Abstract
Chemotherapy is widely used in the treatment of cancer patients, but the cardiotoxicity induced by chemotherapy is still a major concern to most clinicians. Currently, genetic methods have been used to detect patients with high risk of chemotherapy-induced cardiotoxicity (CIC), and our study evaluated the correlation between genomic variants and CIC. The systematic literature search was performed in the PubMed, Cochrane Central Register of Controlled Trials (CENTRAL), China Biology Medicine disc (CBMdisc), the Embase database, China National Knowledge Internet (CNKI) and Wanfang database from inception until June 2020. Forty-one studies were identified that examined the relationship between genetic variations and CIC. And these studies examined 88 different genes and 154 single nucleotide polymorphisms (SNPs). Our study indicated 6 variants obviously associated with the increased risk for CIC, including CYBA rs4673 (pooled odds ratio, 1.93; 95% CI, 1.13–3.30), RAC2 rs13058338 (2.05; 1.11–3.78), CYP3A5 rs776746 (2.15; 1.00–4.62) ABCC1 rs45511401 (1.46; 1.05–2.01), ABCC2 rs8187710 (2.19; 1.38–3.48), and HER2-Ile655Val rs1136201 (2.48; 1.53–4.02). Although further studies are required to validate the diagnostic and prognostic roles of these 6 variants in predicting CIC, our study emphasizes the promising benefits of pharmacogenomic screening before chemotherapy to minimize the CIC.
Keywords: chemotherapy, cardiotoxicity, gene, SNPs, meta-analysis
Introduction
The burgeoning field of cardio-oncology is continuing to grow in step with major scientific developments in oncology that have improved cancer prognosis and survivorship (1). Cardiotoxicity has long been considered one of the main side effects of chemotherapy in cancer patients (2–6). Now, more effective therapies and some forms of radiotherapy may also have multiple cardiovascular (CV) secondary effects, in particularly left ventricular dysfunction (LVD) and heart failure (HF) (7–12). Some of the data from studies on the genetic defects and pharmacological interventions suggested that many molecules, primarily those regulating oxidative stress (OS), autophagy, apoptosis, and metabolism, contributing to the pathogenesis of cardiotoxicity induced by cancer treatment. Treatment with anthracyclines has been reported to increase the risk of cardiotoxicity and death by more than five times (13–15). Early diagnosis and treatment of cardiotoxicity can increase the chances of recovery (16, 17), which highlights the urgent need to develop new technologies and programmes for the early management of cardiotoxicity caused by chemotherapy and for a multidisciplinary clinical approach throughout the chemotherapy process (18–20).
Early identification of the CIC is essential to minimize the harmful side effects of cancer treatment, and could provide oncologists and cardiologists with an ideal choice to allow personalized antitumor treatment strategies or interventions (21). Although some factors may make some patients more susceptible to the severity of toxicity, individual differences in toxicity manifestations still remain large, which will significantly exacerbate these toxic. Genetics, therefore, could provide insights into the development of toxicity induced by some cancer treatments. Identification of the genetic biomarkers that are able to predict whether the patient is at risk of developing cancer therapies-induced cardiac dysfunction will allow for the minimization of cardiotoxicities during cancer treatment by careful monitoring, applying cardioprotective drugs or using optimized cancer therapies. Several recent studies have showed the role of genetic variation as a biomarker for the early detection of CIC (22–25). The aim of this study was to provide an overview of studies focusing on the relationship between polymorphic gene variants and CIC.
Methods
Search Strategy
Our study searched the PubMed, CENTRAL, CBMdisc, the Embase database, Wanfang database and CNKI from the beginning to June 2020. The search terms include chemotherapy, anthracyclines, doxorubicin, daunorubicin, epirubicin, idarubicin, trastuzumab, cyclophosphamide, 5-fluorouracil, methotrexate, adriamycin, cisplatin, cytoxan, cardiotoxicity, HF, cardiomyopathy, arrhythmia, genetic, pharmacogenomics, variant and polymorphism. The search is limited to clinical trials involving human participants. Then, literature titles, abstracts and subject words are carefully analyzed to further identify keywords for document retrieval. If the abstract is relevant to our research, we will read the full text. References in the research were also analyzed to find out some studies that might have been missed in the original search.
Study Selection
Studies that met the criteria were as follows: (i) most of the SNPs were considered as dominant inheritance models unless specifically notified; (ii) original studies that determine the relationship between the genetic polymorphism (including different SNPs in each gene deletions, duplication, and copy-number variants) and cardiotoxicity; (iii) chemotherapy was used regardless of the cycle regimen type, timing, and duration of administration; (iv) in human studies and (vi) in English language. Exclusion criteria: (i) laboratory studies, case series and reports, interim studies; (ii) republication literature; (iii) data with obvious error. The relevance of the article titles and abstract was filtered by two independent reviewers, and the full text was retrieved based on inclusion criteria. Any disagreement will be settled by the third author by decision.
Data Extraction
The authors (GPL and MKG) extracted the data and checked the qualifications and the methodological quality of each included study. Any disagreements will be discussed and if the discussion is not finalized, the disagreements would be resolved by the third author (XYY). This information extracted from each document included the name of the first author, the year of publication, the sample size of the trial, the type of participant, the age of the participant, the type of cancer, the genotyping technique and the definition of cardiotoxicity. At last, we assessed the relationship between different genomic polymorphisms and cardiotoxicity.
Quality Assessment
The STREGA reporting guide was used to assess the quality of each study report (26, 27). STREGA includes five main categories of information: reporting possible genotyping methods and errors, addressing population stratification methods, methods used to inference haplotypes or genotypes, whether the Hardy-Weinberg equilibrium was considered and whether this study is the first to report genetic associations, replication work or both. The quality of the report is assessed independently by two investigators, and differences are resolved through the discussion or through the third author if no consensus is reached between the two investigators.
Statistical Analysis
The meta-analysis was analyzed using the Review Manager 5.3 packages (http://comunity.cochrane.org/tools/review-production-tools/revman-5) (28) and STATA version 13.0. In studies evaluating the same genotype polymorphism, we performed the meta-analysis using the fixed effects (FE) model and the random effects model (29), and this study heterogeneity was evaluated using the I2 statistics. Overall heterogeneity was quantified with the I2, with p < 0.01 used to indicate significance (30). Compared with the sampling error in the study, the true variance ratio of the estimated effects between the included studies was calculated by using the I2 statistic and moderate heterogeneity is considered when I2 is between 50 and 75% and high heterogeneity is considered when I2>75% (30). The sensitivity analysis was conducted to evaluate the stability of this study, namely meta-analysis is performed again after the exclusion of abnormal results, and the results of meta-analysis were compared with those of studies that did not exclude abnormal results.
Result
Study and Patient Characteristics
The study searched 2,277 literatures and 859 that were evaluated. A total of forty-one studies involving in 9,183 patients, were included in this study (Figure 1). Table 1 lists the characteristics of the included studies. Thirteen studies were case-control studies (22, 23, 32, 35, 36, 39, 44, 51, 53, 54, 60, 63, 64), eight were nested case-control studies (24, 25, 31, 34, 37, 48, 52, 59). Another eleven were prospective cohort studies (33, 41–43, 45, 46, 50, 55–57, 62) while eight were retrospective cohort study (38, 40, 48, 58, 61, 65–67). The remaining one was a case report (49). An almost equal number of studies have been performed in children (n = 15) and adults (n = 19). Seven studies included children and adults in our study. The most common cancer types examined include breast cancer (BC) (n = 12), leukemia (n = 9), osteosarcoma (n = 1), lymphoma (n = 3), and hematological neoplasms (n = 3). In eleven other studies, mixed cancers were examined.
Figure 1.

Selection process and criteria for inclusion in this study.
Table 1.
Genetic polymorphisms in chemotherapy-induced cardiotoxicity (CIC).
