Abstract
Conrad Waddington’s epigenetic landscape, a visual metaphor for the development of multicellular organisms, is appropriate to depict the formation of phenotypic variants of bacterial cells. Examples of bacterial differentiation that result in morphological change have been known for decades. In addition, bacterial populations contain phenotypic cell variants that lack morphological change, and the advent of fluorescent protein technology and single-cell analysis has unveiled scores of examples. Cell-specific gene expression patterns can have a random origin or arise as a programmed event. When phenotypic cell-to-cell differences are heritable, bacterial lineages are formed. The mechanisms that transmit epigenetic states to daughter cells can have strikingly different levels of complexity, from the propagation of simple feedback loops to the formation of complex DNA methylation patterns. Game theory predicts that phenotypic heterogeneity can facilitate bacterial adaptation to hostile or unpredictable environments, serving either as a division of labor or as a bet hedging that anticipates future challenges. Experimental observation confirms the existence of both types of strategies in the bacterial world.
Keywords: phenotypic heterogeneity, noise, bistability, lineage formation, DNA methylation
Introduction
During differentiation of tissues in multicellular eukaryotes, genetically identical cells diversify into cell types that differ in both their morphology and their physiology. In the mid-twentieth century, C. H. Waddington envisioned eukaryotic developmental pathways as a series of ridges and valleys traversed by cells on their way to differentiation (Waddington, 1957). Cell differentiation involving change of form is also found in certain prokaryotic species. Well-known examples include the formation of heterocysts in filamentous cyanobacteria (Muro-Pastor and Hess, 2012), sporulation in Bacillus subtilis (Khanna et al., 2020), differentiation of nitrogen-fixing bacteroids in Rhizobium spp. (Kondorosi et al., 2013), asymmetric cell division in Caulobacter (Collier, 2019), and formation of fruiting bodies by myxobacteria (Munoz-Dorado et al., 2016). In other cases, however, cell differentiation occurs without visible morphological change. In the last few decades, the study of bacterial cell variants has been facilitated by growing interest in bacterial multicellularity (Shapiro, 1998) and by technical upturn in single-cell analysis technologies (Bernander et al., 1998; Meyer and Dworkin, 2007; Kreibich and Hardt, 2015; Scheler et al., 2019).
Phenotypic heterogeneity in a bacterial population can be the consequence of chemical communication, leading to a heterogeneous response at the single-cell level. For instance, differentiation of cyanobacterial heterocysts seems to respond to gradients of activator and inhibitor molecules along the cyanobacterial filament (Muro-Pastor and Hess, 2012). In other cases, phenotypic heterogeneity arises without the involvement of environmental cues, and the underlying mechanisms are diverse. Genetic mechanisms include site-specific recombination (Scott and Simon, 1982; Reyes Ruiz et al., 2020), slipped-strand mispairing at tracts of repetitive DNA sequences (Moxon et al., 2006), and amplification of specific genome regions (Belikova et al., 2020; Tomanek et al., 2020). As described below, cell diversification into two or more phenotypic states can also be driven by nongenetic mechanisms, such as propagation of feedback loops (Ferrell, 2002) and formation of DNA methylation patterns (Sanchez-Romero and Casadesus, 2020).
Differentiation of bacterial subpopulations can be interpreted as the manifestation of two different strategies: division of labor and bet hedging (Veening et al., 2008; Lambert and Kussell, 2014). Division of labor is a cooperative activity that increases the fitness of the subpopulations if they coexist (Zhang et al., 2016). Illustrative examples of division of labor have been described in biofilms (van Gestel et al., 2015; Dragos et al., 2018). In bet hedging, a population with more than one phenotype performs better in a changing environment than a population with a homogeneous phenotype, and the variance in offspring numbers across generations is minimized (Gillespie, 1974; de Jong et al., 2011; Schreiber et al., 2016). Bet hedging has been shown to produce subpopulations tolerant to antibacterial agents (Adam et al., 2008; Hernandez et al., 2012; Dewachter et al., 2019) or resistant to bacteriophages (Cota et al., 2015; Turkington et al., 2019).
Natural selection of phenotypic heterogeneity, especially if it involves a bet-hedging strategy, is a controversial notion in classical Darwinism because it involves group selection, which has been traditionally considered a weak evolutionary force (Leigh, 2010). This view is however countered by game theory (Kussell and Leibler, 2005; Wolf et al., 2005; Kussell, 2013).
Sources of Phenotypic Differences in Isogenic Bacterial Cells
Events in cellular physiology involve random encounters between molecules, some of which are present in small numbers. As a consequence, a certain degree of stochasticity exists in many biochemical transactions (Kaern et al., 2005; Sanchez et al., 2013). A physiological event where stochasticity is well known is transcription initiation, which can show differences from one cell to another. As a consequence, cells with distinct transcriptional profiles can be produced in isogenic subpopulations of bacteria (Silva-Rocha and de Lorenzo, 2010). Changes in gene copy number during the bacterial cell cycle and cell-to-cell differences in translation efficiency are additional sources of stochasticity in gene expression (Kaern et al., 2005).
Noise can be sufficient to produce phenotypic heterogeneity in a bacterial population. Because of the finite number effect, a small difference in the number of molecules can produce or not a signal with physiological significance (Kaern et al., 2005). Thresholds are therefore crucial in noisy systems to produce a “meaningful” signal (Anderson, 1972). When distinct gene expression patterns generated by noise are propagated by feedback loops, the bacterial population splits into subpopulations, a phenomenon known as multistability (Thomas and Kaufman, 2001).
Most examples of multistability validated by experimental analysis involve two phenotypic states only (bistability), producing cells with high and low expression of specific genes or gene networks (ON and OFF cells) (Laurent et al., 2005; Dubnau and Losick, 2006). In some cases, formation of cell variants is not stochastic but deterministic, and ON and OFF cells show nonlinear gene expression patterns that do not arise from noise (Casadesus and Low, 2013). Whatever their origin, bistable states can be transmitted to the progeny either by a positive feedback loop or by a double-negative feedback loop (Casadesus and D’Ari, 2002; Ferrell, 2002). When reversion of a bistable state occurs in a programmed manner, bistability is called phase variation (van der Woude, 2011).
Bistable States Propagated by Feedback Loops
Examples of bacterial bistable systems sustained by feedback loops are reviewed below. Some have been chosen because of their historic relevance; other choices may be arbitrary. A rich literature on the subject exists, including comprehensive reviews (Laurent et al., 2005; Dubnau and Losick, 2006; Veening et al., 2008; Casadesus and Low, 2013; Ackermann, 2015; Weigel and Dersch, 2018; Schroter and Dersch, 2019).
Bistability in the lac Operon
An example of bistability propagated by a positive feedback loop was described in the lac operon of Escherichia coli more than six decades ago (Novick and Weiner, 1957). Isopropyl-β-D-1-thio-galactoside (IPTG) is a gratuitous (noncatabolizable) inducer that derepresses the lac operon if added to the bacterial culture at high concentrations. At low concentrations, IPTG is unable to induce a naïve culture. However, if an induced culture is transferred to a culture medium containing a low concentration of IPTG, a subpopulation of cells remains in the induced state (Novick and Weiner, 1957). Maintenance of LacON cells occurs because they have a high level of β-galactoside permease in their membrane. A high level of permease concentrates IPTG inside the cell, and a high concentration of IPTG induces a high level of permease synthesis (Novick and Weiner, 1957; Laurent et al., 2005). In certain cells, however, a decrease in the internal concentration of inducer (which may occur, for instance, during cell elongation or after cell division) reduces permease synthesis, which in turn causes a further reduction in the internal concentration of IPTG, driving the cell toward the LacOFF state (Figure 1). The overall consequence is that a fully induced population bifurcates into two bistable states: LacON and LacOFF (Novick and Weiner, 1957; Casadesus and D’Ari, 2002; Laurent et al., 2005). The positive feedback loop in this system is that a high permease level concentrates inducer inside the cell, and a high internal level of inducer produces a high level of permease. A potential benefit of this loop may be to drain pools of metabolizable β-galactosides by maintaining high levels of permease when the inducer concentration decreases. Otherwise, a certain amount of β-galactoside might be left unused.
Figure 1.
