Abstract
Human organoids are emerging as a valuable resource to investigate human organ development and disease. The applicability of human organoids has been limited, partly due to the oversimplified architecture of the current technology, which generates single-tissue organoids that lack inter-organ structural connections. Thus, engineering organoid systems that incorporate connectivity between neighboring organs is a critical unmet challenge in an evolving organoid field. Here, we describe a protocol for the continuous patterning of hepatic, biliary and pancreatic (HBP) structures from a 3D culture of human pluripotent stem cells (PSCs). After differentiating PSCs into anterior and posterior gut spheroids, the two spheroids are fused together in one well. Subsequently, self-patterning of multi-organ (i.e., HBP) domains occurs within the boundary region of the two spheroids, even in the absence of any extrinsic factors. Long-term culture of HBP structures induces differentiation of the domains into segregated organs complete with developmentally relevant invagination and epithelial branching. This in-a-dish model of human hepato-biliary-pancreatic organogenesis provides a unique platform for studying human development, congenital disorders, drug development and therapeutic transplantation. More broadly, our approach could potentially be used to establish inter-organ connectivity models for other organ systems derived from stem cell cultures.
Introduction
Failure to establish contiguous organ architectures because of developmental defects, infection or drug toxicity leads to critical and, in most cases, lethal organ failures, as seen in biliary atresia, tracheoesophageal fistula and congenital anomalies of the kidneys and urinary tract. Animal models have provided valuable insights into the mechanisms for generating connections between tissues. However, because of profound differences in both metabolism and molecular regulation between animals and humans, translation of these findings from animal models into human biology and disease remains a major challenge1.
Human pluripotent stem cell (hPSC)-derived organoids are increasingly recognized as cutting-edge in vitro model systems for investigating human organ development and diseases, with the potential to also supplant fetal tissue research2–8. However, the applicability of hPSC-derived organoids has been limited thus far, partly because technology has been limited to the generation of single-tissue organoids lacking inter-organ structural connections. It is vital to understand the mechanisms that drive the intricate inter-organ orchestrations that occur during organogenesis, which can then be leveraged for developing appropriate multi-organ 3-D stem cell culture systems that allow for seamless connections to neighboring tissues9–11.
Here, we describe a 3D differentiation approach using hPSCs to grow gut spheroids with distinct regional identities, which are comprised of both endoderm and mesoderm populations. We demonstrate that the anterior and posterior spheroid interactions recapitulate the in vivo foregut and midgut boundary. These interactions generate an organoid composed of several types of organs, called a human hepato-biliary-pancreatic (HBP) organoid (HBPO)12, that can model the inter-coordinated specification and invagination that naturally occur in development. Although this system has limitations, such as requiring further refinement of the later stages of HBP organogenesis (increased tissue maturation would be required for transplant applications), this HBPO system enables modeling of complex multi-organ development and disease in vitro.
Development of the protocol
In early embryogenesis, the foregut endoderm first arises at the early- to mid-primitive streak stages, leading to the formation of the definitive endoderm. This endodermal sheet begins to fold in at the anterior and posterior ends, forming the anterior and caudal intestinal portals, respectively, during early somitogenesis13. During anterior intestinal portal formation and subsequent foregut tubulogenesis, a medial part of the foregut endoderm sheet is roughly defined in the dorsal walls of the primitive gut tubes, and the lateral foregut endoderm cells are relocated into the ventral midline of the gut tube closure14. During foregut closure, the liver, gallbladder, and ventral pancreas primordia are specified within adjacent regions of the ventral foregut endoderm.
The formation of each individual primordium is induced and maintained by distinct signaling factors released from neighboring tissues15,16. In the posterior foregut, mesodermal cues instigate localized proliferation and/or migration of the endoderm, which results in budding of the liver, and dorsal and ventral pancreas by embryonic day 9.5. After embryonic day 9.5, the specified endodermal organ domains of the gut tube undergo morphogenesis and lineage diversification to create the functional architecture connecting neighboring organs.
Well-defined temporal expression of certain genes has been localized to distinct regions during organogenesis, and this gene expression can be exploited to identify particular areas of the embryo. In vivo, the HBP anlage, which is demarcated by HHEX (hematopoietically-expressed homeobox protein) and PDX1 (pancreatic and duodenal homeobox 1) expression17, is first specified at the boundary region between the foregut and midgut, as indicated by expression of the foregut endoderm marker SOX2 (SRY-box2) and the mid/hindgut endoderm marker CDX2 (caudal-type homeobox2), respectively. The HBP anlage forms an epithelial vesicle invaginating ventrally from the primitive gut as a result of epithelial-mesenchymal interactions18,19. Given that genetic perturbation of such boundary-defining genes disrupts integral multi-organ development20, it is clear that continuous, dynamic organogenesis occurs in a complex environment and is probably driven by neighboring factors (such as boundary interactions)21. From this knowledge, we hypothesized that the recapitulation of the foregut-midgut boundary in a stem cell culture acts to induce continuous hepatic, biliary and pancreatic structures.
Therefore, to develop a foregut-midgut boundary, we first exposed definitive endoderm, patterned as previously described4, to fibroblast growth factor and a WNT signaling agonist, which promote gut tube formation and patterning, in the presence or absence of a bone morphogenetic protein antagonist, which induces foregut specification, to direct SOX2 expressing anterior or CDX2 expressing posterior gut fate.
Overview of the procedure
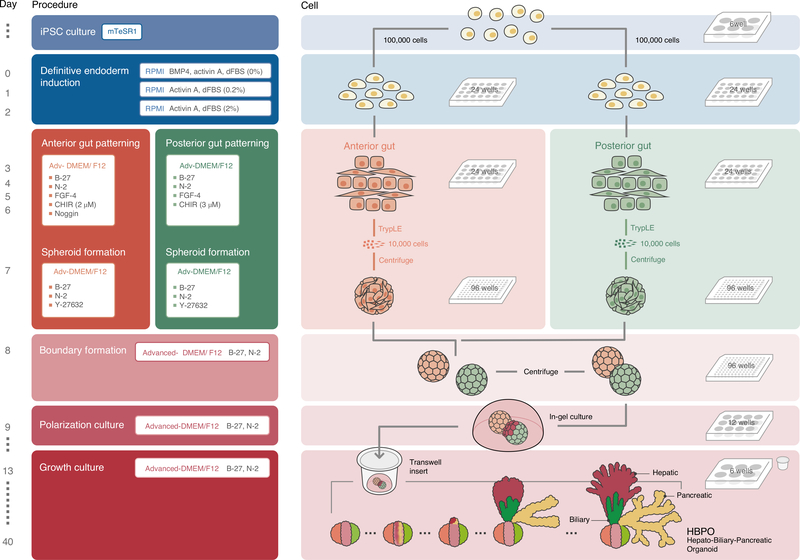
The overall procedure can be separated into six major stages (Fig. 1 and Supplementary Video 1). Stage 1 is PSC culture (Steps 1–19; day −2 to day −1). Stage 2 is definitive endoderm induction (Steps 20–29; day 0–day 2). Stage 3 is anterior/posterior gut patterning (Steps 30–33; day 3–day 6) and anterior/posterior gut spheroid formation (Steps 34–37; day 7). Stage 4 is anterior-posterior boundary formation (Steps 38–41; day 8). Stage 5 is HBPO polarization culture (Steps 42–48; day 9–day 12). Stage 6 is long-term HBPO growth culture (Steps 49–55; day 13 onward).
Fig. 1 |. Overview of the protocol.
Schematic representation of the protocol from passage of PSCs to final HBPO. The image brightness was increased to improve visibility. Refer to Table 1 for the specific details of the different culture media that are required at each stage of the protocol. dFBS, defined FBS.
