Abstract
Exosomes are major contributors in cell to cell communication due to their ability to transfer biological material such as protein, RNA, DNA, and miRNA. Additionally, they play a role in tumor initiation, promotion, and progression, and recently, they have emerged as a potential source of information on tumor detection and may be useful as diagnostic, prognostic, and predictive tools. This review focuses on exosomes from lung cancer with a focus on EGFR mutations. Here, we outline the role of exosomes and their functional effect in carcinogenesis, tumor progression, and metastasis. Finally, we discuss the possibility of exosomes as novel biomarkers in early detection, diagnosis, assessment of prognosis, and prediction of therapeutic response in EGFR-mutated lung cancer.
Keywords: Exosome, EGFR, Biomarker, Lung cancer
Introduction
Lung cancer is one of the primary causes of death from cancer around the world, the prognosis of which is poor (5-year survival rate of 10%). It is possible to categorize lung cancer into two broad groups: small cell lung cancer (SCLC), which accounts for approximately 15% of the total number of cases, and non-small cell lung cancer (NSCLC), which accounts for around 85%. NSCLC is divided into three primary types: adenocarcinoma (representing around 50% of all NSCLC), squamous cell carcinoma (which accounts for approximately 30%), and large-cell undifferentiated carcinoma (comprising around 15%). Surgical resection is the preferred treatment option for a tumor early in its growth, although tumor recurrence and metastasis are commonly used, even after resection, with morphological features such as papillary or solid architecture correlating with poor survival and altered response to chemotherapy [1].
Comprehending the biological mechanisms underlying the development of tumors as well as possible biomarker expression of cancer is critical for precise diagnosis at an early stage, targeted therapy, and the development of medications. Large-scale collaborative works have facilitated the understanding of genomic modifications in tumors, hence enabling possible diagnostic and therapeutic targets to be identified, one of which is the epidermal growth factor receptor (EGFR). For patients with adenocarcinoma, EGFR mutations are now identified when the diagnosis is made. Such a mutation in lung adenocarcinoma has a prevalence that ranges from 10 to 78%, with significant differences according to ethnic origins and geographical locations [2]. The development of EGFR tyrosine kinase inhibitors (EGFR-TKIs) represents considerable progress for the treatment of lung cancer, and they are now recognized first-line treatments for NSCLC patients who have activating EGFR mutations (within exon 19 and the L858R missense mutation within exon 21) [3]. The identification of mutations within the EGFR gene in tumor tissues is acknowledged to be the most effective method of estimating the efficiency of treating NSCLC with EGFR-TKIs. Nevertheless, in up to 20% of patients, tissue biopsies are either not informative or not available due to the lack of sufficient neoplastic tissue or because it is technically infeasible [4]. Additionally, only a limited number of cancers are diagnosed at an early stage, which emphasizes the necessity for early detection and targeted treatment. Consequently, liquid biopsy has the potential to become a complementary/alternative tool for diagnosis and prognosis to conventional tissue biopsy. This method comprises the detection of proteomic and genomic information from body fluids, specifically samples of blood. Moreover, this approach has promise for clinical practice due to its noninvasiveness and easy repeatability. The assessment of a liquid biopsy provides a genetic overview of all cancerous lesions (primary and metastases) [5, 6]. Circulating free DNA (cfDNA), exosomes, and circulating tumor cells (CTCs) can potentially be used for noninvasive screening, diagnosis, and prognosis, as well as evaluating treatment response [7–9]. Additionally, liquid biopsy facilitates the real-time monitoring of disease and the biological responses of the patient undergoing treatment [10].
A growing body of evidence has shown that exosomes can provide important information regarding tumors because of their capability to transfer DNA, RNA, and proteins between cells. Furthermore, in lung cancer harboring EGFR mutations, findings indicate that exosomes represent suitable biomarkers for research into cancer and clinical application while also revealing new pathways in the discipline of nanomedicine. In this review, the function of exosomes in lung cancer is discussed, with a specific focus on lung cancers harboring EGFR mutations. We describe the contribution of exosomes to carcinogenesis, the progression of tumors, and metastasis development. Lastly, we review the potential of exosomes to be used as novel biomarkers for the purpose of diagnosing cancer at an early stage, in addition to assessing prognosis, and as therapeutic predictors in EGFR-mutated lung cancer.
The role of exosomes
In 1987, Johnstone et al. [11] first defined the term “exosomes” for a class of extracellular vesicles (EVs), which Pan et al. [12] initially described in 1985. EVs are grouped into three main categories according to their magnitude and assumed biogenetic pathways; the first is apoptotic bodies that have a diameter ranging from 800 to 5000 nm and are emitted by cells experiencing programmed cell death. The second type is microvesicles, which are large membranous vesicles (with a diameter of 50–1000 nm) that are generated via budding from the membrane of the plasma. The last category is exosomes, which are vesicles with a diameter of 30–100 nm regarded as having endocytic origin and include a lipid bilayer membrane [13].
All types of cells can release exosomes, such as astrocytes, epithelial cells, neurons, hematopoietic stem cells, fibroblasts, adipocytes, melanocytes, mesothelioma cells, and tumor cells [14–16]. Exosomes have high heterogeneity and are likely reflective of the phenotypic condition of the cell from which they are generated. It is possible to detect exosomes in a variety of biological fluids including plasma, serum, semen, breast milk, urine, nasal secretions, saliva, amniotic fluid, cerebrospinal fluid, pleural, and ascitic fluid [17–19]. Exosomes contain proteins, lipids, and RNA (such as mRNA, microRNA [miRNA], and additional noncoding RNA) [20–24]; among those proteins are different human epidermal growth factor receptors [25–27]. Additionally, cytokines, transcription factor receptors, growth factor receptors, and different bioactive molecules are contained in exosomes. In 2019, Jeppesen et al. [28] demonstrated that exosomes do not carry double-stranded DNA (dsDNA) or DNA-binding histones. Exosomal proteins like the tetraspanins (CD63, CD81, CD9), TSG101, Alix, and Heat Shock Protein-70 (HSP-70) are regarded as exosomal markers and are employed for the purpose of identifying the existence of vesicles as verifiable exosomes [20, 28–30]. The lipid components of exosomes include cholesterol, sphingomyelin, hexosylceramides, phosphatidylserine, and saturated fatty acids, which all constitute the plasma membrane [31].
