Abstract
Combination drug therapies have become an integral part of precision oncology, and while evidence of clinical effectiveness continues to grow, the underlying mechanisms supporting synergy are poorly understood. Immortalized human lymphoblastoid cell lines (LCLs) have been proven as a particularly useful, scalable and low-cost model in pharmacogenetics research, and are suitable for elucidating the molecular mechanisms of synergistic combination therapies. In this review, we cover the advantages of LCLs in synergy pharmacogenomics and consider recent studies providing initial evidence of the utility of LCLs in synergy research. We also discuss several opportunities for LCL-based systems to address gaps in the research through the expansion of testing regimens, assessment of new drug classes and higher-order combinations, and utilization of integrated omics technologies.
Keywords: : combination treatment, drug response, LCLs, omics, synergism
Combination drug therapy is the gold standard in modern cancer treatment. While the already large amount of evidence supporting the clinical effectiveness of combination therapy is continually growing, less is known about synergism. Synergism is defined as the effects of two or more agents working in combination that are greater than simply adding together the said agents’ effects. Understanding the underlying mechanisms of synergism, namely how nonadditive interactions occur and why the combined effects of drugs can be larger than individual drugs’ additive effects, is crucial to developing successful new drug cocktails and optimizing treatment regimens. Jia et al. provided an extensive discussion of synergistic mechanisms in 2009, but more recent studies have revealed several issues with the hypothesized mechanisms [1]. For example, the widespread off-target effects demonstrated in high-profile studies, as well as the explanation of synergistic effects as simply the result of patient-to-patient genetic variability, have complicated the already challenging search for a comprehensive understanding of synergism [2,3]. Given the frequent clinical use of combination therapy, the expansion of pharmacogenomics studies to include drug combinations is key for developing a more comprehensive understanding of the mechanisms of synergism and the broad consequences of genetic variability.
Immortalized human lymphoblastoid cell lines (LCLs) [4] created by infecting human lymphocytes with the Epstein-Barr virus [5], provide a scalable, efficient and cost-effective model for pharmacogenomic discovery, and have been successfully used to assay drug response [4,6]. Accordingly, they represent a suitable model for expanding research to include combination drug therapy. Several pharmacogenomics studies have successfully employed the LCL model to further understand the mechanism of action for a range of anticancer drugs, with a growing number of LCL-based pharmacogenomics findings successfully replicated outside the laboratory [7–11]. LCLs have been used to conduct high-throughput screening assays, such as those used to characterize the dose-response relationship of 29 US FDA-approved drugs [12]. Further, genome-wide association analysis of temozolomide response in LCLs identified a clinically relevant association of SNPs in the MGMT gene [13], and a genome-wide meta-analysis of carboplatin- and cisplatin-induced toxicity in LCLs identified SNPs associated with platinum-induced cytotoxicity [14]. These recent successes demonstrate that LCLs provide a valuable screening platform for examining genetic susceptibility and resistance to existing combination therapies and for discovering and testing novel therapeutic combinations [15–17].
In the following, we briefly review the use of LCLs in pharmacogenomic studies seeking to discover synergistic combinations and their mechanisms of action. We subsequently discuss the advantages and disadvantages of using the LCL model as well as opportunities to expand its use to address important challenges in pharmacogenomics.
Advantages of LCLs for synergy pharmacogenomics
The LCL model has several clear advantages. First, genome-wide LCL data for both related and unrelated populations are publicly available, eliminating the cost of genotyping data for use in pharmacogenomic studies and making efficiency a key advantage of LCLs [4]. Second, in contrast to human clinical trials and in vivo model systems, high-throughput in vitro screening technologies, such as those reliant on LCLs, enable easily scaling pharmacogenomic studies to accommodate the large sample sizes needed for gene mapping. This is an important point as sample requirements will continue to grow as increasingly complex study designs are needed for combination drug studies. Third, the LCL model is amenable to high-throughput robotic automation [12]. Fourth, LCLs can be easily manipulated to validate study findings using knock-down or forced gene-expression methods. Fifth, LCLs derived from ovarian cancer patients have shown utility in the putative identification of biomarkers for synergistic response [18] and clinical outcomes [19]. Finally, the LCL model enables study designs that are not possible in vivo. For a comprehensive understanding of the nonadditive components of combination drug response, it is necessary to carefully estimate the additive components, and LCLs enable the assessment of both monotherapy and combination therapy response phenotypes.
