Summary
COVID-19 is a respiratory tract infection that can affect multiple organ systems. Predicting the severity and clinical outcome of individual patients is a major unmet clinical need that remains challenging due to intra- and inter-patient variability. Here, we longitudinally profiled and integrated more than 150 clinical, laboratory, and immunological parameters of 173 patients with mild to fatal COVID-19. Using systems biology, we detected progressive dysregulation of multiple parameters indicative of organ damage that correlated with disease severity, particularly affecting kidneys, hepatobiliary system, and immune landscape. By performing unsupervised clustering and trajectory analysis, we identified T and B cell depletion as early indicators of a complicated disease course. In addition, markers of hepatobiliary damage emerged as robust predictor of lethal outcome in critically ill patients. This allowed us to propose a novel clinical COVID-19 SeveriTy (COST) score that distinguishes complicated disease trajectories and predicts lethal outcome in critically ill patients.
Subject areas: Immunology, Virology, systems biology
Graphical abstract
Highlights
-
•
Unsupervised integration of clinical, laboratory, and immunological data of COVID-19
-
•
Multi-organ dysfunctions are detectable across all disease severities
-
•
Liver damage is an early marker of complicated disease trajectories
-
•
Novel COST score predicts fatal outcome in complicated COVID-19
Immunology; Virology; Systems biology
Introduction
The severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), which was first described in Wuhan, China, is causing coronavirus disease 2019 (COVID-19) and an ongoing pandemic with more than one million confirmed fatalities thus far (cdc.gov). The SARS-CoV-2 virus belongs to the coronavirus family and primarily infects epithelial cells of the respiratory tract and vascular endothelium (Shang et al., 2020; Varga et al., 2020). Patients with COVID-19 exhibit a wide range of symptoms and disease courses (Goyal et al., 2020; Guan et al., 2020). Most patients suffer from mild clinical features such as fatigue, fever, and dry coughs. However, some individuals develop viral pneumonia, or even severe acute respiratory distress syndrome (ARDS), sepsis, and septic shock with an overall case fatality rate of 5%. Factors that determine an unfavorable disease outcome include age, sex, and preconditions such as arterial hypertension, chronic respiratory diseases, or an impaired immune status (Jordan et al., 2020; Williamson et al., 2020). Observational COVID-19 studies have proposed to distinguish the infection into an early phase that is characterized by viral replication in the respiratory system and a later stage of generalized inflammation (Knight et al., 2020). Especially during the later phase, recent studies hypothesize that severe COVID-19 is associated with a dysregulated immune response, hyperactivation, and lymphocyte depletion, yet the underlying disease mechanism and individual patient trajectories remain ill defined (Knight et al., 2020). Analysis of the immune signature of COVID-19 revealed T cell depletion (Lucas et al., 2020) and distinct cytokine profiles (Del Valle et al., 2020; Wang et al., 2020) in patients with complicated disease course. However, more sophisticated integration of clinical, laboratory, and immunological data are challenging owing to inter-individual variability and difficult data acquisition. Although individual risk factors and clinical laboratory values of a complicated disease course have been described (Braun et al., 2020; Dawood et al., 2020), advanced analysis of the multitude of clinical and immunological factors to disentangle different clinical courses of COVID-19 is highly warranted. Further integrative analysis of multidimensional data might help to understand the complex interplay of viral infiltration, immune activation, and organ dysfunction that is crucial to find sensitive biomarkers of poor outcome.
Recent advances in unsupervised systems analysis have led to important milestones in genomics (Stuart and Satija, 2019), epidemiologic studies (Wiemken and Kelley, 2020), and the understanding of complex disease pathogenesis (Tong et al., 2020). For instance, the integration of laboratory and immunological data shed light on the pathogenesis of sepsis and immune responses in patient subgroups that result in different therapeutic and prognostic consequences (Davenport et al., 2016; Seymour et al., 2019). Thus, integrative analysis of multi-dimensional data from patients can lead to individual therapeutic strategies and precision medicine (Alballa and Al-Turaiki, 2021; Haendel et al., 2018; Jung et al., 2021).
Multi-omics approaches of peripheral blood mononuclear cells (PBMCs) and whole blood revealed a wide range of dysregulations of the immunological landscape. COVID-19 is associated with a strong interferon- response across all cell types and severe disease courses, whereas convalescence in patients with moderate COVID-19 was associated with expansion of granulysin+ CD4+ and CD8+ effector T cells (Zhang et al., 2020). Similarly, HLA-DRhighCD11chigh inflammatory monocytes with interferon- responses are already upregulated in mild COVID-19, whereas patients with severe COVID-19 have dysfunctional monocytes and neutrophils (Schulte-Schrepping et al., 2020). Accordingly, single-cell analysis of bronchoalveolar lavage (BAL) and lungs of patients and animal models of COVID-19 revealed clonal expansion of CD8+ cytotoxic T cells and infiltration of pro-inflammatory monocytes from the periphery (Liao et al., 2020; Speranza et al., 2021). However, a systems biology approach that additionally integrates routinely longitudinally assessed laboratory parameters and vital signs with peripheral immunophenotyping of large cohorts offers the chance for fast clinical translation of predictive measures.
Here, we longitudinally integrated biometrical, clinical, and detailed immunological profiles of a large prospective cohort at the University Medical Center Hamburg-Eppendorf. Using the data of a cohort of 173 patients with COVID-19 with a diverse spectrum of outcomes we provide a comprehensive analysis of more than 150 clinical, laboratory, and immunological parameters and demonstrate temporally distinct patterns of organ dysfunction. More importantly, we find immune cell dysregulation as a pivotal determinant of critical and lethal but not uncomplicated COVID-19. Of note, by exploiting unsupervised clustering and trajectory analysis, we identified distinct laboratory and immunological parameters that together robustly predicted lethal COVID-19 outcome. Therefore, our study provides a unique blueprint that allowed us to assemble a novel risk stratification score for patients with COVID-19.
Results
Study cohort
In order to detect prognostic patterns of COVID-19, we unselectively analyzed all consecutive patients who were treated at the University Medical Center Hamburg-Eppendorf (UKE) from 13 February, 2020, until 3 July, 2020. In total, there were 113 male (65%) and 60 female (35%) patients. We graded disease severity by WHO classification (who.int): 48 (28%) patients were classified with mild, 37 patients with moderate (21%), 34 patients with severe (20%), and 28 patients with critical (16%) COVID-19. In 26 patients COVID-19 resulted in a fatal outcome (15%). The median age was 59 (95% confidence interval 54.7–59.9) years, and 148 patients were treated as inpatients, 70 patients (40.5%) were admitted to the intensive care unit (ICU), 27 patients (15.6%) were transferred from other hospitals, and 25 patients (14.5%) were only treated in the emergency room (detailed patient characteristics are provided in Table S1). On average, patients had 2.9 preconditions and were most frequently diagnosed with hypertension (n = 66; 38.2%) or with diabetes mellitus (n = 34; 19.7%). Arterial hypertension, diabetes mellitus, cardiovascular diseases, cerebrovascular diseases, chronic obstructive pulmonary disease, hematologic malignancies, and in particular pre-diagnosed lymphoma had a significant impact on COVID-19 disease severity (Figures S1A–S1C). Moreover, age, number of chronic preconditions, and length of the hospitalization significantly correlated with disease severity (Figures S1D–S1E). Clinical, laboratory, and immunological data were routinely acquired from patients with COVID-19. In total, we analyzed 40 immune cell subsets in these patients with COVID-19 and additionally compared them with 39 healthy individuals (the study cohort is summarized in Figure 1A).
Figure 1.
Systemic differences between early and late COVID-19
(A) Overview of our cohort, including healthy donors and patients with mild, moderate, severe, critical, and lethal COVID-19 according to WHO criteria for classification. Patient cohort is detailed in Table S1.
(B and C) Overrepresentation analysis of clinical themes that drive diseases severity in early (B) and late (C) COVID-19. Color scale represents negative log10 false discovery rate (FDR)-adjusted p value, and size shows the number of parameters in the respective clinical theme. Definitions of clinical themes are provided in Table S2.
(D) Overrepresentation analysis of clinical themes that are enriched in clinical, laboratory, and immunological parameters that define respective COVID-19 severity. Detailed analysis is specified in the STAR Methods section. Color shows negative log10 FDR-adjusted p values. Dashed line represents significance level of negative log10 P = 0.05.
(E) Target plots of clinical themes that define respective early, late, or total COVID-19 severity. Negative adjusted log10 p values are scaled for respective clinical theme between one (outer dashed line) and zero (inner dashed line).
Distinct laboratory signatures during early and late COVID-19
Integration of multiple, high-dimensional longitudinal parametric and non-parametric data can be challenging. Therefore, we first set out to identify laboratory and clinical signatures that distinguish early (less than 6 days from diagnosis to sampling) from late (at least 6 days from diagnosis to sampling) COVID-19 (Lescure et al., 2020). To substantiate pathophysiological changes, we summarized individual laboratory parameters into overarching pathophysiological themes with known organ specificity and biomarker function (definitions of all themes are provided in Table S2). Subsequently, we calculated whether individual parameters that significantly impacted disease severity during early and late COVID-19 were quantitatively overrepresented within these defined themes. Of note, during the early phase of the disease we detected a strong overrepresentation of pathologic serum and urine laboratory parameters that significantly impacted disease severity (Figure 1B). In contrast, late COVID-19 was defined by changes in blood counts and urine laboratory parameters (Figure 1C). These results are in accordance with large observational studies that detected differential pathologic findings during early and late COVID-19 (Chen et al., 2020).