| Study | Participants | Gender: male/female | Age(years): case/control | Drug used | Type of Cancer | Source of DNA sample | Genotyping | Cardiac toxicity |
|---|---|---|---|---|---|---|---|---|
| Wojnowski et al. (31) | NCC; 550 | 50/37; 212/151 | 62.0 ± 10.9; 61.3 ± 11.0 | Doxorubicin | Non-Hodgkin lymphoma | Peripheral blood | Pyrosequencing; PCR | Arrhythmia, CHF, myocarditis-pericarditis |
| Weiss et al. (32) | CC; 197 | ~98/99 | 68 (56–88) | Daunorubicin | AML | Peripheral blood | Multiplex PCR | SWOG toxicity |
| Beauclair et al. (33) | PC; 61 | NR | 50.7 (30.5–83.1) | Trastuzumab | BC | Blood | PCR | Decreased LVEF |
| Blanco et al. (34) | NCC; 145 | 10/20; 57/58 | 10.3 ± 6.5; 9.1 ± 5.8 |
Anthracyclines | Childhood cancer | Buccal cells/saliva | PCR-RFLP; allelic discrimination with specific fluorescent probes | CHF |
| Rossi et al. (35) | CC; 106 | 55/51; 55/51 | 66 (56–75) | Doxorubicin | Large B-cell lymphoma | Peripheral blood | SNP minisequencing | Abnormalities ECG |
| Rajićet al. (36) | CC; 76 | 32/44 | 25.8 ± 5.3 | Anthracyclines | ALL | Bone marrow smears | qPCR; TaqMan genotyping assay | Cardiac damage, SF <30%, LVEF <54% |
| Blanco et al. (37) | NCC; 487 | 76/94; 162/155 | 8.3 ± 6; 8.2 ± 6 | Anthracyclines | Childhood cancer | Peripheral blood/buccal cells/saliva | Allelic discrimination with specific fluorescent probes | Cardiomyopathy, EF <40%, SF <28% |
| Semsei et al. (38) | RC; 235 | 126/109 | 5.7 ± 3.8 | Anthracyclines | ALL | Peripheral blood | Minisequencing; Genome Lab SNP stream genotyping assay | LV dysfunction; reduced LVFS |
| Visscher et al. (39) | CC; 440 | 17/21; 66/52 | 5.5 (0.04–17.0); 3.9 (0.5–16.5) | Anthracyclines | Childhood cancer | NR | Custom Illumina GoldenGate SNP genotyping assay | CHF; SF <26% |
| Cascales et al. (40) | CR; 97 | 37/12; 28/20 | 60 ± 12; 44 ± 18 | Anthracyclines | Hematological neoplasms | Blood | PCR | HF; LVEF decrease; EF <50% |
| Volkan-Salanci et al. (41) | PC; 70 | 7/63 | 49.1 ± 13.6 | Anthracyclines | BC | NR | TaqMan genotyping assay | Cardiac dysfunction; LVEFs <50% |
| Lubieniecka et al. (42) | PC; 185 | 86/99 | 46 (14–74) | Anthracyclines | AML | Blood | Sequenom genotyping assay | LVEF % drop |
| Kitagawa et al. (43) | PC; 34 | 0/34 | 49 (21–71) | Epirubicin, cyclophosphamide, 5-fluorouracil | BC | Whole blood | TaqMan genotyping assay | Arrhythmias; QTc interval prolongation |
| Windsor et al. (44) | CC; 58 | 34/24 | 18 (10–51) | MAP | Osteosarcoma | Peripheral blood | PCR; Illumina microarray | Decreased LVEF |
| Roca et al. (45) | PC; 392 | NR | 48 (24–65) | 5-fluorouracil, epirubicin, cyclophosphamide | BC | Whole blood | PCR | CHF; LVEF <50% |
| Lipshultz et al. (46) | PC; 184 | 101/83 | 15.2 (3.1–31.4) | Doxorubicin | ALL | Peripheral blood | Pyrosequencing; Sequenom genotyping assay; TaqMan genotyping assay | Cardiac dysfunction; LVEF, cTnT, NT-proBNP |
| Armenian et al. (47) | NCC; 255 | 34/43; 119/59 | 49.2 (16–68.8); 51.0 (6.4–72.6) | Anthracyclines | Hematological neoplasms | Peripheral blood | Sequenom MassARRAY | Sign and symptoms |
| Lubieniecka (48) | RC; 91 | 48/43 | 48.4 (19–74) | Daunorubicin | AML | Blood | Sequenom genotyping assay | Decreased LVEF |
| Sachidanandam et al. (49) | CR; 2 | 0/2 | NR | Doxorubicin | Childhood cancer | Whole blood | PCR | Sign and symptoms |
| Vivenza et al. (50) | PC; 48 | 1/47 | 57.5 (28–73) | Anthracyclines | BC | Blood | Allelic discrimination; TaqMan genotyping assay | Decreased LVEF; LVEF <50% |
| Visscher et al. (51) | CC; 218 | 31/25; 75/87 | 21.7 (1.4–33.8); 16.1 (2.3–33.7) | Anthracyclines | Childhood cancer | Blood/saliva/buccal swab | Custom Illumina GoldenGate SNP genotyping assay | SF <24% or symptoms, CTCAE grade 2–4 |
| Wang et al. (52) | NCC; 363 | 40/53; 94/100 | 19.4 (0.4–41.7); 18.5 (3.5–49.2) | Anthracyclines | Children's Oncology | Peripheral blood, buccal cells/ saliva | Illumina IBC cardiovascular SNP array | American Heart Association criteria |
| Wasielewski et al. (53) | CC; 21 | NR | 49 (2–57) | Anthracyclines | NR | NR | Targeted next-generation DNA sequencing | Signs and symptoms; cardiomyopathy |
| Krajinovic et al. (54) | CC; 295 | 134/117; 21/23 | 6.16; 5.27 | Doxorubicin | ALL | Blood, buccal swabs | PCR | Reduction of EF and FS |
| Visscher et al. (23) | CC; 536 | 64/58; 211/187 | 7.4 (0.04–17.6); 4.9 (0.1–17.7) | Anthracyclines | Childhood cancer | Blood, saliva, buccal swabs | Illumina GoldenGate SNP genotyping assay | FS ≤ 26%, LV dysfunction |
| Peña et al. (55) | PC; 78 | NR | 51.72 | Trastuzumab | BC | Saliva | TaqMan allelic discrimination assay | CHF, LVEF <50% |
| Aminkeng et al. (56) | PC; 376 | 27/27; 174/148 | 16.5 (7.5–26); 15 (8–21.5) |
Anthracyclines | Pediatric oncology | NR | Illumina HumanOmniExpress assay | FS ≤ 24%, LVEF <45% |
| Reichwagen et al. (24) | NCC; 520 | 25/31; 46/48 | 68 (61–80); 67 (62–79) |
Doxorubicin | CD20+ B-cell lymphomas | Blood | Pyrosequencing; TaqMan genotyping assays | Arrhythmia, reduced EF |
| Vulsteke et al. (57) | PC; 877 | NR | 50.3 | Epirubicin | BC | Blood | Sequenom MassARRAY | Decrease LVEF, LVEF >10% |
| Stanton et al. (58) | RC; 140 | 0/140 | 56 (32–85) | Trastuzumab | BC | Peripheral blood | PCR | LVEF <55% |
| Hertz et al. (22) | CC; 166 | 0/19; 0/147 | 50 (35–64); 50 (24–80) | Doxorubicin | BC | Blood | Sequenom MassARRAY; TaqMan allelic discrimination assay | EF <55% |
| Reinbolt et al. (59) | NCC; 162 | 0/52; 0/110 | 51.9 ± 11.9; 50.1 ± 9.3 |
Adriamycin, and cytoxan | BC | NR | TaqMan allelic discrimination assay | EF <50% |
| Wang et al. (25) | NCC; 385 | 76/90; 106/113 | 16.1 ± 10.7; 8.3 ± 5.8 |
Anthracyclines | Childhood cancer | Blood, buccal cells, saliva | Illumina HumanOmniExpress assay; Sequenom MassARRAY | Cardiomyopathy, EF <40%, SF <28% |
| Schneider et al. (60) | CC; 102 | NR | NR | Anthracyclines | BC | NR | Illumina Genotyping | CHF; LVEF <50% |
| Ruiz-Pinto et al. (61) | RC; 154 | 0/71; 53/30 | 54.3; 7.8 | Anthracyclines | BC | NR | Illumina HumanExome BeadChip array | Cardiac failure, LVEF decreased |
| Pop-Moldovan et al. (62) | PC; 25 | 13/12 | 59.6 | Doxorubicin | Hematological neoplasms | Blood | qRT-PCR | Diastolic dysfunction; LVEF decreased |
| Ruiz-Pinto et al. (63) | CC; 93 | 33/25; 25/10 | 5.1; 10.4 | Anthracyclines | Pediatric cancer | Peripheral blood | Illumina HumanExome BeadChip array | LV dysfunction |
| Huang et al. (64) | CC; 36 | 22/14 | 7.1 ± 2.3 | Daunorubicin | ALL | Bone marrow | PCR | Abnormal ECG |
| Sági et al. (65) | RC; 680 | NR | 6.6 (± 4.3) | Anthracyclines | ALL | Peripheral blood | TaqMan® Open- Array™ Genotyping | FS ≤ 28%, decreased EF |
| Todorova et al. (66) | RC; 30 | NR | 53.1 (35–76) | Doxorubicin | BC | Peripheral blood | RT-PCR; Illumina HumanOmni BeadChip | Cardiac dysfunction, LVEF <55% |
| Garcia-Pavia et al. (67) | RC; 213 | 33/66; 0/73; 17/24 | 48.7 ± 17.1; 49.6 ± 10.8; 10.8 ± 5.6 |
Anthracyclines | Diverse cancers | Peripheral blood | Illumina TruSight Cardio Sequencing | Cardiomyopathy; LVEF <50% |
The characteristics of included studies. AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; BC, breast cancer; EF, ejection fraction; CC, case-control; LVEF, left ventricular ejection fraction; LVFS, left ventricular fractional shortening; SF, shortening fraction; CHF, congestive heart failure; CTCAE, National Cancer Institute Common Toxicity Criteria; PC, prospective cohort; NCC, nested case control; NR, not reported; RC, retrospective cohort; RFLP, restriction fragment length polymorphism; ECG, echocardiography.
