Abstract
COVID-19 pandemic caused by SARS-CoV-2 globally impacted the humanity causing tragic outcomes; costing millions of lives, destroying economies and demolishing public health infrastructures. The emergence of vaccines using various ingenious approaches in less than a year was deemed the light at the end of the tunnel. However, recent emergence of variants of SARS-CoV-2 in several parts of the world revealed that another hurdle is ahead in the fight against COVID-19. This review will highlight how SARS-CoV-2 mutations, creating different virus variants could potentially impact virus pathogenesis as well as different therapy approaches and vaccine design.
Keywords: COVID-19, SARS-CoV-2, Vaccine, Antibody, Variant, Coronavirus
Introduction
Coronaviruses have a history of causing zoonotic outbreaks that occasionally achieve human-to-human transmission, giving rise to epidemics or pandemics as previously observed with Severe Acute Respiratory Syndrome Coronavirus (SARS) and Middle Eastern Respiratory Syndrome (MERS) coronaviruses [1–3]. While MERS and SARS remained limited to relatively smaller populations, Coronavirus Disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has affected virtually the whole world, wreaking havoc on healthcare systems and costing millions of lives. In December 2019, a SARS-like disease with unknown etiology was initially reported in Wuhan, China, the original epicenter of the pandemic. Later on, a single-stranded RNA virus from patients with severe respiratory illness, the novel betacoronavirus SARS-CoV-2 was determined as the infectious agent [4]. The novel coronavirus SARS-CoV-2, was shown to be capable of human-to-human transmission, leading the whole world to take action, lockdowns being imposed, followed by the World Health Organization (WHO) declaring the outbreak a pandemic [5].
Scientific community demonstrated rigor in investigating the virus itself and its pathogenesis once its genome sequence was revealed. It was quickly shown that the SARS-CoV-2 RNA genome is approximately 30,000 nt in length, including both coding and non-coding regions. The two thirds of its genome encodes non-structural proteins that aid in genome replication and RNA synthesis on the 5′ side. The remainder of its genome codes for several structural proteins including Envelope (E), Spike (S), Nucleocapsid (N) and Membrane (M). Structural proteins help form the virion as well as functioning in various cellular processes [6]. S protein has been widely studied as it is responsible for forming the homotrimeric spike protein that binds to Angiotensin Converting Enzyme 2 (ACE2) promoting virus internalization [7, 8]. Proteolytic cleavage of the S protein by TMPRSS2 is also an important determinant of efficient viral and host cell membrane fusion that ultimately helps viral internalization [9].
RNA viruses are notorious for high mutation rates due in part to the error prone and fast nature of RNA dependent RNA polymerase [10]. Despite the RNA proofreading ability of coronaviruses [11], immune pressure and high mutation rate can eventually lead to protein sequence and structure changes with potential phenotypic impact, creating novel virus variants. This has so far been observed in various instances, namely the early D614G variant, the Danish ‘Cluster 5’ variant, UK (B.1.1.7), South African (B.1.351) and Brazilian (P.1) variants have emerged during the first year of COVID-19 pandemic [12–16] (Table 1). N501Y amino acid change is highlighted as it is at the spike receptor binding domain (RBD), common to several variants, including UK and South African variants, where K417N and E484K substitutions are specific to the South African B.1.351 variant at the ACE2 interface (Fig. 1). These variants are thought to differentially impact ACE2 binding [17] and are thought to potentially have an effect on transmission, effectivity of vaccines and disease progression. Several other variants were reported from the US including, ‘Columbus, Ohio variant’ which contains the N501Y and Q677H substitutions [18], as well as the B.1.429 variant containing L452R substitution that quickly dominated the viral pool in California outbreaks [19] and the fast spreading New York variant [20]. Impact of these and other variants on vaccine efficacy, virulence and pathogenesis is under investigation.
Table 1.
Specifications of current SARS-CoV-2 variants and their transmission/neutralization capacities
| Current SARS-CoV-2 variant lineage nomenclature | Origin of detection | Protein changes of note | Neutralization by natural or vaccine ınduced antibodies | Effect on transmission | References |
|---|---|---|---|---|---|
| D614G | Europe | D614G | Increased | Increased | [95–97] |
| B.1.1.7 | UK | N501Y, ΔH69–V70, P681H | Decreased | Increased | [39, 98] |
| ‘Cluster 5’ | Denmark |
ΔH69–V70 Y453F |
Likely decreased | Likely eliminated | [99, 100] |
| B.1.351 | South Africa | N501Y, K417N, E484K | Decreased | Increased | [38, 101] |
| P.1 | Brazil | E484K, N501Y | Decreased | Increased | [39, 98] |
| B.1.427/B.1.429 | California, USA | S13I, W152C, L452R | Decreased | Increased | [46, 47] |
| B.1.526 | New York, USA | E484K or S447N and D614G, A701V, D253G | Slightly decreased for E484K, no change for S447N | Likely increased | [20, 48] |
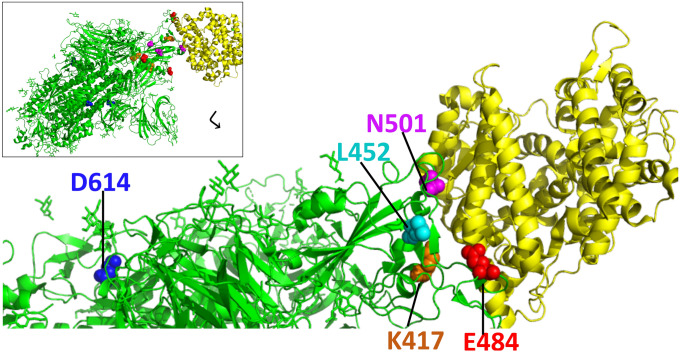
Fig. 1.
Spike glycoprotein of SARS-CoV-2 (green) and angiotensin converting enzyme 2 (ACE2, yellow) receptor complex, zoomed in to highlight variant residues of importance. Modeled using PDB:7DF4. Insert shows the full trimeric spike glycoprotein in complex with an ACE2 molecule. Amino acid residues located at the receptor binding domain (RBD) that changed in several SARS-CoV-2 variants, N501 (magenta spheres), E484 (red spheres), L452 (cyan spheres), K417 (orange spheres), are shown with ACE2. While D614 (blue residue) is not found at the RBD, D to G substitution renders the virus more transmissible and provides ease of entry in ACE2 expressing cells due to allowing the spike RBD to frequently sample the ‘up’ conformation and enhancing ACE2 attachment
Extensive scientific research which accompanied the COVID-19 pandemic led to fast production of both antigen and nucleic acid based diagnostic tests and several vaccines in less than a year which was accompanied, but not yet overshadowed, by the emergence of SARS-CoV-2 variants. This review will highlight the impact of variants on disease management strategies, pathogenesis and vaccines.
SARS-CoV-2 Variants, Their Impact on Disease Outcomes and Vaccine Effectivity
In the light of the high mutation rate associated with RNA viruses, SARS-CoV-2 mutations that create new variants have been expected. The initially dominant variant carried the amino acid change from D to G at the 614th residue creating the D614G variant [12]. This variant, increased in infectivity due to efficient ACE2 binding and replication, quickly took over, dominating the pandemic virus pool globally without causing an increase in disease severity [21].
SARS-CoV-2 was previously shown to cause infection in different animal species and non-human primates [22, 23]. Spillover to-and-fro animals is a real possibility, which may render containment efforts more difficult when compared with a virus that strictly infects humans. SARS-CoV-2 was demonstrated to cause an outbreak in Danish mink farms, where a new variant called ‘Cluster 5’ has emerged [23]. Of note was the Y453F substitution which could increase ACE2 affinity, easing successful introduction to humans. Fast action by Danish authorities prevented this strain spreading to large human populations, kept it limited and helped quickly contain this cluster [13].
