Abstract
Stemona sessilifolia (Miq.) Miq., commonly known as Baibu, is one of the most popular herbal medicines in Asia. In the Chinese Pharmacopoeia, Baibu has multiple authentic sources and there are many similar herbs sold as Baibu in herbal medicine markets. The existence of counterfeits of Baibu brings challenges to its identification. To assist in its accurate identification, we sequenced and analyzed the complete chloroplast genome of S. sessilifolia using next-generation sequencing technology. The genome was found to be 154,037 bp in length, possessing a typical quadripartite structure consisting of a pair of inverted repeats (IRs: 27,090 bp) separated by a large single copy (LSC: 81,949 bp) and a small single copy (SSC: 17,908 bp). A total of 112 unique genes were identified, including 80 protein-coding, 28 transfer RNA and four ribosomal RNA genes. In addition, 45 tandem, 27 forward, 23 palindromic and 104 simple sequence repeats were detected in the genome by repeated analysis. Compared with its counterfeits (Asparagus officinalis and Carludovica palmata) we found that IR expansion and SSC contraction events of S. sessilifolia resulted in two copies of the rpl22 gene in the IR regions and a partial duplication of the ndhF gene in the SSC region. An approximately 3-kb-long inversion was also identified in the LSC region, leading to the petA and cemA genes being presented in the complementary strand of the chloroplast DNA molecule. Comparative analysis revealed some highly variable regions, including trnF-GAA_ndhJ, atpB_rbcL, rps15_ycf1, trnG-UCC_trnR-UCU, ndhF_rpl32, accD_psaI, rps2_rpoC2, trnS-GCU_trnG-UCC, trnT-UGU_trnL-UAA and rps16_trnQ-UUG. Finally, gene loss events were investigated in the context of phylogenetic relationships. In summary, the complete plastome of S. sessilifolia will provide valuable information for the distinction between Baibu and its counterfeits and assist in elucidating the evolution of S. sessilifolia.
Introduction
Radix Stemonae, also known as Baibu, is one of the most popular herbal medicines used in many Asian countries, including China, Korea, Japan, Thailand and Vietnam. It has been used for the treatment of various respiratory diseases such as bronchitis, pertussis and tuberculosis [1, 2]. It was also used for killing cattle parasites, agricultural pests and domestic insects [3, 4]. Stenine B, one of the major chemical ingredients of Baibu, has been considered as a potential drug candidate for use against Alzheimer’s disease due to its significant acetylcholinesterase inhibitory activity [5]. Owing to the important medicinal value, extensive genetic, biochemical and pharmacological studies on Baibu are needed.
According to the Pharmacopoeia of the People’s Republic of China (2015 edition), the root tubers of Stemona tuberosa, S. japonica and S. sessilifolia were all considered to be authentic sources of Baibu. Although these three species have all been employed as the raw materials of Baibu, we cannot ignore their inherent differences. For example, alkaloids from the genus Stemona are the major components responsible for Baibu’s antitussive activities. However, the chemical composition and content of the different members of the genus S. tuberosa, S. japonica and S. sessilifolia vary greatly [6, 7]. These three species differ in their antitussive, anti-bacterial and insecticidal activities [8]. Therefore, it is critical to determine the exact origin of the plant material used as Baibu.
The existence of multiple authentic sources and the similarities between species increase the difficulty for correctly identifying Baibu. In some areas of China, another herbal medicine, Aconitum kusnezoffii Rchb., is also known as Baibu. However, the therapeutic activity of A. kusnezoffii is significantly different from the authentic sources of Baibu described in the Chinese Pharmacopoeia. Research has even reported that this alternative might result in toxicity when A. kusnezoffii is taken in larger quantities [9]. In addition, counterfeits in the herbal market also bring challenges to the correct identification of Baibu. Due to their similar morphologic features to the authentic sources of Baibu, many counterfeits such as Asparagus officinalis, A. filicinus and A. acicularis are often sold as Baibu in the herbal market [10]. Therefore, the distinction between Baibu and its counterfeits is critical for its beneficial usage as a medicinal herb.
Compared to morphological characteristics, a DNA barcode is deemed to be a more efficient and effective method for identifying a particular plant species. Typical barcodes such as ITS, psbA-trnH, matK and rbcL have been used to distinguish different plant species [11–13]. However, these DNA barcodes did not always working effectively, especially when trying to distinguish closely related plant species. Such a phenomenon may be attributed to the fact that a single-locus DNA barcode still lacks adequate variations generally observed in closely related taxa. Compared with a general DNA barcode, the chloroplast genome can provide more abundant genetic information and higher resolutions when identifying plant species. Some researchers have proposed using the chloroplast genome as a species-level DNA barcode [14, 15].
The chloroplast is an organelle which is present in almost all green plants. It is central to photosynthesis and plays a vital role in sustaining life on earth by converting solar energy to carbohydrates. Besides photosynthesis, the chloroplast also plays critical roles in other biological processes, including the synthesis of amino acids, nucleotides, fatty acids and many secondary metabolites. Furthermore, metabolites synthesized in chloroplasts are often involved in plants’ interactions with their environment, such as their response to environmental stress and defense against invading pathogens [16–18]. Due to its essential roles in the cellular processes and its relatively small genome size, the chloroplast genome is a good starting point for resolving phylogenetic ambiguity, discriminating closely between related species and revealing the plants’ evolutionary process [19–21]. To date, over 5000 chloroplast genomes from a variety of land plants are available. Phylogenetic analyses have demonstrated the effectiveness of the chloroplast genome in inferring the phylogenetic identity of plants as well as having the ability to distinguish between closely related plant species [22, 23].
Unfortunately, the taxonomic coverage of the sequenced chloroplast genome is somewhat biased. For example, until now, the chloroplast genome of S. sessilifolia has not been reported. The lack of chloroplast genome information prohibited studies aimed at understanding the evolutionary processes in the family Stemonaceae. Here, we report the full plastid genome of S. sessilifolia. Based on the sequence data, we performed a multi-scale comparative genome analysis among S. sessilifolia, A. officinalis and Carludovica palmata (the major counterfeits of Baibu). We investigated the difference among these three species from three aspects, including general characteristics, repeat sequences and sequence divergence. We also characterized the significant changes, including genome rearrangements, IR expansion and SSC contraction, in the plastid genome of S. sessilifolia, A. officinalis and C. palmata.
Lastly, we investigated the gene loss events in Stemonaceae and its closely related families (Asparagoideae, Velloziaceae, Cyclanthaceae and Pandanaceae). The results reported in this work will provide valuable information for species distinction of herb materials that are used as Baibu. Furthermore, it lays the foundation for elucidating the evolutionary history of plant species in the family Stemonaceae.
Materials and methods
Plant materials and DNA extraction
Fresh young leaves of S. sessilifolia from multiple individual plants were collected from the Institute of Medicinal Plant Development (IMPLAD), Beijing, China, and stored at -80°C for chloroplast DNA extraction. All samples were identified by Professor Zhao Zhang, from IMPLAD, Chinese Academy of Medical Sciences & Peking Union Medical College and voucher specimens were deposited in the herbarium of the institute. S. sessilifolia is not an endangered or protected species and specific permission for the collection of S. sessilifolia was not required. Total DNA was extracted from 100mg of fresh young leaves using a plant genomic DNA kit (Tiangen Biotech, Beijing, Co., Ltd.). Finally, 1.0% agarose gel and a Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific, United States) were used to evaluate the purity and concentration of the extracted DNA, respectively.
Genome sequencing, assembly and annotation
According to the standard protocol, the DNA of Stemona sessilifolia was sequenced using Illumina Hiseq25000 platform, with insert sizes of 500 bases for the library. A total of 5,660,432 paired-end reads (2 × 250bp) were obtained, and low-quality reads were trimmed with Trimmomatic software [24].
