Abstract
In light of the continued threat of antimicrobial-resistant bacteria, new strategies to expand the repertoire of antimicrobial compounds are necessary. Prodrugs are an underexploited strategy in this effort. Here, we report on the enhanced antimicrobial activity of a prodrug toward bacteria having an enzyme capable of its activation. A screen led us to the sulfurol ester of the antibiotic trans-3-(4-chlorobenzoyl)acrylic acid. An endogenous esterase makes Mycolycibacterium smegmatis sensitive to this prodrug. Candidate esterases were identified, and their heterologous production made Escherichia coli sensitive to the ester prodrug. Taken together, these data suggest a new approach to the development of antimicrobial compounds that takes advantage of endogenous enzymatic activities to target specific bacteria.
Keywords: bioinformatics, esterase, mycobacteria, narrow-spectrum antibiotic, prodrug, sulfurol
Graphical Abstract
Antimicrobial resistance represents a growing threat to public health, rendering established therapies ineffective. Changes in the societal use of antibiotics, the discovery of new antibacterial compounds, and the derivatization of known antibacterial compounds all demonstrate great promise for addressing this challenge. Nonetheless, these efforts alone are unlikely to be sufficient to solve this crisis because resistance arises so quickly, even for new classes of antibiotics.1 Rather, new strategies beyond classical broad-spectrum antibacterial compounds are called for.2,3
One potential strategy is the creation of narrow-spectrum antibiotics.4–6 In contrast to broad-spectrum antibiotics, which kill virulent and avirulent bacteria, narrow-spectrum antibiotics are only lethal to particular types of bacteria, ideally, pathogens. The use of broad-spectrum antibiotics has been shown to increase the presence of resistance genes to both the specific antibiotic and other groups of antibiotics.7 In comparison, narrow-spectrum antibiotics apply less selective pressure to the development of resistance because only a few bacterial species present in a treated patient (rather than the entire microbiome) would benefit from resistance. Such drugs could therefore have extended utility.6 In addition, treatment with a narrow-spectrum antibiotic would allow the native fauna of the microbiome to persist, thereby reducing the likelihood of secondary infections and other complications.8
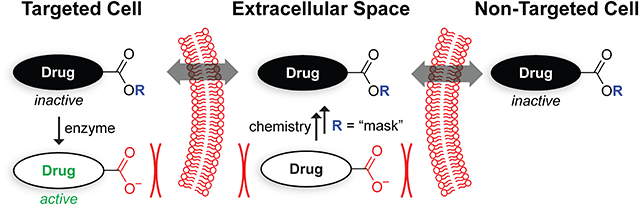
We reasoned that a prodrug approach to antimicrobial compound development could be a novel means of developing targeted antimicrobial compounds if activation were dependent on the activity of an enzyme found only in the target species. Most of the limited work on antimicrobial prodrugs has focused on improving solubility9 or broad-spectrum effectiveness.10 One report has shown, however, that ciprofloxacin can be targeted to pathogenic Escherichia coli when linked as an ester to the siderophore enterobactin, leveraging the receptor for the siderophore to endow specificity.11 Another report has demonstrated the illumination of a fluorogenic probe by an enzyme endogenous in mycobacteria.12 Here, we sought to test the hypothesis that an antibiotic could be targeted through the substrate specificity of an endogenous enzyme.
RESULTS AND DISCUSSION
Screen of Microbial Esterase Activity.
We began by seeking two bacterial species that have vastly different esterase activities. A previous literature report suggested that Escherichia coli contained low esterase activity,13 and the carboxylic ester hydrolase enzymes (CASTLE) database further suggested that E. coli DH10B contained no known carboxylesterases.14 Reports suggest that Mycolycibacterium smegmatis (basionym Mycobacterium smegmatis), on the other hand, has higher levels of esterase activity.15,16 To confirm these reports, we screened a small library of esters for cleavage in Escherichia coli DH10B or Mycolycibacterium smegmatis mc2155 lysates using an esterase-activity assay established in our laboratory (Figures 1A and S1).17 The results suggested that, in line with the literature reports, E. coli cells harbor little esterase activity whereas M. smegmatis has much higher levels. Hence, we explored the esterase activity of M. smegmatis further and established that all of the esters that we tested were cleaved in the presence of its lysate. Together, these literature reports and experimental evidence suggested that E. coli and M. smegmatis possess markedly divergent esterase activity and provided a basis for testing our ester-based prodrug approach.
Figure 1.
