Abstract
As of April 1, 2021, more than 2.8 million people have died of SARS-CoV-2 infection. In addition, the mutation of virus strains that have accompanied the pandemic has brought more severe challenges to pandemic control. Host microRNAs (miRNAs) are widely involved in a variety of biological processes of coronavirus infection, including autophagy in SARS-CoV-2 infection. However, the mechanisms underlying miRNAs involved in autophagy in SARS-CoV-2 infection have not been fully elucidated. In this study, the miRNA and messenger RNA (mRNA) expression profiles of patients with SARS-CoV-2 infection were investigated based on raw data from Gene Expression Omnibus (GEO) datasets, and potential novel biomarkers of autophagy were revealed by bioinformatics analyses. We identified 32 differentially expressed miRNAs and 332 differentially expressed mRNAs in patients with SARS-CoV-2 infection. Cytokine receptor related pathways were the most enriched pathways for differentially expressed miRNAs identified by pathway analysis. Most importantly, an autophagy interaction network, which was associated with the pathological processes of SARS-CoV-2 infection, especially with the cytokine storm, was constructed. In this network, hsa-miR-340–3p, hsa-miR-652–3p, hsa-miR-4772–5p, hsa-miR-192–5p, TP53INP2, and CCR2 may be biomarkers that predict changes in mild SARS-CoV-2 infection. Some molecules, including hsa-miR-1291 and CXCR4, were considered potential targets to predict the emergence of severe symptoms in SARS-CoV-2 infection. To our knowledge, this study provided the first profile analysis of an autophagy interaction network in SARS-CoV-2 infection and revealed several novel autophagy-related biomarkers for understanding the pathogenesis of SARS-CoV-2 infection in vivo.
Keywords: miRNAs, mRNAs, Autophagy, SARS-CoV-2, COVID-19
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in December 2019 in Wuhan, Hubei Province, China [1]. Compared to the infection of other coronaviruses, such as SARS-CoV and MERS-CoV, COVID-19, which is caused by SARS-CoV-2, has a lower fatality rate, but is extremely contagious [2]. SARS-CoV-2, as the seventh member of the family of coronaviruses that can infect humans, rapidly led to a pandemic [3]. SARS-CoV-2 infection shows a variety of symptoms, including fever, acute respiratory distress syndrome, and diarrhea [4,5]. As of April 1, 2021, more than 2.8 million people have died of SARS-CoV-2 [6]. In addition, the mutation of virus strains that has accompanied the pandemic has brought more severe challenges to pandemic control [[7], [8], [9], [10]].
Autophagy has been widely discussed and studied in SARS-CoV-2 infection, and its regulation provides the possibility to produce effective therapies [11,12]. Evidence shows that coronaviruses form double-membrane vesicles via autophagy during infection, which can be used as RNA replication platforms to enhance virus replication efficiency [13]. Tomić et al. reported that downregulated expression of autophagy genes (ULK-1, ATG5, UVRAG, AMBRA, PIK3C3, and LC3) correlated with poor T cell responses in severe COVID-19 patients [14]. On the other hand, previous studies have demonstrated that autophagy targets can combat SARS-CoV-2 infection and affect virus activity [15,16]. Zhu et al. found that the autophagy and the AP-1 signaling pathway activity profiles are significantly correlated with the anti-SARS-CoV-2 activity profile using a high throughput screening database [17].
As a type of non-coding RNA (ncRNA), miRNAs are highly conserved non-coding single-stranded small RNA molecules that combine with target mRNA in the 3′-untranslated region (3′-UTR) through processes, including adenylation and uncoating to regulate gene expression [18]. Studies have shown that miRNAs are widely involved in a variety of biological processes in coronavirus infection, including the immune response, virus replication, and protein interaction [[19], [20], [21]]. Wyler et al. reported that miR-155, a differentiation agent of various immune cells, is upregulated in COVID-19 patients [22]. Sardar et al. revealed nine host miRNAs targeting SARS-CoV2, including hsa-let-7a, hsa-miR 101, and hsa-miR125a-5p, and the expression of these miRNAs is different in SARS-CoV-2 genomes from different geographical sources [23]. Khan et al. revealed the crucial roles of miRNA in SARS-CoV-2 infections and reported that host miRNAs can facilitate viral survival in infected cells by inhibiting autophagy through CXCR4, TGF, and mTOR signaling, and SARS-CoV-2-encoded miRNAs can target autophagy, which was unique compared with other coronaviruses such as SARS-CoV [24]. While miRNAs are known to play an important role in SARS-CoV-2 infection, the functions and underlying mechanisms of most miRNAs in SARS-CoV-2 infection have not been fully elucidated.
Here, we hypothesize that miRNAs participate in the autophagy interaction network and may be useful biomarkers for understanding the pathogenesis of SARS-CoV-2 infection. The current study analyzed the COVID-19 patient dataset from Gene Expression Omnibus (GEO) in NCBI and differentially expressed miRNAs (DEmiRNAs) and differentially expressed mRNAs (DEmRNAs) were identified by the limma package in R language. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were employed to explore the potential functional mechanisms of the differentially expressed genes. Then, bioinformatics analysis was performed to construct competitive endogenous RNA (ceRNA) and protein-protein interaction (PPI) networks. We focused especially on the DEmRNAs associated with autophagy, and constructed an autophagy interaction network in which several novel targets closely associated with SARS-CoV-2 infection were revealed in vivo.
2. Materials and methods
2.1. Raw data
The raw microarray and sequencing data of GSE157859, GSE160351, GSE161918, GSE164805, and GSE166253 were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/), an online public gene data repository for high-throughput sequencing research. In total, miRNA expression data included 17 peripheral mononuclear cell samples of COVID-19 patients and six samples of healthy individuals. The mRNA expression data included 91 peripheral mononuclear cell samples of patients with SARS-CoV-2 infection and 31 healthy samples. The above raw data were extracted from the GEO database. This study did not require ethical review or informed consent because GEO data are publicly available.
2.2. Identification of DEmiRNAs and DEmRNAs
The DEmiRNAs and DEmRNAs were identified by expression differences between COVID-19 patients and healthy samples by analyzing raw data from public GEO databases. The adjusted P-value and absolute log value of fold-change (log|FC|) were analyzed in R language (version 4.0.1) by the limma package. log|FC| > 1.0 and adjusted P-value < 0.05 were the selection criteria to define differentially expressed genes.
