Cancer-prone breast gland exposure to a carcinogen induces immune factors in breast cancer cells and immunity against metastasis.
Abstract
The link between carcinogen exposure and cancer immunogenicity is unclear. Single exposure to 12-dimethylbenz[a]anthracene (DMBA) at puberty accelerated spontaneous breast carcinogenesis in mouse mammary tumor virus-polyoma middle tumor-antigen transgenic (MMTV-PyMTtg or PyMT) and MMTV-Her2/neutg (Her2) mice. Paradoxically, DMBA-treated PyMT and Her2 animals were protected from metastasis. CD8+ T cells significantly infiltrated DMBA-exposed breast cancers. CD8+ T cell depletion resulted in severe lung and liver metastasis in DMBA-treated PyMT mice. Besides increasing tumor mutational burden, DMBA exposure up-regulated Chemokine (C-C motif) ligand 21 (CCL21) in cancer cells and heightened antigen presentation. CCL21 injection suppressed breast cancer growth, and CCL21 receptor deletion attenuated T cell immunity against cancer metastasis in DMBA-treated PyMT animals. CCL21 expression correlated with increased mutational burden and cytolytic activity across human cancers. Higher CCL21 levels correlated with increased CD8+ T cell infiltrates in human breast cancer and predicted lower breast cancer distant recurrence rate. Collectively, carcinogen exposure induces immune-activating factors within cancer cells that promote CD8+ T cell immunity against metastasis.
INTRODUCTION
Cancer immunotherapeutics are effective for the treatment of a subset of cancers such as melanoma, which have large T cell infiltrates at baseline (1). However, most cancers show low response rates to current immunotherapeutics including immune checkpoint blockade (2). This is particularly evident in breast cancer, which has relatively low T cell infiltrate at baseline (3, 4). Even in patients with programmed death ligand 1 (PD-L1)–positive advanced breast cancer, programmed cell death protein 1 (PD1) blockade shows an overall response rate of 18.5% (5). Therefore, it is critical to identify the determinants of cancer-immune recognition to transform the “cold” breast tumors into “hot” immunogenic tumors.
Breast cancer is the most common internal malignancy and a leading cause of cancer-related mortality in women of all ages around the world (6). The presence of tumor-infiltrating CD8+ T cells in breast cancer is associated with a significant reduction in the relative risk of death from disease in both estrogen receptor (ER)–negative (HER2-positive and -negative) and the ER-positive HER2-positive subtypes (7). However, up to 57.6% of primary breast tumors lack tumor-infiltrating CD8+ T cells (8, 9). In addition, immunosuppressive regulatory T cells (Tregs) that are present in 77.8 to 82% of primary breast tumors block antitumor immunity and promote metastasis (9–11). Tregs inhibit effector CD8+ T cells by preventing their differentiation and proliferation or directly blocking CD8+ T cell–mediated cytotoxicity (12). Hence, it is essential to identify the factors that can increase the CD8+ T cells and block Tregs in breast cancer to heighten the antitumor immunity against this disease.
Carcinogen exposure and the ensuing increase in tumor mutational burden (TMB) is a major cancer characteristic that is tightly linked to higher tumor immunogenicity and improved efficacy of immunotherapeutics (2, 13–17). Environmental carcinogens introduce DNA damage and mutations, and drive carcinogenesis (18, 19). In addition to increased TMB, DNA damage and other cellular impacts of carcinogen exposure may result in the release of innate immune-activating signals, which can play an important role in the induction of immunity against cancer cells (20, 21). Thus, it is critical to determine how carcinogens induce immunity against cancer and identify the mediators of the cancer immunogenicity downstream of carcinogen exposure that accompany increased TMB yet have therapeutic potential. Ultimately, these signals can be used to transform cold tumors into hot immunogenic tumors.
Carcinogen exposure such as smoking is associated with a significant increase in the risk of breast cancer (22). To determine the role of carcinogen exposure on the immune response to breast cancer, we investigated the impact of 7,12-dimethylbenz[a]anthracene (DMBA) on breast cancer immunogenicity in oncogene-driven murine models of breast carcinogenesis. We studied mouse mammary tumor virus-polyoma middle tumor-antigen transgenic (MMTV-PyMTtg or PyMT) and MMTV-Her2/neutg (Her2) mice, which are well-established spontaneous breast cancer models and mimic luminal breast cancer in humans (23). DMBA belongs to the family of polycyclic aromatic hydrocarbons, which are the main carcinogens in cigarette smoke and environmental pollutants (24, 25). We treated PyMT and Her2 animals with a single dose of DMBA at puberty when MMTV promoter is turned on in the mammary epithelial cells and before any breast tumor is developed. Long-term monitoring of the animals demonstrated that T cell immunity had no impact on controlling the accelerated rate of primary breast cancer development in DMBA-exposed PyMT and Her2 mice. However, these animals were protected from metastasis in a CD8+ T cell–dependent manner. Furthermore, we found DMBA-treated breast cancer cells to express high levels of CCL21, which is a T cell–recruiting chemokine (26), and to contain significantly more activated antigen-presenting cells (APCs) in addition to increased TMB. CCL21 signaling was essential for the immune-activating effects of DMBA in breast cancer, and CCL21 was sufficient to transform a cold breast tumor into a hot immunogenic tumor. Last, CCL21 expression correlated with increased TMB, immune cytolytic activity (CYT), and T cell infiltrates in human cancers and a lower risk of breast cancer distant recurrence in patients. Collectively, our findings demonstrate that carcinogen exposure induces immune-activating pathways within the cancer cells that subsequently lead to an immunogenic transformation of cancer.
RESULTS
Breast cancer metastasis is blocked in DMBA-exposed animals
To determine the impact of carcinogen exposure on breast cancer development, we treated PyMT mice with a single oral dose of 1 mg of DMBA or olive oil (carrier) at puberty and before the onset of breast tumorigenesis (fig. S1A). After DMBA or olive oil treatment, mice were monitored weekly for any sign of breast cancer development. PyMT-DMBA mice had a significantly shorter breast tumor latency compared with PyMT–olive oil controls (P = 0.0069; Fig. 1A). PyMT-DMBA mice developed significantly more palpable breast tumors compared with their olive oil–treated counterparts (P < 0.05 starting at week 9; fig. S1, B and C). Furthermore, the life spans of PyMT-DMBA mice were significantly shorter than olive oil–treated controls because their primary breast tumors reached the terminal size faster (P < 0.0001; Fig. 1B). A single exposure to DMBA did not induce breast tumors in wild-type (WT) mice (Fig. 1 and fig. S1). When PyMT mice reached their terminal stage, the size of the tumors, tumor histopathological grades, and overall tumor burden in the PyMT-DMBA group were similar to the olive oil–treated controls at their terminal stage (fig. S1, D to F). However, DMBA-treated PyMT mice showed significantly fewer breast cancer metastatic foci in their lung compared with olive oil–treated mice (P = 0.0135; Fig. 1, C and D). Minimal liver metastasis was detected in PyMT-DMBA mice and PyMT–olive oil controls (Fig. 1, E and F, and fig. S2). DMBA-treated Her2 mice also experienced accelerated primary breast tumor development and markedly reduced lung metastasis compared with olive oil–treated controls (figs. S3 and S4). These findings demonstrate the association between DMBA exposure and a block in breast cancer metastasis in genetically predisposed animals.
Fig. 1. Breast cancer metastasis burden is lower in DMBA-treated PyMT mice.
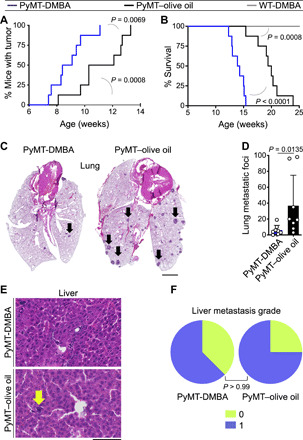
(A) The time to tumor onset in DMBA-treated PyMT mice compared with olive oil–treated PyMT and DMBA-treated WT mice (log-rank test). (B) The survival rate of DMBA-treated PyMT mice compared with olive oil–treated PyMT and DMBA-treated WT mice (log-rank test). The survival rate was determined by the time point at which the animals’ largest primary tumor reached its terminal size. (C and D) Breast cancer metastasis burden in the lung of PyMT mice exposed to DMBA versus olive oil. (C) Representative images of hematoxylin and eosin (H&E)–stained lung (arrows point to metastatic foci) and (D) the number of lung metastatic foci per mouse in each group (graph shows means + SD; Mann-Whitney U test). (E and F) Metastasis burden in the liver of DMBA- and olive oil–treated PyMT mice shown as (E) representative images of H&E-stained liver tissue (the arrow points to a metastatic focus) and (F) liver metastasis grades for PyMT-DMBA and PyMT–olive oil mice (Fisher’s exact test; liver metastasis grades are defined in fig. S2B). n = 8 in each PyMT group and n = 5 in WT group. Scale bars, 2 mm (lung) and 100 μm (liver).