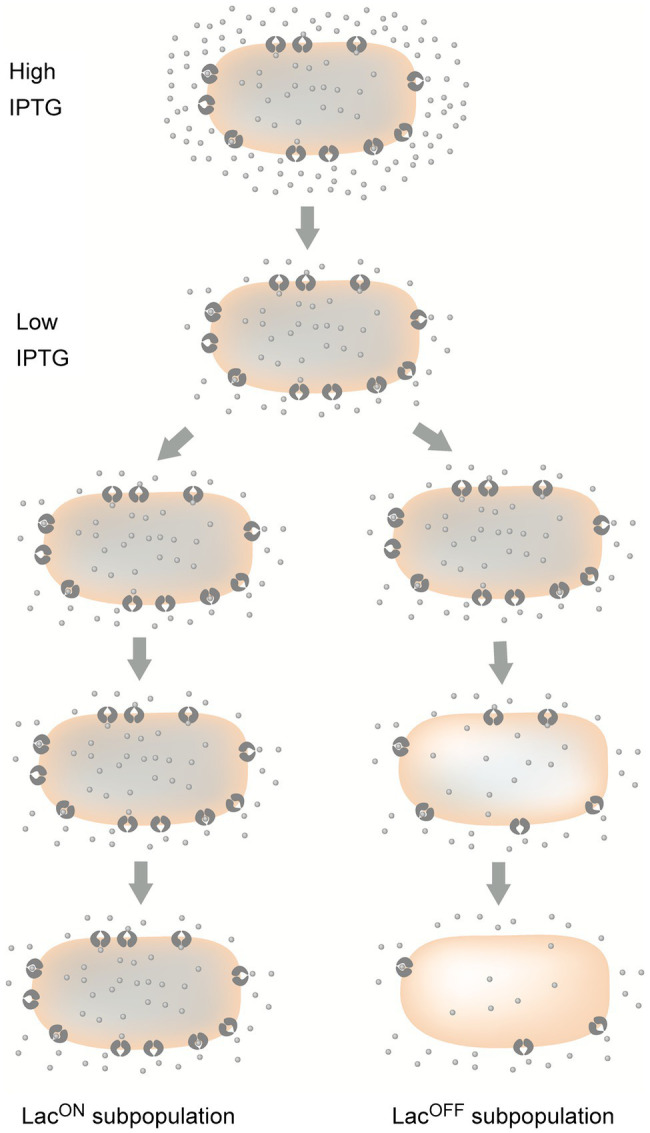
Bifurcation of lac operon expression into ON and OFF states after transfer of an induced culture to lower and intermediate levels of the inducer, IPTG. LacI permease molecules are shown inserted in the cytoplasmic membrane. For simplification, the cell wall and the outer membrane are not shown. Dots represent IPTG molecules.
An increased error rate during transcription, caused by mutations that reduce transcriptional fidelity, can trigger switching of the lac operon from OFF to ON in the presence of suboptimal concentrations of inducer (Gordon et al., 2009). A decrease in the level of functional LacI repressor below a critical threshold permits transcriptional activation in certain cells (Gordon et al., 2009; Satory et al., 2011). Synthesis of permease then creates a positive feedback loop that maintains the ON state. As in other bistable systems, a small number of LacI repressor molecules per cell (~10) is crucial to make the system noisy (Gordon et al., 2009).
Competence Development in Bacillus subtilis
When a B. subtilis culture approaches stationary phase, a fraction of cells acquire competence, a physiological state that enables DNA uptake (Dubnau and Losick, 2006). A crucial factor for the development of competence is accumulation of ComK, a transcription factor that activates genes required for DNA uptake as well as the comK gene itself (van Sinderen et al., 1995). During exponential growth, ComK is synthesized but degraded. When the culture approaches stationary phase, a quorum-sensing-related factor stabilizes ComK (Magnuson et al., 1994; Turgay et al., 1998). At that moment, a competition starts between several repressors and ComK for binding regulatory regions at the comK promoter (Hoa et al., 2002; Hamoen et al., 2003). Binding of ComK initiates a positive feedback loop, leading to increased synthesis of ComK and subsequent transcription of competence genes. Binding of the repressors inhibits comK expression and prevents competence. A crucial property for bifurcation of the population into two subpopulations is that the level of ComK in individual cells fluctuates, generating stochastic noise. When the ComK level reaches a threshold in a B. subtilis cell, a quantitative difference becomes qualitative: the ComK positive feedback loop is activated, and competence is acquired (Smits et al., 2005, 2006; Dandach and Khammash, 2010). Development of competence thus occurs in cells that undergo a small but critical increase in ComK concentration. In turn, comK is repressed in cells in which the ComK level remains below the threshold, and they do not acquire competence (Smits et al., 2006) (Figure 2).
Figure 2.
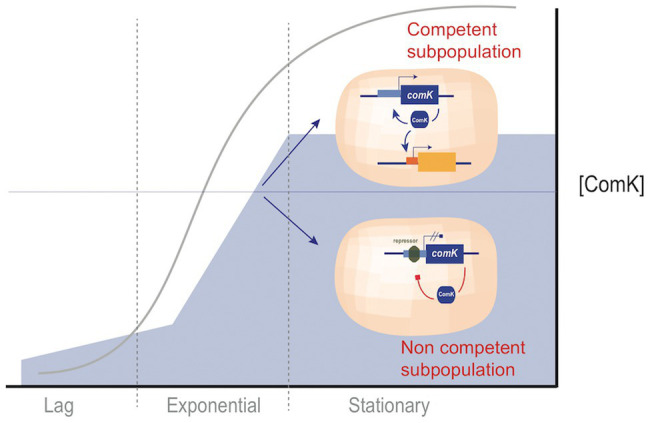
Acquisition of competence by a Bacillus subtilis subpopulation. The fate of individual cells is decided at a critical moment in which ComK levels are intermediate and noisy. Above a threshold level, ComK drives the bacterial cell toward competence. Below the threshold level, repressors prevent ComK synthesis, and the cell does not become competent. Under laboratory conditions, the decision is taken when the culture enters stationary phase.
Lysis and Lysogeny in Bacteriophages
Infection of E. coli by the temperate bacteriophage lambda can follow two developmental programs: lysis of the bacterial cell and lysogeny. Although the decision is influenced by the physiological state of the cell and by environmental cues, the fate of individual infections is unpredictable and may be considered stochastic (Johnson et al., 1981; Casadesus and D’Ari, 2002; Munsky and Khammash, 2010). Phage lambda has two repressors, CI and Cro, each of which represses the expression of the other. At the onset of infection, both repressors are produced, and the lysis-lysogeny decision may be viewed as a race: The repressor that first occupies specific regulatory DNA sites in the lambda genome will repress the synthesis of its antagonist (Johnson et al., 1981). If the winner is Cro, synthesis of CI will be repressed, and lambda will lyse the host cell (Figure 2). If the winner is CI, synthesis of Cro will be repressed, and lambda will lysogenize the cell (Johnson et al., 1981). Note that the outcomes of a positive feedback loop and a double-negative feedback loop are analogous (Casadesus and D’Ari, 2002; Ferrell, 2002). In the case of lambda, preventing the synthesis of Cro by CI is equivalent to positive autoregulation of cI gene expression, and vice versa (Figure 3).
Figure 3.
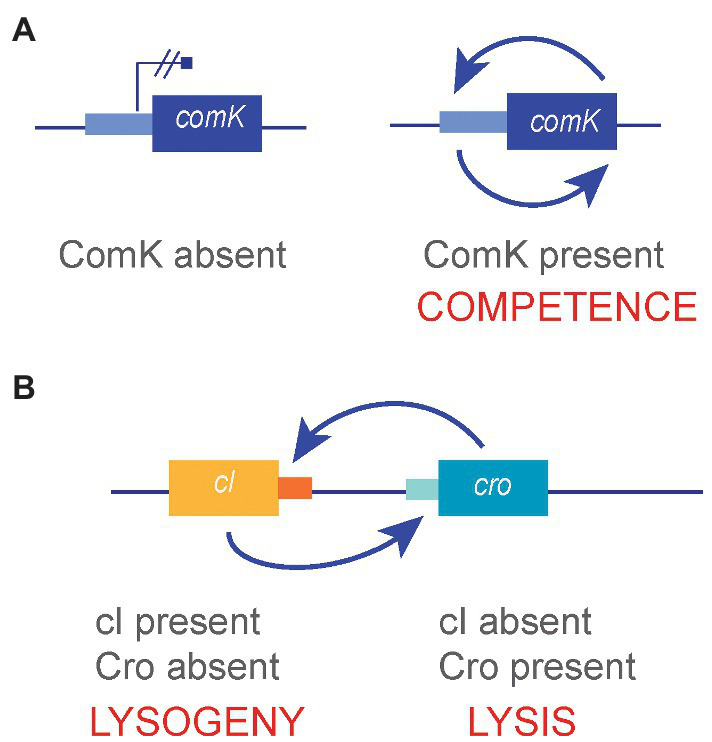
(A) Competence development in B. subtilis, an example of bistability produced by a positive feedback loop. (B) The lysis/lysogeny decision in bacteriophage lambda, an example of bistability produced by a double-negative feedback loop.
In E. coli lysogens for Shiga toxin phages, only a fraction of cells enter the lytic cycle upon prophage induction. This dual strategy may prevent extinction of the bacterial population, at the same time permitting that the phage population generated by induction can introduce the capacity to produce Shiga toxin into new hosts (Imamovic et al., 2016).