The blood and buccal cells were the most commonly biological specimens used for the genotyping. Twenty-six studies used a single biological sample, including blood (24, 31–33, 35, 38, 40, 42–50, 57, 58, 62, 63, 65–67), bone marrow smear (36, 64) or buccal swab (34), while seven researches used more than one bio-specimens (23, 25, 34, 37, 51, 52, 54). Seven studies did not report the bio-specimens used for genotyping (39, 41, 53, 56, 59–61). Thirty studies used a single genotyping assay (24, 32, 33, 35, 37, 39–43, 45, 47–57, 59–65, 67) while the remaining studies used multiple genotyping assays (22, 24, 25, 31, 34, 36, 38, 44, 46, 50, 64). The most common detection technique were Sequenom MassARRAY (22, 25, 47, 57), (n = 4), Sequenom genotyping assay (42, 46, 48) (n = 3), TaqMan genotyping assay (22, 24, 36, 41, 44, 46, 50, 53, 55, 59, 65) (n = 11), pyrosequencing (24, 31, 46) (n = 3) and custom Illumina GoldenGate SNP genotyping assay (23, 39, 51) (n = 3).
The cardiotoxicity definition varied from study to study, with most studies using subjective results (n = 5), objective results (n = 11) or both (n = 24), while one did not define cardiotoxicity. However, most researches using subjective results defined cardiotoxicity as the signs and symptoms that required intervention. Furthermore, some studies have used shortened fraction (SF) or left ventricular ejection fraction (LVEF) as the objective indicators, but the critical points vary. For example, a cut-off value below 40–55% of the LVEF or reduce of more than 10–15% have been used. Five studies (24, 31, 35, 43, 64) also included the definition of electrocardiographic changes in cardiotoxicity, namely arrhythmias and electrocardiogram (ECG) abnormalities, while one study only detected the influences of anthracyclines on the QT intervals and arrhythmias.
The Quality of the Reporting in the Studies
Among the studies reviewed (Table 2), there was only one study met the five main standard for reporting data from the genetic association studies in the STREGA guidelines. Most researches (N = 32) did not report the error rates or call rates related to the genotyping methods. Twenty-six researches did not indicate whether the genotyping was done in the batches or simultaneously. Twenty-nine studies did not provide any information on whether the population stratification was evaluated in our analysis.
Table 2.
The quality assessment of reporting in each study (N = 41).
| Studiesa | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotyping methods and errors | ||||||||||||||
| Describe the laboratory methods: state the source and storage of DNA, the genotyping methods and the platforms | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Describe the laboratory methods: state the error rates and call rates | ✓ | × | × | × | × | × | × | × | × | × | × | × | × | × |
| State the laboratory/center where the genotyping was done | ✓ | × | × | ✓ | × | × | ✓ | ✓ | ✓ | × | × | × | ✓ | × |
| Specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches | ✓ | × | × | × | × | × | × | × | × | × | × | ✓ | ✓ | × |
| Report the numbers of individuals for whom genotyping was attempted and the numbers of individuals for whom genotyping was successful | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | × | ✓ | ✓ |
| Modeling population stratification | ||||||||||||||
| Describe any methods used to assess or address population stratification | ✓ | × | × | × | × | × | × | × | ✓ | × | × | × | × | × |
| Modeling haplotype variation | ||||||||||||||
| Describe any methods used for inferring genotypes or haplotypes | ✓ | NA | NA | NA | NA | NA | NA | ✓ | NA | ✓ | NA | NA | NA | NA |
| Hardy–Weinberg equilibrium | ||||||||||||||
| State whether the Hardy–Weinberg equilibrium was considered | ✓ | × | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | × | ✓ |
| Replication | ||||||||||||||
| State if the study is the first report of a genetic association, a replication effort or both | ✓ | × | ✓ | ✓ | × | × | × | × | × | ✓ | ✓ | × | × | × |
| Studiesa | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 |
| Genotyping methods and errors | ||||||||||||||
| Describe the laboratory methods: state the source and storage of DNA, the genotyping methods and the platforms | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Describe the laboratory methods: state the error rates and call rates | × | × | ✓ | ✓ | × | × | × | × | × | ✓ | ✓ | × | ✓ | × |
| State the laboratory/center where the genotyping was done | × | ✓ | × | × | ✓ | × | × | ✓ | × | × | × | × | ✓ | × |
| Specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches | × | ✓ | × | ✓ | ✓ | × | × | ✓ | × | ✓ | ✓ | × | × | ✓ |
| Report the numbers of individuals for whom genotyping was attempted and the numbers of individuals for whom genotyping was successful | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Modeling population stratification | ||||||||||||||
| Describe any methods used to assess or address population stratification | × | × | × | ✓ | × | × | ✓ | ✓ | × | × | ✓ | × | ✓ | × |
| Modeling haplotype variation | ||||||||||||||
| Describe any methods used for inferring genotypes or haplotypes | NA | NA | NA | ✓ | NA | NA | ✓ | NA | NA | NA | ✓ | NA | ✓ | NA |
| Hardy–Weinberg equilibrium | ||||||||||||||
| State whether the Hardy–Weinberg equilibrium was considered | ✓ | × | ✓ | ✓ | × | ✓ | ✓ | ✓ | × | × | ✓ | ✓ | ✓ | ✓ |
| Replication | ||||||||||||||
| State if the study is the first report of a genetic association, a replication effort or both | × | × | × | × | ✓ | × | × | × | × | × | ✓ | × | × | ✓ |
| Studiesa | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | |
| Genotyping methods and errors | ||||||||||||||
| Describe the laboratory methods: state the source and storage of DNA, the genotyping methods and the platforms | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Describe the laboratory methods: state the error rates and call rates | × | ✓ | × | × | × | × | × | × | × | × | ✓ | ✓ | × | |
| State the laboratory/center where the genotyping was done | ✓ | ✓ | ✓ | ✓ | ✓ | × | ✓ | × | ✓ | ✓ | × | × | × | |
| Specify whether genotypes were assigned using all of the data from the study simultaneously or in smaller batches | × | ✓ | ✓ | × | ✓ | × | ✓ | ✓ | ✓ | × | × | × | × | |
| Report the numbers of individuals for whom genotyping was attempted and the numbers of individuals for whom genotyping was successful | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Modeling population stratification | ||||||||||||||
| Describe any methods used to assess or address population stratification | × | × | × | × | ✓ | ✓ | ✓ | × | ✓ | × | × | × | ✓ | |
| Modeling haplotype variation | ||||||||||||||
| Describe any methods used for inferring genotypes or haplotypes | NA | NA | NA | NA | NA | ✓ | NA | NA | NA | NA | ✓ | NA | NA | |
| Hardy–Weinberg equilibrium | ||||||||||||||
| State whether the Hardy–Weinberg equilibrium was considered | × | ✓ | ✓ | × | ✓ | × | ✓ | × | ✓ | × | ✓ | × | × | |
| Replication | ||||||||||||||
| State if the study is the first report of a genetic association, a replication effort or both | × | × | × | ✓ | ✓ | × | ✓ | × | × | × | × | × | ✓ |
1. Wojnowski et al., 2005; 2. Weiss et al., 2006; 3. Beauclair et al., 2007; 4. Blanco et al., 2008; 5. Rossi et al., 2009; 6. Rajić et al., 2009; 7. Blanco et al., 2012; 8. Semsei et al., 2012; 9. Visscher et al., 2012; 10. Cascales et al., 2012; 11. Volkan-Salanci et al., 2012; 12. Lubieniecka et al., 2012; 13. Kitagawa et al., 2012; 14. Windsor et al., 2012; 15. Roca et al., 2013; 16. Lipshultz et al., 2013; 17. Armenian et al., 2013; 18. Lubieniecka et al., 2013; 19. Sachidanandam et al., 2013; 20. Vivenza et al., 2013; 21. Visscher et al., 2013; 22. Wang et al., 2014; 23. Wasielewski et al., 2014; 24. Krajinovic et al., 2015; 25. Visscher et al., 2015; 26. Peña et al., 2015; 27. Aminkeng et al., 2015; 28. Reichwagen et al., 2015; 29. Vulsteke et al., 2015; 30. Stanton et al., 2015; 31. Hertz et al., 2016; 32. Reinbolt et al., 2016; 33. Wang et al., 2016; 34. Schneider et al., 2017; 35. Ruiz-Pinto et al., 2017; 36. Pop-Moldovan et al., 2017; 37. Ruiz-Pinto et al., 2017; 38. Huang et al., 2017; 39. Sági et al., 2018; 40. Todorova et al., 2018; 41. Garcia-Pavia et al., 2019.
Chemotherapy-Induced Cardiotoxicity (CIC) and Susceptibility Genes
Our study reported a total of 154 SNPs involving eighty-eight genes (Supplementary Table 1). Most of the research has focused on genetic variations linked to chemical synthesis or heart function. Accessable data was found in 17 studies out of the included 41 studies and 14 SNPs were consistently detected. Then these 14 SNPs were subjected to our quantitative analysis (Figures 2–4). And we searched for these six variants in the protein-coding region of the gene in the Pubmed (Table 3). The genes included in the meta-analysis are discussed below.