B.1.1.7: ‘UK Variant’
In late 2020, the UK reported a phylogenetically distinct variant of concern (VOC) termed SARS-CoV-2 VOC 202012/01, lineage B.1.1.7 VOC [24, 25]. Genome wide investigations of this variant revealed a total of 17 variant defining mutations in its genome where 9 of those reside in the Spike protein, including the D614G substitution. These mutations led to the spike amino acid changes N501Y, A570D, D614G, P681H, T716I, S982A and D1118H, deletion of the amino acids Y144_Y145del and H69_V70del [26] (Table 2). Particularly the deletion H69_V70del is of concern as decreased sensitivity for neutralization by SARS-CoV-2 human convalescent serum samples has been reported [27]. N501Y, found in at least 3 variants (Table 2), is another amino acid change of concern as this residue is one of 6 residues that interact with ACE2 receptor [28]. This substitution could cause an increase in ACE2 affinity [29], potentially enhancing viral entry into host cells.
Table 2.
Spike protein amino acid changes and deletions of note found in the variants of concern [20, 26, 47, 48]
| H69_V70del | Y144_Y145del | L242_L245del | L5F | S13I | L18F | T20N | P26S | D80A | T95I | D138Y | W152C | R190S | D215G | R246I | D253G | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B.1.1.7 | * | * | ||||||||||||||
| B.1.351 | * | * | * | * | * | |||||||||||
| P.1 | * | * | * | * | * | |||||||||||
| B.1.427/B.1.429 | * | * | ||||||||||||||
| B.1.526 | * | * | * |
| K417N/T | L452R | E484K | N501Y | A570D | D614G | H655Y | P681H | A701V | T716I | S982A | T1027I | D1118H | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B.1.1.7 | * | * | * | * | * | * | * | ||||||
| B.1.351 | *(N) | * | * | * | * | ||||||||
| P.1 | * (T) | * | * | * | * | * | |||||||
| B.1.427/B.1.429 | * | * | |||||||||||
| B.1.526 | *or S477N | * | * |
*Presence of the amino acid change or deletion in the variant
Initial studies reported high viral load, increased transmissibility but no increased risk of lethality associated with this variant; however, a UK report suggested an increase in fatality of 1.65 fold [30]. Other studies highlighted the increased nasopharyngeal viral load associated with B.1.1.7 [31], despite no association with severe disease in various cohorts [32]. An ecological study out of the UK where B.1.1.7 is the predominant strain, revealed that B.1.1.7 does not alter the presentation of symptoms and does not cause a significant increase in the reinfection rate [32]. It should, however, be noted that this study focused on a small sample of hospitalized patients (n = 484), therefore more generalized cohorts will need to be included in order to gain insight toward the infection fatality rate. Another UK based study of a cohort of 198,420 patients reported an increase in disease severity, critical care admission and 60% increased risk of mortality associated with B.1.1.7 [33]. An increase in the pediatric cases with COVID-19 in the UK during the second wave of the pandemic is attributed to B.1.1.7 prevalence and high transmission nonetheless, no significant increase in morbidity or mortality on these young individuals were observed [34]. Pertaining to vaccine efficacy, despite being 45% more transmissible, prioritized vaccination with an mRNA vaccine coupled with proactive surveillance programs was shown to prevent B.1.1.7 infections especially in the elderly [35].
B.1.351: ‘South Africa Variant’
A variant, lineage B.1.351, was reported by South African authorities, with higher viral load and transmission rates [15]. This variant has more spike amino acid substitutions compared to the UK B.1.1.7 variant including L18F, D80A, D215G, L242-244del, R246I, K417N, E484K, N501Y, D614G, and A701V (Table 2). Three of these amino acid changes are of special significance as they are located in the RBD (K417N, E484K and N501Y) of the spike protein (Fig. 2). SARS-CoV-2 spike RBD and N-terminal domains (NTD) are recognized by various potently neutralizing monoclonal antibodies such that, amino acid changes localized in these domains could interfere with recognition by these antibodies [36]. Furthermore, therapeutic monoclonal antibodies designed to target the virus may lose effectiveness and require updating [37, 38]. This variant, which also includes N501Y present in the UK variant, not only could change disease outcome but it could also interfere with the effectiveness of spike protein based vaccines. For instance, antibody neutralization studies have revealed that, mRNA vaccine induced antibodies from volunteers had reduced (onefold to threefold) neutralization capacity of the South African variant largely due to the E484K substitution [39] (Table 3). E484K amino acid change was demonstrated to interfere with efficient neutralization by convalescent sera and confer resistance to SARS-CoV-2 neutralizing antibodies, potentially preventing therapeutic approaches both by monoclonal antibody therapeutics and via convalescent plasma [37, 38, 40, 41]. While polyclonal antibody response and T-cell response still likely allows mRNA vaccine effectiveness, potential impact of variants on vaccine efficacy and therapeutic approaches can not be ignored.
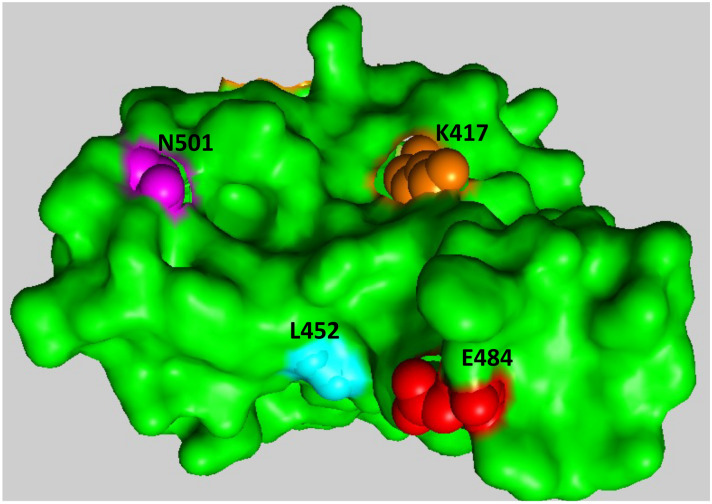
Fig. 2.
Top down view of receptor binding domain of SARS-CoV-2 spike protein demonstrated as surface representation. Modeled using PDB: 6W41. Residues of note, that are observed to change in several VOC, are shown as colored spheres. Red: E484, Orange: K417, Magenta: N501, Cyan: L452
Table 3.
Impact of SARS-CoV-2 variants on neutralization capacities of antibodies elicited by COVID-19 vaccines
| Vaccine ⇒ | BNT162b2 by Pfizer-BioNtech | mRNA-1273 by Moderna | CoronaVac by Sinovac |
|---|---|---|---|
| Variant ⇓ | |||
| B.1.1.7 | Reduced [36] | N/A | Reduced [43] |
| B.1.351 | Reduced ++ [36] | Reduced ++ [76] | Reduced ++ [43] |
| P.1 | Reduced + [36] | N/A | N/A |
| B.1.429 | Reduced + [47] | Reduced [47, 76] | N/A |
N/A data not available
+, relative reduction within the study
P.1: ‘Brazil Variant’
P.1 lineage variant, also known as the Brazilian variant emerged in Manaus in December 2020 [42] and since spread globally. P.1 variant has some of the spike protein amino acid changes observed in other variants including K417N, E484K and N501Y (Fig. 2) [42]. Several in vitro studies revealed that antibody neutralization of P.1 variant is reduced [41, 43, 44]. Specifically, vaccine or infection induced antibodies all had significantly decreased neutralization capacity against P.1, where P.1 demonstrated resistance against two commonly used therapeutic antibodies Casirivimab and Bamlanivimab [44]. Hence, P.1 variant can evade inhibition by neutralizing antibodies and demonstrate resistance against convalescent plasma therapy as well as therapeutic antibodies. Particularly the immune escape of P.1 against convalescent sera may have fueled the pandemic waves in Brazil.