T cells accumulate in breast cancers of mice exposed to DMBA
CD8+ T cells can restrict cancer cell dissemination from the primary tumor site, while Tregs impair the immune-mediated suppression of metastasis (27, 28). To examine the possibility that T cells blocked metastasis of breast cancer exposed to DMBA, we characterized T cell infiltrates in the primary breast tumors of PyMT and Her2 mice treated with DMBA or olive oil. CD8+ T cell frequency and CD8+ T–to–Treg ratio were significantly increased in the tumor-draining lymph nodes of PyMT-DMBA mice compared with controls (Fig. 2A and fig. S5A). Likewise, we observed increased CD8+ T cell frequency and higher CD8+ T–to–Treg ratio in PyMT-DMBA breast tumors compared with their controls (Fig. 2B). Quantification of T cells on histological sections revealed that more T cells and CD8+ T cells accumulated in the stroma surrounding PyMT-DMBA breast tumors than the olive oil–treated controls; however, T cells did not infiltrate into the tumor parenchyma (Fig. 2, C to E). We identified an increased number of lymphoid aggregates (LAs) containing T cells and APCs around the primary breast tumors of DMBA-treated PyMT mice (Fig. 2F and fig. S5B). Increased CD8+ T cell frequency and higher CD8+ T–to–Treg ratio were also found in Her2-DMBA breast tumors compared with controls (fig. S5C). Quantification of the T cells demonstrated more T cells and CD8+ T cells in Her2-DMBA breast tumors compared with olive oil–treated controls (fig. S5, D and E). Collectively, these data indicate the induction of an antitumor T cell immunity against breast cancer progression upon exposure to DMBA.
Fig. 2. Cytotoxic T lymphocytes infiltrate the breast tumors of DMBA-exposed PyMT mice.
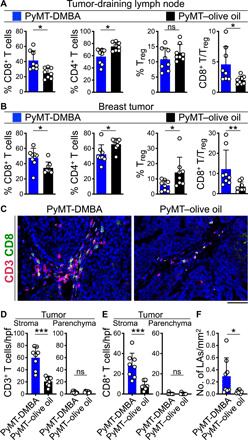
(A and B) T cell subset frequencies and CD8+ T/Treg ratio in the (A) tumor-draining lymph nodes and (B) breast tumor of DMBA- and olive oil–treated PyMT mice. Flow cytometry data were used for this analysis, which were collected when PyMT animals reached the end point. (C to E) CD3+ T cell and CD8+ T cell infiltrates in the stroma and parenchyma of the breast tumors demonstrated by (C) representative immunofluorescence (IF) images of CD3- and CD8-stained tumor tissues and the bar graphs of (D) CD3+ T cell and (E) CD8+ T cell quantifications in high-power field (hpf) images of PyMT-DMBA and PyMT–olive oil breast tumors. (F) LA quantifications per square millimeter of PyMT-DMBA and PyMT–olive oil breast tumors. n = 8 in each group. *P < 0.05, **P < 0.01, and ***P < 0.001, Mann-Whitney U test. Graphs show means + SD. Scale bar, 100 μm. ns, not significant.
CD8+ T cells are required for blocking the metastasis of breast cancer in DMBA-exposed mice
To determine the role of adaptive immunity in primary breast cancer development and metastasis in PyMT-DMBA mice, we generated PyMT mice harboring homozygous null mutations in the recombinase activating gene-1 (Rag1) lacking mature T and B lymphocytes (MMTV-PyMTtg;Rag1−/− or PyMTRag1KO). We treated PyMT and PyMTRag1KO mice with one oral dose of 1 mg of DMBA at puberty (fig. S6A) and monitored their breast cancer development weekly. PyMTRag1KO-DMBA mice had similar tumor latency, palpable breast tumor counts over time, and survival rate compared with PyMT-DMBA mice (fig. S6, B to D). Likewise, the endpoint primary breast tumor sizes, tumor histopathological grades, and breast tumor burden were similar between PyMTRag1KO-DMBA and PyMT-DMBA mice (fig. S6, E to G). Therefore, the depletion of mature T and B cells did not affect the development of primary breast cancer in DMBA-exposed PyMT mice. In contrast, the loss of mature T and B cells in DMBA-treated PyMTRag1KO mice led to a significant increase in breast cancer metastasis to the lung including the size and the number of the metastatic foci (P = 0.0052; Fig. 3, A and B). Notably, we also found a significant metastatic burden in the liver of PyMTRag1KO-DMBA mice, which was not detectable in PyMT-DMBA animals (P = 0.0025; Fig. 3C). To further verify that adaptive immune-mediated restriction of metastasis is specific to DMBA-exposed breast cancer, we treated PyMT and PyMTRag1KO mice with a single dose of olive oil at puberty (fig. S7A) and monitored breast cancer development weekly. The loss of mature T and B cells did not accelerate the primary breast cancer development or metastasis in olive oil–treated PyMTRag1KO mice compared with PyMT controls (figs. S7 and S8).
Fig. 3. CD8+ T cells are required for blocking breast cancer metastasis in DMBA-treated PyMT mice.
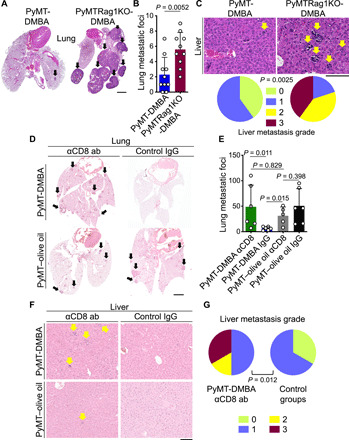
(A and B) Metastasis burden in the lung of DMBA-treated PyMT and PyMTRag1KO mice demonstrated by (A) representative H&E-stained lung images (arrows point to metastatic foci) and (B) a bar graph of the number of lung metastatic foci in each group. (C) Metastasis burden in the liver of DMBA-treated PyMT and PyMTRag1KO mice shown as representative images of H&E-stained liver tissue (arrows point to metastatic foci) and liver metastasis grades. n = 10 in each group for (A) to (C). (D and E) Metastasis burden in the lung of DMBA- and olive oil–treated PyMT mice injected with anti-CD8 antibody (ab) or control IgG demonstrated by (D) representative H&E-stained lung images (arrows point to metastatic foci) and (E) bar graph of the number of lung metastatic foci in each group. (F and G) Metastasis burden in the liver of DMBA- and olive oil–treated PyMT mice injected with anti-CD8 antibody or control IgG shown as (F) representative images of H&E-stained liver tissue (arrows point to metastatic foci) and (G) liver metastasis grades in anti-CD8 antibody–treated PyMT-DMBA mice compared with the control groups combined. n = 5 to 6 in each group for (D) to (G). Mann-Whitney U test was used for lung metastasis, and Fisher’s exact test was used for liver metastasis. Liver metastasis grades are defined in fig. S2. Graphs show means + SD. Scale bars, 2 mm (lung) and 100 μm (liver).
Considering that B cells did not infiltrate PyMT-DMBA or PyMT–olive oil breast tumors (fig. S9) and the previously reported prometastatic role of CD4+ T cells in PyMT mice (29), we examined the role of CD8+ T cells in blocking breast cancer metastasis in PyMT-DMBA mice using anti-CD8α–depleting antibody (fig. S10A). The depletion of CD8+ T cells did not affect the development of primary breast cancer in DMBA- or olive oil–treated PyMT mice (fig. S10, B to G). In contrast, the loss of CD8+ T cells in DMBA-treated PyMT mice led to a significant increase in breast cancer metastasis to the lung compared with control immunoglobulin G (IgG)–treated PyMT-DMBA mice (P = 0.011; Fig. 3, D and E). CD8+ T cell–depleted PyMT-DMBA mice also had a significant metastatic burden in the liver compared with control animals (P = 0.012; Fig. 3, F and G). CD8+ T cell depletion did not affect metastasis outcomes in olive oil–treated PyMT mice (Fig. 3, D to G). These findings demonstrate that CD8+ T cells play an essential and specific role in blocking breast cancer metastasis in DMBA-exposed mice.
Further characterization of CD8+ and CD4+ T cells revealed a trend toward an increase in PD1+ CD8+ T cells in the breast tumors of PyMT-DMBA mice compared with their olive oil–treated controls (P = 0.07; fig. S11, A and B). However, we did not detect any PD-L1 expression on PyMT-DMBA tumor cells (fig. S11C). Tumor-infiltrating CD8+ T cell activation or exhaustion status remains unchanged in breast tumors of mice exposed to DMBA (fig. S11). To determine whether CD8+ T immunity against primary breast tumors of DMBA-exposed PyMT mice could be enhanced by anti-PD1 therapy, we treated PyMT-DMBA mice with anti-PD1 antibody as soon as the first breast tumor reached 5 mm in diameter (fig. S12A). We repeated the intraperitoneal antibody injection every other day for five more doses (fig. S12A). Primary breast cancer development in PyMT-DMBA mice was not affected by anti-PD1 treatment as measured by tumor latency, tumor burden, and survival analysis (figs. S12 and S13). Considering the minimal metastatic burden in PyMT-DMBA mice at the baseline, we were not able to detect any further reduction in lung metastasis in PyMT-DMBA mice treated with anti-PD1 antibody (fig. S13). T cell infiltrates at the sites of the primary breast tumor development in PyMT-DMBA mice did not show any significant change upon anti-PD1 antibody treatment (fig. S14). Similarly, anti- cytotoxic T lymphocyte–associated protein 4 (CTLA4) therapy in PyMT-DMBA mice had no impact on their primary breast cancer or metastasis outcomes (figs. S15 and S16). Together, these results suggest that the immune checkpoint blockade does not affect the primary breast cancer development in mice that are exposed to carcinogen likely due to lack of T cells infiltrating into the tumor parenchyma.