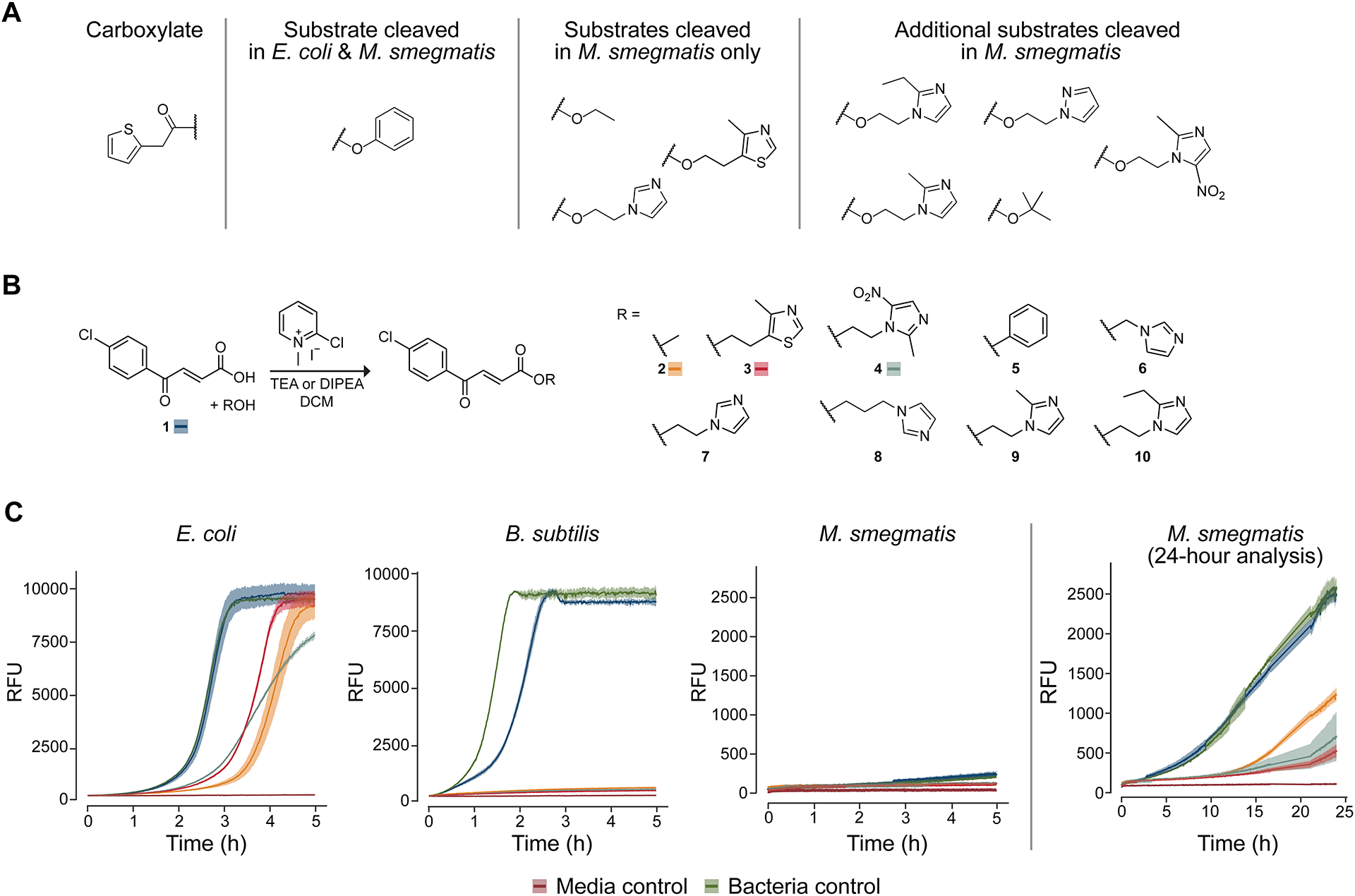
Microbial esterase activity assays. (A) A small panel of esters of 2-thiopheneacetic acid was assessed for cleavability in E. coli DH10B or M. smegmatis mc2155 lysate. Only the phenolic ester was cleaved in E. coli lysate whereas all four esters initially tested were cleaved in M. smegmatis lysate. Consequently, a larger panel of esters was screened in M. smegmatis lysate, and it was found that all esters were subject to cleavage. (B) Synthetic route to esters of trans-3-(4-chlorobenzoyl)acrylic acid. Note that acid 1 and esters 2–4 are labeled with the corresponding color for viability curves in panel C. (C) Viability curves for bacteria incubated with acid 1 and esters 2–4 (100 μM), which have high antimicrobial activity. Viability was assessed through the addition to the medium of resazurin dye, which fluoresces upon reduction in the presence of live, metabolizing cells. Medium: E. coli and B. subtilis, CAMHB; M. smegmatis, 7H9. Lines are the average of triplicate experiments; ribbons depict one standard deviation.
Screen of Ester Prodrugs.