The majority of work on in vitro combination drug exposure has been performed in cancer cell lines [20–22]. While cancer cell lines present important scientific opportunities, LCLs are an important complementary model system [23,24]. Pharmacogenomics must holistically consider the genetics of drug response in both the tumor and host. The tumor genome consists of acquired somatic mutations along with germline variants that represent disease variation and influence drug response, while the host genome consists of inherited germline variants that represent interindividual variability in drug response and influence drug efficacy and toxicity. As immediate surrogates of germline variation, LCLs provide an opportunity for understanding host pharmacogenetics [24]. For instance, several studies have shown that anticancer drug response is a heritable trait and that germline variants influence drug response [4,24–26]. A classic example is the germline variation in the TPMT gene, which affects thiopurine metabolism. The TPMT host genotype is used as a pharmacogenomic biomarker in clinical practice to guide the dosage for various thiopurines such as mercaptopurine and azathioprine [27]. Further, since LCLs represent the entire host genome, they can be directly used to study the impact of germline variation on drug response without confounding by somatic mutations or secondary genomic changes, such as those found in cancer cell lines [5,28,29]. Another important difference is that LCLs are genetically heterogeneous while cancer cell lines are typically clonal [30]. This is an important distinction with subtle yet important consequences. Studying drug response and the evolution of drug resistance in cancer cell lines requires the accumulation and selection of genetic mutations in the original clonal line. In contrast, the heterogeneity available in LCLs allows for the immediate interrogation of population-level variation.
Synergy studies in LCLs
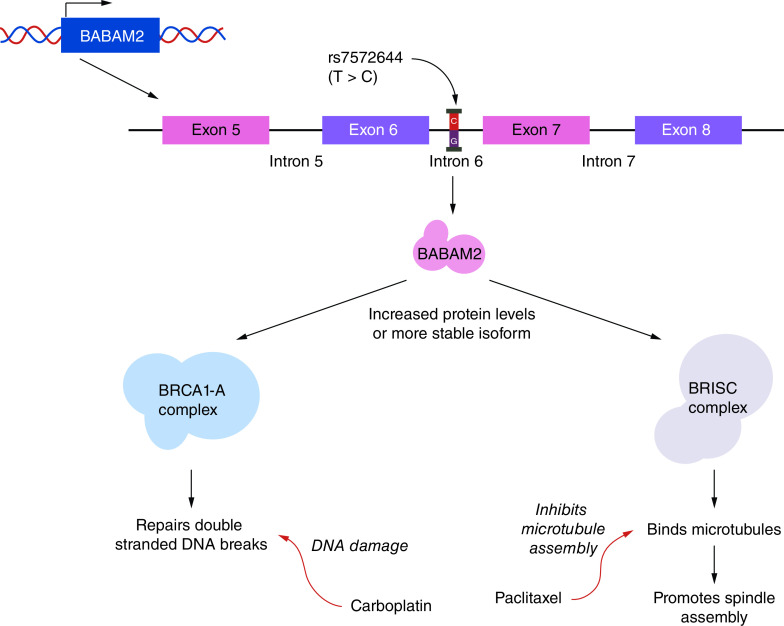
Several recent studies provide proof of principle that the LCL model can be used for pharmacogenomic studies of synergy. In the first genome-wide association study investigating synergy in LCLs [18], LCLs derived from 74 patients with epithelial ovarian cancer were treated with increasing concentrations of carboplatin and/or paclitaxel and assessed for in vitro drug response. Drug response phenotypes, including IC50 (the effective dose at which 50% of cells are viable) and EC50 (the dose resulting in 50% induction of caspase 3/7 activity), were estimated for each patient for paclitaxel and carboplatin treatment, both individually and in combination. In vitro paclitaxel drug-response phenotypes were moderately associated with time to cancer recurrence in vivo. While no pharmacogenomic associations were significant at p < 5 × 10-8, seven genomic loci were associated with drug response at p < 10-6, including 4q21.21 for carboplatin, 4p16.1 and 5q23.2 for paclitaxel, and 3q24, 10q, 1q44 and 13q21 for combination therapy. Importantly, the results revealed regions unique to combination treatment response. In particular, SNP rs7572664 was significantly associated with decreased survival odds and sensitivity to the combination treatment in vitro (Figure 1) [18,19]. This is likely because the SNP either increases BABAM2 protein levels or produces a more stable isoform, enhancing cancer cell survival through the BRCA1-A and BRISC complexes. The effectiveness of treating this mutation with the carboplatin + paclitaxel combination is most likely driven by DNA damage and inhibition of the microtubule assembly that the BRCA1-A and BRISC complexes mitigate.