Applications
Studying mechanisms by which inter-tissue connections are generated in humans remains a major challenge because of limited access to fetal samples and poor manipulability22. Although signaling mechanisms for organ specification have been studied in animal models, the ways in which combinatorial endoderm-mesoderm interactions orchestrate inter-connected and multi-organ patterning in humans remain elusive. Human HBPOs can serve as an in-a-dish model to investigate and interrogate the complex inter-organ segregation process. For example, we have shown that the transient and localized suppression of retinoic acid signaling at the foregut-midgut border is crucial for directing HBP patterning in hPSC-derived organoids12. Interestingly, in agreement with animal studies, the accompanying mesenchyme probably plays an important role in lineage specification into HBP precursors. Therefore, this system provides a novel platform that will facilitate future mechanistic studies about contributions of different cell types in organogenesis. Our integrated multi-organ model system can be used as a tractable, manipulatable and readily achievable model to broaden our knowledge of complex organogenesis and disease mechanisms in humans.
For example, our HBPOs could be useful for mechanistic studies of biliary atresia and evaluation of therapeutic responses to new potential treatments. Biliary atresia, a disease of inter-organ connectivity failure, is the most common indication for pediatric liver transplantation worldwide. Persistent inflammation and obstruction in the bile ducts lead to abnormal accumulation of bile and consequently significant liver damage. Several animal model studies of experimental biliary atresia have investigated the role of a Notch effector, Hes family bHLH transcription factor 1 (HES1), in the initiation and progression of the disease. Deficient Hes1 expression results in induction of experimental biliary atresia in animals23, with ectopic pancreatic endocrine cell generation probably due to the role of HES1 as a general negative regulator of endodermal endocrine differentiation23. We know that HES1 expression is evolutionariy conserved in gallbladder primordium and its derivatives in humans, but it remains unclear how HES1 activity is involved in the segregation of the HBP primordium and subsequent development of the bile duct system. In HBPOs differentiated from PSCs with a genetic knock-out of HES1, biliary specification is abolished, while pancreatic fate commitment is promoted in Hes1-deficient mice12.
Building inter-organ connections in a dish has been a long-standing challenge because of a limited understanding of the developmental processes during organogenesis. Previous studies have also been hindered by lack of efficient programming of the cells to naturally segregate and organize in a spatiotemporally appropriate manner. Many attempts have been made to either developmentally or synthetically solve these issues, including organoid generation, tissue printing, cell-material composite formation and decellularization and recellularization of existing 3D tissue. Each of these methods has shown some success; however, none has provided a durable, persistently functional system1. Recapitulating boundary interactions in 3D stem cell culture is one potential approach to drive an organ segregation program10,24–27. Animal organogenesis studies have revealed that the prospective HBP region arises from a boundary between the foregut and midgut. Co-cultivation of stem cell–derived foregut and midgut is sufficient to induce a boundary-specific transcriptional program that includes the modulation of WNT and retinoic acid pathways, leading to HBPO generation. Thus, in vitro boundary modeling could potentially be used to establish inter-organ connectivity models for other organ systems derived from stem cell cultures.
Experimental design
Differentiation is achieved through several steps, starting with the 2D generation of anterior and posterior gut cells according to previously described methods4,28 with certain key modifications in basal medium and concentration of growth factors. Next, from these 2D cultures, we make single-cell suspensions of the anterior and posterior gut cells, and, in the presence of the Rho kinase inhibitor Y-27632, spheroids are generated on day 7. The successful generation of stable spheroids is absolutely critical to efficiently and consistently form HBPOs. Therefore, the size of the aggregates is crucial, and careful consideration must be given to both the number of cells used and the incubation time for aggregate formation. To generate a boundary organoid, one anterior and one posterior spheroid are carefully mixed in Matrigel in a single well, wherein the two spheroids will fuse overnight (Fig. 1).
For long-term culture, we tested several different culture conditions for the day 13 boundary organoids to evaluate HBP organogenesis12. Among the tested culture conditions, we found that using the air–liquid interface system of excised PROX1+ domains continued the morphogenesis for the longest period, resulting in the formation of a branching structure that we have called HBPO. Time-course imaging showed that the dissected PROX1+ domain changed from an epithelial morphology to a more convoluted structure after 2 d of culture.
Alternative methods
Recently, several groups have described hepato-biliary-like organoids that can be differentiated from hPSCs and fetal liver progenitor cells29,30. However, these methods generate liver tissues composed of intrahepatic biliary epithelial cells and hepatocytes, and do not generate any extrahepatic components or surrounding organs. Tissue decellularization and recellularization methods have shown some success in creating more spatially defined 3D tissues, but the temporal dynamics that naturally occur between organs during biological or pathological processes have not yet been modelled31,32. Our system is potentially compatible with emerging organ-on-chip technologies, provided a measurable readout can be developed33.
Limitations
The early stage of our protocol (up to day 13) can be carried out with high efficiency and reproducibility. However, moving into later stages, morphological variation is observed between experiments because of variation in the efficiency of branching of the pancreas-like tissue. Therefore, the dissection of the HBPO region by using a lineage-specific reporter PSC line is recommended to improve the efficiency of epithelial bifurcation.
Our current HBPO system is less mature than adult tissues, and it is not suitable for transplantation applications. Further HBP organogenesis (e.g., to include the liver budding process, including delamination of the hepatic epithelial sheet by disruption of the laminar layer) would potentially require the addition of stromal cell components, such as septum transversum mesenchyme and/or endothelial progenitors. Nevertheless, the in vitro contiguous specification and early morphogenesis of HBP subdomains provide an opportunity to investigate the complex interactions at the anterior–posterior boundary and the generation of interconnected, multi-organ structures. This model can be used to study personalized human organogenesis and model disease systems in vitro, with the ultimate goal of developing new therapies for human diseases.
Materials
Biological materials
Human PSCs. We have successfully used H1 human embryonic stem cells (RRID: CVCL_9771; provided by the Pluripotent Stem Core Facility, Cincinnati Children’s Hospital Medical Center), induced pluripotent stem cell clone TkDA3–4 (RRID: CVCL_RJ54; provided by University of Tokyo), 317A434 (derived from Riken Cell Bank cat. no. HPS0076; RRID: CVCL_K092), 317D634 (derived from Riken Cell Bank cat. no. HPS0076; RRID: CVCL_K092), 1231A3 (RRID: CVCL_LJ39; provided by CiRA, Kyoto University) and the human 72_3 clone4 (RRID: CVCL_A1BW; generated by the Pluripotent Stem Cell Core Facility, Cincinnati Children’s Hospital Medical Center). TkDA3–4-GFP was a gene-edited clone derived from TkDA3–4 ▲ CRITICAL Use of human tissues must adhere to all relevant ethical guidelines. When appropriate, informed consent must be obtained from donors or their legal guardians. ! CAUTION When handling cells, take the necessary precautions. Use proper personal protective equipment at biosafety level 2 for tissue culture work, and perform the work within a class II biosafety cabinet.
Reagents
! CAUTION When handling reagents, take the necessary precautions. Refer to specific reagent material safety data sheets for additional information.
