This study uses single-cell RNA sequencing to investigate the transcriptional heterogeneity of all cell types known to comprise the acutely injured spinal cord in mice. The authors investigate both cell subtype heterogeneity and potential signaling interactions between myeloid, vascular, and macroglia cells.
Abstract
The wound healing process that occurs after spinal cord injury is critical for maintaining tissue homeostasis and limiting tissue damage, but eventually results in a scar-like environment that is not conducive to regeneration and repair. A better understanding of this dichotomy is critical to developing effective therapeutics that target the appropriate pathobiology, but a major challenge has been the large cellular heterogeneity that results in immensely complex cellular interactions. In this study, we used single-cell RNA sequencing to assess virtually all cell types that comprise the mouse spinal cord injury site. In addition to discovering novel subpopulations, we used expression values of receptor–ligand pairs to identify signaling pathways that are predicted to regulate specific cellular interactions during angiogenesis, gliosis, and fibrosis. Our dataset is a valuable resource that provides novel mechanistic insight into the pathobiology of not only spinal cord injury but also other traumatic disorders of the CNS.
Introduction
In the adult mammalian nervous system, a diverse population of glial and vascular cells are essential for optimal neuronal function. After spinal cord injury (SCI), this cellular diversity becomes even more heterogeneous by infiltrating leukocytes, which, together with neural cells regulate inflammation, cell proliferation, and tissue remodeling events, are collectively termed the wound healing process, which has been studied extensively in the past (Kigerl et al., 2006; Loy et al., 2002; Velardo et al., 2004). Although we currently understand the major cell types that comprise the injury site after SCI, we have a limited knowledge of the phenotypic heterogeneity within each cell type and how these cells interact during the wound healing process.
SCI triggers multiple processes that occur in a temporally defined manner. At 1 d post-injury (dpi), an innate immune response initiated by microglia is amplified by peripheral myeloid cells, mainly neutrophils and monocytes, which migrate to the injury site (Beck et al., 2010; Kigerl et al., 2006; Milich et al., 2019). By 3 dpi, most glial cells, including astrocytes, microglia, and oligodendrocyte progenitor cells (OPCs), are at the peak of their proliferative state, resulting in recovery of cell number and initiating gliosis (Barnabé-Heider et al., 2010; White et al., 2010). Concurrently, monocytes differentiate into macrophages that phagocytose cellular debris, acquire different phenotypic states, and activate perivascular fibroblasts to initiate fibrosis (Kigerl et al., 2009; Zhu et al., 2015). By 7 dpi, the numbers of macrophages and fibroblasts have reached their peak, and the fibrotic scar begins to be surrounded by the astroglial scar, which limits their infiltration into the spinal cord parenchyma (Herrmann et al., 2008; Sofroniew, 2015). In addition, during these first 7 d, hypoxic conditions at the injury site promote angiogenesis and revascularization (Loy et al., 2002; Whetstone et al., 2003). By 14 dpi, gliosis, fibrosis, and angiogenesis have reached a more stable state (Baldwin et al., 1998; Soderblom et al., 2013; Whetstone et al., 2003). Thus, the first 7 d after injury are highly dynamic and provide a window of therapeutic intervention for multiple pathological processes, but we currently have a limited understanding of the cellular interactions that occur during this critical period.
One way to investigate cellular responses to injury is through transcriptomic profiling of cell types that comprise the injury site. Traditional methods of using FACS, or more recent methods using immunoprecipitation of cell type–specific polyribosomes or nuclei, have provided important insight into central nervous system (CNS) pathobiology (Heiman et al., 2008; Sanz et al., 2009). However, these studies have been limited by the focus on a single cell type as well as the lack of information on subpopulation heterogeneity and different cell states. Recent advances in single-cell RNA sequencing (scRNA-seq) technologies have circumvented these limitations by providing a more accurate depiction of the heterogeneous subpopulations that comprise a single cell type (Vanlandewijck et al., 2018; Zeisel et al., 2018). In addition, many different cell types can be obtained and analyzed from a single tissue sample simultaneously, enabling examination of relationships and signaling mechanisms between multiple cell types.
In this study, we used scRNA-seq to generate a single-cell transcriptomics dataset of virtually all cell types that comprise the uninjured and injured spinal cord at 1, 3, and 7 dpi using a mouse mid-thoracic contusion model. We characterized the unique molecular signatures of multiple cell types, as well as their subtypes, and identified the transient appearance of injury-induced subpopulations during this acute period. By assessing expression of ligand–receptor pairs on different cell types, we highlight potential signaling relationships that may mediate angiogenesis, gliosis, and fibrosis. As the first scRNA-seq analysis of all cells that make up the SCI site, our transcriptomic dataset will provide the field with novel and comprehensive insight into early SCI pathology as well as other traumatic injuries to the CNS.
Results
Molecular identification of immune, vascular, and glial cells in the acute injury site
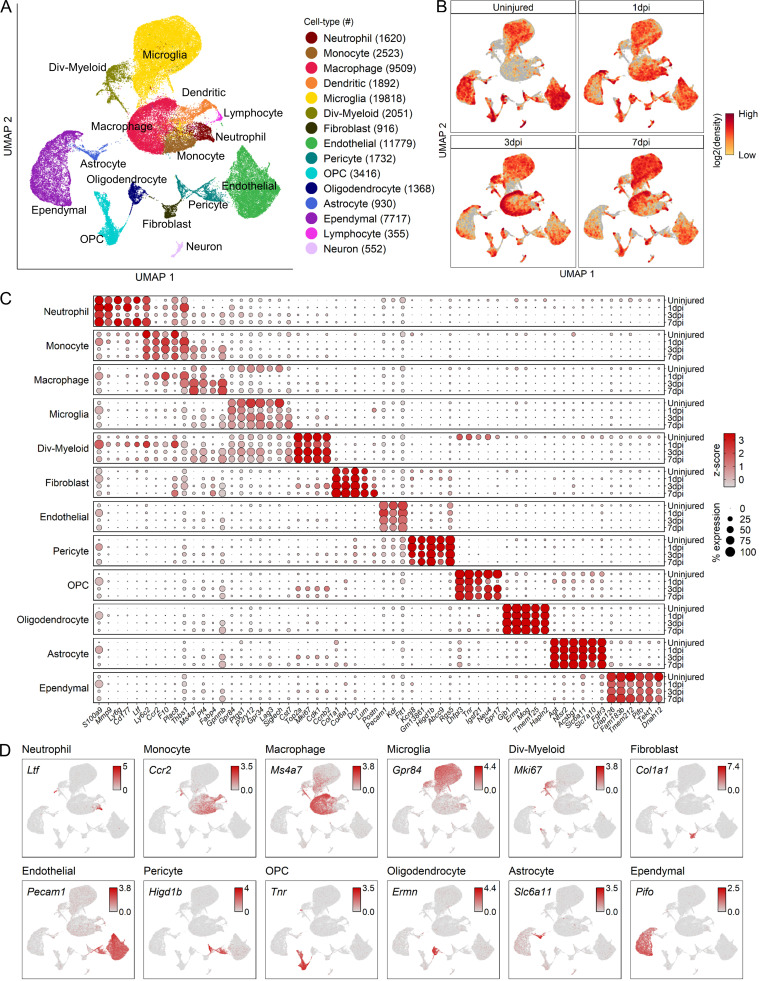
To assess the cellular heterogeneity involved in spinal cord wound healing, we performed mid-thoracic contusion SCI in mice and generated single-cell suspensions for sequencing. After excluding low-quality cells and potential doublets (see Materials and methods), we obtained a total of 66,178 cells from uninjured (12,488 cells across three biological replicates from five animals), 1 dpi (21,010 cells across three biological replicates from five animals), 3 dpi (17,491 cells across two biological replicates from three animals), and 7 dpi tissue (15,189 cells across two biological replicates from three animals). Cluster analysis of these cells resulted in 15 distinct clusters with specific temporal progression when visualized on a uniform manifold approximation and projection (UMAP) plot (Fig. 1, A and B). These 15 clusters represented all major cell types that are known to comprise the SCI site including microglia, monocytes, macrophages, neutrophils, dendritic cells, astrocytes, oligodendrocytes, OPCs, neurons, fibroblasts, pericytes, ependymal cells, and endothelial cells. Principal component analysis (PCA) showed separation between myeloid, vascular, and macroglia cell types (data not shown), and cells were grouped into these categories for further analysis as described below. The distribution of each biological replicate in the UMAP for each cell group category is shown in Fig. S1. Neurons were excluded from the analysis because they were not expected to survive our dissociation protocol and thus were subject to a selection bias. Lymphocytes were also excluded because they are primarily involved in autoimmunity after SCI (Ankeny and Popovich, 2009; Jones, 2014), which is beyond the scope of this study. Cell types pertaining to each cluster were identified using annotated lineage markers and the automatic annotation tool SingleR (Aran et al., 2019; Fig. S2, A and B). These methods combined gave us confidence in the correct identification of the major cell types.
Figure 1.
Transcriptomic identification of major cell types that comprise the mouse mid-thoracic spinal cord contusion site at acute time points.(A) UMAP plot of all cells collected from the uninjured spinal cord and injured spinal cord at 1, 3, and 7 dpi (66,178 cells). Cells are colored and annotated by cell type based on a combination of DEGs, canonical marker genes, and previously published spinal cord scRNA-seq datasets (Fig. S2, A and B). Cell counts for each cell type are displayed in parentheses in the legend on the right. (B) UMAP of all cells split by each time point. Cells are colored by neighborhood density in UMAP space to illustrate shifts over time. Darker red colors indicate greater cell density in the UMAP or gray if not from indicated time point. (C) Dot plot of highest DEGs for each major cell type at each time point (right axis). Dot color intensity represents the z-score of expression values, and dot size represents percent of cells with at least one UMI detected per gene. (D) UMAP of expression pattern of the DEG that best identifies each cell type. Values indicated are log-normalized counts per cell. n = 3 biological replicates for uninjured and 1 dpi; n = 2 biological replicates for 3 and 7 dpi. Div, dividing; OPC, oligodendrocyte progenitor cells.
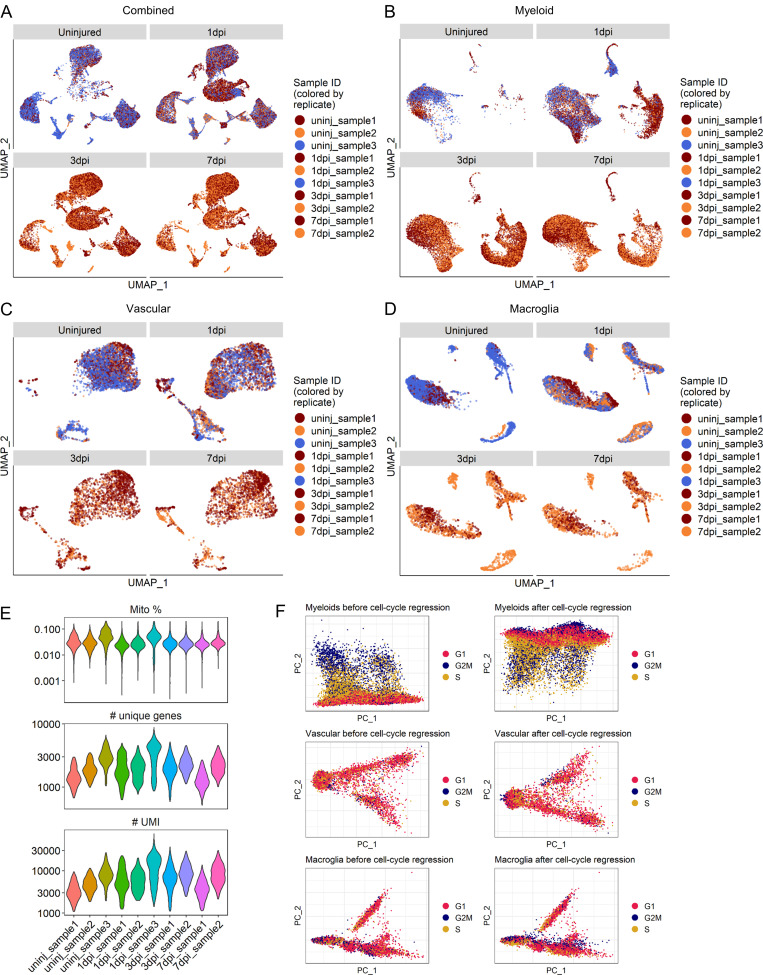
Figure S1.
Sample distribution, quality control metrics, and effect of cell cycle regression.(A–D) UMAP of all cells (A), myeloid cells (B), vascular cells (C), and macroglia cells (D) across all injury time points. Cells are colored by replicate for each injury time point. n = 3 replicates for uninjured and 1 dpi time points and n = 2 for 3 and 7 dpi time points (i.e., uninjured and 1-dpi UMAPs have three colors while 3- and 7-dpi UMAPs have two colors). (E) Sequencing and quality control metrics by sample. Distributions shown are for cells that were retained following removal of low-quality cells. Top: Number of UMIs detected per cell. Middle: Number of unique genes detected per cell. Bottom: Percentage of UMIs that maps to a mitochondrial protein–encoding gene per cell. (F) Plots of first two principal components for each cell group category show separation by cell cycle phase for myeloid cells but not vascular or macroglial cells. Cells are colored by predicted phases using the Seurat CellCycleScoring() function. Cell cycle regression of myeloid cells removed separation between cells in S and G2M phases. Cell cycle regression of vascular and macroglial cells had little to no effect. Mito, mitochondrial; PC, principal component; uninj, uninjured.
Figure S2.
Identification of major cell types using expression pattern of canonical genes and SingleR.(A and B) Cell type identification based on SingleR using the reference data from Rosenberg et al. (2018) (A) or Sathyamurthy et al. (2018) (B). The cell type names used in A and B are those from corresponding references. (C) Expression pattern of previously annotated marker genes used to identify major cell types on the combined UMAP in Fig. 1. Cx3cr1 and Tmem119 identified microglia. Itgam and Ptprc identified leukocytes. Ly6c2 and Ccr1 identified monocytes. Ly6g identified neutrophils. Cd3e identified lymphocytes. Mki67 and Cdk1 identified dividing cells. Pecam1 and Cldn5 identified endothelial cells. Col1a1 identified fibroblasts. Combined expression of Cspg4 and Pdgfrb identified pericytes. Foxj1 identified ependymal cells. Combined expression of Aqp4 and Gfap identified astrocytes. Plp1 and Opalin identified oligodendrocytes. Combined expression of Pdgfra and Olig2 identified OPCs. Snap25 identified neurons. Expression values are log-normalized counts. (D) Immunohistochemical analysis of 7 dpi spinal cord from Postn-CreER/Rosa26-EYFP mice shows genetically labeled EYFP+ fibroblasts exclusively located in the fibrotic area delineated by dense PDGFRβ+ region as well as only in meningeal fibroblasts that overlie the injury site. Arrowheads denote activated fibroblasts that are EYFP+/PDGFRβ+. PDGFRβ+ fibroblasts in the fibrotic scar that do not express EYFP are due to low recombination efficiency in these mice. Arrows indicate perivascular PDGFRβ+ cells in uninjured regions that do not express EYFP. Boxed regions are magnified in the right panels. Scale bars, 100 µm (left) and 30 µm (right). Astro, astrocyte; Endo/endothe, endothelial; Oligo, oligodendrocyte; OPC, oligodendrocyte progenitor cell.