Next, we divided our patient cohort by the WHO classification (WHO reference number: WHO/2019-nCoV/clinical/2020.5) into mild, moderate, severe, critical, and lethal COVID-19 to find pathological patterns of disease progression. We combined immunological, clinical, and laboratory data and calculated dysregulated parameters that were specific for each severity subgroup (summary of specific parameters for each severity are provided in Table S3). Subsequently, we performed an overrepresentation analysis (Yu et al., 2012). Reassuringly, we found that the inflammatory signature was significantly overrepresented across all patients with COVID-19. By contrast, dysregulation of blood counts was only significantly enriched in critical and lethal patients. Changes in the peripheral cellular immunological landscape were overrepresented during the early and late disease course in patients with lethal outcome and thereby, possibly constituting predictive patterns (Figures 1D and 1E). In summary, we found profound differences between early and late COVID-19 reaction patterns that differed according to severity. Complicated COVID-19 correlated with changes of immune cell populations, whereas uncomplicated COVID-19 was predominantly characterized by pathological laboratory parameters.
COVID-19 affects multiple organ systems
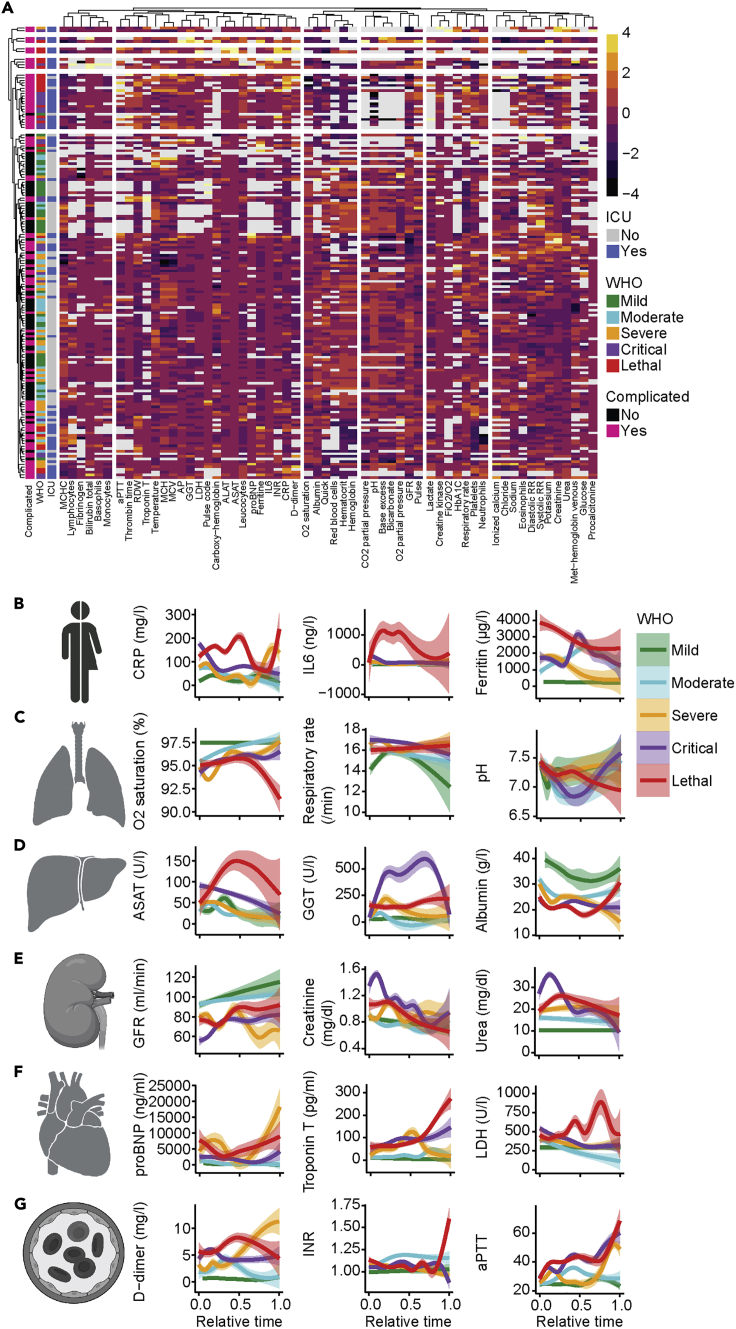
Next, we aimed to further resolve the predictive patterns into their individual determinants. In a first step, we analyzed routinely available laboratory parameters to characterize the organ systems that were affected in association with different COVID-19 severities. Despite inter-individual heterogeneity, k-means clustering of all patients by laboratory parameters already separated different disease severities and in particular patients with ICU admissions (Figure 2A). Of note, in-depth analysis of biomarkers indicative of inflammation (Figures 2B and S2A) and involvement of the respiratory system (Figures 2C and S2B) as well as the hepatobiliary system (Figures 2D and S2C) indicated a severe dysregulation independent of disease severity at early stages of the disease. In comparison with patients with COVID-19 with non-lethal outcomes, patients who died from COVID-19 did not show a recovery of the laboratory dysregulations at later disease stages. In patients with critical and lethal COVID-19, biomarkers of the renal (Figures 2E and S2D), cardiovascular (Figures 2F and S2E), hemostatic (Figures 2G and S2F), and musculoskeletal system (Figure S2G) indicated a dysregulation during later disease stages. Together, our analysis of longitudinal routinely collected laboratory and clinical data revealed systemic multiple organ dysregulations in patients with COVID-19 that clearly correlated with disease severity.
Figure 2.
COVID-19 is a multi-organ disease
(A) Unsupervised clustering of 60 clinical parameters that were available for more than 50 patients. The average value of the respective parameter was used. Rows and columns are arranged by k-means-clustering. WHO severity, ICU admission, and complicated disease course are annotated.
(B–G) Analysis of systemic inflammatory (B; max. CRP, IL-6, ferritin), respiratory (C; min. O2-saturation, max. respiratory rate, min. pH), hepatobiliary (C; max. ASAT, max. GGT, min. albumin), nephrological (D; min. GFR, max. Creatinine, max. Urea), cardiological (E; max. pro-BNP, troponin T, LDH), and coagulation (G; max. D-dimers, INR, aPTT) parameters. The average value of each individual was used. Relative time was adjusted to time of admission (t = 0) and time of discharge (t = 1). The mean and standard error are shown.
In contrast, SARS-CoV-2 viral load as determined by PCR from nasopharyngeal swabs or the presence of SARS-CoV-2-specific IgG antibodies at the day of admission did not correlate with disease severity (Figure S2H) underlining that a complicated disease course is unlikely to be entirely attributable to initial high viral loads.
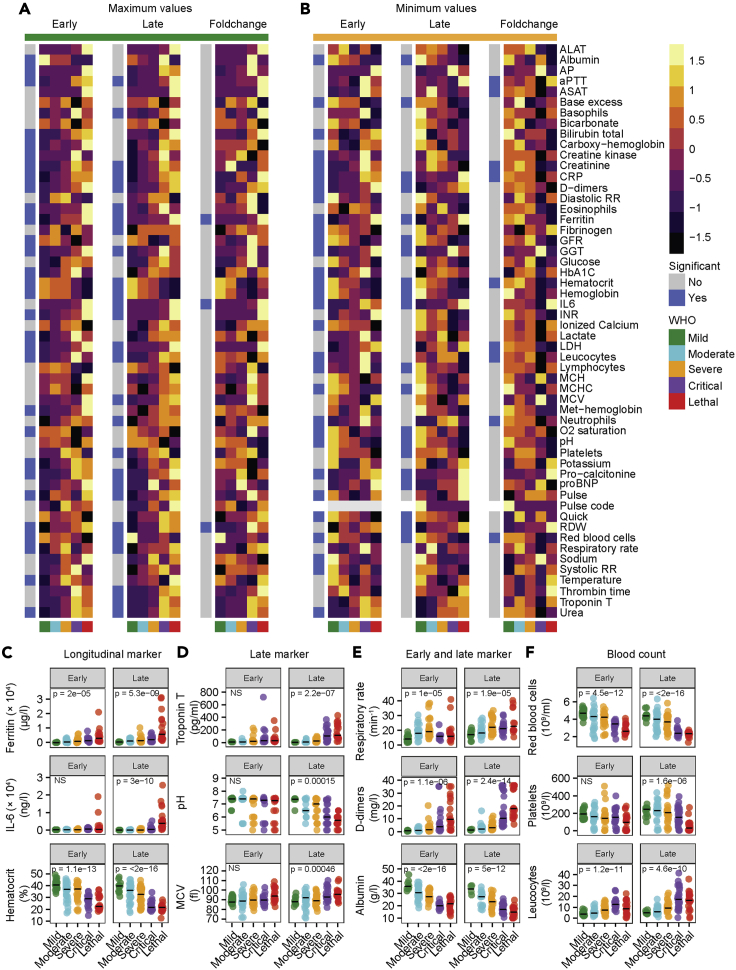
Although we and others (Goyal et al., 2020; Guan et al., 2020) describe multiple organ affections in COVID-19, the temporal sequence of organ-specific manifestations remains elusive. Moreover, the pathogenesis of COVID-19 is determined by different factors during acute and chronic infection (Jordan et al., 2020). To find prognostic laboratory markers we analyzed early and late COVID-19 and compared maximal (Figure 3A) and minimal (Figure 3B) values for each parameter, as well as the longitudinal fold change. Longitudinal markers that showed a significant change during the disease course at early and late time points included an increase of ferritin and decrease of the hematocrit value. Of note, IL-6 levels significantly rose during late but not early time points of the disease and thereby underline the importance of routinely collected laboratory data to estimate disease trajectories (Figure 3C). Furthermore, reduction of the glomerular filtration rate (GFR) and potassium serum levels were related to disease severity (Figure 3B). Together, these important early biomarkers of poor outcome (Huang et al., 2020) might indicate volume loss or redistribution, in addition to generalized inflammation. Higher levels of circulating IL-6 that have been shown to correlate with respiratory failure in other studies (Grifoni et al., 2020) correlated with disease severity during late COVID-19 in our cohort. Other parameters that were significantly changed in late, but not early, disease included troponin T, pH value, and the mean corpuscular volume (Figure 3D). Potentially, these changes display rather the consequences than the cause of earlier pathophysiological events and they might be important determinants of the overall prognosis at later time points. Moreover, we identified laboratory parameters that were significantly dysregulated during early and late disease but did not qualify as longitudinal markers, since we did not detect significant temporal changes during the disease course. These parameters included respiratory rate, raised D-dimers, and abnormal albumin (Figure 3E). As expected, we observed an increase of the maximal respiratory rate in mild, moderate, and severe COVID-19. Of note, the maximal respiratory rate in patients with critical and lethal COVID-19 was comparable with mild disease during early time points. At later stages of the disease the maximal respiratory rate of critical and lethal COVID-19 was increased as well. Hypoxia and acidosis exclusively occurred in critical and lethal disease courses. This might indicate a missing respiratory adaptation to hypoxia in critical and lethal COVID-19 leading to acidosis. In addition, we found strong general dysregulation of blood counts that correlated with poor outcome. In particular, the absolute number of red blood cells was decreased in severe COVID-19 during the early and late phase, whereas platelet numbers were reduced especially during the late phase of the disease. Reduction of platelet counts, together with a pathological increase of D-dimer levels, the international normalized ratio (INR), and the activated partial thromboplastin time (aPTT; Figure 3A) during the late disease phase of complicated COVID-19 underline the potential role of dysregulated coagulation for poor outcome in COVID-19 (Levi et al., 2020). In addition, the absolute leukocyte count was increased at early and late disease stages (Figure 3F).