2.3. Analysis of differentially expressed transcription factors
Based on the DEmiRNAs identified, FunRich (version 3.1.3) was used to analyze and visualize differentially expressed transcription factors. FunRich is a stand-alone software tool used mainly for functional enrichment and interaction network analysis of genes and proteins.
2.4. Pathway enrichment analysis
We used GO annotation (http://www.geneontology.org) and KEGG pathway analysis to determine the functions of the DEmRNAs. DEmRNA functions were classified into three subgroups, namely biological process (BP), cellular component (CC), and molecular function (MF). GO terms with P-value < 0.05 were selected and integrated using the clusterProfiler package in R language (version 4.0.1). The top 10 enriched GO terms were presented. KEGG pathway analysis was performed to determine the involvement of DEmRNAs in different biological pathways. The clusterProfiler package in R language (version 4.0.1) was used to reveal the pathways of the identified DEmRNAs.
2.5. Construction of the ceRNA network
Based on the analysis of raw data from public GEO databases, the DEmiRNA target genes were predicted using the miRDB, miRTarBase and TargetScan databases. In this study, we used the prediction of DEmiRNA target genes to intersect with identified differentially expressed downstream mRNAs to further screen the prediction results. The prediction results were used to construct a ceRNA interaction network.
2.6. PPI network and clustered sub-network construction
The exploration of protein interactions helps reveal the underlying pathological mechanism of SARS-CoV-2 infection. In this study, we used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) to construct a PPI network. The clustered sub-networks and hub genes were identified by employing plugins of Cytoscape, Molecular Complex Detection (MCODE), and cytoHubba (version 3.8.0).
2.7. Construction of an autophagy interaction network
Autophagy genes were obtained from the Human Autophagy Database (http://www.autophagy.lu/). Comparing autophagy genes from the database and the DEmRNAs we identified, the intersecting mRNAs were considered candidate mRNAs in the autophagy interaction network. Upstream miRNAs were predicted using the miRDB, miRTarBase, and TargetScan databases. The candidate miRNAs in the autophagy interaction network were the intersecting mRNAs between the prediction results and the identified DEmRNAs. The autophagy interaction network was constructed based on the above results and the protein-protein interaction relationships between DEmRNAs. The results were visualized using Cytoscape (version 3.8.0).
2.8. Verification of autophagy-related genes using clinical data
The clinical data of patients from previous GEO datasets (GSE157859, GSE161918 and GSE164805, etc) were employed to analyze the relationships between the identified autophagy-related genes and the disease severities using the limma package of R language. The severity of the disease is divided into mild, moderate and severe, according to guidance for COVID-19 pneumonia diagnosis and treatment plan in China. Fold-change > 1.5 and P-value < 0.05 was used as the selection criteria [25,26]. Finally, we analyzed the potential clinical application value of autophagy-related genes identified in this study for evaluating the different disease severities of COVID-19 patients.
2.9. Statistical analysis
COVID-19 patients and healthy individuals were compared to evaluate the statistical significance between the two groups. All data analysis was performed using R software (version 4.0.1), FunRich (version 3.1.3), and Cytoscape (version 3.8.0). A P value of <0.05 was considered statistically significant.
3. Results
3.1. Identification of DEmiRNAs and DEmRNAs
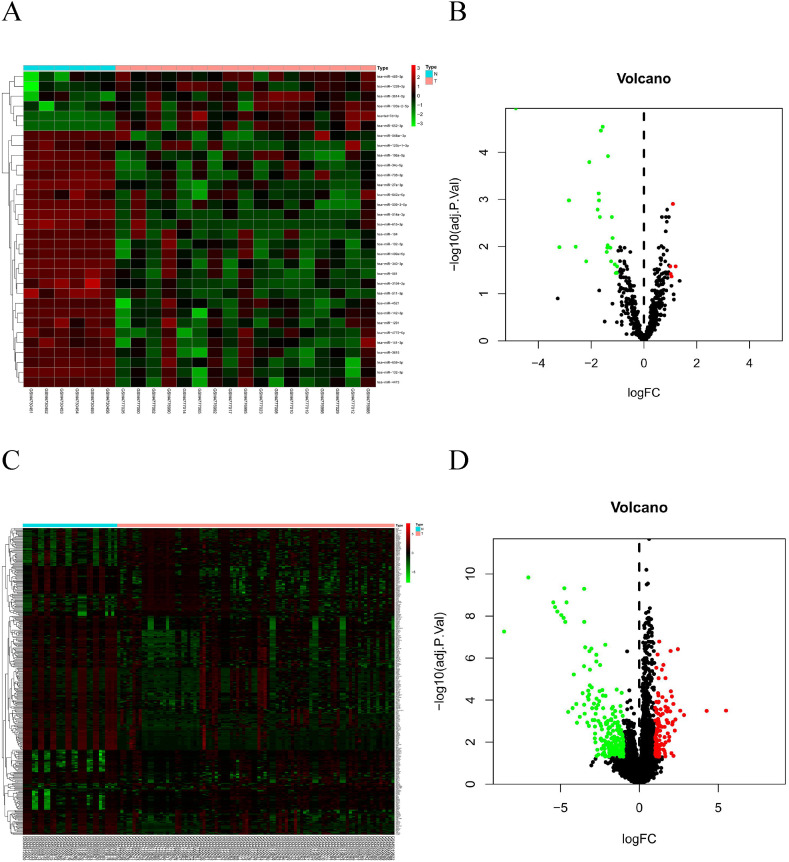
The log|FC| > 1.0 and adj.p < 0.05 were the selection criteria to define differentially expressed genes. In the miRNA dataset, six upregulated miRNAs and 26 downregulated miRNAs were identified (Fig. 1 A and B). In the mRNA dataset, 118 upregulated mRNAs and 214 downregulated mRNA were identified (Fig. 1C and D).
Fig. 1.