Figure 1. . Possible mechanism of synergistic response in lymphoblastoid cell lines from patients with ovarian cancer.
An SNP (rs7572644) found in intron 6 of the BABAM2 gene of ovarian cancer patients was associated with synergistic response to carboplatin + paclitaxel treatment. BABAM2 plays an important role in both the BRCA1-A and BRISC complexes that are responsible for the repair of dsDNA breaks and the promotion of spindle assembly, respectively. As carboplatin and paclitaxel induce DNA damage and inhibit microtubule assembly, respectively, the mechanism for the observed increase in sensitivity to combined treatment could be due to the combination treatment targeting both oncogenic pathways downstream of BABAM2. While the role of rs757266 in this process is unknown, it could result in increased BABAM2 protein levels or the production of a more stable isoform of the protein.
In another important proof-of-principle experiment, researchers used the Chou-Talalay method [31] for the first time to directly quantify synergy for seven drug combinations in LCLs from related pedigrees [32]. Heritability was calculated to understand how much variation in synergism is due to genetic variation, in isolation from the heritable components of monotherapy response. Heritability ranged from 4 to 31%, and linkage analysis results revealed a significant linkage peak unique to the synergistic component of response to epirubicin + paclitaxel treatment. Additional linkage analysis results point to a general trend of regions being uniquely linked to response to individual drugs or combination therapies. Another interesting result of this study is that while it is generally assumed that the tested drug combinations had synergistic effects compared with monotherapies, there was a dramatic level of interindividual variation in both the range of nonadditivity observed, and more importantly, the direction of the effects. While synergy was observed for some cell lines, combination therapies were antagonistic for an unexpected number of cell lines. This preliminary observation highlights the need for further interrogation of the mechanisms of interindividual response heterogeneity.
Opportunities
While these initial studies are exciting, important research questions related to the pharmacogenomics of combination drug response remain unanswered. There are several opportunities to expand work in LCLs to address gaps in the research, including increasing and developing testing regimens, investigating new drug classes and higher-order combinations, and using LCLs in integrative omics and high-resolution phenotyping.
Expansion of testing regimens
The synergy studies described above involve response to simultaneously dosed drug pairs. All current cell-line studies investigating synergy utilize such pair-wise synchronous chemotherapeutic treatment, so it is important to expand the knowledge of differential response to polytherapies and investigate asynchronous treatment [18,22,33–36]. Drug resistance, which can result from tumor genetic heterogeneity, particularly in high-turnover tumors, limits targeted cancer therapies [37,38]. The current approach to addressing this resistance involves the initiation of new drug therapies following clinical/radiographic disease progression. However, polytherapies are more clinically effective than single or dual therapies for addressing tumor heterogeneity and resistance, and computational models suggest asynchronous treatment is more effective than synchronous treatment [37,39–42]. LCLs present opportunities for more extensive, empirical exploration of avenues to expand testing and improve current treatment regimens.
New drug classes & higher-order combinations
Researchers have investigated monotherapies that include chemotherapy, tyrosine kinase inhibitors and monoclonal antibodies in LCLs across a broad range of drugs and drug classes [10,14,18,36,43–54]. Prior work has shown that drug response in LCLs can recapitulate response to drug classes in fully unsupervised clustering [12]. The synergy studies discussed above have focused on platinum agents, microtubule binding agents, nucleosides, fluoropyrimidines and anthracyclines, presenting the opportunity to expand the number of drug combinations across classes in future LCL studies.
