatives for 86%–97% of cases and alternatives with a higher probability of response in 100%.
Table 7.
Summary results for the in silico analyses
| Modelling for all test cases |
Modelling for failures only |
||||||
|---|---|---|---|---|---|---|---|
| Missing baseline data | Model name | Effective alternatives predicted (%) |
Superior alternatives predicted (%) |
Effective alternatives predicted (%) |
Superior alternatives predicted (%) |
||
| S | R | S | S | R | S | ||
| Classifier models: response defined as follow-up plasma viral load of <50 copies/mL HIV RNA | |||||||
|
| |||||||
| None | CG | 91 | 77 | 98 | 87 | 68 | 100 |
| Genotype | CNG | 94 | 80 | 100 | 91 | 71 | 100 |
| Time on therapy | CG(−ToT) | 91 | 76 | 99 | 86 | 65 | 100 |
| Genotype/time on therapy | CNG(−ToT) | 98 | 84 | 100 | 96 | 77 | 100 |
| Genotype/CD4 count | CNG(−CD4) | 95 | 84 | 99 | 91 | 77 | 100 |
| Genotype/time on therapy/CD4 count | CNG(−ToT −CD4) | 98 | 86 | 100 | 97 | 79 | 100 |
|
| |||||||
| Absolute models: response defined as follow-up plasma viral load of <400 copies/mL HIV RNA | |||||||
|
| |||||||
| None | AG | 92 | 84 | 96 | 85 | 73 | 100 |
| Genotype | ANG | 95 | 82 | 98 | 91 | 67 | 100 |
| Time on therapy | AG(−ToT)a | 93 | 86 | 97 | 85 | 74 | 100 |
| Genotype/time on therapy | ANG(−ToT)a | 100 | 96 | 98 | 99 | 92 | 100 |
| Genotype/CD4 count | ANG(−CD4) | 96 | 88 | 99 | 92 | 78 | 100 |
| Genotype/time on therapy/CD4 count | ANG(−ToT −CD4) | 97 | 90 | 98 | 95 | 81 | 100 |
S, standard drug list; R, restricted drug list.
Previously published.15
When the analyses were repeated using the highly restricted list of drugs widely available in LMICs the models identified alternatives that were predicted to be effective in 76%–86% of all cases and for 65%–79% of cases that failed their new regimen in the clinic.
In terms of the absolute models, the percentage of all test cases for which the models found effective alternatives (predicted follow-up viral load <400 copies/mL) from the standard list of regimens in common use ranged from 92% to 100%. They identified alternative regimens with a higher probability of response than the regimen used in the clinic for 96%–99%. For cases that failed in the clinic, the models identified effective alternatives for 85%–99% and alternatives with a higher probability of response in 100%.
When the analyses were repeated using the highly restricted drug list the models identified alternatives that were predicted to be effective in 82%–96% of cases and for 67%–92% for those that failed in the clinic.
Discussion
These latest models, developed using our largest databases to date, produced the most accurate predictions of response to combination ART ever reported. For the first time, to the best of our knowledge, they can factor in total time on therapy, make accurate predictions of response to rilpivirine and dolutegravir, and do so even if substantial baseline data are missing.
Classifier models developed with all baseline variables except time on therapy achieved marginally superior performance to those with all variables, producing an AUC value of 0.90 versus 0.89. This is probably due to them being trained with a larger dataset, the additional 5144 training TCEs more than compensating for the loss of the time-on-therapy information. Both models performed better than any previously published.