Characteristics of differentially-expressed genes (DEGs). (A) Unsupervised clustering analysis of DE micro RNAs (miRNAs). Red dots indicate significantly up-regulated miRNAs, green dots indicate significantly down-regulated miRNAs. (B) Volcano plots of miRNAs. Red dots indicate up-regulated DEmiRNAs, green dots indicate down-regulated DEmiRNAs, black dots indicate non-differentially expressed miRNAs (C) Unsupervised clustering analysis of the DE messenger RNAs (mRNAs). Red dots indicate significantly up-regulated mRNAs, green dots indicate significantly down-regulated mRNAs. (D) Volcano plots of mRNAs. Red dots indicate up-regulated DEmRNAs, green dots indicate down-regulated DEmRNAs, black dots indicate non-differentially expressed mRNAs. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Analysis of differentially expressed transcription factors
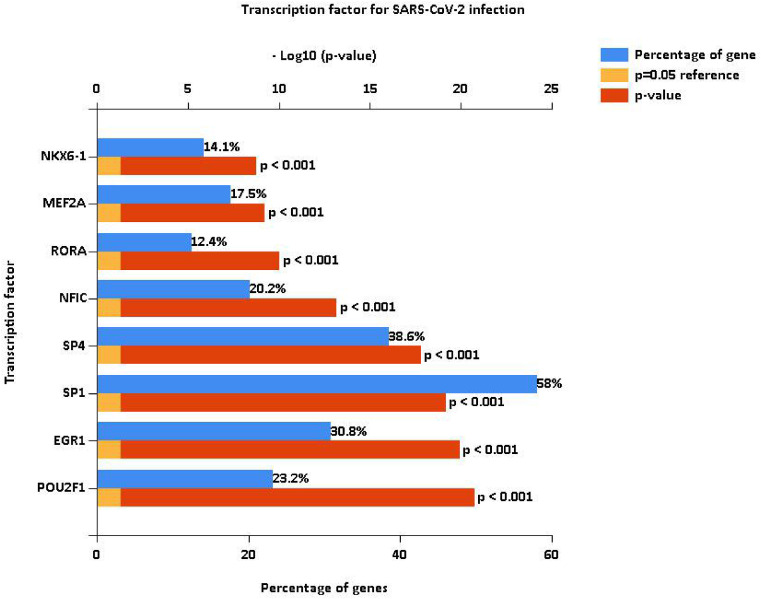
Transcription factors are important molecules that regulate gene expression. We analyzed the differential expression of transcription factors by the 32 identified DEmiRNAs and identified 100 statistically significant transcription factors. Among these, POU2F1, EGR1, SP1, SP4, NFIC, RORA, MEF2A and NKX6-1 were the most significant molecules in COVID-19 patients (Fig. 2 ).
Fig. 2.
Analysis of differentially-expressed transcription factors associated with the identified DEmiRNAs. POU2F1, EGR1, SP1, SP4, NFIC, RORA, MEF2A and NKX6-1 were the most significant differentially-expressed transcription factors in COVID-19 patients.
3.3. Pathway enrichment analysis
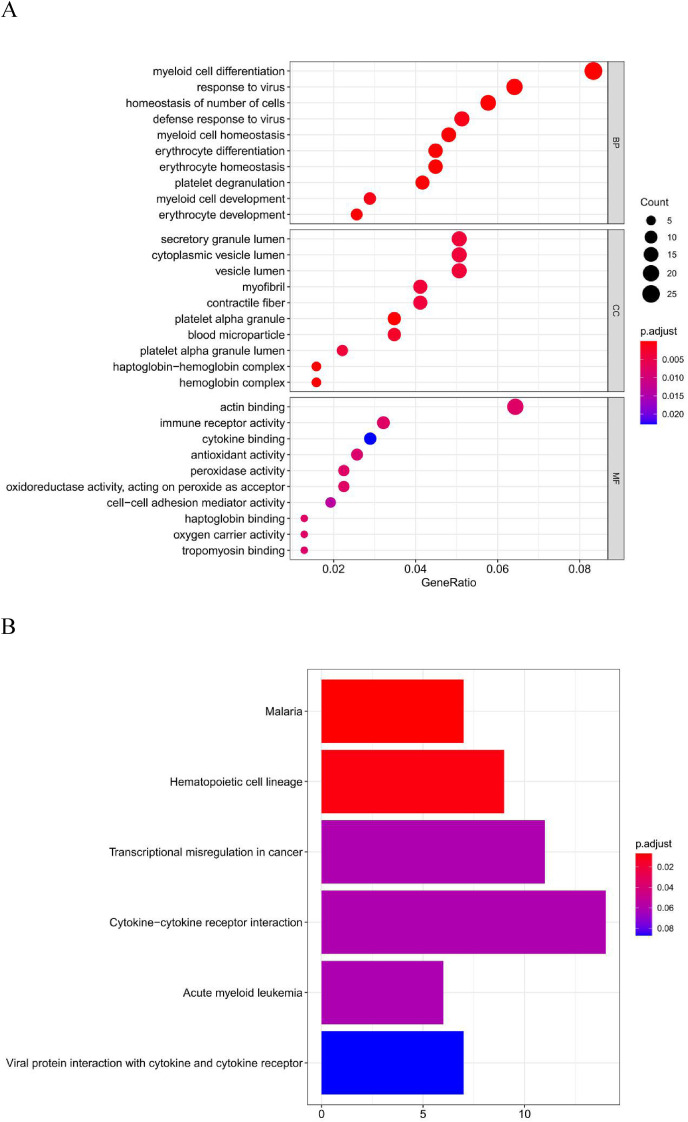
To better understand the biological functions of the identified DEmRNAs in COVID-19 patients, GO annotation and KEGG pathway analyses were performed. In GO annotation analysis, we found that the DEmRNAs were significantly enriched in the terms myeloid cell differentiation, response to virus, and homeostasis of number of cells in the BP subgroup. Secretory granule lumen, cytoplasmic vesicle lumen, and vesicle lumen were the most significant GO terms in the CC subgroup. The top three GO processes included actin binding, immune receptor activity, and cytokine binding in the MF subgroup for DEmRNAs (Fig. 3 A).
Fig. 3.
Pathway enrichment analysis. (A) GO enrichment analysis of the DEmRNAs in biological process (BP), cellular component (CC) and molecular function (MF) subgroups. (B) KEGG enrichment analysis of DEmRNAs.
In KEGG pathway enrichment analysis, malaria and hematopoietic cell lineage, and transcriptional misregulation in cancer were the most significant pathways enriched for DEmRNAs in COVID-19 patients (Fig. 3B).