Figure 3.
Early, late, and longitudinal laboratory patterns of patients with COVID-19
(A and B) Maximal (A) and minimal (B) values of depicted 56 laboratory parameters from early (left) and late (middle) and fold change from early to late (right) COVID-19. The average of all individuals from each disease severity is displayed. Coloring shows Z score. WHO disease severity and significance are annotated. Significance was calculated using one-way ANOVA.
(C–E) Longitudinal marker with significant differences between early and late COVID-19 (C), late marker that reached significance in late but not early COVID-19 (D), and early and late marker (E) for disease severity in COVID-19.
(F) Blood count dysregulation (red blood cells, platelets, leukocytes) in early and late COVID-19. If not stated otherwise, exact p values are provided in the figure and one-way ANOVA was used to test statistical significance. Exact n is provided in Table S4. Individual data points and median are shown.
Together, we detected profound dysregulations of parameters specific for different organ systems and, thereby, defined COVID-19 as a multi-organ disease. In addition, we identified differential organ-specific biomarkers during the early and late stages of the disease. Although renal, respiratory, and hepatobiliary systems were affected early in the disease course, our data indicate that cardiac and coagulative dysfunctions occur only at later stages, possibly reflecting secondary changes.
Longitudinal immune profiling of COVID-19
Since blood count abnormalities were most prominent in patients with COVID-19, we next analyzed how temporal changes of different immune cell populations contribute to the observed leukocytosis. We established four immune cell panels that separately assessed lymphocyte subpopulations, regulatory T cells, subsets of B cells, and myeloid immune cells (panel design and gating strategy provided in Figure S3). Across all time points, we observed a decrease of the absolute number and relative frequency of lymphocytes (Figure 4A) that correlated with poor outcome. In particular, we found a decreased absolute number of T cells (Figure 4B), CD4+ T cells (Figure 4C), CD8+ T cells (Figure 4D), and naive T cells (Figure 4E), whereas their respective relative frequencies did not significantly differ. The relative frequency of regulatory T cells was slightly decreased in moderate, severe, and critical COVID-19 compared with mild disease (Figure S4A). Our B cell panel showed a significant reduction of absolute B cell numbers at early and late time points of patients with COVID-19 with lethal disease course (Figure 4F). We further analyzed B cell subpopulations and found decreased frequencies of naive (Figure 4G) and resting memory B cells (Figure 4H), which was dependent on disease severity. In contrast, we observed increased frequencies of tissue-like (Figure 4I) and activated memory B cells (Figure 4J) in patients with lethal outcome. Tissue-like memory B cells were also significantly upregulated in comparison with healthy donors (Figure S4B).
Figure 4.
Longitudinal profiling of lymphocyte populations in COVID-19
(A–E) Absolute numbers (top row), frequency (middle row), and time course of absolute number (third row) of lymphocytes (A, minimum), T cells (B, mean), CD4+ T cells (C, mean), CD8+ T cells (D, mean), and naive T cells (E, mean) of patients with COVID-19. Relative time was adjusted to time of admission (t = 0) and time of discharge (t = 1). Exact n is provided in Table S4.
(F–J) Absolute number of B cells (F), frequency of naive B cells (G), resting memory B cells (H), tissue-like memory B cells (I), and activated memory B cells (J) of patients with COVID-19. Exact n is provided in Table S4.
(K and L) Heatmap of shown parameters during early (K) and late (L) COVID-19. Color represents Z score.
(M) Heatmap of differences between early and late COVID-19 of shown parameters. Non-significant parameters are colored in gray. Color scale represents signed negative log10 FDR-adjusted p value. If not stated otherwise exact p values are provided in the figure and exact n is provided in Table S4. Individual data points are shown with median.
Recent studies have attributed the immunopathogenesis of COVID-19 to inflammatory cytokine signatures that might lead to disturbances of the immune cell composition (Lucas et al., 2020). However, the predictive value of temporal changes of the immune cell composition in combination with routinely collected data has only been sparsely investigated. Therefore, we compared early and late time points by longitudinal flow cytometry. Our analyses revealed differential patterns of T cells in mild versus lethal COVID-19 (Figures 4K–4M and S4C). Although the average T cell frequency increased over time in mild COVID-19, we observed T cell depletion in longitudinal data of lethal COVID-19. This might be a reflection of T cell exhaustion (Diao et al., 2020) or tissue homing (Yang et al., 2020a) in patients with COVID-19 with lethal outcome. On the other hand, we observed that T cells were only transiently decreased and rapidly recovered in patients with mild COVID-19. Longitudinal profiling of B cells showed a decrease of naive B cells during mild COVID-19 (Figure S4D). Of note, changes in B cell populations associated with complicated COVID-19 already appeared early in the disease course. This could indicate that formation of tissue-like and activated memory B cells is an early event during COVID-19 progression potentially mediating T cell exhaustion (Mathew et al., 2020).
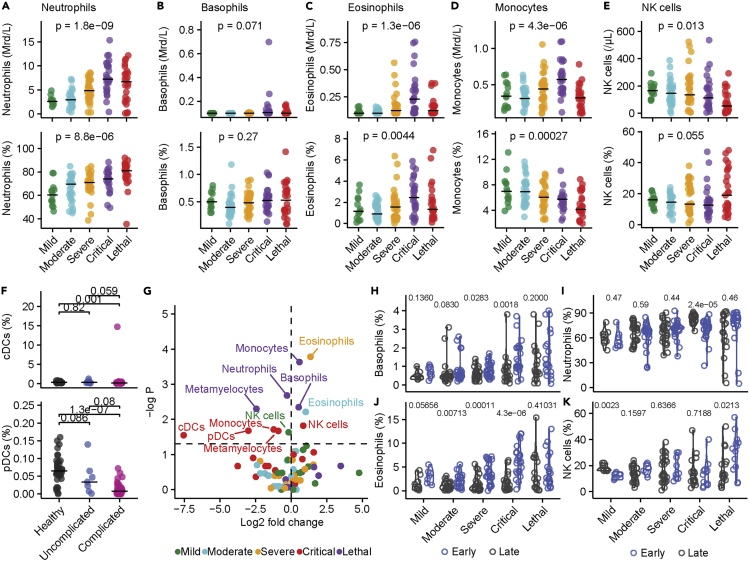
In our myeloid cell panel, we observed an increase in the absolute count and frequency of neutrophils that might explain the general leukocytosis in COVID-19 (Figures 5A and S5A). We did not observe differences in the basophil count (Figures 5B and S5B) but detected a strong increase of peripheral eosinophilic (Figures 5C and S5C) and monocyte cell counts (Figures 5D and S5D) in patients with critical disease. By contrast, natural killer cells (Figures 5E and S5E) as well as conventional and plasmacytoid dendritic cells (Figure 5F) were reduced in patients with COVID-19 in comparison with healthy controls (all significant changes are displayed in Figure 5G). In-depth analysis of myeloid subpopulations revealed an increase of CD16+ non-classical monocytes and CD16+P2X7high monocytes but decreased frequencies of classical monocytes and non-CD2 NK cells in patients with COVID-19 in comparison with healthy donors (Figures S5F–S5J). Longitudinal analysis revealed an increased frequency of basophils in severe and critical COVID-19 as well as a decrease of NK cells in mild COVID-19, whereas patients with lethal disease course showed an increased frequency of NK cells during late disease. Eosinophils were increased during late time points in mild, moderate, severe, and critical COVID-19, which is in accordance with recent findings of a temporary type-2 (anti-helminth) effector response in COVID-19 (Lucas et al., 2020). However, this increase was absent in patients with lethal outcome (Figures 5H–5K), which might indicate an immunomodulatory function of eosinophils during late COVID-19 that potentially counteracts hyperinflammation (Lee and Ashkar, 2018).
Figure 5.
Innate immune dysfunction in COVID-19
(A–E) Absolute count (top row) and frequency (bottom row) of neutrophils (A), basophils (B), eosinophils (C), monocytes (D), and NK cells (E) in patients with COVID-19. Significance was calculated using one-way ANOVA.
(F) Frequency of conventional (cDCs; top) and plasmacytoid dendritic cells (pDCs; bottom) of healthy donors and patients with uncomplicated (mild, moderate) and complicated (severe, critical, lethal) COVID-19. Significance was calculated using one-way ANOVA.
(G) Volcano plot of subset frequencies. The dashed horizonal line represents significance level of negative log10 P = 0.05. Significant populations are labeled in the figure. Significance and fold change were calculated by comparing frequency of the respective populations from each disease severity against each other by Wilcoxon test.