Large, publicly available, combinatorial screens in cancer cell lines by pharmaceutical companies (i.e., Merck and AstraZeneca) and the National Cancer Institute (NCI-ALMANAC) have yielded important information, including oncogenic and nononcogenic drug targets, monotherapeutic utility in a range of cancer types, and alternative combinatorial-drug response prediction models [21,22,35]. To expand this work, the genetic diversity of LCLs can be leveraged in similar combinatorial-drug response studies to assess differential susceptibility.
Integrative omics & higher-resolution phenotyping
Initial drug synergy studies in LCLs focused on linkage or associations of single nucleotide variants with a single phenotypic response such as cell viability or death. However, complex diseases such as cancer are driven by the dynamic interplay of polygenic and nongenetic effects. An ideal approach to quantifying drug synergy in such a complex scenario involves integrating high-dimensional molecular epistatic readouts of omics (e.g., SNPs, copy number variations, gene expression, protein expression and epigenetic modifications) from drug combination assays. Systems biology approaches that include network analysis, protein–protein interaction models, dynamic pathway simulations and reduced network motifs can be used to identify putative mechanisms of synergistic and antagonistic drug–drug and gene–drug interactions [55–57]. Interestingly, network motif studies [58,59] show that synergistic motifs often include serial parallel mixed-type structures or negative feedback loops, while antagonistic motifs are mostly associated with positive feedback loops and a downstream link. These results suggest that genome-wide association study and multi-omics studies conducted in LCLs would provide a rich source of data for these types of mapping studies [6,12,18,32]. They also indicate the potential of gene-editing technologies such as CRISPR-Cas9 for probing human gene combinations and predicting synergistic interactions. Further, pathway-based analysis [60,61] involving gene perturbations is a successful approach to integrating omics data mainly derived from gene-expression profiles from combinations of deletion mutants of protein kinases and phosphatases [62]. The output of this method is a gene-interaction map encoding functional relationships between genes. These network approaches are predicated on the idea that phenotypes from an individual population of tumor cells are driven by a single regulatory network controlled by a limited number of germline or somatic master regulators [63]. While this is a feasible approach to addressing tumor complexity, it does not account for the single-cell variation that gives rise to drug resistance. While no current LCL studies have conducted association mapping of combination therapies at the single-cell level, non-LCL studies using CyTOF [39,64] and scRNA-seq [40,65] have revealed striking cell-to-cell variability for targeted single and combination drug treatments. The results of these non-LCL studies indicate that, on average, the response of individual hormone-treated cells is 30% that of the total expected set of transcriptional glucocorticoid receptor target genes, demonstrating the potential for a single cell to become resistant over time [40].
Addressing this complexity challenges the field of combination therapy association mapping to expand to account for single-cell heterogeneity. Recent efforts to measure the gene expression of LCLs at the single-cell level [66], with the objective of characterizing the function of genetic variants at single-cell resolution or the subcell-type level [67], have significantly improved researchers’ ability to identify gene–gene interactions that account for cell–cell variability in response to combination drug treatment and have been shown to strongly influence the effectiveness of cancer treatment. In an innovative approach for studying the interplay between complementary genetic and epigenetic mechanisms of drug resistance in cancer, researchers have combined lineage tracing using barcoding and scRNA-seq [68]. These studies demonstrate the potential for tracking the epistatic effects of combination treatment at the single-cell level during tumor progression. Recent advancements in cutting-edge omics technologies and gene-editing tools offer new opportunities for studying drug synergy using LCL models at multiple scales.
Limitations & challenges
There are several limitations associated with the LCL model. First, while LCLs have produced clinical successes, translating in vitro results to in vivo biological/clinical relevance remains difficult [5,9,15]. Deficiencies in data on in vivo absorption, metabolism and excretion make extrapolating in vitro test concentrations to in vivo doses challenging [15]. Further, not all biological pathways, and, as with other cell lines used in monoculture, many metabolizing enzymes (e.g., CYP450s) are not represented in LCLs [15], making it difficult to fully test prodrugs and other therapeutics. In addition, in a clinical setting, radiation is often used as a front-line treatment along with chemotherapy [69], and LCLs may not be amenable to this experimental paradigm as studies have shown they have reduced chromosomal radiosensitivity [5]. Second, LCLs are unable to capture somatic variation, the evolution of tumor drug resistance, or the toxic outcomes related to germline variation [37,39–42,54]. Third, as with any cell-line system, drug amenability can be an issue, and chemical properties such as aqueous solubility and light sensitivity need to be considered.
