Abstract
Background
Human leukocyte antigen (HLA)‐DP is much less studied than other HLA class II antigens, that is, HLA‐DR and HLA‐DQ, etc. However, the accumulating data have suggested the important roles of DP‐restricted responses in the context of cancer, allergy, and infectious disease. Lack of animal models expressing these genes as authentic cis‐haplotypes blocks our understanding for the role of HLA‐DP haplotypes in immunity.
Methods
To explore the potential cis‐acting control elements involved in the transcriptional regulation of the HLA‐DPA1/DPB1 gene, we performed the expression analysis using bacterial artificial chromosome (BAC)‐based transgenic humanized mice in the C57BL/6 background, which carried the entire HLA‐DP401 gene locus. We further developed a mouse model of Staphylococcus aureus pneumonia in HLA‐DP401 humanized transgenic mice, and performed the analysis on the expression pattern of HLA‐DP401 and immunological responses in the model.
Results
In this study, we screened and identified a BAC clone spanning the entire HLA‐DP gene locus. DNA from this clone was analyzed for integrity by pulsed‐field gel electrophoresis and then microinjected into fertilized mouse oocytes to produce transgenic founder animals. Nine sets of PCR primers for regional markers with an average distance of 15 kb between each primer were used to confirm the integrity of the transgene in the five transgenic lines carrying the HLA‐DPA1/DPB1 gene. Transgene copy numbers were determined by real‐time PCR analysis. HLA‐DP401 gene expression was analyzed at the mRNA and protein level. Although infection with S aureus Newman did not alter the percentage of immune cells in the spleen and thymus from the HLA‐DP401‐H2‐Aβ1 humanized mice. Increased expression of HLA‐DP401 was observed in the thymus of the humanized mice infected by S aureus.
Conclusions
We generated several BAC transgenic mice, and analyzed the expression of HLA‐DPA1/DPB1 in those mice. A model of S aureus‐induced pneumonia in the HLA‐DP401‐H2‐Aβ1−/− humanized mice was further developed, and S aureus infection upregulated the HLA‐DP401 expression in thymus of those humanized mice. These findings demonstrate the potential of those HLA‐DPA1/DPB1 transgenic humanized mice for developing animal models of infectious diseases and MHC‐associated immunological diseases.
Keywords: bacterial artificial chromosome (BAC), gene expression, HLA‐DP4, humanized mice, Staphylococcus aureus pneumonia
1. INTRODUCTION
In humans, there are three HLA class II molecules, HLA‐DR, HLA‐DQ, and HLA‐DP, which compose of two separate α‐ and β‐chains and present antigenic peptides to CD4+ T lymphocyte. 1 , 2 It has been well known that specific major histocompatibility complex (MHC) II genes are associated with infectious or immunological diseases, such as autoimmune diseases including rheumatoid arthritis (RA), diabetes, and so on. 3 , 4 , 5 , 6 , 7
Compared to HLA‐DR and HLA‐DQ, HLA‐DP is less frequently studied because early study demonstrates that HLA‐DP incompatibility was less likely to cause graft‐vs‐host disease (GVHD). 8 , 9 However, recent data revealed that a single mismatch in this molecules is enough to trigger a specific T cell response after bone marrow transfer. 10 , 11 , 12 Functional studies have also shown that the HLA‐DP molecules are involved in the process of Be presentation to CD4+ T lymphocyte. 13 , 14 , 15 Furthermore, various kind of antigenic peptides originated from viruses, 16 , 17 , 18 fungi, 19 and tumors 20 , 21 can be presented to CD4+ T lymphocyte by HLA‐DP molecules.
In a previous study, FVB/N transgenic mouse strains containing different alleles HLA‐DPB1*0401, HLA‐DPB1*0201, and HLA‐DPB1*1701 were developed, to illustrate that a Be‐induced adaptive immune response is developed by interaction of HLA‐DPB1 molecule and an environmental exposure in mice and human. 22 However, HLA‐DPA1 may participate in disease susceptibility through interactions during positive and negative selection of the T‐cell repertoire or through aspects of their peripheral co‐regulation. 23 , 24 , 25 Furthermore, the role of pseudogenes, intergenic “junk” DNA sequences within HLA‐DP locus cannot be assessed by the single independent gene locus‐based mice. 26 , 27 , 28 Therefore, we generated BAC‐based transgenic humanized mice carrying HLA‐DP haplotype, which contained the linked HLA DOA‐DPA1*0103‐DPB1*0401 (DP401) with the authentic flanking and intervening sequences. The tissue‐specific expression was found for HLA‐DPA1/DPB1 molecules in these transgenic mice, and S aureus infection increased the HLA‐DP401 expression in the thymus, indicated that these mice will be used to set up animal models to clarify the role of HLA‐DP genes in predisposition and development of autoimmunity.
2. METHODS
2.1. Generation of transgenic mice
BAC clone CH501‐138A21 was derived from the BACPAC resources. BAC DNA was prepared by the conventional method as previously described. 29 Briefly, BAC DNA was harvested from 1 L of bacterial culture by alkaline lysis and subsequently purified using NucleoBond BAC 100 Kit (Clontech). Integrity of BAC DNA was confirmed by pulsed field gel electrophoresis in 1% agarose/0.5 × Tris‐Borate‐EDTA buffer for 15 hours at 14°C (6 V/cm, 0.2‐22s switch time) after digestion with restriction endonucleases MluI and XhoI. Purified circular BAC DNA was diluted to 1‐3 ng/μL in microinjection buffer and microinjected into the pronuclei of C57BL/6 mouse zygotes. Microinjection was done at Mouse Genome Editing Lab in Department of Laboratory Animal Science, Shanghai Public Health Clinical Center (SHPHC). Transgenic founder animals were typed by using PCR with the specific primers. Transgenic founders were bred to wild‐type C57BL/6 mice, and offspring were typed by PCR using the following specific primers. For HLA‐DOA, 5′‐CTCCTCTTCCCATCCTG‐3′ (forward) and 5′‐TCATCCACTTTCCTCCC‐3′ (reverse) 1478 bp; for HLA‐DP/LCR7#, 5′‐TGTGGTTCTTGCTTGATGA‐3′ (forward) and 5′‐GTTGCTAAGACAGGAGTGA‐3′ (reverse) 798 bp; for HLA‐DP/LCR8#, 5′‐CTCTTCCTCTTACTGTGGTT‐3′ (forward) and 5′‐TTGATTGCCTGTGCTACTT‐3′ (reverse) 637 bp; for HLA‐DP/LCR9#, 5′‐GAACCTGTGAAGTAGAAGTAAC‐3′ (forward) and 5′‐AGATTGTCCTGAGTGAGTTG‐3′ (reverse) 762 bp; for Hotspot DPA1, 5′‐CAGGAGCCACAGGAGTAT‐3′ (forward) and 5′‐AGCATTAACAGCACATAGGT‐3′ (reverse) 588 bp; for HLA‐DPA1, 5′‐TGTTGCTCCTTCTTCTTCCCC‐3′ (forward) and 5′‐TGGAATAGAGGATGCCAGGAG‐3′ (reverse) 1067 bp; for HLA‐DPB1, 5′‐GAGGATTAGATGAGAGTGGC‐3′ (forward) and 5′‐3′ (reverse) 445 bp; for HLA‐P6, 5′‐GAAGGAAGGAAGGAAGGAAGG‐3′ (forward) and 5′‐GAAGAAAGATGGGGTTTGGAC‐3′ (reverse) 1272 bp; for HLA‐DPA2, 5′‐CCATTCTCCATCTTCTCCTT‐3′ (forward) and 5′‐CCTCCTCTGCTGTCCTAA‐3′ (reverse) 701 bp.
