Abstract
Background
Comprehensive and up-to-date monitoring of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOC) is crucial as these are characterized by their increased transmissibility, immune evasion and virulence.
Objectives
To describe the wide-scale implementation of a reverse transcriptase polymerase chain reaction (RT-PCR) multiple variants assay with melting curve analysis as a routine procedure.
Study design
We prospectively performed multiple variants RT-PCR on consecutive SARS-CoV-2 RT-PCR positive samples from patients, healthcare workers and nursing home residents from our hospital catchment area. This technique was implemented in our automated Roche FLOW system with a turn-around time of 6 h.
Results
Between February 1 and May 2, 2021, 989 samples were tested by the variant RT-PCR. Our method was validated by comparison of variant RT-PCR to whole genome sequencing testing. We observed an increase over time in the proportion of UK variant that became the dominant variant, and the concurrent emergence of the South-African and Brazilian variants. Prompt public health responses for infection control were possible because of this rapid screening method, resulting in early detection and reduction of unnoticed spread of VOC as early as possible.
Conclusion
A variant RT-PCR with additional melting curve analyses is a feasible, rapid and efficient screening strategy that can be implemented in routine microbiological laboratories.
Keywords: SARS-CoV-2, COVID-19, Melting curve, PCR, Variants, screening
1. Introduction
Many countries are experiencing difficulty in effectively reducing the number of the coronavirus disease 2019 (COVID-19) infections despite extensive, prolonged and stringent infection control measures [1]. The implemented rules and restrictions are effective in combatting the originally dominant variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, these seem to be less sufficient for the more transmissible variants, including those labelled as variants of concern (VOC) by the World Health Organization. The first established VOC share the N501Y mutation: UK (B.1.1.7 or 20I/501Y.V1), South-African (B.1.351 or 20H/501Y.V2) and Brazilian P1 variant (B.1.1.28 or 20 J/501Y.V3) (Table 1 ) [2,3]. As of May 11, 2021, the Indian variant (B.1.617.2 or 21A/S:478 K) was added to the WHO list of VOC. These VOC have the highest priority for adequate surveillance, and might have a higher risk for hospital and intensive care unit admissions [4]. The UK variant has expanded rapidly and became the dominant strain in many countries due to increased transmission advantage [5]. The South African variant appears to pose an additional risk because vaccination might be less effective which could be related to the E484K mutation [6]. The Brazilian variant may cause re-infections in individuals who already experienced COVID-19 [7], and seems to be more virulent in young individuals [8]. More importantly, new VOC may emerge over time.
Table 1.
Interpretation schedule for identification of SARS-CoV-2 variants using mutations found in multiple variants RT-PCR.
| Spike protein mutation | Lightmix® kits | No variant of concern (VOC) | United Kingdom B.1.1.7 | South African B.1.351 | Brazil P1 B.1.1.28 |
|---|---|---|---|---|---|
| H69/V70 | VirSNiP SARS-CoV-2 Spike del H69/V70 | Negative | Positive | Negative | Negative |
| N501Y | VirSNiP SARS-CoV-2 Spike del N501Y | Negative | Positive | Positive | Positive |
| E484K | VirSNiP SARS-CoV-2 Spike E484K | Negative | Negative | Positive | Positive |
| K417N | VirSNiP SARS-CoV-2 Spike K417N | Negative | Negative | Positive | Negative |
The current surveillance and detection of SARS-CoV-2 variants is based on whole genome sequencing (WGS), which is an accurate method that generates detailed information, but is time-consuming and only limited available.
We describe our experience after implementing a reverse transcriptase polymerase chain reaction (RT-PCR) variant assay with melting curve analysis as a routine procedure and as a complementary rapid diagnostic screening method in addition to WGS, which can be widely implemented in most laboratories.
2. Methods
As of February 1, 2021, nasopharyngeal samples from patients, healthcare workers and nursing home residents from our hospital catchment area were initially tested for SARS-CoV-2 by our validated in-house RT-qPCR assay according to the national reference method [9], GeneFinder™ Kit on the ELITe InGenius® [10], or GeneXpert (Cepheid Inc, USA).