TMB is increased in breast cancer cells of mice exposed to DMBA
TMB is reported to be positively correlated with increased tumor-infiltrating lymphocytes (TILs) and to be associated with heightened CD8+ T cell CYT in multiple cancer types (30, 31). To determine whether DMBA treatment led to an increase in breast cancer TMB, we performed exome sequencing on DNA extracted from primary breast tumors of DMBA- and olive oil–treated PyMT mice and compared the results to the breast glands of WT littermates. Breast tumors from DMBA-exposed mice had more somatic single-nucleotide variants (SNVs) than controls (Fig. 4A). Furthermore, we detected significantly more missense variants in breast tumors of DMBA-exposed animals (P = 0.038; Fig. 4B), which is a predictor of increased neoantigen load and a higher antitumor T cell immunity against cancer. Signatures of point mutations were represented more in breast tumors of DMBA-treated mice but not in olive oil–treated controls, with a preferential association of DMBA exposure with T → A transversion (Fig. 4C). To examine the impact of increased neoantigens in PyMT-DMBA breast tumors on T cell immunity against breast cancer metastasis, we performed immunoSEQ high-throughput DNA sequencing of the tumor-infiltrating T cell receptor B gene (TCRB). TCRB sequencing demonstrated no differences in the degree of T cell clonality and entropy in the breast tumors of DMBA-treated mice compared with olive oil–treated mice (fig. S17, A to C). Likewise, T cell entropy did not correlate with the number of missense mutations in the breast tumors or the burden of lung metastasis across the animals (fig. S17, D and E). Thus, a single exposure to DMBA before breast tumorigenesis increased the mutational burden in breast tumors and resulted in a potent antitumor immunity found in PyMT-DMBA mice.
Fig. 4. A single exposure to DMBA at puberty increases TMB in PyMT breast tumors.
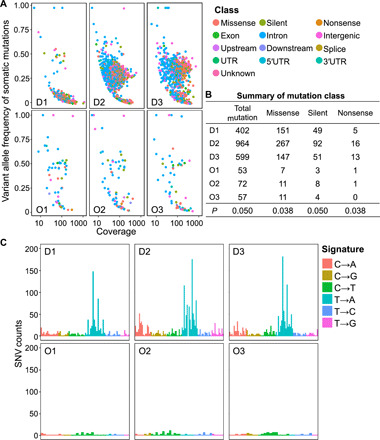
(A and B) Dot plots (A) and the summary of somatic mutations (B) detected in breast tumors of PyMT mice exposed to DMBA versus olive oil determined by exome sequencing. D, DMBA-treated; O, olive oil–treated. n = 3 in each group. P value, one-tailed Mann-Whitney U test comparing DMBA- and olive oil–treated breast tumors. UTR, untranslated region. (C) SNV signatures in breast tumors of PyMT mice exposed to DMBA versus olive oil. SNVs were identified and characterized as C → A, C → G, C → T, T → A, T → C, or T → G.
CCL21 is up-regulated in breast cancer cells of PyMT mice exposed to DMBA
We hypothesized that carcinogens such as DMBA not only increase TMB but also induce the release of immune-activating factors, which can contribute to the immunogenic transformation of cancer cells. To test this hypothesis, we analyzed 113 chemokines and cytokines in protein lysates prepared from primary breast tumors of PyMT mice treated with DMBA or olive oil. Among this large array of immune factors, we found only three factors, CCL21, CCL6, and resistin, to be up-regulated in DMBA-exposed breast tumors compared with controls (Fig. 5, A and B, and fig. S18A). Among them, CCL21 chemokine was expressed in breast cancer cells (Fig. 5C), which was implicated in antigen presentation and T cell priming (26, 32). Consistent with this function of CCL21, we detected an increased number of major histocompatibility complex class II–positive (MHCII+) APCs in breast tumors of PyMT-DMBA mice compared with PyMT–olive oil controls (P = 0.0002; Fig. 5, D and E). Among APCs, we found CD11c+ dendritic cells (DCs) to be significantly increased in PyMT-DMBA tumors (P = 0.0047; fig. S18, B to D). PyMT-DMBA tumor-infiltrating APCs expressed significantly higher levels of MHCI (P = 0.0051; fig. S18, E and F). We found a marked up-regulation of serum CCL21 levels in DMBA-treated mice compared with olive oil–treated mice (P = 0.0007; fig. S19A) and its significant negative correlation with the number of lung metastatic foci across DMBA- and olive oil–treated animals (P = 0.0324; fig. S19B). The assessment of Ccl21 gene expression and CCL21 protein levels in primary breast tumors of DMBA- and olive oil–treated PyMT mice revealed that the DMBA exposure resulted in CCL21 overexpression at the posttranscriptional level (fig. S20). The measurement of CCL21 protein levels in small (<0.25 g) to large (>2 g) primary breast tumors showed its loss as tumors grew in olive oil–treated PyMT mice (fig. S20B). In contrast, breast tumors in DMBA-treated animals expressed high CCL21 levels as they reached their terminal size (fig. S20B). These findings demonstrate that DMBA exposure up-regulates a T cell–recruiting chemokine in the breast cancer cells, which associates with improved antigen presentation within the breast tumor microenvironment and reduced risk of lung metastasis.
Fig. 5. DMBA exposure up-regulates CCL21 in PyMT breast cancer cells.
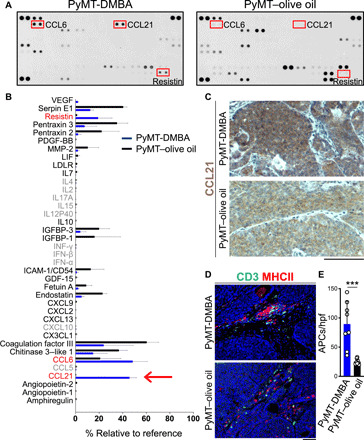
(A) Representative images of cytokine arrays performed on equal amounts of protein lysates from primary breast tumors of DMBA- and olive oil–treated PyMT mice. Red boxes highlight the factors that show significantly higher protein levels in breast tumors of DMBA-exposed PyMT mice. (B) Bar graph of cytokine arrays on breast cancers of DMBA- and olive oil–treated PyMT mice. Significantly up-regulated (red), down-regulated (black), and no change (gray) immune factors in breast tumors of PyMT mice exposed to DMBA compared with carrier control are shown (P < 0.05). The red arrow highlights CCL21 relative levels in the tumor lysates. n = 3 in each group. CCL21 up-regulation was confirmed using enzyme-linked immunosorbent assay (ELISA) on a larger number of samples (fig. S20). VEGF, vascular endothelial growth factor; PDGF-BB, platelet-derived growth factor BB; MMP-2, matrix metalloproteinase 2; LIF, leukemia inhibitory factor; LDLR, low-density lipoprotein receptor; IL7, interleukin-7; IGFBP-3, insulin-like growth factor binding protein 3; ICAM-1; intercellular adhesion molecule–1; GDF-15, growth and differentiation factor 15. (C) Representative images of CCL21 immunohistochemical staining on breast tumors of DMBA- and olive oil–treated PyMT mice. (D and E) Tumor-infiltrating T cells and MHCII+ APCs demonstrated by (D) representative images MHCII- and CD3-stained breast cancer tissues and (E) bar graph of APC counts in PyMT-DMBA and PyMT–olive oil breast tumors. APCs were quantified in 10 randomly selected hpf images per sample. Note the LAs with dense T cell/APC populations that are primarily found in the breast tumors of DMBA-exposed PyMT mice. n = 8 in each group. ***P = 0.0002, Mann-Whitney U test. Graphs show means + SD. Scale bars, 100 μm.
CCL21 is a potent inducer of antitumor CD8+ T cell immunity downstream of DMBA exposure
To assess the impact of CCL21 on enhancing T cell response against breast cancer, we generated a breast cancer cell line using a PyMT tumor on the C57BL/6 background. PyMT cells were injected subcutaneously adjacent to the inguinal mammary fat pads of syngeneic WT mice. Six days later, palpable breast tumors received 1 μg of CCL21 intratumoral injections every other day for six doses. Breast tumors that received CCL21 grew significantly slower compared with phosphate-buffered saline (PBS; carrier)–treated controls (Fig. 6A). We found significantly increased CD3+ and CD8+ T cells infiltrating the PyMT breast tumors in response to CCL21 compared with PBS injections (Fig. 6, B to D). Unlike CCL21, the intratumoral injection of CCL6 had no impact on PyMT tumor growth in WT mice (fig. S21A). Next, we examined whether tumor growth inhibitions by CCL21 relied on adaptive immunity and injected PyMT breast cancer cells into Rag1KO mice, followed by intratumoral injection of CCL21 or PBS. CCL21 had no impact on breast tumor growth in the absence of T and B cells (fig. S21B). To investigate the role of CCL21 in the immunogenic transformation of DMBA-exposed breast cancer, we treated PyMT cells with DMBA or dimethyl sulfoxide (DMSO) (carrier control) in culture and selected a pair of single-cell clones with matched proliferation in vitro (fig. S22, A to C). Similar to spontaneous breast tumors, CCL21 was up-regulated in DMBA-treated PyMT cells compared with DMSO-treated and parental PyMT cells (P = 0.0286; fig. S22D). DMBA-treated PyMT cells formed markedly smaller tumors compared with DMSO-treated PyMT cells in WT mice, while no significant difference in tumor growth was observed between these cell lines in Rag1KO animals (Fig. 6E and fig. S22E). Accordingly, DMBA-treated PyMT cells formed significantly bigger tumors in Rag1KO compared with WT mice (fig. S22F). However, DMSO-treated PyMT cells formed tumors with similar kinetics in WT and Rag1KO mice (fig. S22G), which confirmed the low immunogenicity of parental PyMT breast cancer cells.