It is estimated that normal human blood contains approximately 2000 trillion exosomes, whereas the number of exosomes in the blood of cancer patients is around 4000 trillion [32–34]. Rabinowits et al. [35] conducted a study in which they investigated the extent of circulating tumor exosomes in the plasma of two groups composed of 27 lung adenocarcinoma patients and 9 healthy controls. The findings indicated there were more exosomes in adenocarcinoma patients (mean 2.85 mg/mL) in comparison to the controls (mean 0.77 mg/mL). It was theorized that the higher level of exosomes in diseased patients was caused by alterations to cellular physiology. Studies also revealed that circulating exosomes were more concentrated in patients with breast, ovarian, and pancreatic cancer [36, 37], which could increase their potential to be used as biomarkers for malignancies. Various researchers have concentrated on the underlying mechanism via which exosomes are upregulated in lung cancer. For instance, Yu et al. [38] described that exosome production by cells was found to be regulated by tumor protein p53. In a different study, it was shown that YKT6, which belongs to the soluble N-ethylmaleimide-sensitive-factor attachment receptor (SNARE) family of proteins, is a molecule that plays a critical role in the regulation of exosome release in lung cancer cells [39]. Exosome function depends on their parental cytotypes and contents; exosomes derived from normal cells contribute to the process of sustaining stable homeostasis, while exosomes derived from tumor cells are associated with the progression of the tumor. The identification of specific exosomal markers to discriminate stages and/or histological subtypes in lung cancer is still ongoing. Nonetheless, one study has identified specific miRNA profiles for adenocarcinoma and squamous cell carcinoma [40]. Also, exosomal Tim-3 and Galectin-9, actors of immune surveillance suppression, have been shown to correlate with squamous cell carcinoma and more malignant characteristics [41].
The exact physiological role played by exosomes is still unclear, although it is probable that they have some involvement in the removal of surplus and unneeded cell components, acting as a form of waste disposal; however, recent studies indicate that they also contribute to signaling between cells as a result of their capacity to carry RNA molecules and proteins from one cell to another. It has been recently shown that EVs with the ability to deliver cargos to target cells they meet could eventually cause such recipient cells to be reprogrammed distal from the EV release. Hence, exosomes provide a new means of communication between cells, which could be critical in numerous cellular procedures, including immune responses [42, 43], signal transduction [44], and antigen presentation [45]. Therefore, thanks to their biologically active cargos, exosomes could provide prognostic data on a variety of diseases, including chronic inflammation [46], cardiovascular and renal diseases [47, 48], neurodegenerative diseases [49–51], lipid metabolic diseases [52], and malignancy [53–55].
Exosome contents have a significant influence on how they affect recipient cells. It has been proposed that the miRNA contents of circulating exosomes have similarities to those in cancer cells from which they originated, highlighting the possibility of using miRNA in cancer diagnostics [56–58]. The ability to detect exosomal miRNA in bodily fluids demonstrates the possible benefits of exosomal miRNA as disease biomarkers [59]. Likewise, it is probable that exosome proteins mirror the properties of their origin cells and could indicate the potential for the development of metastasis [60, 61], as communication can occur between parental cells and particular distal or proximal target cells via exosome amplification [62]. Furthermore, the cell tropism of exosomes is specifically related to their cellular origin and properties, characteristics that can be utilized for targeting diseased tissues and/or organs, as suggested by Hoshino et al. [60], who showed that it is possible to use exosomal integrins for the prediction of metastasis in specific organs.
Functional effect of EGFR-mutated exosomes
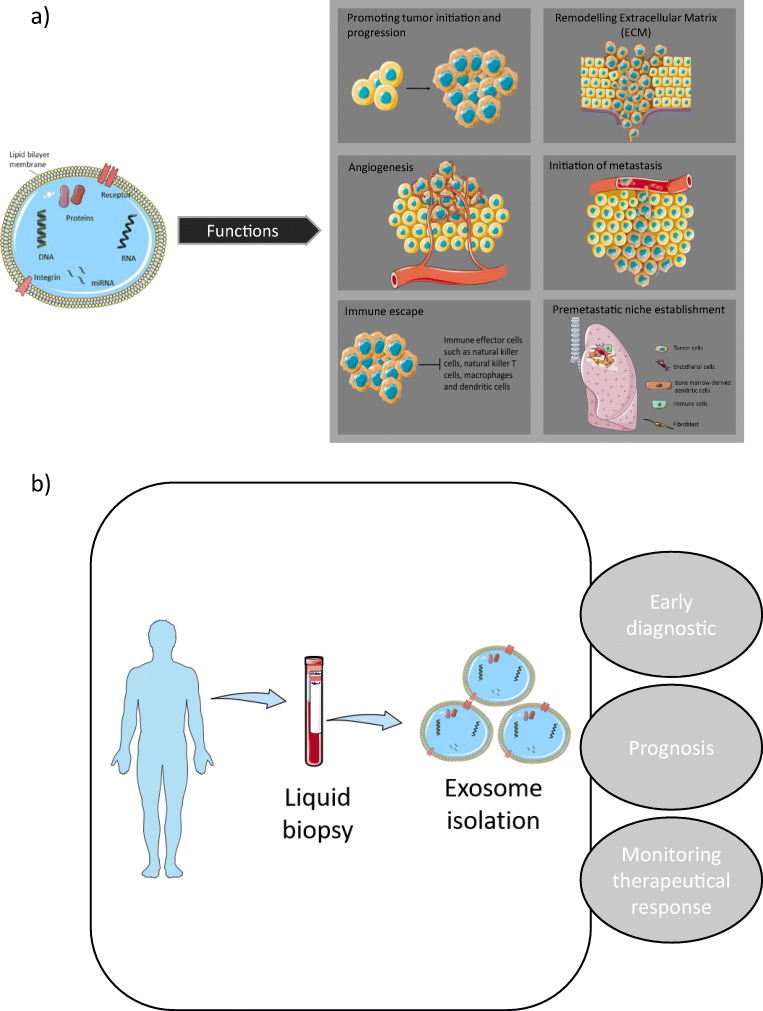
Receptor cells can take up exosomes emitted by donor cells in an autocrine, paracrine, or endocrine manner, thus demonstrating the pivotal role that exosomes play in intercellular communication [63, 64]. Transferring functional exosome content to receptor cells can lead to pathological or physiological effects. Within tumor cells, significant alterations occur to endosomal sorting complexes required for transport, which could cause considerable modifications to the exosomes’ molecular profile as well as the volume of exosomes emitted [29, 65]. An increasing amount of research is being conducted on exosomes as novel mediators of cell-to-cell communication for tumorigenesis as well as progression in the tumor microenvironment in many types of cancer, including lung cancer [66–68] (Fig. 1a). As it is possible to easily obtain and characterize exosomes from the majority of bodily fluids, they have significant potential to be applied in screening programs or as predictive/prognostic biomarkers and can also be used as possible liquid biopsy biomarkers for lung cancer [69]. Exosomes have the ability to transport molecules derived from tumors (DNA and RNA), and the findings of exosomal nucleic acid analysis reveal they have sensitivity in the identification of pertinent mutations [70, 71].
Fig. 1.
Exosomes display different functional roles a allowing potential use in the clinical application b in EGFR-mutated cancer
Zannetti-Domingues et al. [72] depict a major role of EGFR in exosome trafficking (biogenesis and uptake by recipient cells) thereby on its own emission (probably due to the involvement of exosomes in the activation of EGFR recycling mechanisms [73]) and signalization. It was observed that EGFR (as well as its oncogenic mutants) and its signaling network proteins were commonly expressed in lung cancer exosomes from varying sources [74–76]. EGFR mutation relevance in malignancies is well recognized [77]. In so doing, it is important to monitor and further understand the biology of exosomes carrying EGFR mutations.