In order to extract reads belonging to the chloroplast genome, we downloaded 1,688 chloroplast genome sequences from the GenBank and constructed a Basic Local Alignment Search Tool (BLASTn) database. All trimmed reads were mapped to this database using the BLASTN program [25], and reads with an E-value > 1E-5 were extracted. The reads were first assembled using the SPAdes software with default parameters [26]. The contigs were then subjected to gap closures using the Seqman module of DNASTAR (V11.0) [27]. Finally, the quality of the assembled genome was evaluated by mapping the reads to the genome using Bowtie2 (v2.0.1) with default settings [28]. For further evaluation, all the barcode sequences of S. sessilifolia available from the GeneBank were download (S1 File), including matK (1), petD(1), rbcL (1), rpoC1 (1), rps16 (1), rps19-rpl22-psbA (1), trnL (3) and trnL-trnF (2) The numbers enclosed in parentheses represent the number of the barcode sequence. The BLAST program was used to calculate the identity differences between the chloroplast genome sequence of S. sessilifolia and other barcode sequences. As a result, the barcode, rps19-rpl22-psbA, which is located at the boundary of LSC/IRb, was identifies to have a value of 100%. All the other barcode sequences also gave identity values of 100%, indicating the high reliability of the chloroplast genome sequence.
Gene annotation of S. sessilifolia chloroplast genome was conducted using CpGAVAS2, which is an integrated plastome sequence annotator and analyzer [29]. The tRNA genes were confirmed with tRNAscan-SE [30] and ARAGORN (V1.2.38) software packages [31]. Then the gene/intron boundaries were inspected and corrected using the Apollo program (V1.11.8) [32]. The Cusp and Compseq programs from EMBOSS (V6.3.1) were used to calculate the GC content [33]. Finally, OrganellarGenomeDRAW [34] was used to generate the circular chloroplast genome map of S. sessilifolia.
Repeat sequence analysis
Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/) was used to identify simple sequence repeats (SSRs) with the parameters listed as follows: 74 repeat units for mononucleotide SSRs, 20 repeat units for di- and tri-nucleotide repeat SSRs, and 12 repeat units for tetra-, penta-, and hexanucleotide repeat SSRs. Tandem Repeats Finder was used with parameters of 2 for matches and 7 for mismatches and indels [35]. For the minimum alignment score and the maximum period, size was set to 50 and 500, respectively. Palindrome and forward repeats were identified by the REPuter web service [36]. The minimum repeat size and the similarity cut-off were set to 30 bp and 90%, respectively.
Comparative genomic analysis
A total of three species, including S. sessilifolia, A. officinalis (NC_034777), Carludovica palmata (NC_026786), were subjected to multiple sequence alignment using mVISTA with default parameters [37]. Subsequently, 20 introns and 108 intergenic regions shared by S. sessilifolia, A. officinalis, and Carludovica palmata were extracted using custom MatLab scripts and used to perform sequence divergence analysis. Firstly, the sequences of each intergenic-region/intron were aligned individually using the CLUSTALW2 (v2.0.12) [38] program with options "-type = DNA–gapopen = 10 -gapext = 2". Secondly, pairwise distances were calculated with the Distmat program in EMBOSS (v6.3.1) using the Kimura 2-parameters (K2p) evolution model [39]. We attempted to discover highly divergent regions in order to develop novel molecular markers. To identify the occurrence of genome rearrangement events in the chloroplast genome of S. sessilifolia, synteny analysis among the three species mentioned above were performed using Mauve Alignment [40].
Phylogenetic analysis
A total of 11 chloroplast genomes which are distributed into Stemonaceae (3), Cyclanthaceae (1), Pandanaceae (1), Velloziaceae (1) and Asparagoideae (5) were retrieved from the RefSeq database. The protein sequences shared by these chloroplast genomes were used to construct a phylogenetic tree with Veratrum patulum and Paris dunniana as the outgroup taxa (S1 Table). Fifty-eight proteins were involved, and all these protein sequences were aligned using the CLUSTALW2 (v2.0.12) program with options "-gapopen = 10 -gapext = 2 -output = phylip". Then the Maximum Likelihood (ML) method was adopted to infer the evolutionary history of S. sessilifolia and the other closely related species. The detailed parameters were "raxmlHPC-PTHREADS-SSE3 -f a -N 1000 -m PROTGAMMACPREV–x 551314260 -p 551314260-o Nicotiana_tabacum, Solanum_lycopersicum -T 20".
Results
General characteristics of chloroplast genomes
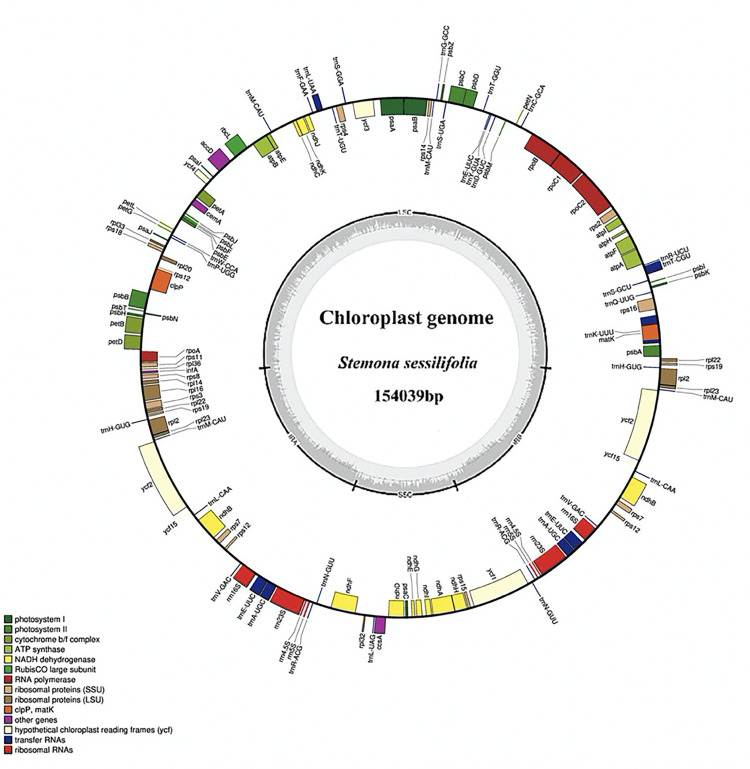
The HiSeq2500 generated about 3.2 GB of data and the average coverage depth of the assembled cp genome was 885×. The gene map of S. sessilifolia is shown in Fig 1. This genome has been deposited in the GenBank (Accession number: MW023922). The chloroplast genomes of S. sessilifolia and two other species share the standard features of possessing a typical quadripartite structure consisting of a pair of inverted repeats (IRs) separating a large single copy (LSC) and a small single copy (SSC). This is similar to other angiosperm chloroplast genomes [41].
Fig 1. Gene maps of the chloroplast genomes of Stemona sessilifolia.
Genes inside and outside the circle were transcribed clockwise and counterclockwise, respectively. The darker gray in the inner circle indicates the GC content. Genes with different functions are characterized with different color bars.
We then carried out a multi-scale comparative genome analysis of these three chloroplast genomes from four aspects, including the size, the guanine-cytosine (GC) content, the number of genes and the gene organization (Table 1). The complete circular chloroplast genomes of S. sessilifolia, A. officinalis and C. palmata were 154,037, 156,699 and 158545 bp, respectively. Compared to A. officinalis and C. palmata, S. sessilifolia showed a relatively short SSC region and a relatively long IR region. We speculated that the chloroplast genome of S. sessilifolia might undertake IR expansion and SSC contraction simultaneously. There was no significant difference between S. sessilifolia, A. officinalis, and C. palmata. Such a result may be attributed to the high conservation of tRNAs and rRNAs. The lengths of the CDS regions of A. officinalis and C. palmata were found to be shorter than S. sessilifolia, indicating that there were probable gene loss events in the chloroplast genome of A. officinalis and C. palmata.