The highest differentially expressed genes (DEGs) provided a unique molecular signature for each cell type (Fig. 1, C and D), which in some cases were different from canonical markers used in prior studies (Fig. S2 C). For example, the highest DEGs in OPCs were noncanonical genes such as tenascin-R (Tnr) and lipoma HMGIG fusion partner (Lhfpl3), which displayed better specificity than canonical OPC markers such as Pdgfra and Cspg4 that were expressed in multiple cell types including fibroblasts and pericytes (Fig. S2 C). Interestingly, while certain marker genes were expressed both before and after injury, others, such as Postn in fibroblasts, changed expression in an injury-dependent manner. While Postn was not expressed in uninjured fibroblasts, there was a graded increase as injury progressed (Fig. 1 C). We validated this by genetic lineage tracing in PostnEYFP mice, which showed enhanced YFP (EYFP) cells present in the fibrotic scar and overlying meninges but absent in surrounding spinal cord tissue (Fig. S2 D). In contrast to marker genes that identify a cell type both before and after SCI, injury-dependent genes offer useful information on cellular pathological states and can serve as potential biomarkers. Taken together, our analysis of DEGs between major cell types uncovers highly specific molecular identifiers, many of which are noncanonical and display temporal specificity.
Myeloid analysis reveals temporal changes in macrophage and microglial subtypes
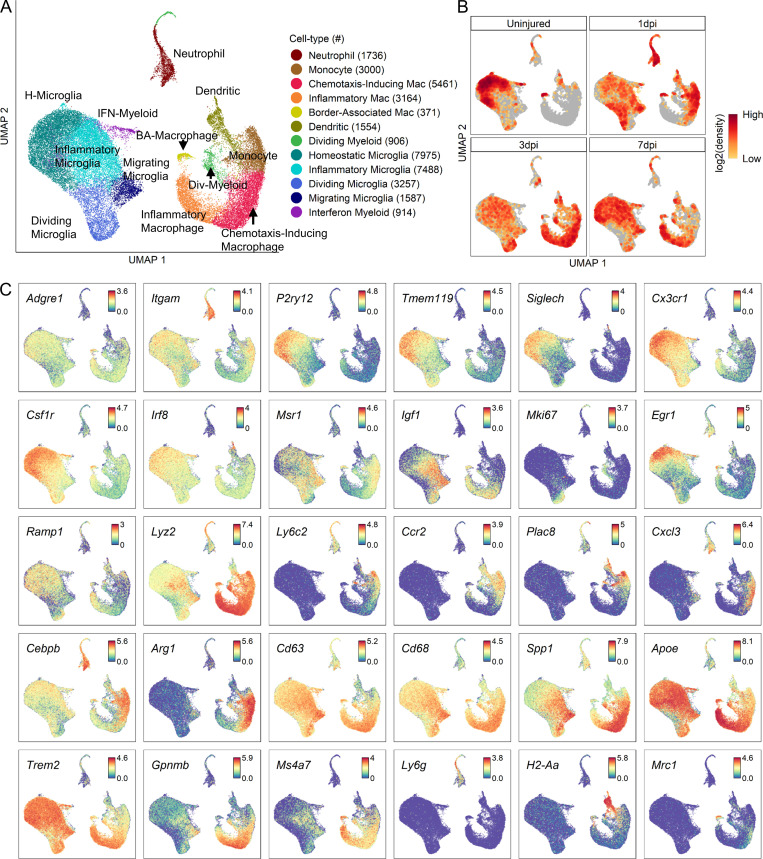
To better understand myeloid heterogeneity, we performed further cluster analysis of neutrophils, monocytes, macrophages, microglia, and dendritic cells and identified 12 distinct subtypes (Fig. 2, A and B). Overall structure of the resulting UMAP revealed two large clusters corresponding to microglia and peripherally derived myeloid cells as identified by annotated marker gene expression (Fig. 2 C). These marker genes displayed expected expression patterns between microglia and monocytes/macrophages for certain genes, but revealed novel insight for others (Fig. 2 C). Of the canonical microglial markers (P2ry12, Siglech, Cx3cr1, and Tmem119), Siglech seemed to be the most specific to microglia (versus monocytes/macrophages), although P2ry12 and Cx3cr1 were expressed at higher levels. Ccr2 and Arg1 were highly specific to monocytes/macrophages (versus microglia) but displayed macrophage subtype specificity.
Figure 2.
Transcriptomic identification of major myeloid subtypes acutely after SCI. (A) UMAP plot of all myeloid cells from uninjured spinal cord and 1, 3, and 7 dpi. Neutrophils, monocytes, macrophages, microglia, div-myeloid, and dendritic cells (Fig. 1 A) were extracted, reclustered, and re-embedded in new UMAP coordinates. Cells are colored by myeloid subtype as shown in legend on right. Cell counts in parentheses. Subtypes were annotated using a combination of DEGs, GO enrichment analyses, and previously published data. (B) UMAP of myeloid cells split by each time point. Cells are colored by neighborhood density in UMAP space to illustrate shifts over time. Darker red colors indicate greater cell density in the UMAP or gray if not from indicated time point. (C) Expression pattern of canonical marker genes, DEGs, and genes implicated in disease from previous studies. Cells are colored by expression value. Values are log-transformed normalized expression counts. The large cluster on the left was identified as microglia by the expression of P2ry12, Tmem119, and Siglech. The large cluster on the right was identified as peripheral myeloid cells by the expression of Ccr2 and Ly6c2. Homeostatic microglia (H-microglia) were identified by high expression of P2ry12, Siglech, and Tmem119. Inflammatory microglia were identified by Igf1, and migrating microglia were identified by Msr1 and Igf1. Dividing myeloid cells were identified by Mki67 and Top2a. Monocytes were identified by Ccr2 and Ly6c2. Macrophages (Mac) were identified by Cd63 and the reduced expression of monocyte markers. BA, border-associated; Div, dividing.
We identified four microglial subtypes. Homeostatic microglia were identified based on its expression of several annotated markers of steady-state microglia, such as P2ry12, Siglech, and Tmem119 (Fig. 2 C; and Fig. 3, B and C). As expected, homeostatic microglia were the predominant myeloid population in the uninjured spinal cord, but by 1 dpi, they were replaced by microglia subtypes that displayed different transcriptional signatures. We identified three nonhomeostatic microglia subtypes, which were labeled inflammatory, dividing, and migrating microglia based on their gene ontology (GO) terms for biological processes (Fig. 3 D). Inflammatory microglia GO terms were mostly associated with cell death and cytokine production. Genes associated with the cell death terms were not self-death pathways (e.g., caspases), but rather mostly cytokines (data not shown). Inflammatory microglia were identified by low expression of the purinergic receptor P2ry12 and increased expression of Igf1. GO terms for dividing microglia were mostly related to cell cycle. Dividing microglia expressed low levels of P2ry12, increased expression of Msr1, and high levels of cell cycle–related genes such as Cdk1. The presence of inflammatory and dividing microglia in uninjured tissue is perhaps due to dissociation-induced stress (Hammond et al., 2019; Wu et al., 2017) and suggests they are the first nonhomeostatic phenotypic states (see Discussion). GO terms for migrating microglia, which represented the smallest subtype, were associated with cell migration and motility. Migrating microglia had low levels of P2ry12, and high levels of Msr1 and the growth factor Igf1.
Figure 3.
Molecular and temporal profile of microglia subtype heterogeneity acutely after SCI. (A) Proportion of each microglia subtype among all microglia at each time point. (B) Dot plot of marker genes that differentiate nonhomeostatic microglia subtypes from homeostatic microglia. Color of dots represents z-scored expression level, and size of dots represents percentage of cells with at least one UMI detected per gene. (C) Heatmap of the highest DEGs per microglia subtype. Color represents z-score expression level. Inset stars in B and C indicate statistically significant greater expression compared with all other microglia combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction; see Materials and methods). (D) GO enrichment analysis of the DEGs per subtype. GO biological process terms displayed along y axis, and –log10 (Bonferroni-corrected P values) displayed along x axis. (E) Scatter plot of flow cytometry analysis of microglia in the uninjured spinal cord and injured spinal cord at 1 and 7 dpi. Y axis and x axis display fluorescence intensities of P2RY12 and MSR1, respectively. After removing nonviable cells and gating out Ly6G+ cells, CD45med/CD11b+ microglia were selected from other leukocytes for further gating along P2RY12 and MSR1 quadrants (see Materials and methods and Fig. 5 B). (F) Quantification of proportion of microglia subtypes in flow cytometry as gated in E. adj., adjusted; H-microglia, homeostatic microglia; Prop., proportion; Uninj, uninjured; Trans, transition; Lo, low; Hi, high.
We used flow cytometry to validate the presence of microglia subtypes in vivo. We focused on dividing/migrating microglia due to their distinct molecular signatures as compared with homeostatic microglia (Fig. 3 C). Microglia were gated on P2RY12 and MSR1 expression based on our sequencing data (Fig. 3, E and F). As expected, >90% of microglia present in the uninjured spinal cord were in the homeostatic state (P2RY12hi/MSR1lo), and this decreased to 20% at 1 dpi (Fig. 3 F). Similar to our sequencing data, uninjured samples also contained a small population of P2RY12lo/MSR1lo inflammatory microglia, which most likely resulted from dissociation-induced stress as mentioned above. At 1 dpi, there was a large increase in MSR1hi microglia, consistent with the appearance of dividing/migrating microglia. However, the majority of MSR1hi microglia at 1 dpi were also P2RY12hi, likely representing a transition state between homeostatic and nonhomeostatic microglia, which are expected to be P2RY12lo. At 7 dpi, we observed a significant decrease in P2RY12hi/MSR1hi transition microglia and a partial return of homeostatic microglia. Taken together, our flow cytometry data support the appearance of several nonhomeostatic microglia subtypes after SCI in vivo, but the temporal effects were more graded than those predicted from our sequencing data, perhaps due to a delay in manifestation of gene expression changes at the protein level.
The peripherally derived myeloid clusters revealed monocytes and two macrophage subtypes in addition to border-associated macrophages (Van Hove et al., 2019) and dendritic cells (Fig. 2 A). The two macrophage subtypes were named chemotaxis-inducing macrophages and inflammatory macrophages based on their GO biological processes terms (Fig. 4 A). GO terms for chemotaxis-inducing macrophages were associated with chemotaxis of other leukocytes such as neutrophils and granulocytes. GO terms for inflammatory macrophages were associated with glial and macrophage activation and inflammatory response. Neutrophils, monocytes, and chemotaxis-inducing macrophages were the predominant populations at 1 dpi, whereas inflammatory and chemotaxis-inducing macrophages were the predominant populations at 7 dpi (Fig. 4 B). Both subtypes expressed the lysosomal gene Cd63 but were distinguished by preferential expression of heme oxygenase Hmox1 in chemotaxis-inducing macrophages and Apoe in inflammatory macrophages (Fig. 4 C). A heatmap of the top DEGs shows both distinct and overlapping molecular signatures in the monocyte-macrophage lineage (Fig. 4 D). The two macrophage subtypes did not correspond to the M1/M2 genetic nomenclature (Murray, 2017). They both expressed low levels of M1 marker genes, and when all myeloid cells were hierarchically clustered based on these marker genes, inflammatory and chemotaxis-inducing macrophages clustered together, indicating that they could not be distinguished from each other based on traditional M1 classification (Fig. 4 E). Although they clustered separately using M2 marker genes, this was driven primarily by higher expression of Chil3 and Arg1 in chemotaxis-inducing macrophages (Fig. 4 E). The higher expression of the anti-inflammatory Arg1 in chemotaxis-inducing macrophages was consistent with GO terms associated with inflammation and glial activation being more prevalent in inflammatory than in chemotaxis-inducing macrophages (Fig. 4 A). In conclusion, our data reveal the presence of multiple cellular states in the monocyte-macrophage lineage that display temporal progression toward a more pro-inflammatory state.
Figure 4.
Molecular and temporal profile of monocyte/macrophage subtype heterogeneity acutely after SCI. (A) GO enrichment analysis of the DEGs distinguishing the two macrophage subtypes. GO biological process terms displayed along y axis and –log10 (Bonferroni-corrected P values) displayed along x axis. (B) Proportion of each peripheral myeloid subtype among all peripheral myeloid cells at each time point. (C) Dot plot of marker genes that differentiate peripheral myeloid subtypes. Color of dots represents z-scored expression level, and size of dots represents percentage of cells with at least one UMI detected per gene. (D) Heatmap of the highest DEGs per peripheral myeloid subtype. Color represents z-score expression level. (E) Heatmaps of M1 marker gene (left) and M2 marker gene (right) expression among all myeloid subtypes show that neither inflammatory nor chemotaxis-inducing subtypes exclusively display M1 or M2 gene signatures. A notable exception is higher expression of M2 markers Chil3 and Arg1 in monocytes and chemotaxis-inducing macrophages compared with inflammatory macrophages. Dendrograms along axes display hierarchical clustering results. Color of tiles represents z-scored expression value. Inset stars in C–E indicate statistically significant greater expression compared with all other cells combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction; see Materials and methods). Prop., proportion; Mac, macrophage.
To validate the presence of chemotaxis-inducing and inflammatory macrophage subtypes in vivo, we performed immunohistochemistry using antibodies against CD63 and APOE to spatially validate our results (Fig. 5 A). While CD11b+ myeloid cells were present at all time points, CD11b+/CD63+ cells started to appear by 3 dpi. By 7 dpi, many CD11b+/CD63+ cells that were either APOElo (chemotaxis-inducing) or APOEhi (inflammatory) were present at the lesion core. Since graded expression based on fluorescence intensity is difficult to quantify using immunohistochemistry, we used flow cytometry to quantify the relative proportion of these two macrophage subtypes. After separating monocytes/macrophages from other leukocytes including microglia (see Materials and methods), we isolated macrophages based on CD63hi expression (Fig. 5 B). Further separation on APOE and CD11b expression revealed two distinct clusters that were consistent with chemotaxis-inducing (CD63hi/CD11bmed/APOElo) and inflammatory (CD63hi/CD11bhi/APOEhi) subtypes (Fig. 5 B). Similar to our sequencing data, the CD63− myeloid cells were the predominant population at 1 dpi, and decreased by 7 dpi, whereas inflammatory macrophages (APOEhi) were the most represented macrophage subtype at 7 dpi (Fig. 5 C). Therefore, both immunohistochemistry and flow cytometry data support the molecular identification of chemotaxis-inducing and inflammatory subtypes and their temporal progression after SCI in vivo. In summary, analysis of myeloid cells reveals novel subtypes of microglia and macrophages during SCI progression.