B.1.427/B.1.429: ‘California Variant’
Viral whole-genome sequencing revealed a variant in California in May, 2020 denoted by two lineages as B.1.427/B.1.429. Carrying the L452R amino acid change, this variant quickly dominated the infection pool in California due to reported increased infectivity and transmissibility [45]. L452R substitution, positioned at the RBD of the spike protein of SARS-CoV-2, could potentially increase affinity of ACE2 interaction as well as mediating escape from neutralizing antibodies (Fig. 2) [46]. Other amino acid changes of note include S13I and W152C where S13I is located at the signal peptide but W152C is found at the NTD [46]. In addition to the increased viral load, B.1.427/B.1.429 was demonstrated to have reduced sensitivity toward neutralization by antibodies obtained from individuals vaccinated with mRNA-based vaccines [47] (Table 3). Considering the increased transmission and infectivity, that is largely attributed to L452R, coupled with a resistance in neutralization by convalescent/vaccinee plasma, close surveillance of this strain along with other preceding VOC is warranted.
B.1.526: ‘New York Variant’
A VOC, lineage B.1.526, spread fast in New York City [48]. Two versions of the variant, both carrying the prevalent D614G substitution of the spike protein, carry either E484K or S477N at their receptor binding domain [20, 48]. Several substitutions of note are D253G which is at the N-terminal supersite that serves as a binding site for neutralizing antibodies and A701V is near the furin processing site [20]. T95I, L5F are also spike protein associated substitutions that are not yet found in other variants [20]. Convalescent sera and vaccine elicited antibodies provided full neutralization profile against the S477N where E484K version elicited decreased neutralization against convalescent and vaccine induced antibodies as well as some therapeutic monoclonal antibodies [20].
Another variant originating from India, B.1.617, carrying L452R and E484Q, P681R key substitutions in spike protein is spreading at an alarming rate and could potentially be involved in immune evasion mechanisms [49].
While various substitutions exist in several variants (Table 2), it should be noted that, these are only the detected variants and it is readily possible that variants keep emerging in different parts of the world. In regions without an efficient research network to study or detect any mutations [50], emergence of variants remain undetected until they spread elsewhere. Therefore, it is likely that we will encounter various variants in the upcoming months and years that may have increased infectivity or antigenicity. Collectively, evidence suggests that precautions should be taken, concerning novel variants and the antigenic drift SARS-CoV-2 is potentially going through. For instance, in vitro studies indicated that further spike variants, N234Q, L452R, A475V, V483A, are capable of immune escape by conferring resistance to monoclonal antibodies [19, 28]. Ensuring globally accessible and adaptable treatment, vaccination, diagnosis strategies is crucial. In order to have the upper hand in containing the pandemic, access to the right tools such as being able to adapt the vaccines to the new variants or manipulating molecular detection kits fast and efficiently, will have critical importance. Biotechnological approaches used during the innovation of mRNA vaccines provide them with the ease of changing the mRNA code to adapt new variants or potentially include various mutations in their formulations, creating multivalent vaccines.
Vaccine Design in the Era of SARS-CoV-2 Variants
While the year 2020 has been marked with the COVID-19 pandemic, it has also been marked by the commendable efforts of the scientific community that worked endlessly to understand the virus, the disease and eventually produce effective vaccines. The challenge now is to ensure the vaccines will be efficient against the current and potential future variants as well.
SARS-CoV-2 spike protein, just like that of SARS-CoV, goes through sequential cleavage, leading to formation of S1 and S2 (S2 is further cleaved to yield S2’) where S1 includes the receptor binding domain and S2 promotes membrane fusion [51]. The prefusion form of the S protein can be recognized by neutralizing monoclonal antibodies, hence is a good vaccine target. Spike RBD can sample between ‘up’ and ‘down’ conformations where the ‘up’ conformation allows it to readily adhere to ACE2 protein [52]. Additionally, previous research has shown that proline substitutions in S2 part of the spike increases expression and immunogenicity of coronavirus spike proteins [53]. Utilizing this knowledge collectively, Pfizer/BioNtech and Moderna used novel mRNA technology coding S protein, and their vaccine proved to be over 90% efficient in Phase III clinical trials [54].
The synthetic mRNA used in both vaccines’ formulation is lipid nanoparticle encapsulated, where upon intramuscular injection, it can gain access into the macrophages in the milieu, causing cellular spike protein expression. Both Moderna and Pfizer/BioNtech have so far reported that their vaccine is effective against the South African and UK variants [40, 55].
While clinical studies will be the ultimate judge of vaccine effectiveness against variants, future variants, carrying multiple changes particularly at antibody recognition sites risk rendering vaccines less effective. Molecular biotechnological tools used in the vaccine design could easily allow manipulating the synthetic mRNA used in vaccine formulation to adapt the mRNA sequence, to include the novel mutations or even potential mutations. Evolutionary projections into the selective pressure toward certain spike residues may help delineate the immune pressure exerted on specific spike residues, helping select for various changes.
Vaccines that can achieve mucosal immunity may increase protection via limiting transmission. In the era of highly transmissible variants, this approach may be useful. One way to induce mucosal immunity may be using a nasal vaccine to help clear the nasal passages from the virus and limit person to person viral spread [6]. Since SARS-CoV-2 is capable of efficient replication in the upper respiratory tract [56], a nasal vaccine interfering with this replication may play a pivotal role in viral elimination. New variants usually take over due to their increased nasopharyngeal viral load leading to high levels of transmission. For instance, the D614G and UK variant B.1.1.7 were shown empirically to have higher viral load in the nasopharyngeal samples [31, 57]. Furthermore, D614G substitution allows the spike RBD to more frequently sample the ‘up’ conformation therefore enhances ACE2 attachment, increasing transmissibility [52]. At the same time since ‘up’ conformation of the RBD is more frequently encountered by D614G substitution, recognition by neutralizing antibodies occur more frequently, rendering this variant neutralization prone to patient sera. To avoid D614G and variants alike with high ACE2 affinity to take a hold of the nasopharynx, utilization of mucosal immunity inducing vaccines could play a role in efficiently limiting viral spread. Considering the most prominent SARS-CoV-2 transmission mode is via respiratory droplets, utilizing a nasal vaccine could also trigger cell mediated responses in addition to mucosal immunity, preventing viral replication in nose and nasopharynx, hence interfering with virus transmission. Efficacy of a single dose adenovirus based intranasal SARS-CoV-2 vaccine was demonstrated in animal models [6].
Inactive virus based vaccines or other viral vector based vaccines may also play an efficient role in achieving herd immunity. Utilizing a vector based approach or inactive virus vaccine could mount a robust T-cell response and prove successful [58]. Preclinical studies on an inactive vaccine were proven successful on animal models on a small-scale study, protecting rhesus macaques against SARS-CoV-2 infection without causing antibody dependent enhancement for 7 days until sacrifice [59]. SARS-CoV-2 that was inactivated in this study despite eliciting neutralizing antibody response and protection in macaques, was inactivated using β-propiolactone that is frequently used in inactive viral vaccine generation [60]. While this vaccine elicited a protective immune response in animal models and was then used in Phase ½ trials [61], it is important to consider spike protein’s structural properties when using an inactivating agent. β-propiolactone has the potential of removing the spike S1 domain and rendering it in a postfusion state where the RBD is not exposed to mount a robust enough immune response [62]. Since spike protein elicits an effective neutralizing antibody and T-cell based immune response, it is important to ensure its structural architecture during vaccine generation.