3.4. Construction of the ceRNA network
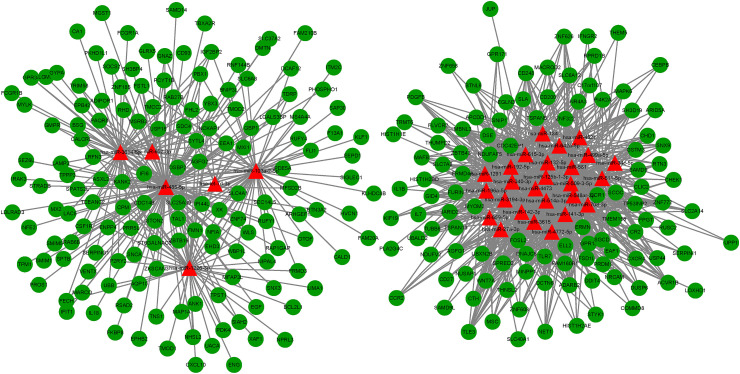
To better understand the endogenous regulatory mechanism of differentially expressed genes (DEGs) in the infection of SARS-CoV-2, we predicted the downstream target genes of DEmiRNAs by employing the miRDB, miRTarBase, and TargetScan databases. At the miRNA level, 32 miRNAs were predicted, and following comparison with DEmRNAs, we identified 257 intersecting mRNAs (Fig. 4 ).
Fig. 4.
The competitive endogenous RNA (ceRNA) network of DEGs in SARS-CoV-2 infection. In total, there were 32 miRNAs and 257 mRNAs in the ceRNA network.
3.5. Construction of the PPI network
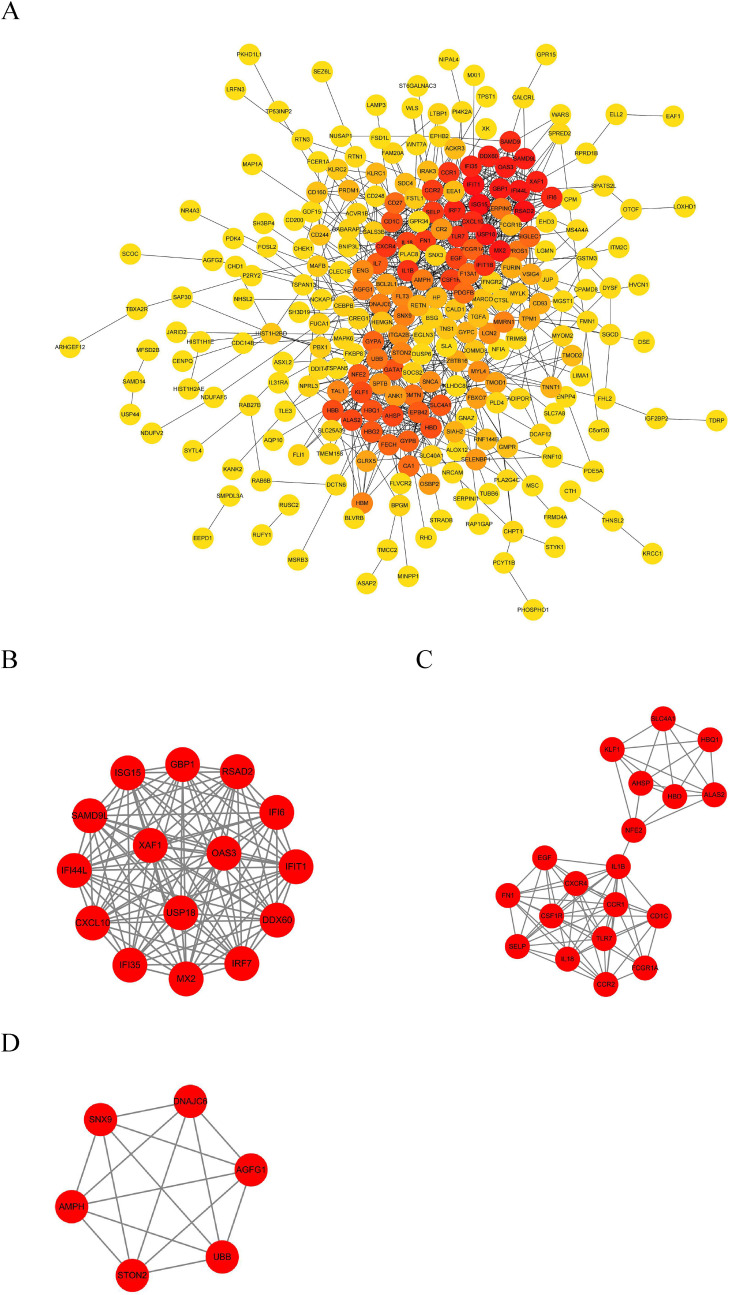
We used the identified DEmRNAs to construct a PPI network, which included 266 nodes and 945 edges, under the condition that the comprehensive Gt score >0.4 and unconnected points were removed. Among the 266 genes, 72 genes had a score >100 analyzed by the maximal clique centrality (MCC) method in Cytohubba (Fig. 5 A). The top five hub genes were ISG15, RSAD2, IFIT1, USP18, and MX2. We also defined the most closely clustered sub-network by employing the MCODE plug-in in Cytoscape, which consisted of 15 nodes and 105 edges (Fig. 5B). In addition, we identified two other clustered sub-networks, with 19 nodes and 76 edges, and six nodes and 15 edges, respectively (Fig. 5C and D).
Fig. 5.