Contribution of Phenotypic Heterogeneity to Antibiotic Tolerance
Stochastic fluctuations in the expression of critical genes can produce bacterial cells that are able to survive in the presence of an antibiotic (Adam et al., 2008). For instance, stochastic activation of the multiple antibiotic resistance activator MarA confers multidrug tolerance in E. coli (El Meouche et al., 2016). In Salmonella enterica, plating of a batch culture on a lethal (but not extremely high) concentration of kanamycin provides two types of kanamycin-resistant isolates. Some are stable, produced by mutation. Other isolates, however, revert to kanamycin sensitivity upon nonselective growth, indicating a nongenetic origin for the kanamycin-tolerant phenotype. A factor that contributes to tolerance is the formation of a subpopulation of cells that contain reduced levels of the OmpC porin in the outer membrane (Sanchez-Romero and Casadesus, 2014). Expression of ompC is noisy, and cells with low OmpC protein in the outer membrane can withstand kanamycin. In the presence of kanamycin, activation of the RpoE-dependent stress response downregulates ompC expression (Woods and McBride, 2017). The resulting feedback loop sustains and/or amplifies the cellular state that initially permitted survival, and a kanamycin-tolerant subpopulation is produced (Sanchez-Romero and Casadesus, 2014).
Resistance to fluoroquinolones also has nongenetic components. For instance, the activity of the AcrAB-TolC efflux pump increases the minimal inhibitory concentration of nalidixic acid in S. enterica isolates that carry gyrase mutations. Because individual Salmonella cells display different levels of acrAB expression, the bacterial population is heterogeneous and includes cells with high AcrAB-mediated efflux (Sanchez-Romero and Casadesus, 2014). These cells have reduced growth rate, which can be regarded as a toll for the acquisition of nongenetic resistance (Motta et al., 2015). An inverse correlation between growth and antibiotic tolerance may be common (Claudi et al., 2014).
Nonmutational tolerance to antibiotics is also found in persisters, subpopulations of cells that adopt a dormant state upon decrease or arrest of growth and metabolism (Balaban, 2011; Fisher et al., 2017). Persistence is a reversible epigenetic state (Balaban et al., 2004). Persisters were first described in Staphylococcus aureus and more recently in other bacterial pathogens. Especially relevant for human health is the role of persisters in asymptomatic carriage of Mycobacterium tuberculosis and other pathogens that cause latent infection (Rhen et al., 2003). Various mechanisms have been proposed to produce persisters (e.g., toxin-antitoxin control, metabolic regulation, and ppGpp-dependent stringent response). The lack of an accepted model may merely reflect the involvement of multiple mechanisms. In fact, persisters of a given species often belong to several phenotypic classes (Dhar and McKinney, 2007; Hofsteenge et al., 2013; Putrins et al., 2015).
DNA Repair Heterogeneity
The SOS regulon, a bacterial gene network responsive to DNA damage, is under the control of the LexA transcriptional repressor (Baharoglu and Mazel, 2014). In the presence of a DNA damaging agent, LexA is degraded and SOS genes are turned on. However, SOS activation is also observed in a subpopulation of E. coli cells during normal growth (McCool et al., 2004; Pennington and Rosenberg, 2007; Kamensek et al., 2010). SOS activation under such conditions is triggered by endogenous DNA-damaging compounds produced by normal metabolism (Xia et al., 2019; Yang et al., 2019). In fact, spontaneous DNA strand breakage is detected in a subset of cells during normal growth (Pennington and Rosenberg, 2007). Heterogeneous activation of the SOS system under apparently optimal growth conditions has been also described in M. tuberculosis (Manina et al., 2019).
Heterogeneous expression may be a common feature of DNA repair systems (Vincent and Uphoff, 2020). As a consequence, repair of DNA lesions by the adaptive response to DNA alkylation damage may be accompanied by an increase in the mutation rate in individual cells (Uphoff et al., 2016). Antibiotic-induced activation of the RpoS-dependent general stress response can likewise increase the mutation rate in an E. coli subpopulation (Pribis et al., 2019). Variation of mutation rates in response to environmental factors is an old prediction of population genetics (Fitch, 1982) and has been validated by experimental studies (Oliver et al., 2000; Saint-Ruf and Matic, 2006; Pribis et al., 2019). Formation of small colony variants of pathogenic bacteria in animal cells and tissues may be also a manifestation of increased mutation rates (Proctor et al., 1994; Cano et al., 2003; Nelson et al., 2010). The notion of stress-induced mutation has raised concerns about mutational burden (Roth et al., 2006). However, such arguments can be lessened if the mutation rate increases only in a subpopulation of cells.
Bistable States in Host-Pathogen Interactions
Formation of cell variants in bacterial pathogens has long been recognized as a strategy for evasion of the immune system (Finlay and McFadden, 2006). In addition, cases of phenotypic heterogeneity whose adaptive value does not seem related to immune evasion have been described.
In the opportunistic pathogen Pseudomonas aeruginosa, a positive feedback loop involving the transcriptional regulator BexR activates the expression of the so-called BexR regulon, which includes the virulence-related aprA gene and other loci of unknown function (Turner et al., 2009). In addition, BexR shows positive autoregulation (Turner et al., 2009). Similar to the B. subtilis ComK system, bistable BexR expression is the consequence of noisy, low-level BexR synthesis, followed by autogenous amplification of the BexR level in cells that produce BexR above a critical threshold (Turner et al., 2009). A difference, however, is that competence is acquired by 10% of B. subtilis cells (Dubnau and Losick, 2006) while the BexR feedback loop is activated in 0.004% cells only (Turner et al., 2009).
In Yersinia pseudotuberculosis, bistable synthesis of the virulence regulator RovA can be viewed as a bet-hedging strategy that preadapts the bacterial population to the changing conditions encountered during early and late stages of infection (Nuss et al., 2016; Weigel and Dersch, 2018). RovA bistability has at least two sources. Activation of rovA transcription by RovA is noisy, and a feedback loop of autogenous activation is triggered in cells where RovA reaches a critical threshold. In addition, posttranscriptional control contributes to bistability: a conformational change in a dimerization domain reduces the RovA DNA-binding capacity and increases proteolytic degradation, thus driving the system toward the OFF state. Control of the ratio of ON/OFF cells can be further modulated by two-component systems and global regulators, adjusting the expression of virulence determinants during different stages of infection and in different tissues (Weigel and Dersch, 2018).
Temporal bistability also modulates virulence in Vibrio cholerae. At a late stage of animal infection, V. cholerae populations bifurcate into two subpopulations, one of which turns off virulence genes while the other remains virulent (Nielsen et al., 2010). The existence of a highly infectious subpopulation in the stools of cholera patients may contribute to V. cholerae dissemination. Bifurcation is reversible, and a bistable switch enables or disables the formation of a feedback loop that controls the expression of ToxT, the master regulator of virulence gene expression (Nielsen et al., 2010).
The Gram positive pathogen S. aureus causes acute and chronic infections, and the infection outcome is controlled by a quorum-sensing system called Agr (Recsei et al., 1986; Benson et al., 2011). This system shows bistability, with concomitant formation of AgrOFF and AgrON subpopulations specialized in planktonic and biofilm-associated lifestyles, respectively (García-Betancur et al., 2017). The AgrON cell lineage, specialized in chronic infection, is produced by a positive feedback loop that activates the expression of biofilm genes in cells where the phosphorylated form of the transcription factor AgrA is present above a critical concentration. Below this threshold, the cells remain AgrOFF and form a subpopulation with acute infection capacity including toxin secretion (García-Betancur et al., 2017).
Another threshold-based decision controls phenotypic heterogeneity in Xenorhabdus nematophila, a Gram-negative bacterium used in biological pest control. X. nematophila lives a double life, as a pathogen of insects and a mutualist of nematodes that transmit the pathogen to insects. Formation of mutualistic and virulent cell variants is under the control of the transcriptional regulator Lrp, which controls transcription of hundreds of genes (Hussa et al., 2015). The level of Lrp shows cell-to-cell variation, and high Lrp levels promote mutualism, while low Lrp levels promote virulence. As infected nematodes age, a decrease in the Lrp level enhances virulence, anticipating exposure to the insect host (Cao and Goodrich-Blair, 2020). Interestingly, lrp mutants, which show a growth advantage at late stages of infection, have reduced virulence and impaired transmission to insects (Cambon et al., 2019), a feature that illustrates how nonmutational variation can be advantageous over mutation.
In S. enterica, expression of pathogenicity island 1 (SPI-1) is bimodal, and virulence determinants are secreted by SPI-1ON cells only (Sturm et al., 2011; Arnoldini et al., 2014). Synthesis and/or activity of the type III secretion system (T3SS) slows down the growth of SPI-1ON cells. However, inflammation triggered by the T3SS generates electron acceptors that provide a growth advantage to Salmonella over the intestinal microbiota (Diard et al., 2013), a benefit for both SPI-1ON and SPI-1OFF cells. Furthermore, fast growth makes SPI-1OFF cells able to outcompete Salmonella avirulent variants (Diard et al., 2013). Epithelial cell invasion by SPI-1OFF cells may extend outcompetition to the intracellular environment, contributing to prevent takeover of the population by avirulent mutants (Sanchez-Romero and Casadesus, 2018).