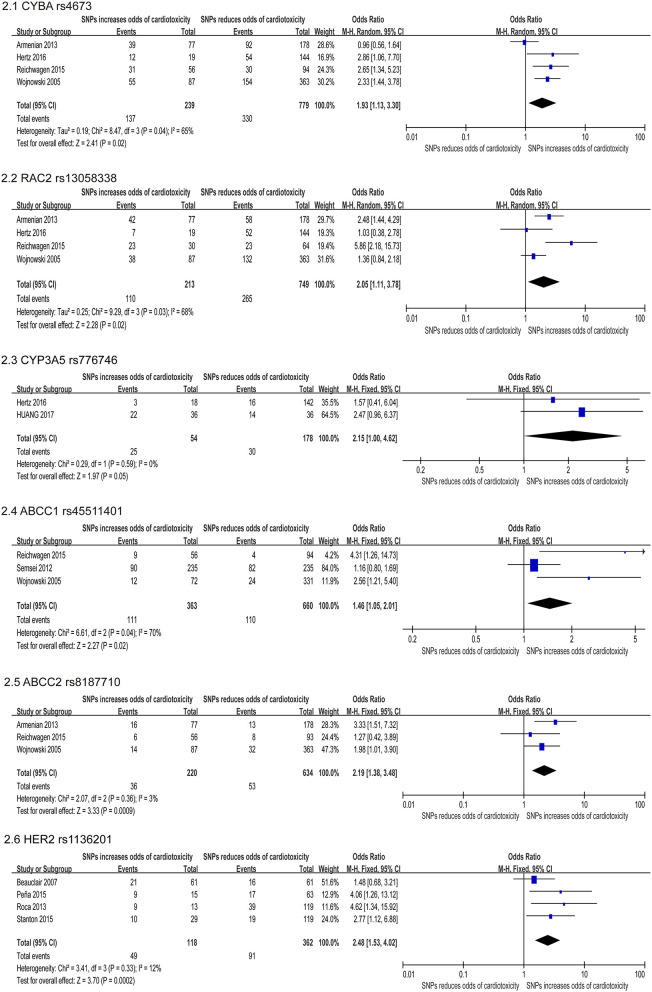
Figure 2.
Forest plot of meta-analysis for 6 SNPs. Six variants, CYBA rs4673, RAC2 rs13058338, CYP3A5 rs776746, ABCC1 rs45511401, ABCC2 rs8187710, and HER2 rs1136201 are significantly increased the odds for chemotherapy induced cardiotoxicity.
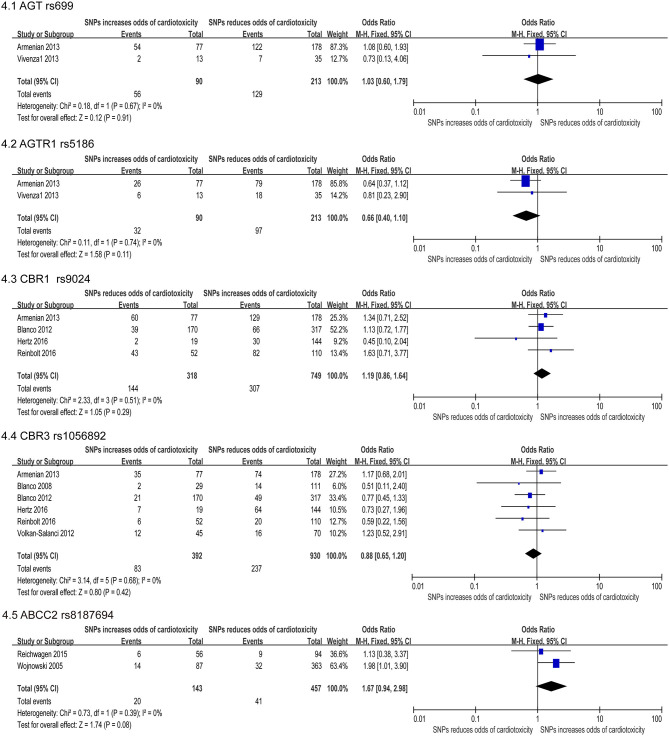
Figure 4.
Forest plot of meta-analysis for five SNPs. Five variants, AGT rs699, AGTR1 rs5186, CBR1 rs9024, CBR3 rs1056892, and ABCC2 rs8187694, are not statistically significant for chemotherapy induced cardiotoxicity.
Table 3.
The 6 variants, annotations and meta-analysis OR and p-value.
| Gene | SNP | Variant type | Alleles | Chromosome | Functional consequence | Clinical significance | Odds ratio | p-value |
|---|---|---|---|---|---|---|---|---|
| CYBA | rs4673 | SNV | A>G,T | 16:88646828 (GRCh38) | Coding sequence variant | Benign, likely-pathogenic | 1.93 | 0.02 |
| RAC2 | rs13058338 | SNV | T>A,G | 22:37236730 (GRCh38) | Intron variant | Not report | 2.05 | 0.02 |
| CYP3A5 | rs776746 | SNV | T>C | 7:99672916 (GRCh38) | Intron variant | Benign, drug-response | 2.15 | 0.05 |
| ABCC1 | rs45511401 | SNV | G>T | 16:16079375 (GRCh38) | Coding sequence variant | Not report | 1.46 | 0.02 |
| ABCC2 | rs8187710 | SNV | G>A | 10:99851537 (GRCh38) | Coding sequence variant | Likely-benign | 2.19 | 0.0009 |
| Her2 | rs1136201 | SNV | A>G,T | 17:39723335 (GRCh38) | Coding sequence variant | Benign, not-provided | 2.48 | 0.0002 |
Cytochrome b-245, Alpha Polypeptide (CYBA) Gene
The CYBA gene encodes a major component of the phagocytes microbicidal oxidase system. Meanwhile, our study confirmed six studies that assessed the correlation between CYBA rs4673 and cardiotoxicity, and four studies (22, 24, 31, 47) were included in quantitative analysis. In our study, this SNP was found to increase the risk of developing cardiotoxicity (OR: 1.93; 95% CI: 1.13–3.30; p = 0.04) with high heterogeneity (I2 = 65%) (Figure 2.1). We used the random effects model and excluded individual studies to conduct sensitivity analysis to account for the heterogeneity, and these results show that excluding the study of Armenian et al. can alter heterogeneity (I2 = 0%).
Ras-Related C3 Botulinum Toxin Substrate 2 (RAC2) Gene
The RAC2 gene encodes proteins that regulate a variety of processes including secretion, phagocytosis, cell polarization, and ROS production. Four of the six studies indicated that the SNP rs13058338 on the RAC2 obviously increased the risk of cardiotoxicity (22, 24, 31, 47). We used the random effects model and the analysis of this variant in four studies suggested that the RAC mutation significantly increased the risk of cardiotoxicity by nearly twice (OR: 2.05; 95% CI: 1.11–3.78; p = 0.02), but with moderate heterogeneity (I2 = 68%) (Figure 2.2). Sensitivity analyses were conducted to explore the potential sources of heterogeneity, but these results did not change substantially [I2 = 74% (47), 74% (22), 60% (24), 66% (31)].
Cytochrome P450 Family 3 Subfamily A Member 5 (CYP3A5) Gene
The CYP3A5 gene is involved in the metabolism and clearance of daunorubicin (DNR), but the activity of CYP3A5 is impacted on the gene polymorphism, which has individual differences (68). Meta-analysis of two studies (22, 64) reported that the rs776746 on CYP3A5 increased cardiotoxicity risk, including a total of 232 patients (54 in the case group and 178 in the control group). Using the FE model, our study suggested that the missense mutation was related to an obviously increase in the risk of cardiotoxicity [OR = 2.15 [95% CI (1.00–4.62)], P = 0.05] (Figure 2.3).
ATP Binding Cassette (ABC) Genes
The ABC transporter gene encodes a superfamily of transmembrane proteins that can use adenosine triphosphate to actively transport substrates including doxorubicin through the membrane (69). Meta-analysis of three studies (24, 31, 47) reported that the ABCC1 rs45511401 increased cardiotoxicity risk, including a total of 1,023 patients (363 in the case group and 660 in the control group). Using the random effects model, we found that missense mutations were related to a significant increase in risk [OR = 1.46 [95% CI (1.05–2.01)], P = 0.02] with moderate heterogeneity (I2 = 70%) (Figure 2.4). The study conducted by Semsei et al. (38) were excluded, changing the results and addressing heterogeneity (I2 = 0%). And three studies (24, 31, 38) indicated that the ABCC2 rs8187710 significantly augmented the risk of cardiotoxicity (OR: 2.19; 95% CI: 1.38–3.48; p = 0.0009) (Figure 2.5).
Human Epidermal Growth Factor Receptor 2 (HER2) Gene
The HER2 is a proto-oncogene that encodes transmembrane proteins that have tyrosine kinase activity, but have not been identified as the physiological ligand. Four included studies (33, 45, 55, 58) revealed Ile655Val rs1136201 on HER2 significantly increase risk for cardiotoxicity, including a total of 480 patients (118 in the case group and 362 in the control group). Using the FE model, the result indicated that the missense mutation was related to a significant increase in the risk of cardiotoxicity [OR = 2.48 [95% CI (1.53–4.02)], P = 0.0002] with low heterogeneity (I2 = 12%) (Figure 2.6).