Next, we established a screen to determine the antimicrobial efficacy of esters of trans-3-(4-chlorobenzoyl)acrylic acid (1) in E. coli versus M. smegmatis. Compound 1 had been reported to have moderate efficacy against M. tuberculosis, and its methyl ester had been reported to have enhanced activity along with low toxicity to mammalian cells.18,19 Still, compound 1 has not been used clinically, and is likely to target more than one species.18,20
We reasoned that different esters of acid 1 would differ in their ability to kill bacteria, depending on both cellular permeability and the rate of esterase-catalyzed hydrolysis. Accordingly, we synthesized an array of nine esters of acid 1 (Figure 1B). Using resazurin dye fluorescence turn-on as a proxy for cell viability, we screened these esters to assess their ability to inhibit the growth of E. coli (Figure S2) or M. smegmatis (Figure S3) at a concentration of 50 μM in Luria–Bertani broth. We found that the free acid form had low activity in both species and that the esters presented differential, species-dependent antimicrobial activity. In E. coli, the methyl ester was highly effective, whereas in M. smegmatis the ester with sulfurol21 showed the most activity, followed closely by the metronidazole ester. Imidazole ester 7, which showed high activity against both species, was observed to undergo rapid hydrolysis in aqueous solution, consistent with previous reports of α-(1H-imidazol-1-yl)alkyl (IMIDA) carboxylic acid esters,22 and was not pursued further.
Intrigued by the differential activity of the esters in M. smegmatis compared to E. coli, we probed for differences in a Gram-positive organism. Specifically, we added Bacillus subtilis OI1085 to our screen because this Gram-positive bacterium, like M. smegmatis, is known to harbor many esterase-encoding genes.23 We chose the three most active compounds identified in our initial screen and increased their concentration to 100 μM. Cation-adjusted Mueller–Hinton broth was used for the screen of E. coli and B. subtilis cells, as is typical for antimicrobial tests in these species, whereas 7H9 medium was used for M. smegmatis. Viability curves confirmed the earlier pattern observed in E. coli and M. smegmatis. In B. subtilis, however, the acid form of the drug showed some activity and all ester variants inhibited bacterial growth for the duration of the viability screen (Figure 1C). We performed a similar screen in the presence of the constituent alcohol moieties alone and found that none of the alcohols influenced the growth of any of the bacteria (Figure S4–S6). MIC values correlated with the trends seen in the viability assays (see: Supporting Information). Specifically, sulfurol ester 3 exhibited a lower MIC for both B. subtilis and M. smegmatis but not for E. coli, consistent with our hypothesis that the presence of esterases capable of cleaving the sulfurol ester led to its enhanced toxicity.
Analysis of Microbial Esterase Activity.
Next, we sought to establish whether the sulfurol ester was indeed hydrolyzed by esterases endogenous in M. smegmatis and B. subtilis but not E. coli. Small cultures of the three bacteria were grown overnight then lysed via sonication. Ester 3 was then added to these lysates, which were incubated at 37 °C for 2 h, extracted with dichloromethane, and analyzed with LC–MS (Figure 2). To provide a semiquantitative assessment of the extent of hydrolysis, standard curves were constructed by spiking known amounts of authentic sulfurol ester, sulfurol, and trans-3-(4-chlorobenzoyl)acrylic acid into lysates of each bacterial species to mimic hydrolysis extents ranging from 0% hydrolyzed to 100% hydrolyzed. A linear relationship was discovered in the range from 25–100% hydrolyzed (Figure S7). The extracted ion chromatograms from the same LC–MS run allowed for integration of the sulfurol ester peak (m/z 336) and the sulfurol ester hydrolysis alcohol peak (m/z 144) using SpectraGryph software (version 1.2.15). Comparison of the ratio in the sample to the ratios determined from an external calibration curve allowed for semiquantitative assessment of the extent of hydrolysis. (The acid itself was excluded from the analysis because its detection was found to be too variable to allow for reliable semiquantitative analysis (Figure S8), perhaps because of its formation of covalent adducts with target enzymes as has been suggested previously.18) In both CAMHB medium and E. coli cell lysate, less hydrolysis was observed than could be determined via the semiquantitative method. In 7H9 medium, however, both partial hydrolysis and partial isomerization were apparent. In the presence of M. smegmatis or B. subtilis lysate, the ester compound was nearly completely hydrolyzed, suggesting that the sulfurol ester is a substrate for one or more esterases in these species.
Figure 2.