Transgenic mice expressing HLA‐DP in the absence of murine endogenous class II molecules was generated as follows. The H2‐Aβ1 neo floxed mutant mice on a C57BL/6 background were crossed with EIIa‐cre transgenic mice. 30 The heterozygotes offspring H2‐Aβ1 ± was crossed with line 3‐3# humanized transgenic mice. The H2‐Aβ1 ± littermate expressing HLA‐DP molecule were selectively intercrossed with the same genotypes to obtain HLA‐DP‐H2‐Aβ1 −/− mice, which did not express any endogenous mouse class II antigens.
2.2. Tail DNA isolation and copy number estimation
DNA was extracted and prepared from mice tail biopsies by conventional method using tissue DNA extract Kit (Trans, Beijing). Genomic DNA samples were quantified on UV spectrophotometer at 260 nm and diluted to 10 ng/μL for real‐time PCR.
A standard curve of real‐time PCR data was generated as previously described. 31 Briefly, BAC DNA were quantified by UV spectrophotometry at 260 nm. Then, A series of standard samples, which contained the given the ratio of BAC molecules ranged from ~1 to ~50 BAC copies, were prepared with the known amounts of BAC template. Two microliters (20 ng) of genomic DNA samples or copy number standards were analyzed in a 10 μL reaction volume with the ABI PRISM® 7900HT sequence detection system. Each experiment was run with no‐template controls. All reactions were carried out in triplicate. Copy number estimates were derived from delta Ct values for standard curve samples. Copy number of samples can be further calculated by the resulting equation (estimated copy number = 2e((deltaCt‐y intercept)/slope)).
The following primer pairs were used: for HLA‐DPA1, 5′‐GGTGTTGCTCCTTCTTCTTCC‐3′ (forward) and 5′‐AACTCCTCCAGATGCCAGAC‐3′ (reverse) 243 bp; HLA‐DPB1, 5′‐GAAGGAAGGAAGGAAGGAAGGA‐3′ (forward) and 5′‐CATTCAGGAACCATCGGACTTG‐3′ (reverse) 217 bp.
2.3. RNA Isolation, cDNA synthesis, and RT‐PCR
RNA was isolated from the PBMC, spleen and lung using Tri‐Reagent (Trans, Beijing) following the manufacturer's protocol. Reverse transcription RNA and cDNA Synthesis with Hifair® Ⅱ 1st Strand cDNA Synthesis Kit (gDNA digester plus) (Yeasen Biotech), PCR amplification with TransTaq DNA Polymerase High Fidelity (Yeasen Biotech), all the steps followed the manufacturer's protocol. Sequences of primers used and the predicted amplification sizes are as follows: for HLA‐DPA1‐V2 E2‐E3, 5′‐GCCTCAGTTCCTCATCAC‐3′ (forward) and 5′‐GAACGCTGGATCAAGGTAT‐3′ (reverse) 381 bp; for HLA‐DPA1‐V2 E4‐E6, 5′‐TTCCACAAGTTCCATTACCT‐3′ (forward) and 5′‐CCTAAGTCCTCTTCTGTTCA‐3′ (reverse) 303 bp; for HLA‐DPB1 E1‐E3, 5′‐CCACTCCAGAGAATTACCTT‐3′ (forward) and 5′‐GGAACCATCGGACTTGAAT‐3′ (reverse) 387 bp; for HLA‐DPB1 E3‐E6, 5′‐GTAATGGAGACTGGACCTTC‐3′ (forward) and 5′‐GACTTCAGAGCAACTTCTTG‐3′ (reverse) 386 bp; for HLA‐DOA E1‐E3, 5′‐CACCAAGGCTGACCACAT‐3′ (forward) and 5′‐CACGATGCAGATGAGGATG‐3′ (reverse) 331 bp; and for HLA‐DOA E3‐E5, 5′‐GTTCCGCAAGTTCCACTA‐3′ (forward) and 5′‐GCACTTAAAGGGCACTGA‐3′ (reverse) 336 bp.
2.4. Expression analysis of transgenic mice with real‐time PCR and immunostaining blotting
Quantitative PCR was conducted in 20 μl reactions containing first‐strand cDNA template, Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Yeasen Biotech), and primer sets using the ABI PRISM® 7900HT sequence detection system (Applied Biosystems). The following oligonucleotide primers for HLA‐DPA1, HLA‐DPB1 and GAPDH were purchased from Sangon Bio, Inc: for HLA‐DPA1, 5′‐GTACAGACGCATAGACCAACAG‐3′ (forward) and 5′‐GAACTTGTCAATGTGGCAGATG‐3′ (reverse) 28 4bp; HLA‐DPB1, and 5′‐GGAACAGCCAGAAGGACATC‐3′ (forward) and 5′‐CAGGAACCATCGGACTTGAAT‐3′ (reverse) 218 bp. Reaction mixtures were incubated for an initial denaturation at 95℃ for 10 minutes, followed by 40 PCR cycles. Each cycle consisted of 95℃ for 15 seconds and 60℃ for 60 seconds. The mRNA levels of all genes were normalized using GAPDH as internal controls.
For immunoblotting, protein extracts (50 μg) were resolved by one‐dimensional SDS‐PAGE (12% acrylamide) under reducing conditions and electrophoretically transferred to polyvinylidene difluoride membranes (GE Amersham). Membranes were probed with primary antibodies against HLA‐DPA1, HLA‐DPB1 or β‐actin (ACTB; 1:10 000 dilution; product sc‐47778; Santa Cruz Biotechnology) diluted in Tris‐buffered saline with 0.1% tween 20 containing 5% bovine serum albumin (Jackson ImmunoResearch Laboratories) or 5% dried milk. Following incubation with a horseradish peroxidase‐conjugated secondary goat anti–rabbit antibody (1:10 000 dilution; product W401B; Promega), the protein bands were visualized by chemiluminescence using a Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
2.5. Histology and immunostaining procedure
Mice were perfused transcardially with 2%‐4% paraformaldehyde (PFA) in phosphate‐buffered saline (PBS; pH 7.2‐7.4). The tissues including spleen, lung, kidney, gut, and thymus was dissected, post‐fixed in 2%‐4% PFA for 2‐12 hours, and cryoprotected in 30% sucrose in PBS at 4°C overnight. After sectioning on a cryostat, 10‐12 µm sections were collected onto Superfrost Plus Microscope slides (Fisher Scientific, Loughborough, UK). The polyclonal primary antibody used was specific for HLA‐DPA1 (rabbit GTX104414; GeneTex Inc, GeneTex, USA), and the mouse monoclonal primary antibody used was specific for HLA‐DPB1 (mouse sc‐134357; Santa Cruz Biotechnology, Santa Cruz, USA).