Fig. 6. CCL21 suppresses breast tumor growth through the induction of antitumor CD8+ T cell immunity in vivo.
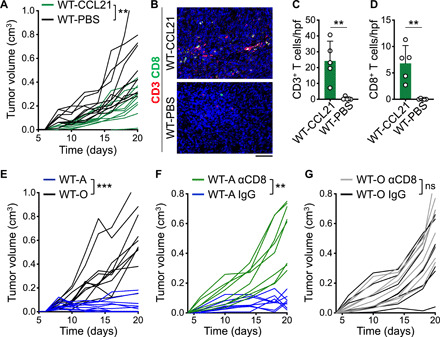
(A) Spider plot of PyMT breast tumor growth in WT mice treated with 1 μg of CCL21 or sterile PBS (carrier only) intratumorally every other day six times starting at day 6 after PyMT cell injection (**P < 0.01 starting at day 8, n = 10 in each group). (B to D) Tumor-infiltrating CD3+ T and CD8+ T cells shown as (B) representative images of CD3- and CD8-stained tumor tissues, (C) CD3+ T cell, and (D) CD8+ T cell counts in CCL21- and PBS-treated breast tumors. Note that T cells were rarely found in PBS-treated breast tumors. n = 5 in each group. (E) Spider plots of DMBA- and DMSO-treated PyMT breast cancer cell growth in WT mice (WT-A and WT-O, respectively; ***P < 0.0001 starting at day 12; n = 10 in each group). (F) Spider plot of DMBA-treated PyMT breast tumor growth in WT mice treated with anti-CD8β versus IgG control antibody (n = 8 per group; **P < 0.01 starting at day 6). (G) Spider plot of DMSO-treated PyMT breast tumor growth in WT mice treated with anti-CD8β versus IgG control antibody (n = 8 per group). Mann-Whitney U test was used. Graphs show means + SD. Scale bar, 100 μm.
To examine the role of CD8+ T cell immunity in suppressing PyMT-DMBA tumor growth, we depleted CD8+ T cells in WT mice that were implanted with PyMT-DMBA versus PyMT-DMSO cells using an anti-CD8β antibody. CD8+ T cell depletion led to a significant increase in PyMT-DMBA tumor growth rate compared with IgG-treated controls (Fig. 6F). However, CD8+ T cell depletion had no impact on the growth of PyMT-DMSO tumors in WT mice (Fig. 6G). Next, we injected DMBA-treated PyMT cells into WT and Ccr7−/− (Ccr7KO) mice, which lack the receptor for CCL21. PyMT-DMBA cells developed significantly bigger tumors in Ccr7KO compared with WT mice, which indicated that blocking the CCL21-CCR7 axis was essential for control of DMBA-exposed breast cancer growth (Fig. 7A). Quantification of the T cells demonstrated a significant decrease in CD3+ T cell and CD8+ T cell infiltrates in DMBA-treated PyMT tumors in Ccr7KO compared with WT mice (P < 0.01; Fig. 7, B to D). The deletion of Ccr7 in DMBA-treated PyMT mice did not affect spontaneous breast tumor development (figs. S23 and S24). However, DMBA-treated PyMTCcr7KO mice developed significantly more breast cancer metastases in the lung compared with DMBA-treated PyMT mice (P = 0.0079; Fig. 7, E and F). Together, our findings demonstrate that CCL21 is sufficient to induce T cell immunity against cold breast tumors and the CCL21-CCR7 axis plays a critical role in the immunogenic transformation of breast cancer in carcinogen-exposed animals.
Fig. 7. CCL21/CCR7 signaling is required for the suppression of DMBA-treated breast tumor growth and metastasis.
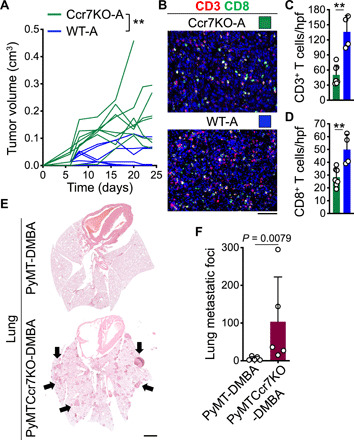
(A) Spider plot of DMBA-treated PyMT breast tumor growth in Ccr7KO (Ccr7KO-A; n = 8) compared with WT (WT-A; n = 10) mice. **P < 0.005 starting at day 8. (B to D) T cell infiltrates in Ccr7KO-A compared with WT-A breast tumors demonstrated by (B) representative images of CD3- and CD8-stained tumor tissues, quantification of (C) CD3+ T cells and (D) CD8+ T cells within hpf images of Ccr7KO-A (n = 7) and WT-A (n = 4) breast tumors (**P < 0.01). (E and F) Metastasis burden in the lung of DMBA-exposed PyMT and PyMTCcr7KO mice demonstrated by (E) representative H&E-stained lung images (arrows point to metastatic foci) and (F) bar graph of the number of lung metastatic foci in each group. n = 5 in each group. Mann-Whitney U test was used. Graphs show means + SD. Scale bars, 100 μm (tumor) and 2 mm (lung).
CCL21 is positively correlated with the mutational burden and T cell infiltration in human cancers and reduced risk of breast cancer distant recurrence
We investigated the correlations of CCL21, CCL23 (homolog of mice Ccl6 in human), and RETN (encoding resistin) that were up-regulated upon DMBA exposure with TMB across The Cancer Genome Atlas (TCGA) cancer types. Median CCL21 expression had positive correlations with median total somatic mutations and missense mutations across TCGA cancer types [P = 0.006 (Rho = 0.47) and P = 0.0099 (Rho = 0.45), respectively; Fig. 8, A and B]. However, no significant correlation between median CCL23 and RETN expressions and TMB was observed across cancer types (fig. S25, A to D). In addition, we did not find any correlations between CCL19 chemokine expression [sharing the same receptor (CCR7) with CCL21] and TMB in TCGA data (fig. S25, E and F). A metric for immune CYT has been validated on the basis of expression levels of granzyme A (GZMA) and perforin (PRF1) across TCGA cancers, which are two key cytolytic effectors and highly up-regulated upon CD8+ T cell activation in the tumors (31, 33). Median CCL21 expression was positively correlated with median CYT across TCGA cancer types [(P = 0.002 (Rho = 0.55); Fig. 8C]. CCL21 was expressed by human breast cancer cells, and breast cancers with high CCL21 levels had significantly higher CD3+ and CD8+ T cell infiltrates compared with CCL21 low breast cancers (P = 0.004 and P = 0.007, respectively; Fig. 9, A to C). In a cohort of 265 hormone receptor–positive (HR+), lymph node–positive primary breast tumors (table S1) (34), high CCL21 expression by tumor cells associated with a significantly lower risk of distant recurrence (P = 0.0174; Fig. 9D). These associations strongly suggest that CCL21 is an important mediator of antitumor immunity in human cancers.
Fig. 8. CCL21 expression is positively associated with TMB and immune CYT across human cancers.
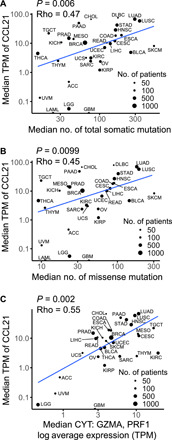
(A to C) Correlations between median TPM of CCL21 and the median number of (A) total somatic mutations, (B) missense mutations, and (C) median immune CYT across 32 TCGA cancer types (Spearman’s rank correlation). As previously described, only solid tumors are included in the CYT correlation in (C) (31). ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Fig. 9. High CCL21 expression by cancer cells is associated with increased TILs and reduced risk of distant recurrence in human breast cancer.
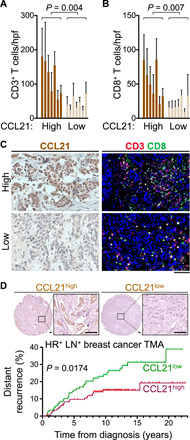
(A and B) T cell infiltrates in human breast cancers with high and low CCL21 levels. Bar graphs show (A) CD3+ T and (B) CD8+ T cell counts in human breast cancers. Each bar represents single breast cancer. Cell counts were based on 8 to 10 randomly selected hpf images. n = 7 in each group. Mann-Whitney U test was used. Graphs show means + SD. (C) Representative images of CCL21 immunohistochemical staining and CD3 and CD8 IF-stained human breast cancer tissues. (D) Representative images of CCL21 immunohistochemical staining on HR+ lymph node–positive (LN+) human breast cancer TMA samples and its correlation with distant recurrence rate. n = 265. clinical characteristics of TMA samples are listed in table S1. Log-rank test was used. Scale bars, 100 μm.