Promotion of tumor initiation and progression
It has been shown that microvesicles originating from glioma cells that express EGFR in its truncated form, EGFRvIII, deliver this oncogenic protein to neighboring cells in which it is not expressed [78]. This causes transforming signaling pathways to be activated as well as the regulation of target gene expression, such as vascular endothelial growth factor (VEGF), anti-apoptotic Bcl-x(L), and cyclin-dependent kinase inhibitor p27. Research has revealed that the transfer of EGFR could occur through exosomes from human carcinoma cell lines containing activated EGFR to endothelial cells, thus causing mitogen-activated protein kinase (MAPK) and AKT pathways to be activated as well as the expression of VEGF, which increases the capacity for anchorage-independent growth and anti-apoptotic gene expression [74]. Testing on EGFR localized to exosome membranes has been performed to evaluate its potential as a marker for diagnosing lung cancer [79]. For instance, EGFR derived from exosomes is a possible differentiating biomarker for diagnosing NSCLC and chronic lung inflammation. Huang et al. [75] determined that around 80% of exosomes from cancer biopsies were positive for EGFR, whereas a mere 2% of exosomes extracted from chronically inflamed lung tissue were EGFR positive. It was shown by Yamashita et al. and Huang et al. [75, 79] that exosomal EGFR protein can potentially be used as a biomarker to characterize lung cancer; nevertheless, the aforementioned studies had comparatively small sample sizes, and therefore, the findings should be corroborated by a larger cohort.
Immune escape
The immune system interacts closely with tumor cells in the body. Immune escape is characterized by the ability of cancer cells to escape immune surveillance and antitumor immunity, which is a key mechanism of metastatic progression.
Other researchers have highlighted that exosomes derived from cancer mediate the interaction among tumor and immune cells to modify antitumor immunity. For instance, EGFR signaling within lung cancer cells can jeopardize antitumor immunity, which is caused by the transferral of EGFR to host macrophages by exosomes, which resultantly suppresses the innate antiviral immunity [80]. Yu et al. [81] investigated the role of the EGFR-containing exosomes on dendritic cells. The findings revealed that lung cancer with the EGFR E746-A750 deletion mutation caused the antitumor immunity to be repressed by dendritic cells via exosomes. It has additionally been demonstrated that stromal-derived exosomal-EGFR is a critical factor in balancing macrophage polarization and prostatic wound healing. Shi et al. [82] revealed that the production and secretion of exosomal-EGFR and other cytokines and chemokines, after shallow heat injury, by prostate stromal fibroblasts caused greater proliferation and migration of prostate epithelial cells as the wound healed. Exosomes derived from cancer cells that contain EGFR contribute to the progression of metastasis; Zhang et al. [83] showed that EGFR-containing exosomes could facilitate the creation of a liver-like microenvironment that promotes metastasis specific to the liver. They demonstrated that exosomes derived from gastric cancer transport EGFR to the liver and such EGFR are ultimately incorporated into liver stromal cells’ plasma membrane. Further in vitro experiments have shown that hepatocyte growth factor (HGF) is effectively activated by such translocated EGFR by the suppression of miR-26a/b, and the upregulated paracrine HGF, which binds the c-MET receptor onto migrated cancer cells, establishes a fertile microenvironment so that metastatic gastric cancer cells can land and proliferate.
Angiogenesis
Angiogenesis is a critical factor in the process of a microscopic tumor expanding into a macroscopic tumor that has clinical relevance. Exosomes derived from cancer have been identified as novel contributors to angiogenesis [84]. The protein cargo and molecular properties of exosomes may be reflective of the hypoxic condition of the tumor and its aggressiveness [85]. Analogous findings were discovered for exosomes derived from lung cancer and leukemia cells. In addition to modified phenotype properties, exosomes derived from hypoxic lung cancer cells have been found to promote angiogenesis and increase endothelial cells’ permeability, thus triggering cancer cell intravasation/extravasation [86, 87]. VEGF is one of the pro-angiogenic proteins that is acknowledged to be an important initiator of angiogenesis [88]. It can be detected within serum exosomes of mice that have recurrent lung cancer in levels that are 213-fold greater than mice without lung tumors. It has been suggested that the poor prognosis and progression of tumors in 60% of NSCLC may be attributed to the overexpression of VEGF [89–91]. Another factor that motivates the exploration of EGFR as a possible target in cancer and angiogenesis is associated with the fact that, while this receptor is predominantly not present in standard endothelial cells, selective overexpression commonly occurs by the tumor-related vasculature [92, 93]. EGFR can be transferred by cancer cell-derived exosomes to vascular endothelial cells (VECs) which enables VEGF to be secreted. Additionally, exosomes can contribute to the promotion of endothelium proliferation by activating the MAPK and Akt signaling pathways, as previously described [74, 94].
Initiation of metastasis and the pre-metastatic niche
Evidence from recent studies suggests that exosomes have a close association with tumor metastasis, and they could cause the epithelial to mesenchymal transition in lung cancer cells. Messages could be transmitted by the exosomes to initiate the epithelial–mesenchymal transition (EMT) and cause tumor organ metastasis [95]. One of the factors that influences the potential for lung cancer cells to metastasize is the tumor microenvironment [96]. The source and progression of the metastasis could be associated with the exosomes and their contents [97]. Exosomes derived from tumors could be possible mediators of EMT in recipient cells. It was shown by Rahman et al. [98] that exosomes derived from lung cancer cells that metastasized induce expression of vimentin and EMT in receptor human bronchial epithelial cells (HBECs). Additionally, it was shown that exosomes acquired from late-stage lung cancer patient serum had increased vimentin expression, and a more metastatic phenotype was induced in recipient cells. It was suggested that exosomes could trigger the epithelial to mesenchymal transition in lung cancer cells. It has been found that exosomes isolated from different tumor cells that express amphiregulin (AREG) can induce the activation of EGFR in recipient cells, which subsequently affects the microenvironment [99] or promotes bone metastases [100]. Tumor metastasis requires the creation of a pre-metastatic niche [101], where hypoxia and extracellular matrix (ECM) remodeling assist with the formation of this niche [102].
Exosomes as potential biomarkers
As previously stated, in comparison to standard cells, it is possible for exosome formation to be significantly changed in tumor cells. Furthermore, while all types of cells release exosomes, they can be found in abundance in tumor cells. The upregulation of exosome levels has been detected in the bodily fluids of patients with lung cancer, suggesting that exosomes play a critical role in the development and progression of lung cancer. A growing body of evidence suggests that key exosomal cargo that has been significantly modified under cancerous conditions can act as a biomarker for the diagnosis, prognosis, and prediction of lung cancer [103] (Fig. 1b). Resistance to EGFR-TKIs has also been linked to exosomal cargo. Liu et al. [104] showed that exosomes from cells carrying T790M mutation can induce resistance to gefitinib in sensitive cells via activation of the PI3K/AKT signaling pathway. Exosomal transfer of wild-type EGFR has been described to confer osimertinib resistance by activating this same pathway [105]. Interestingly, exosome cargo can also play a role in reversing gefitinib resistance as shown by Chen et al. [106]. They described exosomal miR-7 as an actor of chemoresistance to gefitinib through the promotion of YAP phosphorylation, an effector of the Hippo pathway. Those studies imply potential novel resistant mechanisms to EGFR-TKIs in NSCLC via exosomes.