All the remaining classifier models achieved AUC values over 0.80 in cross-validation and independent testing other than those missing three baseline variables (genotype, CD4 count and time on therapy), which achieved 0.78 in independent testing. Nevertheless, it is encouraging that this performance remains highly statistically superior to the use of genotyping with rules-based interpretation, reinforcing previous findings that our models consistently outperform genotyping as a predictor of response.12,14,15
The absolute models produced highly significant correlations between predicted and observed changes in plasma viral load. The mean absolute difference between predicted and observed change ranged from 0.65 to 0.75 log10 copies HIV RNA/mL, which is similar to the typical test-retest error of most commercially available assays. The scatterplots in Figure 2 suggest that the absence of baseline information leads to an increase in incorrect predictions of response, with a cluster of cases with little change in viral load observed in the clinic that were predicted to have decreases of up to 3 log10 copies/mL. The cases in the cluster tended to be older cases with greater use of older drugs.
Both classifier and absolute models identified alternative regimens that were predicted to be effective for the great majority of cases (91%–100%) and, most crucially, for 85%–99% of those cases where the patient failed the new regimen introduced in the clinic. Moreover, this remained the case when the models were restricted to using a highly limited list of 11 relatively old and inexpensive drugs that have been widely used in LMICs, with effective alternatives predicted for 76%–96% of cases and 65%–92% of failures.
The models were able to identify alternative regimens that were predicted to be more effective, i.e. a higher probability of response or a lower predicted follow-up viral load, than the new regimen introduced in the clinic for almost all the cases. These results are a compelling indication of the potential utility of the models to reduce virological failure if used to support treatment decision-making.
It should be noted that while the TCEs used in the modelling all involved virological failure (a baseline viral load >50 copies/mL) they may have been triggered for a variety of reasons including, for example, tolerability. The HIV-TRePS system is designed to provide predictions of response, whatever the reason for switching, and the richness of the real-life data used to train the models underpins this utility.
The study has some limitations. Firstly, a key input variable for these models was the plasma viral load.17 Although viral load monitoring is still not universally available in many LMICs, it is recommended in WHO guidelines for monitoring therapy response.18 A recent study of its scale-up in sub-Saharan Africa showed the percentage of patients with viral loads ranged from 3% in Tanzania to 96% in Namibia. Of 11 million patients on ART in the region, 5 million were estimated to have some access to viral load monitoring.19 As technological advances enable lower costs and point-of-care testing, the use of viral load testing is likely to increase.20–22 Nevertheless, the RDI is currently experimenting with models that do not require a baseline viral load for their predictions.
Secondly, LMICs are somewhat under-represented in the data used in these studies, at 10%. Nevertheless, it could be argued that data from treatment decisions made in well-resourced countries, with the benefit of diagnostics and a wide range of available drugs, are the best data to use in supporting treatment decisions made with less information available and with a limited choice of drugs.
The data used in this study have been collected over many years, including from combinations of drugs no longer in common use. One of the advantages of the modelling methods employed, however, is that these data add useful information on the contribution of the individual drugs within each combination, which improves the accuracy of modelling for different contemporary combinations. This is born out by the high accuracy of the models’ predictions for the most recent regimens and in substudies performed excluding all but the most recent data (data on file). This should not, therefore, be considered a limitation.
There is inevitably a time lag between changes in treatment practice, the monitoring of patients treated according to the new practice, collection of resulting outcome data, their provision to the RDI and the development, testing and release of new models. The relative lack of data involving the very latest drugs and clinical practice will always be a challenge. For example, these studies only involved around 3000 (5%) cases of integrase strand transfer inhibitor (INSTI) use. The RDI is heavily dependent on and grateful to its data contributors for the timely provision of new data to limit this issue as far as possible.
There are significant procedural, cultural, psychological and other ‘soft’ barriers to the use of HIV-TRePS that limit the extent to which it is being used and its potential benefit being realized. For example, most, if not all, countries have set treatment protocols, at least for second-line therapy, and many physicians are reluctant or may not have the agency to depart from these and individualize treatment. At third-line or beyond, protocols generally become less prescriptive, if guidance exists at all, but there can be reluctance to make any further changes, with some patients being left on only partially effective regimens. HIV-TRePS could potentially be of considerable value in such cases and substantially reduce virological failure and clinical progression if these barriers were overcome and the tool was more widely used to individualize salvage therapy.