Additional bifurcations occur during Salmonella infection. Bistable expression of myo-inositol utilization genes may help to overcome nutrient limitation in the intestine and can be viewed as a bet-hedging strategy (Muller et al., 2019). Epithelial cells infected by Salmonella contain either large or small numbers of bacteria, a distribution that suggests bimodality (Garcia-Del Portillo, 2008). During systemic infection, the Salmonella population splits into two subpopulations inside macrophages, and one subpopulation multiplies while the other enters a dormant-like state (Helaine et al., 2010).
Colonization of the gall bladder by Salmonella provides another example of lineage formation. The bile-laden gall bladder is a harsh environment for bacteria because bile salts are bactericidal (Urdaneta and Casadesus, 2017). Salmonella survival in the gall bladder may be facilitated by bifurcation of the population into one lineage that invades the gall bladder epithelium (Menendez et al., 2009), while another lineage remains in the gall bladder lumen. Further diversification occurs if gallstones are present, because Salmonella is able to form biofilms on gallstones (Prouty et al., 2002; Crawford et al., 2010). Survival of planktonic cells in the gall bladder lumen may additionally involve phenotypic heterogeneity associated with noisy activation of the RpoS-dependent general stress response in certain cells (Urdaneta et al., 2019).
Bistable Switches Under DNA Methylation Control
DNA methylation has multiple roles in bacterial physiology, including the control of lineage formation (Casadesus and Torreblanca, 1996; Marinus, 1996; Lobner-Olesen et al., 2005; Wion and Casadesus, 2006; Low and Casadesus, 2008; Vasu and Nagaraja, 2013; Adhikari and Curtis, 2016; Mouammine and Collier, 2018). Formation of cell variants under DNA methylation control may be an especially robust mechanism for subpopulation formation because inheritance of DNA methylation patterns permits faithful transmission of transcriptional states across generations. Furthermore, unlike noise-based switches, the architecture and the DNA methylation state of the regulatory region determine the switching frequencies, thereby producing subpopulations of constant sizes (Casadesus and Low, 2006). In certain cases, the subpopulation sizes can be additionally modulated by cellular regulators responsive to environmental cues (Casadesus and Low, 2006; Sanchez-Romero and Casadesus, 2020).
The widespread involvement of DNA methylation in bacterial pathogenesis (Marinus and Casadesus, 2009; Kumar and Rao, 2013) and the recent development of a DNA sequencing procedure that permits genome-wide detection of N6-methyl-adenine (SMRT sequencing) (Flusberg et al., 2010) has given a fresh impulse to the study of DNA methylation in bacterial genomes (Davis et al., 2013). Among other interesting outcomes, SMRT sequencing has broadened our knowledge of the distribution of DNA methylation in bacterial genomes (Blow et al., 2016) and has provided novel examples of bistable loci under DNA methylation control (Sanchez-Romero et al., 2020).
Control of Lineage Formation by DNA Adenine Methylation
In gammaproteobacteria, formation of DNA adenine methylation patterns (combinations of methylated and nonmethylated GATC sites) provides a mechanism for transmission of epigenetic states to the offspring (Wion and Casadesus, 2006). Nonmethylated sites are often part of clusters of GATCs located within binding sites for transcriptional regulators and are flanked by DNA sequences that reduce the processivity of the Dam methylase (Wion and Casadesus, 2006; Sanchez-Romero and Casadesus, 2020). Binding of the cognate protein hinders Dam methylase activity, generating nonmethylated GATCs after two rounds of DNA replication. Nonmethylation persists as long as the transcription factor remains bound to its cognate sequence, and the methylation pattern can be inherited by daughter cells. However, every DNA replication round provides a window of opportunity to change the DNA methylation pattern of the regulatory region, switching transcription from OFF to ON and vice versa. A paradigm of Dam-dependent epigenetic control is the pap operon of uropathogenic E. coli, whose workings were brilliantly deciphered by David Low and co-workers in the 1990s and early 2000s (van der Woude et al., 1996; Hernday et al., 2004). The pap operon encodes fimbriae that permit adherence to the epithelium of the urinary tract (van der Woude et al., 1996), and formation of PapOFF and PapON lineages may be interpreted as a division of labor: Only the fimbriated subpopulation can colonize the urinary tract, but the nonfimbriated subpopulation makes it possible by avoiding immune system alert. An example of Dam-dependent locus involved in bet hedging is provided by the Salmonella opvAB operon, which produces a lineage of cells resistant to bacteriophages at the expense of reducing virulence (Cota et al., 2015).
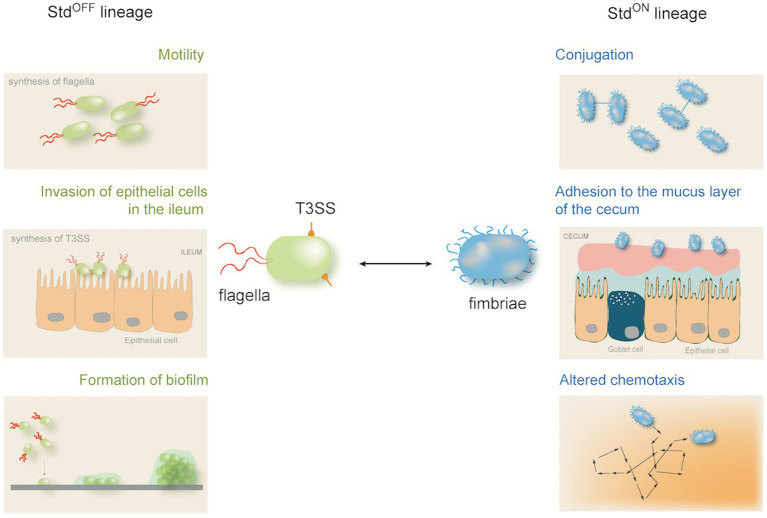
Additional genes and operons under Dam methylation control have been described in E. coli and Salmonella, each with particular traits and switching frequencies (Sanchez-Romero and Casadesus, 2020). An example that stands out because of its unusual pleiotropy is the Salmonella std operon, which encodes fimbriae for adhesion to the mucus layer of the cecum in the large intestine (Suwandi et al., 2019). In addition to fimbrial components, the std operon encodes transcriptional regulators that control the expression of hundreds of genes (Garcia-Pastor et al., 2018). StdOFF and StdON cells thus differ in multiple phenotypic traits, and their formation may be considered a genuine example of bacterial differentiation without visible morphological change (Figure 4).
Figure 4.
Subpopulation formation under the control of the Std pleiotropic switch in Salmonella enterica serovar Typhimurium. In addition to fimbriae, the std operon encodes transcriptional regulators that activate or repress hundreds of genes. As a consequence, the StdOFF and StdON lineages differ in multiple phenotypic traits.
Phase-Variable DNA Methylation
Certain restriction-modification (R-M) systems of types I and III undergo phase variation (De Ste Croix et al., 2017; Seib et al., 2020). Switching between OFF and ON states is caused by alteration of nucleotide repeats in certain systems and by recombination in others. In some such systems, the gene encoding the restriction enzyme is inactivated by mutation, while the DNA methyltransferase gene remains active (Srikhanta et al., 2010). Phase variation of DNA methyltransferase synthesis produces two subpopulations of bacterial cells, one of which contains N6-methyl-adenine in the genome while the other subpopulation does not. As a consequence, each lineage shows a distinct pattern of gene expression in all DNA methylation-sensitive loci. Systems of this kind, known as phasevarions, have been described in human pathogens belonging to the genera Haemophilus, Neisseria, Helicobacter, Moraxella, Mycoplasma, and Streptococcus. Phasevarions have been shown to control envelope structure, as well as virulence and stress responses, and can facilitate immunoevasion (Srikhanta et al., 2010; Phillips et al., 2019; Seib et al., 2020).
Phasevarions are an outstanding evolutionary invention. Most phase variation systems under DNA methylation control (e.g., pap and opvAB) generate heterogeneity of a single phenotypic trait, while the cell lineages under phasevarion control differ in multiple phenotypic traits. An additional tour de force in the capacity of phasevarions to generate cell-to-cell diversity is found in bacterial species that produce DNA methyltransferase variants. A phasevarion of this kind controls lineage formation in the pneumococcus, Streptococcus pneumoniae, an opportunistic pathogen frequently found in the nasopharynx of healthy humans. The pneumococcus also causes several types of acute infection, including pneumonia and meningitis. Pneumococcal populations undergo phase variation between “opaque” and “transparent” colony phenotypes that differ in their virulence properties (Weiser et al., 1994). Subpopulations that combine traits of the two phenotypes are also produced. Formation of such lineages is under the control of a phase-variable DNA adenine methyltransferase of a type I R-M system. Six DNA methyltransferase variants are produced by site-specific recombination, and each variant generates a distinct pattern of genome methylation, which results in the formation of cell types with distinct virulence properties. Formation of such lineages may facilitate adaptation during different stages of the infection, including the crucial passage from the nasopharynx into the lung (Manso et al., 2014; Li et al., 2016; Oliver et al., 2017).