(H–K) Frequency of basophils (H), neutrophils (I), eosinophils (J), and NK cells (K) from early and late time points of patients with mild, moderate, severe, critical, and lethal COVID-19. Significance was calculated by FDR-adjusted Wilcoxon test. If not stated otherwise, exact p values are provided in the figure and exact n is provided in Table S4. Individual data points are shown with median.
In summary, we found a wide range of dysregulated immune phenotypes that were most pronounced in critical COVID-19 and patients with lethal outcome. We observed an increased number of leukocytes, mostly of neutrophils in patients with severe, critical and lethal outcome, and an increase of monocytes and eosinophil counts in patients with critical disease. In contrast, disease severity was associated with generalized lymphocytopenia and decreased T and B cell numbers. However, especially CD4+ and CD8+ T cells were divergently regulated at early and late time points of uncomplicated and complicated COVID-19, possibly indicating a key role for T cell exhaustion in the pathogenesis of COVID-19 (Mahmoudi et al., 2020). Thus, these changes of the immune cell composition might be useful as predictive biomarkers.
Integrative systems analysis identifies specific patterns of complicated COVID-19
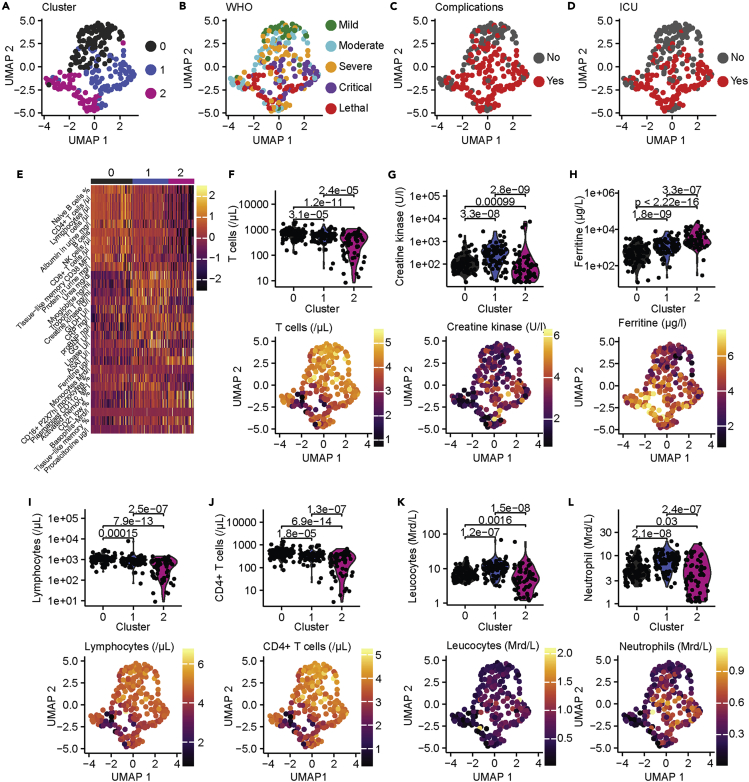
The rapid emergence (Dawood et al., 2020) and the variability of clinical outcomes (Ware, 2020) require early prognostic indicators to robustly discriminate between complicated and uncomplicated COVID-19 for triage and treatment decisions. Thus, to test whether complicated COVID-19 displayed specific early and late signatures, we applied an unbiased machine learning approach with early and late time points from all patients using all available clinical, laboratory, and immunological data. We first filled missing data using random forest modeling that is routinely used for clinical prediction models (Yang et al., 2020b). Subsequently, we performed unsupervised clustering using the UMAP algorithm (Becht et al., 2019). We identified three clusters that robustly separated mild (cluster 0) COVID-19 from patients with critical (cluster 1) and lethal (cluster 2) outcomes (Figure 6A). Reassuringly, ICU admissions were mainly concentrated within clusters 1 and 2 (Figures 6B–6D; distribution of demographic parameters and respective preconditions are displayed in Figures S6A–S6R). Specific markers for cluster 1, which mostly included patients with critical disease courses, consisted of increased levels of urea, troponin T, creatine kinase, proBNP, and gamma-glutamyl transferase (GGT). Cluster 2, which was enriched in lethal COVID-19, was defined by an increase of the laboratory parameters ferritin and aspartate aminotransferase (ASAT) and also an enrichment of activated and tissue-like memory B cells (Figures 6E–6H). Of note, the results of our unsupervised clustering indicated that dysregulation of laboratory markers, indicative of heart and kidney failure was predictive of critical (cluster 1) but not lethal outcome. In addition, the unsupervised clustering was shaped by cluster-dependent depletion of lymphocytes and CD4+ T cell counts (Figures 6I and 6J).Leukocyte and neutrophil counts were increased in cluster 1 in comparison with cluster 0, whereas these populations were decreased in cluster 2 (Figures 6K and 6L). This might reflect divergent immune response patterns in patients with critical COVID-19 and lethal outcome. Specifically, patients with critical COVID-19 developed an immune phenotype that was characterized by increased frequencies of peripheral myeloid immune cell subsets, whereas patients with lethal outcome developed a general leukocytopenia.
Figure 6.
Unsupervised clustering of laboratory, clinical, and immunological data
(A) Unsupervised clustering of early and late time points from our patient cohort. Three clusters were identified.
(B–D) Depiction of WHO disease severity (B), complicative disease courses (C), and patients with ICU admission (D). (E) Heatmap of top 10 defining parameters for each cluster. Color shows Z score.
(F–L) Cluster defining laboratory parameters. T cells (F), creatine kinase (G), ferritin (H), lymphocytes (I), CD4+ T cells (J), leukocytes (K), and neutrophils (L) are shown as violin plots with individuals data points (top row) or mapped onto UMAP plots (bottom row). Statistical significance was calculated using FDR-adjusted Wilcoxon test.
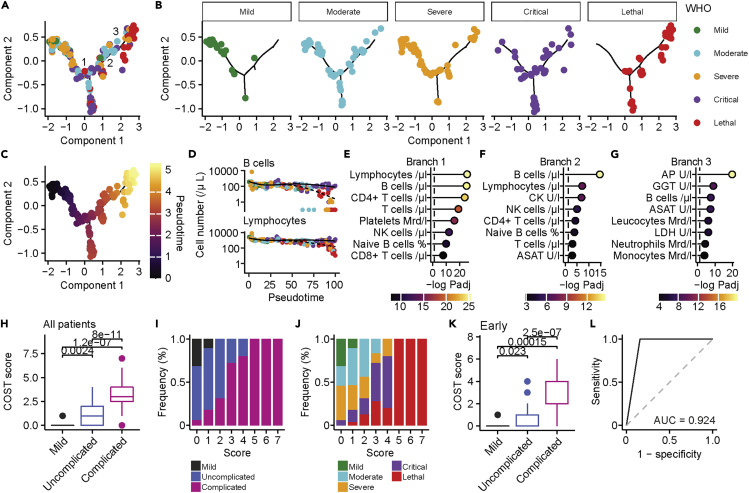
Next, we aimed to identify parameters that predict the progression of disease severity from mild to lethal COVID-19. By using pseudo-time-trajectory analysis we identified positioning of individuals from mild to lethal disease along pseudo-time (Figure 7A). Strikingly, we observed a distinct separation between uncomplicated (mild, moderate, severe) and complicated (critical, lethal) disease trajectories, which allowed us to extract key biomarkers of disease progression (Figures 7B, 7C, S7A, and S7B; distribution of demographic parameters and preconditions are displayed in Figures S7C–S7T). We identified three branches that were enriched in patients with critical (branches 1 and 2) and lethal (branches 2 and 3) COVID-19. The first two branches were characterized by reduction of lymphocytes, B cells, and T cells, whereas damage of the hepatobiliary system, and in particular increased levels of alkaline phosphatase (AP), aspartate amino-transferase, gamma glutamyl-transferase, was the key driver of the third branch (Figures 7D–7G). Our results demonstrate that lymphocyte depletion is a key feature of disease progression. However, signs of organ damage and especially damage of the hepatobiliary system as defined by increased levels of ASAT, alanine aminotransferase (ALAT), GGT, and AP were highly predictive of a lethal outcome. Following this notion, we analyzed ratios of liver enzymes that are routinely used to evaluate hepatocyte destruction. We found that the ASAT/ALAT ratio was increased in the early phase of critical and lethal COVID-19 and further increased over time in patients with lethal outcome (Figure S8A). In contrast, in recovering critically ill patients we observed a decrease of ASAT/ALAT ratio over time. This was mirrored in patients with lethal COVID-19, who showed a decrease of (ASAT + ALAT)/glutamate dehydrogenase (GLDH) ratio (Figure S8B) and an increased GGT/ALAT ratio (Figure S8C), whereas we observed no differences in the ASAT/lactate dehydrogenase (LDH) ratio that is indicative for hemolysis (Figure S8D). Thus, our data support the notion of direct damage of hepatocytes during COVID-19 (Gupta et al., 2020).
Figure 7.
A combined clinical and immunological score robustly classifies complicated COVID-19
(A) Pseudo-time-trajectory analysis of patients with COVID-19. Three branches were identified and are labeled in the figure.
(B) Distribution of patients with mild, moderate, severe, critical, and lethal COVID-19 on the pseudo-time-trajectory.
(C) Pseudo-time is colored on the calculated trajectory.
(D) Changes in cell numbers (cells/μl) of B cells (top) and lymphocytes (bottom) in pseudo-time. Two kinetics from the first branch are shown. Color represents WHO severity.
(E–G) Top eight parameters, ranked by negative log10 FDR-adjusted p values that define the first (E), second (F), and third (G) branch defined by branch expression analysis modeling. Dashed line shows the significance level of negative log10 P = 0.05. Color shows significance level.
(H) Comparison of mild (n = 14), uncomplicated (n = 61; pooled moderate and severe), and complicated (n = 47; pooled critical and lethal) COVID-19. Boxplot with median, interquartile range (IQR), and outliers are displayed.