Subsequently, all consecutive RT-PCR SARS-CoV-2 positive samples with a cycle threshold (Ct) value of <32 were further analyzed by multiple variants RT-PCR and melting curve analyses [11], [12], [13], targeting relevant SARS-CoV-2 mutations to determine the SARS-CoV-2 variant (Table 1). Samples with Ct values >32 did not result in reliable peak profiles. This is in line with the minimal required RNA amount of 50 to 500 copies as specified by the manufacturer. We used four different SARS-CoV-2 Lightmix® kits (TIB Molbiol GmbH, Germany) to detect the presence of the following mutations: N501Y, del H69/V70, E484K and K417N. The selected primers and probes target specific parts of the virus genome that contain these mutations. Samples were diluted with equal volumes lysis buffer, extraction was done on 500 μL of this mixture with the DNA and Viral NA Large Volume kit (Roche, Germany), and 10 μL of the eluate was used in an end volume of 20 μL in each monoplex PCR well. After RT-PCR amplification on the LightCycler® 480 (Roche Diagnostics, Germany) according to manufacturer's instructions, the temperature of the sample was gradually increased while measuring the amount of fluorescence. By analyzing the melting temperatures which is influenced by whether probes are attached to the targets, it can be determined if specific mutations are present. This technique was implemented in our automated Roche FLOW system with a turn-around time of 6 h, but can also be performed with other standard real-time PCR equipment available in most routine diagnostic microbiology laboratories.
As part of the validation and before implementation of our rapid screening method in routine care, 58 samples were sent for validation by WGS to the Department of Viroscience at the Erasmus Medical Center (Rotterdam, The Netherlands), of which 56 identification findings were confirmed by WGS and for two samples sequencing failed probably due to low virus loads.
3. Results
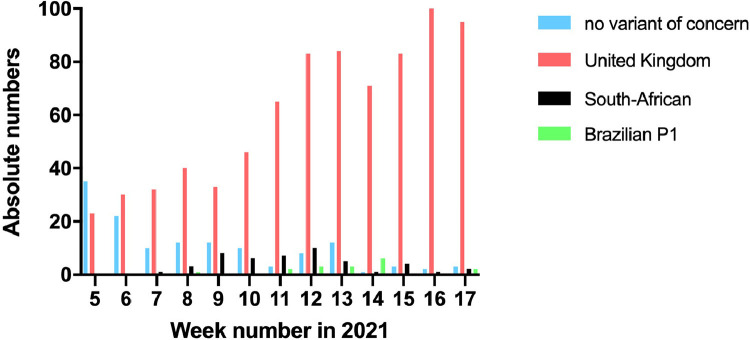
Between February 1 and May 2, 2021, 989 samples were tested by the variant RT-PCR. We observed an increase in the proportion of UK variant that became the dominant variant, followed by the emergence of the South-African and Brazilian variants thereafter (Fig. 1 ). Overall, during a 13-week period we detected 791 (80%) UK, 48 (5%) South-African, 17 (2%) Brazilian P1 SARS-CoV-2 variants, and 133 (13%) no VOC.
Fig. 1.
Number of different SARS-CoV-2 variants over time.
As information regarding variant identification became available within one or two days after sample arrival at our laboratory, quick results were available to clinicians and public health organizations. This allowed a possible prompt public health response for infection control as shown in the following three examples. We detected a cluster of 12 SARS-CoV-2 infections caused by the South African variant in individuals who all had high virus loads, of whom 2 fully vaccinated healthcare workers who did not develop any symptoms, 8 nursing home residents who already received their first vaccination 4 weeks before or earlier and had only mild symptoms, and 2 unvaccinated healthcare workers with severe symptoms. Another South African variant cluster was found among 4 hospitalized crew members of a workboat. In addition, during a short period we observed 14 cases caused by the P1 variant, of which 11 from two adjacent small municipalities.
4. Discussion
By testing of all RT-PCR SARS-CoV-2 positive samples with subsequent variant RT-PCR, we obtained a real-time and complete overview of the circulating SARS-CoV-2 variants in our hospital catchment area. Without any delay in obtaining the results of variant identification the consecutive increase of the UK, and the emergence of South African and Brazilian variants could be observed. In comparison to WGS this PCR-based screening method with melting curve analysis is faster, easy to implement and exceeds in capacity in various settings.
