Abstract
Organoids are multicellular structures that can be derived from adult organs or pluripotent stem cells. Early versions of organoids range from simple epithelial structures to complex, disorganized tissues with large cellular diversity. The current challenge is to engineer cellular complexity into organoids in a controlled manner that results in organized assembly and acquisition of tissue function. These efforts have relied on studies of organ assembly during embryonic development and have resulted in the development of organoids with multilayer tissue complexity and higher-order functions. We discuss how the next generation of organoids can be designed by means of an engineering-based narrative design to control patterning, assembly, morphogenesis, growth, and function.
Organoids are three-dimensional (3D) structures with multicellular complexity and some level of tissue structure and function. For decades, organoids were derived through the deconstruction of adult organs and grown as complex but poorly defined tissues in vitro (1). However, they are also generated from embryonic or adult stem cells. Because organoids are both highly tractable and expandable and can be genetically manipulated, they are well suited to study organ development and pathophysiology in vitro (2). Human organoids have facilitated studies of human birth defects, human-specific pathogens, and screening of experimental drugs for efficacy before testing in patients (3).
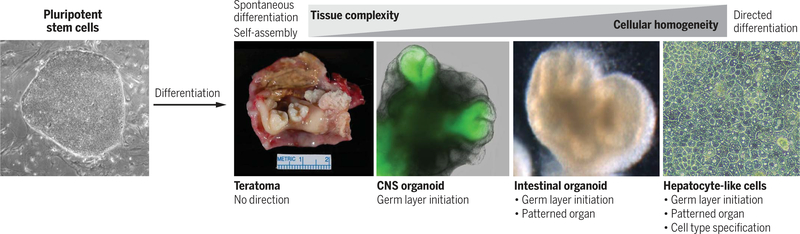
Efforts in adult stem cell research have focused on stem cell identification, isolation, expansion in culture using niche factors (4), and differentiation to specific fates. For the past decade, scientists have sought to harness and control the potential of stem cells to generate organoids with specific tissue-level or organ-level complexity. Unlike adult stem cells, embryonic or induced pluripotent stem cells (PSCs) can form all tissues of the body and will spontaneously differentiate in vivo into a disorganized mass of differentiated tissues called a teratoma (Fig. 1). By manipulating factors that control embryonic organogenesis, methods have been developed to guide the stepwise differentiation of PSCs into embryonic germ layer–restricted organoids, organ-specific organoids, and even specific cell types such as hepatocytes, neurons, and cardiomyocytes (Fig. 1).
Fig. 1. Controlling the chaotic differentiation of pluripotent stem cells.
Pluripotent stem cells (PSCs) stochastically differentiate in vivo into a disordered mix of tissues called teratomas [teratoma image by permission from Sanjay Mukhopadhyay, Cleveland Clinic]. PSC differentiation can be directed in a stepwise manner by controlling the initiation of germ layer formation [CNS organoid with an optic cup shown from (29) by permission from Nature, copyright 2011], organ patterning (intestinal organoids shown), and specification of individual cell types (hepatocytes shown).
A major goal of organoid research is to use in vitro–derived constructs to replace diseased or aging organs. However, cellular complexity, tissue geometry, growth, and function present challenges in moving toward clinical applications. In addition, methods must be developed to generate organoids in a controlled and stereotypic manner to reduce heterogeneity. Below, we explore how the processes of normal organogenesis have been used to improve efforts to generate organoids (“organoidgenesis”) and how engineering-based approaches may help to overcome obstacles and lead to better control over organoid assembly, growth, shape, and function.