Figure 5.
In vivo validation of chemotaxis-inducing and inflammatory macrophages after SCI. (A) Immunohistochemical validation of chemotaxis-inducing and inflammatory macrophages and their temporal changes after SCI. Injured spinal cord tissue shows a steady increase in macrophages (CD11b+/CD63+) over time. APOE expression differentiates between chemotaxis-inducing (CD11b+/CD63+/APOElo) and inflammatory (CD11b+/CD63+/APOEhi) macrophages. Chemotaxis-inducing macrophages are identified by white arrowheads, while inflammatory macrophages are identified by red arrowheads. Boxed regions on the left are shown in higher magnification on the right. Representative images from a total of three biological replicates per each time point. Scale bars in merged images are 300 µm, and scale bars in magnified images are 25 µm. Blue is DAPI to label nuclei. (B) Scatter plot of flow cytometry analysis and gating strategy (see Materials and methods for description) to quantify relative proportions of the two macrophage subtypes. After excluding neutrophils and lymphocytes, monocytes/macrophages were separated from microglia based on CD11b and CD45. After gating for CD63hi macrophages, separation based on CD11b and APOE resulted in CD11bmed/APOElo (chemotaxis-inducing macrophages) and CD11bhi/APOEhi (inflammatory macrophages) clusters. (C) Quantification showed increased proportion of CD11bhi/APOEhi macrophages at 7 dpi compared with 1 dpi. n = 5 biological replicates at each time point for flow cytometry experiment. Macro, macrophage; Mono, monocyte.
Vascular heterogeneity analysis identifies tip cell dynamics
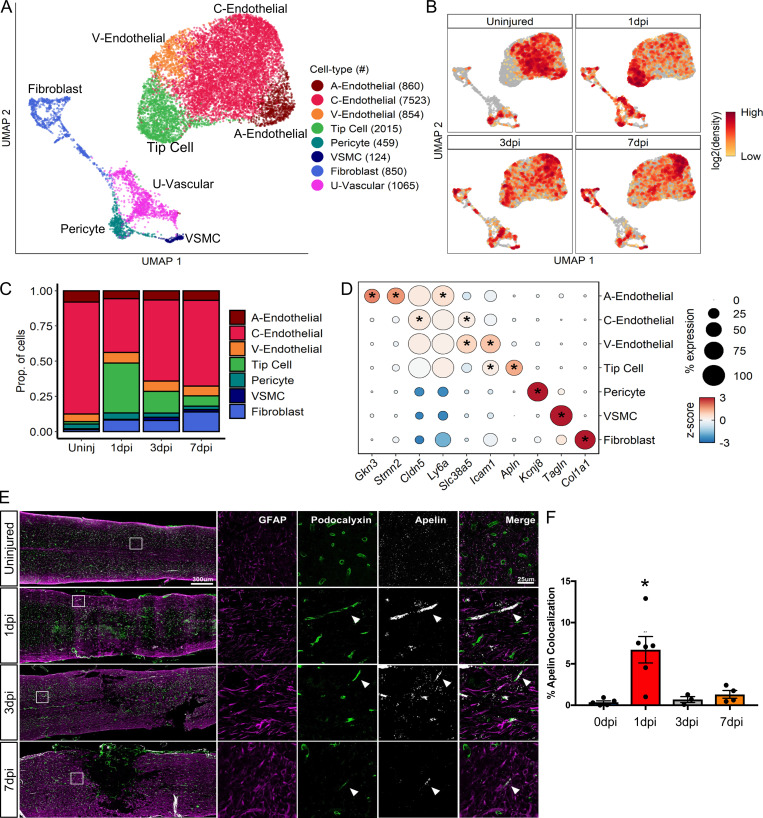
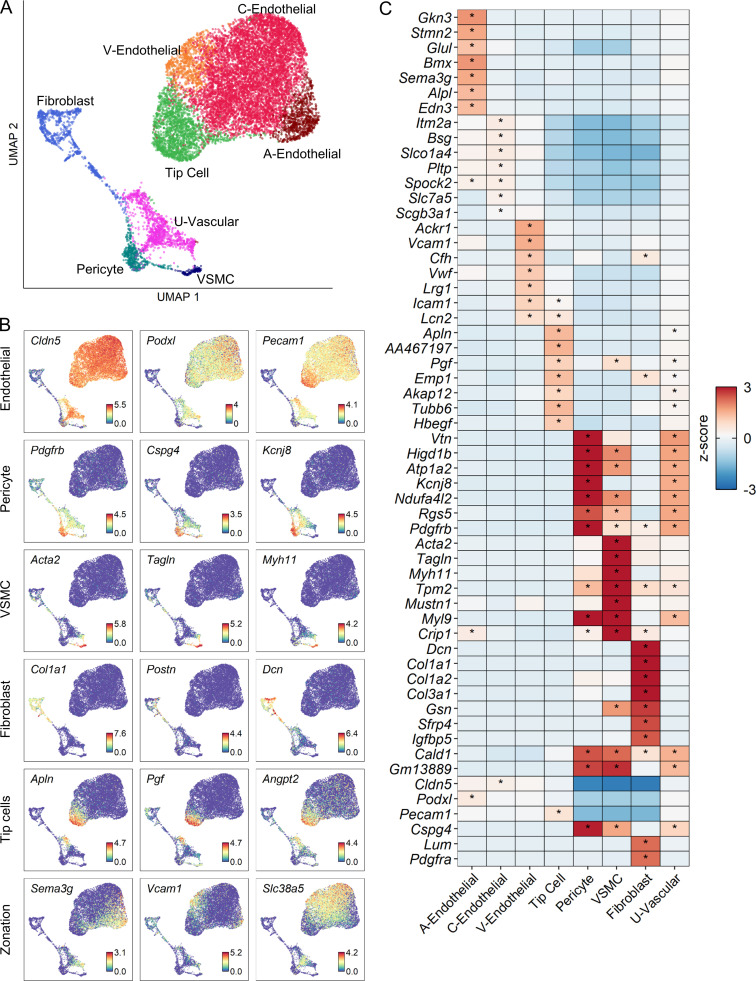
To better understand the heterogeneity of vascular cells, we performed further cluster analysis of endothelial cells, fibroblasts, and pericytes and identified nine distinct subtypes of vascular cells. UMAPs of vascular subtypes revealed a coarse division between endothelial and mural cells and displayed specific temporal progression (Fig. 6, A and B). We annotated each cluster using marker genes from a previous scRNA-seq study of the brain vasculature (Vanlandewijck et al., 2018). We identified fibroblasts, pericytes, and vascular smooth muscle cells (VSMCs) as three distinct populations of perivascular mural cells that were identified based on their expression of Col1a1, Kcnj8, and Acta2, respectively (Fig. S3, A and B). We also identified an unknown vascular subtype (U-vascular) that clustered with mural cells due to its molecular similarity with pericytes, but also expressed endothelial cell markers (Fig. S3, A and C), similar to a cell type described previously (Vanlandewijck et al., 2018; Zeisel et al., 2018). To determine whether the U-vascular cells represented capture of two different cell types during single-cell dissociation (i.e., doublet), we compared doublet likelihood scores (see Materials and methods) and found that this was unlikely.
Figure 6.
Molecular and temporal profile of vascular cell subtype heterogeneity acutely after SCI.(A) UMAP plot of all vascular cells from uninjured spinal cord and 1, 3, and 7 dpi. Fibroblasts, endothelial cells, and pericytes (Fig. 1 A) were extracted, reclustered, and re-embedded in new UMAP coordinates. Cells are colored by vascular subtype as shown in the legend on the right. Cell counts in parentheses. Subtypes were annotated using a combination of DEGs, canonical marker genes, and previously published data (Fig. S3). (B) UMAP of vascular cells split by each time point. Cells are colored by neighborhood density in UMAP space to illustrate shifts over time. Darker red colors indicate greater cell density in the UMAP or gray if not from indicated time point. (C) Proportion of each vascular subtype at each time point among all vascular cells except for a U-vascular subtype. (D) Dot plot of marker genes that differentiate vascular subtypes. Color of dots represents z-scored expression level, and size of dots represents percentage of cells with at least one UMI detected per gene. Inset stars indicate statistically significantly greater expression compared with all other cells combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction; see Materials and methods). (E) Histological validation of the temporal progression of tip cells at the injury site. Uninjured, 1, 3, and 7 dpi spinal cord sections were assessed by immunohistochemistry for podocalyxin (green) to label endothelial cells and for GFAP (purple) to label astrocytes and delineate the injury site. In situ hybridization was used for apelin mRNA (Apln; white puncta) to label tip cells. White box represents the corresponding region of the magnified images on the right. Arrowheads identify tip cells that coexpress podocalyxin and apelin. (F) Quantification of histological analysis represented as the average percent area of apelin and podocalyxin colocalization relative to the entire podocalyxin-positive area. Dots represent individual biological replicates. n = 3–6 mice per time point. Error bars, SEM. *, P < 0.05 compared with all other time points; one-way ANOVA with Bonferroni multiple comparisons. A, arterial; C, capillary; Prop., proportion; Uninj, uninjured; U, unknown; V, venous; VSMC, vascular smooth muscle cell.
Figure S3.
Molecular profile of vascular cells after SCI.(A) UMAP representation of clusters from all vascular cells combining all time points. (B) Expression pattern of annotated marker genes used to identify each vascular cluster. The last row of UMAPs shows the expression of molecular markers used differentiate between arterial, venous, and capillary endothelial cells as previously defined (Vanlandewijck et al., 2018). Endothelial cells on the top row refer to all subtypes except tip cells, which are represented separately. (C) Heatmap of the top DEGs for each vascular subtype. Color of tile indicates z-scored expression and inset stars indicate statistically significant greater expression compared with all other cells combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction). A, arterial; C, capillary; V, venous.
Next, we identified an arterial, a venous, and a capillary subtype based on annotated markers (Vanlandewijck et al., 2018). Arterial endothelial cells were identified by expression of Gkn3 and Stmn2, whereas venous endothelial cells were identified by Vwf and Vcam1. Capillary endothelial cells were identified by the expression of general endothelial cell markers Ly6a and Cldn5, combined with the lack of selective arterial and venous markers (Fig. 6 D and Fig. S3). The fourth endothelial cluster was identified as tip cells based on their expression of the canonical marker Apln. Tip cells are leading cells present during vasculogenesis and angiogenesis, and they have not yet been systematically assessed in SCI. Tip cells were absent in the uninjured spinal cord, but increased considerably at 1 dpi, and gradually decreased by 7 dpi (Fig. 6 C). This tip cell temporal profile was validated using in situ hybridization for Apln mRNA combined with immunostaining for podocalyxin as an endothelial cell marker (Fig. 6 E). In the uninjured cord, Apln transcripts were detected scattered throughout the gray matter, consistent with previous reports of Apln expression in neurons (Reaux et al., 2002). After SCI, dense areas of Apln transcripts were detected in elongated podocalyxin-positive cells near the injury site. These tip cells were most abundant at 1 dpi, and decreased significantly by 3 dpi (Fig. 6 F). Taken together, our analysis identified all known major vascular cell types at the injury site, including previously undescribed tip cell molecular and temporal profiles.
Cellular interactions via angiopoietin (Angpt) and VEGF signaling during angiogenesis
To gain insight into mechanisms of cellular interactions during angiogenesis after SCI, we adapted CellPhoneDB (Vento-Tormo et al., 2018) to calculate “interaction scores” based on the average expression levels of a ligand and its receptor between two cells (Fig. 7 A). Higher interaction scores indicate stronger predicted interactions between two cells via the specified ligand–receptor pathway. We used a database of all known ligand–receptor pairs (Ramilowski et al., 2015) to determine putative interactions between endothelial/tip cells and other cell types at the injury site in an unsupervised manner. Analysis of the uninjured spinal cord showed expected canonical vascular signaling pathways such as Vegfb and Angpt1 (data not shown). We focused on 1 dpi interactions since tip cells were most abundant at this time point, and we included subtype information only for myeloid and oligodendrocyte lineage cells. Interaction plots were generated separately for myeloid, vascular, and macroglia cell types (Fig. 7, B–D). Endothelial and tip cells shared largely similar patterns of interaction scores with other cell types. A notable difference was strong Spp1-Itgav signaling that was specific between chemotaxis-inducing macrophages and tip cells (Fig. 7 B). While the predicted interactions included several canonical angiogenesis pathways such as Vegf, Notch, and Angpt signaling, our analysis revealed several unexpected pathways in the context of CNS injury such as Plgf (placental growth factor), which was present across most vascular and macroglia cell types, and Trf (transferrin), which was highly specific to oligodendrocytes (Fig. 7, C and D). Furthermore, while certain interaction pairs, such as the ADAM–integrins, were shared among all three compartments, others, such as the collagen–integrins, were present only for vascular cells and macroglia but not for myeloid cells.
Figure 7.
Ligand–receptor analysis to assess endothelial and tip cell interactions with other cell types after SCI. (A) Overview of ligand–receptor analysis method. Interaction scores were calculated as the mean of the average log-normalized expression of receptor gene in one cell type and the average log-normalized expression of ligand gene in a second cell type. To test for enrichment in specific pairs, we used a permutation test and compared scores against these random values (see Materials and methods). (B) Dot plot of the interaction scores at 1 dpi between select myeloid subtypes as the ligand-expressing cell (top axis) and endothelial/tip cells as the receptor-expressing cells (bottom axis). Specific ligand–receptor pairs are listed along left axis. Size of the dot indicates –log10(P value). Color of dot indicates interaction score where dark red dots signify stronger predicted interactions. (C) Dot plot of the interaction scores at 1 dpi between vascular subtypes and endothelial/tip cells. (D) Dot plot of interaction scores at 1 dpi between select macroglial subtypes (see Fig. 8) and endothelial/tip cells. (E) Dot plot of expression of Angpts (left of dotted line) and their receptors (right of dotted line) in cell types across all time points. (F) Dot plot of expression of VEGF family member ligands (left of dotted line) and their receptors (right of dotted line) in cell types across all time points. In E and F, color of dots indicates z-scored expression, and size of dot indicates percentage of cells with at least one UMI detected per gene. Inset stars indicate statistically significant greater expression compared with all other cells combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction; see Materials and methods). Div, dividing; Oligo, oligodendrocyte; OPC, oligodendrocyte progenitor cell; VSMC, vascular smooth muscle cell.