Table 1. Chloroplast genome characteristics of Stemona sessilifolia, Asparagus officinalis and Carludovica palmata.
| Plastome | Characteristics | Species | ||
|---|---|---|---|---|
| Stemona sessilifolia | Asparagus officinalis | Carludovica palmata | ||
| Size (bp) | Genome | 154037 | 156699 | 158545 |
| LSC | 81949 | 84999 | 71426 | |
| IR | 27090 | 26531 | 26529 | |
| SSC | 17908 | 18638 | 18364 | |
| tRNA genes | 2877 | 2863 | 2816 | |
| rRNA genes | 9060 | 9052 | 8866 | |
| CDS | 79260 | 77436 | 77802 | |
| GC content (%) | Overall | 38.00 | 37.59 | 37.74 |
| LSC | 36.18 | 35.60 | 35.79 | |
| IR | 42.70 | 42.92 | 42.81 | |
| SSC | 32.13 | 31.50 | 31.51 | |
| tRNA genes | 53.43 | 53.57 | 53.40 | |
| rRNA genes | 55.22 | 55.38 | 55.38 | |
| CDS | 38.29 | 38.1 | 38.41 | |
| 1st position | 45.7 | 45.64 | 45.93 | |
| 2nd position | 38.46 | 38.56 | 38.39 | |
| 3rd position | 30.72 | 30.09 | 30.91 | |
| NO. of genes | Total | 112 | 110 | 112 |
| protein-coding genes | 80 | 78 | 80 | |
| tRNAs | 28 | 28 | 28 | |
| rRNAs | 4 | 4 | 4 | |
| Genes with introns | 18 | 18 | 18 | |
| Genes in IR | 21 | 22 | 19 | |
LSC: large single-copy, IR: inverted repeat, SSC: small single-copy, CDS: coding sequence.
As for the GC content, S. sessilifolia showed a higher value in the regions of the LSC and SSC than A. officinalis and C. palmate, even in the complete chloroplast genome. However, in the IR region, A. officinalis and C. palmata showed a GC content value larger than S. sessilifolia. The GC content decreased markedly from the first position to the third position in the codon position scale. Such a result was in line with the phenomenon observed in most land plant plastomes [42–44].
We identified 112, 110 and 112 genes in the chloroplast genomes of S. sessilifolia, A. officinalis, and C. palmata, respectively. All of these three chloroplast genomes have 28 tRNAs and four rRNAs. The number of genes with introns in each species is 18, similar to reports in prior works [45]. Therefore, we may conclude that there were no intron loss events that occurred in the chloroplast genomes of these three species. Among these 18 genes, 16 of them (10 protein-coding genes and 6 tRNAs) had one intron each, 2 genes (ycf3 and clpP) have two introns each and all of the genes with introns are described in S2 Table. The rps12 gene was divided into 5’-rps12 in the LSC region and 3’-rps12 in IR region. In addition, 20, 22 and 19 genes were predicted for S. sessilifolia, A. officinalis, and C. palmata in the IR regions, respectively.
The gene organization of the three species were compared and the results are presented in Table 2. In the upstream and downstream regions of the A. officinalis chloroplast genome, premature stop codons were discovered in the ycf1 gene, resulting in the loss of this gene. Compared to S. sessilifolia, we found the shorter CDS regions of A. officinalis was directly related to the loss of this gene. We also found a full-length and a pseudogene of the ndhF gene which coexists in the chloroplast genome of S. sessilifolia, which indicated the presence of another SSC contraction event.
Table 2. Genes presented in chloroplast genomes of Stemona sessilifolia, Asparagus officinalis and Carludovica palmata.
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Ribosome RNA genes | rRNA genes | rrn16Sa, rrn23Sa, rrn5Sa, rrn4.5Sa |
| Transfer RNA genes | tRNA genes | trnT-UGU, trnR-ACGa, trnT-GGU, trnS-UGA, trnfM-CAU, trnF-GAA, trnL-UAG, trnV-UAC*, trnL-CAAa, trnM-CAU, trnG-GCC, trnQ-UUG, trnA-UGCa, **, trnD-GUC, trnP-UGG, trnI-CAUa, trnE-UUC**, trnL-UAA**, trnK-UUU**, trnW-CCA, trnY-GUA, trnI-GAUa,*, trnG-UCC*, trnS-GGA, trnR-UCU, trnH-GUGa, trnS-GCU, trnN-GUUa, trnV-GACa, trnC-GCA |
| Others | Large subunit of ribosome | rpl14, rpl16*, rpl2a,*, rpl20, rpl22a, rpl23a, rpl32, rpl33, rpl36 |
| Small subunit of ribosome | rps11, rps12a,b,*, rps14, rps15, rps16*, rps18, rps19a, rps2, rps3, rps4, rps7a, rps8 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Subunits of NADH dehydrogenase | ndhA*, ndhBa,*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, ycf3 | |
| Large subunit of rubisco | rbcL | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| C-type cytochrome synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Protease | clpP** | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Conserved open reading frames | ycf1, ycf2a, ycf15**, ycf4 | |
| Pseudogenes | ycf1ψ, ndhFψ, infAψ, ycf15 a,ψ, ycf68 a,ψ |
*Gene with one intron
**Gene with two introns, a Gene with two copies, b Trans-splicing gene, ψ Pseudo gene. Genes in Bold font were only identified in S. sessilifolia and A. officinalis.Genes with underline were only identified in A. officinalis.
Repeat sequence analysis
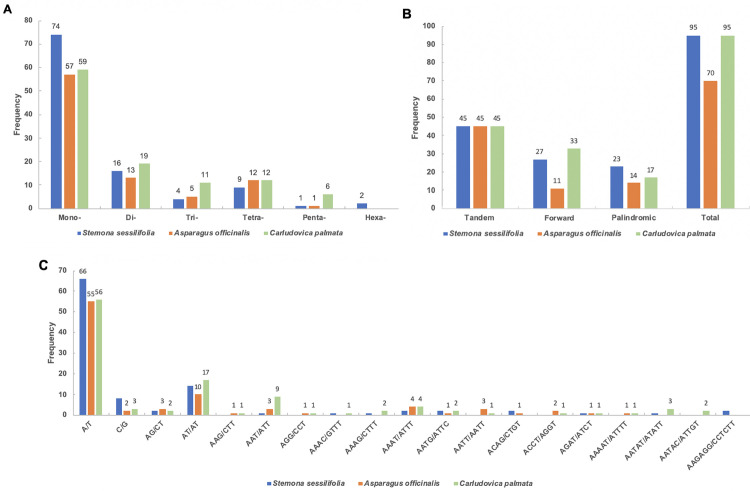
Simple sequence repeats (SSRs), which are tandem repeat sequences consisting of 1–6 repeat nucleotide units, are widely distributed in prokaryotic and eukaryotic genomes. A high degree of polymorphisms of SSRs has been considered to be effective molecular markers when considering species identification, population genetics and phylogenetic research [46, 47]. In the current study, we investigated the distribution of SSRs in the genomes as well as their numbers and types (Fig 2). As a result, a total of 106, 88 and 107 SSRs were detected in S. sessilifolia, A. officinalis and C. palmata, respectively. Mononucleotide motifs showed the highest frequency of SSRs in these species, followed by dinucleotides, tetranucleotides, trinucleotides and pentanucleotides, respectively. However, hexanucleotide repeats were also detected only in S. sessilifolia. As expected, the majority of repeats consisted of A/T and AT/AT repeats which suggest that these chloroplast genomes are rich in short poly-A and poly-T motifs, while poly-C and poly-G ones are relatively rare. These SSRs were highly polymorphic, suggesting they present great potential for the identification of these three species. We then use Tandem Repeats Finder [35] and REPuter [36] to detect long repeats and found 95, 70, and 95 long repeat sequences in S. sessilifolia, A. officinalis and C. palmata, respectively. Tandem, forward and palindromic repeats were present in all these three species with the number of tandem repeats being the same in all of them. In comparison, the number of forward and palindromic repeats were different in the three species. These two types of repeats were most common in S. sessilifolia (27 (54%) and 23 (46%), respectively) and least common in A. officinalis (11 (44%) and 14 (56%), respectively).