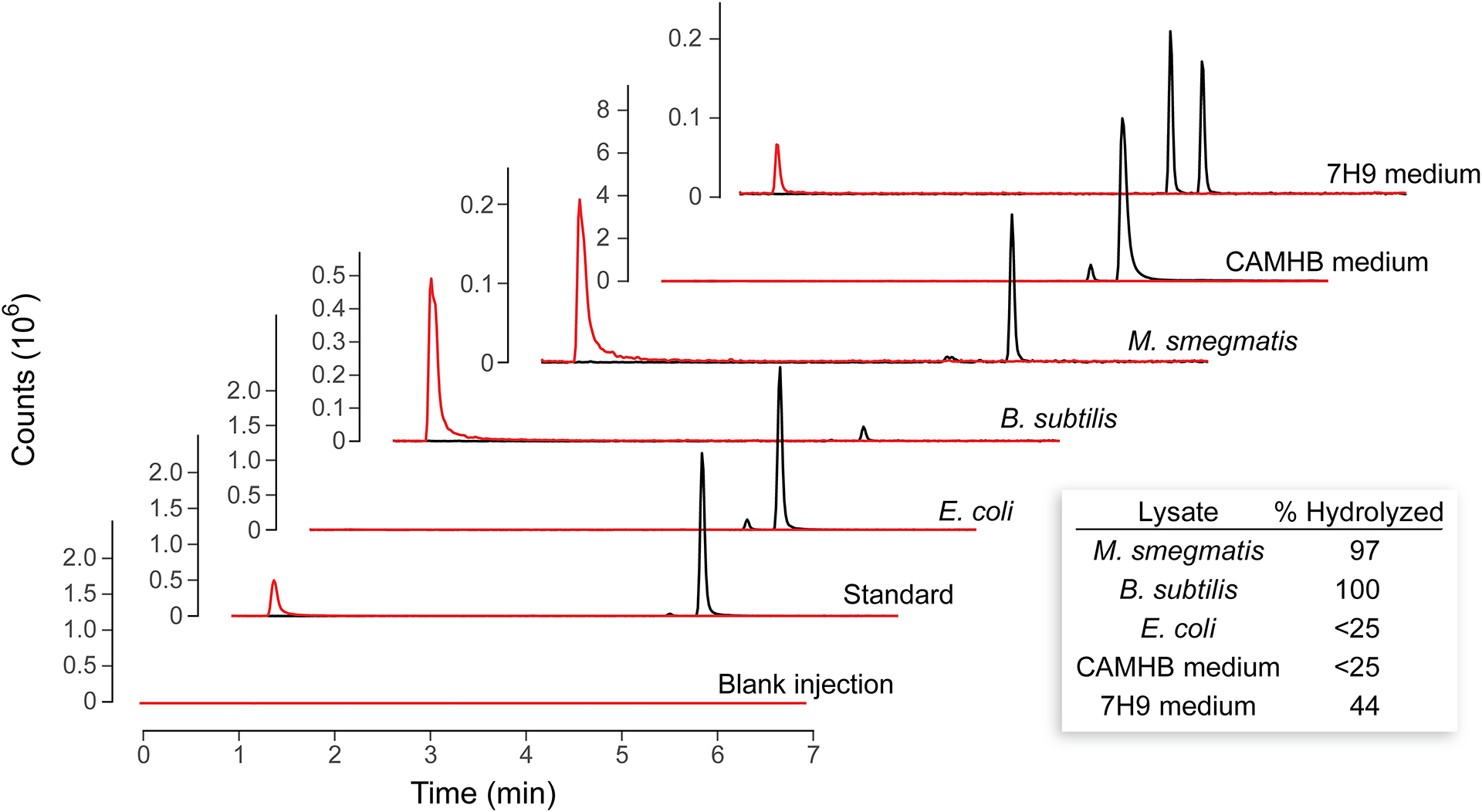
LC–MS extracted ion chromatograms (EICs) of sulfurol and sulfurol ester 3 after incubation with a cell lysate. Red EIC has m/z 144 (sulfurol), and black EIC has m/z 336 (sulfurol ester 3) from the same LC–MS run. The small peak in the black trace is likely the cis isomer of sulfurol ester 3. Standard traces are from a solution containing 200 μM of both sulfurol and sulfurol ester 3. Inset: Sulfurol ester 3 % hydrolyzed was determined from calibration curves as described in the Supporting Information. Note: In addition to hydrolysis, the 7H9 medium led to isomerization.
Bioinformatic Analysis of Bacterial Esterases.
Having demonstrated that M. smegmatis and B. subtilis but not E. coli are capable of hydrolyzing sulfurol ester 3, we used bioinformatics to identify a candidate esterase. We collected the sequences of all proteins annotated as “carboxylesterases” from E. coli DH10B, M. smegmatis mc2155, and B. subtilis subsp. subtilis 168 through searching of the NCBI protein database. B. subtilis subsp. subtilis 168 was used in place of B. subtilis OI1085 because it has a fully sequenced genome and strain OI1085 is descended from strain 168.24–26 Using the tools from the Enzyme Function Institute,27 we uploaded all the sequences thus identified and found 209 with UniProt ID matches, 116 of which were unique (Table S1). We then created a sequence similarity network using a minimum alignment score of 37 (Figure 3A; for determination of minimum alignment score, see the Suppporting Information and Figure S9). Analysis of the network revealed a single cluster that contained esterases from both M. smegmatis and B. subtilis. Because both M. smegmatis and B. subtilis were capable of hydrolyzing ester 3, we concluded that this cluster likely represents carboxylesterases that hydrolyze ester 3 and contribute to its activity.