We focused on Angpt and vascular endothelial growth factor (Vegf) signaling due to their well-known roles in angiogenesis (Lee et al., 2009). Angpt1 is an agonist that promotes stabilization, whereas Angpt2 is an antagonist that promotes destabilization of blood endothelium by binding to the Tie2 receptor (Eklund et al., 2017). Interestingly, Angpt2–Tie2 interaction was only present among vascular cells at 1 dpi (Fig. 7 C). Expression analysis showed that Angpt2 is expressed most highly by tip cells at 1 dpi followed by a gradual decrease over the next 7 d, and Tie2 (and Tie1) is expressed in endothelial cells at all time points (Fig. 7 E). VSMCs and fibroblasts also contributed to this signaling interaction with endothelial cells, albeit at a lower level. Interestingly, Angpt1 expression shifted from VSMC at 1 dpi to astrocytes at 3 and 7 dpi (Fig. 7 E), suggesting that astrocytes are important in vessel stabilization. Collectively, our data provide putative interactions between vascular and neural cells with endothelial cells via Angpt signaling after SCI. These data lead to the potential hypothesis that tip cells promote vessel destabilization during early angiogenesis, and astrocytes mediate vessel stabilization at later periods.
Vegfa binding to Vegfr1 and Vegfr2 on endothelial cells facilitates the proliferation, survival, and directional sprouting of tip cell filipodia during angiogenesis (Apte et al., 2019; Gerhardt et al., 2003; Reinert et al., 2014). High Vegfa interaction scores were present for many cell types across myeloid, vascular, and macroglia categories (Fig. 7, B–D). Vegfa expression was highest in astrocytes before injury and at 1 dpi, and Vegfr1 and Vegfr2 receptors were highly expressed by endothelial cells at all time points. These data may lead to the hypothesis that major cues for new vessel formation are derived from multiple cell types including infiltrating myeloid as well as resident neural cells (Fig. 7 F). Strikingly, the strongest interactions among the Vegf family members after SCI were associated with Plgf binding to Vegfr1 (Fig. 7, C and D). PLGF mediates angiogenesis by binding to VEGFR1 (Nagy et al., 2003; Ribatti, 2008), and our expression analysis showed tip cells and VSMC to be the primary source, although lower levels of Plgf were also expressed by fibroblasts and astrocytes (Fig. 7 F). Similar to our Angpt analysis above, our Vegf analysis may lead to the potential hypothesis that tip cells play a major role in facilitating their own migration and growth, as well as those of other endothelial cells. Collectively, our data provide a rational and temporal framework of how endothelial cells interact with multiple cell types via Angpt and Vegf pathways to revascularize the injured tissue, and highlight the autocrine and paracrine effects of tip cells in this process. However, it is important to note that these interaction scores provide predictive models, rather than final experimental findings, which can be used to generate hypotheses to be tested in the future.
Analysis of macroglia heterogeneity reveals their molecular relationships during gliosis
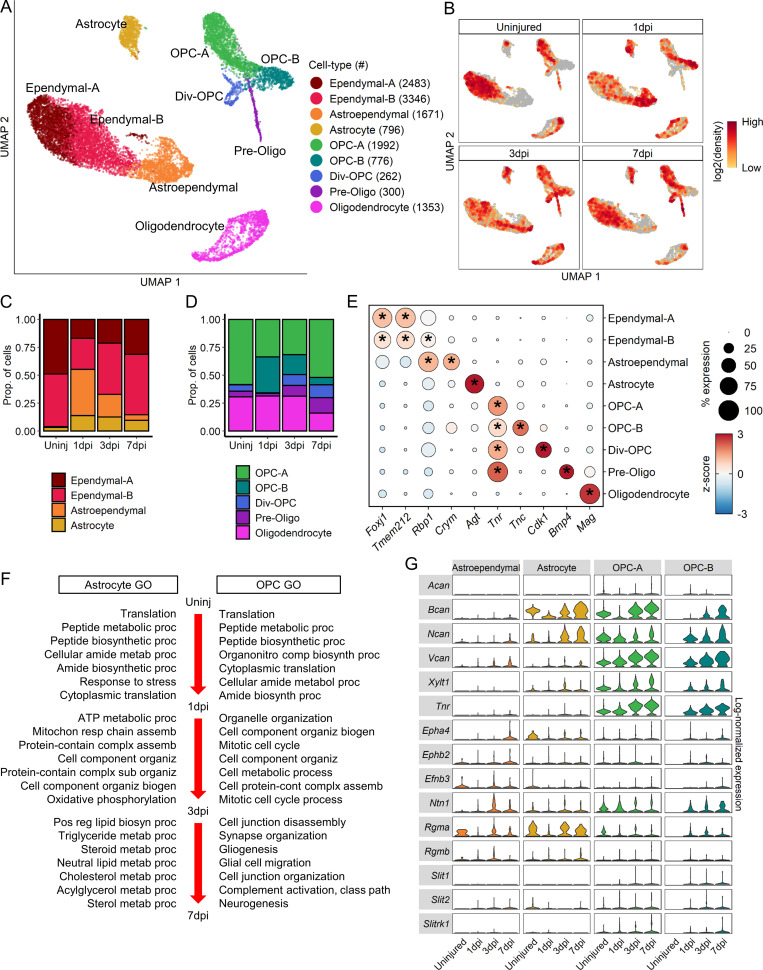
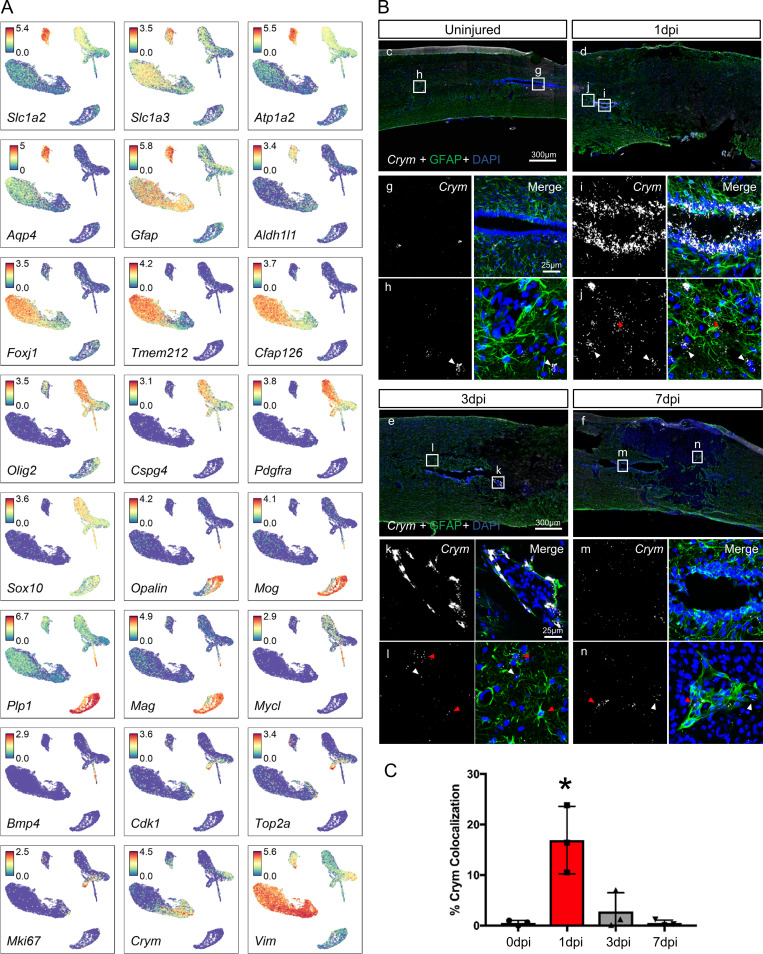
To assess macroglial heterogeneity, oligodendrocyte lineage cells, astrocytes, and ependymal cells were further clustered into nine distinct clusters that were identified based on annotated marker genes (Fig. 8 A and Fig. S4 A). UMAP of macroglia clusters revealed notable temporal progression after injury (Fig. 8 B). We identified two ependymal and an astroependymal subtypes. The two ependymal subtypes did not express molecular markers distinct from each other (Fig. 8 E), and displayed only slight differences in top DEGs (Fig. S5 A) suggesting that they are likely a single population. However, astroependymal cells displayed unique features and a notable temporal progression. Astroependymal cells were absent in the uninjured spinal cord, but increased considerably at 1 dpi and then decreased gradually by 7 dpi (Fig. 8, B and C). They were best identified by expression of the crystallin Crym, and shared common markers with both astrocytes (e.g., Timp1 and Gfap) and ependymal cells (e.g., Vim and Tm4sf1; Fig. 8 E and Fig. S4 A). However, they did not express any cilia-associated genes that are hallmarks of ependymal cells (Fig. S5 A). Consistent with the sequencing data, in situ hybridization for Crym showed highest expression at 1 dpi in central canal cells near the injury site, which decreased significantly by 3 dpi (Fig. S4, B and C). Gliotic regions around the injury site included GFAP+ cells that also expressed Crym (Fig. S4 B).
Figure 8.
Molecular and temporal profile of macroglial heterogeneity acutely after SCI. (A) UMAP plot of all macroglial cells from uninjured spinal cord and 1, 3, and 7 dpi. Ependymal cells, astrocytes, OPCs and oligodendrocytes (Fig. 1 A) were extracted, reclustered, and reembedded in new UMAP coordinates. Cells are colored by macroglia subtype as shown in legend on right. Cell counts in parentheses. Subtypes were annotated using a combination of DEG, GO enrichment analyses, canonical markers, and transcription factor expression (Figs. S4 and S5). (B) UMAP of macroglial cells split by each time point. Cells are colored by neighborhood density in UMAP space to illustrate shifts over time. Darker red colors indicate greater cell density in the UMAP or gray if not from indicated time point. (C) Proportions of subtypes among astrocytes and ependymal cells and (D) proportions of oligodendrocyte-lineage subtypes at each time point. (E) Dot plot of marker genes that differentiate macroglial subtypes. Color of dots represent z-scored expression level and size of dots represent percent of cells with at least one UMI detected per gene. Inset stars indicate statistically significant greater expression compared with all other cells combined (adjusted P value < 10−10, Wilcox rank sum with Bonferroni correction; Materials and methods). (F) GO biological process terms associated with the top DEGs between time points for astrocytes (left) and OPCs (right). OPC-A and OPC-B were combined for this analysis. (G) Violin plot of expression of common axon growth inhibitors between astroependymal cells, astrocytes, and OPC subtypes. Expression levels are log-normalized counts along right axis. Assemb, assembly; Biogen, biogenesis; Div, dividing; Oligo, oligodendrocyte; OPC, oligodendrocyte progenitor cell; Organiz, organization; Proc, process; Prop., proportion; Uninj, uninjured.
Figure S4.
Molecular profile of macroglia cells after SCI.(A) Individual UMAP representations of the expression pattern of previously described marker genes used to annotate each macroglia cluster. (B) In situ hybridization (Crym in white) and immunohistological (GFAP in green) validation of astroependymal cell formation in response to injury. Boxes depict the location of higher magnification images shown below. Images in g, i, k, and m were taken from the central canal. Images shown in h, j, l, and n were taken outside of the central canal. White arrowheads denote DAPI+/Crym+ cells, and red arrowheads denote DAPI+/Crym+/GFAP+ cells. DAPI is in blue labeling cell nuclei. GFAP-negative region in injured tissue represents the lesion. (C) Quantification of Crym+ astroependymal cells in the central canal as defined by the percentage of DAPI+ nuclei that colocalize with Crym+ in situ signal surrounding the central canal. *, P < 0.05 compared with all other groups using one-way ANOVA with Tukey’s post-test. n = 3 biological replicates per time point.
Figure S5.
Molecular comparison of macroglia subtypes. (A) Heatmap of top DEGs in macroglia subtypes that are grouped according to hierarchical clustering. Pairwise differential expression tests between macroglia subtypes were done to identify shared and unique gene signatures. Color represents z-scored expression. (B) Expression levels of transcription factors commonly associated with specific glia point to potential lineage relationships among known and novel macroglia subtypes. Values shown are log-normalized counts. (C) GO biological process terms associated with the top DEGs between OPC-A and OPC-B at 1 dpi. Div, dividing; Oligo, oligodendrocyte.
The oligodendrocyte lineage cells segregated into expected clusters that showed a spatial and temporal progression from OPC to dividing OPC and preoligodendrocytes with mature oligodendrocytes forming their own cluster (Fig. 8, A and B). We identified two OPC clusters (OPC-A and -B) that displayed distinct temporal and molecular features. Whereas OPC-A were present in abundance at all time points, likely representing prototypical OPCs, OPC-B appeared only after injury, with numbers peaking at 1 dpi and decreasing significantly by 7 dpi (Fig. 8 D). OPC-A expressed canonical OPC genes such as Cspg4 and Pdgfra (Fig. S4 A) but was best distinguished from other non-OPC cell types by its expression of Tnr (Fig. 8 E). OPC-B was distinguished from OPC-A by its expression of tenascin-C (Tnc) and lower expression levels of Cspg4 and Pdgfra (Fig. 8 E and Fig. S4 A). GO analysis of DEGs between OPC-A and OPC-B showed biological processes associated with neural development and myelination in OPC-A, and cell growth and proliferation in OPC-B (Fig. S5 C). Interestingly, OPC-B had several features in common with astroependymal cells, including its temporal progression, and expression of Crym and Vim at lower levels (Fig. 8 E and Fig. S4 A). In fact, hierarchical clustering of the top DEGs showed OPC-B, astroependymal cells, and astrocytes forming their own cluster, indicating the shared similarities between these subtypes (Fig. S5 A). In addition, transcription factor expression showed Ascl1 in OPC-A but Sox9 expression in OPC-B (Fig. S5 B). Taken together, our data highlight the heterogeneity and potential lineage relationships between OPCs, astrocytes, and ependymal cells that reflect the effect of SCI on their differentiation state.
Although gliosis has been synonymous with reactive astrocytes, accumulating evidence indicates that OPCs are also an important component of the glial scar (Hackett et al., 2016; Hackett and Lee, 2016). To assess changes in OPC and astrocyte function during injury progression, we tested for DEGs between each sequential time point for each cell type and performed GO enrichment analysis for biological processes. (Fig. 8 F). At 1 dpi, top biological processes for both astrocytes and OPCs pertain to translation and biosynthetic processes. By 3 dpi, astrocytes are defined by processes related to mitochondrial function and oxidative phosphorylation, whereas OPCs are defined by mitosis and cell proliferation. DEGs between 3 and 7 dpi in astrocytes were related to lipid processing, whereas those in OPCs were related to glial cell development, differentiation, and migration. To assess the potential effects of reactive astrocytes and OPCs on axonal growth, we compared the expression levels of axon growth inhibitory molecules using a previously curated list (Anderson et al., 2016; Fig. 8 G). Interestingly, inhibitory proteoglycans such as Acan, Bcan, Ncan, and Vcan were expressed preferentially by OPC-A and -B compared with astrocytes or astroependymal cells. In summary, our data suggest that although astrocytes and OPCs initially display similar generic responses to injury, by 7 dpi, they acquire unique functional identities, and many processes that have traditionally been attributed to reactive astrocytes are also present in OPCs.