The PPI network of the target genes of the identified DEmRNAs. (A) The target genes of the identified DEmRNAs were ranked in the PPI network. The depth of red indicates the importance of genes in the network. (B) The most closely-clustered subnetwork was composed of 15 nodes and 105 edges. (C) The clustered subnetwork identified by the MCODE plug-in in Cytoscape had 19 nodes and 76 edges. (D) The clustered subnetwork identified by the MCODE plug-in in Cytoscape which had six nodes and 15 edges. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Construction of the autophagy interaction network
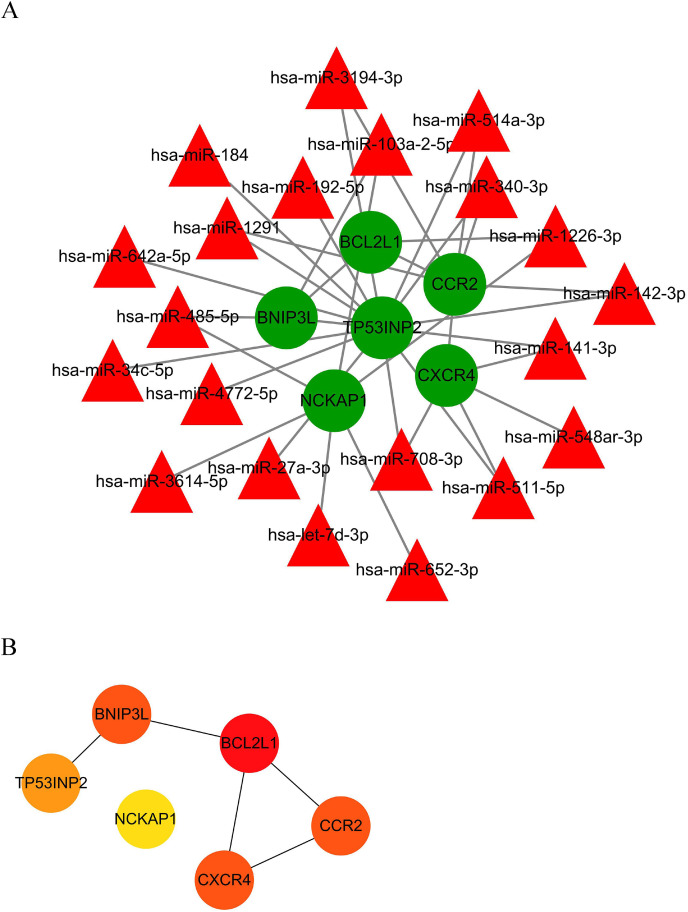
Due to the interesting role of autophagy in SARS-CoV-2 infection, an autophagy interaction network was constructed based on analysis of the ceRNA and PPI networks. Autophagy genes were obtained from the Human Autophagy Database. A total of 21 miRNAs and six mRNAs were involved in network construction (Fig. 6 A). In the autophagy interaction network, BCL2L1 was the most important hub gene among the identified autophagy genes (Fig. 6B).
Fig. 6.
The autophagy interaction network in SARS-CoV-2 infection. (A) Based on the result of the prediction of DEmiRNAs and PPI network, we constructed an autophagy interaction network including 21 miRNAs and six mRNAs. (B) BCL2L1 was the hub gene among autophagy genes in network. The depth of red indicates the importance of genes in the network among mRNAs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.7. Verification of autophagy-related genes using clinical data
The clinical importance of autophagy-related genes identified in this study has been verified to some degree. Most miRNAs in the autophagy interaction network exhibited differential expression between healthy individuals and patients with mild symptoms. Differential expression of hsa-miR-340–3p, hsa-miR-4772–5p, hsa-miR-192–5p, and hsa-miR-652–3p was considered to be statistically significant between patients with mild and moderate symptoms. The expression levels of hsa-miR-3194–3p, hsa-miR-142–3p, hsa-miR-708–3p, hsa-miR-652–3p, hsa-miR-4772–5p, hsa-miR-192–5p, hsa-miR-340–3p, and hsa-miR-1291 showed differences between patients with mild and serious symptoms (p < 0.05). The hsa-miR-103a-2-5p and hsa-miR-141–3p were considered to be differentially expressed in patients with mild and moderate symptoms and in patients with mild and serious symptoms (p < 0.10). The hsa-miR-708–3p also showed differential expressions between patients with moderate and serious symptoms (p < 0.10) (Table 1 ).
Table 1.
The autophagy-related miRNAs by clinical data verification.
| healthy vs mild |
mild vs moderate |
moderate vs serious |
mild vs serious |
|||||
|---|---|---|---|---|---|---|---|---|
| logFC | P.Value | logFC | P.Value | logFC | P.Value | logFC | P.Value | |
| hsa-miR-1226–3p | 1.304 | 0.003 | −0.507 | 0.077 | 0.119 | 0.739 | −0.389 | 0.209 |
| hsa-miR-103a-2-5p | 0.540 | 0.241 | 0.776 | 0.054 | 0.065 | 0.846 | 0.840 | 0.057 |
| hsa-miR-485–5p | 1.423 | 0.008 | −0.322 | 0.315 | −0.012 | 0.978 | −0.347 | 0.388 |
| hsa-miR-3194–3p | −1.263 | <0.001 | −0.411 | 0.088 | −0.231 | 0.357 | −0.613 | 0.030 |
| hsa-miR-514a-3p | −4.862 | <0.001 | 0.100 | 0.905 | −0.358 | 0.700 | −0.254 | 0.754 |
| hsa-miR-1291 | −0.734 | 0.058 | −0.594 | 0.185 | −0.544 | 0.200 | −1.161 | 0.015 |
| hsa-miR-142–3p | −0.729 | 0.004 | −0.611 | 0.114 | −0.121 | 0.766 | −0.720 | 0.031 |
| hsa-miR-340–3p | −1.039 | 0.002 | −0.919 | 0.011 | −0.156 | 0.667 | −1.084 | 0.023 |
| hsa-miR-708–3p | −1.201 | 0.004 | −0.479 | 0.282 | −0.743 | 0.082 | −1.211 | 0.012 |
| hsa-miR-511–5p | −1.165 | 0.001 | −0.350 | 0.205 | 0.051 | 0.843 | −0.313 | 0.308 |
| hsa-miR-141–3p | −0.467 | 0.229 | −0.850 | 0.052 | −0.045 | 0.894 | −0.903 | 0.064 |
| hsa-miR-548ar-3p | −2.071 | 0.009 | −0.506 | 0.578 | −0.624 | 0.466 | −1.158 | 0.181 |
| hsa-miR-652–3p | 0.566 | 0.008 | 0.836 | 0.001 | 0.012 | 0.954 | 0.821 | 0.004 |
| hsa-let-7d-3p | 0.896 | 0.003 | 0.448 | 0.137 | −0.370 | 0.171 | 0.051 | 0.876 |
| hsa-miR-3614–5p | 0.807 | 0.059 | 0.271 | 0.449 | 0.222 | 0.606 | 0.506 | 0.116 |
| hsa-miR-34c-5p | −2.943 | <0.001 | 0.380 | 0.595 | −0.610 | 0.471 | −0.218 | 0.758 |
| hsa-miR-184 | −2.546 | 0.008\ | −0.731 | 0.513 | −0.731 | 0.505 | −1.447 | 0.183 |
| hsa-miR-4772–5p | −0.737 | 0.031 | −0.977 | 0.018 | −0.040 | 0.927 | −1.032 | 0.043 |
| hsa-miR-27a-3p | −1.444 | <0.001 | −0.329 | 0.266 | 0.347 | 0.368 | 0.010 | 0.977 |
| hsa-miR-642a-5p | −0.752 | 0.064 | −0.639 | 0.143 | 0.441 | 0.294 | −0.194 | 0.673 |
| hsa-miR-192–5p | −0.705 | 0.020 | −0.752 | 0.026 | −0.070 | 0.811 | −0.820 | 0.025 |
BCL2L1 and BNIP3L in the autophagy interaction network exhibited differential expressions between healthy individuals and patients with mild symptoms. In the analysis of mRNAs of the autophagy interaction network, differential expression of CCR2 and TP53INP2 was considered to be statistically significant between patients with mild and moderate symptoms (p < 0.05). CXCR4, BCL2L1, and BNIP3L also showed differential expression between patients with mild and serious symptoms (p < 0.05). The expression levels of CCR2 were considered to be significantly different between healthy individuals and patients with mild symptoms, as well as between patients with mild and serious symptoms (p < 0.10). TP53INP2 also showed differential expression between patients with moderate and serious symptoms (p < 0.10) (Table 2 ).