Staining against HLA‐DPA1, HLA‐DPB1 was performed using the standard immunofluorescence procedure. Cryosections were washed in PBS (3 × 10 minutes), placed into blocking solution [1% bovine serum albumin (BSA), 1% normal horse serum, and 0.3% Triton X‐100 in PBS] for 1‐2 hours, and then incubated in a mixture of the polyclonal, primary antisera: rabbit anti‐HLA‐DPA1 (1:100‐500 dilution), mouse anti‐HLA‐DPB1 (1:100‐500 dilution) in blocking solution. Primary antibody incubation lasted for 36‐48 hours at 4 ℃, and then sections were washed in PBS 3 × 10 minutes and incubated for 2‐18 hours in a mixture of secondary antibodies: Alexa633 goat anti‐rabbit or Alexa488 rabbit anti‐mouse (1:400; Molecular Probes, USA). The slides then were washed one time for 10 minutes in 0.1 mol/L PBS and two times for 10 minutes in 0.1 mol/L PBS before coverslipping slides with Fluormount G (Southern Biotechnology Associates, USA). Fluorescent images were captured using a Leica TCS SP2 Spectral Confocal Microscope (Leica Microsystems Inc, Mannheim, Germany).
Staining against HLA‐DPA1, HLA‐DPB1 was performed using the standard immunohistochemical procedure according to the manufacturer's instructions (VECTASTAIN Elite ABC Kit; Vector Laboratories, Burlingame, USA). The standard immunocytochemical procedure used an avidin‐biotin‐peroxidase complex (ABC; Vector Laboratories, Burlingame, CA) and 3,38‐diaminobenzidine (DAB, Sigma). Cryosections were washed in four 4‐minute washes of 0.1 mol/L PBS, pH 7.4, containing 0.3% Triton X‐100. The slides subsequently were incubated in blocking solution [1% bovine serum albumin (BSA), 1% normal horse serum, and 0.3% Triton X‐100 in PBS] for 1‐2 hours, followed by a 24 hours incubation with the primary antibody at 4°C. The slides were washed with PBS four times followed by a 45‐minute application of the biotin‐conjugated secondary antibody. Three additional washes in PBS preceded both the 30‐minute application of ABC and the 10‐minute incubation with a PBS solution containing 0.5 mg/mL DAB, and 0.01% H2O2 to tint the reaction product blue. Counterstaining is carried out with standard hematoxylin staining and bright field images of the sections were captured digitally.
2.6. Bacterial strains, growth conditions, and Staphylococcus aureus pneumonia model
S. aureus stain Newman (a gift from Prof. Minggui Wang at Huashan Hosptital affiliated to Fudan University, Shanghai, China) was used in this study. 32 Bacteria grew aerobically at 37C in tryptic soy broth (TSB) (Difco) with shaking (250 rpm). A microspectrophotometer system (K5600) was used to analyze the bacterial growth curves. Then the bacteria at exponential growth phase were harvested by centrifugation, washed, resuspended in PBS, and adjusted to a final concentration.
The infection murine model of S. aureus was a modification of that described by Lee. 33 Briefly, 8‐week‐old C57BL/6, HLA‐DP‐H2‐Aβ1−/− humanized mice, and H2‐Aβ1−/− mice were intranasally infected with 2.4 × 109 CFU of S aureus in a total volume of 50 µL. After 72 hours post‐infection, mice were sacrificed and the spleen, thymus and lung were aseptically harvested for further analysis.
2.7. Antibodies and flow cytometry
Single‐cell suspensions of splenocytes and thymic cells were incubated with an FcR‐specific blocking mAb (eBioscience) and directly stained with fluorescently conjugated mAbs to murine CD3 (PerCP‐Cy5.5), CD4(Alexa Fluor 700), CD8 (FITC),F4/80 (FITC), CD19 (eFluor 405), CD11b (PE), CD11c (APC‐E780). Then, cells were analyzed by flow cytometer (FACS Aria II, BD Biosciences).
2.8. Statistical analysis
The cytometric bead array data were generated in a graphical and tabular format using the BD cytometric bead array analysis software. The difference between groups was calculated using SPSS statistical software (SPSS Inc, Chicago, USA).
3. RESULTS
3.1. Generation of BAC‐based transgenic humanized mice carrying HLA‐DP4 haplotype and analysis of BAC transgene integrity
Screening human BACs library demonstrate that BAC clone CH501‐138A21, 124 kb in length, contains 12 kb 5′‐UTR of DOA and 27 kb 3′‐UTR DPB1, and spans the entire HLA‐DP gene locus (DOA‐DPA1*0103‐DPB1*0401). These existing flanking regions may keep the cis‐regulatory elements of HLA‐DPA1/DPB1 gene as far as possible (Figure 1A).
FIGURE 1.
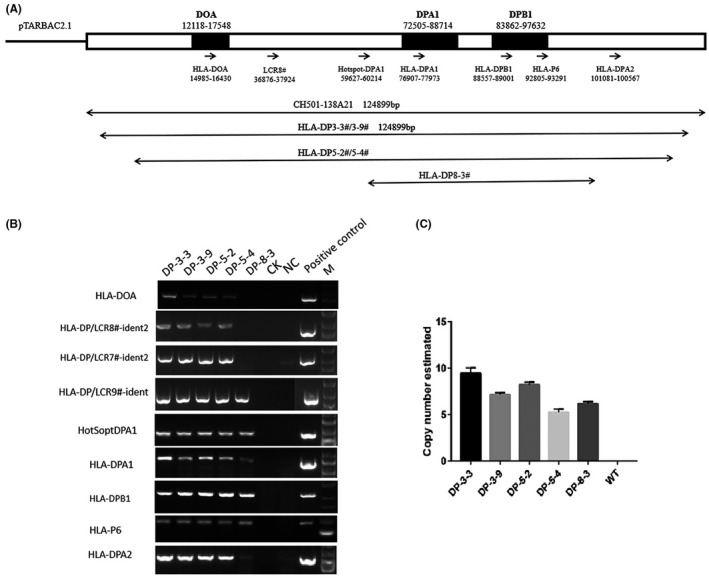
Schematic diagram of the HLA‐DP4‐haplotype‐containing BAC. A, The original BAC CH501‐138A21 is ~124 kb in size, harboring the full HLA‐DP4 locus. Nine sets of PCR primers for regional markers with an average distance of 15 kb between each primer were used to identify the HLA‐DP BAC transgenes. The transgenic lines carrying the different fragments are indicated. BAC transgene integrity assayed by these primers is shown in B. Copy number is estimated by real‐time PCR, and shown in C. M 5Kb DNA marker
BAC DNA constructs were purified and used for pronuclear injections to generate founder lines as previously described. 29 Four founder animals harboring the BAC DNA fragment were generated. Because founder animals are generally considered to be a mosaic of transgenic and non‐transgenic cells, wild‐type animals were crossed with founder animals to get non‐mosaic F1 offspring. Five lines representing five different integration events were established from 3 founders. In order to rule out the insertion of fragmented transgenes, we employed polymorphic marker analysis to monitor integrity of large transgenes in these transgenic lines. Nine sets of PCR primers for regional markers with an average distance of 15 kb between each primer were assessed within the HLA‐DP4 BAC transgenes. Four lines were shown to harbor most of regional markers, suggesting integration of complete BAC molecules (Figure 1A,B). However, one lines lacked regional markers across one portion of the BAC, suggesting this transgene integrated into the mouse genome as partial fragments (Figure 1A,B). Importantly, the LCR8# region, a distal S’‐Y’ modules functioning as transcriptional enhancers, 28 was missing from the “fragmented” transgenic line 8‐3# (Figure 1A,B).