Organogenesis-inspired principles to direct organoidgenesis
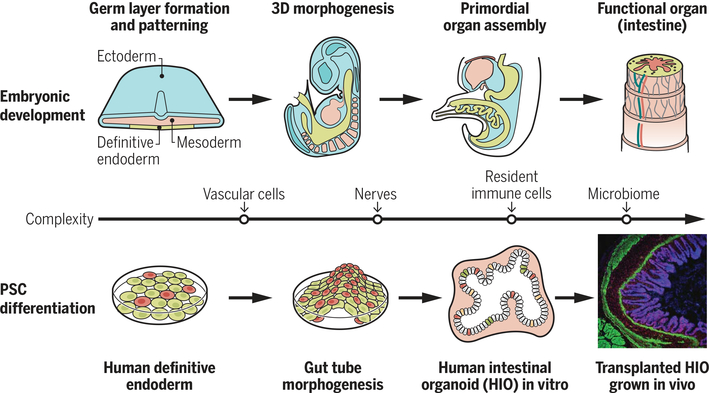
Organoidgenesis efforts have focused on principles learned from studies of organogenesis, the process by which organs form in the developing embryo. Organogenesis can be loosely subdivided into stages (Fig. 2) (5). The first is formation of the three embryonic germ layers during gastrulation: ectoderm, mesoderm, and endoderm. The second stage subdivides (patterns) the germ layers into regional subdomains along the anterior-posterior (A-P) and dorsal-ventral (D-V) axis. The third stage involves a series of morphogenetic processes that drive the formation of 3D organ primordia at precise locations along the A-P and D-V axes. Once organ primordia are established, each organ becomes vascularized and innervated by endothelial precursors and neural crest cells, respectively. Vascularization brings oxygen, nutrients, and circulating factors, as well as hematopoietic cells (including macrophages) that can participate in organ development and persist postnatally.
Fig. 2. Using principles of organogenesis to generate cellular complexity during organoidgenesis.
The upper panel shows some of the main stages that drive assembly of organ primordia. The middle line indicates additional cell types that get incorporated into developing organs (vascular cells, nerves, immune cells) and how these cell types have been experimentally incorporated into developing organoids (lower panel). After transplantation, organoids can become vascularized by the host and continue to grow in size and undergo tissue morphogenesis.
From the onset of gastrulation, within 4 to 5 days in mice and 20 to 30 days in humans, organ primordia contain most of the necessary cellular components that will contribute to the fully functional organ. Much of the remaining development involves reciprocal paracrine interactions between cells and systemic cues delivered by the circulation; these interactions drive tissue growth, morphogenesis, and differentiation. In many respects, these early stages of organ development can be considered “self-assembly,” a process used to describe how organoids form through assembly of a population of tissue progenitors. Therefore, controlling these early stages of organogenesis has been essential in directing organoidgenesis.
Providing direction to chaotic differentiation
Early efforts to control the stochastic differentiation of PSCs focused on guiding their differentiation into one of the three primary germ layers. The first example of this came from experiments with mouse and human PSCs that were differentiated into neural ectoderm aggregates that formed organoids containing a mix of forebrain derivatives such as cerebellum and optical tissues (6). Although ectoderm organoids start as symmetrical structures, uncontrolled symmetry-breaking events result in random pockets of differentiation, resulting in heterogeneous organoids containing a mix of diverse neural tissues. However, by manipulating signaling pathways to uniformly direct the regional pattern of organoids, PSCs can be directed to form specific organoid types representing the midbrain, hypothalamus, cerebellum, retina, cardiac, kidney, lung, esophagus, pancreas, liver, stomach (both fundus and antrum), small intestine, colon, etc. (2). Some of these first-generation organoid systems have remarkable cellular diversity. For example, intestinal organoids contain nearly all of the intestinal epithelial cell types, as well as intestinal mesenchyme that forms fibroblasts, smooth muscle fibers, and interstitial cells of Cajal.
Increasing organoid complexity
The next generation of organoidgenesis focused on incorporating critical cell types that are shared across organs, such as blood vessels, lymphatic vessels, nerves, stromal cells, and immune cells (Fig. 2). During organogenesis, many of these cell types arise in another region of the embryo and are delivered to the developing organ by migration or embryonic morphogenesis. In the case of organoidgenesis, vascular and neuronal cell types can be generated separately and introduced into forming organoids at a time that approximates their normal arrival during embryonic organogenesis. This approach was used to incorporate vascularity into brain and liver organoids (7, 8), interneurons and microglia into brain organoids (9–12), and a functional enteric neuroglial plexus capable of controlling peristalsis in intestinal organoids (13).
A common feature of these studies is the remarkable ability of vascular and neuronal progenitor cells to incorporate into the developing organoid and self-organize into neural and vascular plexi. This supports the notion that stage-matched populations of progenitor cells that are placed together have the intrinsic ability to self-organize (14). As in the self-organization processes that occur during organogenesis in vivo, where different populations of progenitor cells communicate via paracrine factors to coordinate the formation of tissue structure, organoids form tissue structures when cells communicate and coordinate with each other in the micro-environment of the forming organoid.