Growth medium and supplements
▲ CRITICAL We have not tested media or supplements from other vendors.
mTeSR1 (Stemcell Technologies, cat. no. 85850)
Matrigel, Growth Factor Reduced (GFR; Corning Inc., cat. no. 356231)
Matrigel, Phenol Red–Free (Corning Inc., cat. no. 356237)
RPMI 1640 (Thermo Fisher Scientific, cat. no. 22400089)
Defined FBS (dFBS) (Hyclone Laboratories Inc., cat. no. SH3007002)
Advanced DMEM/F-12 (Thermo Fisher Scientific, cat. no. 12634)
B27 Supplement (Thermo Fisher Scientific, cat. no. 17504044)
N2 Supplement (Thermo Fisher Scientific, cat. no. 17502048)
HEPES (Thermo Fisher Scientific, cat. no. 15630080)
l-Glutamine (Thermo Fisher Scientific, cat. no. 25030164)
Penicillin-streptomycin (Thermo Fisher Scientific, cat. no. 15140122)
DMEM/F-12 (e.g., Thermo Fisher Scientific, cat. no. 11330057)
Enzyme and growth factors
▲ CRITICAL We have not tested enzymes and growth factors from other vendors.
Accutase (Thermo Fisher Scientific, cat. no. A1110501)
TrypLE Express (Thermo Fisher Scientific, cat. no. 12604021)
Activin A (R & D Systems, cat. no. 338-AC-01M)
FGF-4 (R & D Systems, cat. no. 235-F4–01M)
CHIR 99021 (Stemgent, cat. no. 04–0004-10)
Y-27632 (Tocris Bioscience, cat. no. 1254)
BMP-4 (R & D Systems, cat. no. 314-BP-050)
Noggin (R & D Systems, cat. no. 6057-NG-100)
Immunostaining reagents
Donkey anti-goat IgG Alexa Fluor 647 (Thermo Fisher Scientific, cat. no. A-21447; RRID: AB_2535864)
Donkey anti-rabbit IgG Alexa Fluor 555 (Thermo Fisher Scientific, cat. no. A-31572; RRID: AB_162543)
Donkey anti-mouse IgG Alexa Fluor 488 (Thermo Fisher Scientific, cat. no. A-21202; RRID: AB_141607)
Donkey anti-guinea pig IgG Alexa Fluor 647 (Jackson ImmunoResearch Labs, cat. no. 706–605-148; RRID: AB_2340476)
DAPI mounting solution (Sigma, cat. no. F6057–20ML)
Rabbit anti-SOX2 (Seven Hills Bioreagents, cat. no. WRAB-1236; RRID: AB_2715498)
Mouse anti-CDX2 (BioGenex, cat. no. AM392; RRID: AB_2650531)
Guinea pig anti-PDX1 (Abcam, cat. no. ab47308; RRID: AB_777178)
Rabbit anti-HHEX (R & D Systems, cat. no. MAB83771)
Rabbit anti-PROX1 (Abcam, cat. no. ab38692; RRID: AB_2170708)
Mouse anti-GATA4 (Santa Cruz Biotechnology, cat. no. sc-25310; RRID: AB_627667)
Mouse anti-SOX17 (R & D Systems, cat. no. MAB1924; RRID: AB_2195646)
Goat anti-SOX17 (R & D Systems, cat. no. AF1924; RRID: AB_355060)
Goat anti-epithelial cell adhesion molecule (EpCAM) (R & D Systems, cat. no. AF960; RRID: AB_355745)
Goat anti-E-cadherin (R & D Systems, cat. no. AF648; RRID: AB_355504)
Mouse anti-alpha-fetoprotein (Thermo Fisher Scientific, cat. no. 14–9499-82; RRID: AB_2572949)
Rabbit anti-HNF4 (Abcam, cat. no. ab200142)
Mouse anti-FOXA2 (Thermo Fisher Scientific, cat. no. 14–4778-82; RRID: AB_10714832)
Rabbit anti-RFP (Rockland, cat. no. 600–401-379S; RRID: AB_11182807)
Mouse anti-albumin (Sigma-Aldrich, cat. no. A6684; RRID: AB_258309)
Fluorescein Labeled Dolichos Biflorus Agglutinin (DBA) (Vector Laboratories, cat. no. FL-1031; RRID: AB_2336394)
Gene-editing reagents
pSpCas9(BB)-2A-GFP(PX458) (Addgene, plasmid no. 48138)
Lipofectamine 3000 Reagent (Thermo Fisher Scientific, cat. no. L3000015)
Opti-MEM (Thermo Fisher Scientific, cat. no. 31985062)
Blood & cell culture DNA mini kit (Qiagen, cat. no. 13323)
Other reagents and chemicals
Dulbecco’s PBS (DPBS) (Thermo Fisher Scientific, cat. no. 14190250)
Paraformaldehyde (PFA) Solution, 4% (wt/vol) in PBS (Affymetrix, cat. no. 19943 1 LT)
Cell Recovery Solution (Corning, cat. no. 354253)
Triton X-100 (Sigma, cat. no. T8787–100ML)
Tween 20 (Sigma, cat. no. P9416–100ML)
BSA Solution in PBS (Sigma, cat. no. SRE0036–250ML)
Dimethyl sulfoxide (Sigma, cat. no. D2650–100ML)
Optimal cutting temperature compound (Tissue-Tek; VWR, cat. no. 25608–930)
Equipment
Cell culture incubator (37 °C, 5% CO2)
Refrigerated centrifuges for 15- and 1.5-ml tubes and multiwell plates
Biosafety cabinet
Mechanical pipettes
Pipette controller
Sterile filter pipette tips
Sterile wide-bore pipette tips (e.g., USA Scientific, 1011–8410)
Sterile 50-ml centrifuge tube (e.g., Corning Inc., 352098)
Sterile 15-ml centrifuge tube (e.g., Corning Inc., 352095)
Sterile 1.5-ml microcentrifuge tubes (e.g., Eppendorf, 0030120086)
96-well Round Bottom Ultra-Low Attachment Microplate (Corning Inc., 7007)
Transwell (Corning Inc., 3470)
6-well multiwell cell culture plate (e.g., Corning Inc., 353046)
24-well multiwell cell culture plate (e.g., Corning Inc., 353047)
24-well multiwell cell culture plates (VWR International, 10062–896)
12-well multiwell cell culture plates (VWR International, 10062–894)
96-well multiwell cell culture plate (e.g., Corning Inc., 353072)
Serological pipettes
Inverted microscope (e.g., Olympus, model CKX53)
Stereomicroscope (e.g., Leica, S8APO)
Cell counter
Cell counter slides
Pasteur pipette
Micro knife (Fine Science Tools, 10315–12)
Spot Plate (Fisher Scientific, 13–748B)
Hydrophobic pen (ImmeEdge pen; Vector Laboratories, cat. no. H-4000)
Disposable base molds (15 × 15 × 5 mm; Fisherbrand, cat. no. 22–363-553)
Nikon A1 single-photon confocal microscope with NIS-Elements Advanced research imaging software
Flow cytometer (BD Biosciences, BD FACS AriaII)
Reagent setup
hPSC lines
Maintain the undifferentiated hPSCs in feeder-free conditions in mTeSR1 medium on plates coated with GFR Matrigel in an incubator with 5% CO2 at 37 °C.
Accutase
Thaw bottle on ice or overnight at 4 °C. Divide accutase into aliquots and store at −20 °C for ≤6 months.
Aliquots of GFR Matrigel for hPSC maintenance and Phenol Red–Free Matrigel for organoid culture
Aliquots of Matrigel should be prepared as detailed by the manufacturer. Briefly, thaw a 10-ml vial of GFR Matrigel or Phenol Red–Free Matrigel in a 4 °C refrigerator overnight. Gently pipette Matrigel by using a pre-cooled pipette to ensure homogeneity, divide into aliquots in 1.5-ml microcentrifuge tubes, quickly transfer filled microcentrifuge tubes onto ice and then store at −20 °C. ▲ CRITICAL Matrigel will rapidly begin to solidify near room temperature, so it is vital to work quickly and keep the Matrigel cold throughout the process of making aliquots.