Conclusions
RF models developed using very large, global datasets are highly accurate predictors of virological response to combination ART, even if a number of baseline input variables are missing. Such models are of greatly enhanced utility in LMICs and any settings with limited and/or infrequent laboratory monitoring.
The gold-standard models reported here are the most accurate developed to date. As more drugs become available in more settings, so many more combinations become possible, only relatively few of which have been subject to clinical trials. Computational modelling using large clinical datasets can enable physicians to expand the options available to them with a level of confidence provided by predictions based on real-life experiences.
Once again, these latest models are better predictors of response to therapy than genotyping with rules-based interpretation, even when these models are missing up to three baseline variables. Since use of these models is free of charge, this again suggests that scarce funds in LMICs would be better spent on antiretroviral drugs and viral load testing than on genotyping. This would enable a greater range of treatments to be offered, failure to be detected earlier and optimal, individualized treatment-change decisions made using the models.
The models described are now available for use through the on-line HIV-TRePS system at http://www.hivrdi.org/treps. The system has the potential to reduce virological failure and improve patient outcomes in all parts of the world, but particularly in LMICs. The use by clinicians of this tool to support optimized treatment decision-making in the absence of resistance tests could also combat the development of drug resistance and its contribution to treatment failure, disease progression and onward viral transmission. The keys to unlocking the potential of this tool lie in the hands of healthcare professionals around the world.
Acknowledgements
RDI data and study group
The RDI wishes to thank all the following individuals and institutions for providing the data used in training and testing its models:
Cohorts: Peter Reiss and Ard van Sighem (ATHENA, the Netherlands); Julio Montaner and Richard Harrigan (BC Center for Excellence in HIV & AIDS, Canada); Tobias Rinke de Wit, Raph Hamers and Kim Sigaloff (PASER-M cohort, The Netherlands); Brian Agan, Vincent Marconi and Scott Wegner (US Department of Defense); Wataru Sugiura (National Institute of Health, Japan); Maurizio Zazzi (MASTER, Italy); Rolf Kaiser and Eugen Schuelter (Arevir Cohort, Köln, Germany); Adrian Streinu-Cercel (National Institute of Infectious Diseases Prof. Dr. Matei Balş, Bucharest, Romania); Gerardo Alvarez-Uria (VFHCS, India), Maria-Jesus Perez-Elias (CORIS, Spain); Federico Garcia (Resistance Work Package of the Spanish HIV Resistance Network; RIS, Spain); Tulio de Oliveira, (SATuRN, South Africa).
Clinics: Jose Gatell and Elisa Lazzari (University Hospital, Barcelona, Spain); Brian Gazzard, Mark Nelson, Anton Pozniak and Sundhiya Mandalia (Chelsea and Westminster Hospital, London, UK); Colette Smith (Royal Free Hospital, London, UK); Lidia Ruiz and Bonaventura Clotet (Fundacion Irsi Caixa, Badelona, Spain); Schlomo Staszewski (Hospital of the Johann Wolfgang Goethe-University, Frankfurt, Germany); Carlo Torti (University of Brescia, Italy); Cliff Lane, Julie Metcalf and Catherine A. Rehm (National Institutes of Health Clinic, Rockville, USA); Maria-Jesus Perez-Elias (Instituto Ramón y Cajal de Investigación Sanitaria, Madrid, Spain); Stefano Vella and Gabrielle Dettorre (Sapienza University, Rome, Italy); Andrew Carr, Richard Norris and Karl Hesse (Immunology B Ambulatory Care Service, St. Vincent’s Hospital, Sydney, NSW, Australia); Dr Emanuel Vlahakis (Taylor’s Square Private Clinic, Darlinghurst, NSW, Australia); Hugo Tempelman and Roos Barth (Ndlovu Care Group, Elandsdoorn, South Africa); Robin Wood, Carl Morrow and Dolphina Cogill (Desmond Tutu HIV Centre, University of Cape Town, South Africa); Chris Hoffmann (Aurum Institute, Johannesburg, South Africa and Johns Hopkins University, Boston, USA); Luminita Ene (‘Dr. Victor Babes’ Hospital for Infectious and Tropical Diseases, Bucharest, Romania); Gordana Dragovic (University of Belgrade, Belgrade, Serbia); Ricardo Diaz and Cecilia Sucupira (Federal University of Sao Paulo, Sao Paulo, Brazil); Omar Sued and Carina Cesar (Fundación Huésped, Buenos Aires, Argentina); Juan Sierra Madero (Instituto Nacional de Ciencias Medicas y Nutricion Salvador Zubiran, Mexico City, Mexico); Pachamuthu Balakrishnan and Shanmugam Saravanan (YRG Care, Chennai, India).