Existing studies on exosome-related biomarkers have largely concentrated on miRNAs and proteins derived from exosomes [107–110]. However, more research is now focusing on exosomal EGFR and the role it plays in the pathogenesis and progression of lung cancer. To improve survival rates and to deliver more effective treatment for patients with early-stage cancer, it is essential that the mutation profile of the tumor is monitored as early as possible.
The majority of studies on noninvasive biomarkers of cancer have focused on circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and circulating free DNA (cfDNA) as well as additional relevant biomarkers, including circulating microRNAs, circulating RNA, platelets, plasma/serum metabolites, or exosomes. A significant amount of cfDNA originates from normal body cells, and only a small proportion (1–3%) of the circulating cfDNA in a patient is associated with tumors, derived from primary tumors, metastatic sites, or CTCs, and is known as ctDNA [111]. Thus far, the detection of EGFR has been primarily based on nucleic acid sourced from ctDNA and is currently being employed in clinical practice [4, 112]. The first liquid biopsy test to receive market approval from the Food and Drug Administration (FDA) is the Cobas EGFR Mutation Test v2® (Roche Diagnostics Inc.) [113, 114]. This test allows the mutations that exist in the cfDNA fraction to be analyzed, such as exon 19 deletions or exon 21 (L858R) substitution mutations in the EGFR gene; however, its ability to detect EGFR-T790M is constrained by 58% sensitivity and 80% specificity. Nevertheless, liquid biopsies using cfDNA can be problematic, largely due to the properties of ctDNA and the detection constraints of the methods, even with assay platforms of the highest sensitivity [115–118]. Moreover, there is increasing evidence of heterogeneity between different metastatic sites within the same patient. In addition, concentrations of cfDNA vary enormously between individuals and their physiopathological conditions, and early-stage cancers display a low amount of cfDNA similar to healthy subjects [119], while the fraction of ctDNA fragments in the total cfDNA augments with tumor burden and multiple metastatic sites [120].
To resolve these problems, three recently conducted studies revealed the advantages of combining the mutations identified exosomal nucleic acids (exoNA) with cfDNA for detecting mutations [121–123]. Moreover, the evidence presented by Fernando et al. [124] showed that a significant proportion (93%) of plasma cfDNA is localized in exosomes. Krug et al. [123] showed the utility of exosomal RNA for identifying somatic mutations originating from tumors. Their study involved the parallel screening of exosomal RNA and cfDNA from a cohort composed of 84 NSCLC patients positive for EGFR (stage IIIB, IV). The research findings indicated that exosomal RNA exhibited considerably more sensitivity with regard to the detection of activating EGFR mutations (98%) and EGFR-T790M (90%) in comparison to corresponding cfDNA (82% and 84% for activating EGFR mutations and EGFR-T790M, respectively).
Furthermore, it was shown by Castellanos-Rizaldos et al. [125] that combining exoRNA/DNA and cfDNA for the purpose of detecting T790M offered increased sensitivity and specificity in comparison to historical cohorts in which only cfDNA was used. The researchers published another study in which they designed a qPCR-based test (ExoDx EGFR) capable of interrogating a panel of 29 mutations in the EGFR gene, which included the activating and resistance mutations utilizing exosomal RNA/DNA and cfDNA obtained from the plasma of a cohort consisting of 110 NSCLC patients According to the assay’s performance results, its general sensitivity was 90% for L858R, 83% for T790M, and 73% for exon 19 deletion with corresponding specificities of 100%, 100%, and 96%, respectively [122]. This could additionally be beneficial for preventing needless tumor biopsies to test for EGFR mutation. Targeting exoNA has increased sensitivity and specificity in the detection of EGFR mutation though the type of detection platform used (Cobas, NGS, ddPCR, ARMS-PCR, etc.) plays a major role in those factors (Table 1).
Table 1.
Detection of EGFR mutation in exosomal nucleic acids
| Techniques | Patients | Mutation | Accuracy | Ref | |
|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | ||||
| ddPCR | 47 NSCLC patients | EGFR activating and T790M mutations | 76.5 | 100 | Kim et al. [126] |
| Cobas® | 84 patients from the TIGER-X trial | EGFR activating and T790M mutations |
98 for EGFR activating 90 for T790M |
94 for L858R 100 for del19 |
Krug et al. [123] |
| ddPCR | 102 NSCLC patients with biopsy-confirmed T790M positive and 108 patients T790M negative | T790 | 92 | 89 | Castellanos-Rizaldos et al. [125] |
| ExoDx EGFR | 110 NSCLC patients | Panel of 29 mutations in EGFR gene |
90 for L858R 83 for T790M 73 for del19 |
100 for L858R 100 for T790M 96 for del19 |
Castellanos-Rizaldos et al. [122] |
| ARMS-PCR | 284 NSCLC patients | EGFR activating and T790M mutations | 27.5 | 96.6 | Wan et al. [127] |
| NGS | 9 patients with EGFR mutation in tissues | T790 | 75 | 100 | Möhrmann et al. [121] |
ddPCR, droplet digital PCR; ARMS-PCR, amplification-refractory mutation system based PCR assay; NGS, next-generation sequencing
As mentioned earlier, miRNAs are highly abundant in exosomes, could regulate the stability or translation of targeted mRNA, and are key factors altering the lung cancer microenvironment, potentially enabling progression, invasion, angiogenesis, metastasis, and drug resistance [56, 110, 128, 129]. Exosome structures prevent miRNAs from being degraded, which is conducive to clinical detection. For lung cancer patients, both circulating exosomes and exosomal miRNA have higher concentrations in comparison to those detected in control patients [35]. Hence, exosomal miRNAs can potentially be employed as a tool for diagnosing, predicting, and prognosing lung cancer and are considered to be possible suitable noninvasive tools for both diagnosing cancer early and as therapeutic targets due to the fact that they include critical information in signaling pathways associated with the biological responses of tumors [130].
Various studies have determined the potential diagnostic ability of tumor biomarkers including miR-139-5p, miR-200b-5p, miR-378a, miR-126, and miR-379 for differentiating between lung cancer and healthy previous smokers. Furthermore, it was determined that six exosomal miRNAs (miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100, and miR-154-3p) were differently expressed in lung adenocarcinoma and granuloma [131–133]. The sensitivity and specificity of these six microRNAs were 96% and 60%, respectively, and they showed high potential for clinical application in terms of differentiating between lung adenocarcinomas and granulomas.
In the current era in which medicine is becoming increasingly personalized, the creation of beneficial and powerful instruments that provide minimal or noninvasive, highly sensitive, simple, quick, and inexpensive diagnosis of cancer is essential. Numerous liquid biopsy biosensors that are intended to detect exosomal proteins, DNA, RNA, and miRNA as biomarkers for the purpose of screening, diagnosing, and prognosticating cancer have been proposed including immunofluorescence biosensor, colorimetric biosensor, surface plasmon resonance (SPR) biosensor, surface-enhanced Raman scattering (SERS) biosensor, electrochemical biosensor, nuclear magnetic resonance (NMR) biosensor, PCR, and next-generation sequencing (NGS) [134]. They could act in parallel or as supplementary tests for screening and diagnosing cancer, personalizing therapy, monitoring response to treatment, and prognosticating.