(I and J) Frequency distribution of mild, uncomplicated, and complicated (I) and WHO severity classified (J) patients with COVID-19 according to our COST score.
(K) Comparison of mild (n = 14), uncomplicated (n = 61; pooled moderate and severe), and complicated (n = 47; pooled critical and lethal) early COVID-19. Boxplot with median, IQR, and outliers are displayed.
(L) ROC analysis of COST score (n = 122). If not stated otherwise, FDR-adjusted Wilcoxon-test was used and exact p values are displayed in the figure.
In the next step, we sought to develop a clinical score that unequivocally differentiates uncomplicated from complicated COVID-19. Therefore, we utilized the key parameters of disease progression that we identified by our unsupervised machine learning approach (COVID-19 SeveriTy score, COST score). In order to establish our COST score, patients received one point for each value across all time points when their maximal levels of (1) AP (U/I), (2) GGT (U/I), (3) or ASAT (U/I) exceeded 1.8 times of the upper reference value or when the minimal counts of (4) lymphocytes (/μL), (5) B cells (/μL), (6) CD4+ T cells (/μL), or (7) platelets (billion/l) fell below 60% of the lower reference value (summary of parameters and cutoffs for COST score are provided in Table S5). By scoring all patients with a maximum COST score of seven, we found a strong separation of patients with mild, uncomplicated (moderate, severe), and complicated (critical, lethal) COVID-19. Of note, we could distinguish patients with lethal outcome from critically ill patients who scored five or more points (Figures 7H–7J and S9A–S9C). Separation by the COST score was independent of pre-existing hematologic malignancies (Figure S9D). This is important since this score relies on the assessment of changes of immune subsets. In confirmation, patients who were admitted to the ICU had significantly higher scores than patients who were treated on a regular ward (Figure S9E). Moreover, we were able to predict different disease trajectories at the early and late phase of the disease (Figures 7K and S9F–S9H). Of note, clinical chemistry signs of hepatobiliary damage emerged in the analysis as specific early predictor of COVID-19 outcome. The COST score was independent of sex and age but, as expected, correlated with demographic variables that predisposed to a complicated disease course (Figures S9I–S9M). Since we faced the constraints that currently no large database with all necessary parameters for validation of our novel score exists, we validated the individual parameters using data from external studies and meta-analyses that partly reported the parameters. By comparing the results from studies that exclusively included non-lethal or lethal COVID-19 (data derived from covidanalytics.io; date of acquisition 09/03/2020) we found decreased lymphocytes and platelets, as well as increased ASAT and GGT (Figures S9N–S9Q) in patients with lethal outcomes, supporting the clinical validity of our newly established scoring system.
Finally, we compared the capability of our COST score to predict lethal disease trajectories with the well-established and clinically widely used sepsis scores Sepsis-related Organ-Failure Assessment score (SOFA) and the Simplified-Acute-Physiology-Score II (SAPSII) in patients with severe, critical, and lethal COVID-19 (Figures S10A and S10B). The COST score showed a statistically significant positive correlation with SAPSII and SOFA scores (Figures S10C and S10D). Although receiver operating characteristic (ROC) curves of the three scores revealed similar area under the curves (AUC) of SAPSII and SOFA scores, our COST score achieved a significantly higher AUC in comparison with the SAPSII and SOFA scores across the whole disease course (Figures 7L and S10E) and the first available time points (Figure S10F). Of note, our COST score achieved similar AUCs with all patients (n = 122, AUC = 0.924; Figure 7L) and half of the patients with available data (n = 61, AUC = 0.913; Figure S10E) underlining its internal validity. In summary, our results indicate a highly predictive accuracy of the COST score to detect lethal disease trajectories in COVID-19.
Discussion
Early and robust identification of critical and lethal disease trajectories is one of the most pressing needs for the clinical care of patients with COVID-19 (Bhatraju et al., 2020). Here, we used an unsupervised systems approach to analyze multidimensional longitudinal data of patients who were treated for COVID-19 at the University Medical Center Hamburg-Eppendorf with diverse outcomes. We identified novel biomarker signatures that distinguish critically ill patients and predict lethal disease trajectories by grading patients with the newly proposed COST score.
Longitudinal analysis of a well-defined cohort by a large number of routine laboratory parameters allowed us to disentangle temporal patterns of biomarkers specific for organ dysfunctions at high resolution. Early changes included laboratory and clinical parameters indicative of kidney damage, generalized volume redistribution, and hypoxia. This supports recent studies that found a high prevalence of SARS-CoV-2 infiltrates in renal glomeruli (Puelles et al., 2020) and observational clinical studies that have correlated kidney damage with multiorgan involvement (Gross et al., 2020). The early renal affection underlines its importance for the pathophysiology of the disease. However, the high prevalence of renal dysfunction in all severity classes of COVID-19 limited its specificity as discriminative biomarker for critical or lethal trajectories in our analysis. In contrast to the early renal involvement, clinical and laboratory biomarkers of cardiovascular involvement, such as pro-BNP and troponin T, were elevated during late COVID-19. Thus, our data indicate that cardiac complications might develop later or be a secondary complication of severe pneumonia or sepsis but not directly due to SARS-CoV-2 infection itself. This is in accordance with postmortem histopathologic studies (Lindner et al., 2020).
We found profound dysregulations of the blood counts during early and late COVID-19 and characterized temporal changes of various immune cell subsets. Although we observed an overall increase of leukocyte counts, we observed depletion of lymphocytes that correlated with disease severity. This is supported by two meta-analyses that have shown that low lymphocyte counts correlated with severe COVID-19, in particular with admission to ICU, ARDS, and mortality (Huang and Pranata, 2020; Zhao et al., 2020). Specifically, we found reduced absolute counts of CD4+ T cells and CD8+ T cells as well as B cells. This is in line with results of large observational studies from the UK (Laing et al., 2020) and China (Lucas et al., 2020) and functional studies that found higher numbers of T cells expressing the exhaustion markers PD-1 and TIM-3 in severe COVID-19 (Diao et al., 2020; Herrmann et al., 2020). Nevertheless, in complicated COVID-19 we observed increased relative frequencies of activated and tissue-like memory B cells, whereas the relative frequencies of naive and resting memory B cells were reduced. Our longitudinal analysis revealed that recovery of T cells predicted a mild disease course underlining its prognostic importance. In contrast, B cell changes developed during early COVID-19. This might indicate that B cell-mediated overstimulation might drive T cell exhaustion (Diao et al., 2020) and that activated memory B cells might be a source of IL-6 (Quinti et al., 2020), thereby fostering a poor disease outcome. We recorded an overall increase of granulocyte counts in the peripheral blood of patients with severe and critical, but not lethal, COVID-19, especially of neutrophils and monocytes. This is in line with multi-omics data of PBMCs of patients with COVID-19 that provided molecular insights into dysregulations of myeloid cells in severe COVID-19 (Schulte-Schrepping et al., 2020). Lineage single-cell and lineage tracing analysis of lungs and BALs of patients with COVID-19 revealed that tissue damage was mostly driven by infiltrating and not tissue-resident monocytes (Wauters et al., 2021). Histopathologic examinations of lung autopsies of deceased patients revealed congestions of micro-vessels by formation of neutrophil extracellular traps (Leppkes et al., 2020). Moreover, neutrophil-induced oxidative stress has been proposed as a key pathophysiological mechanism of tissue damage, thrombosis, and red blood cell pathology (Merad and Martin, 2020). Since the SARS-CoV-2 entry genes ACE2, TMPRSS2, and CTSL are expressed on different cell types across multiple organ systems (Muus et al., 2021) and patients with COVID-19 suffer from a wide range of extra-pulmonary manifestations, further studies are warranted to analyze organ-specific activation of tissue-resident and infiltrating immune cells. Of interest, we observed that eosinophils were sharply increased in patients with COVID-19, who developed severe pneumonia, sepsis, or ARDS. Except for patients with lethal outcome, all patients with COVID-19 showed an increased number of eosinophils in late but not in early phases of the disease, supporting studies that found that severe COVID-19 is associated with a type-2 anti-helminth response (Lucas et al., 2020). Of note, subtropical and tropical countries with a high prevalence of helminth infection have a low prevalence of severe COVID-19 (Hays et al., 2020).
To facilitate clinical translation, we aimed to identify parameters that predict poor and specifically lethal outcome. By exploiting unsupervised clustering of all clinical, laboratory, and immunological data, we identified three clusters that clearly separated uncomplicated (cluster 0) from complicated (cluster 1 and 2) disease trajectories. We found a strong separation between critically ill patients with severe pneumonia, sepsis, or ARDS who survived (cluster 1) and who died (cluster 2). A similar integrative approach using genomic data from peripheral blood leukocytes of patients with sepsis who were admitted to the ICU identified two distinct sepsis response signatures that correlated with mortality and required different treatment strategies (Seymour et al., 2019). Subsequently, we identified key determinants of disease progression by performing pseudo-time-trajectory analysis (Trapnell et al., 2014). We found clear transitions in patients with mild COVID-19 to those with lethal outcomes and identified different branches for critically ill and lethal COVID-19. Key determinants of complicated disease outcomes were depletion of lymphocytes, B cells, CD4+ T cells, and platelets. It is surprising that, among these patients, an increase of the hepatobiliary parameters ASAT, GGT, and AP accurately defined patients with lethal outcome. Detailed analysis of liver enzymes provided further evidence for the hypothesis that hepatocytes are damaged in critical and lethal COVID-19. Of note, hepatobiliary damage has been sparsely described in case series and small observational cohorts (Iavarone et al., 2020). Although histopathological studies could detect SARS-CoV-2 by PCR in livers from deceased patients with COVID-19 (Lagana et al., 2020), it remains elusive whether hepatocytes are directly infected by SARS-CoV-2 or whether they are destroyed by an overly reactive immune response (Yan et al., 2014). Our results point toward a yet underappreciated role of liver involvement for the pathogenesis of COVID-19 and as an important prognostic indicator for disease progression.