Table 2.
The autophagy-related mRNAs by clinical data verification.
| healthy vs mild |
mild vs moderate |
moderate vs serious |
mild vs serious |
|||||
|---|---|---|---|---|---|---|---|---|
| logFC | P.Value | logFC | P.Value | logFC | P.Value | logFC | P.Value | |
| BCL2L1 | −1.018 | 0.001 | 0.504 | 0.458 | 0.462 | 0.552 | 0.979 | 0.016 |
| BNIP3L | −1.118 | <0.001 | 0.892 | 0.170 | 0.141 | 0.846 | 1.047 | 0.002 |
| CCR2 | −0.883 | 0.091 | 1.035 | 0.043 | 0.028 | 0.899 | 1.076 | 0.080 |
| CXCR4 | 0.182 | 0.515 | −0.290 | 0.472 | −0.703 | 0.105 | −1.008 | 0.016 |
| NCKAP1 | 0.127 | 0.382 | 0.111 | 0.258 | 0.381 | 0.004 | 0.496 | 0.002 |
| TP53INP2 | −0.557 | 0.092 | 0.821 | 0.009 | −0.638 | 0.064 | 0.184 | 0.479 |
After comparing the results of autophagy-related miRNAs and mRNAs according to the severity of disease, we identified hsa-miR-340–3p/CCR2, hsa-miR-340–3p/TP53INP2, hsa-miR-4772–5p/TP53INP2, and hsa-miR-192–5p/TP53INP2 as interactive pairs that were statistically significant between patients with mild and moderate symptoms (p < 0.05); hsa-miR-141–3p/TP53INP2 was also considered significant in a relatively wide range (p < 0.10). In the comparison of patients with mild and serious disease, hsa-miR-3194–3p/CCR2, hsa-miR-1291/CCR2, hsa-miR-142–3p/CCR2, and hsa-miR-340–3p/CCR2 showed differences (p < 0.10).
4. Discussion
Coronavirus SARS-CoV-2 caused a widespread and serious pandemic. In the background of the rapid spread of viruses and the rapid mutation of strains, the scientific community is facing severe challenges. Although some studies have described the potential functional mechanisms of miRNAs in SARS-CoV-2 infection, our understanding of functions and complex mechanisms of most miRNAs is still very limited. This study revealed an autophagy interaction network in SARS-CoV-2 infection and identified several novel targets for understanding the pathogenesis of SARS-CoV-2 infection in vivo. Most importantly, the clinical value of these targets has been verified to some degree in this study, which is particularly vital for the current pandemic.
In this study, we used bioinformatics and the genome data of COVID-19 patients in the GEO database to identify differentially expressed miRNAs and mRNAs in SARS-CoV-2 infection in vivo. In the GO annotation analysis, the most enriched terms in the BP, CC, and MF subgroups were myeloid cell differentiation, secretory granule lumen, and actin binding. In KEGG pathway enrichment analysis, cytokine-cytokine receptor interaction was the most enriched pathway of DEmRNAs in SARS-CoV-2 infection. The autophagy interaction network identified 21 miRNAs and six mRNAs involved; BCL2L1, CCR2, and CXCR4 were the top three hub genes.
Some miRNAs in the autophagy network have been reported in other studies; the specific mechanisms of these miRNAs in SARS-CoV-2 infection are worthy of further study. Wang et al. found that hsa-miR-581 can promote the expression of hepatitis B virus surface antigen by targeting Dicer and EDEM1, which could promote the progression of hepatocellular carcinoma [27]. Hsa-miR-511–5p, hsa-miR-141–3p, and hsa-miR-192–5p have been defined as potential biomarkers of hepatitis B virus infection in human serum [28,29]. In the study of Pulati et al., hsa-miR-1291 was considered as a therapeutic target for cervical cancer as it was shown to be upregulated in HPV16 positive cervical cancer [30]. Du et al. reported downregulation of hsa-miR-34c-5p expression in airway epithelial cells with respiratory syncytial virus infection, which promoted MUC5AC production, to increase mucus secretion [31]. We observed that hsa-miR-34c-5p was significantly downregulated in mild patients in our study, which may indicate that hsa-miR-34c-5p is involved in initiating respiratory pathological symptoms of SARS-CoV-2 infection. In our study, hsa-miR-27a-3p was confirmed to be differentially expressed in SARS-CoV-2 infection. Wicik et al. studied ACE2 interaction networks in COVID-19 to predict the outcomes of patients with cardiovascular risk factors; in this study, hsa-miR-27a-3p was considered the main miRNA to regulate the ACE2 network [32]. Further studies will investigate the pathological mechanisms involved in hsa-miR-27a-3p in SARS-CoV-2 infection.
The results of the present study contribute to our understanding of mRNAs in the autophagy interaction network, especially those defined as hub genes, including BCL2L1, CCR2, and CXCR4. Szabo et al. studied the lung tissue transcriptome of 2244 COVID-19 patients and found that high expression of CCR2 was associated with severe disease [33]. The diagnostic potential of CCR2 for COVID-19 with severe disease has also been reported by Kim et al. in immune cells of bronchoalveolar lavage fluids [34]. However, these studies have some limitations, such as lacking different samples from human tissues and from different countries, or lacking a sufficient number of samples. Our study made up for these shortcomings and revealed that even though SARS-CoV-2 infection differs between regions and race, CCR2 was differentially expressed in peripheral blood mononuclear cells of Chinese patients with SARS-CoV-2 infection. In addition, we analyzed COVID-19 patients according to the severity of the disease and found that CCR2 had a more sensitive diagnostic ability. Compared with patients with mild disease, the expression level of CCR2 in patients with moderate disease was differentially upregulated (fold change > 1.5; p < 0.05). Thus, CCR2, as a biological target, can more sensitively identify patients at an early stage. As the upstream target gene of CCR2, hsa-miR-340–3p, compared with the other four upstream target genes, showed a statistically significant difference between patients with mild and moderate disease (fold change > 1.5; p < 0.05). The hsa-miR-1291, hsa-miR-3194–3p, and hsa-miR-142–3p were also upstream miRNAs of CCR2, and showed more significant differences in expression between patients with mild and serious disease (fold change > 1.5; p < 0.05).