Clinical trials: Sean Emery and David Cooper (CREST); Carlo Torti (GenPherex); John Baxter (GART, MDR); Laura Monno and Carlo Torti (PhenGen); Jose Gatell and Bonventura Clotet (HAVANA); Gaston Picchio and Marie-Pierre deBethune (DUET 1 & 2 and POWER 3); Maria-Jesus Perez-Elias (RealVirfen); Sean Emery, Paul Khabo and Lotty Ledwaba (PHIDISA).
Funding
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024, Task Order No. 75N91020F00130. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Transparency declarations
None to declare.
Contributor Information
the RDI study group:
Peter Reiss, Ard van Sighem, Julio Montaner, Richard Harrigan, Tobias Rinke de Wit, Raph Hamers, Kim Sigaloff, Brian Agan, Vincent Marconi, Scott Wegner, Wataru Sugiura, Maurizio Zazzi, Rolf Kaiser, Eugen Schuelter, Adrian Streinu-Cercel, Gerardo Alvarez-Uria, Federico Garcia, Tulio de Oliveira, Jose Gatell, Elisa Lazzari, Brian Gazzard, Mark Nelson, Anton Pozniak, Sundhiya Mandalia, Colette Smith, Lidia Ruiz, Bonaventura Clotet, Schlomo Staszewski, Carlo Torti, Cliff Lane, Julie Metcalf, Catherine A Rehm, Maria-Jesus Perez-Elias, Stefano Vella, Gabrielle Dettorre, Andrew Carr, Richard Norris, Karl Hesse, Emanuel Vlahakis, Hugo Tempelman, Roos Barth, Robin Wood, Carl Morrow, Dolphina Cogill, Chris Hoffmann, Luminita Ene, Gordana Dragovic, Ricardo Diaz, Cecilia Sucupira, Omar Sued, Carina Cesar, Juan Sierra Madero, Pachamuthu Balakrishnan, Shanmugam Saravanan, Sean Emery, David Cooper, Carlo Torti, John Baxter, Laura Monno, Carlo Torti, Jose Gatell, Bonventura Clotet, Gaston Picchio, Marie-Pierre deBethune, Maria-Jesus Perez-Elias, Sean Emery, Paul Khabo, and Lotty Ledwaba
References
- 1.UNAIDS. Seizing the moment. Global AIDS Update 2020. 2020. https://www.unaids.org/en/resources/documents/2020/global-aids-report.
- 2. Montaner JS, Lima VD, Harrigan PR. et al. Expansion of HAART coverage is associated with sustained decreases in HIV/AIDS morbidity, mortality and HIV transmission: the “HIV Treatment as Prevention” experience in a Canadian setting. PLoS One 2014; 9: e87872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. HIV Drug Resistance Report 2019. 2019. https://www.who.int/hiv/pub/drugresistance/hivdr-report-2019/en/.