Matrigel-coated plate for hPSC culture
Coat 6-well or 24-well plates with GFR Matrigel, as detailed by the manufacturer’s instructions. Briefly, thaw the GFR Matrigel aliquot on ice and resuspend in cold DMEM-F12 medium at 1:30 dilution on ice. Transfer sufficient cold medium plus Matrigel to each well of the tissue culture plate so that the entire surface is covered. Usually, 1 ml per well is required for a 6-well dish, and 500 μl per well is required for 24-well dishes. It is vital to incubate all Matrigel-coated dishes at room temperature for ≥1 h before passaging hPSCs. ▲ CRITICAL Matrigel will rapidly begin to solidify and fall out of solution near room temperature, so it is vital to work quickly and add Matrigel to cold medium before coating dishes.
Growth factor and small-molecule reconstitution
Reconstitution information for all growth factors and small molecules can be found in Table 1. After reconstitution, growth factors can be stored at 4 °C for ≤1–2 weeks, whereas small molecules typically can be stored at 4 °C for >2 weeks. Refer to manufacturers’ guidance for storage and stability instructions if any issues arise. For long-term storage (≤6 months), make aliquots and store at −20 or −80 °C. After thawing an aliquot of growth factor or small molecules, store at 4 °C. Avoid freeze–thaw cycles.
Table 1 |.
Reagent reconstitution
| Solvent | Stock concentration | |
|---|---|---|
| Activin A | 4 mM HCl, PBS | 100 μg/ml |
| BMP4 | 0.1% (wt/vol) BSA, 4 mM HCl, PBS | 50 μg/ml |
| Noggin | 0.1% (wt/vol) BSA, PBS | 200 μg/ml |
| FGF4 | 0.1% (wt/vol) BSA, PBS | 100 μg/ml |
| CHIR99021 | Dimethyl sulfoxide | 20 mM |
| Y-27632 | PBS | 10 mM |
Medium preparation
▲ CRITICAL Ideally, make up medium complete with growth factors fresh each day, although this can be stored at 4 °C for ≤3 d. Medium without growth factors can be stored at 4 °C for 2 weeks.
Medium for definitive endoderm induction no. 1 (day 0 medium for first day of differentiation)
Supplement RPMI 1640 with penicillin/streptomycin, 100 ng/ml activin A and 50 ng/ml BMP4.
▲ CRITICAL No serum should be added.
Medium for definitive endoderm induction no. 2 (day 1 medium for second day of differentiation)
Supplement RPMI 1640 with penicillin/streptomycin, 100 ng/ml activin A and 0.2% (vol/vol) dFBS. Medium for definitive endoderm induction no. 3 (day 2 medium for third day of differentiation)
Supplement RPMI 1640 with penicillin/streptomycin and then add 100 ng/ml activin A and 2% (vol/vol) dFBS.
Gut growth medium (for 3D spheroid and organoid growth)
Supplement advanced DMEM-F12 with 1× N2, 1× B27, 2 mM l-glutamine, 100 U/ml (1×) penicillin/streptomycin and 15 mM HEPES. Gut growth medium can be stored at 4 °C for 2 weeks and is largely based on the medium previously described3. For long-term storage (≤6 months), make aliquots and store at −20 °C.
Medium for anterior gut cell induction (anterior day 3–6 medium)
Supplement gut growth medium with 500 ng/ml FGF4, 2 μM CHIR99021 and 200 ng/ml Noggin.
▲ CRITICAL Medium complete with growth factors and small molecules is best if made fresh, but it can be stored at 4 °C for ≤3 d.
Medium for posterior gut cell induction (posterior day 3–6 medium)
Supplement gut growth medium with 500 ng/ml FGF4 and 3 μM CHIR99021. ▲ CRITICAL Medium complete with growth factors and small molecules is best if made fresh, but it can be stored at 4 °C for ≤3 d.
Gut growth medium containing 10 μM Y-27632 (day 7 medium for 3D spheroid and organoid growth)
Supplement gut growth medium with 10 μM Y-27632. ▲ CRITICAL Gut growth medium containing 10 μM Y-27632 can be stored at 4 °C for 2 weeks. For long-term storage (≤6 months), make aliquots and store at −20 °C.
Procedure
Passage of PSCs (day −2) ● Timing 1–2 h
-
1
Pre-coat 24-well plates with GFR Matrigel as described in Reagent setup. The coated plates can be wrapped in Parafilm and kept for ≤2 weeks at 4 °C.
▲ CRITICAL STEP All tissue culture steps in this protocol should take place in a sterile environment and be performed with aseptic technique to prevent culture contamination.
-
2
Take a six-well plate of undifferentiated PSCs and remove the mTeSR1 by aspiration from the wells to be passaged (Fig. 2a).
-
3
Wash the wells with 2 ml of sterile DPBS. Aspirate the DPBS.
-
4
Add 1 ml of accutase to the wells.
-
5
Incubate at 37 °C for 5 min, allowing PSC colonies to dissociate into single cells.
-
6
Add 5 ml of warm DMEM/F12 to each well to sufficiently dilute the accutase.
-
7
Gently triturate each treated well two to three times before transferring the 6 ml of PSCs in medium to a conical tube. Repeat the process with the remaining wells until all cells are collected into the same conical tube.
-
8
Centrifuge the cells at 1,000 r.p.m. for 3 min at 4 °C.
-
9
Aspirate the medium from the conical tube.
-
10
Add warm mTeSR1 to the conical tube.
-
11
Count the cells.
-
12
Aspirate the Matrigel from the prepared Matrigel-coated 24-well plate (from Step 1).
-
13
Plate the cells at a density of 100,000–150,000 cells/well in 500 μl of mTeSR1 including 10 μM Y-27632 on a Matrigel-coated tissue culture 24-well plate. Prepare cells with at least two independent wells to generate anterior and posterior gut spheroids at the same time.
-
14
Place the 24-well plate in a tissue culture incubator and ensure that the single cells are evenly distributed in the wells by gently moving the 24-well plate side to side.
-
15
Incubate overnight in an incubator with 5% CO2 at 37 °C.
Fig. 2 |. Representative images of PSC differentiation into definitive endoderm and anterior/posterior gut cells.