However, a limitation of some of the studies of exosomes as biomarkers includes the retrospective nature of them, a relatively limited follow-up time, and in some instances a small cohort of patients which may not completely reflect real-life clinic situation. Furthermore, exoNA are often studied in combination with cfDNA [121–123, 125], which may interfere with the interpretation of exosomes alone as biomarkers for diagnosing, predicting, and prognosticating in lung cancer. Additional studies in larger cohorts are needed to prove the robustness of exosome application as biomarkers.
There is increasing interest in the therapeutic potential of exosomes. Notably, a multicenter investigation, the EXTRA study [135] is ongoing, which aims to evaluate exosomes and their contribution to EGFR-TKI resistance mechanisms. This trial will utilize genomic, proteomic, epigenomic, and metabolomic data to determine the different mechanisms underlying treatment efficacy and the development of resistance to afatinib of non-small cell lung cancer patients expressing EGFR.
However, the use of exosomes as biomarkers and/or therapeutic agents still faces major challenges, including the lack of standardization in isolation and purification techniques. Establishing a quick, accurate, and low-cost common method is crucial for clinical application. Moreover, the heterogeneity of the exosome population is secreted in body fluids as they originate from a variety of different cell types, making it challenging to determine their tissues of origin. Therefore, a precise understanding of exosome characteristics and profiles is essential.
Conclusion
Exosomes may be used to identify somatic mutations of tumors from liquid biopsies [8, 69], with EGFR and its signaling network proteins frequently detected in exosomes from NSCLC patients, making them potential biomarkers for cancer research and clinical use [75, 76, 78]. New platforms display promising perspectives in their ability to detect EGFR mutations in liquid biopsy. One of the most noteworthy is the recently and first approved FDA cfDNA-based plasma test for EGFR mutations [113]. Promisingly, the measurement of exoNA may expand the utility of exosomes as potential diagnostic and prognostic tools in EGFR-mutated cancers as they could provide a more complete assessment of the evolving tumor and response to targeted therapies.
Funding
Open Access funding provided by the IReL Consortium.
Declarations
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tsao M-S, Marguet S, Le Teuff G, Lantuejoul S, Shepherd FA, Seymour L, et al. Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2015;33(30):3439–3446. doi: 10.1200/JCO.2014.58.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tfayli AH, Fakhri GB, Al Assaad MS. Prevalence of the epidermal growth factor receptor mutations in lung adenocarcinoma patients from the Middle East region. Annals of Thoracic Medicine. 2019;14(3):173–178. doi: 10.4103/atm.ATM_344_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H. Osimertinib making a breakthrough in lung cancer targeted therapy. OncoTargets and therapy. 2016;9:5489–5493. doi: 10.2147/OTT.S114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernabé R, Hickson N, Wallace A, Blackhall FH. What do we need to make circulating tumour DNA (ctDNA) a routine diagnostic test in lung cancer? European Journal of Cancer (Oxford, England: 1990) 2017;81:66–73. doi: 10.1016/j.ejca.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Mayo-de-Las-Casas C, Garzón Ibáñez M, Jordana-Ariza N, García-Peláez B, Balada-Bel A, Villatoro S, et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Review of Molecular Diagnostics. 2018;18(1):35–45. doi: 10.1080/14737159.2018.1407243. [DOI] [PubMed] [Google Scholar]
- 6.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nature Reviews Clinical Oncology. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.Karachaliou N, Mayo-de-las-Casas C, Molina-Vila MA, Rosell R. Real-time liquid biopsies become a reality in cancer treatment. Annals of Translational Medicine. 2015;3(3):8–8. doi: 10.3978/j.issn.2305-5839.2015.01.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantel K, Alix-Panabières C. Liquid biopsy: Potential and challenges. Molecular Oncology. 2016;10(3):371–373. doi: 10.1016/j.molonc.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russano M, Napolitano A, Ribelli G, Iuliani M, Simonetti S, Citarella F, Pantano F, Dell’Aquila E, Anesi C, Silvestris N, Argentiero A, Solimando AG, Vincenzi B, Tonini G, Santini D. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. Journal of Experimental & Clinical Cancer Research. 2020;39(1):95. doi: 10.1186/s13046-020-01601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strijker M, Soer EC, de Pastena M, Creemers A, Balduzzi A, Beagan JJ, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. International Journal of Cancer. 2020;146(5):1445–1456. doi: 10.1002/ijc.32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) Journal of Biological Chemistry. 1987;262(19):9412–9420. doi: 10.1016/S0021-9258(18)48095-7. [DOI] [PubMed] [Google Scholar]
- 12.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. The Journal of Cell Biology. 1985;101(3):942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nature Reviews. Immunology. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics, Proteomics & Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mignot G, Roux S, Thery C, Ségura E, Zitvogel L. Prospects for exosomes in immunotherapy of cancer. Journal of Cellular and Molecular Medicine. 2006;10(2):376–388. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Niel G, Mallegol J, Bevilacqua C, Candalh C, Brugiere S, Tomaskovic-Crook E, et al. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52(12):1690–1697. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinheiro A, Silva AM, Teixeira JH, Gonçalves RM, Almeida MI, Barbosa MA, Santos SG. Extracellular vesicles: intelligent delivery strategies for therapeutic applications. Journal of Controlled Release. 2018;289:56–69. doi: 10.1016/j.jconrel.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Alharbi M, Zuñiga F, Elfeky O, Guanzon D, Lai A, Rice GE, Perrin L, Hooper J, Salomon C. The potential role of miRNAs and exosomes in chemotherapy in ovarian cancer. Endocrine-Related Cancer. 2018;25(12):R663–R685. doi: 10.1530/ERC-18-0019. [DOI] [PubMed] [Google Scholar]
- 19.Vaidyanathan R, Hao Soon R, Zhang P, Jiang K, Teck Lim C. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab on a Chip. 2019;19(1):11–34. doi: 10.1039/C8LC00684A. [DOI] [PubMed] [Google Scholar]
- 20.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 21.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. The Journal of Biological Chemistry. 2014;289(7):3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalluri R. The biology and function of exosomes in cancer. The Journal of Clinical Investigation. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.Adamczyk KA, Klein-Scory S, Tehrani MM, Warnken U, Schmiegel W, Schnölzer M, Schwarte-Waldhoff I. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sciences. 2011;89(9):304–312. doi: 10.1016/j.lfs.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, Czupryna A, Szczepanik A, Zembala M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer immunology, immunotherapy: CII. 2010;59(6):841–850. doi: 10.1007/s00262-009-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, Morelli D, Villa A, Mina PD, Menard S, Filipazzi P, Rivoltini L, Tagliabue E, Pupa SM. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. Journal of Cellular Physiology. 2012;227(2):658–667. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 28.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019;177(2):428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Current Opinion in Cell Biology. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. Journal of Lipid Research. 2019;60(1):9–18. doi: 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caradec J, Kharmate G, Hosseini-Beheshti E, Adomat H, Gleave M, Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clinical Biochemistry. 