Abstract
STUDY QUESTION
Do phytocannabinoids (PCs) affect follicular endocannabinoid signalling and the epigenome in the surrounding granulosa cells (GCs)?
SUMMARY ANSWER
Exposure to PCs increases the expression of endocannabinoid receptors and reduces DNA methylation enzyme expression and global DNA methylation in naïve GCs.
WHAT IS KNOWN ALREADY
Cannabis plant derivatives, known as PCs, are used for medicinal and recreational purposes. The main PC, tetrahydrocannabinol (THC), is the third most commonly used substance by women of childbearing age, hence knowledge of the effect it has on reproduction is of utmost importance. THC exerts its effects via receptors of the endocannabinoid system (ECS) and can interfere with folliculogenesis, oocyte development and ovulation. Endocannabinoids have been measured in follicular fluid (FF) obtained during oocyte retrieval and are implicated in controlling folliculogenesis. It has been established that in the placenta, PCs disrupt endocannabinoid homeostasis via impairment of the synthetic and degrading enzymes, leading to a net increase of endocannabinoid levels. Finally, previous studies have shown that THC alters methylation and histone modifications in sperm, brain and blood cells.
STUDY DESIGN, SIZE, DURATION
This study included an in vivo cohort assessment of cannabis exposure and its effects on the follicle and in vitro assays conducted to validate the in vivo findings and to explore possible mechanisms of action.
PARTICIPANTS/MATERIALS, SETTING, METHODS
A total of 318 FF samples, from 261 patients undergoing IVF treatment at a private fertility clinic who consented for biobanking biological waste material between January 2018 and July 2019, were included in this study. Concentrations of PCs and endocannabinoids were assessed in FF by liquid chromatography-mass spectrometry (LC-MS/MS). Exposure to PCs was determined based on these measured levels. Levels of both endocannabinoid receptors (CB1R, CB2R) and the de novo DNA methylating enzyme, DNMT3b, in GCs were assessed by flow cytometry both in vitro and in vivo and global DNA methylation was assessed in vitro by ELISA. In vivo effects were assessed by comparing samples positive for at least one PC, with samples negative for all measured PCs. In vitro effects were determined in naive GCs, obtained concurrently with FF samples that had tested negative for all PCs. These GCs were treated with different combinations of the main three PCs.
MAIN RESULTS AND THE ROLE OF CHANCE
Overall, 17 patients (6.4%) were positive for cannabis consumption. Furthermore, the prevalence of cannabis positivity in the FF increased from 4% of the tested samples that were collected prior to national legalisation in October 2018 to 12% of those collected following legalisation. Of note, 59% of patients who tested positive for PCs (10 of 17) reported previous or ongoing exposure to cannabis upon their initial intake. Endocannabinoid levels were not affected by the presence of PCs. CB2R was more prevalent than CB1R in GCs and its expression increased following acute and chronic in vitro exposure to PCs. The expression of DNMT3b and global methylation decreased following exposure, suggesting that cannabis may affect the epigenome in the follicular niche. The acute changes were sustained throughout chronic treatment.
LARGE SCALE DATA
N/A.
LIMITATIONS, REASONS FOR CAUTION
Our study is limited by lack of details regarding mode, frequency and timing of PC consumption. Moreover, we were not able to adequately assess the effect of PCs on immediate or long-term clinical outcomes, due to the small sample size and the lack of follow up. Future, large-scale studies should focus on assess the clinical implications of cannabis exposure, validate our findings, and determine to what extent cannabis affects the epigenome ovarian follicle and the developing oocyte.
WIDER IMPLICATIONS OF THE FINDINGS
To our knowledge, this is the first study measuring PCs in FF by LC-MS/MS. We show that consuming cannabis alters the ECS in the developing follicle, and directly affects DNMT expression and global DNA methylation levels. Cannabis legalisation and use is increasing worldwide, therefore further understanding its role in female fertility and folliculogenesis is critical.
STUDY FUNDING/COMPETING INTEREST(S)
All funding was provided by CReATe Fertility Centre through the reinvestment of clinical earnings. The authors declare no competing interests.
Keywords: cannabis, marijuana, endocannabinoid system, epigenetics, female fertility
Introduction
After tobacco and alcohol, cannabis is the most commonly used substance by women of childbearing age, and with increasing legalisation, knowledge of the effect cannabis has on reproduction is of utmost importance (SAMHSA, 2014). In Canada, the average frequency of consumption among the age range of our patient population (25–44 years old) increased significantly following legalisation of cannabis in October 2018, from 21.4% to 24.2% (Rotermann, 2019).
Delta-9-tetrahydrocannabinol (Δ9-THC), the main psychoactive component of cannabis, is a phytocannabinoid (PC) which is used for medicinal and recreational purposes (Sun and Dey, 2012). Other major PCs identified to date include the non-psychotropic cannabidiol (CBD), the degradation byproduct cannabinol (CBN) and the two main metabolites of Δ9-THC metabolism: 11-OH and 11-COOH-THC (ElSohly et al., 2017).
Identification of Δ9-THC led to the discovery of the endocannabinoid system (ECS) (Maccarrone, 2015). Endocannabinoids (eCBs), ligands of the ECS, are essential for folliculogenesis, oocyte maturation and ovulation, among other reproductive functions, and Δ9-THC mimics their mode of action (Piomelli, 2004; Wang et al., 2006; Taylor et al., 2007; Battista et al., 2008). The two main studied eCBs are N-arachidonoylethanolamine (anandamide/AEA) and 2-arachidonoylglycerol (2-AG) (Maccarrone, 2015; Szutorisz and Hurd, 2016) and they exert their actions primarily via two G-protein-coupled receptors (GPCRs), cannabinoid receptors 1 and 2 (CB1R and CB2R), as does Δ9-THC (Howlett, 2002; Fonseca et al., 2013; Yohn et al., 2015). Both cannabinoid receptors (CBRs) are prevalent in the male and female reproductive systems and are essential for the participation of endocannabinoids in oocyte and sperm development as well as uterine preparation for embryo implantation (Battista et al., 2008). It has been established that in the placenta, PCs disrupt endocannabinoid homeostasis via impairment of the synthetic and degrading enzymes, leading to a net increase in endocannabinoid levels (Maia et al., 2019). Furthermore, following exposure, Δ9-THC accumulates in fat which is why the effects could be sustained even after the exposure has been eliminated (Schuel, 2006; Karasu et al., 2011). Clinical studies assessing the effects of cannabis exposure on female fertility performed to date have been based on self-reporting which introduces significant bias. Objective assessment of cannabis exposure can be facilitated by measuring PC levels in the follicular fluid (FF) which represents the immediate microenvironment of the female germ cells (Pichini et al., 2012).