In addition to these limitations, expanding combination therapy study designs introduces several challenges. First, there is no unified approach for detecting and quantifying synergy. Dose-response curves are the bedrock of pharmacological and toxicological studies, and parametric nonlinear least square models are the preferred model for curve fitting [70]. While powerful nonlinear least square models are highly sensitive to starting values, the initial parameter values must be close to the true values because otherwise there may be no convergence [70,71]. These modeling limitations make comparisons across studies challenging, and the addition of combinatorial complexity will likely magnify these effects [72].
Second, the study of synergy in temporally staggered treatments, which are known to affect in vivo efficacy, should be proactively addressed. Such treatments reflect combination therapies that are not commonly given together and are often ignored in the assumptions underlying the standard methods used to predict and quantify synergy, including the Bliss independence [73] and Loewe additivity [74] models.
A third challenge is related to the fact that in recent years researchers have used pharmacological and toxicological molecular end points to study drug synergy through omics and single-cell technologies for a growing number of FDA-approved cancer treatments, including 3D patient-derived cell cultures (e.g., spheroids and organoids) [75]. This challenges the current definition of synergy and the computational methods for predicting it due to the huge number of possible combinations, the complexity of the tissue microenvironment and single-cell heterogeneity. For example, each additional drug considered exponentially increases the number of conditions that must be measured, making an exhaustive search of combinatorial space and events unmanageable. To address this challenge, optimal sampling designs and the development of better computational algorithms that can account for higher-order synergistic interactions beyond pair-wise interactions are needed. Approaches that can predict higher-order synergy using low-order interaction effects (e.g., epistatic nested effects models) [61] can potentially reduce the number of combinations tested, account for high-dimensional hidden epistatic effects and predict higher-order interactions from observed pair-wise interactions.
Conclusion
People with cancers of all types will continue to benefit from more precise multidrug treatments that maximize outcomes and minimize side effects. A better understanding of the underlying mechanisms of successful multidrug treatment will empower efforts to discover new and effective combinations and to optimize individual treatment plans. Patients with brain cancer will especially benefit from additional combination therapies, as monotherapies account for 71% of the recommended first-line treatment options, while in most tissue-specific cancers, they account for 50% or less. Several proof-of-principle studies using the LCL model to evaluate synergy support the use of this model system for such efforts.
Future perspective
Drug combination treatment is a significant and effective clinical method used to treat nearly all cancer types, as administering more than one drug can provide benefits that include higher effectiveness, lower toxicity and delayed onset of acquired drug resistance. Recent large-scale systematic and serendipitous drug discovery efforts have identified synergistic combinations. Considering the vast number of possible drug combinations, a deeper understanding of the molecular mechanisms of synergy and the broad influence of genetic variability is vital for the efficient discovery of effective new cancer treatments. As part of this effort, LCLs have proven utility as a rigorous and high-throughput pharmacogenetics tool. Recent studies have further highlighted their usefulness in interrogating synergy.
Importantly, studies have revealed the ability of LCLs not only to detect in vitro synergy but also to identify genetic response regions unique to combination treatment, the heritability of response, and a surprising mixture of both synergistic and antagonistic responses to cotreatment due to their genetic diversity. These preliminary observations highlight the need for further investigation of the mechanisms of interindividual response heterogeneity. Continued and expanded utilization of LCLs will provide an effective and efficient avenue to discover these mechanisms, test novel drug combinations, and increase the number of options available to clinicians while reducing side effects by taking genetic variability into account, moving us closer to realizing the goals of precision oncology.
Executive summary.
Our understanding of the mechanisms of synergism and the influence of genetic variability on tumor response is limited.