DISCUSSION
To identify the immune-activating factors that can transform cold tumors into hot immunogenic tumors, we focused on carcinogens as the causative agents for increased total and clonal TMB, which is a cardinal cancer characteristic positively associated with cancer immunogenicity and response to immunotherapy (2, 13, 14, 17). Chemical carcinogens such as DMBA are prominent inducers of TMB (35). Therefore, we asked, besides increased TMB, what immune-activating factors are induced by carcinogens in nonimmunogenic cancers such as breast cancer. First, we demonstrate that a single exposure of breast glands to DMBA at puberty accelerates the initiation of primary breast cancer development in spontaneous breast cancer models (18, 19). In stark contrast, carcinogen-exposed mice have a significantly less metastatic burden. CD8+ T cell immunity is responsible for this blockade in metastasis. In addition to increased mutations, carcinogen exposure leads to the up-regulation of CCL21 chemokine in breast cancer cells, which plays a critical role in the immunogenic transformation of breast cancer (36, 37). Last, high CCL21 levels associate with increased TILs in human breast cancers, reduced breast cancer distant recurrence in a large clinical cohort of HR+ lymph node–positive primary breast cancers, and increased TMB and CYT across human cancer types. Therefore, our mechanistic studies on carcinogen-induced cancer immunogenicity have led to the identification of CCL21 as an immune-activating factor expressed by the cancer cells, which can be used for cancer immunotherapy. These outcomes uncover the importance of elucidating the immune-inductive properties of carcinogen exposure besides increased TMB, which can be therapeutically exploited.
Environmental carcinogens initiate and promote cancer development across multiple cancer types (18, 19). However, it has recently become evident that carcinogen-induced cancers have high TMB, contain a high number of TILs at baseline, and show a favorable response to cancer immunotherapy including immune checkpoint blockade (13, 14, 38). The association between increased TMB and cancer immunogenicity has been attributed to the generation of neoantigens in carcinogen-exposed cancers (13, 39, 40). Although neoantigens can provide targets for a tumor-specific T cell response and their recent use as therapeutic vaccines has shown efficacy (41, 42), the induction of mutations in cold tumors to generate neoantigens is not a viable therapeutic option. Accordingly, we find that the cell-autonomous carcinogenic impact of DMBA in the primary breast cancer cells is dominant over any immune-related benefits; a single exposure to DMBA markedly accelerates the primary breast tumor kinetics in PyMT and Her2 mice, which is not altered by the deletion of T cells or the addition of anti-PD1/anti-CTLA4 therapies. However, breast cancer metastasis is blocked in a CD8+ T cell–dependent manner in DMBA-exposed animals. Therefore, our findings suggest that the immune-inductive effect of carcinogen exposure plays a beneficial role against cancer metastasis, where the efficacy of immune checkpoint blockade and its correlation with high TMB have been clinically observed (2).
Metastasis is the leading cause of cancer-related death (43). During metastasis, cancer cells can be exposed, recognized, and killed by the immune system (12). CD8+ cytotoxic T cells play a major role in mediating this immune restriction, and the suppression of CD8+ T cells results in increased cancer metastasis and reduced survival including in patients with breast cancer (7, 27, 44, 45). Despite accelerated primary tumor development, DMBA treatment results in a significantly reduced breast cancer metastasis in PyMT and Her2 mice accompanied by increased tumor-infiltrating CD8+ T cells and CD8+ T cell–to–Treg ratio ratio in the breast tumors and the tumor-draining lymph nodes. In the absence of CD8+ T cells, DMBA exposure results in severe metastatic disease as evidenced by increased metastatic foci in the lung and the appearance of significant liver metastasis. The role of DMBA as the inducer of antitumor T cell immunity against breast cancer metastasis is supported by studies on PyMTRag1KO mice showing a delay in metastasis compared with PyMT animals in the absence of DMBA exposure (29). The similar structure and carcinogenic effects of DMBA to common environmental carcinogens (25) and its ability to increase TMB and induce antitumor T cell immunity in breast cancer models enable the investigation of the immune-activating factors that are up-regulated in the cancer cells in association with carcinogen exposure and increased TMB. These factors can have fundamental roles in the induction of antitumor immunity and be potential candidates for therapeutic intervention.
Among an array of cytokines and chemokines, we have found CCL21 chemokine to be up-regulated in DMBA-exposed breast cancer cells. CCL21 is known to be expressed by high endothelial venules of secondary lymphoid organs such as lymph nodes and spleen, and its signaling through CCR7 recruits and activates T cells during an inflammatory response (46). CCL21 recruits T cells and antigen-stimulated DCs to colocalize into T cell zones of secondary lymphoid organs thereby promoting an early immune activation (26, 47). An intratumoral vaccine of autologous DCs overexpressing CCL21 has shown efficacy in the induction of antitumor T cell immunity in patients with lung cancer (48). Our findings reveal that breast cancer cells themselves express CCL21 in mice and humans, which is up-regulated in cancer cells exposed to carcinogens. Furthermore, CCL21 induction is accompanied by increased APCs and T cells surrounding DMBA-exposed breast tumors in the form of LAs. CCL21 signaling is critical for the immunogenic effects of DMBA on breast cancer cells, and CCL21 is sufficient to promote immunity against cold breast tumors. Therefore, CCL21 intratumoral injection may provide a therapeutic approach to transforming cold breast cancers into hot immunogenic tumors that will be responsive to current immunotherapeutics.
In summary, our results reveal that the immunogenic impact of environmental carcinogens goes beyond the generation of neoantigens and demonstrate how the immune-inductive factors that are up-regulated within the carcinogen-exposed cancer cells can suppress cancer metastasis. Future studies are warranted to identify the full array of cytokines and chemokines that are induced by environmental carcinogens in various cancer types to harness the most potent mediators of antitumor immunity for cancer immunotherapy. Last, our findings indicate that genetically engineered mouse models of cancer, which have very low TMB and immunogenicity at baseline (49–52), can be exposed to carcinogens to generate cancers that more closely resemble human disease. These refined models will be particularly beneficial for mechanistic studies in cancer immunology.
MATERIALS AND METHODS
Mice
All mice were housed under pathogen-free conditions in an animal facility at Massachusetts General Hospital in accordance with animal care regulations. PyMT (a gift of D. DeNardo, Washington University in St. Louis) and PyMTRag1KO mice were maintained on BALB/c background by backcrossing from FVB over 12 generations. PyMTCcr7KO female mice on BALB/c x C57BL/6 background were compared with MMTV-PyMTtg;Ccr7+/− (PyMT) littermates. All other strains of animals were purchased from certified vendors (table S2). Age-matched female mice were used in all the breast cancer studies.
Human tissue samples
Deidentified human breast cancer samples were used in the study. Formalin-fixed paraffin-embedded tissue sections (5 μm) and tissue microarray (TMA) slides were used for manual CCL21 immunohistochemistry (IHC) and CD3/CD8 immunofluorescence (IF) staining. Stained cells were counted blindly.
Spontaneous breast carcinogenesis studies
PyMT and PyMTRag1KO female mice received 1 mg of DMBA (catalog no. D3254, Sigma-Aldrich, St. Louis, MO) dissolved in 100 μl of olive oil or 100 μl of olive oil (carrier alone) by oral gavage at 4 to 6 weeks of age. Her2 mice were treated with 1 mg of DMBA dissolved in 100 μl of olive oil or 100 μl of olive oil by oral gavage at 7 weeks of age. MMTV-PyMTtg;Ccr7+/− (PyMT) and MMTV-PyMTtg;Ccr7−/− (PyMTCcr7KO) female littermates received 1 mg of DMBA dissolved in 100 μl of olive oil by oral gavage three times at 30, 35, and 40 days of age. In the immune checkpoint blockade study, PyMT mice were treated with DMBA as above. When tumors reached 5 mm in diameter, mice received an intraperitoneal injection of 250 μg of anti-PD1 (catalog no. BE0273, Bio X Cell, West Lebanon, NH) or IgG2a isotype (catalog no. BP0089, Bio X Cell) or 200 μg anti-CTLA4 (catalog no. BE0164, Bio X Cell) or IgG2a isotype (catalog no. BE0086, Bio X Cell) every other day for six doses. In the CD8+ T cell depletion study, PyMT mice were treated with DMBA or olive oil as above. Mice received an intraperitoneal injection of 750 μg of anti-CD8α antibody (catalog no. BE0117, Bio X Cell) or IgG isotype (catalog no. BE0090, Bio X Cell) for the first dose delivered 2 days before oral DMBA gavage, and then, the injections were repeated at the dose of 250 μg of antibodies weekly. Tumor onset and growth were monitored every week. The animals were harvested once their tumors reached 2 cm in diameter or the mice showed any sign of distress or weight loss.