It is also important to note that environmental exposures can cause epigenetic modifications, which are heritable changes that do not involve alterations in the DNA sequence itself, and may be sudden or may accumulate overtime (Bird, 2007). Epigenetic modifications include, but are not limited to, DNA methylation and histone modifications (Yohn et al., 2015). While there appear to be some common factors that are epigenetically modified following drug exposure, various parts of the complex epigenomic network may be uniquely mediated by different classes of drugs of abuse (Yohn et al., 2015). It is established that Δ9-THC causes genome-wide histone modifications and altered DNA methylation, in the brain, sperm and blood cells. However, to date, these effects have not been assessed in the reproductive system.
The purpose of this study was to enhance our understanding of the role of PCs in human female reproduction and, specifically, the effects they have on the follicular niche. We aimed to achieve this goal by: (i) developing an objective measurement methodology to assess the level of PCs in FF; (ii) measuring concurrent levels of eCBs and CBRs; and (iii) exploring disruption of the epigenome in human granulosa cells (GCs).
Materials and methods
Ethical approval and licensing
This study had IRB approval (Veritas #16518) for the request of previously biobanked biological waste material. All subjects provided written informed consent for the donation and biobanking of their biological waste material, which included collection of FF and GCs, as well as obtaining de-identified clinical information including age, body mass index, ovarian reserve metrics and treatment regimens (Veritas #16487). The purchase, storage and use of Δ9-THC and its metabolites was approved by Health Canada and all procedures were conducted in accordance with the ‘Cannabis Act’ and ‘Cannabis Regulations’ (License #LIC-A4MUR820SB-2020).
Sample collection
All FF samples biobanked from consenting patients undergoing treatment at the CReATe Fertility Center, between January 2018 and July 2019, were utilised to determine the status of exposure to PCs. Patients were treated using a standard IVF antagonist protocol, with initial gonadotropin dosing and subsequent adjustments at the discretion of the treating physician. Ultrasound-guided oocyte retrieval was performed approximately 36 h following trigger injection. All study personnel were blinded to clinical information associated with the tested samples prior to analysis of PC concentrations. Participants with samples that tested positive for one or more of the PCs in the FF were considered positive for cannabis and were assigned to the case group. For a subset of positive samples, both the dominant and subordinate FF was assayed to determine if there are follicular stage-dependent differences in the concentration of PCs. Case patients were matched by demographic and stimulation parameters with patients whose samples were negative for all PCs (control group), in a 1:1 matching. This matched case-control cohort was used to determine the effect PCs have on the levels of eCB in the FF and the expression of CBRs and DNA methylation enzyme in the GCs. To assess the invivo effects of exposure to PCs, corresponding GCs of the two matched groups were retrieved from the biobank and utilised for functional invitro assessment of PCs on endocannabinoid signalling and epigenetic machinery within the follicular niche.
Measurement of phyto- and endocannabinoids in follicular fluid
All samples included in case and control groups, were assayed for endocannabinoid levels (i.e. palmitoylethanolamide (PEA), alpha-linolenoylethanolamide (ALEA), N‐linoleylethanolamine (LEA), N‐oleoylethanolamine (OEA), eicosapentaenoyl ethanolamide (EPEA), anandamide (AEA), O-arachidonoyl ethanolamine (O-AEA), dehydroepiandrosterone (DHEA), 2‐arachidonoylglycerol (2-AG), and 1‐arachidonoylglycerol (1-AG)) (Lin et al., 2012) and the main PC levels (Δ9-THC, 11-OH-THC, 11-COOH-THC, CBD and CBN) (SensAbues, 2012). Measurements were performed by liquid chromatography-mass spectrometry (LC-MS/MS) on the FF samples, at the Analytical Facility for Bioactive Molecules (Hospital for Sick Children, Toronto, CA, USA). Proteins were precipitated using 1:1 methanol: water (v/v), pelleted, and the supernatants were assessed by LC-MS/MS using a QTRAP 5500 (SCIEX, Concord, CA, USA) and Agilent 1290 HPLC (Agilent, Santa Clara, USA). Calibration curves (0.001–200 ng) were generated using known amounts of all phyto- and endocannabinoids of interest (MilliporeSigma, Oakville, CA, USA) to permit absolute quantification. Any sample above the limit of detection (0.001 ng) was considered positive for the detected compound and assigned to the case group. Data acquisition and quantification were performed with Analyst 1.6.2 software (SCIEX, Concord, CA, USA).
Quantifying in vivo effects of PCs on cannabinoid receptors and DNMT3 levels in the follicular niche
Sample collection and flow cytometry analysis
Previously collected and cryopreserved pooled GCs from all aspirated follicles from patients in both case and control groups were thawed rapidly using a 37°C water bath. Cells were washed in DMEM/F12 + 2.5% FBS to remove contaminating cryoprotectants. The resulting GC pellet was resuspended, and cell number and viability were assessed using the Countess automated cell counter (ThermoFisher Scientific, Mississauga, CA, USA). Equal numbers of cells (250 000 viable cells) were distributed into two polypropylene flow cytometry tubes. The first tube was incubated with anti-human CBR1-APC conjugated primary antibody (R&D Systems, Minneapolis, USA) and anti-human CBR2-Alexa Fluor 488 conjugated primary antibody (R&D Systems, Minneapolis, USA) for 60 min at 4°C. The cells were washed three times with PBS + 3%FBS. The final wash contained 1:1000 of the nuclear stain, Hoechst, to aid identification of live cells (ThermoFisher Scientific, Mississauga, CA, USA). The second tube was fixed using PBS + 4% Formaldehyde for 25 min at 4 °C. Fixed cells were washed three times with PBS + 3%FBS and permeabilised using PBS + 0.5% Triton X-100 for 25 min, at room temperature. The fixed and permeabilised cells were washed three times with PBS + 0.2% Triton X-100. The final wash contained 1:1000 Hoechst in PBS + 3%FBS + 0.2%Triton X-100 to aid in identification of live cells (ThermoFisher Scientific, Mississauga, CA, USA). Finally, cells were stained with anti-human DNMT3b-APC conjugated primary antibodies (Miltenyi Biotec, Bergisch Gladbach, Germany) and incubated at 4°C for 60 min. Samples were analysed on a MACSQuant 10 (Miltenyi Biotec, Bergisch Gladbach, Germany) and median fluorescence intensities were calculated (FlowJo10). Fluorescence cut-offs were established utilising the unstained sample to detect the baseline autofluorescence of the cells.