Neutrophil Cytosolic Factor 4 (NCF4) Gene
The NCF4 gene encode the p40phox subunit of the NAD(P)H oxidase (NOX) (70). The rs1883112 polymorphism of NCF4 promoter blocks the oxidase activation of the enzyme, thus reducing the formation of active oxidant intermediates (71). Three (22, 24, 31, 47) of the six researches (22, 24, 31, 35, 39, 47) studied the roles of SNP rs1883112, and found that this SNP was related to cardiotoxicity, but the combined effect of this synonymous substitution indicated no significant relevancy (OR: 1.13; 95% CI: 0.72–1.77; p = 0.59) (Figure 3.1).
Figure 3.
Forest plot of meta-analysis for 4 SNPs. Four variants, NCF4 rs1883112, SLC28A3 rs7853758, SOD2 rs4880, and NQO1 rs1800566, are not statistically significant for chemotherapy induced cardiotoxicity.
Solute Carrier Family 28 Member 3 (SLC28A3) Gene
It has been previously reported that SLC28A3 (rs7853758) has cardioprotective effects in the multiple patient cohorts with an odds ratio of 0.35–0.42 (39, 51). We identified five studies that assessed the association between rs7853758 missense SNP on SLC28A3 and cardiotoxicity, and three studies (22, 24, 65) were included in the quantitative analysis. We used the random effects model. But the SNP rs7853758 was not statistically significant difference (OR: 1.77; 95% CI: 0.44–7.13; p = 0.42) (Figure 3.2) with high heterogeneity (I2 = 70%). Therefore, we excluded individual studies to conduct sensitivity analyses to account for the heterogeneity, and the result suggested no significant differences between the selected studies.
Superoxide Dismutase II (SOD2) Gene
SOD2 exists in the mitochondria and metabolizes superoxide radicals formed when anthracycline compounds are oxidized to hydrogen peroxide. Meta-analysis of two included studies (36, 47) revealed rs4880 on SOD2 significantly increase risk for cardiotoxicity including a total of 407 participants (153 in the case group and 254 in the control group). But the FE model indicated that the missense mutation was not statistically significant [OR = 1.65 [95% CI (0.94–2.88)], P = 0.08] (Figure 3.3).
NAD(P)H Quinone Dehydrogenase1 (NQO1) Gene
The NQO1 gene is involved in the protection of intracellular OS, and many pro-oxidant drugs induced basic NQO1 activity (34). Two included studies (34, 47) revealed SNP rs1800566 on NQO1 significantly increase risk for cardiotoxicity, including a total of 395 patients (106 in the case group and 289 in the control group). Using the FE model, the result indicated that the missense mutation was not statistically significant [OR = 1.56 [95% CI (0.75–3.24)], P = 0.24] (Figure 3.4).
Angiotensinogen (AGT) Gene
Association between AGT P.m.ET235THr gene polymorphism and cardiovascular disease concluded that there was a positive correlation between essential hypertension (72, 73) and myocardial infarction (74). Two included studies (44, 47) revealed SNP rs699 AGT significantly increase risk for cardiotoxicity, including a total of 303 participants (90 in the case group and 213 in the control group). The FE model indicated that the missense mutation was not statistically significant [OR = 1.93 [95% CI (0.60–1.79)], P = 0.91] (Figure 4.1).
Angiotensin II Type-1 Receptor (AGTR1) Gene
Angiotensin II is the main ligand of AGTR1A, which adjusts the intravascular volume and the blood pressure (75). And functional polymorphisms in the ACE, AGT, and AGTR1 can impact the expression or the function of encoded proteins, and are related to stroke, coronary heart disease, vascular dysfunction and diabetes (76). Meta-analysis of two included studies (47, 50) revealed SNP rs5186 significantly increase risk for cardiotoxicity, including a total of 303 patients (90 in the case group and 213 in the control group). But the FE model indicated that the missense mutation was not statistically significant difference [OR = 0.66 [95% CI (0.40–1.10)], P = 0.11] (Figure 4.2).
Carbonyl Reductases (CBR) Genes
The enzyme encoded by the CBR genes catalyzes the reduction of the endogenous aliphatic aldehydes and ketones as well as various xenobiotic, and thus has a cardioprotective effect against cardiotoxicity. Four SNPs on CBRs were studied, one on carbonyl reductase 1 (CBR1) gene and three on carbonyl reductase 3 (CBR3) gene. However, two SNPs, rs9024 of CBR1 (22, 37, 47, 59) and rs1056892 of CBR3 (22, 34, 37, 41, 47, 59) were not statistically significant (OR: 1.19; 95% CI: 0.86–1.64 and 0.88; 0.65–1.20, respectively) (Figures 4.3,4.4).
ATP Binding Cassette (ABC) Gene
Two studies (24, 31) reported that the ABCC2 rs8187694 increased cardiotoxicity risk, including a total of 600 patients (457 in the case group and 143 in the control group). Using the FE model, we found no statistically significant difference (OR: 1.67; 95% CI: 0.94–2.98; p = 0.08) with low heterogeneity (I2 = 0%) (Figure 4.5).
Sensitivity Analyses
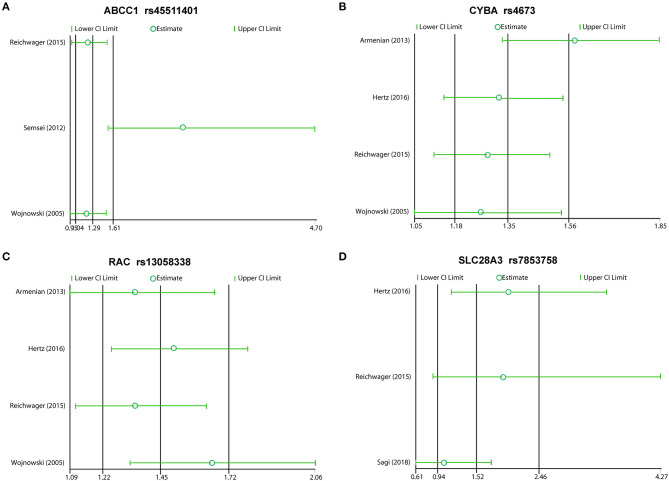
We excluded each study in order, and the rest reported inconsistent results between ABCC1 and CYBA. In this meta-analysis, the studies that included Semsei et al. (38) and Armenia et al. (47) obviously distorted these results, suggesting that the two reports may be statistically unstable (Figures 5A,B). Other results showed consistent results (Figures 5C,D).
Figure 5.
Sensitivity analyses plot of ABCC1 (A), CYBA (B), RAC (C), and SLC28A3 (D).
Discussion
Our study analyzed the association between genetic polymorphism and CIC. We included a total of 41 studies that screened 88 different genes. Gene variation may promote OS, metabolic disorders, mitochondrial dysfunction, calcium overload, myocardial fibrosis, sarcoplasmic reticulum structure and function destruction, cardiomyocyte autophagy and apoptosis in CIC (77, 78). The results of our meta-analysis showed that the polymorphisms in six (6.8%) of eighty-eight genes were obviously associated with the risk of cardiotoxicity in patients receiving chemotherapy.
For genes with positive associations, mechanism studies have indicated that these alleles can alter the encoded proteins expression or activity, leading to cardiotoxicity. The ABCC1 transporter plays an important function in the OS, and is involved in maintaining the adequate levels of glutathione, which is essential for ROS defense. In addition, ABCC1 also requires glutathione to transport anthracycline antibiotics (79–81), which may also affect the OS response induced by anthracycline antibiotics (82). The ABCC2 gene encoded proteins involved in the efflux of substances from cells, and ABCC2 mutation obviously decrease the ATPase-activity, leading to reduce in the efflux activity resulting in the accumulation of anthracyclines in cells (83). CYBA encodes p22phox, which is one of the two subunits of the NOX located in the cell membrane. And the reduced activity of inherited NOX may lead to impaired ROS defense capacity, thus increasing ROS levels under chemotherapy exposure. Similarly, RAC2 encoded by the RAC2 gene is a mitochondrial protein required for the electron transfer reaction of NOX (84) during OS formation (85). Genetic alteration leads to mitochondrial dysfunction, which leads to an increase in ROS production and ultimately cardiomyocyte damage. There is also evidence that the CYP3A5-mediated oxidative metabolism of anthracyclines may induce drug-induced cardiotoxicity by generating OS (86). Taken together, these gene mutations are believed to cause cardiotoxicity on account of the accumulation of chemotherapy and excessive ROS in cardiomyocytes.
Some other genes were also observed to be associated with CIC, such as HER2-Ile655Val rs1136201. The most studied germline polymorphism in the clinical level is associated with the transmembrane domain of HER2 protein 655 A>G Ile/Val (87, 88), which may be related to a high risk of BC (89). The Val allele presence may make cardiomyocytes especially dependent on HER2-Ile655Val rs1136201 signaling and the highly sensitive to trastuzumab (33). This mechanism of cardiotoxicity induced by trastuzumab is unknown, however, HER2/neu has been shown to be critical for cardiomyocytes in animal model. Our meta-analysis also confirmed the role of HER2-Ile655Val polymorphism as a genetic predictor of cardiac toxicity induced by trastuzumab in the BC patients.