Promoting organoid function and tissue maturity
Organoid systems have demonstrated a broad array of functionality, including muscle contractility, epithelial barrier function, neuronal activity, hepatocyte detoxification, gastric acid secretion, and secretion of insulin by beta cells. However, PSC-derived tissues tend to be more fetal in nature and do not have the structure and full functionality of their adult counterparts. In humans, organs are able to support the life of premature infants born as early as 24 weeks of gestation (where full term = 40 weeks). Consistent with this, extending time in culture has improved maturation of brain, intestinal, and kidney organoids in the absence of any other manipulation (15–17). However, organoids in culture do not grow beyond a few millimeters in size because of limitations of passive diffusion of oxygen, nutrients, and the other humoral factors. This can be overcome by engrafting organoids onto a vascular bed in animals, where they become vascularized by the host and can continue to grow and mature (8, 13, 18, 19). Human intestinal organoids continue to grow into centimeter-sized tissues with villi containing mature brush borders, circular and longitudinal muscle layers, and interstitial cells of Cajal. As an alternative to in vivo engraftment, engineered vascular systems could be designed to interface with organoids to promote their continued growth and function in vitro (20).
Tissue development, complexity, function, and maturity are interlinked. As organs develop and acquire early organ function, subsequent development processes can be triggered. For example, in the gut, the epithelium promotes smooth muscle differentiation; in turn, smooth muscle development and contraction can in turn promote epithelial villus formation (21). In the lungs, fetal breathing in the second and third trimester helps the maturation of human lung structure and function (22). Organoids can be used to investigate linkages among elapsed time, cellular complexity, and function, as well as the ways in which each may affect maturation. For example, intestinal organoids have been used to show that innervation, colonization by the microbiome, and mechanical stretch can improve tissue functions including intestinal barrier integrity and peristaltic contractions (23–25).
Organogenesis-inspired approaches have been a valuable driver of organoidgenesis. Organoid platforms can benefit from engineering design principles, synthetic biology (25), and systems biology to improve organoid function, maturation, reproducibility, scale-up, and integration into macrofluidic and microfluidic platforms (26).
Engineering principles to control organoidgenesis: Narrative engineering
“Stigmergy,” a concept first introduced in insect biology to explain eusocial behaviors, is a form of indirect communication: a contextual, environmental, and interdependent coordination between individuals that is indirectly affected by their past actions (27). Dynamic multicellular self-organization in organoids requires the translation of such stigmergic elements (i.e., temporal and self-evolving biological events) into engineering-driven efforts that are not the common goal of the canonical tissue engineering concept (28). Sasai (14) elegantly translated this concept into biological systems of self-organization that tend to be strongly tied to history or memory, whereby the morphogenetic behavior of the group of cells is influenced not only by current conditions but also by preceding events. In other words, biological self-organization arises from progressive local interactions between cells of an initially disorganized system by environmental fluctuations, amplified by positive feedback. Thus, controlling biological history (or “narrative”) in a biological system benefits from an integral design strategy that is founded on multiple evolving engineering-driven principles: tissue engineering, synthetic biology, biofabrication, biomaterials, manufacturing, and computational modeling, to name a few.
The new term narrative engineering applies to the interface between principles of biology and engineering for the controlled development of self-organizing systems (Box 1). Three general design strategies, modeled on how organs are assembled during embryonic development, have been adopted for the robust creation of organoid systems. These strategies comprise spatial, biological, and synthetic considerations (Fig. 3).
Box 1. Narrative engineering.
A new field applies the interface principles of biology and engineering for the controlled development of self-organizing systems. Narrative engineering implements rationalized design to maximize the biological history (stigmergy) dependency that leads to the robust tissue self-organization of cell collectives in both time and space, including patterning, assembly, morphogenesis, and growth processes.
The following are key design considerations of narrative engineering:
Space design: Tissue designed as a spatial default for preparedness toward self-organization.
Biological environmental control: The selection of biology-inspired environmental modulators, which rationally recapitulate the full in vivo complexity of organogenesis, homeostasis, and regeneration.
Synthetic environmental control: The design of synthetic environmental modulators that are studied in multiple cutting-edge, engineer-driven disciplines: tissue engineering, synthetic biology, biofabrication, biomaterials, manufacturing, and computational modeling.
Note that these three elements are not necessarily independent but are interrelated processes during organoidgenesis.