Figure 3.
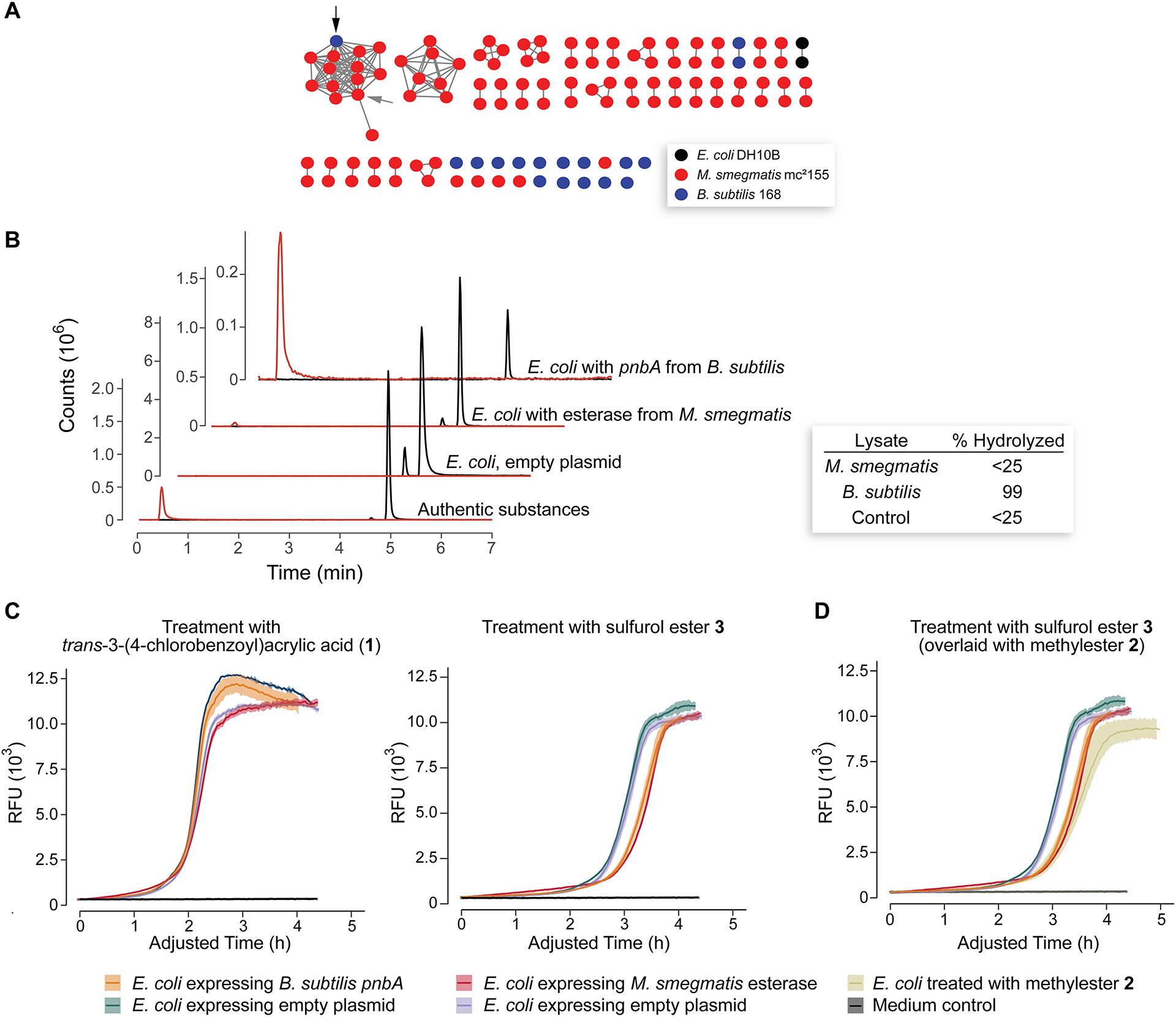
Identification of pnbA and effect of its expression in E. coli DH10B. (A) Sequence similarity network of annotated carboxylesterases from B. subtilis 168, E. coli DH10B, and M. smegmatis mc2155 (Table S1). The black arrow indicates the pnbA carboxylesterase from B. subtilis 168; the gray arrow indicates carboxylesterase A0QYI2 from M. smegmatis. (B) LC–MS extracted ion chromatograms (EICs) of dichloromethane extract from cultures of E. coli exposed to 200 μM of sulfurol ester 3. The “Authentic substances” traces are from a solution containing 200 μM of both sulfurol and sulfurol ester 3. The red EIC has m/z 144 (sulfurol), and the black EIC has m/z 336 (sulfurol ester 3) from the same LC–MS sample. Inset: The sulfurol ester 3 % hydrolyzed was determined from calibration curves as described in the Supporting Information. (C) Viability curves for E. coli expressing the identified carboxylesterase incubated with acid 1 and ester 3 (100 μM). (D) Viability curves for E. coli expressing the identified carboxylesterase incubated with esters 2 and 3 (100 μM). In panels C and D, the ordinate was adjusted such that the bacteria-only growth curves from each gene expression condition have the same inflection point, allowing for a comparison between the antimicrobial compound dose curves. Lines are the average of triplicate experiments; ribbons depict one standard deviation.