Adenovirus based viral-vectored vaccines have previously been approved for various infectious diseases including respiratory syncytial virus and Ebola virus infections, and are generally considered to be safe and immunogenic [63, 64]. Viral vector vaccines produced by Janssen (Johnson & Johnson) and Oxford-AstraZeneca have received approval for emergency use from various independent organizations. Janssen vaccine, Ad26.COV2.S, which encodes full length spike in non-replicating adenovirus type 26 vector is administered as single dose [65]. While the single dose vaccine shows efficacy in clinical trials, studies from animal models highlight the added benefit of a second dose, while also highlighting the vaccine’s effectivity against D614G [66]. AZD1222 vaccine by Oxford-AstraZeneca is based on ChadOx1 vector carrying DNA encoding spike protein administered as a two dose regimen. Both antibody and T-cell responses were detected upon administration of the first dose [67]. While clinical trials for Janssen vaccine, Ad26.COV2.S, were carried out in Belgium and the US, Oxford-AstraZeneca carried out their studies for AZD1222 in South Africa, among other places [68], where the predominant circulating variant was B.1.351; however, a two dose regimen of the Oxford-AstraZeneca vaccine did not elicit protection against B.1.351 variant [69]. Since antibodies elicited against B.1.351 South Africa variant show crossreactivity toward both the original D614G and the P.1 variant, a vaccine eliciting neutralizing antibody response against B.1.351 is thought to provide protection against other variants like D614G and P.1 [70]. While the world is in dire need of vaccines and a large number of them, the reports highlighting the association of these two vaccines with very rare coagulopathies like cerebral venous sinus thrombosis (CVST) [71] and thrombotic thrombocytopenia [72], led regulatory agencies to halt their use. While COVID-19 itself is associated with CVST risk [73, 74] as well as an overall 24% incidence of pulmonary embolism and hypercoagulability [75] the risk benefit ratio of vaccine usage should be carefully evaluated and precautionary measures should be strictly implemented once mechanisms of pathogenesis are delineated.
Neutralization Capacity of Vaccine Induced Antibodies Against SARS-CoV-2 Variants
The emergence of several SARS-CoV-2 variants risks rendering some of the current COVID-19 vaccines less effective. Several studies highlighted the resistance of variants to neutralization by convalescent and vaccine induced antibodies. For instance, vaccine sera from volunteers vaccinated with Pfizer-BioNtech vaccine elicited reduced neutralizing activity against B.1.1.7, B.1.351 South Africa strain and P.1 Brazil strain, with the most prominent reduction observed with B.1.351 relative to the reference D614G strain [36] (Table 3). Another study investigated the neutralization of antibodies elicited by Novavax NVX-CoV2373, a protein subunit vaccine, and that of mRNA-1273 by Moderna against the California variant B.1.429 and B.1.351 pseudoviruses. The small-scale study of 63 volunteers similarly revealed the reduction in neutralization abilities of antibodies elicited by both vaccines. The most drastic reduction, up to 9–14 times decrease in neutralization compared to D614G was observed with B.1.351 pseudovirus where the antibodies were 2–3 times less sensitive against the B.1.429 variant pseudovirus [76]. W152C, and L452R amino acid changes found in B.1.429 NTD and RBD respectively could potentially play a role in decreased sensitivity (Table 3). Vaccines produced using inactivated virus, namely CoronaVac by Sinovac and BBIBP-CorV by Sinopharm were investigated for their neutralization capacity against D614G, B.1.1.7, B.1.351 [43]. Interestingly, both vaccines elicited lower antibody titers compared to convalescent sera. So a comparison group against another well studied vaccine sera would have been useful to assess the comparative baseline neutralization levels. Particularly CoronaVac vaccine revealed a statistically significant decrease in neutralization against B.1.351 and B.1.1.7 compared to the WT pseudovirus (Table 3). For the Sinopharm BBIBP-CorV vaccinee serum samples, 20 serum samples out of 25 showed complete or partial loss of neutralization against B.1.351, despite no statistical significance [43]. While antibody neutralization is crucial, T-cell response elicited particularly by mRNA-based vaccines plays a significant role in early protection against COVID-19, so all branches of the immune system should be concomitantly considered in order to appreciate the real impact of the variants on vaccine mediated protection from COVID-19 [77].
SARS-CoV-2 Variants and Pathogenesis
How variants will impact SARS-CoV-2 pathogenesis is another topic of concern. SARS-CoV-2 has diverse tissue tropism. Due to its higher ACE2 affinity, it can efficiently infect the upper respiratory tract, such as the nasopharynx (also explaining its high transmissibility, along with asymptomatic transmission) unlike SARS-CoV [56]. Following the nasopharynx, it can enter the host via alveolar cells, vascular endothelial cells, alveolar macrophages and vascular endothelial cells. Mammalian gastrointestinal tract is also permissive to SARS-CoV-2 infection [78]. In addition to the classic respiratory symptoms, hyperinflammatory syndrome, gastrointestinal disease, coagulopathies, cardiac pathologies, and neurological problems associated with COVID-19 prompted detailed investigations [79–81].
Autopsy studies aiming at understanding the neurological symptoms, revealed cases of colocalization of SARS-CoV spike protein with neural/neuronal cells in olfactory mucosa of a subset of individuals deceased due to COVID-19. SARS-CoV-2 presence in CNS of a subset of deceased individuals was also detected both via staining and nucleic acid detection methods. These results indicate SARS-CoV-2’s ability in crossing the neural-mucosal interface in olfactory mucosa and entering the nervous system, providing an explanation toward some of the observed neurological symptoms [82]. Considering that novel variants have been causing structural changes in the spike protein, the changes in binding affinity of a variant spike to ACE2 or any processing change by TMPRSS2 or other downstream intracellular processes may affect SARS-CoV-2 variants’ tissue tropism or could change the course of infection. Future work will need to delineate if differential antibody recognition or ACE2 affinity exerted by the variants will be involved in changing viral pathogenesis.
What Determines Disease Severity, Impact of Biological Sex-Specific Traits and How the Variants May Affect It
The large number of people infected with SARS-CoV-2 during COVID-19 pandemic has revealed various outcomes of disease in a relatively short time. For instance, while some people developed no symptoms, others suffered serious, even fatal disease. While a number of people got better in a short time, others, termed ‘long haulers’ or ‘long term COVID patients’ suffered symptoms lasting for many months. In some people, infection led to severe neurological symptoms such as stroke or encephalopathy, whereas other had none or mild neurological symptoms like a state of confusion, loss of taste or smell [82–84].
Over time, gender and age related differences in disease progression became obvious. People over 65 had worse outcomes compared to younger patients and women fared better compared to men [85, 86]. Knowledge on these details became obvious, mainly because of the variety of scientific research carried out in the world on COVID-19, due to the urgency of the situation and the large number of people who got the infection.
Research has focused on how biological sex impacts COVID-19 severity. Initial results highlighted the higher incidence of severe disease in males compared to females. Several mouse studies indicated a higher expression and activity for ACE2 protein in males [87]. In addition to ACE2, TMPRSS2, the serine protease used to cleave the spike protein to aid in viral internalization by allowing fusion of viral and host membranes, was also reported to have a higher expression in males due to higher androgen levels [88]. Furthermore, males may suffer elevated inflammatory immune responses compared to women, leading to fatal lung macrophage-monocyte infiltration and cytokine storm. Previous studies reported a higher C-reactive protein level as well as higher neutrophil to lymphocyte ratio in males, indicating an elevated inflammatory immune response and worse prognosis [86].
