Abstract
The endemicity of several parasitic diseases across the globe and recent evidence of distress among COVID-19 patients with preexisting parasitic infections requires strengthening One Health framework and advanced strategies for parasitic detection. Owing to the greater sensitivity and accuracy, molecular technologies such as conventional polymerase chain reaction (PCR), reverse transcription (RT)-PCR, nested PCR, loop-mediated isothermal amplification (LAMP), and xMAP technology have been extensively studied for parasitic diagnosis. Varieties of genes have been targeted for primer development where 18S rRNA, internal transcribed spacer regions, and mitochondrial DNAs coding for cytochrome, and other enzymes have been widely used. More recent, low-cost sequencing and advances in big data management have resulted in a slow but steady rise of next-generation sequencing-based approaches for parasite diagnosis. However, except for few parasites of global concerns such as Plasmodium and Entamoeba, most of the molecular tools and technologies are yet to witness bench to bedside and field translations. This review looks into some of the advancements in the molecular diagnosis of parasites that have potential relevance to clinical purposes and may pave the way toward disease management in an efficient and timely manner.
Keywords: Molecular diagnosis, parasites, post-COVID-19, zoonotic disease
INTRODUCTION
Parasitic infections in humans are as old as civilization. There are about 300 helminth worms and over 70 protozoal species that have taken shelter of the human body with an ancestral or zoonotic origin.[1] However, among these, some have resulted in serious health consequences, mostly in the tropical regions, and are endemic.[2] As such, the majority of parasitic infections are less studied in comparison with pathogenic virus and bacteria and have prevailed in the sphere of neglected tropical diseases (NTDs) yet have a considerable burden of diseases.[3]
The emergence of zoonosis with pandemic potentials such as the recent SARS-CoV-2 and the past incidences of SARS-CoV, MERS, Ebola, etc., points to the inevitability of future outbreaks from pathogens of zoonotic origin. However, the past outbreaks are mostly viral; the severity of the infection has been correlated to preexisting parasitic infections. Some preliminary investigations on COVID-19 patients coinfected with malaria as well as other NTDs have indicated potential medico-relevant interactions with a higher level and pro-coagulant state, possibly shift of COVID-19 disease dynamics among lower age groups and enhanced chances of morbidity and mortality.[4,5] In contrast, soil-transmitted helminths such as Ascaris, Trichuris, and Ancylostoma, that preform immunomodulation by lowering cytokine and interleukin-12 production and block the TH1 pathway to evade the inflammatory immune response in human for its coexistence, have been reported to lower risk of COVID-19.[6]
Further, climatic changes are mostly expanding the geography of tropical regions with more hot and humid temperatures and paving the way for a greater dispersal of infections that were mostly confined to some tropical regions.[7,8] In combination, management of zoonosis has gained attention worldwide through the One Health approach by integrating cross-sectors of animal-human and environment.
It is, therefore, expected that the parasites, mostly zoonotic, may find their way across the nations including new territories and could threaten the health security of multiple nations.[3,9] Further, recent evidence of the differential influence of parasitic infections to COVID-19 requires that epidemiology of new, emerging, or remerging infectious agents require attention to the agent itself as well as a detailed understanding of the coexisting parasitic infections.[10,11]
Prompt and accurate detection holds the key for early response and management of infections. Pathogen detection strategies that were incepted with simple microscopy have traveled a long journey to molecular regimes.[12] These have shaped human understandings of host-pathogen interactions and have served as the first line of response in developing medical countermeasures as part of health-care services. Irrespective that each technology that was introduced or under study for parasite diagnostics has its own merits and demerits[13] [Figure 1], the selection of a particular strategy has been mostly guided by accuracy, sensitivity, and affordability. In the current scenario, molecular diagnostics that target specific genes or genomes of an organism for direct detection encompassed the benefits of rapid scientific advancements in terms of big data, automation, low-cost sequencing, etc., and are emerging as the preferred choices for parasite detection.[14]
Figure 1.
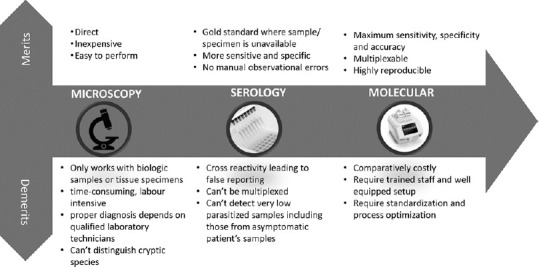
Evolution of parasitic diagnosis technologies with their constraints and benefits. Images inside the circles are for representation purpose and are open source
The reflection of molecular tools and technologies is not well dispersed across categories of microorganisms. Molecular tools are most researched and clinically applied for the diagnosis of bacteria and viruses across the globe. In contrast, ample research has been done on molecular detection of parasites yet used very limited diagnostic purposes except for a few pathogens that have attained a global concern. Even, antimalarial drug resistance of Plasmodium sp. has been tested using molecular approaches by exploring genetic mutations using molecular markers.[15] Sequence analysis of Plasmodium falciparum chloroquine-resistant transporter and multidrug resistance 1 gene fragments from isolates obtained from high and low malaria-endemic areas of India (Pondicherry and Odisha) revealed the varying degree of K76T and N86Y mutations among the isolates of different origin.[16] This warrants large-scale molecular epidemiological studies to identify the nature of antimalarial drug resistance across regions to help develop appropriate regional antimalarial programs.