Measuring the effect of cannabis on the follicular microenvironment in vitro
Granulosa cell culture, phytocannabinoid treatment and flow cytometry
Previously collected and cryopreserved GCs from all aspirated follicles from eight patients who tested negative for all PCs were thawed, washed and resuspended, and their viability was assessed as described above. Cells were seeded on an uncoated 10 cm culture dish and cultured in DMEM/F12 + 2.5%FBS for 4 h to allow for contaminating cells to attach. The floating cells were then distributed equally in 24-well culture plates (ThermoFisher Scientific, Mississauga, CA, USA), (100 000 viable cells per well) and incubated overnight (37°C, 5%CO2, 21% O2) in DMEM/F12 + 2.5%FBS. Cells were treated with one of the following treatments: (i) vehicle control (methanol), (ii) 5-Azacytidine (a demethylating agent) (1uM), (iii) Δ9-THC (100 ng/ml and 500 ng/ml), (iv) 11-COOH-THC (200 ng/ml and 500 ng/ml), (v) combined treatment based on the maximum level measured in FF in our invivo experiment (25 ng/ml Δ9-THC + 5 ng/ml 11-OH-THC + 50 ng/ml 11-COOH-THC), or (vi) combined treatment based on previous invitro studies (100 ng/ml Δ9-THC + 50 ng/ml 11-OH-THC + 200 ng/ml 11-COOH-THC) (López-Cardona et al., 2016). Cells were treated for 24 h to mimic acute exposure to PCs or for 120 h to mimic chronic exposure, with daily media changes and drug treatments. Following treatment, cells were lifted using TrypLE (ThermoFisher Scientific, Mississauga, CA, USA) and the resulting cell suspension was divided in half; the first half of the cells were processed immediately for flow cytometry (as described above), and the second half of cells were pelleted and frozen at −80 C for future DNA extraction and analysis (as described below).
DNA extraction
Genomic DNA was isolated from ∼50 000 cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. Briefly, cells were lysed in Buffer AL and homogenised using a Disruptor Genie for 5 min. The genomic DNA was bound to the supplied column and washed using the supplied buffers. Genomic DNA was eluted in 200 µl of Buffer AE. Total genomic DNA concentration was assayed using Qubit dsDNA HS Assay (ThermoFisher Scientific, Mississauga, CA, USA).
Global DNA methylation quantification
Purified genomic DNA (50 ng) was assayed for global DNA methylation (5-mC) using MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit, according to manufacturer’s instructions (Epigentek, Farmingdale, USA). All samples and standards were assayed in duplicate. DNA was diluted and incubated in the precoated plate for 60 min at 37°C with gentle agitation. Any unbound material was washed away with three consecutive washes. The 5-mC detection complex solution, which binds specifically to methylated cytosine residues, was added and incubated for 50 min at room temperature. Any unbound antibody was washed away with five consecutive washes. Finally, the developer solution was added, and the colourimetric reaction was stopped when the highest concentration positive control turned deep blue. The absorbance was read at 450 nm using FilterMax F5 Plate Reader (Molecular Devices, San Jose, USA). The percentage of 5-mC DNA was calculated based on the standard curve and adjusted for total DNA input.
Statistical analysis
An a priori power analysis was conducted to ensure that a 1:1 matching ratio was sufficient to detect an effect of PC exposure on eCB levels and on CBR and DNMT3b expression. With a sample size of 17 cases, matched 1:1 with controls, we were sufficiently powered (b = 0.8, a = 0.05) to detect a moderate to strong effect (odds ratio 3.5). Categorical variables were represented by n and %, and Student’s t-test or Fisher’s exact tests were used to assess statistical significance. Any P-value below 0.05 was considered significant. For continuous outcomes, average and standard errors are presented by error bars, except where indicated. The differences were considered statistically significant if the 95% confidence intervals were not overlapping and did not cross 1. A Holm-Bonferroni correction was applied for multiple comparisons, to lower the chances of a false positive result. Specific tests utilised for each analysis are also mentioned in the figure legends.