Another determinant of disease severity is associated with the fucosylation profiles of IgG antibodies against SARS-CoV-2. Level of afucosylated IgG antibodies show correlation with disease severity such that patients suffering severe COVID-19 have high levels of afucosylated IgG compared to those with mild symptoms. This increased afucosylated IgG profile amplifies pro-inflammatory cytokine response, leading to acute case manifestation [89]. Interestingly, the level of afucosylated anti-spike RBD IgG was elevated in hospitalized males compared to females [90]. Antibody response elicited in the presence of different SARS-CoV-2 variants in both sexes will be a strong determinant of disease progression. In the light of this information, investigational tools may be adapted to understand the fucosylation levels of therapeutic antibodies. Furthermore, convalescent plasma therapy strategies can be manipulated to include volunteers with high levels of fucosylated IgG. These factors will be particularly important especially if certain variants are indeed causing worse prognosis or if variants give rise to differing antibody or immune profiles.
In addition to IgG fucosylation levels, there are several other variables that affect the success rate of convalescent therapy [91]. Location is thought to be one of the factors contributing to the effectivity of convalescent therapy, as different variants tend to circulate in specific areas. Sera obtained from an individual in a certain area is presumed to have generated an antibody response against a particular variant prevalent in that area, therefore will be most useful for a patient infected with the same variant. Being hospitalized with severe COVID-19, being older and having male sex are associated with having a higher antibody response against SARS-CoV-2, making these individuals better candidates as plasma donors [91]. While males have higher plasma antibody titers, they also have higher levels of afucosylated IgG. Considering the importance of plasma donor in relation to the variants as well as the sex-specific antibody profiles, the selection criteria for convalescent therapy should include sex-specific variations, location and the prevalence of variants.
Not only the SARS-CoV-2 spike variants but also the variants of ACE2 receptor were demonstrated to determine the affinity of host-virus interactions [92, 93]. For instance, ACE2 K31R and E37K substitutions showed decreased affinity where K26R and T92I substitutions showed increased affinity for wild type SARS-CoV-2 spike protein [93]. Rare X-linked alleles, including the ones encoding E37K, are observed twice as frequently in females, suggesting that these missense variants may predominantly affect SARS-CoV-2 affinity in females [94]. Modeling studies coupled with biochemical interaction/affinity evaluation of ACE2 substitutions with spike variants are likely to provide insight toward the potential gender-specific impact of spike variants in host–pathogen interactions as well as susceptibility to SARS-CoV-2 variants.
Conclusions
More variants are expected to emerge especially at the spike RBD as it is under immune selection due to being a major epitope for neutralizing antibodies. Therefore, more variants with differing antibody recognition and ACE2 affinities are expected. Future studies will need to delineate the immune response and pathogenesis associated with novel SARS-CoV-2 variants. Considering varying antibody recognition and neutralization states for different variants, different downstream immune responses can be expected that could eventually affect disease outcome.
Overall, novel SARS-CoV-2 variants run the risk of changing immune response elicited by the host, vaccine and therapeutic antibody efficacy, disease pathogenesis and prognosis. As variants tend to accumulate at the spike protein’s RBD, ACE2 affinity or antibody binding changes can lead to different pathogenesis and immune responses. Monoclonal antibody therapeutics and convalescent plasma therapy options should also be adaptable in the light of current or potential novel variants. Scientific advances achieved by quick and successful vaccine design should now be implemented to keep the vaccines as effective against the current and potentially upcoming SARS-CoV-2 variants.
Declarations
Conflict of interest
The author declares that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, Lui SF, Szeto CC, Chung S, Sung JJ. A major outbreak of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, Lam WK, Seto WH, Yam LY, Cheung TM, Wong PC, Lam B, Ip MS, Chan J, Yuen KY, Lai KN. A cluster of cases of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine. 2003;348(20):1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 3.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio-Medica. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, Kashentseva E, McCune BT, Bailey AL, Zhao H, VanBlargan LA, Dai YN, Ma M, Adams LJ, Shrihari S, Danis JE, Gralinski LE, Hou YJ, Schafer A, Kim AS, Keeler SP, Weiskopf D, Baric RS, Holtzman MJ, Fremont DH, Curiel DT, Diamond MS. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183(1):169–184 e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biology. 2018;16(8):e3000003. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevajol M, Subissi L, Decroly E, Canard B, Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Research. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bulletin of the World Health Organization. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MG, de Rooij MM, Weesendorp E, Engelsma MY, Bruschke CJ, Smit LA, Koopmans M, van der Poel WH, Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. 2020;25(23):2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England, P. H. (2020). Investigation of novel SARS-CoV-2 variant: Variant of concern 202012/01, technical briefing 3.
- 15.Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., Doolabh, D., Pillay, S., San, E. J., Msomi, N., Mlisana, K., von Gottberg, A., Walaza, S., Allam, M., Ismail, A., Mohale, T., Glass, A. J., Engelbrecht, S., Van Zyl, G., … de Oliveira, T. (2020) Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 10.1101/2020.12.21.20248640.
- 16.Voloch, C. M., da Silva Francisco, R., Jr., de Almeida, L. G. P., Cardoso, C. C., Brustolini, O. J., Gerber, A. L., Guimaraes, A. P. C., Mariani, D., da Costa, R. M., Ferreira, O. C., Jr., L.W.A.C.C. Covid19-Ufrj Workgroup, Frauches, T. S., de Mello, C. M. B., Leitao, I. C., Galliez, R. M., Faffe, D. S., Castineiras, T., Tanuri, A., & de Vasconcelos, A. T. R. (2021). Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. Journal of Virology. 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed]
- 17.Santos, J. C., & Passos, G. A. (2021). The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv. 10.1101/2020.12.29.424708.
- 18.Tu, H., Avenarius, M. R., Kubatko, L., Hunt, M., Pan, X., Ru, P., Garee, J., Thomas, K., Mohler, P., Pancholi, P., & Jones, D. (2021). Distinct patterns of emergence of SARS-CoV-2 spike variants including N501Y in clinical samples in Columbus Ohio. bioRxiv. 10.1101/2021.01.12.426407.
- 19.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Zhang L, Li X, Huang W, Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294 e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, H., Dcosta, B. M., Samanovic, M. I., Mulligan, M. J., Landau, N. R., & Tada, T. (2021). B.1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies. bioRxiv. 10.1101/2021.03.24.436620. [DOI] [PMC free article] [PubMed]
- 21.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang RD, Zhang HJ, Gao XX, Peng C, Min J, Chen Y, Si HR, Wu J, Zhou P, Wang YY, Wei HP, Pang W, Hu ZF, Lv LB, Zheng YT, Shi ZL, Yuan ZM. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Research. 2020;30(8):670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koopmans M. SARS-CoV-2 and the human-animal interface: Outbreaks on mink farms. The Lancet Infectious Diseases. 2021;21(1):18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D. L., Volz, E., on behalf of COVID-19 Genomics Consortium UK (CoG-UK). (2020). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org.
- 25.England, P. H. (2020). VOC 202012/01 Technical Briefing 5.
- 26.Public Health England (2021). Technical Briefing 6, Investigation of SARS-CoV-2 Variants of Concern. In P. H. England (Ed.).