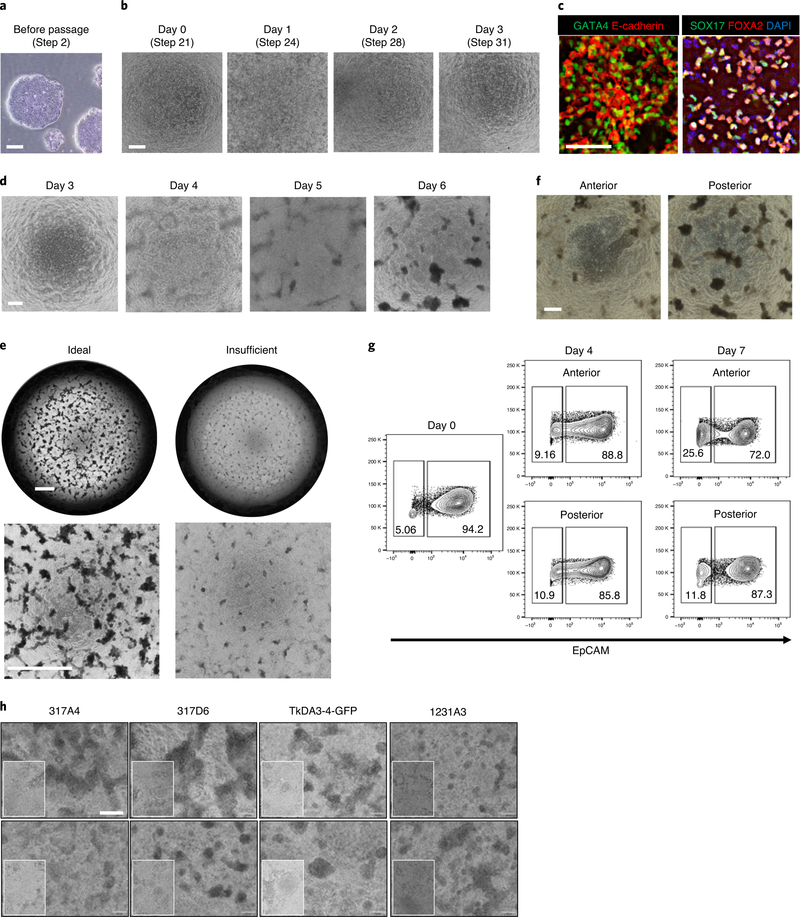
a, Typical morphology of PSC colonies cultured on Matrigel and mTeSR1 medium before passage at day −2. Images are representative of six independent PSC lines with similar results. b, Bright-field images of PSCs from day 0 to day 3 at the definitive endoderm stage. c, Optimal differentiation of PSCs into definitive endoderm displays ~90% of GATA4, E-cadherin positive expression and ~80% SOX17 and FOXA2 double-positive expression by day 3. d, Bright-field image of PSCs from day 3 to day 6. e, Low-magnification images of posterior foregut cells at day 7. The left images are representative of an ideal differentiation; the right images are an example of insufficient differentiation. f, Bright-field images of anterior and posterior gut cells at day 7. g, Flow cytometry analysis of anterior or posterior gut cells. 70–90% of cells were EpCAM positive epithelial cells. h, Confirmation of reproducibility by using various PSC lines. Scale bars, 200 μm (a,b,d,f and h), 100 μm (c) and 500 μm (e).
Change mTeSR medium (day −1) ● Timing 0.5 h
-
16
Warm mTeSR1 at room temperature (15–25 °C).
-
17
Confirm that the plated cells are healthy and are growing at a uniformly high density.
-
18
Aspirate old mTeSR1 with 10 μM Y-27632 to change the medium. Add fresh mTeSR1 without Y-27632 (Step 16), 500 μl/well for a 24-well plate.
-
19
Place the 24-well plate back in an incubator with 5% CO2 at 37 °C overnight.
Differentiation of PSCs into definitive endoderm (days 0–2)
Day 0 ● Timing 0.5 h
-
20
Check the confluency of the cells by using an inverted microscope. Cells should have reached 80–90% confluence (Fig. 2b).
▲ CRITICAL STEP It is imperative that the cell confluence is as close to 80–90% as possible. If the cell density is too high or low, this can result in overgrowth of mesoderm, cell death after activin A treatment and/or incomplete differentiation. Before beginning differentiation, titrate the cell number for each line to ensure 80–90% confluence on day 0. It is possible to wait an additional 24 h until this confluency is reached; aspirate the mTeSR1 and add 500 μl of fresh mTeSR1 to each well.
-
21
Warm an aliquot of day 0 medium to room temperature.
▲ CRITICAL Pre-warming the medium on this day helps to increase cell survival.
-
22
Aspirate the mTeSR1 medium. Add 0.5 ml/well of day 0 medium to the 24-well plate. Place the plate back in the incubator with 5% CO2 at 37 °C overnight.
Day 1 ● Timing 0.5 h
▲ CRITICAL After 1 d of treatment with day 0 medium, there should be evidence of cell death and floating cells in the medium. However, there should still be a layer of cells attached to the plate after treatment.
-
23
View cells under an inverted microscope to determine if cells are still attached to the tissue culture plate (Fig. 2b). If there are few or no cells attached, abandon the protocol and restart.
-
24
Aspirate the day 0 medium.
-
25
Add 0.5 ml/well of fresh day 1 medium to the 24-well plate.
-
26
Place the plate back in the incubator with 5% CO2 at 37 °C overnight.
Day 2 ● Timing 0.5 h
▲ CRITICAL After 1 d of treatment with day 1 medium, there should be evidence of cell death and floating cells in the medium. However, there should still be a layer of cells attached to the plate after treatment. View cells under an inverted microscope to determine if cells are still attached to the tissue culture plate (Fig. 2b). If there are few or no cells attached, abandon the protocol and restart.
-
27
Aspirate the day 1 medium.
-
28
Add 0.5 ml/well of fresh day 2 medium to the 24-well plate.
-
29
Place the plate back in the incubator with 5% CO2 at 37 °C overnight.
Differentiation of PSCs into anterior/posterior gut cells (days 3–6) ● Timing 0.5–1 h every day
-
30
(Optional) If anterior or posterior gut cells cannot be differentiated successfully, confirm that PSCs are induced to definitive endoderm on day 3 by immunostaining5. Endoderm markers, such as GATA binding protein 4 (GATA4), SRY-Box transcription factor 17 (SOX17) and Forkhead box A2 (FOXA2), can be used to determine the efficiency of the definitive endoderm differentiation. Over 70% of cells should express these markers. An example of optimal results is shown in Fig. 2c.
-
31
Each day on days 3–6, warm anterior day 3–6 medium and posterior day 3–6 medium at room temperature and then aspirate the media and replace with 500 μl/well of the prewarmed anterior day 3–6 medium (for anterior gut cells) and prewarmed posterior day 3–6 medium (for posterior gut cells). Owing to the high number of live cells in each well, the phenol red in the medium will turn yellow by 24 h. Confirm the presence of 3D structures forming from the monolayer of cells, including attached and floating spheroids (Fig. 2d–f).
-
32
(Optional) If anterior or posterior gut spheroids cannot be formed successfully, confirm that PSCs are induced to anterior/posterior gut cells by flow cytometry analysis. The epithelial marker EpCAM can be used to determine the efficiency of the gut differentiation. 60–90% of cells in both the anterior and posterior structures should express EpCAM on day 7. An example of optimal results is shown in Fig. 2g.
Anterior/posterior gut spheroid formation (day 7) ● Timing 2–3 h
-
33
On day 7, dissociate the anterior or posterior gut cells to single cells by incubation with TrypLE Express at 37 °C for 5 min (Fig. 3a).
? TROUBLESHOOTING
-
34
Add 500 μl of warm DMEM/F12 to each well to sufficiently dilute the TrypLE.
-
35
Centrifuge the cells at 1,000 r.p.m. for 3 min and, after removing the supernatant, resuspend the pellet in day 7 medium.
-
36
Plate the anterior or posterior gut cell suspensions in 96-well round-bottom ultra-low attachment plates at a density of 10,000 cells with 100 μl day 7 medium per well and incubate at 37 °C overnight to form spheroids. Prepare the same number of anterior and posterior spheroids.
Fig. 3 |. Representative images of anterior or posterior gut cells and boundary organoids.