In light of the parasitic zoonosis, its global spread, and interplay with other pathogens affecting global health security, the present review looks into advancements in the molecular detection of parasites that hold potential clinical relevance to protect and promote One Health safety.
GENE TARGETS FOR PRIMER DESIGNING
Targeting one or multiple highly conserved gene sequences has been widely used as part of the nucleic acid amplification test (NAAT) for the detection of parasites. These sequences act as a signature for particular species wherein a positive amplification guarantees for confirmatory species delineation process. As such, the major attention has been focused on identifying the appropriate target sequences for amplification that holds the key to accurate identification [Table 1]. In this, 18 S rRNA sequences and internal transcribed spacer (ITS) regions were tested extensively. Primers have been developed using partial or full-length 18S rRNA sequence where the 18S rRNA being highly polymorphic and conserved allowed species-level separation and helps to establish phylogenetic relatedness among species.[21,26,30,31] It has been the most preferred choice for malarial diagnostics where multiple NAAT protocols with sensitivity ranging from 1 to 10 parasites/μL have been described.[17,32,33,34] Likewise, 18S rRNA has been preferred most for the detection of different subtypes of Blastocystis, a highly polymorphic parasite, diagnosis of which is very difficult using microscopy.[29,35] In contrast to the protocols used in reverse transcription-polymerase chain reactions (RT-PCRs) that used a single set of primers, the nested PCR protocol used two sets of primers that resulted in greater sensitivity in the detection of four human malaria parasites.[32] Recent findings as a result of genomic data mining of Plasmodium sp. have identified other promising NAAT target sites.[18] In the Plasmodium genome, the 14–41 copies of noncoding subtelomeric repeat sequences Pvr47 and Pfr364 could potentially be targeted for higher sensitivity and detection of mixed infections in the multiplex PCR assay.[36,37]
Table 1.
Examples of primers used for molecular diagnosis of some parasites
| Target parasite | Target gene | Primer sequences | Reference | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Plasmodium genus | 18S | Plasmo 1 5′GTTAAGGGAGTGAAGACGA TCAGA3′ | Plasmo 2 5′AACCCAAAGACTTTGATTTC TCATAA3′ | [17] |
| Plasmodium genus | Pvr47 | 5′TCCGCAGCTCACAAATGTTC3′ | 5′ACATGGGGATTCTAAGCCAATTTA3′ | [18] |
| Plasmodium genus | Pfr364 | 5′ACTCGCAATAACGCTGCAT3′ | 5′TTCCCTGCCCAAAAACGG3′ | [18] |
| Haemosporida | cytb | AE298-EF 5′TGTAATGCCTAGACGTATTCC3′ | AE299-ER 5′GTCAAWCAAACATGAATATAGAC3′ | [19] |
| Haemosporida | co×3 | AE959-F 5′CCATACAATYTCNACRAAATGCC3′ | AE961-R 5′CTGTTATCCCCGGCGAACC3′ | [19] |
| Giardia intestinalis | SSU rRNA | Giardia-80F 5′GACGGCTCAGGACAACGGTT3′ | Giardia-127R 5′TTGCCAGCGGTGT CCG3′ | [20] |
| Cryptosporidium pavaum | SSU rRNA | CrF 5′CGCTTCTCTAGCCTTTCATGA3′ | CrR 5′CTTCACGTGTGTTTGCC AT3′ | [20] |
| Ascaris lumbricoides | 18S | 5′GCTGCGTTCTTCATCGAC3′ 5′CCATGCATGTCTAAGTTCAA3′ | 5′CARAAAWTCGGAGCTTTGGT3′ | [21] |
| Entamoeba genus | 16S-like rRNA gene | E-1 5′TAAGATGCACGAGAGCGAAA3′ | 5′GTACAAAGGGCAGGGACGTA3′ | [22] |
| Entamoeba histolytica | 16S-like rRNA gene | EH-1 5’AAGCATTGTTTCTAGATCTGAG3’ | EH-2 5’AAGAGGTCTAACCGAAATTAG3’ | [22] |
| Entamoeba moshkovskii | 16S-like rRNA gene | Mos-1 5′GAAACCAAGAGTTTCACAAC3′ | Mos-2 5′CAATATAAGGCTTGGATGAT3′ | [22] |
| Entamoeba dispar | 16S-like rRNA gene | ED-1 5’TCTAATTTCGATTAGAACTCT3’ | ED-2 5’TCCCTACCTATTAGACATAGC3’ | [22] |
| Leishmania genus | kDNA | LEISH-1 5′GGTTCCTTTCCTGATTTACG3′ P223 5′AACTTTTCTGGTCCTCCGGGTAG3′ | LEISH-2 5′ACCCCCAGTTTCCCGCC3’ | [23] |
| Leishmania genus | kDNA | P221 5′TCCCATCGCAACCTCGGT3′ | P332 5′GGCCGGTAAAGGCCGAATAG3′ P333 5′AAGCGGGCGCGGTGCTG3′ | [24] |
| Onchocerca volvulus, Mansonella ozzardi, and Mansonella perstans | 18S and ITS | FIL-1F 5′GTGCTGTAACCATTACCGAAAGG3′FIL-2F 5′GGTGAACCTGCGGAAGGATC3′ | FIL-2R (ITS specific) 5′TGCTTATTAAGTCTACTTAA3’ | [25] |
| Strongyloides stercoralis | 18 S rRNA | 5′ACCGTAAACTATGCCTACTAGA3′ | 5′AACCACTAAATCATGAAAGAGCTA3′ | [14] |
| Taenia saginata, Taenia asiatica, and Taenia solium | 18 S rRNA | 5′AGTCGGCGACGGGTCCTT3′ | 5′GAACCTAACGACATAACATAATGA3′ | [21] |
| Enterobius vermicularis | 18 S rRNA | 5′CGCCCTAGTTCTGACCGTAA3′ | 5′GGAGGATTTTCAGGGGGTTA3′ | [26] |
| Trypanosoma genus | ITS | KIN1 5′GCGTTCAAAGATTGGGCAAT3′ | KIN2 5′CGCCCGAAAGTTCACC3′ | [27] |
| Trypanosoma genus | ITS | ITS1 5′CCGGAAGTTCACCGATATTG3′ | ITS1 BR 5′TTGCTGCGTTCTTCAACGAA3′ | [28] |