In the Netherlands, the spread of SARS-CoV-2 virus variants is monitored by a national surveillance system. For this a few selected laboratories send a limited number of positive RT-PCR positive samples to the National Institute for Public Health and the Environment (RIVM) for WGS analysis [14]. The current available sequencing capacity limits the number of variant identifications, is labor-intensive and merely a small selection of samples can be sequenced. As a result, sequence results are only available after a few weeks. A faster and more extensive strategy for detecting VOC, and perhaps also other variants of interest, can have a positive impact on adequate and timely contact tracing, and could facilitate targeted public health measures.
Given the dynamic situation of COVID-19 pandemic, additional diagnostic methods are needed for timely results and sufficient screening capacity on variant identification. Screening by RT-PCR SARS-CoV-2 based tests quickly generates real-life and up-to-date information on the spread of VOC, which enables public health and healthcare institutions to take more targeted measures to prevent further spread of VOC at an earlier stage by means of intensified source and contact tracing. If the variant type would only be determined from a very small proportion of positive samples, the risk remains considerable for delayed awareness because emergence of new variants usually start in small clusters that can be easily missed when using a limited surveillance approach only. Furthermore, it can be important to know the presence of specific mutations which are related to increased transmissibility, immune evasion or virulence as soon as possible.
However, there are also limitations to consider. Similar to WGS the variant PCR assay described here also faced inconclusive results in samples with a low virus load. Therefore, only positive samples with Ct-values <32 were considered for variant testing. Moreover, PCR variant testing cannot replace WGS, which remains crucial to discover and detect new emerging variants that have not yet been detected, and to keep existing PCR-based variant screening methods up to date. PCR variant testing should be used as an additional tool to significantly enhance the extent of epidemiological surveillance and contact tracing.
In conclusion, we share our experience regarding a largescale implementation of PCR-based SARS-CoV-2 variant analysis in routine practice to detect and limit the unnoticed spread of VOC as early as possible. In our setting, the variant RT-PCR with additional melting curve analyses shows how a feasible, rapid and efficient screening strategy can be implemented in microbiological laboratories.
5. Contribution
DSYO, JGHK and PdM contributed to the conception and design of the study. DSYO, SB and PdM acquired and analyzed the data. All authors contributed to the interpretation of the data. DSYO drafted the first manuscript and all other authors revised it critically for important intellectual content. All authors approved this manuscript version to be submitted.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
We would like to thank our laboratory technicians and team managers for their assistance in establishing the diagnostic workflow and performing the tests as part of routine standard diagnostics.
References
- 1.Priesemann V., Balling R., Brinkmann M.M., Ciesek S., Czypionka T., Eckerle I., et al. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021;397:469–470. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021:1–17. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 6.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021:1–6. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 7.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Crispim M.A.E., Fraiji N.A., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M.H.S. de Oliveira, G. Lippi, B.M. Henry, Sudden rise in COVID-19 case fatality among young and middle-aged adults in the south of Brazil after identification of the novel B.1.1.28.1 (P.1) SARS-CoV-2 strain: analysis of data from the state of Parana, medRxiv. (n.d.). https://doi.org/ 10.1101/2021.03.24.21254046.
- 9.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2431. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong D.S.Y., Claas E.C.J., Breijer S., Vaessen N. Comparison of the GeneFinderTM COVID-19 Plus RealAmp Kit on the sample-to-result Platform ELITe InGenius to the national reference method: an added value of N gene target detection? J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan K.-.H., To K.K.W., Chan B.W.K., Li C.P.Y., Chiu S.S., Yuen K.-.Y., et al. Comparison of pyrosequencing, Sanger sequencing, and melting curve analysis for detection of low-frequency macrolide-resistant mycoplasma pneumoniae quasispecies in respiratory specimens. J. Clin. Microbiol. 2013;51:2592–2598. doi: 10.1128/JCM.00785-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheli S., Pietrantonio F., Clementi E., Falvella F.S. LightSNiP assay is a good strategy for pharmacogenetics test. Front. Pharmacol. 2015;6:114. doi: 10.3389/fphar.2015.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon E., Wittwer C.T. LightCycler technology in molecular diagnostics. J. Mol. Diagn. 2009;11:93–101. doi: 10.2353/jmoldx.2009.080094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.https://www.rivm.nl/coronavirus-covid-19/onderzoek/kiemsurveillance (accessed April 30, 2021).