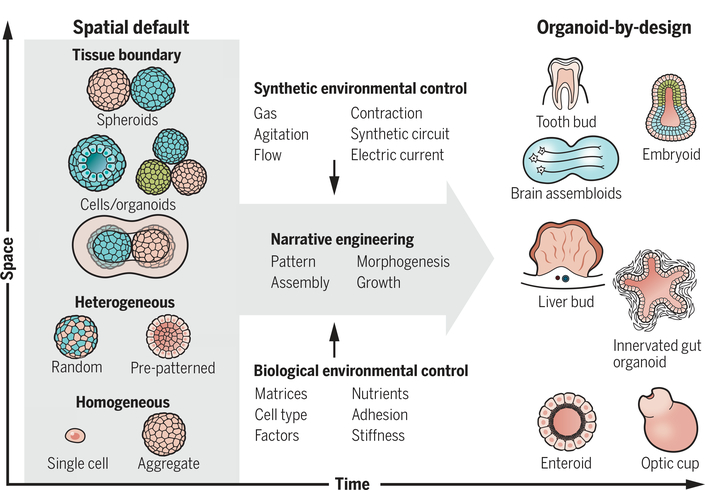
Fig. 3. Concept of narrative engineering.
Starting from the initial default structures, the timed manipulation of environmental factors will facilitate complex and stereotyped organoid formation from stem cells. The right panel explains some examples of terminal products: Enteroids (gut epithelial organoids) or optic cup organoids develop from single or homogeneous stem cell aggregates, whereas vascularized liver bud or innervated gut organoids self-organize by coculturing heterogeneous progenitors. Recent examples of brain organoids and embryoids were self-assembled from the two distinct, preformed tissue aggregates. These self-organization processes are optimized by temporal modulation of biological and/or synthetic parameters.
Space design
Homogeneous aggregate
Default shape and size information is a critical determinant to induce assembly (sorting) and pattern in self-developing systems. Indeed, the aggregation of homogeneous PSCs is most widely used to initiate tissue self-organization for derivation of eye cup (29), brain (6), and blood vessel (30) organoids. This self-organizing process is an aggregate size-sensitive phenomenon. For example, retinal cell differentiation occurs from a small aggregate of 300 mouse embryonic stem cells, and optic cups form from aggregates of 1000 to 2000 cells (14). Precise cell number dependence for asymmetric pattern emergence in part relates to a signaling threshold that will lead to local gradients via reaction-diffusion mechanisms and bistable signaling interaction (14). Therefore, aggregation size controls tissue patterning, which in turn alters subsequent self-organization programs.
Heterogeneous aggregate
Another major approach uses heterogeneous aggregates by coculturing multiple distinct progenitor types. A classical example is the sea sponge self-organization experiment, which uses a heterogeneous aggregate reconstituted from cells from dissociated embryos (31). At early embryonic stages of organogenesis, local intercellular interactions drive a self-assembly program of different regions of developing organs. Recent studies, for example, have cocultured endodermal cells with accessory cell types to reconstitute heterogeneous aggregates from multiple cell types including endothelial (8), neuronal (13), and mesenchymal cells (32). These aggregates self-assemble over time to develop, for instance, primitive vascular networks that augment post-transplant vascular perfusion and engraftment of organoids. Such heterogeneous progenitor interactions aid the integration of accessory structure into 3D organoids to achieve higher-order function.
Tissue boundary
Tissue-tissue interactions have been successfully modeled for the development of complex and interconnected tissues, particularly in the mixed culture of preformed 3D tissues. For example, teeth develop by conjugation of 3D reaggregates of oral ectoderm and tooth mesenchyme cultured in collagen gel to generate a tooth-germ structure, which can grow into a tooth after transplantation into the host (33). Recent experimental approaches have used two distinct stem cell–derived spheroids to model complex tissue-tissue interactions during early embryogenesis (34, 35) and brain development (11, 12). More precise spatial pre-patterning (Fig. 3) to introduce complex boundaries may benefit from evolving bioengineering approaches such as 3D bioprinting and scaffolding methods.
Biological environmental control
Design choice in 3D culture, such as the incorporation of soluble factors or extracellular matrix (ECM), can assist in recapitulating the mechanisms of organogenesis, homeostasis, and regeneration (Fig. 3).
Soluble factors
One of the first examples to incorporate soluble factors was in the derivation of intestinal organoids from adult stem cells [reviewed in (36)]. More recently, timely use of dosed soluble factors has been used to build complex patterning that occurs at the tissue boundary, for example, to model the interface of oral ectoderm and the overlying hypothalamic neuroectoderm to generate adenohypophysis tissues by hedgehog (37), or to balance ureteric epithelium and metanephric mesenchyme fates for kidney organoids by Wnt and fibroblast growth factor (FGF) (17). This provides a time-and dose-specific fluctuation of signaling pathways to trigger pattern formation that is subsequently stabilized by programmed intercellular interactions.