2014;47(13–14):1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican1 identifies cancer exosomes and facilitates early detection of cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: A diagnostic marker for lung cancer. Clinical Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 36.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 37.Huang T, Deng C-X. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. International Journal of Biological Sciences. 2019;15(1):1–11. doi: 10.7150/ijbs.27796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Research. 2006;66(9):4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Martinez M, Navarro A, Marrades RM, Viñolas N, Santasusagna S, Muñoz C, et al. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget. 2016;7(32):51515–51524. doi: 10.18632/oncotarget.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, Su M, Pan H, Shen L, Xie D, Xie C. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clinical Cancer Research. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 41.Gao J, Qiu X, Li X, Fan H, Zhang F, Lv T, Song Y. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochemical and Biophysical Research Communications. 2018;498(3):409–415. doi: 10.1016/j.bbrc.2018.02.114. [DOI] [PubMed] [Google Scholar]
- 42.Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic (Copenhagen, Denmark) 2011;12(12):1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 43.Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Seminars in Cell & Developmental Biology. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Gangoda L, Boukouris S, Liem M, Kalra H, Mathivanan S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics. 2015;15(2–3):260–271. doi: 10.1002/pmic.201400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications. 2011;2(1):282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lässer C, O’Neil SE, Shelke GV, Sihlbom C, Hansson SF, Gho YS, Lundbäck B, Lötvall J. Exosomes in the nose induce immune cell trafficking and harbour an altered protein cargo in chronic airway inflammation. Journal of Translational Medicine. 2016;14(1):181. doi: 10.1186/s12967-016-0927-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez-Calero, L., Martin-Lorenzo, M., & Alvarez-Llamas, G. (2014). Exosomes: A potential key target in cardio-renal syndrome. Frontiers in Immunology, 5. 10.3389/fimmu.2014.00465. [DOI] [PMC free article] [PubMed]
- 48.Kishore R, Garikipati VNS, Gumpert A. Tiny shuttles for information transfer: Exosomes in cardiac health and disease. Journal of Cardiovascular Translational Research. 2016;9(3):169–175. doi: 10.1007/s12265-016-9682-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howitt J, Hill AF. Exosomes in the pathology of neurodegenerative diseases. Journal of Biological Chemistry. 2016;291(52):26589–26597. doi: 10.1074/jbc.R116.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malm T, Loppi S, Kanninen KM. Exosomes in Alzheimer’s disease. Neurochemistry International. 2016;97:193–199. doi: 10.1016/j.neuint.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Jiang L, Dong H, Cao H, Ji X, Luan S, Liu J. Exosomes in pathogenesis, diagnosis, and treatment of Alzheimer’s disease. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2019;25:3329–3335. doi: 10.12659/MSM.914027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Record M, Poirot M, Silvente-Poirot S. Emerging concepts on the role of exosomes in lipid metabolic diseases. Biochimie. 2014;96:67–74. doi: 10.1016/j.biochi.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 53.Salem KZ, Moschetta M, Sacco A, Imberti L, Rossi G, Ghobrial IM, et al. Exosomes in tumor angiogenesis. Methods in Molecular Biology (Clifton, N.J.) 2016;1464:25–34. doi: 10.1007/978-1-4939-3999-2_3. [DOI] [PubMed] [Google Scholar]
- 54.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: Communication from a distance. Developmental Cell. 2019;49(3):347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Advances in Clinical Chemistry. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu C, Meiners S, Lukas C, Stathopoulos GT, Chen J. Role of exosomal microRNAs in lung cancer biology and clinical applications. Cell Proliferation. 2020;53:e12828. doi: 10.1111/cpr.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang S, Yu S, Cheng J, Zhang Y, Huang X. The versatile roles and clinical implications of exosomal mRNAs and microRNAs in cancer. The International Journal of Biological Markers. 2020;35(2):3–19. doi: 10.1177/1724600820920293. [DOI] [PubMed] [Google Scholar]
- 58.Shi Z-Y, Yang X-X, Malichewe C, Li Y-S, Guo X-L. Exosomal microRNAs-mediated intercellular communication and exosome-based cancer treatment. International Journal of Biological Macromolecules. 2020;158:530–541. doi: 10.1016/j.ijbiomac.2020.04.228. [DOI] [PubMed] [Google Scholar]
- 59.Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Molecular Diagnosis & Therapy. 2016;20(6):509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 60.Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Mark MT, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sancho-Albero M, Navascués N, Mendoza G, Sebastián V, Arruebo M, Martín-Duque P, Santamaría J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. Journal of Nanobiotechnology. 2019;17(1):16. doi: 10.1186/s12951-018-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: Biogenesis, biologic function and clinical potential. Cell & Bioscience. 2019;9(1):19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zduriencikova M, Gronesova P, Cholujova D, Sedlak J. Potential biomarkers of exosomal cargo in endocrine signaling. Endocrine Regulations. 2015;49(3):141–150. doi: 10.4149/endo_2015_03_141. [DOI] [PubMed] [Google Scholar]
- 64.Carretero-González A, Otero I, Carril-Ajuria L, de Velasco G, Manso L. Exosomes: Definition, role in tumor development and clinical implications. Cancer Microenvironment: Official Journal of the International Cancer Microenvironment Society. 2018;11(1):13–21. doi: 10.1007/s12307-018-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. Journal of Cell Science. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 66.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 67.Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. Journal of Hematology & Oncology. 2019;12:12. doi: 10.1186/s13045-019-0739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Liao K, Zhou W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells International. 2018;2018:4837370–4837316. doi: 10.1155/2018/4837370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rijavec, E., Coco, S., Genova, C., Rossi, G., Longo, L., & Grossi, F. (2019). Liquid biopsy in non-small cell lung cancer: Highlights and challenges. Cancers, 12(1). 10.3390/cancers12010017. [DOI] [PMC free article] [PubMed]
- 70.Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. Journal of Proteomics. 2016;133:161–169. doi: 10.1016/j.jprot.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 71.Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. Journal of Extracellular Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zanetti-Domingues LC, Bonner SE, Iyer RS, Martin-Fernandez ML, Huber V. Cooperation and interplay between EGFR signalling and extracellular vesicle biogenesis in cancer. Cells. 2020;9(12):2639. doi: 10.3390/cells9122639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montermini L, Meehan B, Garnier D, Lee WJ, Lee TH, Guha A, al-Nedawi K, Rak J. Inhibition of oncogenic epidermal growth factor receptor kinase triggers release of exosome-like extracellular vesicles and impacts their phosphoprotein and DNA content. The Journal of Biological Chemistry. 2015;290(40):24534–24546. doi: 10.1074/jbc.M115.679217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proceedings of the National Academy of Sciences. 2009;106(10):3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang S-H, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Investigation. 2013;31(5):330–335. doi: 10.3109/07357907.2013.789905. [DOI] [PubMed] [Google Scholar]