Results
Following national legalisation, cannabis FF positivity is more prevalent among our patient population
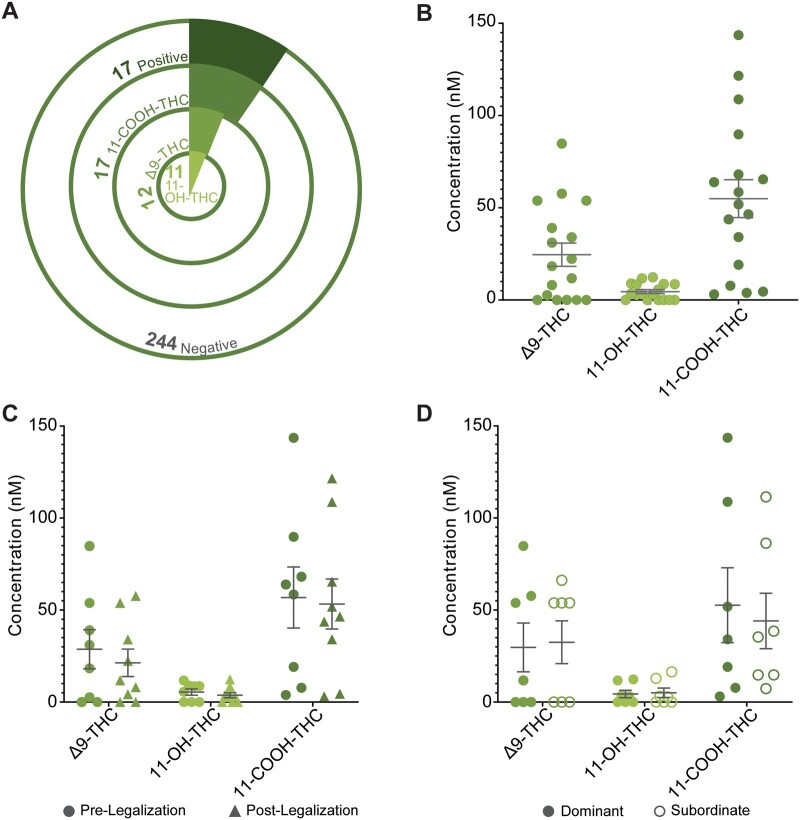
A total of 261 patients (318 individual FF samples) were assessed for exposure to cannabis between January 2018 and July 2019. Patient demographic characteristics are presented in Table I. Ten patients reported previous and/or current exposure to cannabis upon initial intake. Seventeen samples from 17 patients tested positive for at least one PC (6.5% of the tested population) and this group was significantly younger than the patients who tested negative for PCs (Table I). Of these 17 patients, all tested positive for 11-COOH-THC, 11 were positive for 11-OH-THC, and 12 tested positive for Δ9-THC (Fig. 1A). The rate of cannabis FF positivity amongst our patient population increased significantly following national legalisation from 4.3% (8/186) to 12.0% (9/75) (Fisher’s exact test, P < 0.01). The correlation between self-reporting and objective measurements in the FF changed following legalisation as well, from 50% to 67%. The average concentrations of Δ9-THC, 11-OH-THC and 11-COOH-THC in the FF were 32.43 nM, 7.00 nM and 54.95 nM, respectively (Fig. 1B). No patients were positive for CBD or CBN. Despite the frequency of cannabis FF positivity increasing following national legalisation, there was no difference between the FF concentrations of PCs before or after national legalisation (Fig. 1C). For a subset of patients (n = 7 pairs), we assayed both dominant and subordinate follicles, and did not observe a significant difference in concentrations of PCs related to follicular size (Fig. 1D). This study was underpowered to detect a significant difference in ART outcomes between cases who tested positive for PCs and controls (Supplementary Table SI).
Table I.
Patient demographics for all FF assessed for PCs.
| All patients (n = 261) | Patients with PCs in FF (n = 17) | Patients who were negative for all measured PCs in FF (n = 244) | Significance | |
|---|---|---|---|---|
| Age (years) | 32.3 ± 0.4 (21–45) | 28.3 ± 1.3 (21–40) | 32.7 ± 0.4 (22–45) | P = 0.006 |
| BMI (kg/m2) | 24.4 ± 0.3 (14.9–44.6) | 24.9 ± 1.4 (16.8–38.4) | 24.4 ± 0.3 (14.9–44.6) | NS |
| AMH (pmol/L) | 29.6 ± 1.5 (1.2–127.8) | 27.5 ± 5.2 (4.0–94.2) | 29.7 ± 1.7 (1.2–127.8) | NS |
| Day 3 FSH (IU) | 6.4 ± 0.1 (0.2–12.2) | 5.9 ± 0.6 (0.2–8.4) | 6.4 ± 0.2 (0.5–12.2) | NS |
| Peak oestradiol (pmol/L) | 14418.5 ± 530.9 (2363–43 301) | 13595.8 ± 1912.4 (5460–28 472) | 14478.3 ± 612.3 (2363–43 301) | NS |
FF, follicular fluid. NS, not significant. PC, phytocannabinoid.
Figure 1.
Assessment of phytocannabinoids in FF from the general patient population. (A) Frequency of cases in study population divided by type of measured phytocannabinoids, (B) Concentration of individual phytocannabinoids in cases (n = 17), (C) Concentration of phytocannabinoids before (n = 8) and after (n = 9) national legalisation, (D) Concentration of PCs in FF from both dominant and subordinate follicles (n = 7 pairs). Error bars depict standard error. Significance was determined using Student’s t-test assuming equal variance. *P < 0.05, **P < 0.01. FF, follicular fluid; PC, phytocannabinoid.
In vivo implications of cannabis FF positivity
Endocannabinoid levels in FF are not affected by cannabis FF positivity
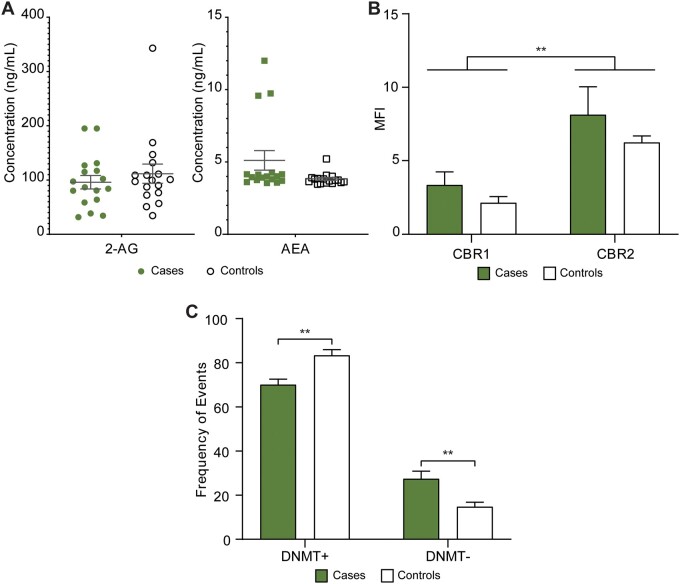
To determine if exogenous PCs alter the delicate internal signalling of the endocannabinoid system in the human follicle, we assayed the levels of endocannabinoids in the 17 cases and in 17 matched controls. The patient characteristics that were the basis for this matching are presented in Table II. All tested endocannabinoids (PEA, ALEA, LEA, OEA, EPEA, AEA, O-AEA, DHEA, 2-AG and 1-AG) were detected except for EPEA and ALEA. None of the measured levels of eCBs differed significantly between the cases and controls. Figure 2A depicts the measured levels of 2-AG and AEA, the primary eCB ligands for CB1R and CB2R.
Table II.