Breast cancer cell lines
PyMT breast cancer cell line derived from spontaneous PyMT breast cancer in C57BL/6 background as previously described (53). PyMT cells were passaged at 70% confluency. In the DMBA-treated PyMT cells experiment, PyMT cells were treated with 20 μmol/liter DMBA dissolved in DMSO (catalog no. 25-950-CQC, Thermo Fisher Scientific, Waltham, MA) or DMSO at 30 to 50% confluency for 24 hours. Cells were then washed and passaged. When cells reached 30 to 50% confluency, they were treated by chemicals as above for another 24 hours. In total, cells were treated six times. Zombie Green (catalog no. 423111, BioLegend, San Diego, CA)–negative live single cells were sorted by fluorescence-activated cell sorter and cultured in three 96-well plates (288 pairs), followed by the selection of 12 growth-matched pairs and then 6 growth-matched pairs based on CellTiter 96 Nonradioactive Cell Proliferation Assay (catalog no. PAG4000, Promega, Madison, WI). Last, one growth-matched cell line pair was selected for further experimentation.
In vivo breast cancer cell line studies
For tumor graft experiments, 1 × 105 breast cancer cells were injected into both flanks adjacent to the inguinal mammary fat pads of WT, Rag1KO, and Ccr7KO mice on the C57BL/6 background. Tumor growth was monitored every day. For chemokine injection experiment, WT and Rag1KO mice were injected with 1 μg of recombinant mouse CCL21 (catalog no. 586408, BioLegend) in 50 μl of sterile PBS or 50 μl of sterile PBS alone intratumorally every other day for six times starting at day 6 after cancer cell injection. WT mice were injected with 1 μg of recombinant mouse CCL6 (catalog no. 758504, BioLegend) in 50 μl of sterile PBS or 50 μl of sterile PBS alone intratumorally every other day for four times starting at day 6 after cancer cell injection. For the CD8+ T cell depletion experiment, WT mice received an intraperitoneal injection of 750 μg of anti-CD8β antibody (catalog no. BE0223, Bio X Cell) or IgG isotype (catalog no. BE0094, Bio X Cell) for the first dose delivered 2 days before tumor injection, and then the injections were repeated at the dose of 250 μg of each antibody weekly. The mice were harvested once their tumors reached 2 cm in diameter or the mice showed any sign of distress or weight loss.
Exome sequencing
Genomic DNA (gDNA) was extracted from fresh-frozen samples stored in −80°C with QIAGEN DNeasy Blood and Tissue Kit (catalog no. 69504, QIAGEN). The gDNA was sequenced by next-generation sequencing system at Novogene Co. Ltd. (Beijing, China). Briefly, sequencing libraries and 180– to 280–base pair gDNA fragments were generated. The gDNA fragments were adenylated at 3′ ends and ligated with adapter oligonucleotides. The library of gDNA fragments was enriched by polymerase chain reaction (PCR) reaction and hybridized with liquid phase of a biotin-labeled probe. Exons were captured by magnetic beads with streptomycin and enriched in a PCR reaction. After products were purified and quantified, sequencing was performed using HiSeq 4000.
Somatic mutation calling
The bioinformatics analysis began using high-quality paired-end reads passing Illumina RTA filter aligned to the National Center for Biotechnology Information (NCBI) mouse reference genome (GRCm38) using Burrows-Wheeler Aligner (BWA) (54). PCR duplicated reads were marked and removed using Picard (http://broadinstitute.github.io/picard/). Putative SNVs and insertions/deletions were identified by running Strelka (v1.0.15) with default parameters (55). The putative somatic mutations were compared to the public mouse germline databases including dbSNV (56) and Mouse Genomes Project by the Wellcome Trust Sanger Institute (57) to further exclude false somatic calls introduced by unapparent germline events. The remaining mutations were annotated using ANNOVAR (58) with the NCBI RefSeq (reference sequence) database. Ambiguous putative mutations were manually inspected to exclude potential false calls introduced by likely sequencing or mapping artifacts. The mutational signatures, in terms of the patterns of substitution types of the mutated base with the immediately 5′ and 3′ bases, were summarized using a customized program for SNVs of each tumor.
TCR sequencing
TCRβ chain Complementarity-determining region (CDR) were sequenced at Adaptive Biotechnologies (Seattle, WA) using gDNA isolated from primary breast tumors of PyMT mice. Multiplex PCR was used to amplify rearranged TCR CDR3 sequences and characterize tens of thousands of TCRβ CDR3 chains. T cell clonality and entropy were analyzed by the immunoSEQ Analyzer platform provided by Adaptive Biotechnologies.
Protein analysis
Lysates of mouse breast tumors or serum were used to quantify cytokines by Proteome Profiler Mouse XL Cytokine Array (catalog no. ARY028, R&D Systems, MN, Canada) and mouse enzyme-linked immunosorbent assay (ELISA) kits for CCL21 (catalog no. LS-F23035, LSBio, Seattle, WA), interferon-α (IFN-α) (catalog no. BMS6027TWO, Thermo Fisher Scientific) and IFN-β (catalog no. 439407, BioLegend). Quantification of the spot intensity in the arrays was conducted by subtracting background in ImageJ (version 2.0.0). The CCL21 protein measurement in breast tumors of various weights was performed by collecting tumor samples from DMBA- and olive oil–treated PyMT mice at postnatal day 90 (for small tumors) and end point.
Quantitative PCR
The mouse Ccl21 mRNA levels were determined by quantitative PCR with an ABI 7500 PCR system (Thermo Fisher Scientific) and SYBR Green Supermix (catalog no. 1725121, Bio-Rad). The primers were as follows: mouse CCL21, 5′-GTGATGGAGGGGGTCAGGA-3′ (forward) and 5′-GGGATGGGACAGCCTAAACT-3′ (reverse); and mouse glyceraldehyde-3-phosphate dehydrogenase, 5′-AATGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-GATGCCTGCTTCACCACCTTCT-3′ (reverse).
Flow cytometry
Tumor drainage lymph nodes were lysed and homogenized to collect single-cell suspensions. Tumor single-cell suspensions were also prepared, and TILs were isolated using CD45+ Microbeads on magnetic columns (Miltenyi Biotec Inc.). Cells were stained with the following surface antibodies: CD3, CD4, CD8α, CD25, CD28, CTLA4, MHCI, MHCII, PD1, and T-Cell Immunoglobulin And Mucin Domain-Containing Protein 3 (TIM-3) (table S3). Next, cells were fixed and permeabilized by the True-Nuclear Transcription Factor Buffer Set (BioLegend). After fixation and permeabilization, intracellular stains were performed using the following antibodies: granzyme B and Forkhead box P3 (FOXP3) (table S3). Stained cells were assayed using a BD LSRFortessa flow cytometer (BD Biosciences), and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Histology, IF, and IHC
TMA of 265 HR+ and lymph node–positive primary human breast cancer samples were from a well-annotated retrospective case series at a single academic institution (Massachusetts General Hospital, Boston, MA) (34). Tissue samples were fixed in 4% paraformaldehyde (catalog no. 441244, Sigma-Aldrich) solution in PBS overnight at 4°C. Samples were processed and embedded in paraffin. Paraffin-embedded tissues were cut at 5 μm and stained for hematoxylin and eosin (H&E). For IF staining, sections were incubated with anti-mouse or anti-human primary antibodies, followed by fluorochrome-conjugated secondary antibodies (table S3). Sections were counterstained with 4′,6-diamidino-2-phenylindole nuclear stain. For IHC staining, sections were incubated with anti-mouse PyMT, anti-mouse PD-L1, anti-mouse CCL21, and anti-human CCL21 antibodies, followed by horseradish peroxidase (HRP)–conjugated secondary antibodies (table S3). Sections were stained with ImmPACT DAB Peroxidase (HRP) Substrate kits (table S3) and counterstained with hematoxylin. IF and IHC images were captured and quantified blindly under ×200 magnification [high-power field (hpf)].
TCGA data analyses
Tumor types selected from TCGA studies were adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS), and uveal melanoma (UVM). TCGA gene expression data of untreated primary tumors samples included the available “Level_3” gene-level data, which were downloaded from the Broad GDAC Firehose (https://gdac.broadinstitute.org). Transcript lengths and transcript IDs to gene symbols alignment from GDAC Firehose were performed using UCSC hg19 version (59). Transcripts per kilobase million (TPM) were determined by rescaling coverage estimates by multiple of 1 × 106 and used as normalization counts for all genes. Point mutation data from PanCancer Atlas were downloaded from cBioPortal for Cancer Genomics (http://cbioportal.org), which supplies the following “.maf” (mutation annotation format). TCGA data were analyzed in R software (version 3.5.1).
Calculation of immune “CYT”
CYT was obtained as previously described (31). Briefly, CYT was calculated as the geometric mean TPM of GZMA and PRF1 genes per sample. For each expression value, 0.01 was added before transformation for avoiding logging zero. Only solid tumor types were included for calculating CYT as previously shown (31).