- 4. Saag MS, Benson CA, Gandhi RT. et al. Antiretroviral drugs for the treatment and prevention of HIV infection in adults: 2018 recommendations of the International Antiviral Society–USA Panel. JAMA 2018; 320: 379–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Writing Group, Williams I, Churchill D. et al. British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2012 (Updated November 2013. All changed text is cast in yellow highlight). HIV Med 2014; 15 Suppl 1: 1–85. [DOI] [PubMed] [Google Scholar]
- 6.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. https://clinicalinfo.hiv.gov/en/guidelines/adult-and-adolescent-arv/.
- 7. Degruttola V, Dix L, D’Aquila R. et al. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther 2000; 5: 41–8. [DOI] [PubMed] [Google Scholar]
- 8. Gupta RK, Hill A, Sawyer AW. et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis 2009; 9: 409–17. [DOI] [PubMed] [Google Scholar]
- 9. Larder B, Wang D, Revell A. et al. The development of artificial neural networks to predict virological response to combination HIV therapy. Antivir Ther 2007; 12: 15–24. [PubMed] [Google Scholar]
- 10. Wang D, Larder B, Revell A. et al. A comparison of three computational modelling methods for the prediction of virological response to combination HIV therapy. Artif Intell Med 2009; 47: 63–74. [DOI] [PubMed] [Google Scholar]
- 11. Larder BA, Revell A, Mican JM. et al. Clinical evaluation of the potential utility of computational modeling as an HIV treatment selection tool by physicians with considerable HIV experience. AIDS Patient Care STDS 2011; 25: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Revell AD, Wang D, Wood R. et al. Computational models can predict response to HIV therapy without a genotype and may reduce treatment failure in different resource-limited settings. J Antimicrob Chemother 2013; 68: 1406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Revell AD, Wang D, Boyd MA. et al. The development of an expert system to predict virological response to HIV therapy as part of an online treatment support tool. AIDS 2011; 25: 1855–63. [DOI] [PubMed] [Google Scholar]
- 14. Revell AD, Wang D, Perez-Elias. et al. 2018 update to the HIV-TRePS system: the development of new computational models to predict HIV treatment outcomes, with or without a genotype with enhanced usability for low-income settings. J Antimicrob Chemother 2018; 73: 2186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Revell AD, Wang D, Perez-Elias. et al. Predicting virological response to HIV treatment over time. A tool for settings with different definitions of virological response. J Acquir Immune Defic Syndr 2019; 81: 207–15. [DOI] [PubMed] [Google Scholar]
- 16. Wensing AM, Calvez V, Ceccherini-Silverstein F. et al. 2019 update of the drug resistance mutations in HIV-1. Top Antivir Med 2019; 27: 111–21. [PMC free article] [PubMed] [Google Scholar]
- 17. Revell AD, Wang D, Harrigan R. et al. Modelling response to HIV therapy without a genotype – an argument for viral load monitoring in resource-limited settings. J Antimicrob Chemother 2010; 65: 605–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. Consolidated Guidelines on the use of Antiretroviral Drugs for Treating and Preventing HIV Infection. Recommendations for a Public Health Approach – Second Edition. 2016. [PubMed] [Google Scholar]
- 19. Lecher S, Ellenberger D, Kim AA. et al. Scale up of HIV viral load monitoring – seven sub-Saharan African countries. MMWR Morb Mortal Wkly Rep 2015; 64: 1281–304. [DOI] [PubMed] [Google Scholar]
- 20. Stevens WS, Scott LE, Crowe SM.. Quantifying HIV for monitoring antiretroviral therapy in resource-poor settings. J Infect Dis 2010; 201: S16–26. [DOI] [PubMed] [Google Scholar]
- 21. Roberts T, Cohn J, Bonner K. et al. Scale up of routine viral load testing in resource-poor settings: current and future implementation challenges. Clin Infect Dis 2016; 62: 1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drain PK, Dorward J, Bender A. et al. Point-of-care HIV viral load testing: an essential tool for a sustainable global HIV/AIDS response. Clin Microbiol Rev 2019; 32: e00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]