Patient demographics of matched case-controls for in vitro assessment of PCs.
| Cases (n = 17) | Controls (n = 17) | Significance | |
|---|---|---|---|
| Age (years) | 28.3 ± 1.3 (21–40) | 31.3 ± 1.2 (23–41) | NS |
| BMI (kg/m2) | 24.9 ± 1.4 (16.8–38.4) | 24.5 ± 1.3 (14.9–38.4) | NS |
| AMH (pmol/L) | 27.5 ± 5.2 (4.0–94.2) | 26.1 ± 5.7 (6.2–80.3) | NS |
| Day 3 FSH (IU) | 5.9 ± 0.6 (0.2–8.4) | 7.0 ± 0.4 (0.1–5.6) | NS |
| Peak oestradiol (pmol/L) | 13595.8 ± 1912.4 (5460–28 472) | 10223.9 ± 974.0 (5864–17 457) | NS |
NS, not significant. PC, phytocannabinoid.
Figure 2.
Measuring the impact cannabis consumption has on the endocannabinoid system and on epigenetic machinery in vivo. (A) Concentration of endocannabinoids in FF from cases (samples that were positive for at least one phytocannabinoid) and matched controls (samples that were negative for at all phytocannabinoids) (n = 17 pairs), (B) median fluorescence intensity (MFI) of cannabinoid receptor 1 (CBR1) and 2 (CBR2) on the cell surface of granulosa cells from cases (n = 8) and matched controls (n = 6), (C) frequency of events either positive (DNMT+) or negative (DNMT−) for DNMT3b expression in granulosa cells from cases (n = 8) and matched controls (n = 6). Error bars depict standard error. Significance was determined using two-way ANOVA with Bonferroni's multiple comparisons test, **P < 0.01. FF, follicular fluid.
Cannabis positivity did not alter CBRs expression in the follicular niche
The overall expression (median fluorescence intensity (MFI)) of CB2R was significantly higher than that of CBR1 (two-way ANOVA, P = 0.002). However, this expression was not significantly altered following exposure to cannabis (Fig. 2B).
Cannabis positivity alters DNA methylation machinery in the follicular niche
DNMT3b, a DNA methylation enzyme involved in denovo methylation, is crucial for epigenetic integrity. There was a significantly higher frequency of DNMT3b negative events in patients who had consumed cannabis when compared with matched controls (1.86-fold, two-way ANOVA, Bonferroni’s multiple comparisons test, P = 0.006) and a significant decrease in the DNMT3b positive events (1.19-fold, two-way ANOVA, Bonferroni’s multiple comparisons test, P = 0.004) (Fig. 2C).
In vitro assessment of the effect of PC exposure on the endocannabinoid system and epigenetic processes within the follicular niche
Cannabis treatment in vitro significantly alters the expression of CB2R but not CB1R
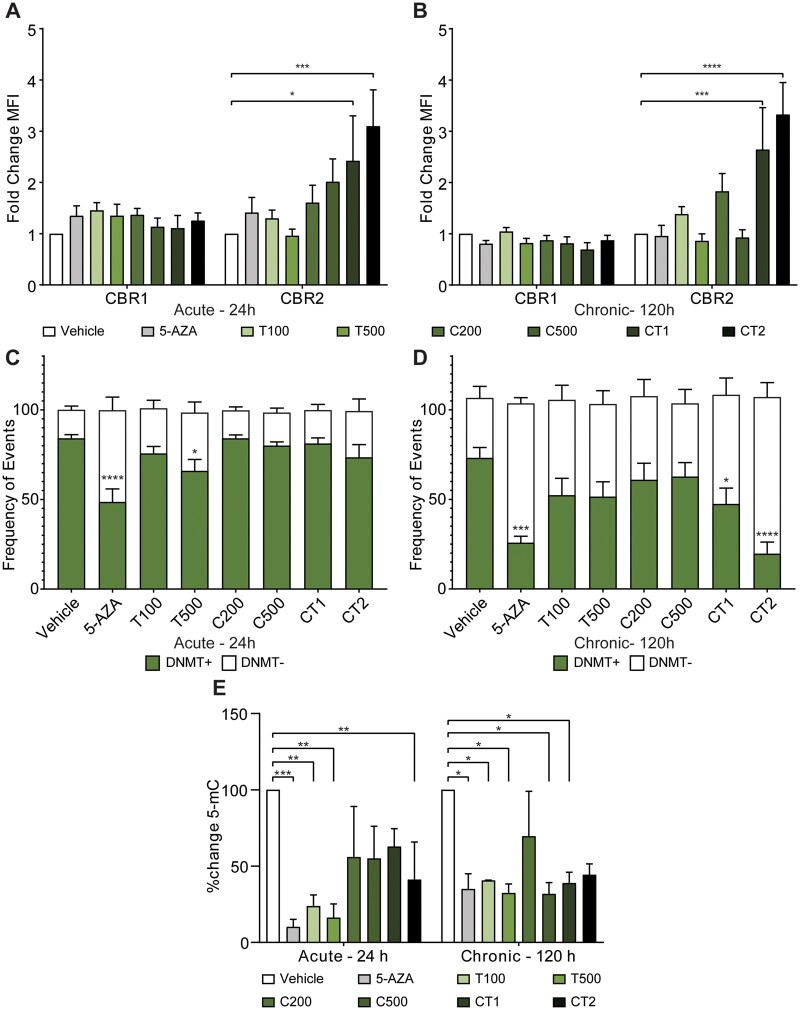
To further explore the potential effect PCs have on CBRs, we assessed their expression in vitro, which allowed us to control for the exposure time and specific metabolite milieu, and explore both the physiological and supraphysiological concentrations. Overall, CB2R was present at higher levels than CB1R on the cell surface (Supplementary Fig. S1). Following different acute and chronic treatment regiments, there was no change in CB1R MFI. The two different acute (24 h) dosing regimens of the combination treatments (CT1 and CT2) caused a significant increase in CB2R expression (2.42-fold (P < 0.05), and 3.10-fold (P < 0.001), respectively) (Fig. 3A). The significant difference between CT1/CT2 and the control was maintained following chronic treatment (120 h) (Fig. 3B).
Figure 3.