Genotyping
PCR was used to genotype genetically engineered mice. The following primer pairs were used for detection of PyMT transgenes and Rag1 WT and mutant alleles (all primers shown are 5′ to 3′): PyMTtg, ATACTGCTGGAAGAAGACGAAATCCTTG (forward) and CTCTGTGAGTAGCTCTCATTCTCTGACTC (reverse); Rag1+, CCAGTAGATACCATTGCGAAGAGG (forward) and CACGTTCTGTGAACCATGCTCTATC (reverse); Rag1−, CCGCTTCCATTGCTCAGCGG (forward) and ACGTTCTGTGAACCATGCTCTATC (reverse); Ccr7+, TAAGGGCATCTTTGGCATCT (forward) and GGTGATCAAGGCCTCCACT (reverse); Ccr7−, TAAGGGCATCTTTGGCATCT (forward) and AGACTGCCTTGGGAAAAGCG (reverse).
Statistical analysis
Tumor onset and survival analyses were performed using the log-rank test. Fisher’s exact test was used for liver metastasis grade outcomes. Mann-Whitney U test was used as the test of significance for tumor counts, tumor volumes, metastasis burden, cell counts, and somatic mutation burden. t test for the Pearson correlation coefficient was used to determine significance in correlations between T cell entropy, serum CCL21 levels, the number of tumor missense mutations, and the number of lung metastatic foci in mice. The correlations between gene expression and mutations and CYT in human tumors were examined by the Spearman rank correlation test. All statistical tests are two-tailed unless otherwise specified. A P value of less than 0.05 was considered significant. All error bars represent SD.
Study approval
All animal studies were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. Massachusetts General Hospital Institutional Review Board approved the deidentified human tissue study.
Acknowledgments
Funding: S.D. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. K.L. was supported by the National Natural Science Foundation of China (81702527), Natural Science Foundation of Guangdong, China (2016A030313185), and China Scholarship Council. Z.F. was supported by the Department of Obstetrics and Gynecology, Peking University First Hospital, Beijing, China. T.L. was supported by the China Scholarship Council and the Department of Otorhinolaryngology–Head and Neck Surgery, Peking University First Hospital, Beijing, China. This work was funded, in part, by a grant from the Breast Cancer Research Foundation (to D.C.S.). K.L., T.L., Z.F., M.H., Z.Y. and S.D. were supported by grants from the Burroughs Wellcome Fund, Breast Cancer Alliance, Sidney Kimmel Foundation, Cancer Research Institute and the NIH (K08AR068619, DP5OD021353, and U01CA233097). L.W., M.L., Q.H., J.W. and S.L. were supported by a grant from the NIH (U24CA232979). Author contributions: K.L., T.L., Z.F. and S.D. conceived and designed the experiments. K.L., T.L., Z.F., M.H., L.W., Z.Y., M.L., Q.H., J.W. and S.L. performed the experiments and analyzed the data. K.L., T.L., D.C.S. and S.D. interpreted the data. K.L. and S.D. wrote the manuscript. D.C.S. contributed breast cancer clinical samples. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The exome sequencing data can be accessed in the NCBI database, SRA accession no.: PRJNA516951. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/25/eabd8936/DC1
REFERENCES AND NOTES
- 1.Hamid O., Robert C., Daud A., Hodi F. S., Hwu W. J., Kefford R., Wolchok J. D., Hersey P., Joseph R. W., Weber J. S., Dronca R., Gangadhar T. C., Patnaik A., Zarour H., Joshua A. M., Gergich K., Elassaiss-Schaap J., Algazi A., Mateus C., Boasberg P., Tumeh P. C., Chmielowski B., Ebbinghaus S. W., Li X. N., Kang S. P., Ribas A., Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarchoan M., Hopkins A., Jaffee E. M., Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P., Allison J. P., The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Vonderheide R. H., Domchek S. M., Clark A. S., Immunotherapy for breast cancer: What are we missing? Clin. Cancer Res. 23, 2640–2646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanda R., Chow L. Q. M., Dees E. C., Berger R., Gupta S., Geva R., Pusztai L., Pathiraja K., Aktan G., Cheng J. D., Karantza V., Buisseret L., Pembrolizumab in patients with advanced triple-negative breast cancer: Phase Ib KEYNOTE-012 study. J. Clin. Oncol. 34, 2460–2467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A., Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Ali H. R., Provenzano E., Dawson S. J., Blows F. M., Liu B., Shah M., Earl H. M., Poole C. J., Hiller L., Dunn J. A., Bowden S. J., Twelves C., Bartlett J. M. S., Mahmoud S. M. A., Rakha E., Ellis I. O., Liu S., Gao D., Nielsen T. O., Pharoah P. D. P., Caldas C., Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 25, 1536–1543 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Mahmoud S. M. A., Paish E. C., Powe D. G., Macmillan R. D., Grainge M. J., Lee A. H. S., Ellis I. O., Green A. R., Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 29, 1949–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Liu S., Foulkes W. D., Leung S., Gao D., Lau S., Kos Z., Nielsen T. O., Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 16, 432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plitas G., Konopacki C., Wu K., Bos P. D., Morrow M., Putintseva E. V., Chudakov D. M., Rudensky A. Y., Regulatory T cells exhibit distinct features in human breast cancer. Immunity 45, 1122–1134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West N. R., Kost S. E., Martin S. D., Milne K., deLeeuw R. J., Nelson B. H., Watson P. H., Tumour-infiltrating FOXP3+ lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br. J. Cancer 108, 155–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen L. M. E., Ramsay E. E., Logsdon C. D., Overwijk W. W., The immune system in cancer metastasis: Friend or foe? J. Immunother. Cancer 5, 79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi N. A., Hellmann M. D., Snyder A., Kvistborg P., Makarov V., Havel J. J., Lee W., Yuan J., Wong P., Ho T. S., Miller M. L., Rekhtman N., Moreira A. L., Ibrahim F., Bruggeman C., Gasmi B., Zappasodi R., Maeda Y., Sander C., Garon E. B., Merghoub T., Wolchok J. D., Schumacher T. N., Chan T. A., Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samstein R. M., Lee C. H., Shoushtari A. N., Hellmann M. D., Shen R., Janjigian Y. Y., Barron D. A., Zehir A., Jordan E. J., Omuro A., Kaley T. J., Kendall S. M., Motzer R. J., Hakimi A. A., Voss M. H., Russo P., Rosenberg J., Iyer G., Bochner B. H., Bajorin D. F., al-Ahmadie H. A., Chaft J. E., Rudin C. M., Riely G. J., Baxi S., Ho A. L., Wong R. J., Pfister D. G., Wolchok J. D., Barker C. A., Gutin P. H., Brennan C. W., Tabar V., Mellinghoff I. K., DeAngelis L. M., Ariyan C. E., Lee N., Tap W. D., Gounder M. M., D’Angelo S. P., Saltz L., Stadler Z. K., Scher H. I., Baselga J., Razavi P., Klebanoff C. A., Yaeger R., Segal N. H., Ku G. Y., DeMatteo R. P., Ladanyi M., Rizvi N. A., Berger M. F., Riaz N., Solit D. B., Chan T. A., Morris L. G. T., Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roh W., Chen P. L., Reuben A., Spencer C. N., Prieto P. A., Miller J. P., Gopalakrishnan V., Wang F., Cooper Z. A., Reddy S. M., Gumbs C., Little L., Chang Q., Chen W. S., Wani K., de Macedo M. P., Chen E., Austin-Breneman J. L., Jiang H., Roszik J., Tetzlaff M. T., Davies M. A., Gershenwald J. E., Tawbi H., Lazar A. J., Hwu P., Hwu W. J., Diab A., Glitza I. C., Patel S. P., Woodman S. E., Amaria R. N., Prieto V. G., Hu J., Sharma P., Allison J. P., Chin L., Zhang J., Wargo J. A., Futreal P. A., Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9, eaah3560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X. Q., Lu H., Nebozhyn M., Zhang C., Lunceford J. K., Joe A., Cheng J., Webber A. L., Ibrahim N., Plimack E. R., Ott P. A., Seiwert T. Y., Ribas A., McClanahan T. K., Tomassini J. E., Loboda A., Kaufman D., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litchfield K., Reading J. L., Puttick C., Thakkar K., Abbosh C., Bentham R., Watkins T. B. K., Rosenthal R., Biswas D., Rowan A., Lim E., al Bakir M., Turati V., Guerra-Assunção J. A., Conde L., Furness A. J. S., Saini S. K., Hadrup S. R., Herrero J., Lee S. H., van Loo P., Enver T., Larkin J., Hellmann M. D., Turajlic S., Quezada S. A., McGranahan N., Swanton C., Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassar D., Latil M., Boeckx B., Lambrechts D., Blanpain C., Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat. Med. 21, 946–954 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Khaled W. T., Choon Lee S., Stingl J., Chen X., Raza Ali H., Rueda O. M., Hadi F., Wang J., Yu Y., Chin S. F., Stratton M., Futreal A., Jenkins N. A., Aparicio S., Copeland N. G., Watson C. J., Caldas C., Liu P., BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat. Commun. 6, 5987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn J., Xia T., Konno H., Konno K., Ruiz P., Barber G. N., Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatzinikolaou G., Karakasilioti I., Garinis G. A., DNA damage and innate immunity: Links and trade-offs. Trends Immunol. 35, 429–435 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Jones M. E., Schoemaker M. J., Wright L. B., Ashworth A., Swerdlow A. J., Smoking and risk of breast cancer in the Generations Study cohort. Breast Cancer Res. 19, 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vargo-Gogola T., Rosen J. M., Modelling breast cancer: One size does not fit all. Nat. Rev. Cancer 7, 659–672 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hecht S. S., Lung carcinogenesis by tobacco smoke. Int. J. Cancer 131, 2724–2732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samanta S. K., Singh O. V., Jain R. K., Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. Trends Biotechnol. 