- 27.Kemp, S. A., Meng, B., Ferriera, I. A., Datir, R., Harvey, W. T., Papa, G., Lytras, S., Collier, D. A., Mohamed, A., Gallo, G., Thakur, N., Carabelli, A. M., Kenyon, J. C. Lever, A. M., De Marco, A., Saliba, C., Culap, K., Cameroni, E., Piccoli, L., … Gupta, R. K. (2021). Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. bioRxiv. 10.1101/2020.12.14.422555.
- 28.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310 e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan, C. E. Z., Seah, S. G. K., Chye, D. H., Massey, S., Torres, M., Lim, A. P. C., Wong, S. K. K., Neo, J. J. Y., Wong, P. S., Lim, J. H., Loh, G. S. L., Wang, D. L., Boyd-Kirkup, J. D., Guan, S., Thakkar, D., Teo, G. H., Purushotorman, K., Hutchinson, P. E., Young, B. E., … Hanson, B. J. (2020). The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. bioRxiv. 10.1101/2020.10.26.355107. [DOI] [PMC free article] [PubMed]
- 30.Horby, P., Bell, I., Breuer, J., Cevik, M., Challen, R., Davies, N., Dabrera, G., Edmunds, J., Ferguson, N., Sebastian, F., Hayward, A., Hippisley-Cox, J., Huntley, C., Humberstone,, B., Huntley, C., McMenamin, J., McKeigue, P., Medley, G., Semple, C. (2021). NERVTAG note on B.1.1.7 severity.
- 31.Calistri P, Amato L, Puglia I, Cito F, Di Giuseppe A, Danzetta ML, Morelli D, Di Domenico M, Caporale M, Scialabba S, Portanti O, Curini V, Perletta F, Camma C, Ancora M, Savini G, Migliorati G, D'Alterio N, Lorusso A. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. International Journal of Infectious Diseases. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, Klaser K, Canas LS, Molteni E, Modat M, Drew DA, Nguyen LH, Polidori L, Selvachandran S, Hu C, Capdevila J, C.-G.U. Consortium. Hammers A, Chan AT, Wolf J, Spector TD, Steves CJ, Ourselin S. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: An ecological study. Lancet Public Health. 2021;6:e335–e345. doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patone, M., Thomas, K., Hatch, R., Tan, P. S., Coupland, C., Liao, W., Mouncey, P., Harrison, D., Rowan, K., Horby, P., Watkinson, P., & Hippisley-Cox, J. (2021). Analysis of severe outcomes associated with the SARS-CoV-2 Variant of Concern 202012/01 in England using ICNARC Case Mix Programme and QResearch databases.medRxiv. 10.1101/2021.03.11.21253364.
- 34.Brookman S, Cook J, Zucherman M, Broughton S, Harman K, Gupta A. Effect of the new SARS-CoV-2 variant B.1.1.7 on children and young people. The Lancet Child & Adolescent Health. 2021;5(4):e9–e10. doi: 10.1016/S2352-4642(21)00030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munitz A, Yechezkel M, Dickstein Y, Yamin D, Gerlic M. BNT162b2 vaccination effectively prevents the rapid rise of SARS-CoV-2 variant B.1.1.7 in high risk populations in Israel. Cell Reports Medicine. 2021;2:100264. doi: 10.1016/j.xcrm.2021.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, Liu J, Errico JM, Xie X, Suryadevara N, Gilchuk P, Zost SJ, Tahan S, Droit L, Turner JS, Kim W, Schmitz AJ, Thapa M, Wang D, Boon ACM, Presti RM, O’Halloran JA, Kim AHJ, Deepak P, Pinto D, Fremont DH, Crowe JE, Jr, Corti D, Virgin HW, Ellebedy AH, Shi PY, Diamond MS. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nature Medicine. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host & Microbe. 2021;29(3):463–476 e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nature Medicine. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P., Nair, M. S., Liu, L., Iketani, S., Luo, Y., Guo, Y., Wang, M., Yu, J., Zhang, B., Kwong, P. D., Graham, B. S., Mascola, J. R., Chang, J. Y., Yin, M. T., Sobieszczyk, M., Kyratsous, C. A., Shapiro, L., Sheng, Z., Huang, Y., & Ho, D. D. (2021). Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature, 593(7857), 130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed]
- 40.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Da Silva J, Xu J, Colbert RA, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faria, N. R., Mellan, T. A., Whittaker, C., Claro, I. M., Candido, D. D. S., Mishra, S., Crispim, M. A. E., Sales, F. C., Hawryluk, I., McCrone, J. T., Hulswit, R. J. G., Franco, L. A. M., Ramundo, M. S., de Jesus, J. G., Andrade, P. S. Coletti, T. M., Ferreira, G. M., Silva, C. A. M., Manuli, E. R., … Sabino, E. C. (2021). Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. Science, 372(6544), 815–821. 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed]
- 43.Wang, G. L., Wang, Z. Y., Duan, L. J., Meng, Q. C., Jiang, M. D., Cao, J., Yao, L., Zhu, K. L., Cao, W. C., & Ma, M. J. (2021). Susceptibility of circulating SARS-CoV-2 variants to neutralization. New England Journal of Medicine. 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed]
- 44.Hoffmann M, Arora P, Gross R, Seidel A, Hornich BF, Hahn AS, Kruger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jack HM, Jahrsdorfer B, Schrezenmeier H, Muller M, Kleger A, Munch J, Pohlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng, X., Garcia-Knight, M. A., Khalid, M. M., Servellita, V., Wang, C., Morris, M. K., Sotomayor-Gonzalez, A., Glasner, D. R., Reyes, K. R., Gliwa, A. S., Reddy, N. P., Sanchez San Martin, C., Federman, S., Cheng, J., Balcerek, J., Taylor, J., Streithorst, J. A., Miller, S., Kumar, G. R., … Chiu, C. Y. (2021). Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant. Cell. 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed]
- 46.Tchesnokova, V., Kulakesara, H., Larson, L., Bowers, V., Rechkina, E., Kisiela, D., Sledneva, Y., Choudhury, D., Maslova, I., Deng, K., Kutumbaka, K., Geng, H., Fowler, C., Greene, D., Ralston, J., Samadpour, M., & Sokurenko, E. (2021). Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv. 10.1101/2021.02.22.432189. [DOI] [PMC free article] [PubMed]
- 47.McCallum, M., Bassi, J., Marco, A., Chen, A., Walls, A. C., Iulio, J. D., Tortorici, M. A., Navarro, M. J., Silacci-Fregni, C., Saliba, C., Agostini, M., Pinto, D., Culap, K., Bianchi, S., Jaconi, S., Cameroni, E., Bowen, J. E., Tilles, S. W., Pizzuto, M. S., … Veesler, D. (2021). SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. 10.1101/2021.03.31.437925.
- 48.Annavajhala, M. K., Mohri, H., Zucker, J. E., Sheng, Z., Wang, P., Gomez-Simmonds, A., Ho, D. D., & Uhlemann, A. C. (2021). A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 10.1101/2021.02.23.21252259.
- 49.Edara, V. V., Lai, L., Sahoo, M. K., Floyd, K., Sibai, M., Solis, D., Flowers, M. W., Hussaini, L., Ciric, C. R., Bechnack. S., Stephens, K., Mokhtari, E. B., Mudvari, P., Creanga, A., Pegu, A., Derrien-Colemyn, A., Henry, A. R., Gagne, M., Graham, B. S., Wrammert, J., Douek, D. C., Boritz, E., Pinsky. B. A., Suthar, M. S. (2021). Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. bioRxiv. 10.1101/2021.05.09.443299.