3.2. Estimating BAC copy number by real‐time PCR
Quantitative PCR can be easily applied to estimate BAC copy number in transgenic mice. 31 Robust standards curves are generated with BAC dilutions in mouse diploid genome. Our data showed the low copy concatamers from 6 to 10 copies in these humanized transgenic mice (Figure 1C), which were consistent with most published reports. 31 , 34 , 35 It has been reported that at least one full‐length monomer is contained in the BAC transgenic mice with multiple‐copy BAC insertions. 31 , 34 After breeding, line 8# founder animals generated close to 50% transgenic littermates, indicating a single site of BAC transgene integration in the founder. At least two distinct, unlinked transgene insertion were found in line 3# founder animals and 5# line founder animals. For example, ten BAC copies were found in 5‐2# line, whereas five copies were found in line 5‐4# (Figure 1C).
3.3. mRNA expression analysis of BAC transgenic mice
Total RNA samples were isolated from spleen tissue of five transgenic mice. Transcript size was determined by RT‐PCR. Primers were specifically designed to amplify the whole transcripts of HLA‐DPA1, HLA‐DPB1 and HLA‐DOA. The expression of HLA‐DPA1 and HLA‐DPB1 was detected in all transgenic lines. But expression of HLA‐DOA was absent from spleen of transgenic lines 8‐3# (Figure S1). PCR products were gel‐recovered, cloned into TA vectors and further verified by sequence (data not shown).
To determine the transgene‐derived transcript levels in five transgenic lines, real‐time PCR was used to analyze the tissue‐specificity of HLA‐DPA1/HLA‐DPB1 expression. Results revealed the lower expression of transgene in peripheral blood mononuclear cell, PBMC (Figure 2A,B), spleen (Figure 2C,D) and lung tissue (Figure 2E,F) from line 8‐3# with the shorter DNA fragments.
FIGURE 2.
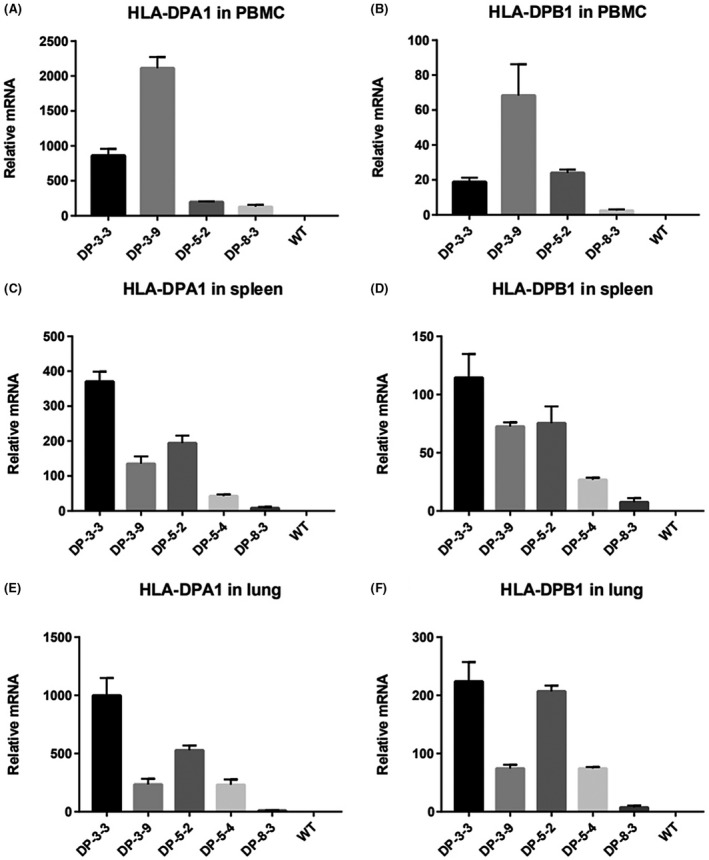
Expression level of the transgene is analyzed by real‐time PCR. A and D PBMC, B and F spleen, C and G lung. Labeling is as shown in Fig
3.4. Distribution pattern of HLA‐DPA1 and HLA‐DPB1 proteins in various tissues from BAC humanized mice
The HLA‐DPA1/HLA‐DPB1 levels in various tissues of transgenic mice were further determined by western blotting. We investigated the HLA‐DPA1 expression in the spleen (Figure 3A) and kidney (Figure 3B). In the kidney, the expression of HLA‐DPA1 was detected in transgenic mice from line 3# and 5#. However, the robust expression of HLA‐DPA1 was only observed from spleen tissue from line 3‐3# (Figure 3A). On the other hand, the expression if HLA‐DPB1 was also observed in various tissues including spleen (Figure 3C), kidney (Figure 3D), lung (Figure 3E), and small intestine (Figure 3F) from all lines. Unexpectedly, we also detected the obvious expression of HLA‐DPB1 in these tissues from “fragmented” transgenic mice, line 8‐3# (Figure 3C‐F).
FIGURE 3.
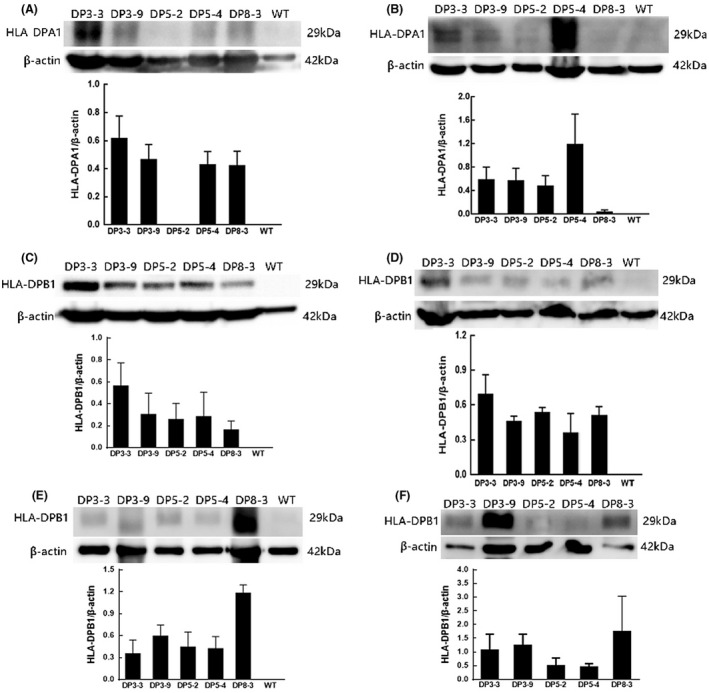
Representative western blot revealed the tissue specificity of transgene expression with antibody against HLA‐DPA1/DPB1. The band at 29 kDa for HLA‐DPA1 is indicated: A, spleen; B, kidney. The band at 29 kDa for HLA‐DPB1 is indicated: C, spleen; D, kidney; E, lung; F, gut
The current results raised a possibility that the differential expression for transgene could be found in five lines. To further verify the distribution of HLA‐DPA1/DPB1, we employed the Immunofluorescence staining to investigate the expression of transgene. Immunofluorescence microscopy revealed the expression of HLA‐DPA1 (Figure 4A‐F) and HLA‐DPB1 (Figure 4G‐L) in the spleen from line 3‐3#. The expression of HLA‐DPB1 was further observed in the lung (Figure 5A‐F) and gut (Figure 5G‐L) from line 3‐3#. Immunohistochemistry staining was also be used to analyze the expression of the transgene. A strong staining of HLA‐DPA1 was detected in the kidney from line 3‐3#, including the proximal convoluted tubule and distal straight tubule (Figure S2A‐D), though no positive signals in the renal corpuscle (Figure S2B). a strong staining of HLA‐DPB1 was observed in the thymus from line 3‐3# (Figure S2E,F).