Assessment of in vitro exposure of phytocannabinoids in naive GCs on cannabinoid receptors, DNA methylating enzymes and global DNA methylation. (A) Median fluorescence intensity (MFI) of cannabinoid receptor 1 (CBR1) and 2 (CBR2) on the cell surface of granulosa cells following treatment with phytocannabinoids in vitro for 24 h (n = 8), (B) MFI of CBR1 and CBR2 following 120 h treatment (n = 8), (C and D) frequency of events either positive (DNMT+) or negative (DNMT−) for DNMT3b expression in granulosa cells following treatment with PCs in vitro for (C) 24 h (n = 8) or (D) following 120 h treatment (n = 8), (E) percent change in global DNA methylation from vehicle treated control following 24 h and 120 h treatments of GCs in vitro (n = 4). Cells were treated with: vehicle control; 5-Azacytidine (5-AZA, 1 µM), a demethylating agent; Δ9-THC (T, 100 ng/ml and 500 ng/ml); 11-COOH-THC (C, 200 ng/ml and 500 ng/ml); CT (combined treatment) 1, 25 ng/ml Δ9-THC + 5 ng/ml 11-OH-THC + 50 ng/ml 11-COOH-THC; or CT2, 100 ng/ml Δ9-THC + 50 ng/ml 11-OH-THC + 200 ng/ml 11-COOH-THC. Error bars depict standard error. Significance was determined using two-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Cannabis treatment in vitro decreases expression of the de novo DNA methylation enzyme; DNMT3b
Figure 3C depicts the frequencies of DNMT3b negative and positive events following acute treatment (24 h) with the different regimens. Higher dosages, specifically of Δ9-THC 500 ng/ml, caused a significant decrease in the prevalence of positive events from 84.1% in the vehicle-treated control to 65.9% (Fig. 3C). Sustained exposure (120 h) further decreased the proportion of positive DNMT3b events across most treatment groups. Specifically, exposure to CT1 (at concentrations measured in our invivo studies) caused a decrease in the proportion of positive DNMT3b events to 52.1% (P < 0.05) and CT2 decreased these events to 19.7% (P < 0.0001) (Fig. 3D). When given as a sole treatment, 11-COOH-THC did not alter expression levels of DNMT3b at any time point or dosage. The flow cytometry histograms for DNMT3b following invitro cannabis exposure are shown in Supplementary Fig. S2.
Cannabis treatment in vitro decreases global DNA methylation
The same patient samples treated with PCs and assessed for CBRs and DNMT3b by flow cytometry were also assessed for global DNA methylation. Figure 3E depicts the percent change in methylated cytosine residues following different treatment regimens. When compared with the vehicle-treated control within each time point (to control for the reduction in methylation following extended culture), both dosages of Δ9-THC (100 ng/ml and 500 ng/ml) decreased DNA methylation significantly following acute exposure (24 h) by 76.2% and 83.8%, respectively. In addition, these changes were also sustained during chronic exposure (120 h). Treatment with 500 ng/ml of 11-COOH-THC decreased DNA methylation significantly only following chronic (120 h) exposure, by 68.2%. A combined treatment with all three PCs, based on concentrations measured during our invivo study (i.e. CT1—25 ng/ml Δ9-THC, 5 ng/ml 11-OH-THC, and 50 ng/ml 11-COOH-THC) caused a 37.2% decrease in 5-mC following acute exposure (24 h); an effect which was further enhanced to a reduction of 61.1% by chronic exposure (120 h). Finally, when treating with a combined regimen based on previous invitro studies (CT2—100 ng/ml Δ9-THC, 50 ng/ml 11-OH-THC, and 200 ng/ml 11-COOH-THC), there was a significant reduction in 5-mC following acute exposure (24 h), by 58.9%; however, this metric did not change any further following chronic exposure (120 h) (Fig. 3E).
Discussion
Legalisation of cannabis in Canada led to increased cannabis positivity by three-fold amongst our patient population and is now aligned with the Canadian national average consumption in females, regardless of age group (12%) (Rotermann, 2019). It is interesting to note that we did not observe an effect of legalisation on the types of consumed products, as the measured metabolites were of similar concentrations. These results are based on a relatively small sample size obtained close to the national legalisation of Cannabis. It would be interesting to explore the dynamics as time from legalisation passes. Several clinical studies have shown that women who use cannabis are at higher risk of ovulatory disorders (Mueller et al., 1990; Jukic et al., 2007; Szutorisz and Hurd, 2016), and when they undergo IVF, they may produce fewer oocytes, with diminished quality, and have lower pregnancy rates (Klonoff-Cohen et al., 2006). However, these studies were based on self-reporting which introduces significant bias. Objective measurements of PCs in humans reported to date include urine testing and hair sampling, but not FF testing which represents the immediate microenvironment of the female germ cells (Pichini et al., 2012).
Herein, we provide the first objective assessment of the prevalence of cannabis consumption in patients undergoing fertility treatments, alongside a description of how it was affected by legalisation. Of note, the concordance between self-reporting and FF positivity showed a trend of improvement following national legalisation (59% to 66%); however, this limited concordance should still be considered when designing clinical studies addressing the implications of cannabis exposure on fertility. Furthermore, we demonstrate the cannabis-related disruption of the endocannabinoid system, and the effects of cannabis on the epigenetic machinery in the follicular niche. This study is novel in the methodology chosen to investigate the relationship between cannabis exposure and human female fertility. First, by modifying an assay to measure concentrations of the active form of cannabis, Δ9-THC and its main metabolites in FF, we were able to provide an accurate assessment of the extent of FF cannabis positivity in our fertility clinic. Other substances have been previously reported to affect follicles in a stage-dependent manner, however we did not observe this effect for PCs (de Angelis et al., 2020).