Fig 2. Simple sequence repeats (SSRs) and long repeat sequences identified in the chloroplast genomes.
(A) Distribution of different types of SSRs in the chloroplast genomes. (B) Distribution of long repeat sequences in the chloroplast genomes. (C) Frequency of SSR motifs in different repeat class types.
In summary, there are significant differences in the types of repeat sequences among S. sessilifolia, A. officinalis, and C. palmata. The occurrence of repeat events in S. sessilifolia was higher than that of A. officinalis and C. palmata. It should be noted that the size of the A. officinalis and C. palmata chloroplast genome is larger than the chloroplast genome of S. sessilifolia. Hence, the relatively larger sizes of the chloroplast genomes of A. officinalis and C. palmata do not result in many repeat sequences.
Sequence divergence analysis
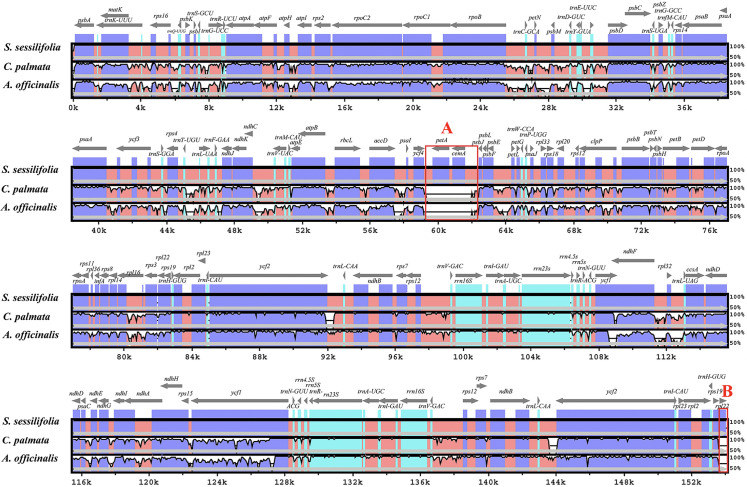
To evaluate the genome sequence divergence, we aligned the sequences from three species using mVISTA [37] (Fig 3). The chloroplast genome of S. sessilifolia was found to be significantly different from A. officinalis and C. palmata. As mycoheterotrophic plants, severe gene loss events always lead to highly reduced plastomes [23, 48]. As expected, the non-coding regions were more divergent than coding regions among these species. The two most divergent regions were the ycf4-psbJ region (red square A) and the rpl22 coding region (red square B). We suspected that such a phenomenon might be attributed to gene loss events or genome rearrangement events, and this will be discussed in detailed later. The Ycf1 gene is also highly divergent, which may be due to the occurrence of pseudogenization. In summary, the LSC region showed the highest divergence, followed by the SSC region and the IR region was less divergent than the LSC and SSC regions. Compared to the coding areas, the intergenic spacers displayed higher divergence areas.
Fig 3. Comparison of the three chloroplast genomes using mVISTA program.
The gray arrows indicate the orientations and positions of genes. Untranslated, conserved non-coding and coding regions were characterized by sky-blue, red and blue blocks, respectively. A cut-off value of 70% was adopted during the process of alignment.
Highly divergent regions can usually assist in the development of molecular markers. Based on the fact that non-coding regions usually evolved more rapidly than coding regions, the intergenic and intron regions are always considered to be ideal candidate regions for molecular markers with high resolution. Therefore, we calculated the Kimura 2-parameter (K2p) distances for each set of the intergenic and intron regions. A relatively higher K2p value between any two species is necessary to distinguish each species from any other two species. Therefore, we calculated the minimal K2p (MK2p) value for each set of intergenic and intron regions. The non-coding regions with higher MK2p values are likely to be the candidate regions for high-resolution molecular markers. Consequently, for introns (S3 Table), the MK2p value ranges from 0.0055 to 0.1096. ClpP_intron2 with the highest MK2p value was followed by rpl16_intron1. The third, fourth and fifth were rps16_intron1, ndhA_intron1 and trnL-UAA_intron1, respectively. For intergenic spacers (S4 Table), five highly conserved intergenic spacers were observed and these were ndhA_ndhH, psaB_psaA, psbL_psbF, rpl2_rpl23 and trnI-GAU_trnA-UGC. The MK2p value of intergenic spacers ranges from 0 to 0.3301, and the top-10 intergenic spacers with higher MK2p values are listed as follows: trnF-GAA_ndhJ, atpB_rbcL, rps15_ycf1, trnG-UCC_trnR-UCU, ndhF_rpl32, accD_psaI, rps2_rpoC2, trnS-GCU_trnG-UCC, trnT-UGU_trnL-UAA and rps16_trnQ-UUG. In conclusion, compared to introns, we observed higher sequence divergence in intergenic spacers. The intergenic spacers with large K2p values represent good candidate molecular markers for distinguishing these three species.
Rearrangement of the chloroplast genome
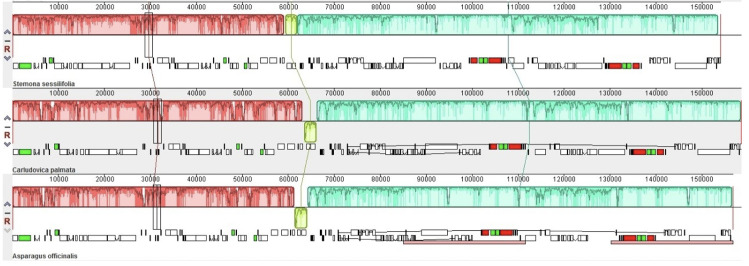
To investigate whether there are significant differences in the yc4-psbJ regions (red square A in Fig 3) and rpl22 coding regions (red square B in Fig 3) between S. sessilifolia and its major counterfeits, we conducted synteny analysis. As plotted in Fig 4, we detected a large inversion of 3 kb long in the LSC region. Interestingly, a similar sequence of an approximately 3-kb long inversion was confirmed to be located in the ycf4-psbJ regions. Therefore, we can conclude that the occurrence of genome rearrangement events leads to a significant difference in the ycf4-psbJ areas between S. sessilifolia and the other two species. To investigate whether the existence of such an inversion in S. sessilifolia is unique occurrence, we conducted synteny analysis between the chloroplast genome of S. sessilifolia and species in Dioscoreales and Liliales, which belong to two closely related orders of Pandanales. Compared to any of the species in Dioscoreales and Liliales, the inversion in the ycf4-psbJ region in S. sessilifolia was always visible (data not shown). Therefore, the inversion in the ycf4-psbJ areas is probably unique to S. sessilifolia.
Fig 4. Comparison of the three chloroplast genomes using the MAUVE algorithm.
Local collinear blocks were colored to indicate syntenic regions, and the histograms within each block indicated the degree of sequence similarity.