| Blastocystis | 18S rRNA | RD5 5’ATCGCCACTTCTCCAAT3’ | BhRDr 5’GAGCTTTTTAACTGCAACAACG3’ | [29] |
Likewise, ITS regions of rRNA have been another popular target choice for the primer construction targeting flanking and/or within the genes. ITS-1 and ITS-2 bearing species-specific signatures and for other properties such as the short length of <800 bp, sequence repetition, and homogenization have been used extensively for the identification of different parasites.[27,28,38] However, ITS-based targeting could not delineate closely related species because of sequence similarities. As such, it is hard to delineate Ascaris lumbricoides (human roundworm) and Ascaris suum (pig roundworm) on ITS-targeted NAAT that also presents difficulties toward estimation of the zoonotic spread of A. suum infection in humans worldwide.[39]
Among other gene targets, mitochondrial DNAs such as genes coding for cytochrome oxidase, cytochrome b, NADH dehydrogenase, etc., have been widely used.[40,41,42,43] This is because most of the parasites except some such as Cryptosporidium parvum contain multiple copies of mitochondria hosting unique gene sequences with comparatively higher evolutionary rates from ITS regions. This has increased chances of amplification and species-specific sensitivity allowing effective separation of cryptic (genetically diverged but morphologically similar) species.[19,43]
MOLECULAR TESTING STRATEGIES
Conventional PCR occupied the initial phases of parasites molecular detection strategies, however, later moved to more refined process such as RT-PCR, nested PCR, loop-mediated isothermal amplification (LAMP), xMAP, etc. Of these, RT-PCR and nested PCR have been explored most both in Research and Development for commercialization purposes in a single or multiplexed format. RT-PCR, a widely used term in clinical pathogen diagnostics, uses fluorescent-tagged primers and allows real-time quantification of the target gene and visualization of amplicons without the need for further processing. Using the optimized protocols, the RT-PCR has detected Plasmodium, Giardia, Cryptosporidium, Leishmania, etc., in a short turnaround time within 3 h.[23,44] In multiplex format, the different fluorophore-tagged primers are used for different target species and allow sensitive detection of mixed species infections in a single reaction. It has been used for the differentiation of species of Plasmodium,[45] mixed infection of diarrhea-causing parasitic protozoa (Giardia intestinalis, Cryptosporidium spp., and Entamoeba histolytica),[20] and even been used to target up to eight parasites, namely C. parvum, G. intestinalis, E. histolytica, Blastocystis hominis, Dientamoeba fragilis, Clonorchis sinensis, Metagonimus yokogawai, and Gymnophalloides seoi in a customized protocol suitable for Korean population.[46] Several commercially approved assays and kits have been developed for the detection of parasites using RT-PCR where some comparative studies on their performances indicated differences in kit-specific sensitivity and specificity.[47,48,49]
In contrast, nested PCR uses two different sets of primers targeting different parts of a gene and offers more sensitive detection, especially delineating mixed infections, yet requires postamplification processing for amplicon visualization and results in interpretation. It has offered a great advantage to detect E. histolytica infection. The parasite has a strong global footprint with major prevalence in developing nations and is one of the top three parasitic causes of mortality across the globe. However, the parasite sometimes remains asymptomatic and morphologically indistinguishable in its cyst and trophozoite stages from nonpathogenic Entamoeba dispar and Entamoeba moshkovskii. These present challenges in microscopic examination of stool and result in very low sensitivity (60%). This was overcome by the use of different molecular diagnosis strategies, some of which have been extensively reviewed earlier.[50] The nested PCR approach has only allowed highly sensitive detection of these three species in stool samples using SSU rDNA as a primer.[51] To understand the genetic basis of the target site for allowing species-specific identification of Entamoeba sp., 16S-like rRNA gene of E. histolytica (439 bp), E. moshkovskii (553 bp), and E. dispar (174 bp) was screed for mutations.[52] The combination of riboprinting and SSCP analysis had revealed several point mutations across the genes that possibly hint toward interspecies genetic signatures for Entamoeba sp.