Evolution of Phenotypic Heterogeneity
Except in obligate parasites, the biochemical machinery of prokaryotes has evolved to facilitate adaptation to changing environments. However, the adaptive capacity of a biological species is restrained by the fact that an organism can only have a limited set of traits (Maynard-Smith, 1982). Production of phenotypic variants can overcome this limitation. Because natural selection acts on phenotypes and not on genotypes, mutational and nonmutational mechanisms can be similarly suitable as sources of cell variation. An advantage of nonmutational heterogeneity is that it avoids the irreversible commitment to a new state imposed by mutation (Veening et al., 2008; Ackermann, 2015; Grimbergen et al., 2015). In fact, game theory analysis predicts that phenotypic heterogeneity can have higher adaptive value than mutation in changing environments (Kussell and Leibler, 2005; Wolf et al., 2005).
Genetic and epigenetic variations are not mutually exclusive. Nonmutational adaptation can provide a temporary window for mutation, a possibility that becomes more likely if the surviving population is large (Baquero, 2013). Hence, bacterial adaptation to new environments can be facilitated by phenotypic adaptation on a short timescale and by tuning via mutations in the long run (Kussell, 2013), and mutations that promote nongenetic variation (e.g., by adjusting the level of noise or the strength of a feedback loop) may be selected in a similar manner as mutations that confer any other adaptive trait.
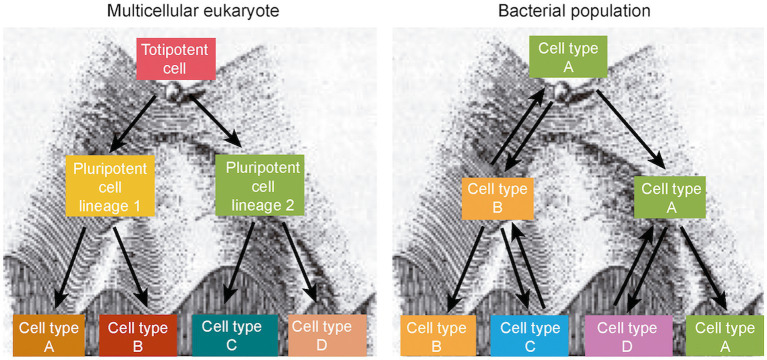
Bacterial Waddington’s landscapes differ from their eukaryotic counterparts in a fundamental aspect, visualized in Figure 5. In a multicellular eukaryote, sequential decisions progressively curtail the cell differentiation capacity. This constraint does not exist in a bacterial population: In principle, a bacterial cell can differentiate into any other cell type. Progression through the Waddington’s landscape is thus orderly in multicellular eukaryotes and chaotic in bacteria. Unrestrained differentiation may be crucial to produce the polymorphism required for bacterial adaptation, and natural selection may tailor the level of cell-to-cell variation to the needs and challenges posed by the lifestyle of the species.
Figure 5.
Adaptation of Waddington’s artistic drawing of an epigenetic landscape as a ball that falls to stable valleys from unstable ridges. In multicellular eukaryotes, the differentiation capacity is narrowed down at each intersection, and development proceeds in an orderly manner (left). In a bacterial epigenetic landscape, loss of differentiation capacity does not occur at any intersection, and formation of cell variants follows an aperiodic behavior typical of deterministic nonlinear systems (right).
Bacterial evolution is speeded up by the fact that bacterial DNA is both somatic and germinal. As a consequence, beneficial mutations are immediately passed to the offspring without the gambles of meiosis and gamete assortment. Furthermore, the absence of canalization in bacteria exposes novel phenotypes to immediate selection (Elena and Lenski, 2001). If an adaptive phenotype is produced, natural selection will instantly act regardless of its genetic or epigenetic origin. Waddington’s landscapes can thus evolve rapidly in the bacterial world.
The capacity of a bacterial population to produce phenotypic cell variants can be appraised if one considers that independent switching of n bistable loci can produce 2n types of cell variants (Sanchez-Romero et al., 2020). This theoretical number may be an overestimation as natural selection can be expected to eliminate lower fitness variants. Anyway, the actual number of phenotypic cell variants in an isogenic population of bacteria may easily surpass the detection capacity of current technologies of single-cell analysis.
Author Contributions
MS-R and JC: conceptualization and investigation. JC: original draft preparation, administration, and funding acquisition. MS-R and JC: review and editing. Both the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to current laboratory members David R. Olivenza, Angela Mérida-Floriano, and Rocío-Fernández-Fernández and to previous laboratory members Marcello Jakomin, Roberto Balbontín, Javier López-Garrido, Ignacio Cota, Elena Puerta-Fernández, Elena Espinosa, Sara B. Hernández, Verónica Urdaneta, and Lucía García-Pastor for their contributions to the study of phenotypic heterogeneity. We also thank colleagues for sharing ideas, criticism, and discussions over the years. The list includes David Low, Martin Marinus, Erik Boye, Marjan van der Woude, Andreas Bäumler, Jay Hinton, Frédéric Barras, Francisco García-del Portillo, Mireille Ansaldi, Eric Cascales, Laurent Aussel, Julien Brillard, and Carmen R. Beuzón.
Footnotes
Funding. Work in our laboratory is currently supported by Grant BIO2016-75235-P from the Ministerio de Ciencia e Innovación of Spain – Agencia Estatal de Investigación and the European Regional Fund.
References
- Ackermann M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508. 10.1038/nrmicro3491, PMID: [DOI] [PubMed] [Google Scholar]
- Adam M., Murali B., Glenn N. O., Potter S. S. (2008). Epigenetic inheritance based evolution of antibiotic resistance in bacteria. BMC Evol. Biol. 8:52. 10.1186/1471-2148-8-52, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari S., Curtis P. D. (2016). DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 40, 575–591. 10.1093/femsre/fuw023, PMID: [DOI] [PubMed] [Google Scholar]
- Anderson P. W. (1972). More is different. Science 177, 393–396. 10.1126/science.177.4047.393, PMID: [DOI] [PubMed] [Google Scholar]
- Arnoldini M., Vizcarra I. A., Pena-Miller R., Stocker N., Diard M., Vogel V., et al. (2014). Bistable expression of virulence genes in salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol. 12:e1001928. 10.1371/journal.pbio.1001928, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z., Mazel D. (2014). SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126–1145. 10.1111/1574-6976.12077, PMID: [DOI] [PubMed] [Google Scholar]
- Balaban N. Q. (2011). Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775. 10.1016/j.gde.2011.10.001, PMID: [DOI] [PubMed] [Google Scholar]
- Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. 10.1126/science.1099390, PMID: [DOI] [PubMed] [Google Scholar]
- Baquero F. (2013). Epigenetics, epistasis and epidemics. Evol. Med. Public Health 2013, 86–88. 10.1093/emph/eot009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikova D., Jochim A., Power J., Holden M. T. G., Heilbronner S. (2020). “Gene accordions” cause genotypic and phenotypic heterogeneity in clonal populations of Staphylococcus aureus. Nat. Commun. 11:3526. 10.1038/s41467-020-17277-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M. A., Lilo S., Wasserman G. A., Thoendel M., Smith A., Horswill A. R., et al. (2011). Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol. Microbiol. 81, 659–675. 10.1111/j.1365-2958.2011.07720.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernander R., Stokke T., Boye E. (1998). Flow cytometry of bacterial cells: comparison between different flow cytometers and different DNA stains. Cytometry 31, 29–36. , PMID: [DOI] [PubMed] [Google Scholar]
- Blow M. J., Clark T. A., Daum C. G., Deutschbauer A. M., Fomenkov A., Fries R., et al. (2016). The epigenomic landscape of prokaryotes. PLoS Genet. 12:e1005854. 10.1371/journal.pgen.1005854, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambon M. C., Parthuisot N., Pages S., Lanois A., Givaudan A., Ferdy J. B. (2019). Selection of bacterial mutants in late infections: when vector transmission trades off against growth advantage in stationary phase. MBio 10:e01437-19. 10.1128/mBio.01437-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano D. A., Pucciarelli M. G., Martinez-Moya M., Casadesus J., Garcia-del Portillo F. (2003). Selection of small-colony variants of Salmonella enterica serovar Typhimurium in nonphagocytic eucaryotic cells. Infect. Immun. 71, 3690–3698. 10.1128/IAI.71.7.3690-3698.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Goodrich-Blair H. (2020). Xenorhabdus nematophila bacteria shift from mutualistic to virulent Lrp-dependent phenotypes within the receptacles of Steinernema carpocapsae insect-infective stage nematodes. Environ. Microbiol. 22, 5433–5449. 10.1111/1462-2920.15286, PMID: [DOI] [PubMed] [Google Scholar]
- Casadesus J., D’Ari R. (2002). Memory in bacteria and phage. BioEssays 24, 512–518. 10.1002/bies.10102, PMID: [DOI] [PubMed] [Google Scholar]
- Casadesus J., Low D. (2006). Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856. 10.1128/MMBR.00016-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus J., Low D. A. (2013). Programmed heterogeneity: epigenetic mechanisms in bacteria. J. Biol. Chem. 288, 13929–13935. 10.1074/jbc.R113.472274, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus J., Torreblanca J. (1996). “Methylation-related epigenetic signals in bacterial DNA” in Epigenetic Mechanisms of Gene Regulation. eds. Russo V. E. A., Martienssen R. A., Riggs A. D. (Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; ), 141–153. [Google Scholar]