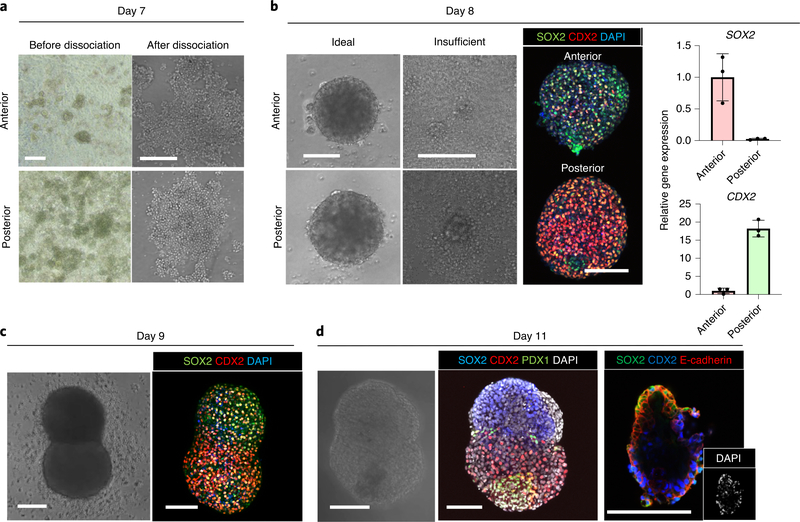
a, Typical morphology of monolayer anterior and posterior gut cells and the image of detached cells seeded on a low-attachment, 96-well plate at day 7. b, Successfully formed spheroids at day 8. Left: the image of spheroids generated under ideal conditions or insufficient culture conditions. Middle: whole-mount immunostaining of successfully formed spheroids for SOX2 and CDX2. Right: qPCR for SOX2 and CDX2. Data are means ± s.d.; n = 3 independent experiments. Unpaired, two-tailed t-test. c, Anterior and posterior boundary organoid at day 9. Right: whole-mount immunostaining for SOX2 and CDX2 of fused organoids. d, Generated boundary organoid at day 11. Whole-mount immunostaining for SOX2, CDX2 and PDX1 shows the PDX1 emergence at the boundary of the fused organoid. Scale bars, 200 μm (a) and 100 μm (b–d).
Anterior-posterior boundary spheroid formation (day 8) ● Timing 1–2 h
-
37
On day 8, pick up one generated single posterior spheroid with entire medium in a well by using a mechanical pipette with a wide-bore 200-μl tip. Pipette up and down first so that the spheroid gets suspended in the medium.
? TROUBLESHOOTING
-
38
Transfer the posterior spheroid to a well containing one anterior spheroid in the same 96-well round-bottom ultra-low attachment plate (Fig. 3b).
-
39
Centrifuge the plate at 1,000 r.p.m. for 1 min to gather spheroids and incubate at 37 °C for 24 h to form fused boundary spheroids (anterior-posterior gut spheroid; A-P spheroids).
? TROUBLESHOOTING
-
40
(Optional) From day 8 to day 11, perform whole-mount immunostaining analysis as a quality-control step (described in Box 1). SOX2, CDX2 and PDX1 expression can be used to determine the efficiency of HBPO formation. SOX2 should be expressed in anterior gut spheroids, CDX2 should be expressed in posterior gut spheroids and PDX1 should be expressed at the boundary after day 11. An example of optimal results is shown in Fig. 3b–d.
Box 1 |. Whole-mount staining of HBPOs.
At day 8, SOX2 positive (~80%) anterior and CDX2 positive (~80%) posterior gut spheroids should form. This can be confirmed by whole-mount immunostaining, as shown in Fig. 3b. At day 11, PDX1-expressing HBP anlage cells should be observed at the boundary of anterior and posterior gut spheroids, as shown in Fig. 3d. HHEX can also be detected at day 11 in the boundary region. In long-term culture conditions, after day 20, PROX1-expressing tissue should start to grow (Fig. 4c). Furthermore, you should be able to see a tissue connection between AFP-expressing liver-like tissue and PDX1-expressing pancreas-like tissue (Fig. 4d,e).
Procedure
Remove as much cell culture medium from the cultures as possible, taking care not to disturb the organoids.
Add 500 μl of pre-chilled Cell Recovery solution.
Pipette up and down gently by using wide-bore tips to carefully break up the Matrigel matrix without damaging the organoids and collect the organoids into a 15-ml conical tube.
Incubate the cultures with Cell Recovery solution at 4 °C for 20 min.
Centrifuge the cultures at 1,000 r.p.m. for 3 min at 4 °C, remove the Cell Recovery solution and repeat steps 2–5.
Visualize the organoids under the microscope to confirm that the Matrigel has been fully depolymerased and that organoids are floating free from the Matrigel. If organoids appear free from the Matrigel, centrifuge the cultures at 1,000 r.p.m. for 3 min at 4 °C to separate the structures from the solution. Remove the Cell Recovery solution.
Add 5 ml of cold DPBS to the tube and gently pipette up and down to wash the Matrigel from the organoids.
Spin down the conical tube at 1,000 r.p.m. for 3 min at 4 °C.
Gently aspirate the DPBS.
Wash a second time with 5 ml of DPBS and spin down at 1,000 r.p.m. for 3 min at 4 °C.
After aspirating DPBS, fix the organoids in 1 ml of 4% (wt/vol) PFA at 4 °C overnight.
Spin down at 1,000 r.p.m. for 3 min. Remove the PFA solution and dispose of properly.
Treat the fixed samples with 500 μl of 4% (wt/vol) PFA with 0.5% (vol/vol) Triton X-100 at room temperature for 15 min.
Spin down at 1,000 r.p.m. for 3 min. Remove the PFA/Triton X-100 solution and dispose of properly.
Add 5 ml of DPBS to the tube to suspend, and spin down the conical tube at 1,000 r.p.m. for 3 min at 4 °C.
Gently aspirate the DPBS.
Wash a second time with 5 ml of DPBS, spin down at 1,000 r.p.m. for 3 min at 4 °C and gently aspirate the DPBS.
Add 1 ml of permeabilization buffer (0.1% (vol/vol) Tween 20/DPBS) to the tube and transfer the organoids in solution to a 1.5-ml centrifuge tube. Incubate for 15 min at room temperature.
After gently removing the permeabilization buffer, add 1 ml of blocking buffer (1% (wt/vol) BSA and 0.3% (vol/vol) Triton X-100 in DPBS) and leave for 1 h at room temperature.
Prepare the primary antibody mixture in blocking buffer. Perform a spin-down at 1,000 r.p.m. for 3 min at 4 °C and aspirate off the blocking buffer. Add the primary antibody mixture and incubate overnight at 4 °C.
Aspirate the primary antibody mixture and perform three washes with 1 ml of DPBS, spinning down and aspirating the DPBS each time.
Prepare the secondary antibody mixture in blocking buffer. Add this mixture to the tube with the organoids and leave at room temperature for 2 h or overnight at 4 °C.
Wash three times with 1 ml of DPBS, spinning down and aspirating the PBS each time.
Add 50 μl of DPBS to each tube of organoids and transfer to one well of an IBIDI glass slide. Add one drop of DAPI mounting medium per well. Gently tilt the slide side to side to mix the DAPI mixture.
Image by using an inverted confocal microscope.
HBPO culture (day 9) ● Timing 2–3 h
-
41
On day 9, embed A–P spheroids in Matrigel drops and culture in gut growth medium to generate multi-organ HBPOs (Fig. 3c). Maintain the spheroid cultures at 37 °C in an atmosphere of 5% CO2, 95% air and replace the gut growth medium every 4 d.
-
42
Pick up a fused A-P spheroid by using a mechanical pipette with a wide-bore 200-μl tip and place the spheroid into a 1.5-ml centrifuge tube.
-
43
Spin down and carefully remove the supernatant with a regular sterile filter pipette tip.
▲ CRITICAL Organoids may be visible but be difficult to see; a dissecting microscope will help with visibility and handling.
-
44
44 Add 20–30 μl/well of 100% Matrigel (Phenol Red–Free) on ice to the fused A-P spheroid in a 1.5-ml tube. Gently re-suspend the single A-P spheroid by using a mechanical pipette with a wide-bore 200-μl tip and slowly plate it on a 24-well tissue culture plate (VWR International) to make a Matrigel drop.
▲ CRITICAL To avoid solidification of Matrigel, this step should be performed quickly. The tissue culture plate does not need to be on ice.