- 50.Volkan E, Volkan E. Under the COVID-19 lockdown: Rapid review about the unique case of North Cyprus. Psychological Trauma. 2020;12(5):539–541. doi: 10.1037/tra0000809. [DOI] [PubMed] [Google Scholar]
- 51.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, Gobeil SMC, Kopp M, Li D, Parks R, Hsu AL, Borgnia MJ, Haynes BF, Acharya P. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nature Structural & Molecular Biology. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proceedings of the National Academy of Sciences USA. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Perez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Jr, Hammitt LL, Tureci O, Nell H, Schaefer A, Unal S, Tresnan DB, Mather S, Dormitzer PR, Sahin U, Jansen KU, Gruber WC, C.C.T. Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New England Journal of Medicine. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muik A, Wallisch AK, Sanger B, Swanson KA, Muhl J, Chen W, Cai H, Maurus D, Sarkar R, Tureci O, Dormitzer PR, Sahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 57.Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, 3rd, Leist SR, Schafer A, Nakajima N, Takahashi K, Lee RE, Mascenik TM, Graham R, Edwards CE, Tse LV, Okuda K, Markmann AJ, Bartelt L, de Silva A, Margolis DM, Boucher RC, Randell SH, Suzuki T, Gralinski LE, Kawaoka Y, Baric RS. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert SC. T-cell-inducing vaccines—What’s the future. Immunology. 2012;135(1):19–26. doi: 10.1111/j.1365-2567.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, Li J, Gong X, Lou X, Shi W, Wu D, Zhang H, Zhu L, Deng W, Li Y, Lu J, Li C, Wang X, Yin W, Zhang Y, Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonnafous P, Nicolai MC, Taveau JC, Chevalier M, Barriere F, Medina J, Le Bihan O, Adam O, Ronzon F, Lambert O. Treatment of influenza virus with beta-propiolactone alters viral membrane fusion. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2014;1838(1 Pt B):355–363. doi: 10.1016/j.bbamem.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, Han W, Chen Z, Tang R, Yin W, Chen X, Hu Y, Liu X, Jiang C, Li J, Yang M, Song Y, Wang X, Gao Q, Zhu F. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. The Lancet Infectious Diseases. 2021;21(2):181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Mendonca L, Yang Y, Gao Y, Shen C, Liu J, Ni T, Ju B, Liu C, Tang X, Wei J, Ma X, Zhu Y, Liu W, Xu S, Liu Y, Yuan J, Wu J, Liu Z, Zhang Z, Liu L, Wang P, Zhang P. The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by Cryo-EM and Cryo-ET. Structure. 2020;28(11):1218–1224 e4. doi: 10.1016/j.str.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anywaine Z, Whitworth H, Kaleebu P, Praygod G, Shukarev G, Manno D, Kapiga S, Grosskurth H, Kalluvya S, Bockstal V, Anumendem D, Luhn K, Robinson C, Douoguih M, Watson-Jones D. Safety and immunogenicity of a 2-dose heterologous vaccination regimen with Ad26.ZEBOV and MVA-BN-Filo Ebola vaccines: 12-month data from a phase 1 randomized clinical trial in Uganda and Tanzania. The Journal of Infectious Diseases. 2019;220(1):46–56. doi: 10.1093/infdis/jiz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams K, Bastian AR, Feldman RA, Omoruyi E, de Paepe E, Hendriks J, van Zeeburg H, Godeaux O, Langedijk JPM, Schuitemaker H, Sadoff J, Callendret B. Phase 1 safety and immunogenicity study of a respiratory syncytial virus vaccine with an adenovirus 26 vector encoding prefusion F (Ad26.RSV.preF) in adults aged >/=60 years. The Journal of Infectious Diseases. 2020;222(6):979–988. doi: 10.1093/infdis/jiaa193. [DOI] [PubMed] [Google Scholar]
- 65.Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, Stoop J, Tete S, Van Damme W, Leroux-Roels I, Berghmans PJ, Kimmel M, Van Damme P, de Hoon J, Smith W, Stephenson KE, De Rosa SC, Cohen KW, McElrath MJ, Cormier E, Scheper G, Barouch DH, Hendriks J, Struyf F, Douoguih M, Van Hoof J, Schuitemaker H. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid-19 vaccine. New England Journal of Medicine. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Lubbe JEM, Rosendahl Huber SK, Vijayan A, Dekking L, van Huizen E, Vreugdenhil J, Choi Y, Baert MRM, Feddes-de Boer K, Izquierdo Gil A, van Heerden M, Dalebout TJ, Myeni SK, Kikkert M, Snijder EJ, de Waal L, Stittelaar KJ, Tolboom J, Serroyen J, Muchene L, van der Fits L, Rutten L, Langedijk JPM, Barouch DH, Schuitemaker H, Zahn RC, Wegmann F. Ad26.COV2.S protects Syrian hamsters against G614 spike variant SARS-CoV-2 and does not enhance respiratory disease. NPJ Vaccines. 2021;6(1):39. doi: 10.1038/s41541-021-00301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, Flaxman A, Wright D, Bellamy D, Bittaye M, Dold C, Provine NM, Aboagye J, Fowler J, Silk SE, Alderson J, Aley PK, Angus B, Berrie E, Bibi S, Cicconi P, Clutterbuck EA, Chelysheva I, Folegatti PM, Fuskova M, Green CM, Jenkin D, Kerridge S, Lawrie A, Minassian AM, Moore M, Mujadidi Y, Plested E, Poulton I, Ramasamy MN, Robinson H, Song R, Snape MD, Tarrant R, Voysey M, Watson MEE, Douglas AD, Hill AVS, Gilbert SC, Pollard AJ, Lambe T, C.V.T.G. Oxford T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nature Medicine. 2021;27(2):270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 68.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, Morrison H, Mujadidi YF, Nana A, O'Reilly PJ, Padayachee SD, Pittella A, Plested E, Pollock KM, Ramasamy MN, Rhead S, Schwarzbold AV, Singh N, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Tarrant R, Thomson EC, Torok ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Watson MEE, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ, C.V.T.G. Oxford Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, Padayachee SD, Dheda K, Barnabas SL, Bhorat QE, Briner C, Kwatra G, Ahmed K, Aley P, Bhikha S, Bhiman JN, Bhorat AE, du Plessis J, Esmail A, Groenewald M, Horne E, Hwa SH, Jose A, Lambe T, Laubscher M, Malahleha M, Masenya M, Masilela M, McKenzie S, Molapo K, Moultrie A, Oelofse S, Patel F, Pillay S, Rhead S, Rodel H, Rossouw L, Taoushanis C, Tegally H, Thombrayil A, van Eck S, Wibmer CK, Durham NM, Kelly EJ, Villafana TL, Gilbert S, Pollard AJ, de Oliveira T, Moore PL, Sigal A, Izu A, N.-S.G.W.-V.C. Group Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. New England Journal of Medicine. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moyo-Gwete, T., Madzivhandila, M., Makhado, Z., Ayres, F., Mhlanga, D., Oosthuysen, B., Lambson, B. E., Kgagudi, P., Tegally, H., Iranzadeh, A., Doolabh, D., Tyers, L., Chinhoyi, L. R., Mennen, M., Skelem, S., Marais, G., Wibmer, C. K., Bhiman, J. N., Ueckermann, V., … Moore, P. L. (2021). Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351). New England Journal of Medicine, 384(22), 2161–2163. 10.1056/NEJMc2104192. [DOI] [PMC free article] [PubMed]