Myeloid-mediated mechanisms of gliosis and fibrosis
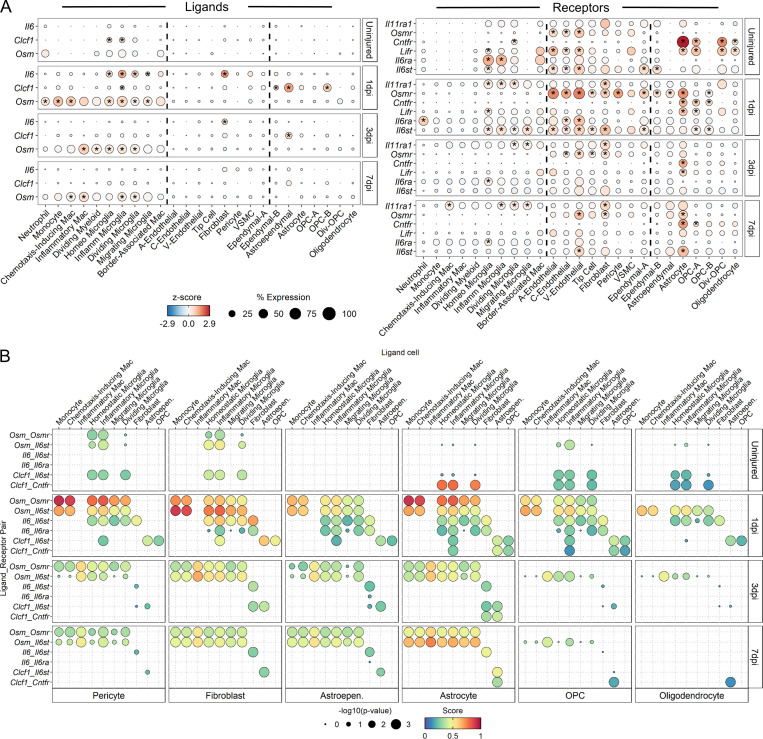
Previous studies have shown that STAT3 plays an important role in astrogliosis (Bonni et al., 1997; Herrmann et al., 2008; Wanner et al., 2013), but its upstream activators have yet to be clearly defined. Since IL-6 cytokine family members are the main STAT3 activators, we assessed their expression across all cell types and found oncostatin M (Osm) expressed at highest levels in myeloid cells, Il6 expressed at highest levels in fibroblasts, and Clcf1 expressed at highest levels in astroependymal cells and OPC-B (Fig. 9 A). Other IL6 cytokines were either not expressed highly or dropped out of our sequencing analysis. Osm receptor (Osmr) and the signaling coreceptor gp130 (Il6st) were expressed highly in fibroblasts, pericytes, and astrocytes, and interaction scores for IL-6 cytokine family members were highest for OSM signaling between myeloid cells and these three cell types (Fig. 9 B). To validate the sequencing data, we used 7 dpi injury site tissue to perform double in situ hybridization for Osm and Itgam (i.e., CD11b) and found a significant increase in Osm in Itgam+ myeloid cells compared with Itgam−non-myeloid cells (Fig. 10, A and C). To assess OSMR expression, we used immunohistochemistry in adjacent sections and found increasing expression of OSMR in both PDGFRβ+ fibroblasts and GFAP+ astrocytes in the fibrotic and glial scars, respectively (Fig. 10, B, D, and E). Taken together, our results lead to the hypothesis that the gp130 signaling pathway induced by OSM is a common mechanism by which astrocytes and fibroblasts are preferentially activated by myeloid cells after SCI.
Figure 9.
Ligand–receptor analysis of IL-6 cytokine family members identify oncostatin M signaling between myeloid cells and astrocytes/fibroblasts. (A) Dot plot of expression of IL-6 family member ligands (left) and their receptors (right) in previously identified cell subtypes across injury time points. In each panel, dotted lines demarcate myeloid (left column), vascular (middle column), or macroglial (right column) subtypes. Color of dots represents z-scored expression, and size of dots represents percentage of cells with at least one UMI detected per gene. Gene members of the IL-6 cytokine family that were not expressed in at least 10% of any subtype at any time point were excluded. Inset stars indicate statistically significant greater expression compared with all other cells combined (adjusted P value < 10−10; Wilcoxon rank-sum with Bonferroni correction; see Materials and methods). (B) Dot plot of ligand–receptor interaction scores of IL-6 family member signaling shows highest predicted interactions between myeloid cells and astrocytes/fibroblasts via onctostatin M signaling. Size of dot represents –log10(P value), and color represents interaction score where dark red colors indicate greater predicted interactions. Specific IL-6 family ligand–receptor pairs are listed along left axis. Ligand-expressing cells are listed across top axis. Receptor-expressing subtypes are listed across bottom axis. Select subtypes with greatest number of significant scores are shown. A, arterial; Astroepen., astroependymal; C, capillary; Div, dividing; Homeo, homeostatic; Inflamm, inflammatory; Mac, macrophage; OPC, oligodendrocyte progenitor cell; V, venous.
Figure 10.
Validation of Osm and Osmr expression and ligand–receptor analysis of other potential interactions between macrophages and astrocytes/fibroblasts.(A) Left column: Histological validation of Osm expression in myeloid cells at all time points after injury. Spinal cord sections were assessed by in situ hybridization for Itgam mRNA (also known as CD11b; green puncta) to label myeloid cells and Osm mRNA (white puncta). DAPI (blue) denotes cell nuclei. White arrowheads indicate CD11b+/Osm+ nuclei. Dotted white box represents the corresponding region of the magnified images on the right. Right column: Histological validation of oncostatin M receptor expression in astrocytes and fibroblasts at all time points after injury. Spinal cord sections were assessed by immunohistochemistry for GFAP (red) to label astrocytes, PDGFRβ (green) to label fibroblasts, and OSMR (white). Scale bar, 15 µm. (B) Quantification of Osm expression in myeloid (DAPI+/Itgam+/Osm+) and nonmyeloid (DAPI+/Itgam−/Osm+) cells shows significantly higher Osm expression in myeloid cells. (C and D) Quantification showed increasing colocalization of OSMR with PDGFRβ (C) and GFAP (D) immunoreactivity over time. Error bars represent SEM. *, P < 0.05 compared with uninjured tissue. One-way ANOVA and Tukey’s post hoc test. Each data point represents a biological replicate. (E) Dot plot of interaction scores for select ligand–receptor pairs at 3 and 7 dpi. Data are segregated to show interactions unique between macrophages and astrocytes (red bracket on top), unique between macrophages and fibroblasts (green bracket on bottom), or common between astrocytes and fibroblasts with macrophages (blue bracket). Macrophage interactions were split further by chemotaxis-inducing (Chemo) or Inflammatory (Inflam) macrophage subtypes. Astro, astrocyte; Fibro, fibroblast; Macro, macrophage; uninj, uninjured.
To identify other putative pathways by which macrophage subtypes mediate astrogliosis and fibrosis, we used a database of all known ligand–receptor pairs (Ramilowski et al., 2015) to calculate interaction scores between chemotaxis-inducing/inflammatory macrophages and astrocytes/fibroblasts at 3 and 7 dpi (Fig. 10 E). While many ligands expressed by macrophages signaled to receptors on both astrocytes and fibroblasts (i.e., common pathways), there were many more ligand–receptor pairs unique to macrophage–fibroblast interactions than macrophage–astrocyte interactions (i.e., distinct pathways). These unique macrophage–fibroblast interactions included signaling related to IL1α/β, Vegfa/b, Pdgfa, and Tgfβ1. Overall, the highest interaction scores were associated with Spp1 and Apoe signaling, which were common to both astrocytes and fibroblasts. In general, chemotaxis-inducing and inflammatory macrophage subtypes displayed interaction patterns similar to those of astrocytes and fibroblasts. In summary, our analysis highlights the utility of our scRNA-seq dataset in identifying putative signaling mechanisms that can be hypothesized to mediate astrogliosis and fibrosis after SCI.
Discussion
The overall goal of this study was to generate a detailed transcriptomic profile at the single-cell level in order to gain insight into the complex cellular heterogeneity and interactions that occur at the SCI site. We were successful in isolating every major cell type known to be present at the injury site, which provided an opportunity to assess the wound healing process with much less bias than prior studies that focused on specific cell types and molecules. In this regard, we found that myeloid subtypes display distinct temporal regulation of angiogenesis, gliosis, and fibrosis. These results support previous studies showing that myeloid depletion leads to reduced angiogenesis, astrogliosis, and fibrosis after CNS injury (Amankulor et al., 2009; Bellver-Landete et al., 2019; Zhu et al., 2015), and provide new molecular and cellular details by specific myeloid subtypes. Although there are species-specific features of these pathological processes (e.g., mice have more fibrosis and less cavitation compared with rats and humans), these differences appear at time points after those investigated in this study. During the first 7 d after SCI, mice and rat injury sites are mostly similar in terms of histopathology, which is one of the reasons we chose these particular time points.
It is important to note that while we were successful in isolating all major cell types present at the injury site, the cell recovery numbers do not accurately reflect the true number of cells that are present at the injury site (e.g., 930 astrocytes versus 7,717 ependymal cells). This is likely due to the dissociation process being more optimal for certain cell types than others, and is in line with our expectation that cells such as leukocytes, endothelial cells, and ependymal cells would survive the dissociation procedure better than more vulnerable neural cell types such as oligodendrocytes, OPCs, astrocytes, and, most of all, neurons. However, within a cell population, our data seem to portray a more accurate representation of the subpopulation proportions. For example, the percentages of microglia subpopulations are similar between our single-cell and flow cytometry data (Fig. 3, A and E). To consider potential confounds from tissue dissociation, we compared our data to previous SCI bulk RNA-seq data (Chen et al., 2013) and found that temporal changes in gene expression were in agreement with the appearance of all the injured microglia subsets and their molecular markers. For example, in the bulk RNA-seq data, expression of P2ry12 was decreased by 2 d post-injury and restored to uninjured levels by 7 dpi, whereas activated microglia genes such as Igf1, Spp1, Osm, and Msr1 remained elevated at 2 and 7 dpi compared with uninjured controls (data not shown). However, using bulk RNA-seq data to assess the effects of dissociation-induced stress in uninjured tissue is difficult because we do not know whether low FPKM values are due to basal expression levels that are not physiologically relevant or whether stress-induced gene expression was diluted by the bulk processing technique. Thus, we tried to address this issue using expression data from the Allen Brain Atlas and found very low expression of Igf1 (mostly in neurons), and almost undetectable expression of Il1b. Since Igf1 and Il1b were markers of the inflammatory microglia subcluster, this suggests that their presence in our uninjured samples may be due to tissue dissociation. However, it is important to note that the single-cell analysis allowed us to distinguish the true homeostatic microglia subcluster in the uninjured tissue, which was used for downstream analyses. This provides confidence that our analyses were not confounded by dissociation-induced artifacts. In addition, using in situ hybridization, a recent study has shown Igf1 as a molecular marker of activated microglia in the injured spinal cord (Bellver-Landete et al., 2019), suggesting that inflammatory microglia present in our injured samples are likely due to SCI rather than dissociation-induced stress. Last, although a very recent study reported expression of many immediate early genes due to dissociation-induced stress in microglia (Hammond et al., 2019), we detected only Fos among the DEGs in the inflammatory microglia.
We can begin to infer putative functions of the two macrophage subtypes by placing their temporal progression in context of prior studies. Our finding that inflammatory and chemotaxis-inducing macrophage subtypes do not correspond to the M1/M2 nomenclature (Fig. 4 E) is consistent with findings from other scRNA-seq studies (Lin et al., 2019; Müller et al., 2017; Ydens et al., 2020). In fact, we found that the commonly used M2 marker CD206 (Mrc1) was almost exclusively expressed by border-associated macrophages (Fig. 4 C), which adds complexity to the M1/M2 debate. However, there are parallels with the generally accepted view that macrophages shift to a more pro-inflammatory state over time after SCI (Kigerl et al., 2009; Kroner et al., 2014; Wang et al., 2015). As the major peripheral myeloid composition at the injury site shifts from monocytes to chemotaxis-inducing macrophages and then to inflammatory macrophages over time, there is a progressive decrease in the classic anti-inflammatory enzyme arginase 1 that is associated with increased pro-inflammatory biological processes in inflammatory macrophages. Despite these pro-inflammatory functions, our data identified several mechanisms by which macrophages may have beneficial effects on the wound healing process. For example, inflammatory macrophages express Vegfa, which is well-known to be necessary for growth and differentiation of endothelial cells during angiogenesis (Neufeld et al., 1999). In addition, macrophages are a major source of osteopontin (Spp1) and Apoe, both of which have neuroprotective roles after SCI. Spp1 and APOE knockout mice both display worse histopathology and behavioral recovery after SCI, whereas administration of APOE mimetic improves recovery (Hashimoto et al., 2007; Wang et al., 2014; Yang et al., 2018). Thus, a future challenge is to limit the pro-inflammatory macrophage state without interfering with macrophages’ beneficial effects on wound healing.
Microglia were notable for their increasing expression of Msr1 from inflammatory to migrating subtypes, as well as their expression of the neuroprotective growth factor Igf1 (Guan et al., 2015). We noted that dividing microglia expressed only moderate levels of Igf1. It may be that their identity as dividing cells makes them unique compared with the other two subtypes. Interestingly, a recent microglia depletion study demonstrated that they have a neuroprotective role by inducing astrogliosis via Igf1, and that Igf1-expressing microglia are located between the astroglial and fibrotic scars (Bellver-Landete et al., 2019). Although the three nonhomeostatic microglia subtypes showed a transcriptional decrease in P2ry12, there was some divergence in our flow cytometry data, which showed a large population of P2RY12hi/MSR1hi microglia at 1 dpi. Based on increased MSR1 expression, these are most likely dividing and migrating microglia, whose transcriptional decrease of P2ry12 has not yet manifested at the protein level. Indeed, by 7 dpi, these transitional dividing/migrating microglia are significantly decreased. However, this model does not account for the decreased P2ry12 expression in inflammatory microglia, which seem to be the earliest nonhomeostatic microglia state since they are present in uninjured tissue (most likely due to dissociation-induced stress; Wu et al., 2017). Therefore, it is possible that rather than a linear progression from inflammatory to dividing to migrating microglia, inflammatory and dividing/migrating microglia follow a divergent path of activation from homeostatic microglia.