- Claudi B., Sprote P., Chirkova A., Personnic N., Zankl J., Schurmann N., et al. (2014). Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158, 722–733. 10.1016/j.cell.2014.06.045, PMID: [DOI] [PubMed] [Google Scholar]
- Collier J. (2019). Cell division control in Caulobacter crescentus. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 685–690. 10.1016/j.bbagrm.2018.04.005, PMID: [DOI] [PubMed] [Google Scholar]
- Cota I., Sanchez-Romero M. A., Hernandez S. B., Pucciarelli M. G., Garcia-Del Portillo F., Casadesus J. (2015). Epigenetic control of Salmonella enterica O-antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genet. 11:e1005667. 10.1371/journal.pgen.1005667, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. W., Rosales-Reyes R., Ramirez-Aguilar Mde L., Chapa-Azuela O., Alpuche-Aranda C., Gunn J. S. (2010). Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U. S. A. 107, 4353–4358. 10.1073/pnas.1000862107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandach S. H., Khammash M. (2010). Analysis of stochastic strategies in bacterial competence: a master equation approach. PLoS Comp. Biol. 6:e1000985. 10.1371/journal.pcbi.1000985, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. M., Chao M. C., Waldor M. K. (2013). Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr. Opin. Microbiol. 16, 192–198. 10.1016/j.mib.2013.01.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong I. G., Haccou P., Kuipers O. P. (2011). Bet hedging or not? A guide to proper classification of microbial survival strategies. BioEssays 33, 215–223. 10.1002/bies.201000127, PMID: [DOI] [PubMed] [Google Scholar]
- De Ste Croix M., Vacca I., Kwun M. J., Ralph J. D., Bentley S. D., Haigh R., et al. (2017). Phase-variable methylation and epigenetic regulation by type I restriction-modification systems. FEMS Microbiol. Rev. 41(Suppl. 1), S3–S15. 10.1093/femsre/fux025, PMID: [DOI] [PubMed] [Google Scholar]
- Dewachter L., Fauvart M., Michiels J. (2019). Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol. Cell 76, 255–267. 10.1016/j.molcel.2019.09.028, PMID: [DOI] [PubMed] [Google Scholar]
- Dhar N., McKinney J. D. (2007). Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 10, 30–38. 10.1016/j.mib.2006.12.007, PMID: [DOI] [PubMed] [Google Scholar]
- Diard M., Garcia V., Maier L., Remus-Emsermann M. N., Regoes R. R., Ackermann M., et al. (2013). Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494, 353–356. 10.1038/nature11913, PMID: [DOI] [PubMed] [Google Scholar]
- Dragos A., Kiesewalter H., Martin M., Hsu C. Y., Hartmann R., Wechsler T., et al. (2018). Division of labor during biofilm matrix production. Curr. Biol. 28, 1903–1913. 10.1016/j.cub.2018.04.046, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Losick R. (2006). Bistability in bacteria. Mol. Microbiol. 61, 564–572. 10.1111/j.1365-2958.2006.05249.x, PMID: [DOI] [PubMed] [Google Scholar]
- El Meouche I., Siu Y., Dunlop M. J. (2016). Stochastic expression of a multiple antibiotic resistance activator confers transient resistance in single cells. Sci. Rep. 6:19538. 10.1038/srep19538, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S. F., Lenski R. E. (2001). Epistasis between new mutations and genetic background and a test of genetic canalization. Evolution 55, 1746–1752. 10.1111/j.0014-3820.2001.tb00824.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ferrell J. E., Jr. (2002). Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148. 10.1016/S0955-0674(02)00314-9, PMID: [DOI] [PubMed] [Google Scholar]
- Finlay B. B., McFadden G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. 10.1016/j.cell.2006.01.034, PMID: [DOI] [PubMed] [Google Scholar]
- Fisher R. A., Gollan B., Helaine S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. 10.1038/nrmicro.2017.42, PMID: [DOI] [PubMed] [Google Scholar]
- Fitch W. M. (1982). The challenges to Darwinism since the last centennial and the impact of molecular studies. Evolution 36, 1133–1143. 10.1111/j.1558-5646.1982.tb05484.x, PMID: [DOI] [PubMed] [Google Scholar]
- Flusberg B. A., Webster D. R., Lee J. H., Travers K. J., Olivares E. C., Clark T. A., et al. (2010). Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465. 10.1038/nmeth.1459, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Betancur J. C., Goñi-Moreno A., Horger T., Schott M., Sharan M., Eikmeier J., et al. (2017). Cell differentiation defines acute and chronic infection cell types in Staphylococcus aureus. eLife 6:e28023. 10.7554/eLife.28023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Del Portillo F. (2008). Heterogeneity in tissue culture infection models: a source of novel host-pathogen interactions? Microbes Infect. 10, 1063–1066. 10.1016/j.micinf.2008.07.004, PMID: [DOI] [PubMed] [Google Scholar]
- Garcia-Pastor L., Sanchez-Romero M. A., Gutierrez G., Puerta-Fernandez E., Casadesus J. (2018). Formation of phenotypic lineages in Salmonella enterica by a pleiotropic fimbrial switch. PLoS Genet. 14:e1007677. 10.1371/journal.pgen.1007677, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H. (1974). Natural selection for within-generation variance in offspring number. Genetics 76, 601–606. 10.1093/genetics/76.3.601, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. J., Halliday J. A., Blankschien M. D., Burns P. A., Yatagai F., Herman C. (2009). Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 7:e44. 10.1371/journal.pbio.1000044, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbergen A. J., Siebring J., Solopova A., Kuipers O. P. (2015). Microbial bet-hedging: the power of being different. Curr. Opin. Microbiol. 25, 67–72. 10.1016/j.mib.2015.04.008, PMID: [DOI] [PubMed] [Google Scholar]
- Hamoen L. W., Kausche D., Marahiel M. A., van Sinderen D., Venema G., Serror P. (2003). The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol. Lett. 218, 299–304. 10.1111/j.1574-6968.2003.tb11532.x, PMID: [DOI] [PubMed] [Google Scholar]
- Helaine S., Thompson J. A., Watson K. G., Liu M., Boyle C., Holden D. W. (2010). Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U. S. A. 107, 3746–3751. 10.1073/pnas.1000041107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez S. B., Cota I., Ducret A., Aussel L., Casadesus J. (2012). Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet. 8:e1002459. 10.1371/journal.pgen.1002459, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday A., Braaten B., Low D. (2004). The intricate workings of a bacterial epigenetic switch. Adv. Exp. Med. Biol. 547, 83–89. 10.1007/978-1-4419-8861-4_7 [DOI] [PubMed] [Google Scholar]
- Hoa T. T., Tortosa P., Albano M., Dubnau D. (2002). Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43, 15–26. 10.1046/j.1365-2958.2002.02727.x, PMID: [DOI] [PubMed] [Google Scholar]
- Hofsteenge N., van Nimwegen E., Silander O. K. (2013). Quantitative analysis of persister fractions suggests different mechanisms of formation among environmental isolates of E. coli. BMC Microbiol. 13:25. 10.1186/1471-2180-13-25, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa E. A., Casanova-Torres A. M., Goodrich-Blair H. (2015). The global transcription factor Lrp controls virulence modulation in Xenorhabdus nematophila. J. Bacteriol. 197, 3015–3025. 10.1128/JB.00272-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamovic L., Balleste E., Martinez-Castillo A., Garcia-Aljaro C., Muniesa M. (2016). Heterogeneity in phage induction enables the survival of the lysogenic population. Environ. Microbiol. 18, 957–969. 10.1111/1462-2920.13151, PMID: [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. (1981). Lambda repressor and cro – components of an efficient molecular switch. Nature 294, 217–223. 10.1038/294217a0, PMID: [DOI] [PubMed] [Google Scholar]
- Kaern M., Elston T. C., Blake W. J., Collins J. J. (2005). Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451–464. 10.1038/nrg1615, PMID: [DOI] [PubMed] [Google Scholar]
- Kamensek S., Podlesek Z., Gillor O., Zgur-Bertok D. (2010). Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogeneous expression. BMC Microbiol. 10:283. 10.1186/1471-2180-10-283, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K., Lopez-Garrido J., Pogliano K. (2020). Shaping an endospore: architectural transformations during Bacillus subtilis sporulation. Annu. Rev. Microbiol. 74, 361–386. 10.1146/annurev-micro-022520-074650, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E., Mergaert P., Kereszt A. (2013). A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628. 10.1146/annurev-micro-092412-155630, PMID: [DOI] [PubMed] [Google Scholar]
- Kreibich S., Hardt W. D. (2015). Experimental approaches to phenotypic diversity in infection. Curr. Opin. Microbiol. 27, 25–36. 10.1016/j.mib.2015.06.007, PMID: [DOI] [PubMed] [Google Scholar]