20, 243–248 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Friedman R. S., Jacobelli J., Krummel M. F., Surface-bound chemokines capture and prime T cells for synapse formation. Nat. Immunol. 7, 1101–1108 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Eyles J., Puaux A. L., Wang X., Toh B., Prakash C., Hong M., Tan T. G., Zheng L., Ong L. C., Jin Y., Kato M., Prévost-Blondel A., Chow P., Yang H., Abastado J. P., Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J. Clin. Invest. 120, 2030–2039 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P., Li Q. J., Feng Y., Zhang Y., Markowitz G. J., Ning S., Deng Y., Zhao J., Jiang S., Yuan Y., Wang H. Y., Cheng S. Q., Xie D., Wang X. F., TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 22, 291–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNardo D. G., Barreto J. B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., Coussens L. M., CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadara H., Choi M., Zhang J., Parra E. R., Rodriguez-Canales J., Gaffney S. G., Zhao Z., Behrens C., Fujimoto J., Chow C., Yoo Y., Kalhor N., Moran C., Rimm D., Swisher S., Gibbons D. L., Heymach J., Kaftan E., Townsend J. P., Lynch T. J., Schlessinger J., Lee J., Lifton R. P., Wistuba I. I., Herbst R. S., Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann. Oncol. 28, 75–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooney M. S., Shukla S. A., Wu C. J., Getz G., Hacohen N., Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Johnson B. J., Costelloe E. O., Fitzpatrick D. R., Haanen J. B. A. G., Schumacher T. N. M., Brown L. E., Kelso A., Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc. Natl. Acad. Sci. U.S.A. 100, 2657–2662 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Schroeder B. E., Jerevall P. L., Ly A., Nolan H., Schnabel C. A., Sgroi D. C., A novel breast cancer index for prediction of distant recurrence in HR+ early-stage breast cancer with one to three positive nodes. Clin. Cancer Res. 23, 7217–7224 (2017). [DOI] [PubMed] [Google Scholar]
- 35.McCreery M. Q., Halliwill K. D., Chin D., Delrosario R., Hirst G., Vuong P., Jen K. Y., Hewinson J., Adams D. J., Balmain A., Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat. Med. 21, 1514–1520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizeq B., Malki M. I., The role of CCL21/CCR7 chemokine axis in breast cancer progression. Cancers 12, 1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S., Lu X., Zhang Z. L., Lei P., Hu P., Wang M., Huang B., Xing W., Jiang X. T., Liu H. J., Zhu Z. G., Li W. H., Zhu H. F., Fu N., Shen G. X., CC chemokine ligand 21 enhances the immunogenicity of the breast cancer cell line MCF-7 upon assistance of TLR2. Carcinogenesis 32, 296–304 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Lauss M., Donia M., Harbst K., Andersen R., Mitra S., Rosengren F., Salim M., Vallon-Christersson J., Törngren T., Kvist A., Ringnér M., Svane I. M., Jönsson G., Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 8, 1738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdegaal E. M. E., de Miranda N. F. C. C., Visser M., Harryvan T., van Buuren M. M., Andersen R. S., Hadrup S. R., van der Minne C. E., Schotte R., Spits H., Haanen J. B. A. G., Kapiteijn E. H. W., Schumacher T. N., van der Burg S. H., Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 536, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Segal N. H., Parsons D. W., Peggs K. S., Velculescu V., Kinzler K. W., Vogelstein B., Allison J. P., Epitope landscape in breast and colorectal cancer. Cancer Res. 68, 889–892 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Keskin D. B., Anandappa A. J., Sun J., Tirosh I., Mathewson N. D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E., Shukla S. A., Hu Z., Li L., le P. M., Allesøe R. L., Richman A. R., Kowalczyk M. S., Abdelrahman S., Geduldig J. E., Charbonneau S., Pelton K., Iorgulescu J. B., Elagina L., Zhang W., Olive O., McCluskey C., Olsen L. R., Stevens J., Lane W. J., Salazar A. M., Daley H., Wen P. Y., Chiocca E. A., Harden M., Lennon N. J., Gabriel S., Getz G., Lander E. S., Regev A., Ritz J., Neuberg D., Rodig S. J., Ligon K. L., Suvà M. L., Wucherpfennig K. W., Hacohen N., Fritsch E. F., Livak K. J., Ott P. A., Wu C. J., Reardon D. A., Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott P. A., Hu Z., Keskin D. B., Shukla S. A., Sun J., Bozym D. J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., Chen C., Olive O., Carter T. A., Li S., Lieb D. J., Eisenhaure T., Gjini E., Stevens J., Lane W. J., Javeri I., Nellaiappan K., Salazar A. M., Daley H., Seaman M., Buchbinder E. I., Yoon C. H., Harden M., Lennon N., Gabriel S., Rodig S. J., Barouch D. H., Aster J. C., Getz G., Wucherpfennig K., Neuberg D., Ritz J., Lander E. S., Fritsch E. F., Hacohen N., Wu C. J., An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehlen P., Puisieux A., Metastasis: A question of life or death. Nat. Rev. Cancer 6, 449–458 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Bidwell B. N., Slaney C. Y., Withana N. P., Forster S., Cao Y., Loi S., Andrews D., Mikeska T., Mangan N. E., Samarajiwa S. A., de Weerd N. A., Gould J., Argani P., Möller A., Smyth M. J., Anderson R. L., Hertzog P. J., Parker B. S., Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 18, 1224–1231 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Coffelt S. B., Kersten K., Doornebal C. W., Weiden J., Vrijland K., Hau C. S., Verstegen N. J. M., Ciampricotti M., Hawinkels L. J. A. C., Jonkers J., de Visser K. E., IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willimann K., Legler D. F., Loetscher M., S. Roos R., Belen Delgado M., Clark-Lewis I., Baggiolini M., Moser B., The chemokine SLC is expressed in T cell areas of lymph nodes and mucosal lymphoid tissues and attracts activated T cells via CCR7. Eur. J. Immunol. 28, 2025–2034 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Cyster J. G., Chemokines and the homing of dendritic cells to the T cell areas of lymphoid organs. J. Exp. Med. 189, 447–450 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J. M., Lee M. H., Garon E., Goldman J. W., Salehi-Rad R., Baratelli F. E., Schaue D., Wang G., Rosen F., Yanagawa J., Walser T. C., Lin Y., Park S. J., Adams S., Marincola F. M., Tumeh P. C., Abtin F., Suh R., Reckamp K. L., Lee G., Wallace W. D., Lee S., Zeng G., Elashoff D. A., Sharma S., Dubinett S. M., Phase I trial of intratumoral injection of CCL21 gene–modified dendritic cells in lung cancer elicits tumor-specific immune responses and CD8+T-cell infiltration. Clin. Cancer Res. 23, 4556–4568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DuPage M., Jacks T., Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr. Opin. Immunol. 25, 192–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McFadden D. G., Politi K., Bhutkar A., Chen F. K., Song X., Pirun M., Santiago P. M., Kim-Kiselak C., Platt J. T., Lee E., Hodges E., Rosebrock A. P., Bronson R. T., Socci N. D., Hannon G. J., Jacks T., Varmus H., Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 113, E6409–E6417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gross E. T., Han S., Vemu P., Peinado C. D., Marsala M., Ellies L. G., Bui J. D., Immunosurveillance and immunoediting in MMTV-PyMT-induced mammary oncogenesis. Onco. Targets. Ther. 6, e1268310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rautela J., Baschuk N., Slaney C. Y., Jayatilleke K. M., Xiao K., Bidwell B. N., Lucas E. C., Hawkins E. D., Lock P., Wong C. S., Chen W., Anderson R. L., Hertzog P. J., Andrews D. M., Möller A., Parker B. S., Loss of host type-I IFN signaling accelerates metastasis and impairs NK-cell antitumor function in multiple models of breast cancer. Cancer Immunol. Res. 3, 1207–1217 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Demehri S., Cunningham T. J., Manivasagam S., Ngo K. H., M. Tuchayi S., Reddy R., Meyers M. A., DeNardo D. G., Yokoyama W. M., Thymic stromal lymphopoietin blocks early stages of breast carcinogenesis. J. Clin. Invest. 126, 1458–1470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saunders C. T., Wong W. S. W., Swamy S., Becq J., Murray L. J., Cheetham R. K., Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., Sirotkin K., dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., Furlotte N. A., Eskin E., Nellåker C., Whitley H., Cleak J., Janowitz D., Hernandez-Pliego P., Edwards A., Belgard T. G., Oliver P. L., McIntyre R. E., Bhomra A., Nicod J., Gan X., Yuan W., van der Weyden L., Steward C. A., Bala S., Stalker J., Mott R., Durbin R., Jackson I. J., Czechanski A., Guerra-Assunção J. A., Donahue L. R., Reinholdt L. G., Payseur B. A., Ponting C. P., Birney E., Flint J., Adams D. J., Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K., Li M., Hakonarson H., ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W., Haussler D., Kent W. J., The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/25/eabd8936/DC1