- 76.Park JO, Choi D-Y, Choi D-S, Kim HJ, Kang JW, Jung JH, Lee JH, Kim J, Freeman MR, Lee KY, Gho YS, Kim KP. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics. 2013;13(14):2125–2134. doi: 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- 77.Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Critical Reviews in Oncology/Hematology. 2021;157:103194. doi: 10.1016/j.critrevonc.2020.103194. [DOI] [PubMed] [Google Scholar]
- 78.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 79.Yamashita T, Kamada H, Kanasaki S, Maeda Y, Nagano K, Abe Y, Inoue M, Yoshioka Y, Tsutsumi Y, Katayama S, Inoue M, Tsunoda S. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Die Pharmazie. 2013;68(12):969–973. [PubMed] [Google Scholar]
- 80.Gao L, Wang L, Dai T, Jin K, Zhang Z, Wang S, Xie F, Fang P, Yang B, Huang H, van Dam H, Zhou F, Zhang L. Tumor-derived exosomes antagonize innate antiviral immunity. Nature Immunology. 2018;19(3):233–245. doi: 10.1038/s41590-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 81.Yu S, Sha H, Qin X, Chen Y, Li X, Shi M, Feng J. EGFR E746-A750 deletion in lung cancer represses antitumor immunity through the exosome-mediated inhibition of dendritic cells. Oncogene. 2020;39(13):2643–2657. doi: 10.1038/s41388-020-1182-y. [DOI] [PubMed] [Google Scholar]
- 82.Shi F, Deng Z, Zhou Z, Jiang B, Jiang C-Y, Zhao R-Z, Sun F, Cui D, Sun MH, Sun Q, Wang XJ, Wu Q, Xia SJ, Han B-M. Heat injured stromal cells-derived exosomal EGFR enhances prostatic wound healing after thulium laser resection through EMT and NF-κB signaling. The Prostate. 2019;79(11):1238–1255. doi: 10.1002/pros.23827. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Wang X, Yang H, Li J, Ning T, Huang D, Li H, Zhang L, Ying G, Ba Y. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nature Communications. 2017;8(1):1–11. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ludwig N, Whiteside TL. Potential roles of tumor-derived exosomes in angiogenesis. Expert opinion on therapeutic targets. 2018;22(5):409–417. doi: 10.1080/14728222.2018.1464141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsu Y-L, Hung J-Y, Chang W-A, Lin Y-S, Pan Y-C, Tsai P-H, Wu CY, Kuo P-L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 87.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. Journal of Biological Chemistry. 2013;288(48):34343–34351. doi: 10.1074/jbc.M113.480822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferrara N. Vascular endothelial growth factor: Basic science and clinical progress. Endocrine Reviews. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 89.Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(8):10703–10714. doi: 10.1007/s13277-016-4939-8. [DOI] [PubMed] [Google Scholar]
- 90.Zheng C-L, Qiu C, Shen M-X, Qu X, Zhang T-H, Zhang J-H, Du J-J. Prognostic impact of elevation of vascular endothelial growth factor family expression in patients with non-small cell lung cancer: An updated meta-analysis. Asian Pacific journal of cancer prevention: APJCP. 2015;16(5):1881–1895. doi: 10.7314/apjcp.2015.16.5.1881. [DOI] [PubMed] [Google Scholar]
- 91.Amiri A, Pourhanifeh MH, Mirzaei HR, Nahand JS, Moghoofei M, Sahebnasagh R, Mirzaei H, Hamblin MR. Exosomes and lung cancer: Roles in pathophysiology, diagnosis and therapeutic applications. Current Medicinal Chemistry. 2021;28(2):308–328. doi: 10.2174/0929867327666200204141952. [DOI] [PubMed] [Google Scholar]
- 92.Kuwai T, Nakamura T, Sasaki T, Kim S-J, Fan D, Villares GJ, et al. Phosphorylated epidermal growth factor receptor on tumor-associated endothelial cells is a primary target for therapy with tyrosine kinase inhibitors. Neoplasia (New York, N.Y.) 2008;10(5):489–500. doi: 10.1593/neo.08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baker CH, Kedar D, McCarty MF, Tsan R, Weber KL, Bucana CD, Fidler IJ. Blockade of epidermal growth factor receptor signaling on tumor cells and tumor-associated endothelial cells for therapy of human carcinomas. The American Journal of Pathology. 2002;161(3):929–938. doi: 10.1016/S0002-9440(10)64253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu S, Zhan Y, Luo J, Feng J, Lu J, Zheng H, Wen Q, Fan S. Roles of exosomes in the carcinogenesis and clinical therapy of non-small cell lung cancer. Biomedicine & Pharmacotherapy. 2019;111:338–346. doi: 10.1016/j.biopha.2018.12.088. [DOI] [PubMed] [Google Scholar]
- 95.Syn N, Wang L, Sethi G, Thiery J-P, Goh B-C. Exosome-mediated metastasis: From epithelial–mesenchymal transition to escape from immunosurveillance. Trends in Pharmacological Sciences. 2016;37(7):606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treatment Reviews. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 97.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Reviews. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7(34):54852–54866. doi: 10.18632/oncotarget.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corrado C, Saieva L, Raimondo S, Santoro A, De Leo G, Alessandro R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. Journal of Cellular and Molecular Medicine. 2016;20(10):1829–1839. doi: 10.1111/jcmm.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taverna S, Pucci M, Giallombardo M, Di Bella MA, Santarpia M, Reclusa P, et al. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Scientific Reports. 2017;7:7. doi: 10.1038/s41598-017-03460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. Journal of Hematology & Oncology. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lobb RJ, Lima LG, Möller A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Seminars in Cell & Developmental Biology. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 103.Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018;116:46–54. doi: 10.1016/j.lungcan.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 104.Liu X, Jiang T, Li X, Zhao C, Li J, Zhou F, Zhang L, Zhao S, Jia Y, Shi J, Gao G, Li W, Zhao J, Chen X, Su C, Ren S, Zhou C. Exosomes transmit T790M mutation-induced resistance in EGFR-mutant NSCLC by activating PI3K/AKT signalling pathway. Journal of Cellular and Molecular Medicine. 2020;24(2):1529–1540. doi: 10.1111/jcmm.14838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu S, Luo M, To KKW, Zhang J, Su C, Zhang H, et al. Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Molecular Cancer. 2021;20(1):17. doi: 10.1186/s12943-021-01307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen, R., Qian, Z., Xu, X., Zhang, C., Niu, Y., Wang, Z., et al. (2021). Exosomes-transmitted miR-7 reverses gefitinib resistance by targeting YAP in non-small-cell lung cancer. Pharmacological Research, 105442. 10.1016/j.phrs.2021.105442. [DOI] [PubMed]
- 107.Rolfo C, Chacartegui J, Giallombardo M, Alessandro R, Peeters M. 71P Exosomes isolated in plasma of non-small cell lung cancer patients contain microRNA related to the EGFR pathway: Proof of concept. Journal of Thoracic Oncology. 2016;11(4):S85. doi: 10.1016/S1556-0864(16)30184-8. [DOI] [Google Scholar]
- 108.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2016;8(4):6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Molecular Oncology. 2016;10(10):1595–1602. doi: 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ingenito, F., Roscigno, G., Affinito, A., Nuzzo, S., Scognamiglio, I., Quintavalle, C., & Condorelli, G. (2019). The role of exo-miRNAs in cancer: A focus on therapeutic and diagnostic applications. International Journal of Molecular Sciences, 20(19). 10.3390/ijms20194687. [DOI] [PMC free article] [PubMed]
- 111.Chen Q, Zhang Z-H, Wang S, Lang J-H. Circulating cell-free DNA or circulating tumor DNA in the management of ovarian and endometrial cancer. OncoTargets and therapy. 2019;12:11517–11530. doi: 10.2147/OTT.S227156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer—Which one will win? Translational Lung Cancer Research. 2016;5(5):466–482. doi: 10.21037/tlcr.2016.10.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Research, C. for D. E. and. (2018). Cobas EGFR mutation test v2. FDA. Retrieved from https://www.fda.gov/drugs/resources-information-approved-drugs/cobas-egfr-mutation-test-v2. Accessed 29 May 2020.