FIGURE 4.
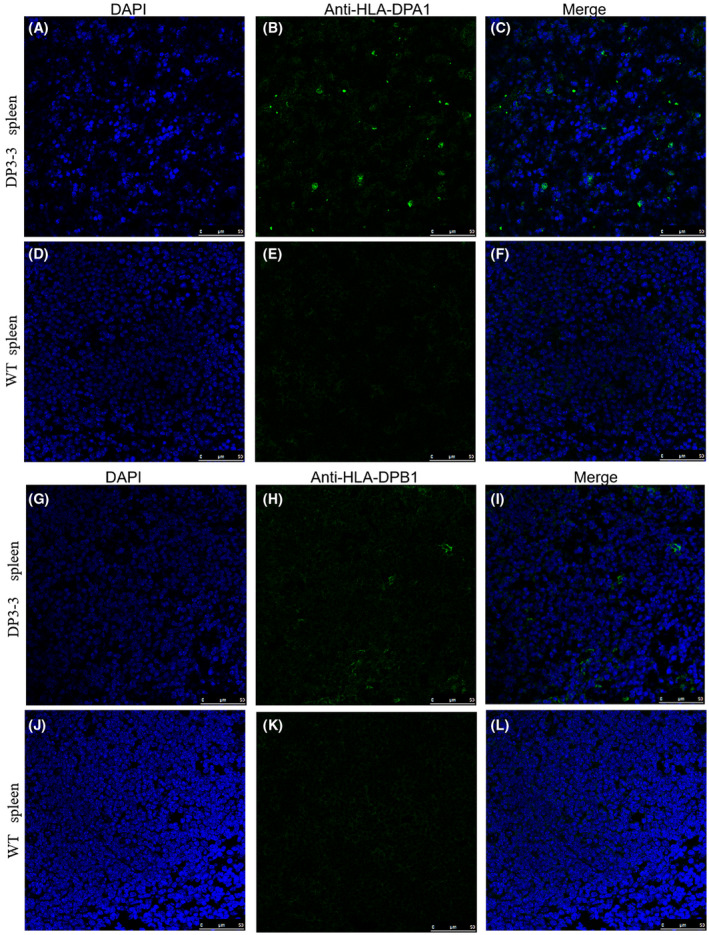
Immunofluorescence staining reveal the expression of HLA‐DPA1/DPB1 in transgenic mice. A‐C HLA‐DPA1 in spleen from transgenic mice DP3‐3#, D‐F spleen from WT mice, G‐I HLA‐DPB1 in spleen from transgenic mice DP3‐3#, J‐L spleen from WT mice. Scale bar 50 µm
FIGURE 5.
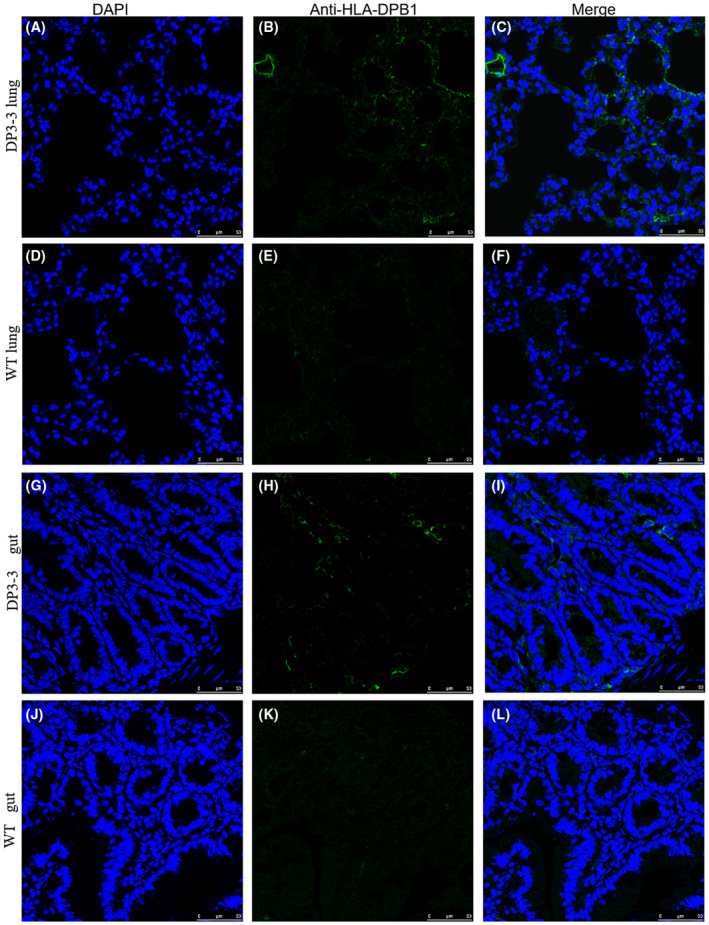
Immunofluorescence staining reveal the expression of HLA‐DPB1 in transgenic mice. A‐C HLA‐DPB1 in lung from transgenic mice DP3‐3#, D‐F lung from WT mice, G‐I HLA‐DPB1 in gut from transgenic mice DP3‐3#, D‐F gut from WT mice. Scale bar 50 µm
3.5. Positive and negative selection of CD4+ T cells in BAC humanized mice
To comprehend whether the introduction of the HLA‐DP transgene affect the CD4 T lymphocyte compartment in the murine class II knockout Aβ mice, T‐cell subpopulations were analyzed by flow cytometry in thymocytes and splenocytes from transgenic and control mice. CD3+ and CD4+ T cells significantly decreased in the spleen from the H2Aβ1−/− mice and BAC‐transgenic homozygotes HLA‐DP‐H2Aβ1−/− (Figure 6A‐C). It has been reported that there was a significant decrease in the percentage of CD4+ T cells in the H2‐Aβ‐deficient DQ8 transgenic mice, indicating that the class II molecules expressed may differentially affect CD4+ T‐cell development in the H2‐M‐deficient background. 36 The percentage of CD19+ B cells (Figure 6D), CD11c+ dendritic cells (Figure 6E) and F4/80+ macrophage (Figure 6F) did not significantly differ among these groups. We also analyzed the T cell development in thymus (Figure 6G‐I). The percentage of CD4+ T cells significantly reduced in thymus from BAC‐transgenic homozygotes HLA‐DP‐H2Aβ1−/− compared with the H2‐Aβ1−/− mice (Figure 6H), indicating inefficient interaction between murine and human class II. 37
FIGURE 6.