Nested PCR has also been tested to distinguish Plasmodium sp.,[53] Leishmania sp.,[24] Onchocerca volvulus, Mansonella ozzardi, Mansonella perstans,[25] etc. Multiplexing in nested PCR was also introduced where multiple primer sets for different species have been used in a single tube run that saved time and offered equal sensitivity as monospecies targeted nested PCR. Using this technique, Hu et al. (2015) had simultaneously detected Ancylostoma ceylanicum, A. caninum, and G. intestinalis.54 Likewise, Saito et al. (2018) had identified five human Plasmodium species in a single PCR tube multiplex assay.55 This techniques was also useful for accurate differentiation of pathogenic E. histolytica, from nonpathogenic E. moshkovskii and E. dispar in stool sample.22 Considering that morphometric differentiation between these Entamoeba species is impossible, such multiplex nested PCR has offered potential diagnostic benefits including accuracy and sensitivity. It has also been used to detect E. histolytica DNA in liver abscess pus and urine among patients with amoebic liver abscess (ALA).[56] Likewise, the same group also detected E. histolytica DNA in saliva using 16S-like rRNA gene-based nested multiplex PCR.[57] These studies, for the first-time established presence of E. histolytica DNA in the urine and saliva of ALA patients. This suggests potential clinical use of the technology being non-invasive in nature. Also, it offers edges toward differentiation from pyogenic liver abscesses that appears identical in available imaging techniques.
Different other forms such as semi-nested PCR have also been used mostly for identification of Plasmodium infection.[58,59] Irrespective of the type of nested PCR protocol used, some studies have indicated possibilities of false amplification leading to error-prone reporting. For example, in a study on avian model infected with Haemoproteus, Plasmodium, and Leucocytozoon sp., the nested PCR had amplified larger Leucocytozoon fragments and reported a false positive.[60] This emphasizes a cautious use of nested PCR with multiple checkpoints including an appropriate selection of primers having different annealing temperatures.
Parasitic endemic regions expect analysis of large quantities of samples and demand short turnaround time where RT and nested PCRs mostly struggle. Procedural complexity and data interpretation strategies involved in these PCRs make it difficult to manage higher sample load effortless. For malarial diagnosis, the challenges have been addressed by the introduction of “direct PCR,” both nested and single format using mitochondrial COXIII gene as a primer, and showed higher sensitivity over the previously used techniques.[61] This direct PCR eliminates DNA extraction and purification steps that result in a more rapid turnaround time (1 h), prevents chances of cross-contamination, and eliminates procedural complexity making it highly suitable for commercial purposes.
LAMP, a non-PCR-based DNA amplification strategy, has gained importance in parasite detection. It is rapidly evolving as a promising point-of-care (POC) technology as it is thermal cycler independent, rapid, inexpensive, and easy to perform. Several commercial kits are now available for molecular diagnosis of human African trypanosomiasis, malaria, and leishmaniasis.[62] Recently, as a proof-of-concept portable system for the molecular diagnosis of tropical diseases, the Q3-Plus system has been used.[63] It used a disposable, silicon-based cartridge, preloaded with quantitative PCR reagents for parasite detection in a portable Q3-Plus instrument that integrates PCR technology, electrical temperature control, illumination, and optical detection.[64] For the detection of parasites such as Trypanosoma cruzi and Plasmodium sp., the Q3-Plus system has shown excellent sensitivity (limit of detection of 1.5 parasites/μL) and 100% specificity, thereby offering great POC application both for the field and bedside use.[63]
Unlike other PCR-based molecular techniques that require maintaining different temperature profile during the reaction, LAMP is typically performed at a constant temperature (60°C–65°C) where a Bst DNA polymerase with its strand displacement activity amplifies target genes in the presence of six primer sets and results and accumulates almost 109 copies of target DNA within an hour.[65] The use of a larger number of primer sets and conditional amplification subject to all primer recognition of target makes the process highly specific and sensitive. Amplification results in the production of pyrophosphate that increases turbidity and confirms a positive test. It has been used for the detection of helminths, Plasmodium, Entamoeba, Trypanosoma, Cryptosporidium, etc., and has been extensively reviewed.[62,66,67] It has also been multiplexed LAMP to detect multiple species in mixed infection[68,69,70] and was modified further to enhance sensitivity.[71] In a study, Mohon et al. applied ultrasensitive LAMP and had reported a sensitivity of 97.0% and specificity of 99.1% for the detection of Plasmodium species in asymptomatic individuals from Africa and Asia.[71] In general, LAMP offers sensitivity and specificities above 80% and 94%, respectively, for human leishmaniasis and sensitivity below 0.01 ng of DNA/reaction with 100% specificity for helminth diagnosis.