There are some limitations to this study that deserve discussion. Firstly, the sample size of the study was small. This is particularly important for genome-wide association study (GWAS), with some literature advocating 10,000 cases to gain sufficient statistical power to detect causality through meta-analysis and data aggregation. Sixty one percentage of the included studies had fewer than 200 people. Secondly, there is selection bias in this study. Most literature used a retrospective method, usually by convenience sampling to recruit patients who were still in the hospital system, thereby further limiting the choice to participants who are still alive. In addition, the analysis was based on previous reports, which may not be complete or accurate. Finally, due to the lack of raw data, studies on drug dosage protocols and study periods varied widely. In addition, patient cohorts were often heterogeneous in terms of disease, drug dose, drug route, and administration, all of which may confuse toxicity associated with the target drug.
Conclusions
This study suggests that the polymorphisms in multiple pharmacogenetic in the biochemical and cardiotoxicity pathways may be predictors of CIC. However, for limited quantitative analysis, the evidences are limited and too diverse. Further researches are needed to produce reliable genetic predictors of CIC in order to achieve the goal of individualized chemotherapy.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YX, HS, and SD designed the idea for drafting this review. XY, GL, MG, and AB collected the documents and wrote the paper. TY, QD, CZ, WL, and NA contributed to discussion. FY, HP, PW, CL, YGo, and YGa revised and edited the review. All authors commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Key R&D Program of China (grants 2017YFC1700400 and 2017YFC1700402), National Key R&D Program of China (grants 2018YFC1704900 and 2018YFC1704901), National high-level talent special support plan (No. W02020052), and China Scholarship Council financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2021.651269/full#supplementary-material
References
- 1.Zaha VG, Meijers WC, Moslehi J. Cardio-immuno-oncology. Circulation. (2020) 141:87–9. 10.1161/CIRCULATIONAHA.119.042276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang HM, Moudgil R, Scarabelli T, Okwuosa TM, Yeh ETM. Cardiovascular complications of cancer therapy best practices in diagnosis, prevention, and management: part 1. J Am Coll Cardiol. (2017) 70:2536–51. 10.1016/j.jacc.2017.09.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang XY, Li XY, Yuan MC, Tian C, Yang YH, Wang XF, et al. Anticancer therapy-induced atrial fibrillation: electrophysiology and related mechanisms. Front Pharmacol. (2018) 9:1058. 10.3389/fphar.2018.01058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suter TM, Ewer MS. Cancer drugs and the heart: importance and management. Eur Heart J. (2013) 34:1102–11. 10.1093/eurheartj/ehs181 [DOI] [PubMed] [Google Scholar]
- 5.Farmakis D, Parissis J, Filippatos G. Insights into onco-cardiology: atrial fibrillation in cancer. J Am Coll Cardiol. (2014) 63:945–53. 10.1016/j.jacc.2013.11.026 [DOI] [PubMed] [Google Scholar]
- 6.López-Sendón J, Álvarez-Ortega C, Auñon PZ, Soto AB, Lyon AR, Farmakis D, et al. Classification, prevalence, and outcomes of anticancer therapy-induced cardiotoxicity: the CARDIOTOX registry. Eur Heart J. (2020) 41:1720–9. 10.1093/eurheartj/ehaa006 [DOI] [PubMed] [Google Scholar]
- 7.Ameri P, Canepa M, Anker MS, Belenkov Y, Bergler-Klein J, Cohen-Solal A, et al. Cancer diagnosis in patients with heart failure: epidemiology, clinical implications and gaps in knowledge. Eur J Heart Fail. (2018) 20:879–87. 10.1002/ejhf.1165 [DOI] [PubMed] [Google Scholar]
- 8.Kenigsberg B, Wellstein A, Barac A. Left ventricular dysfunction in cancer treatment. Is it relevant? J Am Coll Cardiol HF. (2018) 6:87–95. 10.1016/j.jchf.2017.08.024 [DOI] [PubMed] [Google Scholar]
- 9.Mehta LS, Watson KE, Barac A, Beckie TM, Bittner V, Cruz-Flores S, et al. Cardiovascular disease and breast cancer: where these entities intersect: a scientific statement from the American Heart Association. Circulation. (2018) 137:e30–66. 10.1161/CIR.0000000000000556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai MY, Jellis CL, Kotecha R, Johnston DR, Griffin BP. Radiation-associated cardiac disease: a practical approach to diagnosis and management. JACC Cardiovasc Imaging. (2018) 11:1132–49. 10.1016/j.jcmg.2018.04.028 [DOI] [PubMed] [Google Scholar]
- 11.Bonaca MP, Olenchock BA, Salem JE, Wiviott SD, Ederhy S, Cohen A, et al. Myocarditis in the setting of cancer therapeutics: proposed case definitions for emerging clinical syndromes in cardio-oncology. Circulation. (2019) 140:80–91. 10.1161/CIRCULATIONAHA.118.034497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banke A, Fosbøl ML, Ewertz M, Videbæk L, Dahl JS, Poulsen MK, et al. Long-term risk of heart failure in breast cancer patients after adjuvant chemotherapy with or without trastuzumab. J Am Coll Cardiol HF. (2019) 7:217–24. 10.1016/j.jchf.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Von Hoff DD, Layard MW, Basa P, Davis HL, Jr, Von Hoff AL, Rozencweig M, et al. Risk factors for doxorubicin-induced congestive heart failure. Ann Intern Med. (1979) 91:710–7. 10.7326/0003-4819-91-5-710 [DOI] [PubMed] [Google Scholar]
- 14.Benjamin RS, Wiernik PH, Bachur NR. Adriamycin chemotherapy-efficacy, safety, and pharmacologic basis of an intermittent single high-dosage schedule. Cancer. (1974) 33:19–27. [DOI] [PubMed] [Google Scholar]
- 15.Smith LA, Cornelius VR, Plummer CJ, Levitt G, Verrill M, Canney P, et al. Cardiotoxicity of anthracycline agents for the treatment of cancer: systematic review and meta-analysis of randomised controlled trials. BMC Cancer. (2010) 10:337. 10.1186/1471-2407-10-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardinale D, Colombo A, Lamantia G, Colombo N, Civelli M, De Giacomi G, et al. Anthracycline-induced cardiomyopathy. Clinical relevance and response to pharmacologic therapy. J Am Coll Cardiol. (2010) 55:213–20. 10.1016/j.jacc.2009.03.095 [DOI] [PubMed] [Google Scholar]
- 17.Cardinale D, Colombo A, Bacchiani G, Tedeschi I, Meroni CA, Veglia F, et al. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. (2015) 131:1981–8. 10.1161/CIRCULATIONAHA.114.013777 [DOI] [PubMed] [Google Scholar]
- 18.Lancellotti P, Suter TM, Lo'pez-Ferna'ndez T, Galderisi M, Lyon AR, Van der Meer P, et al. Cardio-oncology services: rationale, organization, and implementation: a report from the ESC Cardio-Oncology council. Eur Heart J. (2019) 40:1756–63. 10.1093/eurheartj/ehy453 [DOI] [PubMed] [Google Scholar]
- 19.Farmakis D, Keramida K, Filippatos G. How to build a cardio-oncology service? Eur J Heart Fail. (2018) 12:1732–4. 10.1002/ejhf.1336 [DOI] [PubMed] [Google Scholar]
- 20.Pareek N, Cevallos J, Moliner P, Shah M, Tan LL, Chambers V, et al. Activity and outcomes of a cardio-oncology service in the United Kingdom-a five-year experience. Eur J Heart Fail. (2018) 20:1721–31. 10.1002/ejhf.1292 [DOI] [PubMed] [Google Scholar]
- 21.Wouters KA, Kremer LC, Miller TL, Herman EH, Lipshultz SE. Protecting against anthracycline-induced myocardial damage: a review of the most promising strategies. Br J Haematol. (2005) 131:561–78. 10.1111/j.1365-2141.2005.05759.x [DOI] [PubMed] [Google Scholar]
- 22.Hertz DL, Caram MV, Kidwell KM, Thibert JN, Gersch C, Seewald J. Evidence for association of SNPs in ABCB1 and CBR3, but not RAC2, NCF4, SLC28A3 or TOP2B, with chronic cardiotoxicity in a cohort of breast cancer patients treated with anthracyclines. Pharmacogenomics. (2016) 17:231–40. 10.2217/pgs.15.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visscher H, Rassekh SR, Sandor GS, Caron HN, van Dalen EC, Kremer LC, et al. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics. (2015) 16:1065–76. 10.2217/pgs.15.61 [DOI] [PubMed] [Google Scholar]
- 24.Reichwagen A, Ziepert M, Kreuz M, Gödtel-Armbrust U, Rixecker T, Poeschel V, et al. Association of NADPH oxidase polymorphisms with anthracycline-induced cardiotoxicity in the RICOVER-60 trial of patients with aggressive CD20(+) B-cell lymphoma. Pharmacogenomics. (2015) 16:361–72. 10.2217/pgs.14.179 [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Can-Lan Sun C, Quiñones-Lombraña A, Singh P, Landier W, Hageman L, et al. CELF4 variant and anthracycline-related cardiomyopathy: a children's oncology group genome-wide association study. J Clin Oncol. (2016) 34:863–70. 10.1200/JCO.2015.63.4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Elm E, Moher D, Little J. Reporting genetic association studies: the STREGA statement. Lancet. (2009) 374:98–100. 10.1016/S0140-6736(09)61265-4 [DOI] [PubMed] [Google Scholar]
- 27.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement. Genet Epidemiol. (2009) 33:581–98. 10.1002/gepi.20410 [DOI] [PubMed] [Google Scholar]
- 28.Review Manager (RevMan) [Computer program] . Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration (2014). [Google Scholar]
- 29.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. (1986) 7:177–88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 30.Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al. Cochrane Handbook for Systematic Reviews of Interventions (ver5.1.0). (2011). Available online at: http://handbook. cochrane.org/front_page.htm (accessed January 1, 2016).