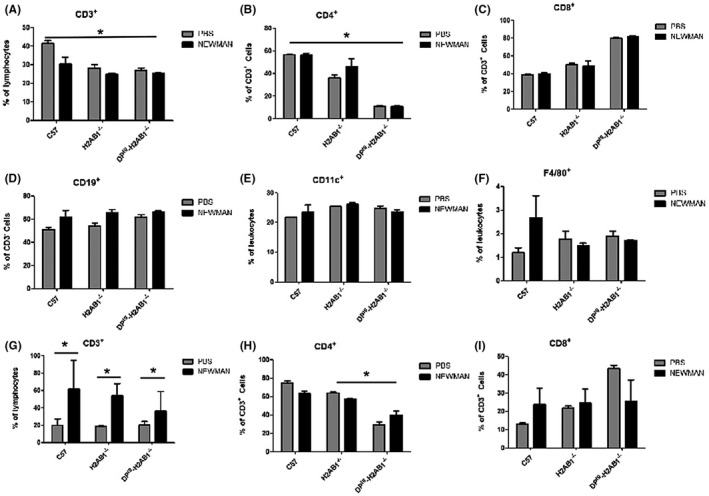
Positive and negative selection of CD4+ T cells in HLA‐DP401 humanized mice. A‐C, Splenic T cells from mice with the indicated genotypes were double‐stained with CD3, CD4 and CD8. D, Splenic B cells from mice with the indicated genotypes were CD19. Splenic cells were stained with antibodies against CD11c, CD11b, F4/80. Subsequently, cells were gated on CD11b+CD11c+ dendritic cells (E) or F4/80+ CD11b+ macrophage (F). (G), (H) and (I) thymic T cells from mice with the indicated genotypes were double‐stained with CD3, CD4 and CD8. *P < .05
3.6. Expression of HLA‐DP significantly increased in thymus from Staphylococcus aureus pneumonia of BAC humanized mice
To further investigate the regulation of human class II molecular expression in the BAC transgenic humanized mice, cell surface expression of DP molecules was examined on splenocyte and thymic cells from HLA‐DP‐H2Aβ1−/−. Line 3‐3# humanized transgenic mice were bred onto the H2‐Aβ1−/− background, in which inactivation of the H2‐Aβ1−/− gene results in the loss of cell surface murine MHC‐II expression. S aureus infection failed to change the percentage of CD3+ T cells in spleen from HLA‐DP‐H2Aβ1−/− and H2Aβ1−/− (Figure 6A). But, the percentage of CD3+ T cells significantly increased in thymus from various genotypes including the C57BL/6, HLA‐DP‐H2Aβ1−/− and H2Aβ1−/− after S aureus infection (Figure 6G). on the other hand, S aureus infection did not result in the change of CD4+ T cells percentage in thymus from the HLA‐DP‐H2Aβ1−/− humanized mice (Figure 6H).
As expected, the expression of HLA‐DP molecule was found in thymic cells (Figure 7A). Interestingly, the percentage of DP+CD4+ (Figure 7B), and DP+CD8+ (Figure 7C) T‐cell lymphocytes significantly increased in the thymus from S aureus pneumonia of BAC transgenic humanized mice. We also found the expression of HLA‐DP molecule in splenocyte cells (Figure 7D). The expression of the HLA‐DP molecules had no deleterious effect on B‐cell development in spleen from Staphylococcus aureus pneumonia of BAC humanized mice (Figures 6D and 7E). The percentage of DP+CD4+ (Figure 7F), DP+CD8+ (Figure 7G), DP+CD11c+ dendritic cells (Figure 7H) and DP+F4/80+ macrophage (Figure 7I) did not significantly differ among these groups. In short, the current results show that bacterial infection can upregulate the expression of HLA‐DP molecule in the thymus, not in spleen of S aureus pneumonia.
FIGURE 7.
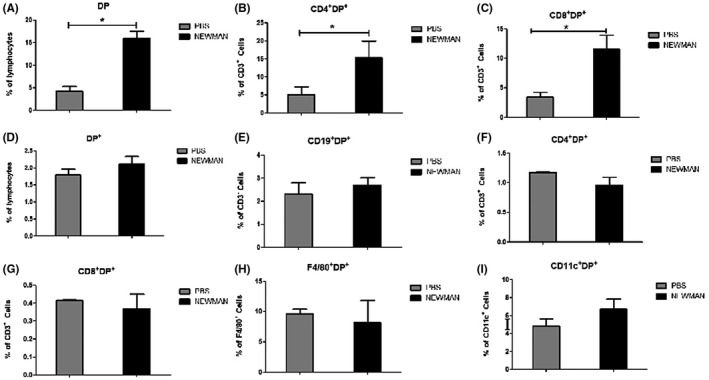
HLA‐DP expression on spleen and thymus cells from HLA‐DP401 humanized mice. HLA‐DP‐H2Aβ1−/− and H2Aβ1−/− mice were intranasally infected with Staphylococcus aureus. Spleen and thymus were collected 3 d later, HLA‐DP expression were quantified by FACS analysis. Y‐axis represents percentages in total spleen and thymus, each bar represents mean ± SD from three mice. The expression of HLA‐DP in thymus significantly increased in Staphylococcus aureus pneumonia of HLA‐DP humanized mice. A, The percentage of DP+ cells in total thymus. B, The percentage of DP+CD4+ T cells in total thymus. C, The percentage of DP+CD8+ T cells in total thymus. D, The percentage of DP+ cells in total spleen. E, The percentage of DP+CD19+ B cells in total spleen. F, The percentage of DP+CD4+ T cells in total spleen. G, The percentage of DP+CD8+ T cells in total spleen. H, The percentage of DP+CD11c+ dendritic cells. I, The percentage of DP+F4/80+ macrophage. *P < .05
4. DISCUSSION
To the best of our knowledge, this is the first report describing expression analysis in humanized transgenic mice contained the intact HLA‐DPA1/DPB1 gene locus (DOA‐DPA1*0103‐DPB1*0401). Our data showed that intact 5′ upstream and 3 kb of 3′ downstream sequences was insufficient to establish consistent expression of HLA‐DPA1/DPB1 in the “fragmented” DP8‐3# transgenic mice. In contrast, inclusion of 60‐kb upstream sequences gave reproducible expression of HLA‐DPA1/DPB1 in the tissues tested. These data suggest that sequences required for efficient expression of the HLA‐DPA1/DPB1 genes, such as enhancers, are not closely linked to these genes and are situated elsewhere in the HLA‐DP locus, which supports the notion that dominant cis‐acting control elements might be involved in the expression of the entire HLA‐DP locus, possibly in analogy to the locus control region (LCR) described for the HLA‐DRA locus. 28 , 38 , 39 Indeed, we found that the transgene in the “intact” transgenic lines including genomic DNA spanning HLA‐DOA and HLA‐DPA1 was expressed at high levels, while the transgene in the “fragmented” line missing LCR8# region was expressed at extremely low levels. LCR8# region contains a distal S’‐Y’ modules 8 situated >30 kb away from the DP region can induces global histone hyperacetylation extending up to 5 kb upstream of the HLA‐DRA. 27 , 28 Another study has also revealed that XL9 site, located in the intergenic region between HLA‐DRB1 (−29 kb downstream) and HLA‐DQA1 (−15 kb upstream), play a role in determining a global increase in chromatin accessibility. 27 , 40 , 41 , 42 Detailed molecular mechanism needs to be further clarified in future study.