- 114.Wu Y-L, Lee V, Liam C-K, Lu S, Park K, Srimuninnimit V, et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer. 2018;126:1–8. doi: 10.1016/j.lungcan.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 115.Karlovich C, Goldman JW, Sun J-M, Mann E, Sequist LV, Konopa K, Wen W, Angenendt P, Horn L, Spigel D, Soria JC, Solomon B, Camidge DR, Gadgeel S, Paweletz C, Wu L, Chien S, O'Donnell P, Matheny S, Despain D, Rolfe L, Raponi M, Allen AR, Park K, Wakelee H. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686) Clinical Cancer Research. 2016;22(10):2386–2395. doi: 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 117.O’Donnell P, May T, DeMartin K, Ferguson J, Halait H, Wei W, Yu K, Scudder S. Performance characteristics of a real-time polymerase chain reaction assay for the detection of epidermal growth factor receptor (EGFR) mutations in plasma samples of non-small cell lung cancer (NSCLC) patients. Molecular Diagnosis & Therapy. 2020;24:451–460. doi: 10.1007/s40291-020-00458-7. [DOI] [PubMed] [Google Scholar]
- 118.Minari R, Mazzaschi G, Bordi P, Gnetti L, Alberti G, Altimari A, et al. Detection of EGFR-activating and T790M mutations using liquid biopsy in patients with EGFR-mutated non-small-cell lung cancer whose disease has progressed during treatment with first- and second-generation tyrosine kinase inhibitors: A multicenter real-life retrospective study. Clinical Lung Cancer. 2020;21:e464–e473. doi: 10.1016/j.cllc.2020.02.021. [DOI] [PubMed] [Google Scholar]
- 119.Bettegowda C, Sausen M, Leary R, Kinde I, Agrawal N, Bartlett B, et al. Detection of circulating tumor DNA in early and late stage human malignancies. Neuro-Oncology. 2014;16(suppl_3):iii7–iii7. doi: 10.1093/neuonc/nou206.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biology & Therapy. 2019;20(8):1057–1067. doi: 10.1080/15384047.2019.1598759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Möhrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, Subbiah V, Karp DD, Naing A, Krug A, Enderle D, Priewasser T, Noerholm M, Eitan E, Coticchia C, Stoll G, Jordan LM, Eng C, Kopetz ES, Skog J, Meric-Bernstam F, Janku F. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clinical Cancer Research. 2018;24(1):181–188. doi: 10.1158/1078-0432.CCR-17-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Castellanos-Rizaldos E, Zhang X, Tadigotla VR, Grimm DG, Karlovich C, Raez LE, Skog JK. Exosome-based detection of activating and resistance EGFR mutations from plasma of non-small cell lung cancer patients. Oncotarget. 2019;10(30):2911–2920. doi: 10.18632/oncotarget.26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, Brinkmann K, Emenegger J, Grimm DG, Castellanos-Rizaldos E, Goldman JW, Sequist LV, Soria JC, Camidge DR, Gadgeel SM, Wakelee HA, Raponi M, Noerholm M, Skog J. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Annals of Oncology. 2018;29(3):700–706. doi: 10.1093/annonc/mdx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLOS ONE. 2017;12(8):e0183915. doi: 10.1371/journal.pone.0183915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Castellanos-Rizaldos E, Grimm DG, Tadigotla V, Hurley J, Healy J, Neal PL, Sher M, Venkatesan R, Karlovich C, Raponi M, Krug A, Noerholm M, Tannous J, Tannous BA, Raez LE, Skog JK. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2018;24(12):2944–2950. doi: 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 126.Kim Y, Shin S, Kim B, Lee K-A. Selecting short length nucleic acids localized in exosomes improves plasma EGFR mutation detection in NSCLC patients. Cancer Cell International. 2019;19(1):251. doi: 10.1186/s12935-019-0978-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wan Y, Liu B, Lei H, Zhang B, Wang Y, Huang H, Chen S, Feng Y, Zhu L, Gu Y, Zhang Q, Ma H, Zheng S-Y. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Annals of Oncology. 2018;29(12):2379–2383. doi: 10.1093/annonc/mdy458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu Z, Wang Z, Sun H, Xin H. Evaluation of exosomal miRNA in blood as a potential diagnostic biomarker for human non-small cell lung cancer. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2020;26:e924721. doi: 10.12659/MSM.924721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peng X-X, Yu R-Y, Wu X, Wu S-Y, Pi C, Chen Z-H, et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. Journal for Immunotherapy of Cancer. 2020;8(1):e000376. doi: 10.1136/jitc-2019-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo-miRNAs as a new tool for liquid biopsy in lung cancer. Cancers. 2019;11(6):888. doi: 10.3390/cancers11060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qin, X., Xu, H., Gong, W., & Deng, W. (2015). The tumor cytosol miRNAs, fluid miRNAs, and exosome miRNAs in lung cancer. Frontiers in Oncology, 4. 10.3389/fonc.2014.00357. [DOI] [PMC free article] [PubMed]
- 132.Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer. 2013;8(9):1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monaco F, Gaetani S, Alessandrini F, Tagliabracci A, Bracci M, Valentino M, Neuzil J, Amati M, Bovenzi M, Tomasetti M, Santarelli L. Exosomal transfer of miR-126 promotes the anti-tumour response in malignant mesothelioma: Role of miR-126 in cancer-stroma communication. Cancer Letters. 2019;463:27–36. doi: 10.1016/j.canlet.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 134.Liu C, Yang Y, Wu Y. Recent advances in exosomal protein detection via liquid biopsy biosensors for cancer screening, diagnosis, and prognosis. The AAPS journal. 2018;20(2):41. doi: 10.1208/s12248-018-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okuma Y, Morikawa K, Tanaka H, Yokoyama T, Itani H, Horiuchi K, Nakagawa H, Takahashi N, Bessho A, Soejima K, Kishi K, Togashi A, Kanai Y, Ueda K, Horimoto K, Matsutani N, Seki N. Prospective exosome-focused translational research for afatinib study of non-small cell lung cancer patients expressing EGFR (EXTRA study) Thoracic Cancer. 2019;10(2):395–400. doi: 10.1111/1759-7714.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]