xMAP technology, a registered technology of Luminex Corporation that was invented in the 1990s, uses different sets of fluorophore-tagged microspheres in a liquid suspension. This has enabled high-throughput multiplex detection of pathogens when microspheres were covalently bound to target nucleic acid, thereby serving as a probe.[72] However, the nucleic acid-based xMAP analysis requires PCR amplification of target genes followed by DNA-DNA hybridization. As such, the process is strictly labor-intensive and complex yet capable of identifying single-nucleotide differences. Using the ML-2 regions of each species as oligonucleotide probes, xMAP was used successfully to separate C. hominis and C. parvum that no other traditional assay could perform.[73] Likewise, it was also used to correctly identify E. histolytica, E. dispar, Entamoeba hartmanni, Entamoeba coli, and E. moshkovskii in mixed infection with 100% accuracy.[74]
The fourth industrial revolution has accumulated more technical advances in big data, sequencing, low operational costs, and more simplified instrument and analytic interfaces. This has already popularized metagenomic investigations through next-generation sequencing (NGS) across any sources including nonculturable species. Compared to the NGS analysis of bacteria, and virus populations, the use of NGS in parasite biology is new and is facing challenges due to the limited availability of primers suitable for NGS and limited whole-genome sequences of parasites in the database to compare.[75,76] However, the future looks promising with greater attention to it and is offering new opportunities in parasite diagnostics. NGS has been used for the detection of Leishmania in the bone marrow of visceral leishmaniasis,[77] accurate diagnosis of neurocysticercosis infection in patients with atypical clinical manifestations,[78] diarrheal infections,[74] and detection of blood-borne helminths and protozoa.[79,80]
CONCLUSION
The large varieties of parasites, their global footprint, asymptomatic presence, and related influences on disease dynamics due to coinfection mandate the commissioning of an efficient mechanism for parasitic detection where the molecular diagnosis could play the catalytic roles. However, most clinical laboratories, especially in developing and underdeveloped nations, still prefer serological and/or morphological tests either due to limited resources, comparatively higher cost, and technical constraints. Most of these molecular tests today require sophisticated laboratory facilities and trained workforce that always presented a challenge toward POC application and applicability in endemic nations, most of which have very limited health-care support infrastructure and funds. As such, molecular tests of parasites are not rewarded to the extent proportional to the benefits it offers in detection.
The molecular diagnostics technologies can identify cryptic species, differentiate mixed infection, and detect sub-microscopic parasitic load in a highly sensitive and accurate manner. Technologies such as direct PCR, LAMP, and the recent chip-based prototype Q3-Plus platform offer easy to perform rapid turnaround tools for parasitic diagnosis with good sensitivity and specificity. Therefore, it is critically needed to integrate and introduce molecular diagnostics for patient care in a phased manner, especially when the contribution of parasitic infection to pandemic severity has been established. Although the ongoing R and Ds and steady scientific advancements may soon outperform the current challenges, an increase in funding for capacity building in parasitic molecular diagnostics is highly recommended.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Cox FE. History of human parasitology. Clin Microbiol Rev. 2002;15:595–612. doi: 10.1128/CMR.15.4.595-612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotez PJ. Ten global “Hotspots” for the neglected tropical diseases. PLoS Negl Trop Dis. 2014;8:e2496. doi: 10.1371/journal.pntd.0002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisarski K. The global burden of disease of zoonotic parasitic diseases: Top 5 contenders for priority consideration. Trop Med Infect Dis. 2019;4:44. doi: 10.3390/tropicalmed4010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutman JR, Lucchi NW, Cantey PT, Steinhardt LC, Samuels AM, Kamb ML, et al. Malaria and parasitic neglected tropical diseases: Potential syndemics with COVID-19? Am J Trop Med Hyg. 2020;103:572–7. doi: 10.4269/ajtmh.20-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdoli A. Helminths and COVID-19 co-infections: A neglected critical challenge. ACS Pharmacol Transl Sci. 2020;3:1039–41. doi: 10.1021/acsptsci.0c00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cepon-Robins TJ, Gildner TE. Old friends meet a new foe: A potential role for immune-priming parasites in mitigating COVID-19 morbidity and mortality. Evol Med Public Health. 2020;2020:234–48. doi: 10.1093/emph/eoaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen MK, Reinemeyer CR, Leathwick D, Sauermann C. Environmental factors affecting parasite transmission. In: Nielsen MK, Reinemeyer CR, editors. Handbook of Equine Parasite Control. New Jersey, United States: Wiley; 2018. pp. 45–5. 3. [Google Scholar]
- 8.Yan C, Liang LJ, Zheng KY, Zhu XQ. Impact of environmental factors on the emergence, transmission and distribution of Toxoplasma gondii. Parasit Vectors. 2016;9:137. doi: 10.1186/s13071-016-1432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kołodziej-Sobocińska M. Factors affecting the spread of parasites in populations of wild European terrestrial mammals. Mammal Res. 2019;64:301–18. [Google Scholar]