- 31.Wojnowski L, Kulle B, Schirmer M, Schlüter G, Schmidt A, Rosenberger A, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. (2005) 112:3754–62. 10.1161/CIRCULATIONAHA.105.576850 [DOI] [PubMed] [Google Scholar]
- 32.Weiss JR, Kopecky KJ, Godwin J, Anderson J, Willman CL, Moysich KB. Glutathione S-transferase (GSTM1, GSTT1 and GSTA1) polymorphisms and outcomes after treatment for acute myeloid leukemia: pharmacogenetics in Southwest Oncology Group (SWOG) clinical trials. Leukemia. (2006) 20:2169–71. 10.1038/sj.leu.2404421 [DOI] [PubMed] [Google Scholar]
- 33.Beauclair S, Formento P, Fischel JL, Lescaut W, Largillier R, Chamorey E, et al. Role of the HER2 [Ile655Val] genetic polymorphism in tumorogenesis and in the risk of trastuzumab-related cardiotoxicity. Ann Oncol. (2007) 18:1335–41. 10.1093/annonc/mdm181 [DOI] [PubMed] [Google Scholar]
- 34.Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, Kawashima TI, Davies SM, Relling MV, et al. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer. (2008) 112:2789–95. 10.1002/cncr.23534 [DOI] [PubMed] [Google Scholar]
- 35.Rossi D, Rasi S, Franceschetti S, Capello D, Castelli A, De Paoli L, et al. Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP21. Leukemia. (2009) 23:1118–26. 10.1038/leu.2008.398 [DOI] [PubMed] [Google Scholar]
- 36.Rajić V, Aplenc R, Debeljak M, Prestor VV, Karas-Kuzelicki N, Mlinaric-Rascan I, et al. Influence of the polymorphism in candidate genes on late cardiac damage in patients treated due to acute leukemia in childhood. Leuk Lymphoma. (2009) 50:1693–8. 10.1080/10428190903177212 [DOI] [PubMed] [Google Scholar]
- 37.Blanco JG, Sun CL, Landier W, Chen L, Esparza-Duran D, Leisenring W, et al. Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes—a report from the children's oncology group. J Clin Oncol. (2012) 30:1415–21. 10.1200/JCO.2011.34.8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semsei AF, Erdelyi DJ, Ungvari I, Csagoly E, Hegyi MZ, Kiszel PS, et al. ABCC1 polymorphisms in anthracycline-induced cardiotoxicity in childhood acute lymphoblastic leukaemia. Cell Biol Int. (2012) 36:79–86. 10.1042/CBI20110264 [DOI] [PubMed] [Google Scholar]
- 39.Visscher H, Ross CJD, Rassekh SR, Barhdadi A, Marie-Pierre Dubé M, Al-Saloos H. Pharmacogenomic Prediction of Anthracycline-Induced Cardiotoxicity in Children. J Clin Oncol. (2012) 30:1422–8. 10.1200/JCO.2010.34.3467 [DOI] [PubMed] [Google Scholar]
- 40.Cascales A, Sánchez-Vega B, Navarro N, Pastor-Quirante F, Corral J, Vicente V, et al. Clinical and genetic determinants of anthracycline-induced cardiac iron accumulation. Int J Cardiol. (2012) 154:282–6. 10.1016/j.ijcard.2010.09.046 [DOI] [PubMed] [Google Scholar]
- 41.Volkan-Salanci B, Aksoy H, Kiratli PÖ, Tülümen E, Güler N, Öksüzoglu B, et al. The relationship between changes in functional cardiac parameters following anthracycline therapy and carbonyl reductase 3 and glutathione S transferase Pi polymorphisms. J Chemother. (2012) 24:285–91. 10.1179/1973947812Y.0000000037 [DOI] [PubMed] [Google Scholar]
- 42.Lubieniecka JM, Liu J, Heffner D, Graham J, Reid R, Hogge D, et al. Single-nucleotide polymorphisms in aldo-keto and carbonyl reductase genes are not associated with acute cardiotoxicity after daunorubicin chemotherapy. Cancer Epidemiol Biomarkers Prev. (2012) 21:2118–20. 10.1158/1055-9965.EPI-12-1037 [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa K, Kawada K, Morita S, Inada M, Mitsuma A, Sawaki M, et al. Prospective evaluation of corrected QT intervals and arrhythmias after exposure to epirubicin, cyclophosphamide, and 5-fluorouracil in women with breast cancer. Ann Oncol. (2012) 23:743–7. 10.1093/annonc/mdr296 [DOI] [PubMed] [Google Scholar]
- 44.Windsor RE, Strauss SJ, Kallis C, Wood NE, Whelan JS. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma. Cancer. (2012) 118:1856–67. 10.1002/cncr.26472 [DOI] [PubMed] [Google Scholar]
- 45.Roca L, Diéras V, Roché H, Lappartient E, Kerbrat P, Cany L, et al. Correlation of HER2, FCGR2A, and FCGR3A gene polymorphisms with trastuzumab related cardiac toxicity and efficacy in a subgroup of patients from UNICANCERPACS04 trial. Breast Cancer Res Treat. (2013) 139:789–800. 10.1007/s10549-013-2587-x [DOI] [PubMed] [Google Scholar]
- 46.Lipshultz SE, Lipsitz SR, Kutok JL, Miller TL, Colan SD, Neuberg DS, et al. Impact of hemochromatosis gene mutations on cardiac status in doxorubicin-treated survivors of childhood high-risk leukemia. Cancer. (2013) 119:3555–62. 10.1002/cncr.28256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armenian SH, Ding Y, Mills G, Sun C, Venkataraman K, Wong FL, et al. Genetic susceptibility to anthracycline-related congestive heart failure in survivors of haematopoietic cell transplantation. Br J Haematol. (2013) 163:205–13. 10.1111/bjh.12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lubieniecka JM, Graham J, Heffner D, Mottus R, Reid R, Hogge D, et al. A discovery study of daunorubicin induced cardiotoxicity in a sample of acute myeloid leukemia patients prioritizes P450 oxidoreductase polymorphisms as a potential risk factor. Front Genet. (2013) 4:231. 10.3389/fgene.2013.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachidanandam K, Gayle AA, Robins HI, Kolesar JM. Unexpected doxorubicin-mediated cardiotoxicity in sisters-possible role of polymorphisms in histamine N-methyl transferase. J Oncol Pharm Pract. (2013) 19:269–72. 10.1177/1078155212461022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vivenza D, Feola M, Garrone O, Monteverde M, Merlano M, Nigro CL, et al. Role of the renin-angiotensin-aldosterone system and the glutathione S-transferase Mu, Pi and Theta gene polymorphisms in cardiotoxicity after anthracycline chemotherapy for breast carcinoma. Int J Biol Markers. (2013) 28:e336–47. 10.5301/JBM.5000041 [DOI] [PubMed] [Google Scholar]
- 51.Visscher H, Ross CJ, Rassekh SR, Sandor GS, Caron HN, van Dalen EC, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. (2013) 60:1375–81. 10.1002/pbc.24505 [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Liu W, Sun CL, Armenian SH, Hakonarson H, Hageman L, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children's oncology group. J Clin Oncol. (2014) 32:647–53. 10.1200/JCO.2013.50.3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasielewski M, van Spaendonck-Zwarts KY, Westerink NL, Jongbloed JDH, Postma A, Gietema JA, et al. Potential genetic predisposition for anthracycline-associated cardiomyopathy in families with dilated cardiomyopathy. Open Heart. (2014) 1:e000116. 10.1136/openhrt-2014-000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krajinovic M, Elbared J, Drouin S, Bertout L, Rezgui A, Ansari M, et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics J. (2016) 16:530–5. 10.1038/tpj.2015.63 [DOI] [PubMed] [Google Scholar]
- 55.Peña CG, Dávila-Fajardo CL, Martínez-González LJ, Carmona-Sáez P, Pino MJS, Ramos JS, et al. Influence of the HER2 Ile655Val polymorphism on trastuzumab-induced cardiotoxicity in HER2-positive breast cancer patients: a meta-analysis. Pharmacogenet Genomics. (2015) 25:388–93. 10.1097/FPC.0000000000000149 [DOI] [PubMed] [Google Scholar]
- 56.Aminkeng F, Bhavsar AP, Visscher H, Rassekh SR, Li Y, Lee JW, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. (2015) 47:1079–84. 10.1038/ng.3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vulsteke C, Pfeil AM, Maggen C, Schwenkglenks M, Pettengell R, Szucs TD, et al. Clinical and genetic risk factors for epirubicin-induced cardiac toxicity in early breast cancer patients. Breast Cancer Res Treat. (2015) 152:67–76. 10.1007/s10549-015-3437-9 [DOI] [PubMed] [Google Scholar]