The vascular analysis revealed novel insight into the contribution of tip cells and astrocytes during angiogenesis after SCI. The data indicate that tip cells are highly dynamic; they appear quickly at 1 dpi and are mostly gone by 7 dpi. They are a major source of Angpt-2, which destabilizes vessels, as well as Plgf, which promotes vessel formation, suggesting that tip cells mediate vessel remodeling during the few days after SCI. Interestingly, their disappearance overlaps with increased expression of Angpt-1 in astrocytes. Angpt-1 typically promotes vessel stabilization, suggesting that astrocytes contribute to vessel maturation after the initial phase of vascular remodeling by tip cells. This is corroborated by a previous study demonstrating that when the blood–spinal cord barrier is restored by 21 dpi, blood vessels closely associated with astrocytes in the glial scar region express Glut1, whereas vessels in the astrocyte-devoid fibrotic region do not express this mature blood–spinal cord barrier marker (Whetstone et al., 2003). These data highlight an understudied role of astrogliosis in vessel maturation after SCI, and provide new insight into prior astrocyte ablation studies that suggested “corralling” of infiltrating leukocytes by the astroglial scar (Faulkner et al., 2004; Wanner et al., 2013). Our data provide mechanistic insight into previous astrocyte ablation studies showing that reducing astrogliosis prevents maturation of blood vessels, which leads to increased infiltration of leukocytes into the spinal cord parenchyma surrounding the injury site (Sofroniew, 2015). Other possible mechanisms of interactions between astrocytes, vasculature, and axons need to be explored in the future.
The appearance of astroependymal cells in our data is notable due to previous studies showing that ependymal cells can differentiate into astrocytes and oligodendrocytes after SCI (Barnabé-Heider et al., 2010), although the extent of this differentiation is debated (Ren et al., 2017). Consistent with these previous findings, astroependymal cells were present only after injury, and expressed astrocyte markers such as Gfap as well as some overlapping markers with OPC-B. Hierarchical clustering of the macroglial subtypes showed astrocytes, astroependymal cells, and OPC-B in the same cluster (Fig. S5 A). In addition, GO analysis of DEG between OPC-A and OPC-B showed epithelial development genes enriched with OPC-B, which is notable since ependymal cells are the epithelial lining of the central canal. The transient appearance of astroependymal cells after SCI (1–3 dpi) as well as their lack of proliferation may explain their limited contribution to the glia population at the injury site, and raise the possibility of whether experimentally extending their presence would enhance the regenerative response after SCI.
Our scRNA-seq dataset is the first comprehensive transcriptomic analysis that captures virtually all cells that contribute to the injury site pathology acutely after SCI. This dataset can be used to assess not only heterogeneity of the cells that comprise the injury site but also signaling pathways that underlie cellular interactions at the injury site. Our own analysis revealed novel insight into myeloid cell heterogeneity, and specific signaling pathways by which unique myeloid subtypes contribute to the wound healing process, including angiogenesis and scar formation. These new insights can help decode the complex processes that underlie SCI pathobiology.
Materials and methods
Mice and SCIs
For sequencing, flow cytometry, and histological validation of tip cells, female C57BL/6J mice 8–10 wk of age were purchased from Jackson Laboratory (stock 000664). Injury sites (or corresponding segments in uninjured controls) were dissected for dissociation as described below. One sample from each time point consisted of cells from a single spinal cord. In all other samples, cells were pooled from two animals. In total, we sequenced three uninjured biological replicates (from five animals), three biological replicates at 1 dpi (from five animals), two biological replicates at 3 dpi (from three animals), and two biological replicates at 7 dpi (from three animals). For histological validation of activated fibroblasts, Postn-CreER mice (Jackson Laboratory; stock 029645) were bred to Rosa26-EYFP mice (Jackson Laboratory; stock 006148) to generate PostnEYFP mice. All PostnEYFP mice were of the C57BL/6 background. To induce EYFP expression, PostnEYFP mice were injected i.p. with 0.124 mg/g body weight of tamoxifen (MP Biomedicals; dissolved in 90% sunflower oil and 10% ethanol) for five consecutive days. Contusive SCIs were performed 7 d after the last tamoxifen injection.
Contusive SCIs for all mice were performed as previously described (Zhu et al., 2015). Briefly, mice were anesthetized using ketamine/xylazine (100 mg/15 mg/kg i.p.) and received a 65-kilodyne mid-thoracic (T8) contusion injury. Laminectomies were performed at the T8 vertebral level followed by stabilization via a spinal frame clamping the T7/T9 spinous processes. A computer-controlled contusion injury was delivered using the Infinite Horizon Impactor device (Precision Systems and Instrumentation, LLC). Injured mice were given post-operative fluids subcutaneously consisting of lactated Ringer’s solution (1 ml), gentamycin (5 mg/kg), and buprenorphine (0.05 mg/kg) twice daily for 7 d. Bladders were manually expressed twice daily until the end of the experiment. All procedures were in accordance with University of Miami Institutional Animal Care and Use Committee and National Institutes of Health guidelines.
Tissue dissociation
Mice were anesthetized with Avertin (250 mg/kg i.p) before transcardial perfusion with artificial cerebrospinal fluid (CSF) solution (87 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 75 mM sucrose, 20 mM glucose, 1.0 mM CaCl2, and 2 mM MgSO4). The solution was oxygenated on ice for 10 min before use. An 8-mm section of spinal cord centered at the injury site (or the corresponding location in uninjured tissue) was dissected. After removal of the meninges, the tissue was chopped using a razor blade, washed with 5 ml CSF solution, and centrifuged at 300 g for 5 min at 4°C. The pelleted cells were processed using the Miltenyi Neural Tissue Dissociation Kit-P (cat #130–092-628) following the manufacturer’s suggested protocol to obtain a single-cell suspension. In brief, the pellets were incubated in 2 ml of enzyme mixture 1 for 30 min at 37°C (50 µl Enzyme P in 1.9 ml Buffer X) with gentle shaking by hand every 5 min. 30 µl of enzyme mixture 2 (10 µl Enzyme A in 20 µl Buffer Y) was added to the cell suspensions and manually triturated (slowly, 10 times per sample) with a large-opening (1,000 µm diameter) fire-polished pipette. After being incubated for 10 min at 37°C, suspensions were triturated with a medium- and then small-opening (750 µm and 500 µm diameter, respectively) fire-polished pipette to produce single-cell suspensions. Suspensions were strained through a 70-µM cell strainer and washed with 10 ml CSF solution. Suspensions were centrifuged at 300 g for 5 min, and supernatants were discarded. Cell pellets were incubated in 10 µl Miltenyi Myelin Removal Beads II (cat #130–096-733) and 90 µl MACS buffer (0.5% BSA in HBSS without [w/o] Ca2+/Mg2+) for 15 min at 4°C. Cells were washed with 10 ml MACS buffer and centrifuged at 300 g for 5 min. Supernatants were discarded, and pellets were resuspended in 1 ml MACS buffer. Suspensions were then applied onto equilibrated LS Miltenyi MACS Magnetic Bead Columns (cat #130–042-401). Columns were washed with 2 ml MACS buffer, flow-through was centrifuged at 300 g for 5 min, and supernatants were discarded.
We enriched our second set (and third set for uninjured and 1-dpi samples) of replicates with astrocytes by pooling from two spinal cord samples. We processed two spinal cords from each injury time point in parallel as described above to yield two suspensions per time point. One suspension from each time point was kept on ice while the other was further processed with the Miltenyi Anti-ACSA-2 MicroBead kit according to the manufacturer’s instructions. In brief, suspensions were incubated with Miltenyi ACSA2 Beads for 15 min at 4°C and subsequently washed with 1 ml of MACS buffer. Suspensions were applied to LS Miltenyi MACS Magnetic Bead Columns, and the remaining cells were flushed from each column using LS Miltenyi column syringes and collected in separate tubes. ACSA-2–enriched flow-throughs were then pooled with the single-cell suspension from the other tissue.
Combined suspensions were centrifuged at 300 g for 5 min, and the pellets were resuspended in Red Blood Cell Lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.09 mM Na4-EDTA) and incubated at room temperature (RT) for 1 min. After washing with 5 ml MACS Buffer, suspensions were centrifuged at 300 g for 5 min, and supernatants were discarded. Pellets were resuspended in 50 µl FACS buffer (1% BSA + 0.05% sodium azide in HBSS w/o Ca2+/Mg2+) and processed for scRNA-seq. Before submitting for sequencing, each sample underwent a quality control assessment that included cell viability and number. Briefly, cell suspensions were incubated with ViaStain AOPI Staining Solution, and cell concentration/viability was assessed using the Nexcelom Cellometer K2. Each submitted sample met a viability cutoff of 80% or higher. Each sample was diluted accordingly to achieve a target cell capture of 10,000 cells, with our actual capture for the replicates totaling 12,488 for uninjured (five animals for three biological replicates), 21,010 for 1 dpi (five animals for three biological replicates, 17,491 for 3 dpi (three animals for two biological replicates), and 15,189 for 7 dpi (three animals for two biological replicates).
Histology
Mice were anesthetized with Avertin as described above and transcardially perfused with cold 4% paraformaldehyde (PFA) in PBS. Whole spinal cords were dissected, post-fixed in 4% PFA for 2 h, and transferred to 30% sucrose in PBS solution for overnight incubation on a shaker. An 8-mm segment of spinal cord centered at the injury site (or corresponding region in uninjured samples) was embedded in O.C.T. compound (Tissue-Tek) and sectioned sagittally at 16-µm thickness on a cryostat. Serial tissue sections were mounted onto Superfrost Plus slides and were stored at −20°C.
Immunohistochemistry
After drying at RT, tissue sections were washed once with PBS, permeabilized with 0.3% Triton X-100 in PBS (PBS-T) for 10 min at RT, and incubated in blocking solution (5% normal donkey serum in PBS-T) for 1 h at RT. Sections were then incubated in primary antibody diluted in 5% normal donkey serum overnight at 4°C (primary antibodies are listed below). After primary antibody incubation, slides were washed two times with PBS for 2 min each. Slides were incubated with appropriate Alexa Fluor secondary antibodies (1:500 in PBS-T) for 1 h at RT. Slides were washed twice with PBS for 2 min each and incubated with DAPI (1:15,000) for 3 min at RT in the dark. Slides were washed with PBS, and glass coverslips were mounted with Fluoromount-G (Southern Biotech; cat #0100–01). Images were taken on an Olympus Fluoview1000 Confocal Microscope or on an Olympus VSI Slide Scanner. Primary antibodies used were: chicken anti-GFAP (Abcam; ab4674/AB-304558; 1:500), rat anti-GFAP (Life Technologies; 13–0300/AB-86543; 1:500), rabbit anti-PDGFRβ (Abcam; ab32570/AB-777165; 1:200), goat anti-OSMR (R&D Systems; AF-662-SP/AB-355511; 1:100), goat anti-Podocalyxin (R&D Systems; AF1556/AB-354858; 1:200), rabbit anti-APOE (Abcam; ab183597/AB-2832971; 1:100), CD63 (PE/Dazzle 594; Biolegend; 143913/2565503; 1:100), and rat anti-CD11b (BioRad; MCA74GA/AB-324660; 1:500).
In situ hybridization
Tissue sections were prepared as described above. In situ hybridization was performed using the RNAscope Multiplex Fluorescent Kit v2 (Advanced Cell Diagnostics; cat #323100) with a few modifications to the manufacturer’s protocol. All incubations were at RT unless otherwise noted. Briefly, slides were incubated in hydrogen peroxide for 10 min. After washing with distilled water, slides were incubated in 1× Target Retrieval Solution at 95°C for 1 min. After washing with distilled water, slides were incubated in 100% EtOH for 3 min. Slides were left to dry before incubation in RNAscope Protease III solution at 40°C for 30 min (all 40°C incubations were in the Advanced Cell Diagnostics HybEZ Oven). After incubation, slides were washed with distilled water and incubated in probe solution for 2 h at 40°C. The following probes were purchased from Advanced Cell Diagnostics: C2 Mm-OSM (cat #427071-C2; 1:50), C1 Mm-ITGAM (cat #311491; no dilution, used working solution), C2 Mm-Apln (cat #415371-C2; 1:50), and C3 Mm-Crym (cat #466131-C3; 1:50). Following probe hybridization, each amplifier was individually incubated with slides for 30 min at 40°C. Before each fluorophore incubation, slides were incubated in RNAscope HRP solution for 15 min at 40°C. Fluorophores for each channel were prepared at 1:750 in RNAscope Diluent Solution, and individually incubated with slides for 30 min at 40°C. Different combinations of TSA Plus Cyanine 5 (Perkin Elmer; cat #NEL745001KT) and TSA Plus Fluorescein (Perkin Elmer; cat #NEL74100KT) were used with different probe channels. After each individual fluorophore incubation, slides were washed with RNAscope 1× Wash Buffer and incubated with RNAscope HRP Blocker solution for 15 min at 40°C. After in situ hybridization, slides were washed with PBS and immunohistochemically stained with appropriate primary antibodies as described above.
Tissue quantifications
For each sagittal tissue section of the OSMR/GFAP/PDGFR-β and OSM/GFAP/CD11b stain (immunohistochemistry and in situ hybridization, respectively), three 40× confocal images were taken surrounding the injury site (three serial tissue sections were analyzed per animal). Images were analyzed using the Cellomics High-Content Screening software (Thermo Fisher Scientific). To quantify total signal colocalization between GFAP/PDGFR-β and OSMR, we used the Target Activation V3 algorithm, with a minimum intensity threshold set to 350 for OSMR fluorescence. To count the number of OSM+/CD11b+ nuclei within the injury site, we used the Colocalization V4 algorithm. For each sagittal section of the Apln/podocalyxin/GFAP stain, a 1,000-µm2 box was drawn surrounding the injury site. Within this region, cells were analyzed for colocalization of Apln and podocalyxin using the Colocalization Threshold function in ImageJ. Areas were reported as the average area of all individual colocalizing particles within the 1,000-µm2 box. Individual particle areas were restricted to an area of 2 µm2 or larger using the Analyze Particles function in ImageJ. For each sagittal section of the Crym/GFAP stain, a 200-µm2 area of central canal was isolated for analysis. To analyze the colocalization of Crym particles within DAPI+ nuclei, the Analyze Particles function was used in ImageJ.