The mechanism by which cannabis exposure alters female fertility is yet to be determined. In the current study, we chose to focus on two possible mechanisms: the first being disruption to the endocannabinoid system, and the second being possible epigenetic modifications caused by exposure to PCs. The eCBs exert their actions through two GPCRs (CB1R and CB2R), and are known to be important for human female fertility (Wang et al., 2006; Taylor et al., 2007; Battista et al., 2008; El-Talatini et al., 2009; Bagavandoss and Grimshaw, 2010; Cacciola et al., 2010). By virtue of our study design, we were able to tease out how PCs alter the ECS. While we managed to measure eCBs in FF, reiterating existing literature (Schuel, 2006; Wang et al., 2006; Brents, 2016), none of these seemed to be affected by the presence of PCs. As described earlier (El-Talatini et al., 2009; Cacciola et al., 2010), we observed significantly higher expression of CB2R than of CB1R in GCs, both in vivo and in vitro. Interestingly, while the expression levels of both eCB receptors in the surrounding GCs were not affected by the presence of PCs in our invivo measurements, we did observe a significant increase of CB2R expression following combined treatment with Δ9-THC and its two main metabolites in vitro. The different findings could be explained by the different samples. In their study, De Domenico et al. (2017) was able to show in mice that invitro pharmacological stimulation of the CB2R in oocytes accelerated meiosis and apoptosis and caused a significant reduction in primordial and primary follicles, with a consequent depletion of ovarian reserve. This study used the compound JWH133, which is a selective agonist of CB2R, and has a similar binding affinity to this receptor as Δ9-THC (Pertwee, 2008; Huffman et al., 2010). This raises the question as to whether Δ9-THC could have a similar effect. Taken together, these findings warrant future exploration regarding the role of CB2R in the effects of cannabis consumption on female fertility, and more specifically, on the follicular niche on growing oocytes.
Next, we explored epigenetic modifications as a possible mechanism for the effect PCs exert on female fertility. While it has been established that cannabis can cause epigenetic modifications to the brain, sperm and blood cells (Yang et al., 2014; Watson et al., 2015; Yohn et al., 2015; Santoro et al., 2017; Murphy et al., 2018), this is the first study to explore these effects in the follicular niche. In our invivo experiments, we observed a significantly reduced expression of the denovo methylating enzyme DNMT3b in FF samples from patients who had consumed cannabis. These findings were further validated in our invitro studies. These changes were demonstrated by treating GCs either with Δ9-THC alone or by treating GCs with a combined treatment of all three metabolites, thereby mimicking a real-life scenario following cannabis consumption. The observed effect was substantial, dose dependent and cumulative up to 120 h of treatment. In Canada, 4.5% of female cannabis consumers identify as frequent consumers of cannabis, indicating a significant proportion of the population who are susceptible to these potentially harmful epigenetic changes (Rotermann, 2019).
To further provide evidence that DNA methylation in GCs is affected by cannabis exposure, we performed an ELISA for global DNA methylation. This experiment further demonstrated a meaningful and significant decrease in global DNA methylation following treatment with Δ9-THC alone, with 11-COOH-THC alone and with the combined regimens. These effects were once again, substantial, dose dependent and cumulative up to 120 h of exposure. These findings reinforce what was observed at the DNMT3b level.
Here, we propose a potential model for the mechanism of action of cannabis in the follicular niche: exposure of GCs to PCs increases CB2R cell surface expression which, through an unknown mechanism, reduces DNMT expression, and in turn inhibits the cells ability to maintain epigenetic integrity, thus resulting in loss of DNA methylation.
In this study, invitro experiments were based directly on findings from invivo measurements in the same patient population, and under similar laboratory conditions. This further corroborates our findings and represents a strength of our study design. Furthermore, our invitro studies help mitigate the potential sampling biases in our invivo cohort of samples. Our ability to associate the above concentrations with actual ART outcomes was hindered by the small sample size and should be further explored in larger-scale studies currently underway in our laboratory.
Future studies should focus on: (i) the direct effects PC exposure has on the developing oocyte, (ii) the receptor activity of PCs on CBRs in the developing follicle, (iii) the mechanism of action of PCs on the dysregulation of epigenetic machinery in the cell, (iv) other epigenetic modifications that may be disrupted by PCs, (v) the functional consequences of the observed epigenetic modifications, and (vi) associations between PC concentrations in FF with ART outcomes in larger-scale studies.
Conclusions
To our knowledge, this is the first study to explore the epigenetic implications of cannabis consumption on the follicular niche. As cannabis legalisation increases worldwide, it is critical that we increase our understanding of its role in female fertility, and its potential to cause epigenetic modifications that could be transmitted from parent to offspring.
Supplementary data
Supplementary data are available at Human Reproduction online.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
Acknowledgements
The authors would like to thank: CReATe Fertility Centre patients, for donation of their material to this study, the CReATe BioBank and its personnel, for the provision of samples and associated de-identified data for this study, Ms. Ashley St. Pierre at the Analytical Facility for Bioactive Molecules, for conducting the LC-MS/MS and analysis, and finally, all embryologists, nurses and staff at the clinic, for helping with data collection.
Authors’ roles
N.F.W. and B.A.W. designed this study, analysed the data, interpreted the results and drafted the manuscript. B.A.W. performed the majority of the experiments, with technical guidance and support from P.S. and M.D. performed clinical data analysis. S.J. collected, processed and released samples and raw deidentified clinical data. C.L. critically reviewed and revised the manuscript. All authors read and approved the final version of the paper.
Funding
All funding was provided by CReATe Fertility Centre through the reinvestment of clinical earnings.
Conflict of interest
The authors declare no conflicts of interest.