- 58.Stanton SE, Ward MM, Christos P, Sanford R, Lam C, Cobham MV, et al. Pro1170 Ala polymorphism in HER2-neu is associated with risk of trastuzumab cardiotoxicity. BMC Cancer. (2015) 15:267. 10.1186/s12885-015-1298-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinbolt RE, Patel R, Pan X, Timmers CD, Pilarski R, Shapiro CL, et al. Risk factors for anthracycline-associated cardiotoxicity. Support Care Cancer. (2016) 24:2173–80. 10.1007/s00520-015-3008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider BP, Shen F, Gardner L, Radovich M, Li L, Miller KD, et al. Genome wide association study for anthracycline-induced congestive heart failure. Clin Cancer Res. (2017) 23:43–51. 10.1158/1078-0432.CCR-16-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz-Pinto S, Pita G, Miguel Martín M, Alonso-Gordoa T, Barnes DR, Alonso MR, et al. Exome array analysis identifies ETFB as a novel susceptibility gene for anthracycline-induced cardiotoxicity in cancer patients. Breast Cancer Res Treat. (2018) 167:249–56. 10.1007/s10549-017-4497-9 [DOI] [PubMed] [Google Scholar]
- 62.Pop-Moldovan AL, Trofenciuc NM, Dărăbantiu DA, Precup C, Branea H, Christodorescu R, et al. Customized laboratory TLR4 and TLR2 detection method from peripheral human blood for early detection of doxorubicin-induced cardiotoxicity. Cancer Gene Ther. (2017) 24:203–7. 10.1038/cgt.2017.4 [DOI] [PubMed] [Google Scholar]
- 63.Ruiz-Pinto S, Pita G, Patiño-García A, Alonso J, Pérez-Martínez A, Cartón AJ, et al. Exome array analysis identifies GPR35 as a novel susceptibility gene for anthracycline-induced cardiotoxicity in childhood cancer. Pharmacogenet Genomics. (2017) 27:445–53. 10.1097/FPC.0000000000000309 [DOI] [PubMed] [Google Scholar]
- 64.Huang Z, Wang J, Qian J, Li Y, Xu Z, Chen M, et al. Effects of cytochrome P450 family 3 subfamily A member 5 gene polymorphisms on daunorubicin metabolism and adverse reactions in patients with acute leukemia. Mol Med Rep. (2017) 15:3493–8. 10.3892/mmr.2017.6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sági JC, Egyed B, Kelemen A, Kutszegi N, Hegyi M, Gézsi A, et al. Possible roles of genetic variations in chemotherapy related cardiotoxicity in pediatric acute lymphoblastic leukemia and osteosarcoma. BMC Cancer. (2018) 18:704. 10.1186/s12885-018-4629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Todorova VK, Makhoul I, Dhakal I, Wei J, Stone A, Carter W, et al. Polymorphic variations associated with doxorubicin-induced cardiotoxicity in breast cancer patients. Oncol Res. (2017) 25:1223–9. 10.3727/096504017X14876245096439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Pavia P, Kim Y, Restrepo-Cordoba MA, Lunde IG, Wakimoto H, Smith AM, et al. Genetic variants associated with cancer therapy-induced cardiomyopathy. Circulation. (2019) 140:31–41. 10.1161/CIRCULATIONAHA.118.037934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fetterly GJ, Aras U, Lal D, Murphy M, Meholick PD, Wang ES. Development of a preclinical PK-PD model to assess antitumor response of a sequential aflibercept and doxorubicin-dosing strategy in acute myeloid leukemia. AAPS J. (2013) 15:662–73. 10.1208/s12248-013-9480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cascorbi I. Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol Ther. (2006) 112:457–73. 10.1016/j.pharmthera.2006.04.009 [DOI] [PubMed] [Google Scholar]
- 70.Lopes LR, Dagher MC, Gutierrez A, Young B, Bouin AP, Fuchs A, et al. Phosphorylated p40PHOX as a negative regulator of NADPH oxidase. Biochemistry. (2004) 43:3723–30. 10.1021/bi035636s [DOI] [PubMed] [Google Scholar]
- 71.Vulsteke C, Lambrechts D, Dieudonné A, Hatse S, Brouwers B, van Brussel T, et al. Genetic variability in the multidrug resistance associated protein-1 (ABCC1/MRP1) predicts hematological toxicity in breast cancer patients receiving (neo-)adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide (FEC). Ann Oncol. (2013) 24:1513–25. 10.1093/annonc/mdt008 [DOI] [PubMed] [Google Scholar]
- 72.Sethi AA, Nordestgaard BG, Tybjaerg-Hansen A. Angiotensinogen gene polymorphism, plasma angiotensinogen, and risk of hypertension and ischemic heart disease: a metaanalysis. Arterioscler Thromb Vasc Biol. (2003) 23:1269–75. 10.1161/01.ATV.0000079007.40884.5C [DOI] [PubMed] [Google Scholar]
- 73.Sethi AA, Nordestgaard BG, Gronholdt ML, Steffensen R, Jensen G, Tybjaerg-Hansen A. Angiotensinogen single nucleotide polymorphisms, elevated blood pressure, and risk of cardiovascular disease. Hypertension. (2003) 41:1202–11. 10.1161/01.HYP.0000072334.34433.17 [DOI] [PubMed] [Google Scholar]
- 74.Freitas AI, Mendonça I, Brión M, Sequeira MM, Reis RP, Carracedo A, et al. RAS gene polymorphisms, classical risk factors and the advent of coronary artery disease in the Portuguese population. BMC Cardiovasc Disord. (2008) 8:15. 10.1186/1471-2261-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gard PR. Implications of the angiotensin converting enzyme gene insertion/deletion polymorphism in health and disease: a snapshot review. Int J Mol Epidemiol Genet. (2010) 1:145–57. [PMC free article] [PubMed] [Google Scholar]
- 76.Ruggenenti P, Cravedi P, Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol. (2010) 6:319–30. 10.1038/nrneph.2010.58 [DOI] [PubMed] [Google Scholar]
- 77.Liu B, An T, Li M, Yi Z, Li C, Sun X, et al. The association between early-onset cardiac events caused by neoadjuvant or adjuvant chemotherapy in triple-negative breast cancer patients and some novel autophapy-related polymorphisms in their genomic DNA: a real-world study. Cancer Commun. (2018) 38:71. 10.1186/s40880-018-0343-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Linschoten M, Teske AJ, Cramer MJ, Wall E, Asselbergs FW. Chemotherapy-related cardiac dysfunction: a systematic review of genetic variants modulating individual risk. Circ Genom Precis Med. (2018) 11:e001753. 10.1161/CIRCGEN.117.001753 [DOI] [PubMed] [Google Scholar]
- 79.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. (2000) 92:1295–302. 10.1093/jnci/92.16.1295 [DOI] [PubMed] [Google Scholar]
- 80.Kruh GD, Belinsky MG. The MRP family of drug efflux pumps. Oncogene. (2003) 22:7537–52. 10.1038/sj.onc.1206953 [DOI] [PubMed] [Google Scholar]
- 81.Bakos E, Homolya L. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1). Pflugers Arch. (2007) 453:621–41. 10.1007/s00424-006-0160-8 [DOI] [PubMed] [Google Scholar]
- 82.Wojtacki J, Lewicka-Nowak E, Lesniewski-Kmak K. Anthracycline induced cardiotoxicity: clinical course, risk factors, pathogenesis, detection and prevention – review of the literature. Med Sci Monit. (2000) 6:411–20. [PubMed] [Google Scholar]
- 83.Elens L, Tyteca D, Panin N, Courtoy P, Lison D, Demoulin JB, et al. Functional defect caused by the 4544G>A SNP in ABCC2: potential impact for drug cellular disposition. Pharmacogenet Genomics. (2011) 21:884–93. 10.1097/FPC.0b013e32834d672b [DOI] [PubMed] [Google Scholar]
- 84.Diebold BA, Bokoch GM. Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat Immunol. (2001) 2:211–5. 10.1038/85259 [DOI] [PubMed] [Google Scholar]
- 85.Dorseuil O, Vazquez A, Lang P, Bertoglio J, Gacon G, Leca G. Inhibition of superoxide production in B lymphocytes by rac antisense oligonucleotides. J Biol Chem. (1992) 267:20540–2. 10.1016/S0021-9258(19)36716-X [DOI] [PubMed] [Google Scholar]
- 86.Zordoky BN, Anwar-Mohamed A, Aboutabl ME, El-Kadi AO. Acute doxorubicin cardiotoxicity alters cardiac cytochrome P450 expression and arachidonic acid metabolismin rats. Toxicol Appl Pharmacol. (2010) 242:38–46. 10.1016/j.taap.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 87.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci USA. (2002) 99:15937–40. 10.1073/pnas.252640799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han W, Kang D, Lee JE, Park IA, Choi JY, Lee KM, et al. A haplotype analysis of HER-2 gene polymorphisms: association with breast cancer risk, HER-2 protein expression in the tumor, and disease recurrence in Korea. Clin Cancer Res. (2005) 11:4775–8. 10.1158/1078-0432.CCR-04-2208 [DOI] [PubMed] [Google Scholar]
- 89.Xie D, Shu XO, Deng Z, Wen WQ, Creek KE, Dai Q, et al. Population-based, case-control study of HER2 genetic polymorphism and breast cancer risk. J Natl Cancer Inst. (2000) 92:412–7. 10.1093/jnci/92.5.412 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.