Flow cytometry
Mice received SCI as described above, and injury sites were processed for flow cytometry. Mice were anesthetized with Avertin followed by transcardial perfusion with ice-cold PBS. An 8-mm segment of the spinal cord centered at the injury site was dissected, and the meningeal layer was removed. Spinal cords were manually dissociated by mechanical grinding against a 70-µm cell strainer (the plunger of a 3-ml syringe was used for mechanical dissociation). Strainers were washed with 10 ml HBSS (w/o Ca2+/Mg2+) and centrifuged at 300 g for 10 min. Supernatant was aspirated, and the pellet was resuspended in 10 µl Miltenyi Myelin Removal Beads II (Miltenyi Biotec; cat #130–096-733) and 90 µl MACS buffer (0.5% BSA in HBSS w/o Ca2+/Mg2+) and incubated for 15 min at 4°C. Cells were washed with 10 ml MACS buffer and centrifuged at 300 g for 10 min. The pellet was resuspended in 1 ml MACS buffer and applied onto equilibrated LS Miltenyi MACS Magnetic Bead Columns (Miltenyi Biotec; cat #130–042-401). Columns were washed with 2 ml MACS buffer, and the suspension was centrifuged at 300 g for 5 min. The pellet was resuspended in RBC lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.09 mM Na4-EDTA) and kept at RT for 1 min. To identify live cells, the pellet was resuspended in Zombie Aqua Viability Dye and incubated for 30 min at RT (Biolegend; cat #423101). Following a wash in PBS and spin at 300 g, cells were resuspended in Fix/Perm Solution for 20 min at 4°C to fix and permeabilize the cells (BD Bioscience Cytofix/Cytoperm; cat #554714). After incubation, cells were washed with 1 ml of Perm/Wash solution. After centrifugation at 300 g for 10 min, supernatant was aspirated, and the cells were incubated in blocking buffer mixture for 5 min at 4°C (1 μl anti-mouse CD16/32 TruStain Fcx; RRID: AB-1574975; Biolegend; cat #101320; and 99 μl FACS buffer [1% BSA + 0.05% sodium azide in HBSS w/o Ca2+/Mg2+]). Antibody mixture was added directly to the cell suspension and incubated for 20 min at 4°C. Cells were washed with 5 ml FACS buffer and centrifuged at 300 g for 10 min. For fixation, cells were resuspended in 4% PFA and incubated for 30 min at 4°C. After centrifugation at 300 g for 10 min, cells were resuspended in 250 µl FACS buffer and transferred into a flat bottom 96-well plate. 10 µl of 123 eBeads Counting Beads (Thermo Fisher Scientific; cat #01–1234-42) were added to each sample well, and cells were kept at 4°C until analysis on a Cytoflex Flow Cytometer (Beckman Coulter). Antibody mixture for macrophages was as follows: CD16/32 TruStain Fcx (Biolegend; 101320/AB-1574975; 1:100), Zombie Aqua Viability Dye (Biolegend; 423101/NA; 1:2.5), CD45.2 (APC-Cy7; Biolegend; 103116/312981; 1:200), CD11b (APC; Biolegend; 101212/312795; 1:200), Ly6G (PerCP Cy5.5; Biolegend; 127616/AB-1877271; 1:200), CD63 (PE/Dazzle 594; Biolegend; 143913/2565503; 1:100), and APOE (Dylight 680; Novus Biologicals; NB110-60531FR/NA; 1:100). Antibody mixture for microglia was as follows: CD16/32, Zombie Aqua Dye, CD45.2, CD11b, and Ly6G were the same as above; P2RY12 (PE; Biolegend; 848003/2721644; 1:100); and MSR1 (Alexa Fluor 700; BioRad; MCA1322A700/2251086; 1:50). After selecting for viable CD45+ leukocytes, neutrophils were excluded based on Ly6G expression. Ly6G− cells were separated based on CD11b and CD45, which identified lymphocytes (CD11b−), monocyte/macrophages (CD11b+/CD45hi), and microglia (CD11b+/CD45lo). The microglia cluster was gated on P2RY12 and MSR1 to differentiate between homeostatic and non-homeostatic microglia. The monocyte/macrophage cluster was first gated on CD63+ cells to identify macrophages, and then gated on APOE and CD11b to separate the two macrophage subtypes. Spinal cord tissues rostral and caudal to the injury site as well as injury site from MSR1 knockout mice were used as negative controls for gating.
scRNA-seq and bioinformatics
scRNA-seq using 10X Genomics platform
Single-cell suspensions were prepared as described above. A total of 10 samples were sequenced with a median of 8,583 cells per sample. The mean value of mean reads per cell was 87,860 with a mean of 5,819 unique molecular identifiers (UMIs) per cell across all samples. Sample libraries were prepared according to Chromium Single Cell 3′ Library and Gel Bead Kit instructions (v2-v3). All samples were indexed with Chromium i7 Multiplex Kits (10X Genomics; PN-120262), and single-cell suspensions were processed according to the manufacturer’s instructions to construct final libraries for Illumina sequencing. For sequencing, libraries were loaded at recommended loading concentrations onto an Illumina NextSeq 500 flow cell and paired-end sequenced under recommended settings (R1: 28 cycles; i7 index: 8 cycles; i5 index: none; and R2: 91 cycles). After sequencing, Illumina output was processed using CellRanger’s (v2-4) recommended pipeline to generate gene-barcode count matrices. In brief, base call files for each sample were demultiplexed into FASTQ reads and then aligned to the mouse mm10 reference genome using the STAR splice-aware aligner. Reads that confidently intersected at least 50% with an exon were considered exonic and further aligned with annotated transcripts. Reads were then filtered to remove UMIs and barcodes with single base substitution errors and finally used for UMI counting. The output was a count matrix containing all UMI counts for every droplet. Sequence alignment and transcript counts were performed using CellRanger.
Preprocessing and quality control
To distinguish cell-containing droplets from empty droplets, we performed cell calling on the unfiltered UMI count matrices using a combination of barcode-ranking and empty-droplet detection algorithms. First, cell barcodes were ranked according to UMI count and visualized in a log-total UMI count versus log-rank plot. A spline curve was fit to the data to identify knee and inflection points. All cells with a UMI count above the knee were considered cell-containing droplets. To further distinguish cells from data below the knee, we used the emptyDrops function from the DropletUtils R package (Lun et al., 2019) using the following fixed parameters for all samples: lower = 250; maximum fit.bounds = 1e06; false discovery rate = 0.001; ignore = 10. Some parameters varied between samples accordingly: “retain” was set to knee point values, and “lower” was set to inflection point values. The result was a filtered count matrix of all putative cell-containing droplets. To distinguish low-quality cells, we considered cell-level metrics such as total UMI count, number of unique genes, percentage of UMIs mapping to mitochondrial genes, percentage of UMIs mapping to ribosomal proteins, and doublet detection algorithm outputs. For each sample, using all metrics except doublet scores, thresholds were computed by determining the median absolute deviation and removing all cells with values three median absolute deviations above or below the median. We observed that removing cells in the lowest quantiles of total UMI counts preferentially selected against endothelial cells, while removing cells in the highest quantiles of mitochondrial percentage preferentially selected against astrocytes. Finally, we removed potential red blood cells by removing any cell with >1 in 5,000 UMIs mapping to hemoglobin genes.
To remove potential doublets, we applied the Scrublet Python package (Wolock et al., 2019) for each individual sample. In brief, the Scrublet simulates multiplets by sampling from the data and builds a nearest-neighbor classifier. Cells from the data that have high local densities of simulated doublets are flagged and removed. We set the expected_doublet_rate for each sample according to the estimated doublet rate per cells sequenced as published by 10X, and default values for all other parameters. The percentage of cells that had to be removed because of doublet detection using Scrublet ranged from 1–5% from each sample. The doublet rates were approximately equal to the expected doublet rates as outlined in the 10X Genomics Single Cell 3′ Gene Expression v3.1 assay user guide (∼0.8% per 1,000 cells). Most samples had estimated rates within 1% of the 10X expected rates, demonstrating that cells were not loaded at excessively high concentrations. In downstream analyses of each of the three compartments, we further removed small clusters of putative doublets (based on shared expression of distinct cell-type markers). In the analysis of the vascular cells, we observed a cluster of cells (U-vascular) that showed both pericyte-like and endothelial-like characteristics and whose numbers were greater than that of pericytes. We ruled out the possibility of U-vascular cells being doublets by comparing the Scrublet scores among all cells and found that scores were not unusually high.
Analysis of all SCI cells
To generate the full SCI dataset, all samples were processed and combined using Seurat v3 (Stuart et al., 2019). After filtering each sample count matrix for genes that were expressed in at least 10 cells, each dataset was independently normalized and scaled using the SCTransform() function, which performs a variance-stabilizing transformation using negative binomial regression (Hafemeister and Satija, 2019). To remove cell cycle genes as a confounding source of variation, cell cycle scores based on the expression of canonical G2M and S phase markers were computed for each cell. Cell cycle genes were provided through the Seurat tutorial. These score values were then used as input for the “vars.to.regress” argument in the SCTransfrom() function. PCA showing separation of cells by cell cycle phase before and after regression is shown in Fig. S1 F. To identify shared and unique molecular cell types across datasets and time points, sample expression matrices were batch-corrected using Seurat’s Data Integration workflow (Butler et al., 2018), which uses a mutual-nearest-neighbor–based method. For the full SCI dataset, the 3,000 most variable genes were used as input for the “anchor.features” argument of the FindIntegrationAnchors() function. This resulted in a single batch-corrected expression matrix for containing all cells.
For clustering, we performed Seurat clustering as recommended. Briefly, we performed PCA on the 3,000 variable genes and selected the top 20 principal components based on the elbow plot heuristic, which measures the contribution of variance by each component. We used the FindNeighbors() and FindClusters() functions with default parameters to perform graph-based clustering on a shared-nearest-neighbor graph. To visualize the data, we generated UMAP plots using the RunUMAP function() on the top 20 principal components. We then annotated the clusters using previously described marker genes (Fig. S1 C). To verify our cell-type annotations, we used the SingleR package (Aran et al., 2019) to compare our data with previously published datasets (Fig. S1 B). In brief, the SingleR package performs a Spearman rank correlation between each query cell and assigned cell-type labels of a reference dataset. For references, we used scRNA-seq data produced from spinal cord tissue by Rosenberg et al. (2018) and Sathyamurthy et al. (2018).
Analysis of myeloid, vascular, and macroglial cells
For downstream analysis of the three compartments, we took subsets of the full SCI count matrix and repeated batch correction and clustering. We performed batch correction in two steps. In the first step, we corrected between replicates of each time point using the rescaleBatches() function from the batchelor package to yield a single count matrix for each time point (Amezquita et al., 2020). This function performs gene-wise linear regression between replicates and scales expression to the lowest mean. In the second step, we performed the Seurat data integration procedure on the log-normalized expression values. Integration was performed using the Seurat data integration procedure on the top 2,000 variable genes, and cell cycle scores were regressed out using the ScaleData() function on the resulting expression matrix. For cluster analysis, we sought to identify reproducible clusters by iterating through a range of parameter values in PCA and graph-based clustering. Specifically, we tested different numbers of components and resolution values in the FindNeighbors() and FindClusters() functions, respectively, and inspected cluster memberships for stable configurations. For the myeloid, vascular, and macroglia, we took the top 11, 11, and 8 principal components and set resolutions to 0.35, 0.4, and 0.4, respectively.
Differential expression testing and GO
To identify marker genes for each cluster, we used the FindAllMarkers() function using default parameters, which implements a Wilcoxon rank-sum test comparing gene expression of cells within a given cluster versus all other cells. We repeated this test for each of the three compartments to identify marker genes for each subtype. To identify DEGs for GO analysis, we used the MAST framework as implemented in the Seurat FindMarkers() function and set the “latent.vars” argument to “nFeature_RNA,” which is equivalent to adjusting for the cellular detection rate (Finak et al., 2015). For GO analysis, we used Fisher’s exact test as implemented in the topGO package (Rahnenfuhrer, 2020). For the chemotaxis-inducing versus inflammatory GO analysis, we took all genes with an average log-fold change >0.5 and adjusted P values <0.001. For GO analysis of OPCs and astrocytes over time, we compared cells from each sequential time point and took all DEGs with an average log fold-change >0.25 and adjusted P values <0.001. We excluded OPC-B cells from all OPCs in these comparisons because OPC-Bs were not present in the uninjured spinal cord, and their lineage may be different from canonical OPCs. Instead, we compared GO terms between OPC-A and OPC-B at 1 dpi, the peak of OPC-B numbers, using the DEGs with adjusted P values <0.001. We show all GO results with at least four significant genes mapping to a term.
Ligand–receptor interaction analysis
To infer potential ligand–receptor interactions between two cell types, we adapted the method used in CellPhoneDB (Vento-Tormo et al., 2018). We first pulled a reference list of human ligand–receptor pairs published previously (Ramilowski et al., 2015) and converted the genes into mouse orthologues using the Ensembl biomaRt package (Durinck et al., 2009). We defined the ligand–receptor score as the mean of the average log-normalized expression of the receptor gene in one cell type and the average log-normalized expression of the ligand gene in a second cell type. To identify enriched ligand–receptor interactions, we applied a permutation test to identify interactions scores that are enriched in a specific <ligand cell A, receptor cell B, time point> combination. For each of 1,000 permutations, we randomly shuffled the cell-type and time point labels and calculated an interaction scores for all possible <ligand cell, receptor cell, time point> combinations. Repeating this 1,000 times generated a null distribution of interaction scores for each ligand–receptor pair. We compared the interaction scores of the actual (ligand cell A, receptor cell B, time point) labels to the null distribution and calculated P values as the proportion of null scores, which are equal to or greater than the actual interaction score.
Online supplemental material
Fig. S1 shows the distribution of each sample, quality control metrics data, and the PCA before and after cell cycle regression. Fig. S2 shows the feature plots of the annotated genes and SingleR analysis used to identify the major cell clusters. Genetic lineage tracing of POSTNEYFP mice is also included here. Fig. S3 shows the feature plot and heatmap of the top molecular markers of vascular cells. Fig. S4 shows the feature plot of the annotated genes used to identify macroglia clusters, and histological assessment of Crym as a marker of astroependymal cells. Fig. S5 shows the comparison of different macroglia cell types.
Acknowledgments
The authors thank the Sylvester Comprehensive Cancer Center Onco-Genomics Shared Resource and Biostatistics and Bioinformatics Shared Resource, Division of Veterinary Resources, Diabetes Research Institute Flow Cytometry Core, and Yan Shi at the Miami Project to Cure Paralysis Microscopy Core. The authors thank Dr. Lina Shehadeh at the University of Miami School of Medicine, Miami, FL, for generously donating the periostin-CreER mice. The authors thank Dr. Nagi Ayad and Robert Suter at the University of Miami School of Medicine for their technical assistance with data processing. Special thanks to Shaffiat Karmally and Yadira Salgueiro for their assistance in multiple aspects of this study, including animal care and logistics.
This study was funded by National Institute of Neurological Disorders and Stroke grant R01NS081040 (to J.K. Lee), University of Miami SAC Award UM SIP 2019-2 (to J.K. Lee), National Institute of Neurological Disorders and Stroke grant F31NS115225 (to C. Ryan), the Miami Project to Cure Paralysis, the Buoniconti Fund to Cure Paralysis, a generous gift from the Craig H. Nielsen Foundation (J.K. Lee), and National Institutes of Health grant 1S10OD023579-01 for the VS120 Slide Scanner housed at the University of Miami Miller School of Medicine Analytical Imaging Core Facility.