Heterologous Esterase Production.
Two nodes from this cluster were cloned for further analysis. The first, a node from B. subtilis annotated as a para-nitrobenzylesterase A (pnbA, UniProtKB P37967) and having a known three-dimensional structure,28 had been produced previously in E. coli.29 The second, a node from M. smegmatis that is annotated, was annotated as a carboxylic ester hydrolase and to the best of our knowledge had not been characterized experimentally (UniProtKB A0QYI2). To confirm that proteins from this cluster were capable of hydrolyzing ester 3, we constructed two pET22b plasmids, each harboring one of the esterase genes identified above under the control of a T7 promoter. The indicated esterase gene and a gene encoding T7 RNA polymerase on plasmid pCS630 were coexpressed overnight in E. coli DH10B cells, the culture was lysed, and the sulfurol ester was added to the lysate. As a control, an empty pET22b plasmid and plasmid pCS6 were treated in the same fashion. LC–MS analysis of the extracts following 2 h at 37 °C demonstrated that the heterologous production of the pnbA esterase from B. subtilis enabled E. coli DH10B cells to hydrolyze sulfurol ester 3 nearly to completion using our semiquantitative method, whereas the esterase from M. smegmatis hydrolyzed sufficient quantities to produce a visible peak in the hydrolysis product extracted ion chromatogram (EIC) but that was less than our method could quantify. No hydrolysis was detectable in the control culture (Figure 3B). Encouraged by this confirmatory activity, we also sought to determine whether production of the esterases in E. coli enhances the antimicrobial activity of the sulfurol ester. We reasoned that even a low degree of esterase activity in the lysate might be sufficient to enhance the activity of the ester prodrug in vivo. Because the control strain harboring an empty plasmid and the strain expressing the esterase experienced a different lag time in the viability assay (Figure S10), we first had to adjust the time course such that the untreated growth curves overlapped. Hence, we fitted the growth curve of the bacterial strains growing in medium alone to Richards’s logistic growth curve31,32 to determine the inflection time point, which is the point at which the growth rate has hit a maximum. We then adjusted the time of the curves such that the untreated bacteria growth curves had identical inflection points, thereby ensuring that they overlapped. Fits were conducted with R software,33 and they are shown in the Supporting Information. This experiment was additionally complicated by the fact that hydrolytic activity was observed in the culture supernatant of E. coli cells expressing pnbA from B. subtilis (Figure S11); nonetheless, we observed that if pnbA expression was held to a low level, thereby holding extracellular pnbA to a minimum, a significant increase in sensitization could be observed in pnbA-expressing E. coli versus E. coli expressing an empty plasmid (Figure 3C). Additionally, a similar significant increase in sensitization was observed in the E. coli expressing the esterase from M. smegmatis. In both cases, this increase in sensitization drove the activity of E. coli cells for the sulfurol ester (3) to match that for the methyl ester (2), which was the most active variant of trans-3-(4-chlorobenzoyl)acrylic acid for parent E. coli cells (Figures 3D and S10).
In conclusion, we have demonstrated that a sulfurol ester prodrug of trans-3-(4-chlorobenzoyl)acrylic acid is more effective against M. smegmatis than are other esters. Importantly, the sulfurol ester is less active against E. coli than is the methyl ester, but this gap is bridged by the heterologous production of an esterase that hydrolyzes the sulfurol ester in E. coli cells. These data indicate that esterase activity profiles can be used to craft an ester prodrug of a carboxylic acid-containing antimicrobial agent. Hence, the alcohol moiety in an ester prodrug should be viewed as more than merely a mask of a negative charge—different esters can confer markedly different activity profiles.
Our findings point to a new strategy for developing prodrugs in which a unique enzymatic activity within a target species is harnessed to unmask an antimicrobial agent, thereby providing a narrow spectrum of action that can be identifiable with a bioinformatic analysis. In this work, we relied on esterase specificity to achieve this goal. We are aware, however, that bacteria are fantastic chemists. Their enzymes catalyze a wide range of unique reactions that are only beginning to be mapped effectively.34–37 Future work could harness a range of bacterial enzymes to develop an array of narrow-spectrum antibiotics.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Dr. Eric M. Gordon for his encouragement of this work and Dr. Paul A. Clemons (Broad Institute of MIT and Harvard) for his helpful suggestions regarding our bioinformatic analysis.