- 10.de Souza W. COVID-19 and parasitology. Parasitol Res. 2020;119:2369–70. doi: 10.1007/s00436-020-06719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradbury RS, Piedrafita D, Greenhill A, Mahanty S. Will helminth co-infection modulate COVID-19 severity in endemic regions? Nat Rev Immunol. 2020;20:342. doi: 10.1038/s41577-020-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smolinski MS, Hamburg MA, Lederberg J. New Jersey, United States: National Academies Press (US); 2003. Pathogen discovery, detection, and diagnostics. In: Microbial Threats to Health: Emergence, Detection, and Response. [PubMed] [Google Scholar]
- 13.Pritt BS. Molecular diagnostics in the diagnosis of parasitic infection. Methods Microbiol. 2015;42:111–60. [Google Scholar]
- 14.Wong SS, Fung KS, Chau S, Poon RW, Wong SC, Yuen KY. Molecular diagnosis in clinical parasitology: When and why? Exp Biol Med (Maywood) 2014;239:1443–60. doi: 10.1177/1535370214523880. [DOI] [PubMed] [Google Scholar]
- 15.Parija SC, Praharaj I. Drug resistance in malaria. Indian J Med Microbiol. 2011;29:243–8. doi: 10.4103/0255-0857.83906. [DOI] [PubMed] [Google Scholar]
- 16.Antony HA, Das S, Parija SC, Padhi S. Sequence analysis of pfcrt and pfmdr1 genes and its association with chloroquine resistance in Southeast Indian Plasmodium falciparum isolates. Genom Data. 2016;8:85–90. doi: 10.1016/j.gdata.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–43. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaral LC, Robortella DR, Guimarães LF, Limongi JE, Fontes CJ, Pereira DB, et al. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of Platyhelminth parasites. Malar J. 2019;18:154. [Google Scholar]
- 19.Vilas R, Criscione CD, Blouin MS. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of Platyhelminth parasites. Parasitology. 2005;131:839–46. doi: 10.1017/S0031182005008437. [DOI] [PubMed] [Google Scholar]
- 20.Nazeer JT, El Sayed Khalifa K, von Thien H, El-Sibaei MM, Abdel-Hamid MY, Tawfik RA, et al. Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol Res. 2013;112:595–601. doi: 10.1007/s00436-012-3171-8. [DOI] [PubMed] [Google Scholar]
- 21.Leles D, Araújo A, Vicente AC, Iñiguez AM. Molecular diagnosis of ascariasis from human feces and description of a new Ascaris sp.genotype in Brazil. Vet Parasitol. 2009;163:167–70. doi: 10.1016/j.vetpar.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 22.Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7:47. doi: 10.1186/1471-2180-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sales KG, Miranda DE, Paiva MH, Figueredo LA, Otranto D, Dantas-Torres F. Fast multiplex real-time PCR assay for simultaneous detection of dog and human blood and Leishmania parasites in sand flies. Parasit Vectors. 2020;13:131. doi: 10.1186/s13071-020-3994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deepachandi B, Weerasinghe S, Soysa P, Karunaweera N, Siriwardana Y. A highly sensitive modified nested PCR to enhance case detection in leishmaniasis. BMC Infect Dis. 2019;19:623. doi: 10.1186/s12879-019-4180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang TH, López-Vélez R, Lanza M, Shelley AJ, Rubio JM, Luz SL. Nested PCR to detect and distinguish the sympatric filarial species Onchocerca volvulus, Mansonella ozzardi and Mansonella perstans in the Amazon Region. Mem Inst Oswaldo Cruz. 2010;105:823–8. doi: 10.1590/s0074-02762010000600016. [DOI] [PubMed] [Google Scholar]
- 26.Zelck UE, Bialek R, Weiss M. Molecular phylogenetic analysis of Enterobius vermicularis and development of an 18S ribosomal DNA-targeted diagnostic PCR. J Clin Microbiol. 2011;49:1602–4. doi: 10.1128/JCM.02454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin GL, Ssenyonga SS, Nanteza E, Rubaire-Akiki, Wafula O, Hansen RD, et al. PCR based detection and typing of parasites. In: Zcel MA, Alkan MZ, editors. Parasitology for the 20th Century. Wallingford: CAB International; 1996. pp. 261–87. [Google Scholar]
- 28.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–92. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 29.Padukone S, Mandal J, Rajkumari N, Bhat BV, Swaminathan RP, Parija SC. Detection of Blastocystis in clinical stool specimens using three different methods and morphological examination in Jones' medium. Trop Parasitol. 2018;8:33–40. doi: 10.4103/tp.TP_4_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters AP. The ribosomal RNA genes of Plasmodium. Adv Parasitol. 1994;34:33–79. doi: 10.1016/s0065-308x(08)60136-0. [DOI] [PubMed] [Google Scholar]
- 31.Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP, et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–7. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 32.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–20. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 33.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, et al. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43:2435–40. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy SC, Prentice JL, Williamson K, Wallis CK, Fang FC, Fried M, et al. Real-time quantitative reverse transcription PCR for monitoring of blood-stage Plasmodium falciparum infections in malaria human challenge trials. Am J Trop Med Hyg. 2012;86:383–94. doi: 10.4269/ajtmh.2012.10-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padukone S, Mandal J, Parija SC. Severe Blastocystis subtype 3 infection in a patient with colorectal cancer. Trop Parasitol. 2017;7:122–4. doi: 10.4103/tp.TP_87_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demas A, Oberstaller J, DeBarry J, Lucchi NW, Srinivasamoorthy G, Sumari D, et al. Applied genomics: Data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA. J Clin Microbiol. 2011;49:2411–8. doi: 10.1128/JCM.02603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med. 2015;12:e1001788. doi: 10.1371/journal.pmed.1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roeber F, Jex AR, Gasser RB. Next-generation molecular-diagnostic tools for gastrointestinal nematodes of livestock, with an emphasis on small ruminants: A turning point? Adv Parasitol. 2013;83:267–333. doi: 10.1016/B978-0-12-407705-8.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parasites – Ascariasis. [[Last accessed on 2021 Feb 04]]. Available from: https://www.cdc.gov/parasites/ascariasis/index.html .