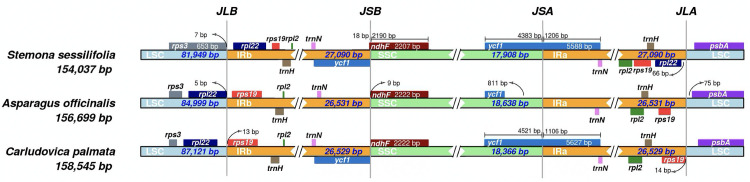
IR expansion and SSC contraction
IR contractions and expansions are common evolutionary events contributing to chloroplast genomes size variation [49]. Here, the JL (LSC/IR) and JS (IR/SSC) boundary comparison analysis was performed by which we attempted to identify IR contraction and expansion events (Fig 5). Compared to A. officinalis and C. palmata, the relatively larger IR regions indicated the occurrence of IR expansion events in S. sessilifolia. Simultaneously, the SSC region was shorter than A. officinalis and C. palmate by 465-737bp, suggesting the occurrence of SSC contraction events in S. sessilifolia. For A. officinalis and C. palmata, one copy of the rpl22 gene is located at the LSC region. However, the IR regions of S. sessilifolia span to the intergenic spacers between the rpl22 and rps3 genes, resulting in the presence of two copies of the rpl22 gene. Therefore, we can claim that the significant differences in rpl22 coding regions between S. sessilifolia and its major counterfeits can be attributed to the occurrence of IR expansion events.
Fig 5. Comparison of IR, LSC and SSC regions among Stemona sessilifolia, Carludovica palmata and Asparagus officinalis.
The numbers above, below or adjacent to genes represent the length of genes or the distances from the front or end of genes to the boundary sites. It should be pointed out that the figure features are not to scale.
Furthermore, the ndhF gene located at SSC regions in A. officinalis and C. palmata, ranges from 9-40bp away from the SSC/IRb junction. However, in S. sessilifolia, the shortening of the SSC region leads to the ndhF gene extending into the IRb region by 18bp. The occurrence of the ndhF gene located at the SSC/IRb junction resulted in partial duplication of this gene at the corresponding region. The ycf1 gene is found at the IRb/SSC junction, creating a ycf1 pseudogene in S. sessilifolia and C. palmata. Considering that premature stop codons were discovered in the ycf1 gene, only one ycf1 gene was annotated in the SSC region in A. officinalis. An overlap of 18bp between the ndhF gene and the ycf1 pseudogene was also observed in S. sessilifolia. In summary, compared to A. officinalis and C. palmata, significant junction expansion and contraction events were observed in S. sessilifolia simultaneously, which were probably responsible for the length variations of these three cp genomes sequences.
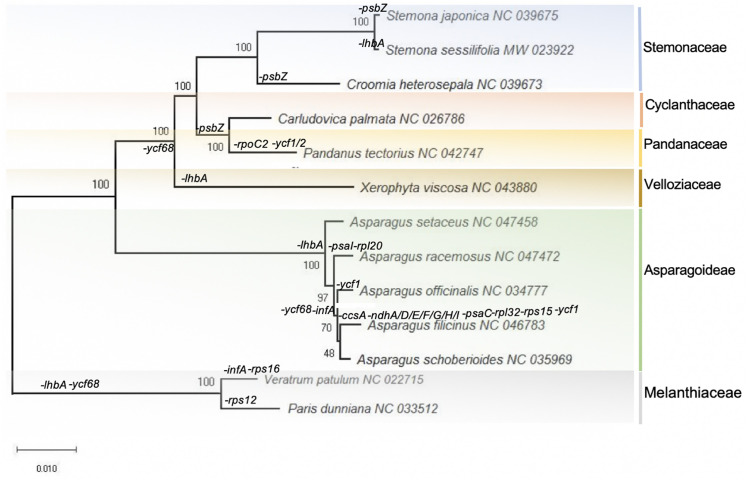
Phylogenetic analysis
The chloroplast genome has been successfully used to determine plant categories and reveal plant phylogenetic relationships [50, 51]. To determine the phylogenetic position of S. sessilifolia, we constructed a phylogenetic tree with species in Stemonaceae and its closely related families (Asparagoideae, Velloziaceae, Cyclanthaceae and Pandanaceae). A total of 13 chloroplast genomes were retrieved from the RefSeq database, and 58 protein sequences shared by these species were used to construct a phylogenetic tree with Veratrum patulum and Paris dunniana serving as the outgroups (Fig 6). As a result, the species in Stemonaceae, Asparagoideae and Velloziaceae formed a cluster, respectively. In addition, S. sessilifolia and S. japonica formed a cluster within Stemonaceae with a bootstrap value of 100%, indicating a sister relationship between these two species.
Fig 6. Molecular phylogenetic analysis of Pandanales and its closely related orders.
The tree was constructed with 58 protein sequences presented in 116 species using the Maximum Likelihood method and implemented in RAxML with Nicotiana tabacum and Solanum lycopersicum serving as the outgroups. The numbers associated with the nodes indicate bootstrap values tested with 1000 replicates. The orders and families to which each species belongs are marked beside the branches as well as the occurrence of gene loss events.
As showed in Fig 6, a series of gene loss events were observed throughout Stemonaceae and its closely related families (Asparagoideae, Velloziaceae, Cyclanthaceae and Pandanaceae). A total of 21 genes are lost in these species, including ycf68 (11), lhbA (9), infA (4), psbZ (4), ycf1 (3), ccsA (1), ndhA (1), ndhD (1), ndhE (1), ndhF (1), ndhG (1), ndhH (1), ndhI (1), psaC (1), psaI (1), ycf2 (1), rps16 (1), rpl20 (1), rpoC2 (1), rps12 (1) and rps15 (1). The numbers enclosed in parentheses represent the frequency of gene loss events. As expected, closely related species always tend to undergo the same gene loss events. A series of clusters formed by species which undergo the same gene deletion events further confirmed such a phenomenon. C. palmata and P. tectorius formed a cluster, and both of these species lack the psbZ gene. The species from Pandanales (Steminaceae, Cyclanthaceae, Pandanaceae and Velloziaceae) formed a cluster without the ycf68 gene. The species from Asparagoideae formed a cluster without the lhbA gene.
The Ycf68 gene has the highest frequency of gene deletions, and the second was the lhbA gene. The following three genes were infA, psbZ and ycf1, respectively. Actually, the ycf68 gene was only found in two species (A. racemosus and A. setaceus), and the lhbA gene was only found in four species (C. palmata, C. heterosepala, P. tectorius and S. japonica). The functions of the ycf68, lhbA and ycf1 genes remain unknown. The occurrence of premature stop codons may account for the rare existence of these three genes in chloroplast genomes [41, 52, 53]. As one of the most active genes in the chloroplast genome, the infA gene plays an essential role in protein synthesis. The frequent absence of the infA gene may be attributed to the transfer of this gene between the cytoplasm and nucleus [41, 54]. The absence of the subunits of the photosystem II gene, psbZ, was frequently observed in Pandanales (Steminaceae, Cyclanthaceae and Pandanaceae). For each of the remaining 16 genes, only one gene loss event was observed, respectively. There was a variety of gene absences in the chloroplast genome of each species, indicating the diversity of variations in the chloroplast genomes. However, for 16 out of 21 genes, the frequency of gene loss events was limited to only one, suggesting that the chloroplast genome is highly conserved at the scale of gene content. Such a phenomenon is consistent with the highly conserved nature of the chloroplast genome as well as its feature of rich variations.
Discussion
Chloroplast genome is frequently used for species identification and plant phylogenetics at generic level [42, 50]. It has also been used at the family level to infer family level phylogenetic relationships and species identification events [55]. Counterfeits medicine are a type of crude drug preparation with a similar morphology but having lower effective components when compared to the authentic medicine. Therefore, it is an important task to distinguish traditional Chinese medicine and its counterfeits. In this study, we sequenced and analyzed the chloroplast genome of S. sessilifolia and performed multi-scale comparative genomics of S. sessilifolia and the major counterfeits of Baibu, A. officinalis, and C. palmata. We also characterized the major changes in the chloroplast genome of these three species, including genome rearrangements, IR expansions and SSC contractions, and investigated the occurrence of gene loss events in Dioscoreales, Liliales, Pandanales and Asparagaceae. Such chloroplast genome analyses can broaden the knowledge regarding the genome organization and phylogenetics of S. sessilifolia and its counterfeits. In addition, two divergence hotspots and 10 intergenic spacers with large K2p values were found in the current study and these might be used for the development of molecular markers.