- Kumar R., Rao D. N. (2013). Role of DNA methyltransferases in epigenetic regulation in bacteria. Subcell. Biochem. 61, 81–102. 10.1007/978-94-007-4525-4_4 [DOI] [PubMed] [Google Scholar]
- Kussell E. (2013). Evolution in microbes. Annu. Rev. Biophys. 42, 493–514. 10.1146/annurev-biophys-083012-130320, PMID: [DOI] [PubMed] [Google Scholar]
- Kussell E., Leibler S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078. 10.1126/science.1114383, PMID: [DOI] [PubMed] [Google Scholar]
- Lambert G., Kussell E. (2014). Memory and fitness optimization of bacteria under fluctuating environments. PLoS Genet. 10:e1004556. 10.1371/journal.pgen.1004556, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent M., Charvin G., Guespin-Michel J. (2005). Bistability and hysteresis in epigenetic regulation of the lactose operon. Since Delbruck, a long series of ignored models. Cell. Mol. Biol. 51, 583–594. [PubMed] [Google Scholar]
- Leigh E. G., Jr. (2010). The group selection controversy. J. Evol. Biol. 23, 6–19. 10.1111/j.1420-9101.2009.01876.x, PMID: [DOI] [PubMed] [Google Scholar]
- Li J., Li J. W., Feng Z., Wang J., An H., Liu Y., et al. (2016). Epigenetic switch driven by DNA inversions dictates phase variation in Streptococcus pneumoniae. PLoS Pathog. 12:e1005762. 10.1371/journal.ppat.1005762, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobner-Olesen A., Skovgaard O., Marinus M. G. (2005). Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8, 154–160. 10.1016/j.mib.2005.02.009, PMID: [DOI] [PubMed] [Google Scholar]
- Low D. A., Casadesus J. (2008). Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11, 106–112. 10.1016/j.mib.2008.02.012, PMID: [DOI] [PubMed] [Google Scholar]
- Magnuson R., Solomon J., Grossman A. D. (1994). Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77, 207–216. 10.1016/0092-8674(94)90313-1, PMID: [DOI] [PubMed] [Google Scholar]
- Manina G., Griego A., Singh L. K., McKinney J. D., Dhar N. (2019). Preexisting variation in DNA damage response predicts the fate of single mycobacteria under stress. EMBO J. 38:e101876. 10.15252/embj.2019101876, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso A. S., Chai M. H., Atack J. M., Furi L., De Ste Croix M., Haigh R., et al. (2014). A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 5:6055. 10.1038/ncomms6055, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. (1996). “Methylation of DNA” in Escherichia coli and Salmonella: Cellular and Molecular Biology. eds. Neidhardt F. C., Curtiss R., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., et al. (Washington, DC: ASM Press; ), 782–791. [Google Scholar]
- Marinus M. G., Casadesus J. (2009). Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33, 488–503. 10.1111/j.1574-6976.2008.00159.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J. (1982). Evolution and the Theory of Games. Cambridge, England: Cambridge University Press. [Google Scholar]
- McCool J. D., Long E., Petrosino J. F., Sandler H. A., Rosenberg S. M., Sandler S. J. (2004). Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53, 1343–1357. 10.1111/j.1365-2958.2004.04225.x, PMID: [DOI] [PubMed] [Google Scholar]
- Menendez A., Arena E. T., Guttman J. A., Thorson L., Vallance B. A., Vogl W., et al. (2009). Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200, 1703–1713. 10.1086/646608, PMID: [DOI] [PubMed] [Google Scholar]
- Meyer P., Dworkin J. (2007). Applications of fluorescence microscopy to single bacterial cells. Res. Microbiol. 158, 187–194. 10.1016/j.resmic.2006.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Motta S. S., Cluzel P., Aldana M. (2015). Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS One 10:e0118464. 10.1371/journal.pone.0118464, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouammine A., Collier J. (2018). The impact of DNA methylation in Alphaproteobacteria. Mol. Microbiol. 110, 1–10. 10.1111/mmi.14079, PMID: [DOI] [PubMed] [Google Scholar]
- Moxon R., Bayliss C., Hood D. (2006). Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40, 307–333. 10.1146/annurev.genet.40.110405.090442, PMID: [DOI] [PubMed] [Google Scholar]
- Muller J., Spriewald S., Stecher B., Stadler E., Fuchs T. M. (2019). Evolutionary stability of Salmonella competition with the gut microbiota: how the environment fosters heterogeneity in exploitative and interference competition. J. Mol. Biol. 431, 4732–4748. 10.1016/j.jmb.2019.06.027, PMID: [DOI] [PubMed] [Google Scholar]
- Munoz-Dorado J., Marcos-Torres F. J., Garcia-Bravo E., Moraleda-Munoz A., Perez J. (2016). Myxobacteria: moving, killing, feeding, and surviving together. Front. Microbiol. 7:781. 10.3389/fmicb.2016.00781, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B., Khammash M. (2010). Identification from stochastic cell-to-cell variation: a genetic switch case study. IET Syst. Biol. 4, 356–366. 10.1049/iet-syb.2010.0013, PMID: [DOI] [PubMed] [Google Scholar]
- Muro-Pastor A. M., Hess W. R. (2012). Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol. 20, 548–557. 10.1016/j.tim.2012.07.005, PMID: [DOI] [PubMed] [Google Scholar]
- Nelson L. K., Stanton M. M., Elphinstone R. E. A., Helwerda J., Turner R. J., Ceri H. (2010). Phenotypic diversification in vivo: Pseudomonas aeruginosa gacS- strains generate small colony variants in vivo that are distinct from in vitro variants. Microbiology 156, 3699–3709. 10.1099/mic.0.040824-0 [DOI] [PubMed] [Google Scholar]
- Nielsen A. T., Dolganov N. A., Rasmussen T., Otto G., Miller M. C., Felt S. A., et al. (2010). A bistable switch and anatomical site control vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 6:e1001102. 10.1371/journal.ppat.1001102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick A., Weiner M. (1957). Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. U. S. A. 43, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss A. M., Schuster F., Roselius L., Klein J., Bucker R., Herbst K., et al. (2016). A precise temperature-responsive bistable switch controlling Yersinia virulence. PLoS Pathog. 12:e1006091. 10.1371/journal.ppat.1006091, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Canton R., Campo P., Baquero F., Blazquez J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254. 10.1126/science.288.5469.1251, PMID: [DOI] [PubMed] [Google Scholar]
- Oliver M. B., Basu Roy A., Kumar R., Lefkowitz E. J., Swords W. E. (2017). Streptococcus pneumoniae TIGR4 phase-locked opacity variants differ in virulence phenotypes. mSphere 2:e00386-17. 10.1128/mSphere.00386-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington J. M., Rosenberg S. M. (2007). Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39, 797–802. 10.1038/ng2051, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips Z. N., Husna A. U., Jennings M. P., Seib K. L., Atack J. M. (2019). Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiology 165, 917–928. 10.1099/mic.0.000805, PMID: [DOI] [PubMed] [Google Scholar]
- Pribis J. P., Garcia-Villada L., Zhai Y., Lewin-Epstein O., Wang A. Z., Liu J., et al. (2019). Gamblers: an antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol. Cell 74, 785–800. 10.1016/j.molcel.2019.02.037, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. A., Balwit J. M., Vesga O. (1994). Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 3, 302–312. PMID: [PubMed] [Google Scholar]
- Prouty A. M., Schwesinger W. H., Gunn J. S. (2002). Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70, 2640–2649. 10.1128/IAI.70.5.2640-2649.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putrins M., Kogermann K., Lukk E., Lippus M., Varik V., Tenson T. (2015). Phenotypic heterogeneity enables uropathogenic Escherichia coli to evade killing by antibiotics and serum complement. Infect. Immun. 83, 1056–1067. 10.1128/IAI.02725-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P., Kreiswirth B., O’Reilly M., Schlievert P., Gruss A., Novick R. P. (1986). Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202, 58–61. 10.1007/BF00330517, PMID: [DOI] [PubMed] [Google Scholar]
- Reyes Ruiz L. M., Williams C. L., Tamayo R. (2020). Enhancing bacterial survival through phenotypic heterogeneity. PLoS Pathog. 16:e1008439. 10.1371/journal.ppat.1008439, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen M., Eriksson S., Clements M., Bergstrom S., Normark S. J. (2003). The basis of persistent bacterial infections. Trends Microbiol. 11, 80–86. 10.1016/S0966-842X(02)00038-0, PMID: [DOI] [PubMed] [Google Scholar]
- Roth J. R., Kugelberg E., Reams A. B., Kofoid E., Andersson D. I. (2006). Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 60, 477–501. 10.1146/annurev.micro.60.080805.142045, PMID: [DOI] [PubMed] [Google Scholar]