Lymphoblastoid cell lines (LCLs) are a suitable model for expanding research to include combination drug therapy through publicly available genome data. They are amenable to high-throughput robotic automation, are easily scalable and provide molecular tractability.
The heterogeneity available in LCLs allows for the immediate interrogation of population-level variation. Recent results have revealed genomic regions of response unique to combination treatment as well as interindividual response heterogeneity ranging from synergism to antagonism.
LCLs present opportunities for more extensive, empirical exploration of both combination and asynchronous treatment regimens, expanded assessment of differential susceptibility, and investigation of combination therapies at the single-cell level.
LCLs provide an effective avenue to discover the underlying mechanisms of synergy, test novel drug combinations and bring us closer to the goal of precision oncology.
Acknowledgments
The authors would like to thank T Wiltshire, H McLeod and B Small for discussion on synergy in the LCL model.
Footnotes
Author contributions
All authors contributed to the writing of the manuscript.
Financial & competing interests disclosure
This work was supported by R01CA161608 from the National Cancer Institute and by intramural funds from the National Institute of Environmental Health Sciences. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Jia J, Zhu F, Ma X, Cao ZW, Li YX, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discov. 8(2), 111–128 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Lin A, Giuliano CJ, Palladino A et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11(509), 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171(7), 1678–1691.e1613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack J, Rotroff D, Motsinger-Reif A. Lymphoblastoid cell lines models of drug response: successes and lessons from this pharmacogenomic model. Curr. Mol. Med. 14(7), 833–840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain T, Mulherkar R. Lymphoblastoid cell lines: a continuous in vitro source of cells to study carcinogen sensitivity and DNA repair. Int. J. Mol. Cell. Med. 1(2), 75–87 (2012). [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler HE, Dolan ME. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics 13(1), 55–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellsworth KA, Eckloff BW, Li L et al. Contribution of FKBP5 genetic variation to gemcitabine treatment and survival in pancreatic adenocarcinoma. PLoS ONE 8(8), e70216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang RS, Johnatty SE, Gamazon ER et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin. Cancer Res. 17(16), 5490–5500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon J-P. Chapter 6 – Human Lymphoblastoid Cell Lines in Pharmacogenomics. : Handbook of Pharmacogenomics and Stratified Medicine. Padmanabhan S (). Academic Press, CA, USA, 89–110 (2014). [Google Scholar]
- 10.Li L, Fridley B, Kalari K et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 68(17), 7050–7058 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation 118(4), 355–362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown CC, Havener TM, Medina MW et al. Genome-wide association and pharmacological profiling of 29 anticancer agents using lymphoblastoid cell lines. Pharmacogenomics 15(2), 137–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CC, Havener TM, Medina MW et al. A genome-wide association analysis of temozolomide response using lymphoblastoid cell lines shows a clinically relevant association with MGMT. Pharmacogenet. Genomics 22(11), 796–802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler HE, Gamazon ER, Stark AL et al. Genome-wide meta-analysis identifies variants associated with platinating agent susceptibility across populations. Pharmacogenomics J. 13(1), 35–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motsinger-Reif AA, Rotroff DM. Leveraging lymphoblastoid cell lines for drug response modeling. J. Data Min. Genomics Proteomics 6(3), 1 (2015). [Google Scholar]
- 16.Niu N, Wang L. In vitro human cell line models to predict clinical response to anticancer drugs. Pharmacogenomics 16(3), 273–285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark AL, Dolan ME. Lymphoblastoid cell lines in pharmacogenomics: how applicable are they to clinical outcomes? Pharmacogenomics 14(5), 447–450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridley BL, Ghosh TM, Wang A et al. Genome-wide study of response to platinum, taxane, and combination therapy in ovarian cancer: in vitro phenotypes, inherited variation, and disease recurrence. Front Genet 7, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto R, Assis J, Nogueira A et al. Pharmacogenomics in epithelial ovarian cancer first-line treatment outcome: validation of GWAS-associated NRG3 rs1649942 and BRE rs7572644 variants in an independent cohort. Pharmacogenomics J. 19(1), 25–32 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Bansal M, Yang J, Karan C et al. A community computational challenge to predict the activity of pairs of compounds. Nat. Biotechnol. 