Extracellular matrix
The ECM has important signaling roles and is one of the most commonly manipulated parameters in tissue engineering (1). Matrigel-embedded cultures enabled the study of branching morphogenesis. In addition, a collagen I matrix was shown to change epithelial cell behavior, reducing the kinetics of cell polarization. Manipulating matrix composition is being leveraged to drive interconnected, fused intestinal organoid structure formation (38). Stiffness of the ECM, involving cell adhesion and contraction, is also an important modifier of biological properties. Larger (millimeter-scale) aggregates, termed condensates, can be stimulated by modulation of the mesenchyme-driven actomyosin pathway (32). Mesenchyme-driven condensation coupled with collagenous ECM has recently been used to design various tissue folding (39). Future advancements in spatiotemporal ECM manipulation, such as photodegradable or photoactivatable materials, are pivotal for engineering more complex context in vitro to achieve higher-order functions (40).
Synthetic environmental control
Historically, biologists have attempted to understand how mammalian organs can be cultured ex vivo. Organoid cultures based on such culture techniques include air-liquid interface, on-gel surface, gel-embedding, and roller ball cultures (41). These experimental systems feature the modulation of variables such as cell-intrinsic properties, perfusion, and mechanical properties (Fig. 3).
Cell-intrinsic properties
Recent synthetic biology–inspired processes such as chemical and genetic programming of tissue assembly represent powerful means to control symmetry breaking and facilitate programmed sorting (40). For example, hybridization of complementary DNA sequences coated on a cellular surface enabled the assembly of multicellular structures with defined cell-cell contacts (42). More recently, engineering cadherin-based adhesion in multicellular systems through the synthetic Notch system was shown to promote collective assembly from a random pattern (43). Thus, generation of pattern and shape can be triggered by a synthetic program.
Perfusion
Engineering approaches enable the precise control of the geometry input and output flow conditions, nutrient supply, and shear stress stimulation, as well as the local mechanical properties of the growing 3D tissues (44). Indeed, several fluidic culture systems containing a perfusible vascular system have been developed, one of which was proven to drive the maturation of PSC-derived kidney organoids (45). Designing vascularized systems to control in vitro growth, morphogenesis, and maturity of organoids will be an essential component of any narrative engineering approach.
Other mechanical and electrical stimuli
Mechanical forces (for example, fluid shear stress, contraction, hydrostatic pressure, and tissue distortion) can have a substantial impact on self-driven behaviors, including mechanochemical coupling and force sensing through the YAP/TAZ pathway (46), durotaxic collective migration (47), tissue-specific control of stiffness and mechanical anisotropy (48), and apoptosis-related force (49). Thus, mechanical force directs another signaling dimension in the regulation of tissue self-organization. Evolving organ-on-a-chip–based approaches, coupled with PSC tissue–based approaches, enable advanced mechanical modeling in organoids to stimulate maturation, for example, by mini-scale agitation in brain organoids (50), turbulence in megakaryocytes (51), and contraction in cardiac tissues (52). Cell behaviors can be electrochemically controlled using optogenetics, as in the control of neuronal activity in brain organoids (7, 8). In biological self-organization, interaction rules of elements are not constant but generally evolve in time and space; therefore, timely strategic integration of engineering principles will complement the limitations of biological approaches to devise a superior, synergistic strategy to construct elaborate organoid architecture.
Conclusion
By applying the principles of organogenesis and narrative engineering, it should be possible to design organoids that are simple but highly defined, or highly complex and functional organoids. This approach acts as an interface between biology and engineering to drive robust, reliable, and effective organoidgenesis. New organoid systems can be designed to interrogate complex organogenesis that is otherwise inaccessible. For example, optic cup organoids coupled with 4D measurements, theoretical modeling, and experimental perturbation resolved a controversy associated with eye cup organogenesis (29). Emerging tools such as gene editing, single-cell analysis, optogenetics, chemogenetics, and superresolution/macroresolution imaging can be combined with in silico tools [e.g., agent-based (53) and force-based (14) modeling] to supervise better organoidgenesis using the three key design elements of narrative engineering. A more holistic approach may prove essential for improving the robustness of organoidgenesis to drive the growth and maturation that is required for better organ modeling and eventual transplantation-based therapies.
ACKNOWLEDGMENTS
There are many excellent recent studies in organoid engineering that cannot be introduced because of space constraints. We apologize to those authors whose work we did not reference.