- Saint-Ruf C., Matic I. (2006). Environmental tuning of mutation rates. Environ. Microbiol. 8, 193–199. 10.1046/j.1462-2920.2003.00397.x-i1, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez A., Choubey S., Kondev J. (2013). Regulation of noise in gene expression. Annu. Rev. Biophys. 42, 469–491. 10.1146/annurev-biophys-083012-130401, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez-Romero M. A., Casadesus J. (2014). Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc. Natl. Acad. Sci. U. S. A. 111, 355–360. 10.1073/pnas.1316084111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Romero M. A., Casadesus J. (2018). Contribution of SPI-1 bistability to Salmonella enterica cooperative virulence: insights from single cell analysis. Sci. Rep. 8:14875. 10.1038/s41598-018-33137-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Romero M. A., Casadesus J. (2020). The bacterial epigenome. Nat. Rev. Microbiol. 18, 7–20. 10.1038/s41579-019-0286-2, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez-Romero M. A., Olivenza D. R., Gutierrez G., Casadesus J. (2020). Contribution of DNA adenine methylation to gene expression heterogeneity in Salmonella enterica. Nucleic Acids Res. 48, 11857–11867. 10.1093/nar/gkaa730, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satory D., Gordon A. J., Halliday J. A., Herman C. (2011). Epigenetic switches: can infidelity govern fate in microbes? Curr. Opin. Microbiol. 14, 212–217. 10.1016/j.mib.2010.12.004, PMID: [DOI] [PubMed] [Google Scholar]
- Scheler O., Postek W., Garstecki P. (2019). Recent developments of microfluidics as a tool for biotechnology and microbiology. Curr. Opin. Biotechnol. 55, 60–67. 10.1016/j.copbio.2018.08.004, PMID: [DOI] [PubMed] [Google Scholar]
- Schreiber F., Littmann S., Lavik G., Escrig S., Meibom A., Kuypers M. M., et al. (2016). Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat. Microbiol. 1:16055. 10.1038/nmicrobiol.2016.55, PMID: [DOI] [PubMed] [Google Scholar]
- Schroter L., Dersch P. (2019). Phenotypic diversification of microbial pathogens -cooperating and preparing for the future. J. Mol. Biol. 431, 4645–4655. 10.1016/j.jmb.2019.06.024, PMID: [DOI] [PubMed] [Google Scholar]
- Scott T. N., Simon M. I. (1982). Genetic analysis of the mechanism of the Salmonella phase variation site specific recombination system. Mol. Gen. Genet. 188, 313–321. 10.1007/BF00332694, PMID: [DOI] [PubMed] [Google Scholar]
- Seib K. L., Srikhanta Y. N., Atack J. M., Jennings M. P. (2020). Epigenetic regulation of virulence and immunoevasion by phase-variable restriction-modification systems in bacterial pathogens. Annu. Rev. Microbiol. 74, 655–671. 10.1146/annurev-micro-090817-062346, PMID: [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. (1998). Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104. 10.1146/annurev.micro.52.1.81, PMID: [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R., de Lorenzo V. (2010). Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64, 257–275. 10.1146/annurev.micro.091208.073229, PMID: [DOI] [PubMed] [Google Scholar]
- Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. (2005). Stripping bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56, 604–614. 10.1111/j.1365-2958.2005.04488.x, PMID: [DOI] [PubMed] [Google Scholar]
- Smits W. K., Kuipers O. P., Veening J. W. (2006). Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4, 259–271. 10.1038/nrmicro1381, PMID: [DOI] [PubMed] [Google Scholar]
- Srikhanta Y. N., Fox K. L., Jennings M. P. (2010). The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 8, 196–206. 10.1038/nrmicro2283, PMID: [DOI] [PubMed] [Google Scholar]
- Sturm A., Heinemann M., Arnoldini M., Benecke A., Ackermann M., Benz M., et al. (2011). The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7:e1002143. 10.1371/journal.ppat.1002143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwandi A., Galeev A., Riedel R., Sharma S., Seeger K., Sterzenbach T., et al. (2019). Std fimbriae-fucose interaction increases Salmonella-induced intestinal inflammation and prolongs colonization. PLoS Pathog. 15:e1007915. 10.1371/journal.ppat.1007915, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Kaufman M. (2001). Multistationarity, the basis of cell differentiation and memory. II. Logical analysis of regulatory networks in terms of feedback circuits. Chaos 11, 180–195. 10.1063/1.1349893, PMID: [DOI] [PubMed] [Google Scholar]
- Tomanek I., Grah R., Lagator M., Andersson A. M. C., Bollback J. P., Tkacik G., et al. (2020). Gene amplification as a form of population-level gene expression regulation. Nat. Ecol. Evol. 4, 612–625. 10.1038/s41559-020-1132-7, PMID: [DOI] [PubMed] [Google Scholar]
- Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998). Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738. 10.1093/emboj/17.22.6730, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkington C. J. R., Morozov A., Clokie M. R. J., Bayliss C. D. (2019). Phage-resistant phase-variant sub-populations mediate herd immunity against bacteriophage invasion of bacterial meta-populations. Front. Microbiol. 10:1473. 10.3389/fmicb.2019.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. H., Vallet-Gely I., Dove S. L. (2009). Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR-type transcription regulator. PLoS Genet. 5:e1000779. 10.1371/journal.pgen.1000779, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff S., Lord N. D., Okumus B., Potvin-Trottier L., Sherratt D. J., Paulsson J. (2016). Stochastic activation of a DNA damage response causes cell-to-cell mutation rate variation. Science 351, 1094–1097. 10.1126/science.aac9786, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta V., Casadesus J. (2017). Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4:163. 10.3389/fmed.2017.00163, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta V., Hernandez S. B., Casadesus J. (2019). Mutational and non mutational adaptation of Salmonella enterica to the gall bladder. Sci. Rep. 9:5203. 10.1038/s41598-019-41600-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude M., Braaten B., Low D. (1996). Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4, 5–9. 10.1016/0966-842X(96)81498-3, PMID: [DOI] [PubMed] [Google Scholar]
- van der Woude M. W. (2011). Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14, 205–211. 10.1016/j.mib.2011.01.002, PMID: [DOI] [PubMed] [Google Scholar]
- van Gestel J., Vlamakis H., Kolter R. (2015). Division of labor in biofilms: the ecology of cell differentiation. Microbiol. Spectr. 3:MB-0002-2014. 10.1128/microbiolspec.MB-0002-2014, PMID: [DOI] [PubMed] [Google Scholar]
- van Sinderen D., Luttinger A., Kong L., Dubnau D., Venema G., Hamoen L. (1995). comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15, 455–462. 10.1111/j.1365-2958.1995.tb02259.x, PMID: [DOI] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72. 10.1128/MMBR.00044-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J. W., Smits W. K., Kuipers O. P. (2008). Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210. 10.1146/annurev.micro.62.081307.163002, PMID: [DOI] [PubMed] [Google Scholar]
- Vincent M. S., Uphoff S. (2020). Bacterial phenotypic heterogeneity in DNA repair and mutagenesis. Biochem. Soc. Trans. 48, 451–462. 10.1042/BST20190364, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H. (1957). The Strategy of the Genes. London: George Allen and Unwin. [Google Scholar]
- Weigel W. A., Dersch P. (2018). Phenotypic heterogeneity: a bacterial virulence strategy. Microbes Infect. 20, 570–577. 10.1016/j.micinf.2018.01.008, PMID: [DOI] [PubMed] [Google Scholar]
- Weiser J. N., Austrian R., Sreenivasan P. K., Masure H. R. (1994). Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62, 2582–2589. 10.1128/IAI.62.6.2582-2589.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion D., Casadesus J. (2006). N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192. 10.1038/nrmicro1350, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. M., Vazirani V. V., Arkin A. P. (2005). Diversity in times of adversity: probabilistic strategies in microbial survival games. J. Theor. Biol. 234, 227–253. 10.1016/j.jtbi.2004.11.020, PMID: [DOI] [PubMed] [Google Scholar]
- Woods E. C., McBride S. M. (2017). Regulation of antimicrobial resistance by extracytoplasmic function (ECF) sigma factors. Microbes Infect. 19, 238–248. 10.1016/j.micinf.2017.01.007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Chiu L. Y., Nehring R. B., Bravo Nunez M. A., Mei Q., Perez M., et al. (2019). Bacteria-to-human protein networks reveal origins of endogenous DNA damage. Cell 176, 127–143.e124. 10.1016/j.cell.2018.12.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Mih N., Anand A., Park J. H., Tan J., Yurkovich J. T., et al. (2019). Cellular responses to reactive oxygen species are predicted from molecular mechanisms. Proc. Natl. Acad. Sci. U. S. A. 116, 14368–14373. 10.1073/pnas.1905039116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Claessen D., Rozen D. E. (2016). Understanding microbial divisions of labor. Front. Microbiol. 7:2070. 10.3389/fmicb.2016.02070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]