The present data may also provide new insights into the tissue‐specificity of HLA‐DPA1 and HLA‐DPB1 expression. The current study only observed the expression of HLA‐DPB1 in the kidney from line 8‐3#. Several functional genes encoding the α‐and β‐chains of the classical MHC‐II isotypes are generally co‐regulated. 43 , 44 , 45 The encoded surface heterodimers are affected by the expression levels of the MHC II alleles. The immune status varies depending on the MHC II gene transcription and the mRNA processing. 42 , 43 , 45 , 46 Growing evidence suggests the existence of discoordinate regulation in MHC II gene expression. A mutant lymphoblastoid cell line (LCL), 45.EM2, express HLA‐DR and ‐DQ but fail to express HLA‐DP, indicating that the specific mechanisms is involved in the regulation of HLA‐DP expression and a specific trans‐acting factor may be involved in the control of DPB1 gene expression. 2 , 47
Previous study has shown that particular HLA class II alleles contribute to various autoimmune disease, 48 , 49 and S. aureus infection MHCII molecules. 32 HLA‐DR4, play an important role during the S. aureus infection for superantigens mediate interactions between MHC class II and the CDR2 loop of the variable chain of the T‐cell receptor. 50 , 51 , 52 But the role of HLA‐DP during the S. aureus infection is still unclear. Although both the α‐ and β‐chains of HLA‐DP molecules are polymorphic molecules, but HLA‐DP4, including the molecule DPA1*0103/DPB1*0401 (DP401) and DPA1*0103/DPB1*0402 (DP402), is the most prevalent HLA class II antigen in the worldwide population. 53
We developed a mouse model of S aureus pneumonias by HLA‐DP401 transgenic humanized mice. The percentage of immune cells was significantly not altered in spleen and thymus after infection. However, S aureus infection results in the rising of HLA‐DP expression in thymus. It has reported that a number of viruses could downregulate MHC I expression such as influenza. 54 S aureus has also been reported to interfere with host immune function by a lot of molecular mechanism. 55 , 56 , 57 , 58 In vitro studies have shown that S aureus can reduce the expression of major histocompatibility complex (MHC) class II on antigen presenting cells. 59 The expression of the MHC class II‐related genes HLA‐DRA and CD74 is significantly reduced in complicated S aureus bacteremia (SAB) than in uncomplicated SAB, indicating that there is an association between complicated SAB and reduced MHC class II expression. 60 It is not clear yet why the HLA‐DP expression upregulated in thymus after S aureus infection. The future study will be devoted to understand the exact mechanisms underlying HLA‐DP upregulation in thymus by S aureus.
In this study, we report a novel transgenic mouse containing the intact HLA‐DP401 molecules, which are the prevalent MHCII molecules in the worldwide population. These mice can not only be used to establish valuable humanized animal models for the study in human infectious and immunological diseases, but also would contribute to identifying universal epitopes to be used in peptide‐based vaccines for cancer, as well as for allergic or infectious diseases.
Supporting information
Fig S1
Fig S2
ACKNOWLEDGMENTS
This work is supported by the Mouse Genome Editing Lab and the followed project: National Science and Technology Major Project (2017ZX10304402‐001‐006, 2017ZX10304402‐001‐012, 2016YFD0500208); Shanghai Professional Platform for High‐level Biosafety Pathogenic Microorganism Detection (18DZ2293000); Scientific Research Projects of Shanghai Science and Technology Commission (19140905300), Shanghai Public Health Clinical Center General Program and the Start‐on Funding (KY‐GW‐2017‐06, KY‐GW‐2018‐11, KY‐GW‐2018‐04, KY‐GW‐2019‐11, and KY‐GW‐2019‐19).
Li F, Zhu M, Niu B, et al. Generation and expression analysis of BAC humanized mice carrying HLA‐DP401 haplotype. Anim Models Exp Med. 2021;4:116–128. 10.1002/ame2.12158
Contributor Information
Feng Li, Email: lifeng30286@shphc.org.cn.
Xiaohui Zhou, Email: zhouxiaohui@shphc.org.cn.
REFERENCES
- 1. Jardetzky TS, Brown JH, Gorga JC, et al. Crystallographic analysis of endogenous peptides associated with HLA‐DR1 suggests a common, polyproline II‐like conformation for bound peptides. Proc Natl Acad Sci USA. 1996;93:734‐738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kelly A, Trowsdale J. Genetics of antigen processing and presentation. Immunogenetics. 2019;71:161‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luckey D, Weaver EA, Osborne DG, Billadeau DD, Taneja V. Immunity to Influenza is dependent on MHC II polymorphism: study with 2 HLA transgenic strains. Sci Rep. 2019;9:19061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mangalam AK, Rajagopalan G, Taneja V, David CS. HLA class II transgenic mice mimic human inflammatory diseases. Adv Immunol. 2008;97:65‐147. [DOI] [PubMed] [Google Scholar]
- 5. Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol. 2013;190:513‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taneja V, David CS. HLA class II transgenic mice as models of human diseases. Immunol Rev. 1999;169:67‐79. [DOI] [PubMed] [Google Scholar]
- 7. Taneja V, David CS. Role of HLA class II genes in susceptibility/resistance to inflammatory arthritis: studies with humanized mice. Immunol Rev. 2010;233:62‐78. [DOI] [PubMed] [Google Scholar]
- 8. Moreau P, Cesbron A. HLA‐DP and allogeneic bone marrow transplantation. Bone Marrow Transplant. 1994;13:675‐681. [PubMed] [Google Scholar]
- 9. Petersdorf EW, Smith AG, Mickelson EM, et al. The role of HLA‐DPB1 disparity in the development of acute graft‐versus‐host disease following unrelated donor marrow transplantation. Blood. 1993;81:1923‐1932. [PubMed] [Google Scholar]
- 10. Meurer T, Arrieta‐Bolanos E, Metzing M, et al. Dissecting genetic control of HLA‐DPB1 expression and its relation to structural mismatch models in hematopoietic stem cell transplantation. Front Immunol. 2018;9:2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petersdorf EW, Malkki M, O’hUigin C, et al. High HLA‐DP expression and graft‐versus‐host disease. N Engl J Med. 2015;373:599‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schone B, Bergmann S, Lang K, et al. Predicting an HLA‐DPB1 expression marker based on standard DPB1 genotyping: linkage analysis of over 32,000 samples. Hum Immunol. 2018;79:20‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falta MT, Tinega AN, Mack DG, et al. Metal‐specific CD4+ T‐cell responses induced by beryllium exposure in HLA‐DP2 transgenic mice. Mucosal Immunol. 2016;9:218‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontenot AP, Keizer TS, McCleskey M, et al. Recombinant HLA‐DP2 binds beryllium and tolerizes beryllium‐specific pathogenic CD4+ T cells. J Immunol. 2006;177:3874‐3883. [DOI] [PubMed] [Google Scholar]
- 15. Mack DG, Falta MT, McKee AS, et al. Regulatory T cells modulate granulomatous inflammation in an HLA‐DP2 transgenic murine model of beryllium‐induced disease. Proc Natl Acad Sci USA. 2014;111:8553‐8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Graaff PM, Heidema J, Poelen MC, et al. HLA‐DP4 presents an immunodominant peptide from the RSV G protein to CD4 T cells. Virology. 2004;326:220‐230. [DOI] [PubMed] [Google Scholar]