Finally, we chose the most significantly regulated branch-defining parameters (lymphocyte count, CD4+ T cells, B cells, platelet count, ASAT, GGT, AP) and evaluated their dysregulation in our COVID-19 cohort by creating the COST score. We could robustly separate mild, uncomplicated, and complicated disease outcomes at early and late phases of the disease using this score. Of note, within the subgroup of patients who suffered from sepsis, severe pneumonia, or ARDS, our score accurately separated patients with lethal outcome from patients who survived. The prognostic separation of critically ill from lethal patients was mainly dependent on the increase in laboratory parameters, indicative of hepatobiliary damage. Therefore, our score might have to be adapted for these patient groups. However, recent studies have shown that pre-existing liver pathologies, for example, liver cirrhosis, are a priori associated with worse disease outcome (Iavarone et al., 2020). Thus, our score is likely not applicable to these patient groups.
In summary, we provide a comprehensive and longitudinal systems analysis of clinical, laboratory, and immunological parameters of a large European cohort with COVID-19. This multi-parametric dataset vividly illustrates that COVID-19 evolves in different phases, affects multiple organs, and is associated with immunological dysregulations that qualify as key drivers of complicated disease outcomes. By unsupervised analysis we identified that a combination of lymphocyte depletion was able to separate mild from uncomplicated COVID-19 and that early hepatobiliary damage predicted later lethal outcome of critically ill patients. We anticipate that our findings will help to understand the sequence of pathophysiological events on different disease trajectories and may have direct therapeutic implications. Further functional analysis and histopathologic examinations are needed to understand the exact role of the liver in the pathophysiology of COVID-19.
Limitations of the study
Limitations of our study are inherent to real-world single center studies. Although we were able to analyze longitudinal data from 156 of 173 affected patients, samples from patients with mild disease who were only seen at our emergency room were gained only once during the duration of this study. In addition, 27 patients were transferred from external hospitals; therefore, initial laboratory values were not available for most of these patients for our analysis and they were only analyzed in the late COVID-19 group. Furthermore, not all parameters were available for each patient at all time points. Therefore, we chose to only analyze parameters that were gained from more than 50 patients in order to obtain representative and robust results. Furthermore, the effect of anti-infectives, such as remdesivir (Grein et al., 2020), or immunosuppressive agents, such as glucocorticoid treatments (Horby et al., 2021), was not taken into account for our systems analysis. However, only a small subset of patients received specific treatments in this cohort. Moreover, we did not find overrepresentation of therapy groups in our unsupervised clustering analysis, therefore excluding systemic bias. Furthermore, another limitation of the COST score is the potential need for acquisition of additional laboratory and immunological data. Other scores, as for example, the c4 mortality score (Hundt et al., 2020), that are based on vital signs might be more feasible for immediate assessment of patients with COVID-19 regardless of the setting. However, the laboratory parameters of the COST score are part of routinely collected panels. Since we aimed to establish a set of biomarkers for clinical routine, we were limited in the number of parameters and cellular subsets that could be analyzed. For example, we did not include Th17 and T follicular helper cells in our panel that each have been hypothesized to play an important role in SARS-CoV-2 immunopathogenesis (Orlov et al., 2020). Since leukopenia is a widely described diagnostic feature of COVID-19 many laboratories and commercial vendors offer different immunological panels that can be integrated into diagnostic procedures. Prospectively, functional analysis of additional cohorts for distribution, differentiation, phenotype, and cytokine secretion of additional immune cell populations is required to refine biomarkers that can be established for clinical testing. In addition, our COST score needs to be validated and compared against other COVID-19-specific and other clinically widely used scores. Thus, further prospective studies of independent larger cohorts are warranted for validation and refinement of the COST score, in terms of timing of laboratory data acquisition, validation in patients infected with SARS-CoV-2 variants, and special patient groups like transplant patients, patients treated with monoclonal antibodies, or patients with partial immunity after vaccination or previous COVID-19 infection.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD16/CD56 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD45 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD4 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD19 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD8 | BD Bioscience | cat. #644611; RRID:AB_2870318 |
| CD25 | BioLegend | cat. #356106; RRID:AB_2561863 |
| CD127 | BD Biosciences | cat. #557938; RRID:AB_2296056 |
| CD4 | BD Biosciences | cat. #345771; RRID:AB_2868799 |
| CXCR3 | BioLegend | cat. #353704; RRID:AB_10983066 |
| HLA-DR | BioLegend | cat. #307606; RRID:AB_2339602 |
| CD38 | BioLegend | cat. #303522; RRID:AB_10953960 |
| CD21 | BioLegend | cat. #354912; RRID:AB_2561577 |
| CD10 | BioLegend | cat. #312210; RRID:AB_314921 |
| CD19 | BioLegend | cat. #302218; RRID:AB_314248 |
| CD73 | BD Biosciences | cat. #562430; RRID:AB_11153119 |
| CD27 | BioLegend | cat. #302835; RRID:AB_2561382 |
| CD303 | BioLegend | cat. #354208; RRID:AB_2561364 |
| CD123 | BioLegend | cat. #306006; RRID:AB_314580 |
| CD16 | BioLegend | cat. #360712; RRID:AB_2562955 |
| HLA-DR | BioLegend | cat. #307616; RRID:AB_493588 |
| CD2 | BioLegend | cat. #300220; RRID:AB_2571989 |
| CD11c | BD Biosciences | cat. #565806; RRID:AB_2869718 |
| CD14 | BioLegend | cat. #301842; RRID:AB_2561946 |
| Software and algorithms | ||
| R studio version 1.2.5.002 | R studio | https://www.rstudio.com/ |
| R version 3.6.3 | R | https://www.r-project.org/ |
| Seurat (v.3.2.3) | Stuart et al. (2019) | https://satijalab.org/seurat/ |
| Monocle (v.2.18.0) | Trapnell et al. (2014) | https://github.com/cole-trapnell-lab/monocle-release |
| clusterProfiler (v.3.18.0) | Yu et al. (2012) | https://guangchuangyu.github.io/software/clusterProfiler/ |
| pROC (v.1.17.0.1) | Robin et al. (2011) | https://rdrr.io/cran/pROC/man/roc.html |
| tidyverse (v.1.3.0) | R environment | https://www.tidyverse.org/ |
| patchwork (v.1.1.1) | R environment | https://patchwork.data-imaginist.com/ |
| outliers (v.0.14) | R environment | https://cran.r-project.org/web/packages/outliers/index.html |
| pheatmap (v.1.0.12) | R environment | https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12/topics/pheatmap |
| ggpubr (v.0.4.0) | R environment | https://cran.r-project.org/web/packages/ggpubr/index.html |
| ggsignif (v.0.6.0) | R environment | https://github.com/const-ae/ggsignif |
| ggcorrplot (v.0.1.3) | R environment | https://cran.r-project.org/web/packages/ggcorrplot/readme/README.html |
| openxlsx (v.4.2.3) | R environment | https://cran.r-project.org/web/packages/openxlsx/index.html |
| randomForest (v.4.6-14) | R environment | https://www.rdocumentation.org/packages/randomForest/versions/4.6-14/topics/randomForest |
| flashclust (v.1.01-2) | R environment | https://cran.r-project.org/web/packages/flashClust/index.html |
Resource availability
Lead contact
Further information and requests for resources and code should be directed to and will be fulfilled by the Lead Contact, Julian Schulze zur Wiesch (j.schulze-zur-wiesch@uke.de)
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data are not publicly available due to ethical restrictions because their containing information could compromise the privacy of the reported patients. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. All packages used are publicly available and are listed in the key resource table. Scripts for analysis are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Study approval
All participants gave written consent in this study that was approved by the local ethic board. Patient characteristics are provided in Figure S1 and Table S1.
Study design
This was a prospective study of consecutive patients with COVID-19 who were admitted to the University Medical Center Hamburg-Eppendorf from 13th February 2020 until 3rd July 2020. All samples were centrally stored. All participants gave written consent in this study that was approved by the local ethic board. All patients with COVID-19 where positive for SARS-CoV-2 by PCR or had SARS-CoV-2 specific antibody titers. The severity of COVID-19 into mild, moderate, severe, critical, lethal disease courses was classified using the WHO criteria (WHO reference number: WHO/2019-nCoV/clinical/2020.5; date of acquisition 08/10/2020). The WHO classification was based on the most affected pathologies per day during the longitudinal disease trajectories. We included 113 male (65%) and 60 female (35%) patients. The mean age was 57.3 years. Patient characteristics and impact of sex and age on disease severity are provided in Figure S1 and Table S1. In order to simplify the visualization of the trajectories, we summarized moderate and severe to uncomplicated and critical and lethal to complicated COVID-19 for some of the visualizations as detailed in the respective figure legends (Figures 6C, 7H, 7I, S7A, S7B, S9A, S9D, and S9F). For deep immune profiling of myeloid subsets, we summarized mild and moderate as uncomplicated and severe, critical and lethal as complicated COVID-19 (Figures 5F, S4B, and S5A). Infection with SARS-CoV-2 was confirmed in all patients by PCR. We defined early disease as less than six days from day of diagnosis to time point of sampling and late disease as at least six days from day of diagnosis to time point of sampling. Data from covidanalytics.io was acquired at 09/03/2020.