Funding
This work was supported by the Merkin Institute for Transformative Technologies in Healthcare. Additional support was provided by Grants R21 GM135780 and R01 GM044783 (NIH). M.A.A.R. was supported by the Undergraduate Research Opportunities Program at the Massachusetts Institute of Technology.
ABBREVIATIONS
- CAMHB
cation-adjusted Mueller–Hinton broth
- EIC
extracted-ion chromatogram
- LB
Luria–Bertani lysogeny broth
- LC–MS
liquid chromatography–mass spectrometry
- MIC
minimum inhibitory concentration
- NCBI
National Center for Biotechnology Information
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at https://pubs.acs.org/doi/10.1021/acschembio.0c00894.
Procedures for the synthesis of esters 2–10 and cognate esters with 2-thiophenacetic acid, biological methods, annotated carboxylesterases in the sequence similarity network; esterase activity of bacterial cell lysates; screen of antimicrobial esters against E. coli and M. smegmatis; screen of alcohol compounds against E. coli, B. subtilis, and M. smegmatis; calibration of sulfurol ester:sulfurol peak-area ratio versus % hydrolyzed; LC−MS spectra and calibration curves of trans-3-(4-chlorobenzoyl)acrylic acid; determination of the e-value threshold for SSN; viability curves of E. coli DH10B cells; extracted ion chromatograms of sulfurol ester 3; NMR spectra (PDF)
Accession Code
Bacillus subtilis, P37967
Mycolicibacterium smegmatis, A0QYI2
The authors declare no competing financial interest.
REFERENCES
- (1).Kolter R; van Wezel GP Goodbye to brute force in antibiotic discovery? Nat. Microbiol 2016, 1, 1–2. [DOI] [PubMed] [Google Scholar]
- (2).Lakemeyer M; Zhao W; Mandl FA; Hammann P; Sieber SA Thinking outside the box—novel antibacterials to tackle the resistance crisis. Angew. Chem., Int. Ed 2018, 57, 14440–14475. [DOI] [PubMed] [Google Scholar]
- (3).Theuretzbacher U; Outterson K; Engel A; Karlen A The global preclinical antibacterial pipeline. Nat. Rev. Microbiol 2020, 18, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Clatworthy AE; Pierson E; Hung DT Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol 2007, 3, 541–548. [DOI] [PubMed] [Google Scholar]
- (5).Brown ED; Wright GD Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [DOI] [PubMed] [Google Scholar]
- (6).Melander RJ; Zurawski DV; Melander C Narrow-spectrum antibacterial agents. MedChemComm 2018, 9, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Willmann M; El-Hadidi M; Huson DH; Schutz M; Weidenmaier C; Autenrieth IB; Peter S Antibiotic selection pressure determination through sequence-based metagenomics. Antimicrob. Agents Chemother 2015, 59, 7335–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Blaser MJ The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol 2017, 17, 461–463. [DOI] [PubMed] [Google Scholar]
- (9).Gordon EM; Duncton MAJ; Gallop MA Orally absorbed derivatives of the β-lactamase inhibitor avibactam. Design of novel prodrugs of sulfate containing drugs. J. Med. Chem 2018, 61, 10340–10344. [DOI] [PubMed] [Google Scholar]
- (10).Fleck LE; North EJ; Lee RE; Mulcahy LR; Casadei G; Lewis K A screen for and validation of prodrug antimicrobials. Antimicrob. Agents Chemother 2014, 58, 1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Neumann W; Sassone-Corsi M; Raffatellu M; Nolan EM Esterase-catalyzed siderophore hydrolysis activates an enterobactin–ciprofloxacin conjugate and confers targeted antibacterial activity. J. Am. Chem. Soc 2018, 140, 5193–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hodges HL; Brown RA; Crooks JA; Weibel DB; Kiessling LL Imaging mycobacterial growth and division with a fluorogenic probe. Proc. Natl. Acad. Sci. USA 2018, 115, 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Antonczak AK; Simova Z; Tippmann EM A critical examination of Escherichia coli esterase activity. J. Biol. Chem 2009, 284, 28795–28800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chen Y; Black DS; Reilly PJ Carboxylic ester hydrolases: Classification and database derived from their primary, secondary, and tertiary structures. Protein Sci. 2016, 25, 1942–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tallman KR; Levine SR; Beatty KE Profiling esterases in Mycobacterium tuberculosis using far-red fluorogenic substrates. ACS Chem. Biol 2016, 11, 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bassett B; Waibel B; White A; Hansen H; Stephens D; Koelper A; Larsen EM; Kim C; Glanzer A; Lavis LD; Hoops GC; Johnson