? TROUBLESHOOTING
-
45
Incubate the drop in a CO2 incubator at 37 °C for 5 min.
-
46
Flip the plate upside down and incubate for a further 5 min to prevent the spheroid from attaching to the bottom.
-
47
Add enough gut growth medium to cover the entire drop and culture for 4 d to generate HBPOs. Cultures for spheroids should be maintained at 37 °C in an atmosphere of 5% CO2, 95% air, and the gut growth medium should be replaced every 4 d.
? TROUBLESHOOTING
Air–liquid interface culture (day 13) (Optional) ● Timing 20–80 d
▲ CRITICAL For longer- term culture, HBPOs can be transferred to six-well Transwells for air–liquid interface culture at day 13.
-
48
Aspirate the medium from the well. Using a mechanical pipette with a wide-bore 200-μl tip, pick up the entire Matrigel drop with embedded HBPO. The Matrigel-embedded HBPO should dissociate easily from the bottom of the well.
-
49
(Optional) If using an HBP anlage specific reporter line (such as a PROX1 reporter) before transferring to Transwells, perform dissection of the boundary region of the HBPO to increase epithelial bifurcation rate. Transfer the HBPO to a spot plate containing DPBS placed on a fluorescent microscope. Using two clean micro knives in either hand, carefully dissect the HBPO by studying the fluorescent activity under the microscope. Using a Pasteur pipette, pick up the dissected HBPO.
-
50
Transfer the (dissected or undissected) HBPO to a six-well Transwell. Add one HBPO per Transwell.
-
51
Add 10 μl of fresh pre-chilled 100% Matrigel over the organoid to cover it.
-
52
Incubate the organoid in a CO2 incubator at 37 °C for 3 min.
-
53
Carefully add 500 μl–1 ml of gut growth medium to the bottom of the Transwell.
▲ CRITICAL For air-liquid interface culture, do not wet the upper side of the Transwell membrane.
-
54
Maintain organoid cultures at 37 °C in an atmosphere of 5% CO2, 95% air and replace the gut growth medium every 2 d. The HBPO on the Transwell can be cultured for ≤50 d without passaging.
▲ CRITICAL Do not wet the upper side of the Transwell membrane during medium exchange.
? TROUBLESHOOTING
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 33 | Gut cells are not differentiated | Low quality of PSCs | Check the pluripotency of the undifferentiated state of PSCs by general undifferentiated marker expression |
| Low number of input cells | Confirm that the cell confluency at day 0 is ~90% | ||
| Insufficient storage of aliquots of small molecules and proteins | Confirm that each aliquot of a small molecule or recombinant protein is reconstituted and stored appropriately according to the manufacturer's document | ||
| Insufficient storage of medium | Do not store the medium in the refrigerator for >1 week after adding small molecules, recombinants, serum or supplements | ||
| Wrong composition of medium | Check the concentration of each component in the differentiation medium | ||
| Insufficient timing of medium change | Confirm that all medium changes were performed at the same hour each day | ||
| 37 | Gut spheroids do not form | Using wrong cell detachment solution | Use TrypLE Express for dissociation on day 7, not accutase, ethylenediaminetetraacetic acid or trypsin |
| Lack of Y-27632 | Confirm that 10 μM Y-27632 was added in the medium on day 7 | ||
| Insufficient seeding cell number | Confirm that the seeding cell number was appropriate on day 7 | ||
| Lack of spin-down | Centrifuge the plate at 1,000 r.p.m. for 1 min before starting culture on day 7 | ||
| Low quality of differentiation | Consider the solutions described above for issues encountered on day 7 | ||
| 39 | A-P spheroids do not form | Lack of spin-down | Centrifuge the plate at 1,000 r.p.m. for 1 min before starting culture on day 8 |
| 44 | The Matrigel drop does not form appropriately | Low concentration of Matrigel | Remove the supernatant before adding Matrigel |
| Low concentration of Matrigel | Do not dilute the Matrigel | ||
| Using an insufficient cell culture plate | The type of tissue culture plate strongly affects the efficiency of Matrigel drop formation. From our comparison of plates from different companies, we recommended using the Multiwell Cell Culture Plates (VWR International, 10062–896) for Matrigel drop formation | ||
| 47, 54 | The HBP domain is not observed | Low differentiation quality | Consider the solutions described above for issues encountered on day 7 |
| Insufficient embedding in Matrigel drop | Confirm that the spheroid was completely covered by the Matrigel drop | ||
| Low volume of medium | Confirm that the Matrigel drop was completely covered by medium | ||
| Presence of Y-27632 | Do not add Y-27632 to the medium on day 9 | ||
Timing
Steps 1–15, passage of PSCs: 1–2 h
Steps 16–19, change mTeSR medium: 1 d
Steps 20–29, differentiation of PSCs into definitive endoderm: 3 d
Steps 30–32, differentiation of PSCs into anterior/posterior gut cells: 4 d
Steps 33–36, anterior/posterior gut spheroid formation: 1 d
Steps 37–40, anterior-posterior boundary spheroid formation: 1 d
Steps 41–47, HBPO culture: 4 d (20–80 d for long-term culture)
Steps 48–54, (optional) air–liquid interface culture: 20–80 d
Anticipated results
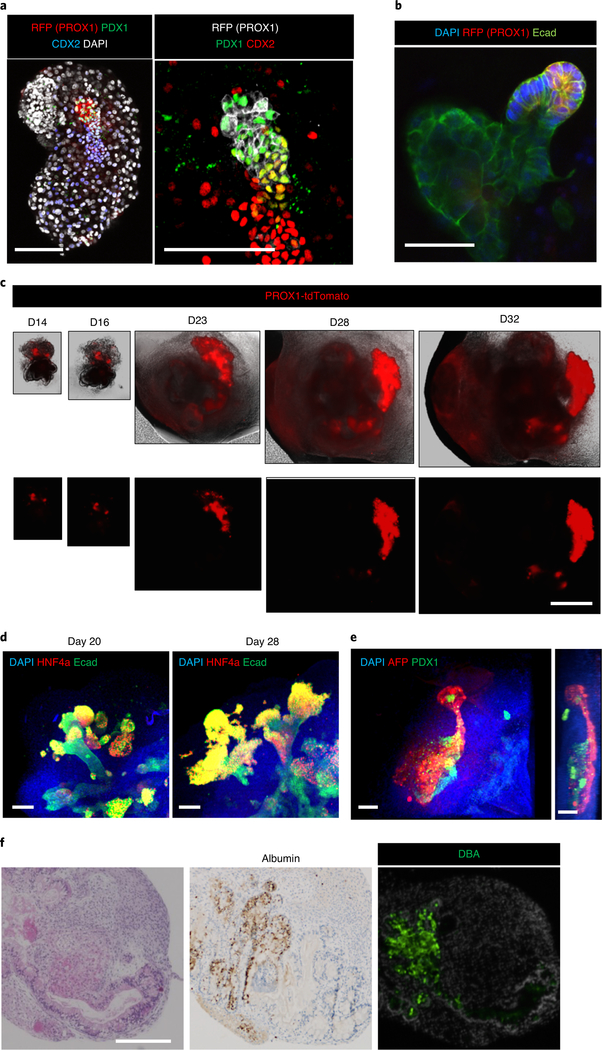
The boundary interactions between anterior and posterior gut spheroids that have been differentiated from PSCs enable autonomous emergence of HBP organ domains specified at the foregut-midgut boundary in the absence of any added extrinsic factors. Definitive endoderm differentiation (step 30; day 3; Fig. 2b–d)) yields a population of cells over 70% pure by SOX17 and FOXA2 double-positive staining (Fig. 2c)4,28,35. During the gut cell induction (Steps 31 and 32; days 3–7), 3D structures gradually form in a highly reproducible manner in various lines of PSCs (Fig. 2h). At day 7 (Step 33), we separately dissociate anterior gut cells and posterior gut cells to reaggregate them in low-attachment 96-well round-bottom plates. At day 8, SOX2 positive (~80%) anterior and CDX2 positive (~80%) posterior gut spheroids should form, and this can be confirmed by whole-mount immunostaining as in Fig. 3b and FACS12. After mixing each gut spheroid in a low-attachment, 96-well round-bottom plate, fused spheroids form with a high rate of reproducibility12. At day 9, PDX1 or PROX1 expression is not observed; however, during polarization culture in Matrigel, both proteins began to be expressed at the boundary domain of the fused spheroids (Fig. 3d). We find that morphogenesis continues in these PDX1 and PROX1 positive domains, which form a branching structure called an HBPO (Fig. 4a,b). The efficiency of HBPO production was increased approximately two-fold by air–liquid interface culture in our validation study12. Time-course imaging shows that the PROX1 positive epithelium in the HBPO starts to invaginate and grow outward in multiple directions, progressively forming a branching structure around day 23 (Fig. 4c). Immunofluorescent staining with the liver markers alpha fetoprotein (AFP), hepatocyte nuclear factor 4 alpha (HNF4A) and albumin, the pancreatic marker PDX1 and the bile duct marker DBA confirms that each lineage of primitive tissues segregates in the HBPOs after 30 d of culture (Fig. 4d–f). In conclusion, this HBPO system allows a detailed study of the early morphogenesis of HBP subdomains, and the generation of inter-connected, multi-organ structures in vitro from healthy and diseased PSC lines that can be reversibly manipulated by emerging technology such as genome editing (Box 2).