UKE COVID-19 standard operation diagnostic procedures
Patients with positive SARS-CoV-2 RT-PCR were hospitalized when they showed clinical symptoms requiring hospitalization or we found radiological evidence of infiltrates (CT, chest radiograph) and the patient suffered from pre-existing risk factors. Patients were admitted to the ICU when oxygen saturation was below 90% during oxygen insufflation of maximum of 4 liters/minute, respiratory rate above 22/min, systolic blood pressure below 100 mmHg or we found elevated lactate levels. On day of admission, all patients received oropharyngeal swab for SARS-CoV-2 RT-PCR and if necessary, for multiplex-PCR for other respiratory pathogens, 12-lead ECG, chest radiograph for all patients, low-dose chest computed tomography scan (CT) if clinically necessary and blood cultures. Furthermore, they we draw blood to analyze laboratory parameters, including complete blood count, urea, creatinine, glomerular filtration rate, ASAT, ALAT, total bilirubin, gamma-GT, alkaline phosphatase, LDH, C-reactive protein (CRP), procalcitonin, venous blood gases, blood sugar level, HbA1C (in patients > 45 years), coagulation screen, D-dimers, fibrinogen, proBNP, creatine kinase, albumin, ferritin, interleukin 6. At the day of admission vital signs (oxygen saturation, respiratory rate, blood pressure, heart rate), body temperature, Glasgow Coma Scale, clinical risk factors and symptoms were documented. During the inpatient stay vital signs were assessed daily and oxygen saturation was measured thrice per day. Depending on the disease course further diagnostics such as radiologic imaging, echocardiography or laboratory parameters were acquired.
Method details
Laboratory parameters
Laboratory parameters were recorded by standard procedures of the UKE. We only included parameters for subsequent analysis that were available for at least 50 patients. In total, we analyzed 159 parameters that consisted of 67 immunological, 92 laboratory and vital parameters. For every patient, laboratory parameters of at least one time point were available. Longitudinal laboratory data were available or 156 patients. The SARS-CoV-2 antibody status was available for 42 patients. In total, standard immunological profiling (Leukocytes counts and relative percentage of T cells, B cells, CD4+ T cells, CD8+ T cells, monocytes, neutrophiles, eosinophiles, basophiles) were available for 131 patients for at least one time point, and from 101 patients for early as well as late time points. Deep immune subset-, and immune cell activation profiling was available for 76 patients for at least one time point and of 52 patients for early as well as late time points. In depth profiling of myeloid subsets was available of 16 patients for early, and for 21 patients for late time points. For all healthy patients were laboratory data available and for 18 healthy patients deep immune profiling. In average laboratory parameters from 25 different time points were available for each patient. All analyzed parameters and respective number of patients in total and divided into WHO disease severities, or early and late time points are provided in Table S4. Antibody titers against SARS-CoV-2 and PCR cycle threshold values from nasopharyngeal swabs described in (Pflüger et al., 2020) , were available for 74 patients. Detailed number of individuals used for each analysis are mentioned in the respective figure legend and are listed in Table S4. The “Relative time” in Figures 2 and 4 were calculated by aligning the time points of data acquisition to the time of admission (t = 0) and time of discharge (t = 1). Thus, all individuals of each WHO severity were visualized for the depicted parameters. The fold change in Figure 3 was calculated by dividing the maximal (Figure 3A) or minimal (Figure 3B) values of the late time point by the maximal or minimal values of the early time point. Thus, we calculated the relative change of maximal and minimal values of each parameter of late time points in comparison to early time points.
Flow cytometry phenotyping
Staining was performed on 50 μl (lymphocyte subsets) or 100 μl (regulatory T cell, B cell and myeloid differentiation panels) whole blood samples containing EDTA as an anticoagulant. The whole blood samples were incubated for 15 min at room temperature in the dark with the antibody cocktails listed in Table S6 and in the Key Resource Table. Erythrocytes were lysed by addition of 500 μl FACS Lysing Solution (BD Biosciences) and incubated for a further 15 min before analysis on a FACS Canto II flow cytometer (BD Biosciences). Analysis of lymphocyte subsets and regulatory T cells was performed with Diva software (BD Biosciences), panels for B cell and myeloid differentiation were analyzed using FlowJo software (TreeStar).
Overrepresentation tests
To analyze biological, immunological and diagnostic themes over time in our COVID-19 cohort, we compiled 18 groups (immune system, immune cells, immune activation, blood count, electrolytes, vital parameter, coagulation, inflammation, organ damage, red blood cell pathology, urine pathology, laboratory parameter, respiration, destruction parameter, endocrinology, hepatobiliary, cardiologic, nephrological parameters) that consisted of well-known and widely established parameters. To define the themes, we only used parameters that were acquired for at least three individuals for each WHO severity group to obtain statistical meaningful results. The themes and respective defining parameters are provided in Table S2. First, we performed 1-way-ANOVA with all parameters for early (Figure 1B), and late (Figure 1C) time points to identify dynamic parameters across all disease severities. P-values were FDR-adjusted. For each time point, the parameters were ranked by negative log10 adjusted P values (most significant parameter at the top, least significant parameter at the end/bottom). Subsequently, we tested overrepresentation of our themes in ranked lists of parameters by hypergeometric testing using the GSEA (gene set enrichment analysis) function of clusterProfiler (v.3.18.0) (Yu et al., 2012). P-values were FDR-adjusted for multiple comparisons. Results were visualized leveraging non-circular cnet-plots.
Disease severity specific parameters
To find parameters that were specific for respective COVID-19 WHO disease severity, we compared average, maximal and minimal values of early and late time points of each parameter from each WHO group with each other by Wilcoxon-test with FDR-adjustment for multiple comparisons. Significant results were considered an adjusted P-value < 0.05. Subsequently, we only took into account the most significant statistical values (maximal or minimal or average) for each parameter. Thus, every parameter is only represented once per disease severity. Therefore, for each disease severity, we filtered out parameters that were significant in comparison to every other severity group and specific for respective disease severity. The computational analysis was performed with tidyverse (v.1.3.0) and stats (v.4.2.0) packages. overrepresentation tests of our themes with disease severity defining parameters by hypergeometric testing were performed as described above with clusterProfiler (v.3.18.0). P-values were FDR-adjusted for multiple comparisons. Results were visualized as lollipop-plots.
Unsupervised clustering analysis
Unsupervised clustering and heatmap visualization of laboratory parameters that is shown in Figure 2A was performed using the pheatmap (v.1.0.12) package. We only included parameters that were available for more than 50 patients. Pearson correlation was applied to calculate the relative distance and dendrograms were calculated using the hclust function from the flashclust (v.1.01-2) package with default parameters. Data was linearly column-wise scaled to the 99th percentile to exclude outliers. To identify markers of severe COVID-19 we used an unsupervised systems approach. We used early (less than six days from symptom onset to sampling) and late (at least six days from symptom onset to sampling) time points from each patient as input (173 patients with COVID-19 × 2 time points = 246 column variables). We excluded patients with more than 170 missing parameters. We included all laboratory, immunological and vital parameters but excluded parameters that were absent in at least 200 individuals. After filtering, we analyzed 233 patients from two different time points and 170 parameters. We used the random forest algorithm of the randomForest (v.4.6-14) package with 300 trees and 1000 iteration with WHO severity as classifier to impute missing data. Otherwise default parameters were used. Next, we performed logarithmic scaling and performed unsupervised clustering with 10 principal components using the UMAP algorithm according to the Seurat (v.3.2.3) pipeline using default parameters. Cluster-defining parameters were calculated using FDR-adjusted Wilcoxon-test. Subsequent pseudo-time-trajectory analysis was performed using the monocle (v.2.18.0) pipeline with default parameters. Parameters that uniquely define late-lethal COVID-19 were provided as endpoint-input for pseudo-time-calculations. Branched expression analysis modeling was used to identify branch-dependent parameters. We defined three branches as number of the output.
Score
We included the following seven parameters for our predictive COVID-19 score that were significant in the resulting three branches of our pseudo-time trajectory analysis (see above): Lymphocytes (/ μL), CD4+ T cells (/ μL), B cells (/ μL), platelets (billion/l), alkaline phosphatase (U/l), GGT (U/l) and ASAT (U/l). For 122 patients all necessary parameters were available and thus, were used to validate our score (for patients with mild n = 14, uncomplicated n = 61, complicated n = 47 disease course). For comparison with SAPS II and SOFA score, we included 61 patients with parameters available for calculating all three scores. Patients received a point for each value when during the disease they exceeded 1.8 times the upper reference of AP (U/l), GGT (U/l) and ASAT (U/l) or fell below 0.6 times the lower reference of lymphocytes (/ μL), CD4+ T cells (/ μL), B cells (/ μL), platelets (billion/l). The components and respective cutoffs we used to calculate the COST score are available in Table S5. SOFA and SAPS II and our score were available for 61 patients. Correlation was calculated using Pearson’s R. We calculated ROC curves of COST, SOFA and SAPS II score using pROC package (v.1.17.0.1) using default parameters. Significance was calculated using the bootstrapping method with 10000 iterations.
Quantification and statistical analysis
Data was analyzed within the R environment (Version 1.2.5.002) on a Mac OS X. The following R packages were used for analysis and visualization: tidyverse (v.1.3.0), patchwork (v.1.1.1), outliers (v.0.14), pheatmap (v.1.0.12), ggpubr (v.0.4.0), ggsignif (v.0.6.0), ggcorrplot (v.0.1.3), openxlsx (v.4.2.3), randomForest (v.4.6-14) , flashclust (v.1.01-2), Seurat (v.3.2.3) (Stuart et al., 2019), clusterprofiler (v.3.18.0) (Yu et al., 2012), monocle (v.2.18.0) (Trapnell et al., 2014), pROC (v.1.17.0.1) (Robin et al., 2011). Detailed analysis is specified in the respective section of the results and in the figure legends. Unless stated otherwise comparisons between two experimental groups are presented as violin plot or dots with median and differences were determined using unpaired Wilcoxon-Mann-Whitney-test and were FDR-corrected for multiple comparisons. Unless stated otherwise comparisons between more than two groups were analyzed by 1-way-ANOVA. Correlations were analyzed using Pearson’s correlation. We excluded values as outlier that exceeded the 99th percentile based on chi-square approximation. Overrepresentation of diagnostic themes was calculated by hypergeometric testing.
Additional resources
COVID-19 disease severity was classified by WHO (WHO reference number: WHO/2019-nCoV/clinical/2020.5; date of acquisition 08/10/2020). Data of external cohorts for validation were derived from covidanalytics.io (date of acquisition 09/03/2020).