- 40.Vasoo S, Pritt BS. Molecular diagnostics and parasitic disease. Clin Lab Med. 2013;33:461–503. doi: 10.1016/j.cll.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Singh B. Molecular methods for diagnosis and epidemiological studies of parasitic infections. Int J Parasitol. 1997;27:1135–45. doi: 10.1016/s0020-7519(97)00111-2. [DOI] [PubMed] [Google Scholar]
- 42.Polley SD, Mori Y, Watson J, Perkins MD, González IJ, Notomi T, et al. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–71. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacheco MA, Cepeda AS, Bernotienė R, Lotta IA, Matta NE, Valkiūnas G, et al. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int J Parasitol. 2018;48:657–70. doi: 10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madison-Antenucci S, Relich RF, Doyle L, Espina N, Fuller D, Karchmer T, et al. Multicenter evaluation of BD max enteric parasite real-time PCR assay for detection of Giardia duodenalis, Cryptosporidium hominis, Cryptosporidium parvum, and Entamoeba histolytica. J Clin Microbiol. 2016;54:2681–8. doi: 10.1128/JCM.00765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shokoples SE, Ndao M, Kowalewska-Grochowska K, Yanow SK. Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J Clin Microbiol. 2009;47:975–80. doi: 10.1128/JCM.01858-08. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Won EJ, Kim SH, Kee SJ, Shin JH, Suh SP, Chai JY, et al. Multiplex real-time PCR assay targeting eight parasites customized to the Korean population: Potential use for detection in diarrheal stool samples from gastroenteritis patients. PLoS One. 2016;11:e0166957. doi: 10.1371/journal.pone.0166957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuin NA, Tan AF, Lew YL, Piera KA, William T, Rajahram GS, et al. Comparative evaluation of two commercial real-time PCR kits (QuantiFast™ and abTES™) for the detection of Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. Malar J. 2020;19:306. doi: 10.1186/s12936-020-03379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulos S, Saugar JM, de Lucio A, Fuentes I, Mateo M, Carmena D. Comparative performance evaluation of four commercial multiplex real-time PCR assays for the detection of the diarrhoea-causing protozoa Cryptosporidium hominis/parvum, Giardia duodenalis and Entamoeba histolytica. PLoS One. 2019;14:e0215068. doi: 10.1371/journal.pone.0215068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartmeyer GN, Hoegh SV, Skov MN, Dessau RB, Kemp M. Selecting PCR for the diagnosis of intestinal parasitosis: Choice of targets, evaluation of in-house assays, and comparison with commercial kits. J Parasitol Res. 2017;2017:6205257. doi: 10.1155/2017/6205257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parija SC, Mandal J, Ponnambath DK. Laboratory methods of identification of Entamoeba histolytica and its differentiation from look-alike Entamoeba spp. Trop Parasitol. 2014;4:90–5. doi: 10.4103/2229-5070.138535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parija SC, Khairnar K. Entamoeba moshkovskii and Entamoeba dispar-associated infections in Pondicherry, India. J Health Popul Nutr. 2005;23:292–5. [PubMed] [Google Scholar]
- 52.Parija SC, Khairnar K. Mutation detection analysis of a region of 16S-like ribosomal RNA gene of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii. BMC Infect Dis. 2008;8:131. doi: 10.1186/1471-2334-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snounou G, Singh B. Vol. 72. New Jersey, United States: Methods in Molecular Medicine™; 2002. Malaria Methods and Protocols; p. 189. [Google Scholar]
- 54.Hu W, Wu S, Yu X, Abullahi AY, Song M, Tan L, et al. A Multiplex PCR for Simultaneous Detection of Three Zoonotic Parasites Ancylostoma ceylanicum, A. caninum, and Giardia lamblia Assemblage A. Biomed Res Int. 2015;2015:406168. doi: 10.1155/2015/406168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito T, Kikuchi A, Kaneko A, Isozumi R, Teramoto I, Kimura M, et al. Rapid and sensitive multiplex single-tube nested PCR for the identification of five human Plasmodium species. Parasitol Int. 2018;67:277–83. doi: 10.1016/j.parint.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Parija SC, Khairnar K. Detection of excretory Entamoeba histolytica DNA in the urine, and detection of E. histolytica DNA and lectin antigen in the liver abscess pus for the diagnosis of amoebic liver abscess. BMC Microbiol. 2007;7:41. doi: 10.1186/1471-2180-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khairnar K, Parija SC. Detection of Entamoeba histolytica DNA in the saliva of amoebic liver abscess patients who received prior treatment with metronidazole. J Health Popul Nutr. 2008;26:418–25. doi: 10.3329/jhpn.v26i4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ongagna-Yhombi SY, Corstjens P, Geva E, Abrams WR, Barber CA, Malamud D, et al. Improved assay to detect Plasmodium falciparum using an uninterrupted, semi-nested PCR and quantitative lateral flow analysis. Malar J. 2013;12:74. doi: 10.1186/1475-2875-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubio JM, Benito A, Roche J, Berzosa PJ, García ML, Micó M, et al. Semi-nested, multiplex polymerase chain reaction for detection of human malaria parasites and evidence of Plasmodium vivax infection in Equatorial Guinea. Am J Trop Med Hyg. 1999;60:183–7. doi: 10.4269/ajtmh.1999.60.183. [DOI] [PubMed] [Google Scholar]