CXCR4, another hub gene in the autophagy interaction network, was first reported in SARS-CoV-2 as a differentially expressed gene in peripheral blood mononuclear cells. It was considered to participate in the innate myeloid response of patients with SARS-CoV-2 infection; this mRNA may be a potential biological target to predict the deterioration of disease by calprotectin plasma level and routine flow cytometry [35]. In a study of serious COVID-19 patients by Taniguchi-Ponciano et al., it was found that CXCR4 was expressed in peripheral blood cells and bronch
Oalveolar cells, and participated in processes such as inflammation and immunometabolism, which make it a potential molecular marker for COVID-19 severity and molecular therapy [36]. Our research promoted CXCR4 as a biological target that could simplify detection methods. Our study also suggests that CXCR4 may have potential as a target for predicting changes in patients with serious disease according to expression level (fold change > 1.5; p < 0.05).
As BCL2L1 is the most important hub gene in the autophagy interaction network, the importance of BCL2L1 was further highlighted in this study. Baralić et al. evaluated the safety of drug combinations for COVID-19 treatment using an in silico toxicogenomic data-mining approach, and found that the drug combination of chloroquine and azithromycin affected the expression of BCL2L1 in vivo [37]. Moreover, Liu et al. reported that matrine could reduce COVID-19 with liver damage by increasing the expression of BCL2L1 [38]. Thus, BCL2L1 shows great potential as a drug target. The results of this study in terms of CXCR4 and CCR2 have been confirmed by other studies to varying degrees. We have reason to believe that BCL2L1 plays an important role in SARS-CoV-2 infection, although this has not yet been confirmed by other studies. Given the significant impact of a large number of COVID-19 patients on the public health system and the emergence of severe forms by strong cytokine storms in individual patients within a short time, these targets are of great significance for screening and predicting changes in patients [39].
In functional analyses, the most enriched terms of the mRNAs in the autophagy interaction network were dendritic cell chemotaxis, autophagosome, and C–C chemokine receptor activity in the BP, CC, and MF subgroups, respectively, based on GO annotations. CCR2 and CXCR4 were annotated in terms related to cytokine activity. Strong cytokine storms are widely present in patients with severe COVID-19. The elimination of cytokine storms can improve clinical results, including acute respiratory distress syndrome, acute heart injury and acute renal failure [40,41]. These results indicated that the autophagy interaction network established based on the ceRNA and protein interaction mechanisms may play an important role in the cytokine storm. This was also verified in the KEGG analysis of mRNAs in the autophagy interaction network; the most significant pathways were mitophagy, autophagy, and viral protein interaction with cytokine and cytokine receptor.
Taken together, this study is the first report of the autophagy interaction network in SARS-CoV-2 infections with a relatively large number of peripheral mononuclear cell samples. The importance of these autophagy-related targets was highlighted with the verification of clinical value in this study. A larger number and different types of samples will help further verify the potential function of the autophagy network. However, on the basis of a limited number of patients with individual characteristics, biased results may be included in our study. Experimental verification would make our study more convincing. The autophagy-related targets reported in this study will be helpful for further research in terms of SARS-CoV-2 infection despite experimental condition and sample acquisition limitations.
5. Conclusions
The first autophagy interaction network associated with the SARS-CoV-2 infection was constructed and verified by the analysis of COVID-19 patients and healthy individuals. Several potential novel biomarkers were revealed for understanding the pathogenesis of SARS-CoV-2 infections in vivo. This study supported previous studies that suggested BCL2L1, CCR2, CXCR4, hsa-miR-34c-5p, and hsa-miR-27a-3p as important factors and promising biomarkers of pathogenesis and molecular therapy in SARS-CoV-2 infection. Moreover, this study showed that the autophagy interaction network we constructed participates extensively in the different stages of the cytokine storm of SARS-CoV-2 infection, highlighting the value of this network. More importantly, the clinical characteristics of autophagy-related genes has been verified; thus, these targets could be a focus in the current pandemic. Although this study lacks experimental validation, it undoubtedly provides a new perspective for future studies regarding SARS-CoV-2 infection.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgement
This study was supported by the Natural Science Foundation of Jiangsu province, China (BK20181173), Gusu health youth talent of Suzhou (GSWS2019039, GSWS2020030), Jiangsu youth medical talents program (QNRC-866, 867, 877), Discipline Construction of The Second Affiliated Hospital of Soochow University (XKTJ-TD202001) and the Science and Technology Program of Suzhou (SYSD2018101, SZS201715, SLT201934, SYS2020023, SS201764), the National Natural Science Foundation of China (3100006, 81572032, 81803266), Key research and development project of Jiangsu provincial science and Technology Department (BE2017654).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2021.105051.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanche S., Lin Y.T., Xu C., Romero-Severson E., Hengartner N., Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7):1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hick J.L., Biddinger P.D. Novel coronavirus and old lessons - preparing the health system for the pandemic. N. Engl. J. Med. 2020;382(20):e55. doi: 10.1056/NEJMp2005118. [DOI] [PubMed] [Google Scholar]
- 4.Pan L., Mu M., Yang P., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115(5):766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO WHO Coronavirus Disease (COVID-19) Dashboard 31 December. 2020 https://covid19.who.int/ Available from: [Google Scholar]
- 7.Ogawa J., Zhu W., Tonnu N., et al. bioRxiv; 2020. The D614G Mutation in the SARS-CoV2 Spike Protein Increases Infectivity in an ACE2 Receptor Dependent Manner. [Google Scholar]