- 17. de Waal L, Yuksel S, Brandenburg AH, et al. Identification of a common HLA‐DP4‐restricted T‐cell epitope in the conserved region of the respiratory syncytial virus G protein. J Virol. 2004;78:1775‐1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ru Z, Xiao W, Pajot A, et al. Development of a humanized HLA‐A2.1/DP4 transgenic mouse model and the use of this model to map HLA‐DP4‐restricted epitopes of HBV envelope protein. PLoS One. 2012;7:e32247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang WL, Audet RG, Aizenstein BD, Hogan LH, DeMars RI, Klein BS. T‐Cell epitopes and human leukocyte antigen restriction elements of an immunodominant antigen of Blastomyces dermatitidis. Infect Immun. 2000;68:502‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laheurte C, Galaine J, Beziaud L, et al. Immunoprevalence and magnitude of HLA‐DP4 versus HLA‐DR‐restricted spontaneous CD4(+) Th1 responses against telomerase in cancer patients. Oncoimmunology. 2016;5:e1137416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schultz ES, Lethe B, Cambiaso CL, et al. A MAGE‐A3 peptide presented by HLA‐DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Can Res. 2000;60:6272‐6275. [PubMed] [Google Scholar]
- 22. Tarantino‐Hutchison LM, Sorrentino C, Nadas A, et al. Genetic determinants of sensitivity to beryllium in mice. Journal of immunotoxicology. 2009;6:130‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hilhorst M, Arndt F, Joseph Kemna M, et al. HLA‐DPB1 as a risk factor for relapse in antineutrophil cytoplasmic antibody‐associated vasculitis: a cohort study. Arthritis Rheumatol. 2016;68:1721‐1730. [DOI] [PubMed] [Google Scholar]
- 24. Lauterbach N, Crivello P, Wieten L, et al. Allorecognition of HLA‐DP by CD4+ T cells is affected by polymorphism in its alpha chain. Mol Immunol. 2014;59:19‐29. [DOI] [PubMed] [Google Scholar]
- 25. Yamazaki T, Umemura T, Joshita S, Yoshizawa K, Tanaka E, Ota M. A cis‐eQTL of HLA‐DPB1 affects susceptibility to type 1 autoimmune hepatitis. Sci Rep. 2018;8:11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dawkins R, Leelayuwat C, Gaudieri S, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275‐304. [DOI] [PubMed] [Google Scholar]
- 27. Gianfrani C, Pisapia L, Picascia S, Strazzullo M, Del Pozzo G. Expression level of risk genes of MHC class II is a susceptibility factor for autoimmunity: new insights. J Autoimmun. 2018;89:1‐10. [DOI] [PubMed] [Google Scholar]
- 28. Krawczyk M, Peyraud N, Rybtsova N, et al. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J Immunol. 2004;173:6200‐6210. [DOI] [PubMed] [Google Scholar]
- 29. Li F, Zhou M. Depletion of bitter taste transduction leads to massive spermatid loss in transgenic mice. Mol Hum Reprod. 2012;18:289‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hashimoto K, Joshi SK, Koni PA. A conditional null allele of the major histocompatibility IA‐beta chain gene. Genesis. 2002;32:152‐153. [DOI] [PubMed] [Google Scholar]
- 31. Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard‐Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mammalian Genome. 2007a;18:693‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu SX, Gilmore KJ, Szabo PA, et al. Superantigens subvert the neutrophil response to promote abscess formation and enhance Staphylococcus aureus survival in vivo. Infect Immun. 2014;82:3588‐3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee MH, Arrecubieta C, Martin FJ, Prince A, Borczuk AC, Lowy FD. A postinfluenza model of Staphylococcus aureus pneumonia. J Infect Dis. 2010;201:508‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007b;27:2934‐2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard‐Smith EM. Distant regulatory elements in a Sox10‐beta GEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest‐derived tissues. Dev Dyn. 2006;235:1413‐1432. [DOI] [PubMed] [Google Scholar]
- 36. Rajagopalan G, Smart MK, Cheng S, Krco CJ, Johnson KL, David CS. Expression and function of HLA‐DR3 and DQ8 in transgenic mice lacking functional H2‐M. Tissue Antigens. 2003;62:149‐161. [DOI] [PubMed] [Google Scholar]
- 37. Patel SD, Cope AP, Congia M, et al. Identification of immunodominant T cell epitopes of human glutamic acid decarboxylase 65 by using HLA‐DR(alpha1*0101, beta1*0401) transgenic mice. Proc Natl Acad Sci USA. 1997;94:8082‐8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol. 2003;4:132‐137. [DOI] [PubMed] [Google Scholar]
- 39. Masternak K, Reith W. Promoter‐specific functions of CIITA and the MHC class II enhanceosome in transcriptional activation. EMBO J. 2002;21:1379‐1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA‐DRB1 and HLA‐DQA1 genes are separated by a CTCF‐binding enhancer‐blocking element. J Biol Chem. 2006;281:18435‐18443. [DOI] [PubMed] [Google Scholar]
- 41. Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long‐distance chromatin interactions. J Exp Med. 2008;205:785‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raj P, Rai E, Song R, et al. (2016). Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. eLife;5:e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Curr Opin Immunol. 2011;23:81‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Handunnetthi L, Ramagopalan SV, Ebers GC, Knight JC. Regulation of major histocompatibility complex class II gene expression, genetic variation and disease. Genes Immun. 2010;11:99‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017;18:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21‐33. [DOI] [PubMed] [Google Scholar]
- 47. Coiras MT, Alvarez‐Barrientos AM, Diaz G, Arroyo J, Sanchez‐Perez M. Evidence for discoordinate regulation of the HLA‐DPB1 gene. Tissue Antigens. 2002;60:505‐514. [DOI] [PubMed] [Google Scholar]
- 48. Ito K, Bian HJ, Molina M, et al. HLA‐DR4‐IE chimeric class II transgenic, murine class II‐deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635‐2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maier LA, McGrath DS, Sato H, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171:6910‐6918. [DOI] [PubMed] [Google Scholar]
- 50. Arad G, Levy R, Nasie I, et al. Binding of superantigen toxins into the CD28 homodimer interface is essential for induction of cytokine genes that mediate lethal shock. PLoS Biol. 2011;9:e1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levy R, Rotfogel Z, Hillman D, et al. Superantigens hyperinduce inflammatory cytokines by enhancing the B7–2/CD28 costimulatory receptor interaction. Proc Natl Acad Sci USA. 2016;113:E6437‐E6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tuffs SW, Haeryfar SMM, McCormick JK. Manipulation of innate and adaptive immunity by Staphylococcal superantigens. Pathogens. 2018;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Castelli FA, Buhot C, Sanson A, et al. HLA‐DP4, the most frequent HLA II molecule, defines a new supertype of peptide‐binding specificity. J Immunol. 2002;169:6928‐6934. [DOI] [PubMed] [Google Scholar]
- 54. Koutsakos M, McWilliam HEG, Aktepe TE, et al. Downregulation of MHC class I expression by influenza A and B viruses. Front Immunol. 2019;10:1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol. 2012;15:92‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med. 2009;206:2417‐2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13:529‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342:863‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang J, Roderiquez G, Norcross MA. Control of adaptive immune responses by Staphylococcus aureus through IL‐10, PD‐L1, and TLR2. Sci Rep. 2012;2:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rasmussen G, Cajander S, Backman A, Kallman J, Soderquist B, Stralin K. Expression of HLA‐DRA and CD74 mRNA in whole blood during the course of complicated and uncomplicated Staphylococcus aureus bacteremia. Microbiol Immunol. 2017;61:442‐451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2