Our results show that the genome organization and content as well as the synteny characteristics were similar among S. sessilifolia, A. officinalis, and C. palmata. This could be attributed to the fact that chloroplast genomes of land plants have conserved features [56–58]. Nevertheless, previous studies have shown that different regions of the chloroplast genome have different GC content [56, 58], while the IR region has high GC content due to the existence of rRNAs which have high GC content. These three species also have similarities in genes content and genome organization. Interestingly, a large inversion was found in S. sessilifolia. The reverse orientation of the SSC region has also been reported in a wide variety of plant species, such as S. japonica, Croomia heterosepala and C. japonica, which all belong to Stemonaceae [59]. By constrast, A. officinalis and other species such as Salvia miltiorrhiza [60] and Cornales [61] do not have this inversion of the SSC region. This phenomenon is sometimes interpreted as a major inversion existing within the species [62–64]. In fact, the two orientations of the SSC region have been found to occur regularly during the course of chloroplast DNA replication within individual plant cells [65, 66]. Therefore, the reverse orientation of the SSC region in S. sessilifolia and other Stemona species may represent a form of chloroplast heteroplasmy.
SSRs have been widely used as molecular markers in studies of species identification, population genetics and phylogenetic investigations based on their high-degree of variations [67]. The SSRs consisting of mononucleotide A/Ts are the most abundant types in S. sessilifolia, A. officinalis and C. palmata. A similar trend of SSRs was also reported in the chloroplast genomes of not only Stemona species but also in other families of angiosperms [56, 59, 68]. These SSRs sequences are often composed of simple repeating units such as polyadenine (PolyA) or polythymine (Poly T) repeats, which have a significant impact on the overall G/C content of the genomes [69]. With the length of polymorphisms in S. sessilifolia, A. officinalis and C. palmata, they suggest great potential for use in the identification of these three species.
Previous studies have shown that highly divergent regions identified by comparative genomics can reveal sites that can be used for DNA barcoding [68, 70]. Such divergent sites in the chloroplast can be applied to DNA barcoding [43, 44, 71–73]. Here, we determined a 3-kb long inversion in the chloroplast genome of S. sessilifolia which might result from a genome rearrangement event. This unique inversion phenomenon led to significant differences in the ycf4-psbJ region among S. sessilifolia, A. officinalis and C. palmata, which can be used as a candidate region to identify S. sessilifolia from counterfeits. Furthermore, the 10 intergenic spacers with large MK2p values in our study could be applied to DNA barcoding. Fan et al. reported the nucleotide sequences of chloroplast DNA trnL-trnF, trnH-psbA, petB-petD and trnK-rps16 regions which can provide useful information in order to discriminate the Stemona species (S. sessilifolia, S. japonica and S. tuberosa), as well as the common counterfeits such as the Asparagus species [10]. Among them, nucleotide variations were found in the partial sequences of the rps16 and trnL genes. In our study, trnT-UGU_trnL-UAA and rps16_trnQ-UUG were also among the top 10 intergenic spacers with higher MK2p values. Whether there are variable sites in the intergenic regions that we found lead to large MK2p values still needs to be further elucidated. Lu et al. compared the pairwise sequence divergence values across all introns and intergenic spacers in two Stemona species (S. japonica and S. mairei) revealed that the ndhF–rpl32 and trnS–trnG regions were the fastest-evolving regions. These findings agreed with our results. These regions are therefore likely to be the choices for molecular evolutionary and systematic studies between S. sessilifolia and its counterfeits.
In addition, there were significant differences in the IR contractions and expansions between S. sessilifolia and the other two species. As far as the JLB (IRb/LSC) boundary is concerned, we found that S. sessilifolia was significantly expanded in the IR region, which led to the presence of two copies of the rpl22 gene. Besides, SSC region contraction resulted in partial duplication of the ndhF gene at the corresponding region and the ycf1 pseudogene in S. sessilifolia. It was reported that the ndhF gene is involved in photosynthesis, and it was often detected during the positive selection that occurs during the evolutionary process of the species [44, 74]. The function of the ycf1 genes is mostly unknown, but it is known to evolve rapidly [21]. The contraction and expansion of the IR region in S. sessilifolia suggests that there is a significant difference in gene sequences between S. sessilifolia and its counterfeits, and it is important to understand the genome structure and evolutionary process of the chloroplast genome.
Phylogenetic analyses showed that S. sessilifolia and S. japonica (both of which are authentic sources of Baibu according to Pharmacopoeia of the People’s Republic of China (2015 edition) were placed close to each other with bootstrap values of 100%, while A. officinalis and C. palmata were on the other branches. When we investigated the gene loss events in the context of phylogenetic relationships, we also found that the cp genomes of S. sessilifolia and S. japonica have similar gene loss patterns. These findings support the pharmaceutical use of S. sessilifolia and S. japonica as genuine Baibu, and also suggest the urgent need for finding new molecular markers for the identification of genuine Baibu. This study will be of value in determining genome evolution and understanding the phylogenetic relationships within Pandanales and other closely related species.
Conclusions
In summary, the complete plastome of S. sessilifolia (Miq.) Miq. is provided in the current study. We believe it will be of benefit as a reference for further complete chloroplast genome sequencing within the family. Based on sequence data provided, a multi-scale comparative genome analysis of S. sessilifolia and the major counterfeits of Baibu, A. officinalis and C. palmata, was performed. Comparative analysis of these three species revealed the existence of a unique inversion in the ycf4-psbJ regions. Interestingly, IR expansion and SSC contraction were observed in S. sessilifolia simultaneously, resulting in a rare boundary pattern. Some highly variable regions were screened as potential DNA barcodes for identification of these three species, including trnF-GAA_ndhJ, atpB_rbcL, rps15_ycf1, trnG-UCC_trnR-UCU, ndhF_rpl32, accD_psaI, rps2_rpoC2, trnS-GCU_trnG-UCC, trnT-UGU_trnL-UAA and rps16_trnQ-UUG. Phylogenetic analyses showed that the two Stemona species were placed close to each other with a bootstrap value of 100%. Finally, we investigated the gene loss events in the context of the phylogenetic relationship. It is obvious that closely related species always tend to share similar gene loss patterns, consistent with those observed previously. This study will be of value in determining genome structure differences, which can be utilized to identify S. sessilifolia and its counterfeits and understanding the phylogenetic relationships within Stemonaceae and its closely related families.
Supporting information
(FASTA)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
Acknowledgments
The authors are grateful to Dr. Dev Sooranna of Imperial College London for English language editing of the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-3-016, 2017-I2M-1-013) from the Chinese Academy of Medical Science, National Natural Science Foundation of China (31460237),Guangxi science and technology program (AD17292003), Guangxi R&D project of medical and health technology (S201529). The funders were not involved in the study design, data collection, and analysis, decision to publish, or manuscript preparation.