- 8.Sun Y.S., Xu F., An Q., et al. A SARS-CoV-2 variant with the 12-bp deletion at E gene. Emerg. Microb. Infect. 2020;9(1):2361–2367. doi: 10.1080/22221751.2020.1837017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eskier D., Suner A., Karakülah G., Oktay Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ. 2020;8 doi: 10.7717/peerj.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong Y.N., Tsao K.C., Hsiao M.J., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microb. Infect. 2020;9(1):1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domdom M.A., Brest P., Grosjean I., et al. A multifactorial score including autophagy for prognosis and care of COVID-19 patients. Autophagy. 2020;16(12):2276–2281. doi: 10.1080/15548627.2020.1844433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Pérez B.E., González-Rojas J.A., Salazar M.I., Torres-Torres C., Castrejón-Jiménez N.S. Taming the autophagy as a strategy for treating COVID-19. Cells. 2020;9(12) doi: 10.3390/cells9122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomić S., Đokić J., Stevanović D., et al. Reduced expression of autophagy markers and expansion of myeloid-derived suppressor cells correlate with poor T cell response in severe COVID-19 patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.614599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halfon P., Bestion E., Zandi K., et al. bioRxiv; 2020. GNS561 Exhibits Potent in Vitro Antiviral Activity against SARS-CoV-2 through Autophagy Inhibition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorshkov K., Chen C.Z., Bostwick R., et al. bioRxiv; 2020. The SARS-CoV-2 Cytopathic Effect Is Blocked with Autophagy Modulators. [Google Scholar]
- 17.Zhu H., Chen C.Z., Sakamuru S., et al. Mining of high throughput screening database reveals AP-1 and autophagy pathways as potential targets for COVID-19 therapeutics. Sci. Rep. 2021;11(1):6725. doi: 10.1038/s41598-021-86110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 19.Wu W., Choi E.J., Lee I., Lee Y.S., Bao X. Non-coding RNAs and their role in respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) infections. Viruses. 2020;12(3) doi: 10.3390/v12030345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouda R., Onomoto K., Takahasi K., et al. Retinoic acid-inducible gene I-inducible miR-23b inhibits infections by minor group rhinoviruses through down-regulation of the very low density lipoprotein receptor. J. Biol. Chem. 2011;286(29):26210–26219. doi: 10.1074/jbc.M111.229856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondanese V.P., Francisco-Garcia A., Bedke N., Davies D.E., Sanchez-Elsner T. Identification of host miRNAs that may limit human rhinovirus replication. World J. Biol. Chem. 2014;5(4):437–456. doi: 10.4331/wjbc.v5.i4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyler Km E., Franke Ad V., Gottula Ra L.T., Klironomos F. bioRxiv; 2020. Bulk and Single-Cell Gene Expression Profiling of SARS-CoV-2 Infected Human Cell Lines Identifies Molecular Targets for Therapeutic Intervention. [Google Scholar]
- 23.Sardar Ds R., Birla Dg S. bioRxiv; 2020. Comparative Analyses of SAR-CoV2 Genomes from Different Geographical Locations and Other Coronavirus Family Genomes Reveals Unique Features Potentially Consequential to Host-Virus Interaction and Pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan M.A., Sany M., Islam M.S., Islam A. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W., Liu Y., Pan Z., et al. Systematic analysis of tRNA-derived small RNAs discloses new therapeutic targets of caloric restriction in myocardial ischemic rats. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.568116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timbergen M., Boers R., Vriends A., et al. Differentially methylated regions in desmoid-type fibromatosis: a comparison between CTNNB1 S45F and T41A tumors. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.565031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y.Q., Ren Y.F., Song Y.J., et al. MicroRNA-581 promotes hepatitis B virus surface antigen expression by targeting Dicer and EDEM1. Carcinogenesis. 2014;35(9):2127–2133. doi: 10.1093/carcin/bgu128. [DOI] [PubMed] [Google Scholar]
- 28.Tan Y., Ge G., Pan T., et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PloS One. 2014;9(9) doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan Y., Ge G., Pan T., Wen D., Gan J. Serum MiRNA panel as potential biomarkers for chronic hepatitis B with persistently normal alanine aminotransferase. Clin. Chim. Acta. 2015;451(Pt B):232–239. doi: 10.1016/j.cca.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Pulati N., Zhang Z., Gulimilamu A., Qi X., Yang J. HPV16+ -miRNAs in cervical cancer and the anti-tumor role played by miR-5701. J. Gene Med. 2019;21(11):e3126. doi: 10.1002/jgm.3126. [DOI] [PubMed] [Google Scholar]
- 31.Du X., Yang Y., Xiao G., et al. Respiratory syncytial virus infection-induced mucus secretion by down-regulation of miR-34b/c-5p expression in airway epithelial cells. J. Cell Mol. Med. 2020;24(21):12694–12705. doi: 10.1111/jcmm.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wicik Z., Eyileten C., Jakubik D., et al. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 2020;9(11) doi: 10.3390/jcm9113743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pairo-Castineira E., Clohisey S., Klaric L., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 34.Kim C.W., Oh J.E., Lee H.K. Single cell transcriptomic Re-analysis of immune cells in bronchoalveolar lavage fluids reveals the correlation of B cell characteristics and disease severity of patients with SARS-CoV-2 infection. Immune Netw. 2021;21(1) doi: 10.4110/in.2021.21.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvin A., Chapuis N., Dunsmore G., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418. doi: 10.1016/j.cell.2020.08.002. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniguchi-Ponciano K., Vadillo E., Mayani H., et al. Increased expression of hypoxia-induced factor 1α mRNA and its related genes in myeloid blood cells from critically ill COVID-19 patients. Ann. Med. 2021;53(1):197–207. doi: 10.1080/07853890.2020.1858234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baralić K., Jorgovanović D., Živančević K., et al. Safety assessment of drug combinations used in COVID-19 treatment: in silico toxicogenomic data-mining approach. Toxicol. Appl. Pharmacol. 2020;406 doi: 10.1016/j.taap.2020.115237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F., Li Y., Yang Y., et al. Study on mechanism of matrine in treatment of COVID-19 combined with liver injury by network pharmacology and molecular docking technology. Drug Deliv. 2021;28(1):325–342. doi: 10.1080/10717544.2021.1879313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhaskar S., Sinha A., Banach M., et al. Cytokine storm in COVID-19-immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper. Front. Immunol. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minakshi R., Jan A.T., Rahman S., Kim J. A testimony of the surgent SARS-CoV-2 in the immunological panorama of the human host. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.575404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stasi A., Castellano G., Ranieri E., et al. SARS-CoV-2 and viral sepsis: immune dysfunction and implications in kidney failure. J. Clin. Med. 2020;9(12) doi: 10.3390/jcm9124057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.