Acknowledgments
We thank the UKE COVID-19 study team for their support. We thank members of the Friese and Schulze zur Wiesch laboratories for discussions. We thank Marc Lütgehetmann for providing PCR and antibody titers. We thank BioRender.com for providing illustrations that were used in Figures 2 and S2. J.S.Z.W., F.H., and M.A.F. receive funding from DFG SFB1328; J.S.Z.W. and S.H. additionally get funding from DFG SFB841 (A6); M.A.F. receives DFG Focus funding COVID-19 FR1720/18-1. J.S.Z.W. and M.M.A. receive funding from DZIF.
Author contributions
Conceptualization, M.S.W., J.S.Z.W., and M.A.F.; methodology, M.S.W. and F.H.; formal analysis, M.S.W.; investigation, A.N., D.J., K.R., C.M., T.T.B., M.V.M., M.S.W., A.H., M.C., C.B., W.F., P.K., A.S., S.H., M.M.A., S.S., S.K., and J.S.Z.W.; resources, M.A.F., S.H., S.K., and J.S.Z.W.; data curation, A.N., D.J., K.R., C.M., T.T.B., M.V.M., M.S.W., A.H., M.C., C.B., W.F., P.K., A.S., S.H., M.M.A., S.S., S.K., and J.S.Z.W.; writing – original draft, M.S.W., J.S.Z.W., and M.A.F.; writing – review & editing, M.S.W., J.S.Z.W., and M.A.F.; visualization, M.S.W.; supervision, M.S.W., J.S.Z.W., M.A.F., and S.K.; project administration, M.S.W., J.S.Z.W., M.A.F., and S.K.; funding acquisition, M.A.F., S.H., S.K., and J.S.Z.W.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102752.
Contributor Information
Manuel A. Friese, Email: manuel.friese@zmnh.uni-hamburg.de.
Julian Schulze zur Wiesch, Email: j.schulze-zur-wiesch@uke.de.
Supplemental information
References
- Alballa N., Al-Turaiki I. Machine learning approaches in COVID-19 diagnosis, mortality, and severity risk prediction: a review. Inform. Med. Unlocked. 2021;24:100564. doi: 10.1016/j.imu.2021.100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., Greninger A.L., Pipavath S., Wurfel M.M., Evans L. COVID-19 in critically ill patients in the Seattle region — case series. N. Engl. J. Med. 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F., Lütgehetmann M., Pfefferle S., Wong M.N., Carsten A., Lindenmeyer M.T., Nörz D., Heinrich F., Meißner K., Wichmann D. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396:597–598. doi: 10.1016/S0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport E.E., Burnham K.L., Radhakrishnan J., Humburg P., Hutton P., Mills T.C., Rautanen A., Gordon A.C., Garrard C., Hill A.V.S. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir. Med. 2016;4:259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood F.S., Ricks P., Njie G.J., Daugherty M., Davis W., Fuller J.A., Winstead A., McCarron M., Scott L.C., Chen D. Observations of the global epidemiology of COVID-19 from the prepandemic period using web-based surveillance: a cross-sectional analysis. Lancet Infect. Dis. 2020;20:1255–1262. doi: 10.1016/S1473-3099(20)30581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., Satlin M.J., Campion T.R., Nahid M., Ringel J.B. Clinical characteristics of covid-19 in New York city. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni E., Valoriani A., Cei F., Lamanna R., Gelli A.M.G., Ciambotti B., Vannucchi V., Moroni F., Pelagatti L., Tarquini R. Interleukin-6 as prognosticator in patients with COVID-19. J. Infect. 2020;81:452–482. doi: 10.1016/j.jinf.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O., Moerer O., Weber M., Huber T.B., Scheithauer S. COVID-19-associated nephritis: early warning for disease severity and complications? Lancet. 2020;395:e87–e88. doi: 10.1016/S0140-6736(20)31041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- Haendel M.A., Chute C.G., Robinson P.N. Classification, ontology, and precision medicine. N. Engl. J. Med. 2018;379:1452–1462. doi: 10.1056/NEJMra1615014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays R., Pierce D., Giacomin P., Loukas A., Bourke P., McDermott R. Helminth coinfection and COVID-19: an alternate hypothesis. PLoS Negl. Trop. Dis. 2020;14:e0008628. doi: 10.1371/journal.pntd.0008628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M., Schulte S., Wildner N.H., Wittner M., Brehm T.T., Ramharter M., Woost R., Lohse A.W., Jacobs T., Schulze zur Wiesch J. Analysis of Co-inhibitory receptor expression in COVID-19 infection compared to acute plasmodium falciparum malaria: LAG-3 and TIM-3 correlate with T cell activation and course of disease. Front. Immunol. 2020;11:1870. doi: 10.3389/fimmu.2020.01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19. NEJM. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J. Intensive Care. 2020;8:36. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt M.A., Deng Y., Ciarleglio M.M., Nathanson M.H., Lim J.K. Abnormal liver tests in COVID-19: a retrospective observational cohort study of 1827 patients in a major U.S. Hospital network. Hepatology. 2020;72:1169–1176. doi: 10.1002/hep.31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone M., D’Ambrosio R., Soria A., Triolo M., Pugliese N., Del Poggio P., Perricone G., Massironi S., Spinetti A., Buscarini E. High rates of 30-day mortality in patients with cirrhosis and COVID-19. J. Hepatol. 2020;73:1063–1071. doi: 10.1016/j.jhep.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R.E., Adab P., Cheng K.K. Covid-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- Jung S., Potapov I., Chillara S., del Sol A. Leveraging systems biology for predicting modulators of inflammation in patients with COVID-19. Sci. Adv. 2021;7:eabe5735. doi: 10.1126/sciadv.abe5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Stephen R., Ho Antonia, Pius Riinu, Buchan Iain, Carson Gail, Drake Thomas M., Dunning Jake, Fairfield Cameron J., Gamble Carrol, Green Christopher A., Gupta Rishi K. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana S.M., Kudose S., Iuga A.C., Lee M.J., Fazlollahi L., Remotti H.E., Del Portillo A., De Michele S., de Gonzalez A.K., Saqi A. Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod. Pathol. 2020;33:2147–2155. doi: 10.1038/s41379-020-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Lee A.J., Ashkar A.A. The dual nature of type I and type II interferons. Front. Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M., Thachil J., Iba T., Levy J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7:e438–e440. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lindner D., Fitzek A., Bräuninger H., Aleshcheva G., Edler C., Meissner K., Scherschel K., Kirchhof P., Escher F., Schultheiss H.P. Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol. 2020;5:1281–1285. doi: 10.1001/jamacardio.2020.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi S., Rezaei M., Mansouri N., Marjani M., Mansouri D. Immunologic features in coronavirus disease 2019: functional exhaustion of T cells and cytokine storm. J. Clin. Immunol. 2020;40:974–976. doi: 10.1007/s10875-020-00824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C., Luecken M.D., Eraslan G., Sikkema L., Waghray A., Heimberg G., Kobayashi Y., Vaishnav E.D., Subramanian A., Smillie C. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 2021;27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlov M., Wander P.L., Morrell E.D., Mikacenic C., Wurfel M.M. A case for targeting Th17 cells and IL-17A in SARS-CoV-2 infections. J. Immunol. 2020;205:892–898. doi: 10.4049/jimmunol.2000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflüger L.S., Bannasch J.H., Brehm T.T., Pfefferle S., Hoffmann A., Nörz D., van der Meirschen M., Kluge S., Haddad M., Pischke S. Clinical evaluation of five different automated SARS-CoV-2 serology assays in a cohort of hospitalized COVID-19 patients. J. Clin. Virol. 2020;130:104549. doi: 10.1016/j.jcv.2020.104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinti I., Lougaris V., Milito C., Cinetto F., Pecoraro A., Mezzaroma I., Mastroianni C.M., Turriziani O., Bondioni M.P., Filippini M. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J. Allergy Clin. Immunol. 2020;146:211–213.e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour C.W., Kennedy J.N., Wang S., Chang C.-C.H., Elliott C.F., Xu Z., Berry S., Clermont G., Cooper G., Gomez H. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321:2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Pérez- Pérez L., Meade-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci. Transl. Med. 2021;13:eabe8146. doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong D.L., Kempsell K.E., Szakmany T., Ball G. Development of a bioinformatics framework for identification and validation of genomic biomarkers and key immunopathology processes and controllers in infectious and non-infectious severe inflammatory response syndrome. Front. Immunol. 2020;11:380. doi: 10.3389/fimmu.2020.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware L.B. Physiological and biological heterogeneity in COVID-19-associated acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:1163–1165. doi: 10.1016/S2213-2600(20)30369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters E., Van Mol P., Garg A.D., Jansen S., Van Herck Y., Vanderbeke L., Bassez A., Boeckx B., Malengier-Devlies B., Timmerman A. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021;31:272–290. doi: 10.1038/s41422-020-00455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemken T.L., Kelley R.R. Machine learning in epidemiology and health outcomes research. Annu. Rev. Public Health. 2020;41:21–36. doi: 10.1146/annurev-publhealth-040119-094437. [DOI] [PubMed] [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Li S., Li S. The role of the liver in sepsis. Int. Rev. Immunol. 2014;33:498–510. doi: 10.3109/08830185.2014.889129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal. Transduct. Target. Ther. 2020;5:1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wu H., Jin X., Zheng P., Hu S., Xu X., Yu W., Yan J. Study of cardiovascular disease prediction model based on random forest in eastern China. Sci. Rep. 2020;10:5245. doi: 10.1038/s41598-020-62133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Meng M., Kumar R., Wu Y., Huang J., Deng Y., Weng Z., Yang L. Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a systemic review and meta-analysis. Int. J. Infect. Dis. 2020;96:131–135. doi: 10.1016/j.ijid.2020.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data are not publicly available due to ethical restrictions because their containing information could compromise the privacy of the reported patients. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code. All packages used are publicly available and are listed in the key resource table. Scripts for analysis are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.