32(12), 1213–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menden MP, Wang D, Mason MJ et al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat. Commun. 10(1), 2674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Neil J, Benita Y, Feldman I et al. An unbiased oncology compound screen to identify novel combination strategies. Mol. Cancer Ther. 15(6), 1155–1162 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Menden MP, Casale FP, Stephan J et al. The germline genetic component of drug sensitivity in cancer cell lines. Nat. Commun. 9(1), 3385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moen EL, Godley LA, Zhang W, Dolan ME. Pharmacogenomics of chemotherapeutic susceptibility and toxicity. Genome Med. 4(11), 90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolan ME, Newbold KG, Nagasubramanian R et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 64(12), 4353–4356 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat. Rev. Genet. 14(1), 23–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relling MV, Schwab M, Whirl-Carrillo M et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT 15 genotypes: 2018 update. Clin. Pharmacol. Ther. 105(5), 1095–1105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J. Natl Cancer Inst. 105(7), 452–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain T, Kotnis A, Sarin R, Mulherkar R. Establishment & characterization of lymphoblastoid cell lines from patients with multiple primary neoplasms in the upper aero-digestive tract & healthy individuals. Indian J. Med. Res. 135(6), 820–829 (2012). [PMC free article] [PubMed] [Google Scholar]
- 30.Hynds RE, Vladimirou E, Janes SM. The secret lives of cancer cell lines. Dis. Model Mech. 11(11), dmm037366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70(2), 440–446 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Roell KR, Havener TM, Reif DM et al. Synergistic chemotherapy drug response is a genetic trait in lymphoblastoid cell lines. Front. Genet. 10, 829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crystal AS, Shaw AT, Sequist LV et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346(6216), 1480–1486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gayvert KM, Aly O, Platt J, Bosenberg MW, Stern DF, Elemento O. A computational approach for identifying synergistic drug combinations. PLoS Comput. Biol. 13(1), e1005308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holbeck SL, Camalier R, Crowell JA et al. The National Cancer Institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res. 77(13), 3564–3576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roell KR, Reif DM, Motsinger-Reif AA. An introduction to terminology and methodology of chemical synergy-perspectives from across disciplines. Front. Pharmacol. 8, 158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonsson VD, Blakely CM, Lin L et al. Novel computational method for predicting polytherapy switching strategies to overcome tumor heterogeneity and evolution. Sci. Rep. 7, 44206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komarova NL, Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc. Natl Acad. Sci. USA 102(27), 9714–9719 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anchang B, Davis KL, Fienberg HG et al. DRUG-NEM: optimizing drug combinations using single-cell perturbation response to account for intratumoral heterogeneity. Proc. Natl Acad. Sci. USA 115(18), E4294–E4303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman JA, Papas BN, Trotter KW, Archer TK. Single-cell RNA sequencing reveals a heterogeneous response to glucocorticoids in breast cancer cells. Commun. Biol. 3(1), 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B, Hemann MT, Lauffenburger DA. Intratumor heterogeneity alters most effective drugs in designed combinations. Proc. Natl Acad. Sci. USA 111(29), 10773–10778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B, Pritchard JR, Lauffenburger DA, Hemann MT. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov. 4(2), 166–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SH, Yang W, Fan Y et al. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia 25(1), 66–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dzamko N, Deak M, Hentati F et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J. 430(3), 405–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA 107(20), 9287–9292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartford CM, Duan S, Delaney SM et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood 113(10), 2145–2153 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotaki R, Kawashima M, Yamamoto Y et al. Dasatinib exacerbates splenomegaly of mice inoculated with Epstein-Barr virus-infected lymphoblastoid cell lines. Sci. Rep. 10(1), 4355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loomis R, Carbone R, Reiss M, Lacy J. Bcl-2 antisense (G3139, Genasense) enhances the in vitro and in vivo response of Epstein-Barr virus-associated lymphoproliferative disease to rituximab. Clin. Cancer Res. 9(5), 1931–1939 (2003). [PubMed] [Google Scholar]