- 60.Szöllsi E, Hellgren O, Hasselquist D. A cautionary note on the use of nested PCR for parasite screening – An example from avian blood parasites. J Parasitol. 2008;94:562–4. doi: 10.1645/GE-1286.1. [DOI] [PubMed] [Google Scholar]
- 61.Echeverry DF, Deason NA, Davidson J, Makuru V, Xiao H, Niedbalski J, et al. Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene. Malar J. 2016;15:128. doi: 10.1186/s12936-016-1185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nzelu CO, Kato H, Peters NC. Loop-mediated isothermal amplification (LAMP): An advanced molecular point-of-care technique for the detection of Leishmania infection. PLoS Negl Trop Dis. 2019;13:e0007698. doi: 10.1371/journal.pntd.0007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rampazzo RC, Graziani AC, Leite KK, Surdi JA, Biondo CA, Costa ML, et al. Proof of concept for a portable platform for molecular diagnosis of tropical diseases: On-chip ready-to-use real-time quantitative PCR for detection of Trypanosoma cruzi or Plasmodium spp. J Mol Diagn. 2019;21:839–51. doi: 10.1016/j.jmoldx.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 64.Cereda M, Cocci A, Cucchi D, Raia L, Pirola D, Bruno L, et al. Q3: A compact device for quick, high precision qPCR. Sensors (Basel) 2018;18:2583. doi: 10.3390/s18082583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sirichaisinthop J, Buates S, Watanabe R, Han ET, Suktawonjaroenpon W, Krasaesub S, et al. Evaluation of loop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am J Trop Med Hyg. 2011;85:594–6. doi: 10.4269/ajtmh.2011.10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng MH, Zhong LY, Kamolnetr O, Limpanont Y, Lv ZY. Detection of helminths by loop-mediated isothermal amplification assay: A review of updated technology and future outlook. Infect Dis Poverty. 2019;8:20. doi: 10.1186/s40249-019-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, et al. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–8. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue N, et al. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods. 2007;71:281–7. doi: 10.1016/j.mimet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 70.Moonga LC, Hayashida K, Kawai N, Nakao R, Sugimoto C, Namangala B, et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) method for simultaneous detection of spotted fever group Rickettsiae and malaria parasites by dipstick DNA chromatography. Diagnostics (Basel) 2020;10:897. doi: 10.3390/diagnostics10110897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohon AN, Getie S, Jahan N, Alam MS, Pillai DR. Ultrasensitive loop mediated isothermal amplification (US-LAMP) to detect malaria for elimination. Malar J. 2019;18:350. doi: 10.1186/s12936-019-2979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reslova N, Michna V, Kasny M, Mikel P, Kralik P. xMAP technology: Applications in detection of pathogens. Front Microbiol. 2017;8:55. doi: 10.3389/fmicb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandyopadhyay K, Kellar KL, Moura I, Casaqui Carollo MC, Graczyk TK, Slemenda S, et al. Rapid microsphere assay for identification of Cryptosporidium hominis and Cryptosporidium parvum in stool and environmental samples. J Clin Microbiol. 2007;45:2835–40. doi: 10.1128/JCM.00138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santos HL, Bandyopadhyay K, Bandea R, Peralta RH, Peralta JM, Da Silva AJ. LUMINEX®: A new technology for the simultaneous identification of five Entamoeba spp.commonly found in human stools. Parasit Vectors. 2013;6:69. doi: 10.1186/1756-3305-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kounosu A, Murase K, Yoshida A, Maruyama H, Kikuchi T. Improved 18S and 28S rDNA primer sets for NGS-based parasite detection. Sci Rep. 2019;9:15789. doi: 10.1038/s41598-019-52422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joensen KG, Engsbro AL, Lukjancenko O, Kaas RS, Lund O, Westh H, Aarestrup FM. Evaluating next-generation sequencing for direct clinical diagnostics in diarrhoeal disease. Eur J Clin Microbiol Infect Dis. 2017;36:1325–38. doi: 10.1007/s10096-017-2947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen H, Fan C, Gao H, Yin Y, Wang X, Zhang Y, et al. Leishmaniasis diagnosis via metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2020;10:528884. doi: 10.3389/fcimb.2020.528884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu P, Weng X, Zhou J, Xu X, He F, Du Y, et al. Next generation sequencing based pathogen analysis in a patient with neurocysticercosis: A case report. BMC Infect Dis. 2018;18:113. doi: 10.1186/s12879-018-3015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flaherty BR, Talundzic E, Barratt J, Kines KJ, Olsen C, Lane M, et al. Restriction enzyme digestion of host DNA enhances universal detection of parasitic pathogens in blood via targeted amplicon deep sequencing. Microbiome. 2018;6:164. doi: 10.1186/s40168-018-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flaherty BR, Barratt J, Lane M, Talundzic E, Bradbury RS. Sensitive universal detection of blood parasites by selective pathogen-DNA enrichment and deep amplicon sequencing. Microbiome. 2021;9:1. doi: 10.1186/s40168-020-00939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