- 71.Castelli GP, Pognani C, Sozzi C, Franchini M, Vivona L. Cerebral venous sinus thrombosis associated with thrombocytopenia post-vaccination for COVID-19. Critical Care. 2021;25(1):137. doi: 10.1186/s13054-021-03572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muir KL, Kallam A, Koepsell SA, Gundabolu K. Thrombotic thrombocytopenia after Ad26.COV2.S vaccination. New England Journal of Medicine. 2021;384:1964–1965. doi: 10.1056/NEJMc2105869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medicherla CB, Pauley RA, de Havenon A, Yaghi S, Ishida K, Torres JL. Cerebral venous sinus thrombosis in the COVID-19 pandemic. Journal of Neuro-Ophthalmology. 2020;40(4):457–462. doi: 10.1097/WNO.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 74.Hughes C, Nichols T, Pike M, Subbe C, Elghenzai S. Cerebral venous sinus thrombosis as a presentation of COVID-19. European Journal of Case Reports in Internal Medicine. 2020;7(5):001691. doi: 10.12890/2020_001691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bompard F, Monnier H, Saab I, Tordjman M, Abdoul H, Fournier L, Sanchez O, Lorut C, Chassagnon G, Revel MP. Pulmonary embolism in patients with COVID-19 pneumonia. European Respiratory Journal. 2020;56(1):2001365. doi: 10.1183/13993003.01365-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen, X., Tang, H., Pajon, R., Smith, G., Glenn, G. M., Shi, W., Korber, B., & Montefiori, D. C. (2021). Neutralization of SARS-CoV-2 variants B.1.429 and B.1.351. New England Journal of Medicine. 10.1056/NEJMc2103740.. [DOI] [PMC free article] [PubMed]
- 77.Kalimuddin, S., Tham, C. Y., Qui, M., de Alwis, R., Sim, J. X., Lim, J. M., Tan, H. C., Syenina, A., Zhang, S. L., Le Bert, N., Tan, A. T., Leong, Y. S., Yee, J. X., Ong, E. Z., Ooi, E. E., Bertoletti, A., & Low, J. G. (2021). Early T cell and binding antibody responses are associated with Covid-19 RNA vaccine efficacy onset. Med (N Y). 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed]
- 78.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833 e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheung EW, Zachariah P, Gorelik M, Boneparth A, Kernie SG, Orange JS, Milner JD. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA. 2020;324(3):294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang J, Shen J, Zhu LR, Chen Y, Iacucci M, Ng SC, Ghosh S, Chen MH. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. The Lancet Gastroenterology & Hepatology. 2020;5(7):667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A, Heinrich F, Mushumba H, Kniep I, Schroder AS, Burdelski C, de Heer G, Nierhaus A, Frings D, Pfefferle S, Becker H, Bredereke-Wiedling H, de Weerth A, Paschen HR, Sheikhzadeh-Eggers S, Stang A, Schmiedel S, Bokemeyer C, Addo MM, Aepfelbacher M, Puschel K, Kluge S. Autopsy findings and venous thromboembolism in patients with COVID-19: A prospective cohort study. Annals of Internal Medicine. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conde Cardona G, Quintana Pajaro LD, Quintero Marzola ID, Ramos Villegas Y, Moscote Salazar LR. Neurotropism of SARS-CoV 2: Mechanisms and manifestations. Journal of the Neurological Sciences. 2020;412:116824. doi: 10.1016/j.jns.2020.116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurology. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klein SL, Dhakal S, Ursin RL, Deshpande S, Sandberg K, Mauvais-Jarvis F. Biological sex impacts COVID-19 outcomes. PLoS Pathogens. 2020;16(6):e1008570. doi: 10.1371/journal.ppat.1008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng Y, Wu P, Lu W, Liu K, Ma K, Huang L, Cai J, Zhang H, Qin Y, Sun H, Ding W, Gui L, Wu P. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: A retrospective study of 168 severe patients. PLoS Pathogens. 2020;16(4):e1008520. doi: 10.1371/journal.ppat.1008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J, Ji H, Zheng W, Wu X, Zhu JJ, Arnold AP, Sandberg K. Sex differences in renal angiotensin converting enzyme 2 (ACE2) activity are 17beta-oestradiol-dependent and sex chromosome-independent. Biology of Sex Differences. 2010;1(1):6. doi: 10.1186/2042-6410-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mikkonen L, Pihlajamaa P, Sahu B, Zhang FP, Janne OA. Androgen receptor and androgen-dependent gene expression in lung. Molecular and Cellular Endocrinology. 2010;317(1–2):14–24. doi: 10.1016/j.mce.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 89.Larsen, M. D., de Graaf, E. L., Sonneveld, M. E., Plomp, H. R., Nouta, J., Hoepel, W., Chen, H. J., Linty, F., Visser, R., Brinkhaus, M., Sustic, T., de Taeye, S. W., Bentlage, A. E. H., Toivonen, S., Koeleman, C. A. M., Sainio, S., Kootstra, N. A., Brouwer, P. J. M., Geyer, C. E., … Vidarsson, G. (2021). Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science,371(6532), eabc8378. 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed]
- 90.Chakraborty S, Gonzalez J, Edwards K, Mallajosyula V, Buzzanco AS, Sherwood R, Buffone C, Kathale N, Providenza S, Xie MM, Andrews JR, Blish CA, Singh U, Dugan H, Wilson PC, Pham TD, Boyd SD, Nadeau KC, Pinsky BA, Zhang S, Memoli MJ, Taubenberger JK, Morales T, Schapiro JM, Tan GS, Jagannathan P, Wang TT. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nature Immunology. 2021;22(1):67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klein SL, Pekosz A, Park HS, Ursin RL, Shapiro JR, Benner SE, Littlefield K, Kumar S, Naik HM, Betenbaugh MJ, Shrestha R, Wu AA, Hughes RM, Burgess I, Caturegli P, Laeyendecker O, Quinn TC, Sullivan D, Shoham S, Redd AD, Bloch EM, Casadevall A, Tobian AA. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. The Journal of Clinical Investigation. 2020;130(11):6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY) 2020;12(11):10087–10098. doi: 10.18632/aging.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suryamohan K, Diwanji D, Stawiski EW, Gupta R, Miersch S, Liu J, Chen C, Jiang YP, Fellouse FA, Sathirapongsasuti JF, Albers PK, Deepak T, Saberianfar R, Ratan A, Washburn G, Mis M, Santhosh D, Somasekar S, Hiranjith GH, Vargas D, Mohan S, Phalke S, Kuriakose B, Antony A, Ustav M, Jr, Schuster SC, Sidhu S, Junutula JR, Jura N, Seshagiri S. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Communications Biology. 2021;4(1):475. doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gibson, W. T., Evans, D. M. An, J., & Jones, S. J. (2020). ACE 2 coding variants: A potential X-linked risk factor for COVID-19 disease. bioRxiv. 10.1101/2020.04.05.026633.
- 95.Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, LaBranche CC, Edwards RJ, Sutherland L, Santra S, Mansouri K, Gobeil S, McDanal C, Pardi N, Hengartner N, Lin PJC, Tam Y, Shaw PA, Lewis MG, Boesler C, Sahin U, Acharya P, Haynes BF, Korber B, Montefiori DC. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host & Microbe. 2021;29(1):23–31 e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole A, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, Connor TR. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75 e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield C-GG, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827 e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance. 2021;26(1):2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McCarthy, K. R., Rennick, L. J., Nambulli, S., Robinson-McCarthy, L. R., Bain, W. G., Haidar, G., & Duprex, W. P. (2021). Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. bioRxiv,371(6534), 1139–1142. 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed]
- 101.Eurosurveillance editorial team Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA—First update. Eurosurveillance. 2021;26(3):2101211. doi: 10.2807/1560-7917.ES.2021.26.3.2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]