- 49.Mitra AK, Crews K, Pounds S et al. Impact of genetic variation in FKBP5 on clinical response in pediatric acute myeloid leukemia patients: a pilot study. Leukemia 25(8), 1354–1356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donnell PH, Gamazon E, Zhang W et al. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenet. Genomics 20(5), 327–337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters EJ, Kraja AT, Lin SJ et al. Association of thymidylate synthase variants with 5-fluorouracil cytotoxicity. Pharmacogenet. Genomics 19(5), 399–401 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Shan D, Ledbetter JA, Press OW. Apoptosis of malignant human B cells by ligation of CD20 with monoclonal antibodies. Blood 91(5), 1644–1652 (1998). [PubMed] [Google Scholar]
- 53.Tan XL, Moyer AM, Fridley BL et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin. Cancer Res. 17(17), 5801–5811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziliak D, O'Donnell PH, Im HK et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl. Res. 157(5), 265–272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen D, Liu X, Yang Y, Yang H, Lu P. Systematic synergy modeling: understanding drug synergy from a systems biology perspective. BMC Syst. Biol. 9, 56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regan-Fendt KE, Xu J, Divincenzo M et al. Synergy from gene expression and network mining (SynGeNet) method predicts synergistic drug combinations for diverse melanoma genomic subtypes. NPJ Syst. Biol. Appl. 5, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7(6), 460–466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Smolen P, Baxter DA, Byrne JH. Computational analyses of synergism in small molecular network motifs. PLoS Comput. Biol. 10(3), e1003524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin N, Ma W, Pei J, Ouyang Q, Tang C, Lai L. Synergistic and antagonistic drug combinations depend on network topology. PLoS ONE 9(4), e93960 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8(2), e1002375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pirkl M, Diekmann M, Van Der Wees M, Beerenwinkel N, Fröhlich H, Markowetz F. Inferring modulators of genetic interactions with epistatic nested effects models. PLoS Comput. Biol. 13(4), e1005496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Wageningen S, Kemmeren P, Lijnzaad P et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143(6), 991–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez MJ, Subramaniam PS, Tang LH et al. A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors. Nat. Genet. 50(7), 979–989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baskar R, Fienberg HG, Khair Z et al. TRAIL-induced variation of cell signaling states provides nonheritable resistance to apoptosis. Life Sci. Alliance 2(6), e201900554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong SP, Chan TE, Lombardo Y et al. Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat. Commun. 10(1), 3840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osorio D, Yu X, Yu P, Serpedin E, Cai JJ. Single-cell RNA sequencing of a European and an African lymphoblastoid cell line. Sci. Data 6(1), 112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borel C, Ferreira PG, Santoni F et al. Biased allelic expression in human primary fibroblast single cells. Am. J. Hum. Genet. 96(1), 70–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eyler CE, Matsunaga H, Hovestadt V, Vantine SJ, Van Galen P, Bernstein BE. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 21(1), 174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Therapy Advisor. Breast, skin, prostate, lung cancer treatment regimens. Haymarket Media, Inc., NY, USA. https://www.cancertherapyadvisor.com/home/cancer-treatment-regimens/ [Google Scholar]
- 70.Ma J, Bair E, Motsinger-Reif A. Nonlinear dose-response modeling of high-throughput screening data using an evolutionary algorithm. Dose Response 18(2), 1559325820926734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bates D, Watts D. Nonlinear regression: iterative estimation and linear approximations. : Nonlinear Regression Analysis and Its Applications. John Wiley & Sons, Inc., NJ, USA, 32–66 (2008). [Google Scholar]
- 72.Meyer CT, Wooten DJ, Lopez CF, Quaranta V. Charting the fragmented landscape of drug synergy. Trends Pharmacol. Sci. 41(4), 266–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bliss CI. The toxicity of poisons applied jointly 1. Ann. Appl. Biol. 26(3), 585–615 (1939). [Google Scholar]
- 74.Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3(6), 285–290 (1953). [PubMed] [Google Scholar]
- 75.Folkesson E, Niederdorfer B, Nakstad VT et al. High-throughput screening reveals higher synergistic effect of MEK inhibitor combinations in colon cancer spheroids. Sci. Rep. 10(1), 11574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]