Funding: Supported by NIH grants R01DK092456, U19 AI116491, P01 HD093363-01, and UG3 DK119982 (J.M.W.) and by NIH grant UG3 DK119982, PHS grant P30 DK078392 of the Digestive Disease Research Core Center in Cincinnati, the NY Stem Cell Foundation, JSPS, Takeda Science Foundation, and AMED grants JP18fk0210037h0001 and JP18bm0704025h0001 (T.T.).
Competing interests: J.M.W. is an inventor on the following patent/patent applications: 9,719,068, 10,174,289, 16/084,599, 15/515,840, PCT/US2018/054635, PCT/US2018/029083, and PCT/US2017/064600 that were held/submitted by Cincinnati Children’s Hospital Medical Center and cover Organoid Based Technologies. T.T. serves on the scientific advisory board of Healios and granted licenses to Healios through Yokohama City University over their inventions that relate to the subject of this manuscript.
REFERENCES AND NOTES
- 1.Simian M, Bissell MJ, J. Cell Biol. 216, 31–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCauley HA, Wells JM, Development 144, 958–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekkers JF et al. , Sci. Transl. Med 8, 344ra84 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Wells JM, Watt FM, Nature 557, 322–328 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Zorn AM, Wells JM, Annu. Rev. Cell Dev. Biol. 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiraku M et al. , Cell Stem Cell 3, 519–532 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Mansour AA et al. , Nat. Biotechnol. 36, 432–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takebe T et al. , Nature 499, 481–484 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Ormel PR et al. , Nat. Commun. 9, 4167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abreu CM et al. , Front. Microbiol. 9, 2766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y et al. , Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA, Nat. Methods 14, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workman MJ et al. , Nat. Med. 23, 49–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasai Y, Nature 493, 318–326 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Sloan SA et al. , Neuron 95, 779–790.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spence JR et al. , Nature 470, 105–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takasato M et al. , Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Daviaud N, Friedel RH, Zou H, eNeuro 5, ENEURO.0219–18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson CL et al. , Nat. Med. 20, 1310–1314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase K, Kamm RD, Regen. Med. 12, 285–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walton KD, Mishkind D, Riddle MR, Tabin CJ, Gumucio DL, Wiley Interdiscip. Rev. Dev. Biol. 7, e317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warburton D, Neonatology 111, 398–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill DR et al. , eLife 6, e29132 (2017).29110754 [Google Scholar]
- 24.Poling HM et al. , Nat. Biomed. Eng. 2, 429–442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitada T, DiAndreth B, Teague B, Weiss R, Science 359, eaad1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SE, Georgescu A, Huh D, Science 364, 960–965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theraulaz G, Bonabeau E, Artif. Life 5, 97–116 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Langer R, Vacanti JP, Science 260, 920–926 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Eiraku M et al. , Nature 472, 51–56 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wimmer RA et al. , Nature 565, 505–510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeichi M, Dev. Cell 21, 24–26 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Takebe T et al. , Cell Stem Cell 16, 556–565 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Nakao K et al. , Nat. Methods 4, 227–230 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M, Science 356, eaal1810 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Rivron NC et al. , Nature 557, 106–111 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Sato T, Clevers H, Science 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Suga H et al. , Nature 480, 57–62 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Sachs N, Tsukamoto Y, Kujala P, Peters PJ, Clevers H, Development 144, 1107–1112 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Hughes AJ et al. , Dev. Cell 44, 165–178.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murrow LM, Weber RJ, Gartner ZJ, Development 144, 998–1007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamir ER, Ewald AJ, Nat. Rev. Mol. Cell Biol. 15, 647–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gartner ZJ, Bertozzi CR, Proc. Natl. Acad. Sci. U.S.A. 106, 4606–4610 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA, Science 361, 156–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sontheimer-Phelps A, Hassell BA, Ingber DE, Nat. Rev. Cancer 19, 65–81 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Homan KA et al. , Nat. Methods 16, 255–262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupont S et al. , Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Sunyer R et al. , Science 353, 1157–1161 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Nerurkar NL, Lee C, Mahadevan L, Tabin CJ, Nature 565, 480–484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS, Science 321, 1683–1686 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian X et al. , Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito Y et al. , Cell 174, 636–648.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Ronaldson-Bouchard K et al. , Nature 556, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaul H, Ventikos Y, Brief. Bioinform. 16, 137–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]