RJ Measuring the global substrate specificity of Mycobacterial serine hydrolases using a library of fluorogenic ester substrates. ACS Infect. Dis 2018, 4, 904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hetrick KJ; Aguilar Ramos MA; Raines RT Terbium(III) luminescence-based assay for esterase activity. Anal. Chem 2019, 91, 8615–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Li X; Liu N; Zhang H; Knudson SE; Li HJ; Lai CT; Simmerling C; Slayden RA; Tonge PJ CoA adducts of 4-oxo-4-phenylbut-2-enoates: Inhibitors of MenB from the M. tuberculosis menaquinone biosynthesis pathway. ACS Med. Chem. Lett 2011, 2, 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ren JF; Xu J; Zhang GN; Xu CL; Zhao LL; You XF; Wang YC; Lu Y; Yu LY; Wang JX Design, synthesis, and bioevaluation of a novel class of (E)-4-oxocrotonamide derivatives as potent antituberculosis agents. Bioorg. Med. Chem. Lett 2019, 29, 539–543. [DOI] [PubMed] [Google Scholar]
- (20).Xu CL; Bai XG; Xu J; Ren JF; Xing Y; Li ZQ; Wang JX; Shi JJ; Yu LY; Wang YC Substituted 4-oxo-crotonic acid derivatives as a new class of protein kinase B (PknB) inhibitors: Synthesis and SAR study. RSC Adv. 2017, 7, 4763–4775. [Google Scholar]
- (21).Sulfurol is a natural catabolite of thiamine pyrophosphate (vitamin B1) that is used as an industrial flavorant and fragrance.
- (22).Majumdar S; Spaeth MM; Sivendran S; Juntunen J; Thomas JD; Sloan KB α-(1H-Imidazol-1-yl)alkyl (IMIDA) carboxylic acid esters as prodrugs of carboxylic acid containing drugs. Tetrahedron Lett. 2007, 48, 4609–4611. [Google Scholar]
- (23).Eggert T; Pencreac’h G; Douchet I; Verger R; Jaeger K-E A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase. Eur. J. Biochem 2000, 267, 6459–6469. [DOI] [PubMed] [Google Scholar]
- (24).Barat M; Anagnostopoulos C; Schneider AM Linkage relationships of genes controlling isoleucine, valine, and leucine biosynthesis in Bacillus subtilis. J .Bacteriol 1965, 90, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ordal GW; Goldman DJ Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science 1975, 189, 802–805. [DOI] [PubMed] [Google Scholar]
- (26).Ullah AH; Ordal GW In vivo and in vitro chemotactic methylation in Bacillus subtilis. J. Bacteriol 1981, 145, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gerlt JA; Bouvier JT; Davidson DB; Imker HJ; Sadkhin B; Slater DR; Whalen KL Enzyme function initiative–enzyme similarity tool (EFI–EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta Proteins Proteomics 2015, 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Spiller B; Gershenson A; Arnold FH; Stevens RC A structural view of evolutionary divergence. Proc. Natl. Acad. Sci. USA 1999, 96, 12305–12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zock J; Cantwell C; Swartling J; Hodges R; Pohl T; Sutton K; Rosteck P Jr.; McGilvray D; Queener S The Bacillus subtilis pnbA gene encoding p-nitrobenzyl esterase: Cloning, sequence and high-level expression in Escherichia coli. Gene 1994, 151, 37–43. [DOI] [PubMed] [Google Scholar]
- (30).Schmidt CM; Shis DL; Nguyen-Huu TD; Bennett MR Stable maintenance of multiple plasmids in E. coli using a single selective marker. ACS Synth. Biol 2012, 1, 445–450. [DOI] [PubMed] [Google Scholar]
- (31).Richards FJ A flexible growth function for empirical use. J. Exp. Bot 1959, 10, 290–300. [Google Scholar]
- (32).Zwietering MH; Jongenburger I; Rombouts FM; van’t Riet K Modeling of the bacterial growth curve. Appl. Environ. Microbiol 1990, 56, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- (34).Veprinskiy V; Heizinger L; Plach MG; Merkl R Assessing in silico the recruitment and functional spectrum of bacterial enzymes from secondary metabolism. BMC Evol. Biol 2017, 17, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Rodríguez Benítez A; Narayan ARH Frontiers in biocatalysis: Profiling function across sequence space. ACS Cent. Sci 2019, 5, 1747–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Fisher BF; Snodgrass HM; Jones KA; Andorfer CM; Lewis JC Site-selective C–H halogenation using flavin-dependent halogenases identified via family-wide activity profiling. ACS Cent. Sci 2019, 5, 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pyser JB; Baker Dockrey SA; Benítez AR; Joyce LA; Wiscons RA; Smith JL; Narayan ARH Stereodivergent, chemoenzymatic synthesis of azaphilone natural products. J. Am. Chem. Soc 2019, 141, 18551–18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