Author contributions: L.M. Milich, J.S. Choi, J.K. Lee, and C. Ryan designed and performed the experiments, analyzed the data, and wrote the manuscript. S.L. Yahn, S.R. Cerqueira, and S. Benavides performed the experiments and wrote the manuscript. P. Tsoulfas designed the experiments, analyzed the data, and wrote the manuscript.
Data availability
The RNA sequencing data generated in this study are available at NCBI GEO accession no. GSE162610. Any other data can be provided upon request.
References
- Amankulor, N.M., Hambardzumyan D., Pyonteck S.M., Becher O.J., Joyce J.A., and Holland E.C.. 2009. Sonic hedgehog pathway activation is induced by acute brain injury and regulated by injury-related inflammation. J. Neurosci. 29:10299–10308. 10.1523/JNEUROSCI.2500-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezquita, R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. 2020. Orchestrating single-cell analysis with Bioconductor. Nat. Methods. 17:137–145. 10.1038/s41592-019-0654-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.A., Burda J.E., Ren Y., Ao Y., O’Shea T.M., Kawaguchi R., Coppola G., Khakh B.S., Deming T.J., and Sofroniew M.V.. 2016. Astrocyte scar formation aids central nervous system axon regeneration. Nature. 532:195–200. 10.1038/nature17623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankeny, D.P., and Popovich P.G.. 2009. Mechanisms and implications of adaptive immune responses after traumatic spinal cord injury. Neuroscience. 158:1112–1121. 10.1016/j.neuroscience.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte, R.S., Chen D.S., and Ferrara N.. 2019. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell. 176:1248–1264. 10.1016/j.cell.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran, D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20:163–172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, S.A., Broderick R., Blades D.A., and Scheff S.W.. 1998. Alterations in temporal/spatial distribution of GFAP- and vimentin-positive astrocytes after spinal cord contusion with the New York University spinal cord injury device. J. Neurotrauma. 15:1015–1026. 10.1089/neu.1998.15.1015 [DOI] [PubMed] [Google Scholar]
- Barnabé-Heider, F., Göritz C., Sabelström H., Takebayashi H., Pfrieger F.W., Meletis K., and Frisén J.. 2010. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell. 7:470–482. 10.1016/j.stem.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Beck, K.D., Nguyen H.X., Galvan M.D., Salazar D.L., Woodruff T.M., and Anderson A.J.. 2010. Quantitative analysis of cellular inflammation after traumatic spinal cord injury: evidence for a multiphasic inflammatory response in the acute to chronic environment. Brain. 133:433–447. 10.1093/brain/awp322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellver-Landete, V., Bretheau F., Mailhot B., Vallières N., Lessard M., Janelle M.E., Vernoux N., Tremblay M.E., Fuehrmann T., Shoichet M.S., and Lacroix S.. 2019. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 10:518. 10.1038/s41467-019-08446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni, A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D.A., Rozovsky I., Stahl N., Yancopoulos G.D., and Greenberg M.E.. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 278:477–483. 10.1126/science.278.5337.477 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K., Deng S., Lu H., Zheng Y., Yang G., Kim D., Cao Q., and Wu J.Q.. 2013. RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: a resource for understanding the pathology at the systems level. PLoS One. 8:e72567. 10.1371/journal.pone.0072567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck, S., Spellman P.T., Birney E., and Huber W.. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4:1184–1191. 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, L., Kangas J., and Saharinen P.. 2017. Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin. Sci. (Lond.). 131:87–103. 10.1042/CS20160129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner, J.R., Herrmann J.E., Woo M.J., Tansey K.E., Doan N.B., and Sofroniew M.V.. 2004. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J. Neurosci. 24:2143–2155. 10.1523/JNEUROSCI.3547-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak, G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16:278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt, H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., and Betsholtz C.. 2003. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 161:1163–1177. 10.1083/jcb.200302047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J., Harris P., Brimble M., Lei Y., Lu J., Yang Y., and Gunn A.J.. 2015. The role for IGF-1-derived small neuropeptides as a therapeutic target for neurological disorders. Expert Opin. Ther. Targets. 19:785–793. 10.1517/14728222.2015.1010514 [DOI] [PubMed] [Google Scholar]
- Hackett, A.R., and Lee J.K.. 2016. Understanding the NG2 Glial Scar after Spinal Cord Injury. Front. Neurol. 7:199. 10.3389/fneur.2016.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett, A.R., Lee D.H., Dawood A., Rodriguez M., Funk L., Tsoulfas P., and Lee J.K.. 2016. STAT3 and SOCS3 regulate NG2 cell proliferation and differentiation after contusive spinal cord injury. Neurobiol. Dis. 89:10–22. 10.1016/j.nbd.2016.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C., and Satija R.. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20:296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. 2019. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 50:253–271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Sun D., Rittling S.R., Denhardt D.T., and Young W.. 2007. Osteopontin-deficient mice exhibit less inflammation, greater tissue damage, and impaired locomotor recovery from spinal cord injury compared with wild-type controls. J. Neurosci. 27:3603–3611. 10.1523/JNEUROSCI.4805-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman, M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 135:738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, J.E., Imura T., Song B., Qi J., Ao Y., Nguyen T.K., Korsak R.A., Takeda K., Akira S., and Sofroniew M.V.. 2008. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J. Neurosci. 28:7231–7243. 10.1523/JNEUROSCI.1709-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.B.2014. Lymphocytes and autoimmunity after spinal cord injury. Exp. Neurol. 258:78–90. 10.1016/j.expneurol.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Kigerl, K.A., McGaughy V.M., and Popovich P.G.. 2006. Comparative analysis of lesion development and intraspinal inflammation in four strains of mice following spinal contusion injury. J. Comp. Neurol. 494:578–594. 10.1002/cne.20827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigerl, K.A., Gensel J.C., Ankeny D.P., Alexander J.K., Donnelly D.J., and Popovich P.G.. 2009. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J. Neurosci. 29:13435–13444. 10.1523/JNEUROSCI.3257-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner, A., Greenhalgh A.D., Zarruk J.G., Passos Dos Santos R., Gaestel M., and David S.. 2014. TNF and increased intracellular iron alter macrophage polarization to a detrimental M1 phenotype in the injured spinal cord. Neuron. 83:1098–1116. 10.1016/j.neuron.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Lee, H.S., Han J., Bai H.J., and Kim K.W.. 2009. Brain angiogenesis in developmental and pathological processes: regulation, molecular and cellular communication at the neurovascular interface. FEBS J. 276:4622–4635. 10.1111/j.1742-4658.2009.07174.x [DOI] [PubMed] [Google Scholar]
- Lin, J.D., Nishi H., Poles J., Niu X., Mccauley C., Rahman K., Brown E.J., Yeung S.T., Vozhilla N., Weinstock A., et al. 2019. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight. 4:e124574. 10.1172/jci.insight.124574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy, D.N., Crawford C.H., Darnall J.B., Burke D.A., Onifer S.M., and Whittemore S.R.. 2002. Temporal progression of angiogenesis and basal lamina deposition after contusive spinal cord injury in the adult rat. J. Comp. Neurol. 445:308–324. 10.1002/cne.10168 [DOI] [PubMed] [Google Scholar]
- Lun, A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., and Marioni J.C.. participants in the 1st Human Cell Atlas Jamboree . 2019. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20:63. 10.1186/s13059-019-1662-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich, L.M., Ryan C.B., and Lee J.K.. 2019. The origin, fate, and contribution of macrophages to spinal cord injury pathology. Acta Neuropathol. 137:785–797. 10.1007/s00401-019-01992-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Kohanbash G., Liu S.J., Alvarado B., Carrera D., Bhaduri A., Watchmaker P.B., Yagnik G., Di Lullo E., Malatesta M., et al. 2017. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 18:234. 10.1186/s13059-017-1362-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, P.J.2017. Macrophage Polarization. Annu. Rev. Physiol. 79:541–566. 10.1146/annurev-physiol-022516-034339 [DOI] [PubMed] [Google Scholar]
- Nagy, J.A., Dvorak A.M., and Dvorak H.F.. 2003. VEGF-A(164/165) and PlGF: roles in angiogenesis and arteriogenesis. Trends Cardiovasc. Med. 13:169–175. 10.1016/S1050-1738(03)00056-2 [DOI] [PubMed] [Google Scholar]
- Neufeld, G., Cohen T., Gengrinovitch S., and Poltorak Z.. 1999. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 13:9–22. 10.1096/fasebj.13.1.9 [DOI] [PubMed] [Google Scholar]
- Rahnenfuhrer, A.A.2020. topGO: Enrichment Analysis for Gene Ontology. R package. Version 2.42.0. https://rdrr.io/bioc/topGO/
- Ramilowski, J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., and Forrest A.R.. 2015. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6:7866. 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaux, A., Gallatz K., Palkovits M., and Llorens-Cortes C.. 2002. Distribution of apelin-synthesizing neurons in the adult rat brain. Neuroscience. 113:653–662. 10.1016/S0306-4522(02)00192-6 [DOI] [PubMed] [Google Scholar]
- Reinert, R.B., Cai Q., Hong J.Y., Plank J.L., Aamodt K., Prasad N., Aramandla R., Dai C., Levy S.E., Pozzi A., et al. 2014. Vascular endothelial growth factor coordinates islet innervation via vascular scaffolding. Development. 141:1480–1491. 10.1242/dev.098657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y., Ao Y., O’Shea T.M., Burda J.E., Bernstein A.M., Brumm A.J., Muthusamy N., Ghashghaei H.T., Carmichael S.T., Cheng L., and Sofroniew M.V.. 2017. Ependymal cell contribution to scar formation after spinal cord injury is minimal, local and dependent on direct ependymal injury. Sci. Rep. 7:41122. 10.1038/srep41122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribatti, D.2008. The discovery of the placental growth factor and its role in angiogenesis: a historical review. Angiogenesis. 11:215–221. 10.1007/s10456-008-9114-4 [DOI] [PubMed] [Google Scholar]
- Rosenberg, A.B., Roco C.M., Muscat R.A., Kuchina A., Sample P., Yao Z., Graybuck L.T., Peeler D.J., Mukherjee S., Chen W., et al. 2018. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 360:176–182. 10.1126/science.aam8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, E., Yang L., Su T., Morris D.R., McKnight G.S., and Amieux P.S.. 2009. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA. 106:13939–13944. 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamurthy, A., Johnson K.R., Matson K.J.E., Dobrott C.I., Li L., Ryba A.R., Bergman T.B., Kelly M.C., Kelley M.W., and Levine A.J.. 2018. Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep. 22:2216–2225. 10.1016/j.celrep.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderblom, C., Luo X., Blumenthal E., Bray E., Lyapichev K., Ramos J., Krishnan V., Lai-Hsu C., Park K.K., Tsoulfas P., and Lee J.K.. 2013. Perivascular fibroblasts form the fibrotic scar after contusive spinal cord injury. J. Neurosci. 33:13882–13887. 10.1523/JNEUROSCI.2524-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew, M.V.2015. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16:249–263. 10.1038/nrn3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove, H., Martens L., Scheyltjens I., De Vlaminck K., Pombo Antunes A.R., De Prijck S., Vandamme N., De Schepper S., Van Isterdael G., Scott C.L., et al. 2019. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22:1021–1035. 10.1038/s41593-019-0393-4 [DOI] [PubMed] [Google Scholar]
- Vanlandewijck, M., He L., Mäe M.A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., et al. 2018. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 554:475–480. 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- Velardo, M.J., Burger C., Williams P.R., Baker H.V., López M.C., Mareci T.H., White T.E., Muzyczka N., and Reier P.J.. 2004. Patterns of gene expression reveal a temporally orchestrated wound healing response in the injured spinal cord. J. Neurosci. 24:8562–8576. 10.1523/JNEUROSCI.3316-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo, R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. 2018. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 563:347–353. 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Hong J., Lu M., Neil J.E., Vitek M.P., Liu X., Warner D.S., Li F., and Sheng H.. 2014. ApoE mimetic ameliorates motor deficit and tissue damage in rat spinal cord injury. J. Neurosci. Res. 92:884–892. 10.1002/jnr.23371 [DOI] [PubMed] [Google Scholar]
- Wang, X., Cao K., Sun X., Chen Y., Duan Z., Sun L., Guo L., Bai P., Sun D., Fan J., et al. 2015. Macrophages in spinal cord injury: phenotypic and functional change from exposure to myelin debris. Glia. 63:635–651. 10.1002/glia.22774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner, I.B., Anderson M.A., Song B., Levine J., Fernandez A., Gray-Thompson Z., Ao Y., and Sofroniew M.V.. 2013. Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J. Neurosci. 33:12870–12886. 10.1523/JNEUROSCI.2121-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstone, W.D., Hsu J.Y., Eisenberg M., Werb Z., and Noble-Haeusslein L.J.. 2003. Blood-spinal cord barrier after spinal cord injury: relation to revascularization and wound healing. J. Neurosci. Res. 74:227–239. 10.1002/jnr.10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R.E., McTigue D.M., and Jakeman L.B.. 2010. Regional heterogeneity in astrocyte responses following contusive spinal cord injury in mice. J. Comp. Neurol. 518:1370–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock, S.L., Lopez R., and Klein A.M.. 2019. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 8:281–291.e9. 10.1016/j.cels.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.E., Pan L., Zuo Y., Li X., and Hong W.. 2017. Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron. 96:313–329.e6. 10.1016/j.neuron.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Yang, X., Chen S., Shao Z., Li Y., Wu H., Li X., Mao L., Zhou Z., Bai L., Mei X., and Liu C.. 2018. Apolipoprotein E Deficiency Exacerbates Spinal Cord Injury in Mice: Inflammatory Response and Oxidative Stress Mediated by NF-κB Signaling Pathway. Front. Cell. Neurosci. 12:142. 10.3389/fncel.2018.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydens, E., Amann L., Asselbergh B., Scott C.L., Martens L., Sichien D., Mossad O., Blank T., De Prijck S., Low D., et al. 2020. Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat. Neurosci. 23:676–689. 10.1038/s41593-020-0618-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel, A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G., et al. 2018. Molecular Architecture of the Mouse Nervous System. Cell. 174:999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Soderblom C., Krishnan V., Ashbaugh J., Bethea J.R., and Lee J.K.. 2015. Hematogenous macrophage depletion reduces the fibrotic scar and increases axonal growth after spinal cord injury. Neurobiol. Dis. 74:114–125. 10.1016/j.nbd.2014.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA sequencing data generated in this study are available at NCBI GEO accession no. GSE162610. Any other data can be provided upon request.