References
- 1.Pilli RA, Rosso GB, Oliveira MdCFd. The chemistry of Stemona alkaloids: An update. Natural Product Reports. 2010;27. doi: 10.1039/c005018k [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Yang W, Yang P, Gao B, Luo L. Effect of Radix Stemonae concentrated decoction on the lung tissue pathology and inflammatory mediators in COPD rats. BMC Complementary and Alternative Medicine. 2016;16(1):1–7. doi: 10.1186/s12906-016-1444-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brem B, Seger C, Pacher T, Hofer O, Greger H. Feeding deterrence and contact toxicity of Stemona alkaloids-a source of potent natural insecticides. Journal of Agricultural & Food Chemistry. 2002;50(22):6383–8. doi: 10.1021/jf0205615 [DOI] [PubMed] [Google Scholar]
- 4.Liu ZL, Goh SH, Ho SH. Screening of Chinese medicinal herbs for bioactivity against Sitophilus zeamais Motschulsky and Tribolium castaneum (Herbst). Journal of Stored Products Research. 2007;43(3):290–6. [Google Scholar]
- 5.Lai DH, Yang ZD, Xue WW, Sheng J, Shi Y, Yao XJ. Isolation, characterization and acetylcholinesterase inhibitory activity of alkaloids from roots of Stemona sessilifolia. Fitoterapia. 2013;89(Complete):257–64. doi: 10.1016/j.fitote.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 6.Fan LL, Xu F, Hu JP, Yang DH, Chen HB, Komatsu K, et al. Binary chromatographic fingerprint analysis of Stemonae Radix from three Stemona plants and its applications. Journal of Natural Medicines. 2015. doi: 10.1007/s11418-015-0887-7 [DOI] [PubMed] [Google Scholar]
- 7.Li SL, Jiang RW, Hon PM, Cheng L, Shaw PC. Quality evaluation of Radix Stemonae through simultaneous quantification of bioactive alkaloids by high-performance liquid chromatography coupled with diode array and evaporative light scattering detectors. Biomedical Chromatography Bmc. 2010;21(10):1088–94. [DOI] [PubMed] [Google Scholar]
- 8.Xu YT, Hon PM, Jiang RW, Cheng L, Li SH, Chan YP, et al. Antitussive effects of Stemona tuberosa with different chemical profiles. Journal of Ethnopharmacology. 2006;108(1):46–53. doi: 10.1016/j.jep.2006.04.022 [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, Zhang A, Dong H, Yan G, Wang X. Toxicity and Detoxification Effects of Herbal Caowu via Ultra Performance Liquid Chromatography/Mass Spectrometry Metabolomics Analyzed using Pattern Recognition Method. Pharmacognosy Magazine. 2017;13(52):683–92. doi: 10.4103/pm.pm_475_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan LL, Zhu S, Chen HB, Yang DH, Cai SQ, Komatsu K. Molecular analysis of Stemona plants in China based on sequences of four chloroplast DNA regions. Biological & Pharmaceutical Bulletin. 2009;32(8):1439. [DOI] [PubMed] [Google Scholar]
- 11.Vere ND, Rich TCG, Trinder SA, Long C. DNA Barcoding for Plants. Methods in Molecular Biology. 2015;1245:101–18. doi: 10.1007/978-1-4939-1966-6_8 [DOI] [PubMed] [Google Scholar]
- 12.Penikar ZF, Buzan EV. 20 years since the introduction of DNA barcoding: From theory to application. Journal of Applied Genetics. 2013;55(1):43–52. doi: 10.1007/s13353-013-0180-y [DOI] [PubMed] [Google Scholar]
- 13.John W., Kress, Carlos, García-Robledo, Maria, et al. DNA barcodes for ecology, evolution, and conservation. Trends in Ecology & Evolution. 2015. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. Plant DNA barcoding: from gene to genome. Biological Reviews. 2015. [DOI] [PubMed] [Google Scholar]
- 15.Hollingsworth PM, Li DZ, Michelle VDB, Twyford AD. Telling plant species apart with DNA: from barcodes to genomes. Philosophical Transactions of the Royal Society B Biological Sciences. 2016;371(1702):20150338. doi: 10.1098/rstb.2015.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuhaus H., E.,, and, M., J., et al. NONPHOTOSYNTHETIC METABOLISM IN PLASTIDS. Annual Review of Plant Physiology & Plant Molecular Biology. 2000. doi: 10.1146/annurev.arplant.51.1.111 [DOI] [PubMed] [Google Scholar]
- 17.Krzysztof B, Burch-Smith TM. Chloroplast signaling within, between and beyond cells. Frontiers in Plant Science. 2015;6(781):781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya D, Chakraborty S. Chloroplast: the Trojan horse in plant–virus interaction. Molecular Plant Pathology. 2018. doi: 10.1111/mpp.12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xinlian Chen, Yingxian Cui, Liping Nie, et al. Identification and Phylogenetic Analysis of the Complete Chloroplast Genomes of Three Ephedra Herbs Containing Ephedrine. Biomed Research International. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Chen X, Nie L, Sun W, Hu H, Lin Y, et al. Comparison and Phylogenetic Analysis of Chloroplast Genomes of Three Medicinal and Edible Amomum Species. Int J Mol Sci. 2019;20(16). Epub 2019/08/23. doi: 10.3390/ijms20164040 ; PubMed Central PMCID: PMC6720276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Zhang H, Jiang M, Chen H, Huang L, Liu C. Complete plastome sequence of Iodes cirrhosa Turcz., the first in the Icacinaceae, comparative genomic analyses and possible split of Idoes species in response to climate changes. PeerJ. 2019;7:e6663. Epub 2019/04/12. doi: 10.7717/peerj.6663 ; PubMed Central PMCID: PMC6448556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubuka T, Tsutsui K. Comparative and Evolutionary Aspects of Gonadotropin-Inhibitory Hormone and FMRFamide-Like Peptide Systems. Frontiers in Neuroscience. 2018;12. doi: 10.3389/fnins.2018.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam VKY, Marybel SG, Graham SW. The Highly Reduced Plastome of Mycoheterotrophic Sciaphila (Triuridaceae) Is Colinear with Its Green Relatives and Is under Strong Purifying Selection. Genome Biology and Evolution. 2015;(8):8. doi: 10.1093/gbe/evv134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Marc L, Bjoern U. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;(15):2114–20. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camacho C, Coulouris G, Avagyan V, Ning M, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 10:421. Bmc Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. Epub 2012/04/18. doi: 10.1089/cmb.2012.0021 ; PubMed Central PMCID: PMC3342519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burland TG. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71 [DOI] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. Epub 2009/03/06. doi: 10.1186/gb-2009-10-3-r25 ; PubMed Central PMCID: PMC2690996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, et al. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47(W1):W65–w73. Epub 2019/05/09. doi: 10.1093/nar/gkz345 ; PubMed Central PMCID: PMC6602467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(Web Server issue):W686-9. Epub 2005/06/28. doi: 10.1093/nar/gki366 ; PubMed Central PMCID: PMC1160127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–6. Epub 2004/01/06. doi: 10.1093/nar/gkh152 ; PubMed Central PMCID: PMC373265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra S, Harris N. Using Apollo to browse and edit genome annotations. 2006. [DOI] [PubMed] [Google Scholar]
- 33.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–7. Epub 2000/05/29. doi: 10.1016/s0168-9525(00)02024-2 . [DOI] [PubMed] [Google Scholar]
- 34.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 2007;52(5–6):267–74. doi: 10.1007/s00294-007-0161-y [DOI] [PubMed] [Google Scholar]
- 35.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. Epub 1998/12/24. doi: 10.1093/nar/27.2.573 ; PubMed Central PMCID: PMC148217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42. Epub 2001/11/20. doi: 10.1093/nar/29.22.4633 ; PubMed Central PMCID: PMC92531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–9. Epub 2004/06/25. doi: 10.1093/nar/gkh458 ; PubMed Central PMCID: PMC441596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. Epub 2007/09/12. doi: 10.1093/bioinformatics/btm404 . [DOI] [PubMed] [Google Scholar]
- 39.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. Epub 1980/12/01. doi: 10.1007/BF01731581 . [DOI] [PubMed] [Google Scholar]