References
- Bagavandoss P, Grimshaw S.. Temporal and spatial distribution of the cannabinoid receptors (CB1, CB2) and fatty acid amide hydroxylase in the rat ovary. Anat Rec 2010;293:1425–1432. [DOI] [PubMed] [Google Scholar]
- Battista N, Pasquariello N, Di Tommaso M, Maccarrone M.. Interplay between endocannabinoids, steroids and cytokines in the control of human reproduction. J Neuroendocrinol 2008;20(Suppl 1):82–89. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature 2007;447:396–398. [DOI] [PubMed] [Google Scholar]
- Brents LK. Marijuana, the endocannabinoid system and the female reproductive system. Yale J Biol Med 2016;89:175–191. [PMC free article] [PubMed] [Google Scholar]
- Cacciola G, Chianese R, Chioccarelli T, Ciaramella V, Fasano S, Pierantoni R, Meccariello R, Cobellis G.. Cannabinoids and reproduction: a lasting and intriguing history. Pharmaceuticals 2010;3:3275–3323. [Google Scholar]
- de Angelis C, Nardone A, Garifalos F, Pivonello C, Sansone A, Conforti A, Di Dato C, Sirico F, Alviggi C, Isidori A. et al. Smoke, alcohol and drug addiction and female fertility. Reprod Biol Endocrinol 2020;18:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico E, Todaro F, Rossi G, Dolci S, Geremia R, Rossi P, Grimaldi P.. Overactive type 2 cannabinoid receptor induces meiosis in fetal gonads and impairs ovarian reserve. Cell Death Dis 2017;8:e3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElSohly MA, Radwan MM, Gul W, Chandra S, Galal A.. Phytochemistry of Cannabis sativa L. Prog Chem Org Nat Prod 2017;103:1–36. [DOI] [PubMed] [Google Scholar]
- El-Talatini MR, Taylor AH, Elson JC, Brown L, Davidson AC, Konje JC.. Localisation and function of the endocannabinoid system in the human ovary. PLoS One 2009;4:e4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca BM, Correia-Da-Silva G, Almada M, Costa MA, Teixeira NA (2013) The endocannabinoid system in the postimplantation period: a role during decidualization and placentation. Int J Endocrinol 2013;2013:510540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC. The cannabinoid receptors. Prostaglandins Other Lipid Mediat 2002;68–69:619–631. [DOI] [PubMed] [Google Scholar]
- Huffman JW, Hepburn SA, Lyutenko N, Thompson AL, Wiley JL, Selley DE, Martin BR.. 1-Bromo-3-(1',1'-dimethylalkyl)-1-deoxy-Δ(8)-tetrahydrocannabinols: new selective ligands for the cannabinoid CB(2) receptor. Bioorg Med Chem 2010;18:7809–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic AM, Weinberg CR, Baird DD, Wilcox AJ.. Lifestyle and reproductive factors associated with follicular phase length. J Womens Health (Larchmt) 2007;16:1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasu T, Marczylo TH, Maccarrone M, Konje JC.. The role of sex steroid hormones, cytokines and the endocannabinoid system in female fertility. Hum Reprod Update 2011;17:347–361. [DOI] [PubMed] [Google Scholar]
- Klonoff-Cohen HS, Natarajan L, Chen RV.. A prospective study of the effects of female and male marijuana use on in vitro fertilization (IVF) and gamete intrafallopian transfer (GIFT) outcomes. Am J Obstet Gynecol 2006;194:369–376. [DOI] [PubMed] [Google Scholar]
- Lin L, Yang H, Jones PJH. Quantitative analysis of multiple fatty acid ethanolamides using ultra-performance liquid chromatography-tandem mass spectrometry. Prostaglandins Leukot Essent Fatty Acids 2012;87:189–195. [DOI] [PubMed] [Google Scholar]
- López-Cardona AP, Sánchez-Calabuig MJ, Beltran-Breña P, Agirregoitia N, Rizos D, Agirregoitia E, Gutierrez-Adán A. Exocannabinoids effect on in vitro bovine oocyte maturation via activation of AKT and ERK1/2. Reproduction 2016;152:603–612. [DOI] [PubMed] [Google Scholar]
- Maccarrone M. Endocannabinoid signaling in female reproductive events: a potential therapeutic target? Expert Opin Ther Targets 2015;19:1423–1427. [DOI] [PubMed] [Google Scholar]
- Maia J, Midao L, Cunha SC, Almada M, Fonseca BM, Braga J, Goncalves D, Teixeira N, Correia-da-Silva G.. Effects of cannabis tetrahydrocannabinol on endocannabinoid homeostasis in human placenta. Arch Toxicol 2019;93:649–658. [DOI] [PubMed] [Google Scholar]
- Mueller BA, Daling JR, Weiss NS, Moore DE.. 'Recreational drug use and the risk of primary infertility. Epidemiology 1990;1:195–200. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau MH, Price TM, Raburn DJ. et al. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 2018;13:1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol 2008;153:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichini S, De Luca R, Pellegrini M, Marchei E, Rotolo MC, Spoletini R, D'Aloja P, Pacifici R, Mortali C, Scaravelli G.. Hair and urine testing to assess drugs of abuse consumption in couples undergoing assisted reproductive technology (ART). Forensic Sci Int 2012;218:57–61. [DOI] [PubMed] [Google Scholar]
- Piomelli D. The endogenous cannabinoid system and the treatment of marijuana dependence. Neuropharmacology 2004;47:359–367. [DOI] [PubMed] [Google Scholar]
- Rotermann M. (2019) Analysis of Trends in the Prevalence of Cannabis use and Related Metrics in Canada, Ottawa. Ontario, Canada: Statistics Canada82-003-X. [DOI] [PubMed]
- SAMHSA (2014) Results from the 2013 National Survey on Drug Use and Health: Summary of National Findings. Rockville, MD: Substance Abuse and Mental Health Services Administration HHS Publication No. (SMA) 14-4863).
- Santoro M, Mirabella M, De Fino C, Bianco A, Lucchini M, Losavio F, Sabino A, Nociti V.. Sativex® effects on promoter methylation and on CNR1/CNR2 expression in peripheral blood mononuclear cells of progressive multiple sclerosis patients. J Neurol Sci 2017;379:298–303. [DOI] [PubMed] [Google Scholar]
- Schuel H. Tuning the oviduct to the anandamide tone. J Clin Invest 2006;116:2087–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SensAbues (2012) SOP&9;Drug&9;Breath&9;Testing&9;v2.0. Solna, Sweden: SensAbues—Karolinska Institutet Science Park. [Google Scholar]
- Sun X, Dey SK.. Endocannabinoid signaling in female reproduction. ACS Chem Neurosci 2012;3:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Hurd YL.. Epigenetic effects of cannabis exposure. Biol Psychiatry 2016;79:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AH, Ang C, Bell SC, Konje JC.. The role of the endocannabinoid system in gametogenesis, implantation and early pregnancy. Hum Reprod Update 2007;13:501–513. [DOI] [PubMed] [Google Scholar]
- Wang H, Xie H, Dey SK.. Endocannabinoid signaling directs periimplantation events. AAPS J 2006;8:E425–E432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, Hurd YL.. Genome-wide DNA methylation profiling reveals epigenetic changes in the rat nucleus accumbens associated with cross-generational effects of adolescent THC exposure. Neuropsychopharmacology 2015;40:2993–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hegde VL, Rao R, Zhang J, Nagarkatti PS, Nagarkatti M.. Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J Biol Chem 2014;289:18707–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn NL, Bartolomei MS, Blendy JA.. Multigenerational and transgenerational inheritance of drug exposure: the effects of alcohol, opiates, cocaine, marijuana, and nicotine. Prog Biophys Mol Biol 2015;118:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.