Fig. 4 |. Long-term culture–induced HBPO maturation.
a, Whole-mount immunostaining of an HBPO organoid at day (D) 12. Left: blue, CDX2; green, PDX1; red, RFP; white, DAPI. Right: a magnified image of the left sample. Green, PDX1; red, CDX2; white, RFP. b, Whole-mount staining of an HBPO organoid at day 13. Green, E-cadherin (Ecad); red, RFP; blue, DAPI. c, Morphogenic change of the PROX1+ domain of the air–liquid interface culture. Red, PROX1-tdTomato. d, Whole-mount immunostaining of a long-term cultured HBPO. Blue, DAPI; green, E-cadherin; red, HNF4a. e, Whole-mount immunostaining of a long-term cultured HBPO. Blue, DAPI; green, PDX1; red, AFP. Right: lateral view. f, Histological analysis of a long-term cultured HBPO. Left: H&E staining; middle: immunostaining for albumin; right: DBA staining. Scale bars, 100 μm (a and e), 50 μm (b and d) and 200 μm (c and f).
Box 2 |. CRISPR editing for HES1-knockout PSC line.
Candidate guide RNAs (gRNAs) targeting the start codon of HES1 transcript were designed by using online tools available at https://www.benchling.com. The sequence of our gRNA is listed in our original article. The candidate gRNA oligo was cloned into a pSpCas9(BB)-2A-GFP vector (Addgene, 48138).
Procedure
Seed PSCs onto a Matrigel-coated 24-well plate.
The next day, transfect with 2 μg of the vector plasmid into the PSCs by using Lipofectamine 3000 (following the manufacturer’s instructions).
After 24 h of transfection, coat a 96-well plate with Matrigel GFR. Add mTeSR supplemented with 10 μM Y-27632 into each well. Sort the GFP-expressing single cells into the prepared 96-well plate by flow cytometer.
After 2 d of sorting, change the medium to mTeSR without Y-27632.
Repeat daily medium changes until a colony is observed.
Passage and collect the colony. Isolate genomic DNA from part of the collected colony by using a blood and cell culture DNA mini kit (following the manufacturer’s instructions) and store the remaining cells.
Confirm the sequence of the target region of the gRNA.
Expand the resulting clones for the organoid experiment.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data to show validation of this protocol are included in the main article, the Supplementary Information files and the supporting primary research article12. The underlying raw datasets will be provided from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We express sincere gratitude to Hiro Nakazawa for helping with the audio in the video abstract, and the other Takebe laboratory members and the Wells-Zorn laboratory members for their support and excellent technical assistance. This work was supported by a Cincinnati Children’s Research Foundation grant, an NIH Director’s New Innovator Award (DP2 DK128799-01) and a PRESTO grant from Japan Science and Technology Agency (JST) to T.T. This work was also supported by NIH grant UG3 DK119982, a Cincinnati Center for Autoimmune Liver Disease Fellowship Award, PHS Grant P30 DK078392 (Integrative Morphology Core and Pluripotent Stem Cell and Organoid Core) of the Digestive Disease Research Core Center in Cincinnati, a Takeda Science Foundation Award, a Mitsubishi Foundation award, AMED JP19fk0210037, JP19bm0704025, JP19fk0210060, 20ta0127003h0001 and JP19bm0404045, and JSPS JP18H02800 and 19K22416. T.T. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-020-00441-w.
Peer review information Nature Protocols thanks Hans Clevers, Kenneth S. Zaret and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takebe T & Wells JM Organoids by design. Science 364, 956–959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 4.McCracken KW et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouchi R et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takebe T et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Sato T & Clevers H Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Takebe T et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 16, 556–565 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Xiang Y et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24, 487–497.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang Y et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song L et al. Assembly of human stem cell-derived cortical spheroids and vascular spheroids to model 3-D brain-like tissues. Sci. Rep. 9, 5977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike H et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 574, 112–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin V et al. Regionalisation of the endoderm progenitors and morphogenesis of the gut portals of the mouse embryo. Mech. Dev. 125, 587–600 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Molkentin JD, Lin Q, Duncan SA & Olson EN Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11, 1061–1072 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Wells JM & Watt FM Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Zorn AM & Wells JM Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udager A, Prakash A & Gumucio DL Dividing the tubular gut: generation of organ boundaries at the pylorus. Prog. Mol. Biol. Transl. Sci. 96, 35–62 (2010). [DOI] [PubMed] [Google Scholar]
- 18.McLin VA, Rankin SA & Zorn AM Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207–2217 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Nissim S et al. Iterative use of nuclear receptor Nr5a2 regulates multiple stages of liver and pancreas development. Dev. Biol. 418, 108–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spence JR et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev. Cell 17, 62–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bort R, Martinez-Barbera JP, Beddington RS & Zaret KS Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 131, 797–806 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Bredenoord AL, Clevers H & Knoblich JA Human tissues in a dish: the research and ethical implications of organoid technology. Science 355, eaaf9414 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Sumazaki R et al. Conversion of biliary system to pancreatic tissue in Hes1-deficient mice. Nat. Genet. 36, 83–87 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Bagley JA, Reumann D, Bian S, Levi-Strauss J & Knoblich JA Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison SE, Sozen B, Christodoulou N, Kyprianou C & Zernicka-Goetz M Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Rivron NC et al. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Spence JR et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu F et al. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 70, 1145–1158 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Vyas D et al. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 67, 750–761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeishi K et al. Assembly and function of a bioengineered human liver for transplantation generated solely from induced pluripotent stem cells. Cell Rep. 31, 107711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl Acad. Sci. USA 113, 2206–2211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer S et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 7, 14620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okita K et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011). [DOI] [PubMed] [Google Scholar]
- 35.McCracken KW, Howell JC, Wells JM & Spence JR Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 6, 1920–1928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data to show validation of this protocol are included in the main article, the Supplementary Information files and the supporting primary research article12. The underlying raw datasets will be provided from the corresponding author upon reasonable request.