- 40.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. Epub 2010/07/02. doi: 10.1371/journal.pone.0011147 ; PubMed Central PMCID: PMC2892488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76(3–5):273–97. Epub 2011/03/23. doi: 10.1007/s11103-011-9762-4 ; PubMed Central PMCID: PMC3104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehmood F, Abdullah, Shahzadi I, Ahmed I, Waheed MT, Mirza B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 2020;112(2):1522–30. Epub 2019/08/31. doi: 10.1016/j.ygeno.2019.08.024 . [DOI] [PubMed] [Google Scholar]
- 43.Mehmood F, Abdullah, Ubaid Z, Bao Y, Poczai P, Mirza B. Comparative Plastomics of Ashwagandha (Withania, Solanaceae) and Identification of Mutational Hotspots for Barcoding Medicinal Plants. Plants (Basel). 2020;9(6). Epub 2020/06/19. doi: 10.3390/plants9060752 ; PubMed Central PMCID: PMC7355740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehmood F, Abdullah, Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, et al. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 2020;8:e9552. Epub 2020/08/11. doi: 10.7717/peerj.9552 ; PubMed Central PMCID: PMC7382938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Shao J, Zhang H, Jiang M, Huang L, Zhang Z, et al. Sequencing and Analysis of Strobilanthes cusia (Nees) Kuntze Chloroplast Genome Revealed the Rare Simultaneous Contraction and Expansion of the Inverted Repeat Region in Angiosperm. Front Plant Sci. 2018;9:324. Epub 2018/03/30. doi: 10.3389/fpls.2018.00324 ; PubMed Central PMCID: PMC5861152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue J, Wang S, Zhou SL. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae). Am J Bot. 2012;99(6):e240–4. Epub 2012/05/23. doi: 10.3732/ajb.1100547 . [DOI] [PubMed] [Google Scholar]
- 47.Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci U S A. 1995;92(17):7759–63. Epub 1995/08/15. doi: 10.1073/pnas.92.17.7759 ; PubMed Central PMCID: PMC41225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25(10):3711–25. Epub 2013/10/22. doi: 10.1105/tpc.113.113373 ; PubMed Central PMCID: PMC3877813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8:36. Epub 2008/02/02. doi: 10.1186/1471-2148-8-36 ; PubMed Central PMCID: PMC2275221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104(49):19369–74. Epub 2007/12/01. doi: 10.1073/pnas.0709121104 ; PubMed Central PMCID: PMC2148296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A. 2007;104(49):19363–8. Epub 2007/12/01. doi: 10.1073/pnas.0708072104 ; PubMed Central PMCID: PMC2148295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang M, Chen H, He S, Wang L, Chen AJ, Liu C. Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea. Int J Mol Sci. 2018;19(5). Epub 2018/05/12. doi: 10.3390/ijms19051419 ; PubMed Central PMCID: PMC5983699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen PA, Kim JS, Kim JH. The complete chloroplast genome of colchicine plants (Colchicum autumnale L. and Gloriosa superba L.) and its application for identifying the genus. Planta. 2015;242(1):223–37. Epub 2015/04/24. doi: 10.1007/s00425-015-2303-7 . [DOI] [PubMed] [Google Scholar]
- 54.Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13(3):645–58. Epub 2001/03/17. doi: 10.1105/tpc.13.3.645 ; PubMed Central PMCID: PMC135507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li CJ, Wang RN, Li DZ. Comparative analysis of plastid genomes within the Campanulaceae and phylogenetic implications. PLoS One. 2020;15(5):e0233167. Epub 2020/05/15. doi: 10.1371/journal.pone.0233167 ; PubMed Central PMCID: PMC7224561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdullah, Mehmood F, Shahzadi I, Waseem S, Mirza B, Ahmed I, et al. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics. 2020;112(1):581–91. Epub 2019/04/19. doi: 10.1016/j.ygeno.2019.04.010 . [DOI] [PubMed] [Google Scholar]
- 57.Amiryousefi A, Hyvönen J, Poczai P. The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae. PLoS One. 2018;13(4):e0196069. Epub 2018/04/26. doi: 10.1371/journal.pone.0196069 ; PubMed Central PMCID: PMC5919006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. Epub 2006/10/13. doi: 10.1186/1471-2105-7-439 ; PubMed Central PMCID: PMC1615880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Q, Ye W, Lu R, Xu W, Qiu Y. Phylogenomic and Comparative Analyses of Complete Plastomes of Croomia and Stemona (Stemonaceae). Int J Mol Sci. 2018;19(8). Epub 2018/08/15. doi: 10.3390/ijms19082383 ; PubMed Central PMCID: PMC6122011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One. 2013;8(2):e57607. Epub 2013/03/06. doi: 10.1371/journal.pone.0057607 ; PubMed Central PMCID: PMC3584094 “Guangzhou Pharmaceutical Holding Limited”. All the authors are cooperating partners in this study about sequencing and analyzing the chloroplast genome of Salvia miltiorrhiza. There are competing interests in employment, consultancy, patents, products in development or marketed products, etc. All the authors confirm that this does not alter their adherence to all the PLOS ONE policies on sharing data and materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu CN, Li HT, Milne R, Zhang T, Ma PF, Yang J, et al. Comparative analyses of plastid genomes from fourteen Cornales species: inferences for phylogenetic relationships and genome evolution. BMC Genomics. 2017;18(1):956. Epub 2017/12/09. doi: 10.1186/s12864-017-4319-9 ; PubMed Central PMCID: PMC5721659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 2007;45(2):547–63. Epub 2007/07/24. doi: 10.1016/j.ympev.2007.06.004 . [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, et al. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 2013;8(2):e57533. Epub 2013/03/06. doi: 10.1371/journal.pone.0057533 ; PubMed Central PMCID: PMC3583863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker JF, Zanis MJ, Emery NC. Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am J Bot. 2014;101(4):722–9. Epub 2014/04/05. doi: 10.3732/ajb.1400049 . [DOI] [PubMed] [Google Scholar]
- 65.Wang W, Lanfear R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol Evol. 2019;11(12):3372–81. Epub 2019/11/22. doi: 10.1093/gbe/evz256 ; PubMed Central PMCID: PMC7145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palmer, Jeffrey D. Chloroplast DNA exists in two orientations. Nature. 1983;301(5895):92–3. [Google Scholar]
- 67.Provan J, Corbett G, McNicol JW, Powell W. Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome. 1997;40(1):104–10. Epub 1997/02/01. doi: 10.1139/g97-014 . [DOI] [PubMed] [Google Scholar]
- 68.Choi KS, Chung MG, Park S. The Complete Chloroplast Genome Sequences of Three Veroniceae Species (Plantaginaceae): Comparative Analysis and Highly Divergent Regions. Front Plant Sci. 2016;7:355. Epub 2016/04/06. doi: 10.3389/fpls.2016.00355 ; PubMed Central PMCID: PMC4804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gichira AW, Avoga S, Li Z, Hu G, Wang Q, Chen J. Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species: DNA barcodes and phylogenetics. Bot Stud. 2019;60(1):17. Epub 2019/08/24. doi: 10.1186/s40529-019-0265-y ; PubMed Central PMCID: PMC6706487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henriquez CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, et al. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae). Planta. 2020;251(3):72. Epub 2020/03/01. doi: 10.1007/s00425-020-03365-7 . [DOI] [PubMed] [Google Scholar]
- 71.Ahmed I, Matthews PJ, Biggs PJ, Naeem M, McLenachan PA, Lockhart PJ. Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol Ecol Resour. 2013;13(5):929–37. Epub 2013/05/31. doi: 10.1111/1755-0998.12128 . [DOI] [PubMed] [Google Scholar]
- 72.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7(4):e35071. Epub 2012/04/19. doi: 10.1371/journal.pone.0035071 ; PubMed Central PMCID: PMC3325284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen VB, Park HS, Lee SC, Lee J, Park JY, Yang TJ. Authentication Markers for Five Major Panax Species Developed via Comparative Analysis of Complete Chloroplast Genome Sequences. J Agric Food Chem. 2017;65(30):6298–306. Epub 2017/05/23. doi: 10.1021/acs.jafc.7b00925 . [DOI] [PubMed] [Google Scholar]
- 74.Peng L, Shikanai T. Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 2011;155(4):1629–39. Epub 2011/02/01. doi: 10.1104/pp.110.171264 ; PubMed Central PMCID: PMC3091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(FASTA)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.